ABSTRACT
Nuclear egress of herpesvirus capsids is mediated by the conserved nuclear egress complex (NEC), composed of the membrane-anchored pUL34 and its nucleoplasmic interaction partner, pUL31. The recently solved crystal structures of the NECs from different herpesviruses show a high structural similarity, with the pUL34 homologs building a platform recruiting pUL31 to the inner nuclear membrane. Both proteins possess a central globular fold, while the conserved N-terminal portion of pUL31 forms an extension reaching around the core of pUL34. However, the extreme N terminus of the pUL31 homologs, which is highly variable in length and amino acid composition, had to be removed for crystallization. Several pUL31 homologs contain a classical nuclear localization signal (NLS) within this part mediating efficient nuclear import. In addition, membrane-binding activity, blocking premature interaction with pUL34, nucleocapsid trafficking, and regulation of NEC assembly and disassembly via phosphorylation were assigned to the extreme pUL31 N terminus. To test the functional importance in the alphaherpesvirus pseudorabies virus (PrV) pUL31, N-terminal truncations and site-specific mutations were generated, and the resulting proteins were tested for intracellular localization, interaction with pUL34, and functional complementation of PrV-ΔUL31. Our data show that neither the bipartite NLS nor the predicted phosphorylation sites are essential for pUL31 function during nuclear egress. Moreover, nearly the complete variable N-terminal part was dispensable for function as long as a stretch of basic amino acids was retained. Phosphorylation of this domain controls efficient nucleocapsid release from the perinuclear space.
IMPORTANCE Nuclear egress of herpesvirus capsids is a unique vesicle-mediated nucleocytoplasmic transport. Crystal structures of the heterodimeric NECs from different herpesviruses provided important details of this viral nuclear membrane deformation and scission machinery but excluded the highly variable N terminus of the pUL31 component. We present here a detailed mutagenesis study of this important portion of pUL31 and show that basic amino acid residues within this domain play an essential role for proper targeting, complex formation, and function during nuclear egress, while phosphorylation modulates efficient release from the perinuclear space. Thus, our data complement previous structure-function assignments of the nucleocapsid-interacting component of the NEC.
KEYWORDS: herpesvirus, pseudorabies virus, nuclear egress complex (NEC), nuclear envelope, nuclear localization signal (NLS), nuclear export signal (NES)
INTRODUCTION
Nuclear egress of newly assembled herpesvirus nucleocapsids is mediated by the nuclear egress complex (NEC), a heterodimeric structure consisting of homologs of the membrane anchored pUL34 and the nucleoplasmic pUL31 (reviewed in references 1–3). Crystal structures of the NECs from different herpesviruses (4–7) are surprisingly similar given the only moderate conservation of amino acid sequences. The membrane-anchored pUL34 homologs form a platform at the inner nuclear membrane, with the globular pUL31 homologs sitting on top of it and projecting into the nucleoplasm or toward the nucleocapsid in the primary enveloped virions. Only the N-terminal arm of the conserved core region of the pUL31 homologs, consisting of two alpha-helices arranged in a V-shape, form a hook-like extension, which wraps around and inserts into the core of the pUL34 homolog. However, to generate high-quality structural information, the C terminus of the pUL34 homologs including the transmembrane domain, as well as the flexible N terminus of the pUL31 homologs had to be removed. Thus, no structural data are available for these elements in any of the studied herpesvirus NECs.
Comparative analyses of the amino acid sequences of pUL31 homologs from various herpesviruses allowed the identification of four conserved regions (CR1 to CR4), while the very N terminus was found to be highly variable in length and amino acid composition (8). A classical nuclear localization signal (NLS) for efficient nuclear targeting was predicted in several but not all pUL31 homologs (8, 9). An additional PY motif-containing NLS, located between amino acids (aa) 21 and 34, was suggested for PrV pUL31 (10). While in pUL31 of herpes simplex virus 1 (HSV-1) the N terminus is required for full function (11), the predicted bipartite NLS is not (12). Nevertheless, basic patches identified within the N-terminal 44 aa were shown to be critical for virus propagation. It was suggested that this part of pUL31 is important to prevent premature interaction with pUL34 in the cytoplasm and to traffic mature capsids in the nucleus toward the inner nuclear membrane for subsequent interaction with pUL34, thereby providing a link between capsid maturation and primary envelopment (12). In addition, for HSV-1 pUL31, aa 41 to 50 were shown to mediate membrane interaction when tested in giant unilamellar vesicles in complex with soluble pUL34, but the specific amino acids involved in this membrane targeting could not be identified (13).
Phosphorylation plays an important role in the function of this part of pUL31. Serine residues present within the N-terminal 50 aa in HSV-1 pUL31 (aa 11, 24, 26, 27, 40, and 43) were shown to be phosphorylated by the alphaherpesviral pUS3 kinase (11). Simultaneous replacement of all six residues by alanine resulted in an accumulation of primary enveloped virions in the perinuclear cleft, similar to those found after deletion or inactivation of the alphaherpesviral protein kinase pUS3, while mimicking phosphorylation by replacement with glutamic acid impaired efficient primary envelopment. These data indicated that only nonphosphorylated pUL31 assembles into the NEC coat, while disassembly may be induced by phosphorylation within the primary virions during deenvelopment (11). Very recently, a poly(ADP-ribose) binding motif has been identified in HSV-2 pUL31 which might target pUL31 to sites of DNA damage (14) and overlaps a functionally uncharacterized nucleotidylylation signal (15).
For pseudorabies virus (PrV) pUL31, a classical bipartite NLS was predicted between aa 5 and 20 (9, 16) and shown to be functional in nuclear targeting (10, 17, 18). pUL31-ΔNLS, lacking the variable N terminus completely, was nonfunctional, and the protein localized nearly exclusively in the cytoplasm. This was surprising, since the approximately 28-kDa protein should be able to enter the nucleus by passive diffusion. This observation led to the identification of a nuclear export signal (NES) present in the C terminus of PrV pUL31 (17). Deletion or site-specific mutation of both signals resulted in the expected diffuse pancellular distribution. Nonfunctional pUL31-ΔNLS exerted a dominant-negative effect on replication of wild-type PrV, which was attributed to a mislocalization of the complex partner pUL34 from the nuclear envelope to cytoplasmic membranes (17). While deletion of either the nuclear import or export signal resulted in nonfunctional proteins, site-specifically mutated pUL31-NLSPM carrying alanine substitutions at arginine (R) positions 5, 6, 16, and 17 retained the ability to partially complement the defect of a mutant lacking pUL31, indicating that an intact NLS motif in pUL31 is not required for NEC function. In contrast, two leucine residues (L252/254A) located in the NES motif are essential. In RK13 cells expressing UL31-NESPM infected with PrV-ΔUL31, nucleocapsids were formed but remained in the nucleus, many of them closely attached to the INM, indicating that the NES-containing region is required for the transition of the flat NEC sheet into a curved envelope coat (17).
To characterize the function of the N-terminal part of PrV pUL31 in more detail, we generated different truncated versions by deleting aa 2 to 6 (pUL31-N7), 2 to 10 (pUL31-N11), 2 to 13 (pUL31-N14), or 2 to 17 (pUL31-N18). In addition, basic residues and predicted phosphorylation sites were altered to alanine. To generate an artificial basic patch, alanine codons at positions 23 and 24 were replaced by arginine codons. Mutated proteins were tested for localization, complex formation after transient transfection, and functional complementation of PrV-ΔUL31.
Our data show that the major part of the PrV pUL31 N terminus is not required for function if one stretch of basic residues is retained. This basic patch is required for efficient nuclear localization of pUL31 and interaction with pUL34 at the INM. A major effect of the predicted phosphorylation sites within the very N-terminal part on viral titers could not be found. However, in their absence primary virions accumulated in the perinuclear space, similar to the phenotype described for mutants lacking the pUS3 kinase (19, 20), indicating that phosphorylation of the pUL31-N-terminal part is relevant for efficient release of nucleocapsids from the perinuclear space.
RESULTS
The bipartite NLS is dispensable for PrV pUL31 function.
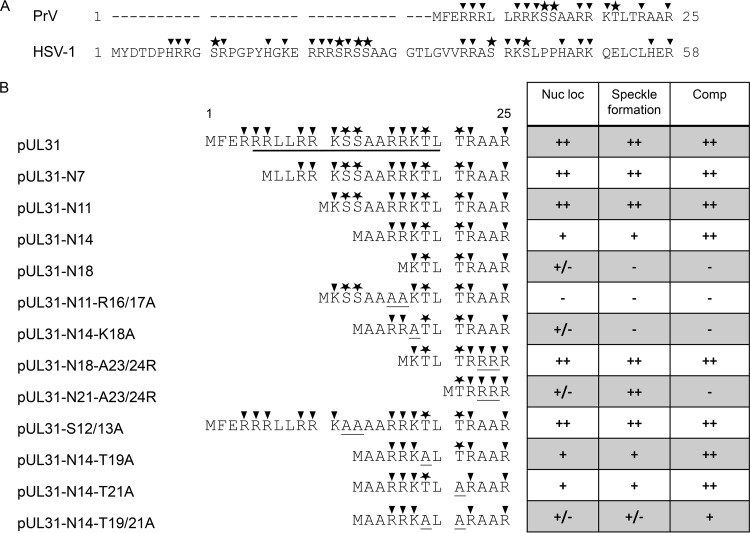
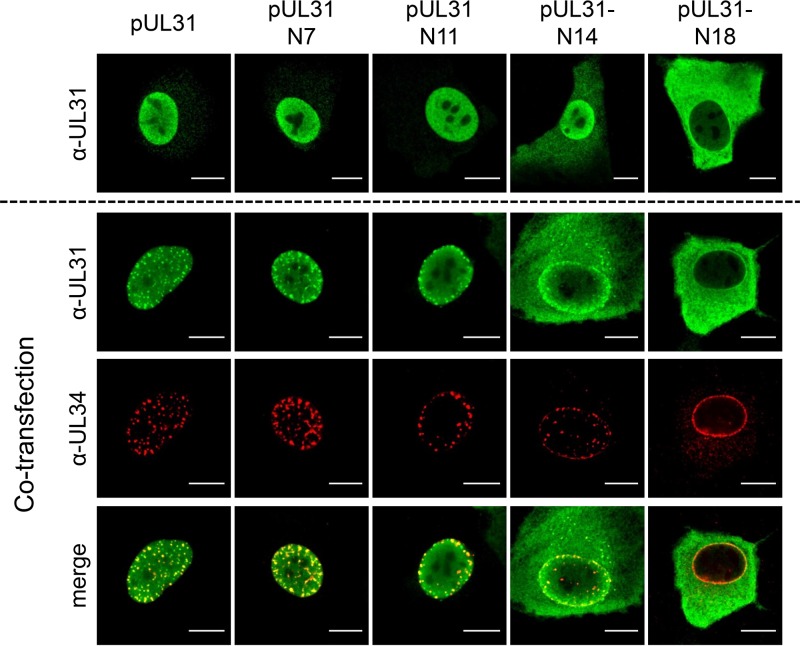
The nonconserved N terminus of PrV pUL31 comprises only 25 amino acids (aa) and is significantly shorter than the 58-aa N terminus of HSV-1 pUL31 (Fig. 1A). Nevertheless, both sequences contain a predicted bipartite NLS composed of several clusters of basic residues (9, 12, 16, 17) (Fig. 1B). To test whether this region is important for protein localization and function, we generated N-terminally truncated PrV pUL31 mutants starting at aa 7 (pUL31-N7), 11 (pUL31-N11), 14 (pUL31-N14), or 18 (pUL31-N18) (Fig. 1B). Corresponding expression plasmids were transfected into rabbit kidney (RK13) cells, and the mutant proteins were detected by indirect immunofluorescence using a monospecific anti-pUL31 rabbit serum (Fig. 2, upper panel). Truncation mutants pUL31-N7 and pUL31-N11 localized like wild-type pUL31 in the nucleus. pUL31-N14, which lacks more than half of the predicted bipartite NLS, still showed a predominant nuclear localization in addition to a diffuse cytoplasmic staining, pointing to impaired nuclear targeting. In contrast, pUL31-N18, lacking all clusters of basic amino acids, was found nearly exclusively in the cytoplasm, as was the previously described pUL31-ΔNLS lacking the complete variable N terminus (aa 2 to 25) (17).
FIG 1.
Summary of PrV pUL31 mutants. (A) The variable N termini of PrV and HSV-1 pUL31 are aligned. Basic amino acids are marked by triangles, and predicted phosphorylation sites are shown by asterisks. (B) The N-terminal amino acid sequences of the full-length, N-terminally truncated, and site-specifically mutated pUL31 are shown. Basic amino acids as well as predicted phosphorylation sites are highlighted as shown for panel A. The black bar indicates location of the predicted bipartite NLS; amino acids substituted in this study are underlined. A summary of the results is presented in the table on the right, with nuclear localization (Nuc loc) after transfection, colocalization in punctate pattern after cotransfection with pcDNA-UL34, and complementation (Comp) of PrV-ΔUL31 by stably expressing cell lines (++, wild-type; +, slightly reduced; +/−, significantly reduced; −, negative).
FIG 2.
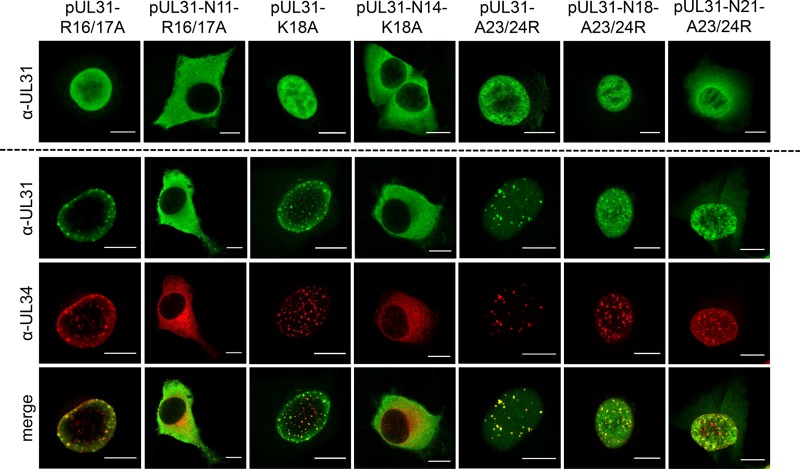
Localization of N-terminally truncated pUL31 and colocalization with pUL34. The pUL31 expression constructs were transfected (upper row) or cotransfected with pcDNA-UL34 (lower rows) into RK13 cells. Two days posttransfection, cells were fixed, permeabilized, and stained with the monospecific pUL31 antiserum (green) and a monoclonal anti-pUL34 antibody (red). Fluorescent signals were recorded with a confocal laser-scanning microscope (Leica SP5). Bars, 10 μm.
Interaction with pUL34 was tested after cotransfection of plasmids expressing the truncated pUL31 constructs together with pcDNA-UL34. Colocalization and speckle formation indicative for interaction and formation of primary vesicles (21) indistinguishable from wild-type pUL31 transfected cells was observed after cotransfection with plasmids expressing pUL31-N7 and pUL31-N11 (Fig. 2, lower panel). Despite increased cytoplasmic presence of pUL31-N14, the typical speckle pattern, where both proteins colocalize, was evident after coexpression with pUL34. However, after cotransfection of pcDNA-UL31-N18 with pcDNA-UL34, no cofluorescing punctae were evident at the nuclear rim, and pUL31-N18 could be detected in the cytoplasm while pUL34 was found with an undisturbed smooth nuclear rim staining, as observed after single transfection (21).
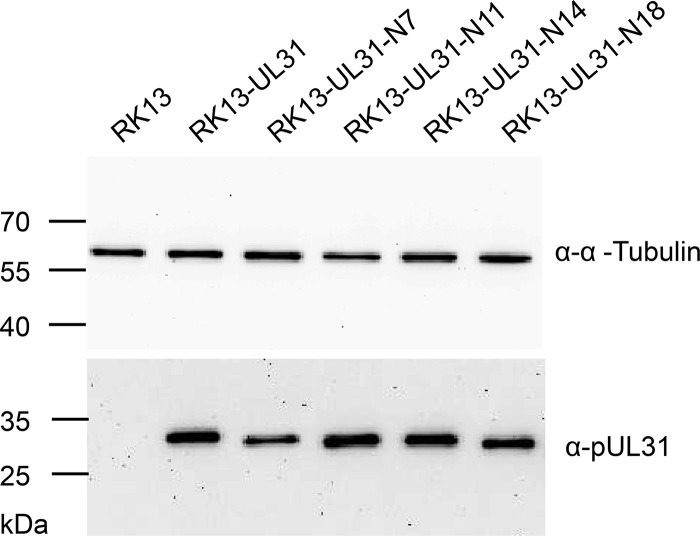
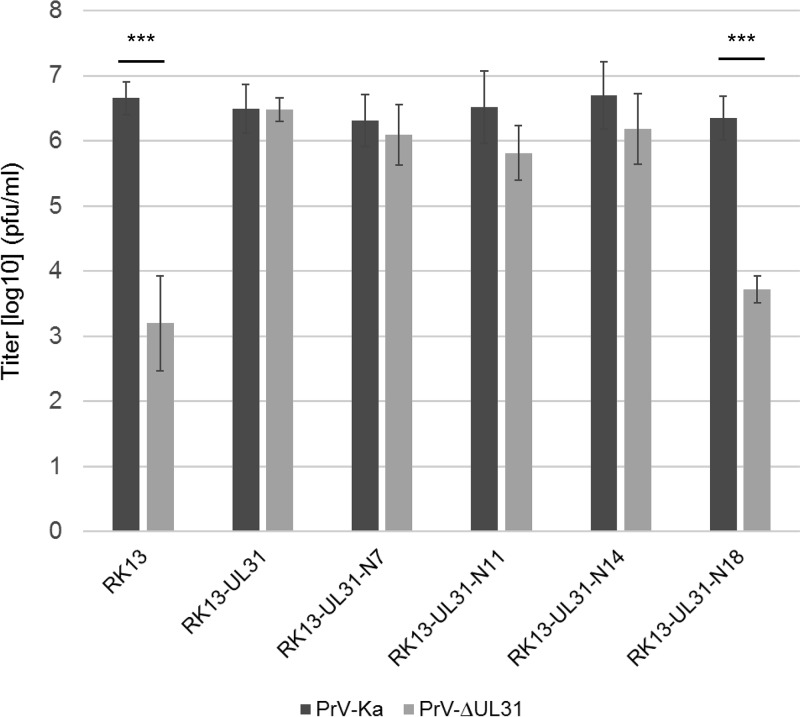
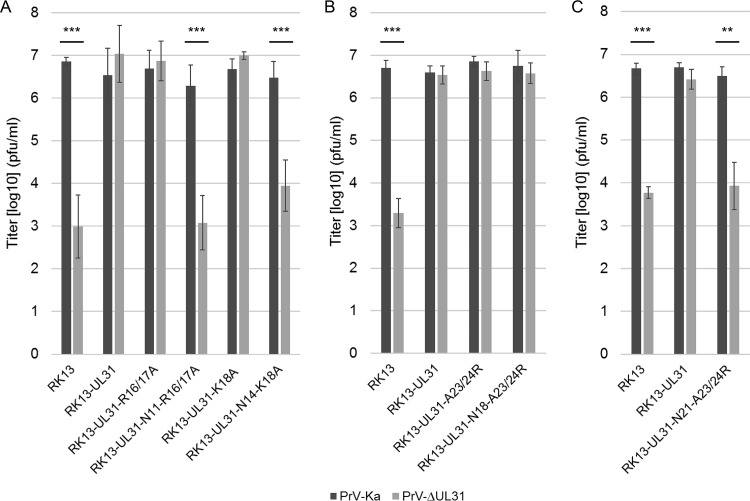
To test whether the truncated pUL31 constructs are able to complement the defect of PrV-ΔUL31, stably expressing RK13 cell lines were generated. Expression of truncated pUL31 variants with the expected, slightly decreasing molecular mass could be detected by Western blotting (Fig. 3). The stably expressing cell lines were infected with the PrV laboratory strain Kaplan (PrV-Ka) (22) or PrV-ΔUL31 (16) at a multiplicity of infection (MOI) of 5. Cells and supernatants were harvested 24 h postinfection (p.i.), and progeny virus titers were determined on RK13-UL31 cells (Fig. 4). PrV-Ka titers from all cell lines, including nontransgenic RK13 cells, were within the same range and reached values above 106 PFU per ml, showing that the expressed proteins exerted no significant dominant-negative effect. RK13-UL31-N7 and RK13-UL31-N14 also efficiently complemented the defect of PrV-ΔUL31, while titers from RK13-UL31-N11 were slightly lower than those from PrV-Ka. In contrast, PrV-ΔUL31 titers derived from RK13-UL31-N18 resembled those obtained from nontransfected cells (Fig. 4).
FIG 3.
Western blot analysis of the pUL31-expressing cell lines. Stably pUL31-expressing or nontransgenic RK13 cells were harvested and lysed. Proteins were separated on an SDS-10% polyacrylamide gel. After transfer to nitrocellulose, membranes were incubated with the monospecific anti-pUL31 rabbit serum or murine anti-alpha tubulin as a loading control. Molecular masses of marker proteins are given on the left (in kDa).
FIG 4.
In vitro replication. RK13, RK13-UL31, and cells expressing truncated pUL31 were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 5 and harvested 24 h p.i. Shown are mean values from four independent experiments with the corresponding standard deviations. Statistically significant differences between the titers of infection with PrV-Ka and PrV-ΔUL31 of each transgenic cell line are indicated: ***, P < 0.001.
One stretch of basic amino acids in the pUL31 N terminus is required for function during nuclear egress.
In contrast to the nonfunctional pUL31-N18, pUL31-N11 and pUL31-N14 retained three consecutive basic amino acids. To test whether this basic patch plays an important role, codons for two arginine residues (R) at positions 16 and 17 were replaced by alanine residues in pUL31-N11 and lysine at position 18 in pUL31-N14 was altered to alanine, resulting in pUL31-N11-R16/17A and pUL31-N14-K18A, respectively. In addition, to restore a basic patch, in the nonfunctional pUL31-N18 two alanine residues were replaced by arginine, giving rise to pUL31-N18-A23/24R (Fig. 1B). For a control, corresponding amino acids were also replaced in full-length pUL31, yielding pUL31-R16/17A, pUL31-K18A, and pUL31-A23/24R. All full-length pUL31 mutants with these amino acid substitutions showed a distribution similar to that of the wild-type protein after transfection or cotransfection with pcDNA-UL34 (Fig. 5). In contrast, pUL31-N11-R16/17A and pUL31-N14-K18A showed a nearly exclusive cytoplasmic localization. In addition, in the presence of pUL31-N11-R16/17A and pUL31-N14-K18A, coexpressed pUL34 was also predominantly located in the cytoplasm. Reconstitution of a basic patch in pUL31-N18 (pUL31-N18-A23/24R) restored efficient nuclear targeting and speckle formation in the presence of pUL34 (Fig. 5).
FIG 5.
Localization of site-specifically mutated pUL31 and colocalization with pUL34. Transfections, cotransfections with pcDNA-UL34, and processing for confocal microscopy were done as described in the legend to Fig. 2. Bars, 10 μm.
In complementation assays on stably expressing cell lines (Fig. 6A), RK13-UL31-R16/17A and RK13-UL31-K18A fully complemented the defect of PrV-ΔUL31, while titers from RK13-N11-R16/17 and RK13-N14-K18A were similar to those from nontransgenic RK13 cells, indicating that a stretch of basic amino acids in the N terminus of pUL31 is required for functional complementation. This was supported by the fact that pUL31-N18 regained functionality after reconstitution of a basic patch in pUL31-N18-A23/24R (Fig. 6B). However, further truncation of the protein, as in pUL31-N21-A23/24R, resulted in a noncomplementing phenotype (Fig. 6C) despite partial nuclear localization and punctate colocalization with pUL34 in the nuclear envelope (Fig. 5).
FIG 6.
In vitro replication. RK13, RK13-UL31, and cells expressing site-specifically mutated pUL31 as indicated were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 5 (A to C). After 24 h p.i. cells and supernatant were harvested and infectious virus titers were determined on RK13-UL31 cells. Given are mean values from at least three independent experiments with the corresponding standard deviations. Statistically significant differences are indicated: **, P < 0.01; ***, P < 0.001.
Predicted phosphorylation sites in the PrV pUL31 N terminus are not essential for nuclear egress.
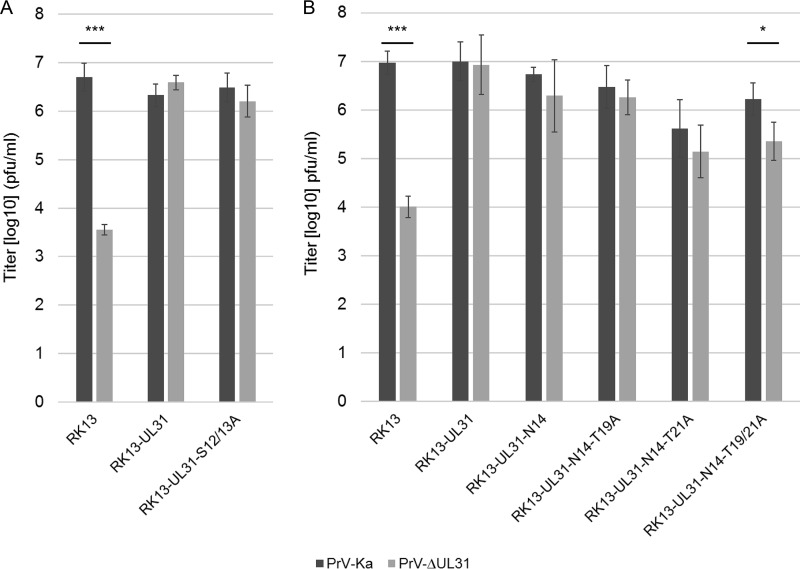
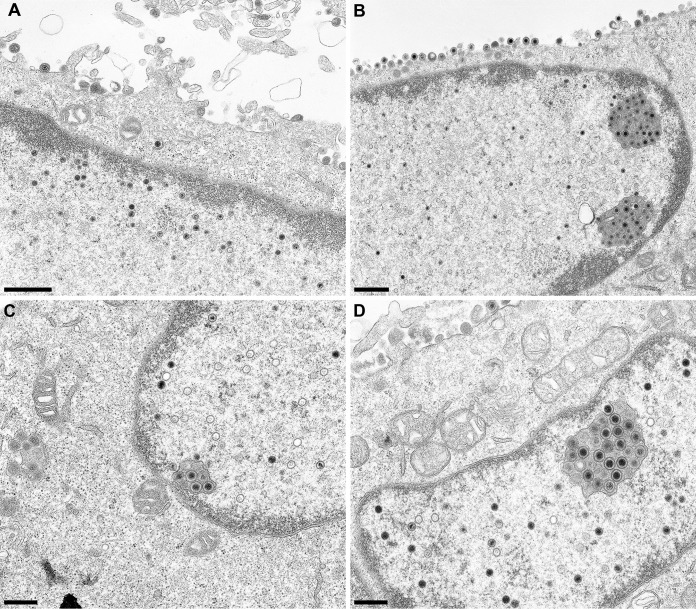
For HSV-1 pUL31, six phosphorylation sites had been predicted within the N-terminal 50 amino acids which were shown to be phosphorylated by pUS3 in vitro (11). Simultaneous mutation of all six serine residues to alanine resulted in a phenotype similar to that of a mutant lacking the protein kinase pUS3. The short N terminus of PrV pUL31 comprises only four predicted phosphorylation sites (NetPhos; http://www.cbs.dtu.dk/services/NetPhos [23]) at positions S12, S13, T19, and T21 (Fig. 1). The functional complementation of PrV-ΔUL31 by pUL31-N14, lacking two of the four predicted phosphorylation sites, indicated that these sites are either not used or not important for nuclear egress of PrV. To support this assumption, we generated pUL31-S12/13A, in which both serine residues were replaced by alanine, pUL31-N14-T19A and pUL31-N14-T21A, which code for alanine instead of threonine at wild-type position 19 or 21, and pUL31-N14-T19/21A, lacking all predicted potential phosphorylation sites. Localization of the mutated proteins and speckle formation after cotransfection with pcDNA-UL34 was comparable to that of wild-type pUL31 or pUL31-N14, respectively (data not shown). Titers derived from stably expressing cell lines documented efficient complementation of PrV-ΔUL31 for most of the mutant proteins (Fig. 7). Only RK13-UL31-N14-T19/21A cells complemented to a ca. 10-fold lower level. However, ultrastructural analyses revealed accumulations of primary virions in the perinuclear space in the PrV-ΔUL31-infected cell lines RK13-UL31-N14-T19A (Fig. 8B), RK13-UL31-N14-T21A (Fig. 8C), and RK13-UL31-N14-T19/21A (Fig. 8D), while they were not obvious in RK13-UL31-N14 (Fig. 8A).
FIG 7.
Site-specific mutation of predicted phosphorylation sites. The predicted phosphorylation sites at positions 12 and 13 were changed in full-length pUL31, giving rise to pUL31-S12/13A (A), or the threonine residues at position 19 and 21 were replaced by alanine in the N-terminally truncated pUL31-N14 mutant singly or in combination (B). Stably expressing cell lines were infected as described in the legend to Fig. 6. Shown are the mean values for three independent experiments with the corresponding standard deviations (*, P < 0.05; ***, P < 0.001).
FIG 8.
Ultrastructural analyses. RK13-UL31-N14 (A), RK13-UL31-N14-T19A (B), RK13-UL31-N14-T21A (C), and RK13-UL31-N14-T19/21A (D) cells were infected with PrV-ΔUL31 at an MOI of 1. Cells were processed for electron microscopy 14 h p.i. Bars indicate 1 μm (A and B) and 500 nM (C and D).
DISCUSSION
Herpesvirus nuclear egress, i.e., the vesicle-mediated translocation of nucleocapsids from the nucleus into the cytoplasm, is still not completely understood. However, the recently published crystal structures for several NECs from different herpesvirus subfamilies (4–7), high-resolution imaging approaches (24, 25), as well as in vitro studies (13, 26) provided detailed insights into how membrane vesicles are formed at and constricted from lipid bilayers. These data showed that the NEC alone drives vesicle formation and scission from lipid membranes by dimerization as well as oligomerization, forming a curved protein coat at the inner nuclear membrane. Major portions of the NEC components, pUL31 and pUL34, are dedicated to dimerization, and stable complexes are formed when coexpressed in bacteria (4). However, for two domains of the NEC components structural information is missing, since they interfered with crystallization, the nonconserved C terminus of the pUL34 homologs, including the transmembrane domain, and the variable, apparently highly flexible N terminus of the pUL31 homologs. Both regions are predicted to be positioned in or close to the inner nuclear membrane and are anticipated to play important regulatory roles.
The PrV pUL31 nonconserved N terminus comprises only 25 amino acids, which is significantly shorter than, e.g., 58 aa in HSV-1 pUL31 and HCMV pUL53 or 108 aa in murine cytomegalovirus (MCMV) M53. More than one-third is composed of basic residues arranged in three stretches of three consecutive basic amino acids (aa 4 to 6, aa 9 to 11, and aa 16 to 18) (Fig. 1A). In silico analyses predicted a bipartite NLS (aa 5 to 20), and efficient nuclear localization for pUL31 was observed after transfection but also infection (16, 17). To test the relevance of this domain for localization and function during nuclear egress, we generated and tested different N-terminally truncated as well as site-specifically mutated variants of PrV pUL31.
pUL31-N11, lacking aa 2 to 10 including two of the three basic patches, was efficiently targeted to the nucleus comparably to the wild-type protein and fully complemented the defect of PrV-ΔUL31. Deletion of an additional three residues (pUL31-N14), including a single basic residue (K11) and two predicted phosphorylation sites (S12/S13) but still retaining three consecutive basic residues, also had no detectable effect on nuclear egress or formation of infectious progeny, indicating that the predicted bipartite NLS is not essential for pUL31 nuclear targeting or function. However, nuclear localization of the protein was not as strict as with the wild-type pUL31, and a significant portion of pUL31-N14 remained cytoplasmic. However, even the reduced amount of nuclear pUL31-N14 was sufficient for interaction with INM-localized pUL34 forming the typical costaining speckles. It also efficiently complemented capsid translocation, as deduced from wild-type-like infectious titers of trans-complemented PrV-ΔUL31. In contrast, pUL31-N18, which lacks any N-terminal basic patch, was nonfunctional and localized nearly exclusively in the cytoplasm. This can be attributed to the leucine-rich nuclear export sequence present in the C terminus (aa 246 to 254), as suggested for PrV pUL31-ΔNLS (pUL31-N26) (17). Mutation of this NES sequence (L252/254A) resulted in the expected diffuse pancellular staining (data not shown).
Neutralization of the remaining basic patch in the functional pUL31-N11 (pUL31-N11-R16/17A) or pUL31-N14 (pUL31-N14-K18A) also resulted in predominant cytoplasmic localization of the proteins and a defect in complementation of PrV-ΔUL31, corresponding to the phenotype found for pUL31-N18. In contrast, restoration of a basic patch in the nonfunctional pUL31-N18 by altering two alanine residues to arginine (pUL31-N18-A23/24R) led to efficient nuclear localization, speckle formation with pUL34 in the nucleus, as well as full complementation of PrV-ΔUL31 with no obvious impairment in nuclear egress or production of infectious progeny. These data highlight the importance of the basic patch in the pUL31 N terminus for proper localization and functional complementation, as has also been observed for HSV-1 pUL31 (12). Further, N-terminal truncation (pUL31-N21-A23/24R) resulted in a noncomplementing phenotype despite partial nuclear localization as well as wild-type-like speckle formation after cotransfection with pcDNA-UL34.
Basic residues, especially lysine and arginine, are frequently found to interact electrostatically with membranes (13). Thus, in addition to the predicted transmembrane domain in pUL34, the basic amino acids in the pUL31 N terminus might support membrane attachment and promote membrane curvature. In line with this, artificially membrane-tethered PrV pUL31 alone is sufficient to form intralumenal vesicles in the giant unilamellar vesicle system (26). It remains to be tested whether the N-terminally truncated and mutated pUL31 variants will also support vesicle formation from synthetic membranes.
For HSV-1 pUL31 it was suggested that the N-terminal domain blocks premature interaction with pUL34 in the cytoplasm and in the nucleus. In the nucleus, pUL31 was shown to be targeted to nucleocapsids, resulting in structural rearrangements allowing pUL34 interaction and, thus, NEC formation at the INM. The basic patch in the N-terminal part seems to be critical for promoting capsid translocation to the budding sites (12). In our studies, however, we did not detect PrV pUL31 on nuclear capsids despite efficient demonstration of the protein at the nuclear envelope and in primary enveloped virions (16).
Besides other functions, activity of the alphaherpesviral protein kinase pUS3 is required for efficient release of capsids from the perinuclear space. In the absence of pUS3, primary virions accumulate in large invaginations of the inner nuclear membrane, but viral titers are only approximately 10-fold reduced, indicating that pUS3 kinase function is not essential for nuclear egress (19, 20, 27). For HSV-1 it was shown that only mutation of all six predicted phosphorylation sites in the nonconserved N-terminal domain of pUL31 resulted in a phenotype similar to that in a pUS3 protein kinase knockout (11). Mutation of all predicted phosphorylation sites in the PrV pUL31 nonconserved region (pUL31-N14-T19/21A) also resulted in accumulations of primary virions in the perinuclear space, while replacement of two serine residues (S12/13) in full-length pUL31 had no detectable effect. Changing the threonine at position 19 or threonine at position 21 or both in pUL31-N14-T19/21A resulted in huge accumulations of primary virions in the perinuclear space. Mutation of all predicted phosphorylation sites resulted in ca. 10-fold-reduced viral titers. Thus, modification of the pUL31 N terminus by phosphorylation is beneficial but not absolutely essential for nuclear egress.
In summary, we show that most of the flexible N terminus of PrV pUL31 that is absent from the NEC crystal structure can be deleted without detectable loss of function during nuclear egress. One basic patch of amino acids is sufficient to efficiently target the protein to and maintain it in the nucleus. Phosphorylation of this domain supports but is not essential for release of the nucleocapsids from the perinuclear space into the cytoplasm.
MATERIALS AND METHODS
Viruses and cells.
PrV laboratory strains Kaplan (PrV-Ka) (22) and PrV-ΔUL31 (16) were used. Viruses were propagated in rabbit kidney (RK13) or RK13-UL31 cells, respectively (16).
Generation of N-terminal truncations and site-specific mutagenesis.
N-terminal truncations were generated by PCR using primers listed in Table 1, with pcDNA-UL31 as the template using Platinum Pfx polymerase (Invitrogen) and with UL31-R (16) as the reverse primer. PCR products were cloned into pcDNA3 via EcoRI sites provided with the primer sequences. Correct cloning was tested by restriction enzyme digestion and sequencing. For site-specific mutations, the QuikChange site-directed mutagenesis kit (Agilent) was used as described previously (17) with primers listed in Table 1 and pcDNA-UL31 as the template. Correct mutagenesis was verified by sequencing.
TABLE 1.
Primers used in this study
| Primera | Sequenceb (5′–3′) |
|---|---|
| UL31-N7 | CACA GAATTC ACA ATG CTC CTG CGG CGC |
| UL31-N11 | CACA GAATTC ACA ATG AAG TCG TCG GCC |
| UL31-N14 | CACA GAATTC ACA ATG GCC GCG CGG CGC |
| UL31-N18 | CACA GAATTC ACA ATG AAG ACG CTG ACG |
| PUL31-R | CTG AAT TCT TCG CGG CGC TCA CGG |
| S12/13A | CTC CTG CGG CGC AAG GCG GCG GCC GCG CGG CGC AAG |
| R16/17A | AAG TCG TCG GCC GCG GCG GCC AAG ACG CTG ACG CGC |
| K18A | GCC GCG CGG CGC GCG ACG CTG ACG CGC |
| T19A | GCG CGG CGC AAG GCG CTG ACG CGC GCG |
| T21A | GC AAG ACG CTG GCG CGC GCG GCC CG |
| T19/21A | GC AAG GCG CTG GCG CGC GCG GCC CG |
| A23/24R | G ACG CTG ACG CGC CGG CGC CGC GAT CGC TAC G |
Only one primer used for each mutagenesis reaction is shown.
Restriction enzyme recognition motifs are shown in italics, artificial start and stop codons are shown in boldface, and altered nucleotides are underlined.
In vitro characterization.
Localization was tested after transfection of the corresponding pUL31 expression vectors with or without pcDNA-UL34 (28) using calcium phosphate coprecipitation (29). Cells were fixed 2 days posttransfection with 3% paraformaldehyde, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline, and incubated with the polyclonal rabbit anti-pUL31 (dilution, 1:500) (16) and an anti-pUL34 monoclonal antibody (4B10C; dilution, 1:20) (30). Bound antibody was detected by Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa 555-conjugated goat anti-mouse IgG (Invitrogen). Images were recorded with a confocal laser scanning microscope (Leica SP5; Germany) and processed by ImageJ (31).
Generation of stably expressing RK13 cell lines.
Stably expressing cell lines were generated by transfection of RK13 cells with the corresponding expression vectors. Two days posttransfection cells were split and selected in medium containing G418 (500 μg/ml). Cell colonies were picked by aspiration and tested by indirect immunofluorescence and Western blotting for pUL31 expression with the monospecific anti-pUL31 serum. For Western blot analyses cell lysates were prepared as described previously and incubated with monospecific anti-pUL31 serum (16) and a monoclonal anti-alpha tubulin antibody (Sigma) as a loading control. Bound antibody was detected by peroxidase-coupled goat anti-rabbit and anti-mouse antibodies and visualized by enhanced chemiluminescence (Clarity Western ECL blotting substrate; Bio-Rad) and recorded in an image analyzer (Bio-Rad, Germany).
In vitro replication studies.
To test for functional complementation, stably expressing cell lines were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 5 and harvested 24 h later. Titers were determined on RK13-UL31 cells. Shown are mean values of at least three independent experiments with corresponding standard deviations. The statistical significance of the differences between infection with PrV-Ka and PrV-ΔUL31 was evaluated by Student's t test.
Ultrastructural analyses.
Cell lines were infected with PrV-ΔUL31 at an MOI of 1 and processed for transmission electron microscopy as described previously (28).
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG ME 854/12-2).
We thank Diana Sydow, Petra Gawol, Karla Günther, Cindy Meinke, and Petra Meyer for technical help and Mandy Jörn for photographic assistance.
REFERENCES
- 1.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 4.Zeev-Ben-Mordehai T, Weberruss M, Lorenz M, Cheleski J, Hellberg T, Whittle C, El Omari K, Vasishtan D, Dent KC, Harlos K, Franzke K, Hagen C, Klupp BG, Antonin W, Mettenleiter TC, Grunewald K. 2015. Crystal structure of the herpesvirus nuclear egress complex provides insights into inner nuclear membrane remodeling. Cell Rep 13:2645–2652. doi: 10.1016/j.celrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walzer SA, Egerer-Sieber C, Sticht H, Sevvana M, Hohl K, Milbradt J, Muller YA, Marschall M. 2015. Crystal structure of the human cytomegalovirus pUL50-pUL53 core nuclear egress complex provides insight into a unique assembly scaffold for virus-host protein interactions. J Biol Chem 290:27452–27458. doi: 10.1074/jbc.C115.686527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lye MF, Sharma M, El Omari K, Filman DJ, Schuermann JP, Hogle JM, Coen DM. 2015. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J 34:2937–2952. doi: 10.15252/embj.201592651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lotzerich M, Ruzsics Z, Koszinowski UH. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J Virol 80:73–84. doi: 10.1128/JVI.80.1.73-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmeiser C, Borst E, Sticht H, Marschall M, Milbradt J. 2013. The cytomegalovirus egress proteins pUL50 and pUL53 are translocated to the nuclear envelope through two distinct modes of nuclear import. J Gen Virol 94:2056–2069. doi: 10.1099/vir.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Jiang S, Wang J, Mo C, Zeng Z, Yang Y, Chen C, Li X, Cui W, Huang J, Peng T, Cai M. 2015. Characterization of the nuclear import and export signals of pseudorabies virus UL31. Arch Virol 160:2591–2594. doi: 10.1007/s00705-015-2527-7. [DOI] [PubMed] [Google Scholar]
- 11.Mou F, Wills E, Baines JD. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J Virol 83:5181–5191. doi: 10.1128/JVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funk C, Ott M, Raschbichler V, Nagel CH, Binz A, Sodeik B, Bauerfeind R, Bailer SM. 2015. The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog 11:e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherry MR, Hay TJM, Gulak MA, Nassiri A, Finnen RL, Banfield BW. 2017. The herpesvirus nuclear egress complex component, UL31, can be recruited to sites of DNA damage through poly-ADP ribose binding. Sci Rep 7:1882. doi: 10.1038/s41598-017-02109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaho JA, Mitchell C, Roizman B. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem 269:17401–17410. [PubMed] [Google Scholar]
- 16.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol 76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paßvogel L, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. 2015. Functional characterization of nuclear trafficking signals in pseudorabies virus pUL31. J Virol 89:2002–2012. doi: 10.1128/JVI.03143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai M, Chen D, Zeng Z, Yang H, Jiang S, Li X, Mai J, Peng T, Li M. 2016. Characterization of the nuclear import signal of herpes simplex virus 1 UL31. Arch Virol 161:2379–2385. doi: 10.1007/s00705-016-2910-z. [DOI] [PubMed] [Google Scholar]
- 19.Wagenaar F, Pol JM, Peeters B, Gielkens AL, de Wind N, Kimman TG. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol 76(Part 7):1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 20.Klupp BG, Granzow H, Mettenleiter TC. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J Gen Virol 82:2363–2371. doi: 10.1099/0022-1317-82-10-2363. [DOI] [PubMed] [Google Scholar]
- 21.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A 104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 23.Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 24.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bauerlein FJ, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grunewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb WW, Fontana J, Winkler DC, Cheng N, Heymann JB, Steven AC. 2017. The primary enveloped virion of herpes simplex virus 1: its role in nuclear egress. mBio 8:e00825-17. doi: 10.1128/mBio.00825-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz M, Vollmer B, Unsay JD, Klupp BG, Garcia-Saez AJ, Mettenleiter TC, Antonin W. 2015. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J Biol Chem 290:6962–6974. doi: 10.1074/jbc.M114.627521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol 76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. [DOI] [PubMed] [Google Scholar]
- 30.Ronfeldt S, Klupp BG, Franzke K, Mettenleiter TC. 6 September 2017. Lysine 242 within helix 10 of the pseudorabies virus nuclear egress complex pUL31 component is critical for primary envelopment of nucleocapsids. J Virol doi: 10.1128/JVI.01182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]