Several members of the family Flaviviridae, including HCV, have adapted cap-independent translation strategies to overcome canonical eukaryotic translation pathways and use cis-acting RNA-elements, designated viral internal ribosome entry sites (IRES), to initiate translation. Although novel hepaciviruses have been identified in different animal species, only limited information is available on their biology on molecular level. Therefore, our aim was a fundamental analysis of BovHepV IRES functions. The findings which show that functional IRES elements are also crucial for BovHepV translation expand our knowledge on molecular mechanism of hepacivirus propagation. We also studied the possible effects of one major host factor implicated in HCV pathogenesis, miR-122. The results of mutational analyses suggested that miR-122 enhances virus translation mediated by BovHepV IRES.
KEYWORDS: bovine hepacivirus, 5′ nontranslated region, internal ribosome entry site, microRNA-122
ABSTRACT
In recent years, hepatitis C virus (HCV)-related viruses were identified in several species, including dogs, horses, bats, and rodents. In addition, a novel virus of the genus Hepacivirus has been discovered in bovine samples and was termed bovine hepacivirus (BovHepV). Prediction of the BovHepV internal ribosome entry site (IRES) structure revealed strong similarities to the HCV IRES structure comprising domains II, IIIabcde, pseudoknot IIIf, and IV with the initiation codon AUG. Unlike HCV, only one microRNA-122 (miR-122) binding site could be identified in the BovHepV 5′ nontranslated region. In this study, we analyzed the necessity of BovHepV IRES domains to initiate translation and investigated possible interactions between the IRES and core coding sequences by using a dual luciferase reporter assay. Our results suggest that such long-range interactions within the viral genome can affect IRES-driven translation. Moreover, the significance of a possible miR-122 binding to the BovHepV IRES was investigated. When analyzing translation in human Huh-7 cells with large amounts of endogenous miR-122, introduction of point mutations to the miR-122 binding site resulted in reduced translation efficiency. Similar results were observed in HeLa cells after substitution of miR-122. Nevertheless, the absence of pronounced effects in a bovine hepatocyte cell line expressing hardly any miR-122 as well suggests additional functions of this host factor in virus replication.
IMPORTANCE Several members of the family Flaviviridae, including HCV, have adapted cap-independent translation strategies to overcome canonical eukaryotic translation pathways and use cis-acting RNA-elements, designated viral internal ribosome entry sites (IRES), to initiate translation. Although novel hepaciviruses have been identified in different animal species, only limited information is available on their biology on molecular level. Therefore, our aim was a fundamental analysis of BovHepV IRES functions. The findings which show that functional IRES elements are also crucial for BovHepV translation expand our knowledge on molecular mechanism of hepacivirus propagation. We also studied the possible effects of one major host factor implicated in HCV pathogenesis, miR-122. The results of mutational analyses suggested that miR-122 enhances virus translation mediated by BovHepV IRES.
INTRODUCTION
Recently, a novel HCV related virus was identified in bovine serum samples and was termed bovine hepacivirus (BovHepV). Like HCV, it belongs to the family Flaviviridae and is capable of establishing persistent infections (1). Taking into account the identification of a growing number of HCV related viruses in various animal species, including dogs, horses, rodents, bats, and cattle, an update of the taxonomy of hepaciviruses has been recently proposed which included the suggestion to allocate the BovHepVs to the species Hepacivirus N (2, 3). HCV is one of the leading causes for chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (4). Although no indications for clinical implication of BovHepV infections have been noticed so far, high levels of BovHepV RNA in the bovine liver are suggestive of a liver tropism (1). Like HCV and other members of the genus Hepacivirus, the genome of BovHepV consists of a single-stranded positively orientated RNA organized as one large open reading frame of 8,340 nucleotides (nt) with a 5′ and a 3′ nontranslated region (NTR) (1). It is assumed that mature structural and nonstructural proteins are a result of cleavage through host signal peptidases and viral proteases. The 5′ NTR of pestiviruses, pegiviruses, and hepaciviruses, including hepatitis C virus (HCV), GBV-B, and nonprimate hepacivirus (NPHV), contains an internal ribosome entry site (IRES) enabling 5′ cap-independent translation of viral proteins (5–7). The first step of eukaryotic translation initiation involves recognition of the 5′ cap structure by the eukaryotic translation initiation factors (eIFs) elF4F and elF4A (8). However, the translation initiation pathway mediated by HCV is eIFs independent as the ribosomal subunit 40S can directly bind the IRES through a high-affinity interaction (9). Regarding HCV and NPHV, translation of viral RNA is further regulated by host factors, especially miR-122 (10, 11). MicroRNAs (miRNAs) are short, single-stranded RNAs, generally 19 to 24 bases in length, which negatively regulate translation of cellular messenger RNAs (mRNAs) (12). miR-122 is highly expressed in the human liver and, contrary to its usual function, increases the translation efficiency of HCV by binding at two seed match sites in the IRES sequence. This interaction protects the noncapped viral RNA against degradation by host exoribonucleases (13). Other functions of miR-122 include stabilization of the viral genome, as well as enhancement of viral replication and support in production of infectious particles (14–16). Moreover, miR-122 interferes with a long-range interaction of the HCV 5′ NTR and an inhibitory core sequence leading to an open conformation of the 5′ NTR which stimulates viral translation. In the absence of miR-122, this long-range interaction would have a negative effect on translation (17). Double mutations within the two HCV miR-122 seed match sites lead to decreased RNA accumulation. More evidence that miR-122 stimulates translation was provided through depletion of Ago2, resulting in decreased HCV translation (18–20). Among vertebrates, the sequence of mature miR-122 is fully conserved (21). Considering the hepatotropism of BovHepV (1) and the fact that miR-122 is also highly expressed in the bovine liver, it is reasonable to hypothesize a crucial involvement of miR-122 in the BovHepV life cycle. Additional nucleotide interactions involved in regulating HCV translation were identified. Stable RNA structures directly downstream of the initiation codon have an adverse impact on translation, whereas destabilization of domain IV was shown to be essential for interactions of 40S subunits with viral RNA. Thereby, an adenosine-rich part of hepacivirus core encoding sequences is capable of mediating destabilization of domain IV (22). BovHepV shares strong similarities with other hepaciviruses regarding genomic organization, but functions of the viral 5′ NTR with respect to translation of the viral polyprotein have not been investigated so far. Therefore, the aim of this study was to identify essential parts of the bovine hepacivirus IRES sequence required for efficient translation. Moreover, the impact of potential interactions between the BovHepV 5′ NTR and core coding sequences, as well as consequences of miR-122 binding for viral translation, was investigated here.
RESULTS
Sequence analyses and modeling of BovHepV IRES secondary structure.
The results of the RACE (rapid amplification of cDNA ends)-PCR revealed that the BovHepV strains investigated here comprise a 5′ NTR that is 294 nt in length. The 5′ NTR sequences showed 98.6% to 100% nucleotide identity but only 51.0% to 52.6% identity to the 5′ NTR of HCV genotype 1 (GenBank accession number AF009606) (23).
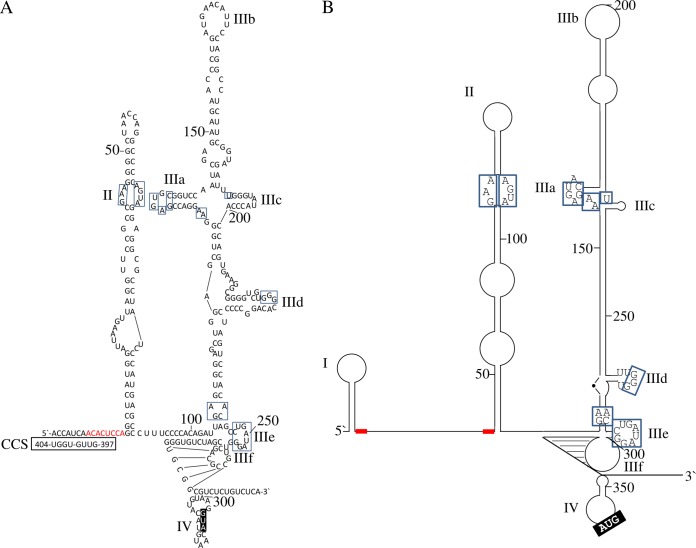
By using the mfold algorithm to determine the highest free enthalpy of folded IRES structures, three major domains strongly resembling HCV IRES domains II to IV were identified in the BovHepV 5′ NTR (Fig. 1). Compared to HCV, the BovHepV 5′ NTR lacks domain I. Also, domain II of BovHepV does not comprise the basal loop characteristic for domain II of HCV, but apart from this they are similar in structure and share several conserved nucleotides. Furthermore, the structure of BovHepV domain III resembles domain III of the HCV IRES comprising six subdomains IIIa to IIIf, including conserved nucleotides at corresponding positions. According to the predicted RNA secondary structure model, BovHepV domain IIIf forms a pseudoknot, while the initiation codon (AUG) is situated in domain IV. At least four 5′-terminal nucleotides of the core protein coding sequence contribute to formation of domain IV. In contrast to HCV, the IRES of BovHepV contains only one putative miR-122 binding site (nt 8 to 15), which corresponds to the first miR-122 binding site in the HCV IRES.
FIG 1.
Overview of BovHepV (A) and HCV (B) 5′ NTR predicted RNA secondary structures (29, 32, 42). Blue, highly conserved nucleotides among BovHepV and HCV; red, miR-122 match sites; black boxed nucleotides, translation initiating codon (AUG). Potential long-range interaction between BovHepV 5′ NTR nt 1 to 9 and nt 397 to 404 of the core protein coding sequence are depicted. CCS, core coding sequence. NTR, nontranslated region.
All predicted domains of BovHepV IRES are important for translation initiation, whereas incomplete domain IV enhances translation efficiency.
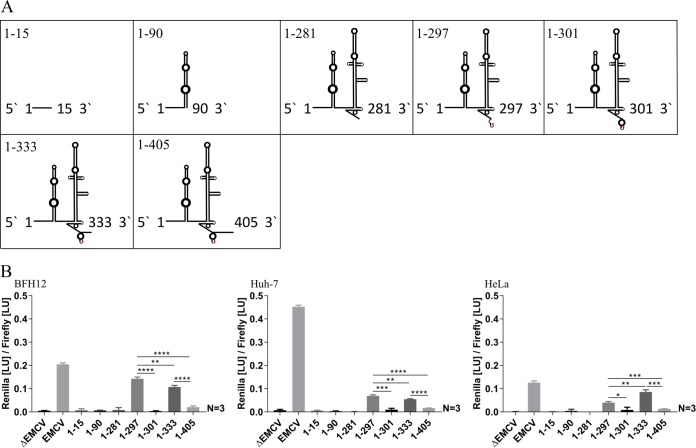
In the dual luciferase reporter system used here, renilla light units are directly correlated to the ability and efficiency of partial and complete IRES elements to initiate translation and are normalized to firefly luciferase light units. All constructs analyzed in this study are described in detail in Fig. 2. First of all, proof of principle was given. Three different hepatic/nonhepatic cell lines of either bovine or human origin were utilized to resolve differences in translation ability depending on host or tissue. The wild-type plasmid containing the type II encephalomyocarditis virus IRES sequence (EMCV) was able to initiate translation best in Huh-7 cells. In contrast, no translation was observed in any of the cell lines following transfection with vectors not containing any IRES structure (ΔEMCV) (Fig. 3). Regarding BovHepV, it could be shown that all domains of the IRES structure are needed for efficient translation, since only very little renilla luciferase activity was observed after transfection of cells with constructs 1-15 (lacking domains II to IV), 1-90 (lacking domains III and IV), and 1-281 (lacking domain IV), which do not encompass the complete BovHepV IRES structure. Remarkably, transfection with the construct 1-297, where complete formation of domain IV was abrogated, significantly increased translation in all three cell lines. Thereby, this construct was most effective to initiate translation in BFH12 cells (Fig. 3). Moreover, two constructs, 1-333 and 1-405, containing partial core coding sequence were analyzed. Construct 1-333 comprises the complete BovHepV 5′ NTR, followed by the 5′ terminal 39 nt of the open reading frame. Measurement of luciferase light units following transfection with this plasmid, which also includes an adenosine-rich core protein coding sequence (nt 318 to 331), demonstrated similar translation activation capacities as construct 1-297. Furthermore, a possible negative influence of a downstream core coding sequence on translation was analyzed. The construct 1-405 which contains a part of the core coding sequence (nt 397 to 404) predicted to interact with the IRES sequence at nt 1 to 9 (see Fig. 1A) showed significantly decreased translation efficiency in all three cell lines in comparison to constructs 1-333 and 1-297 (Fig. 3).
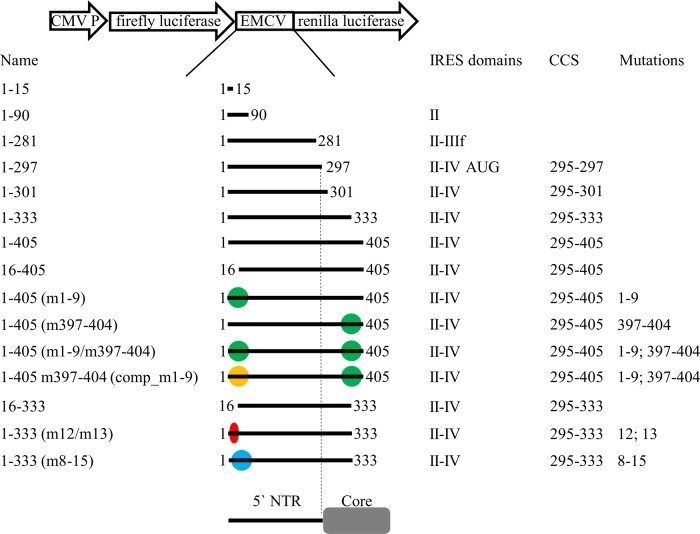
FIG 2.
Illustration of the essential parts of plasmid pF/R_EMCV. EMCV sequence flanked by coding sequences of firefly and renilla luciferases was replaced by different BovHepV sequences. The names of the resulting constructs and the lengths of the BovHepV genome sequences are given. Colored dots indicate mutations introduced in the BovHepV genomic sequence. Green, mutations of complementary sequences in the 5′ NTR (nt 1 to 9), respectively, core coding sequence (nt 397 to 404); yellow, mutations in the 5′ NTR (nt 1 to 9) complementary to the mutated sequence in the core coding sequence (nt 397 to 404); red, point mutations in the 5′ NTR miR-122 match site (nt 12 and 13); blue, mutation of the complete miR-122 match site in the 5′ NTR. Enclosed IRES domains, the lengths of included partial core coding sequences, and the positions of mutations are displayed. CMV P, cytomegalovirus promoter; EMCV, encephalomyocarditis virus IRES sequence; m, mutation; comp, compensation; CCS, core coding sequence. NTR, nontranslated region.
FIG 3.
(A) Schematic diagrams of investigated BovHepV 5′ NTR secondary structures. Red, translation initiation codon (AUG). (B) Luciferase activities at 48 h posttransfection in BFH12, Huh-7, and HeLa cells. Plasmid pF/R_EMCV (EMCV) was used as a positive control, and plasmid lacking the EMCV IRES sequence (ΔEMCV) was used as a negative control. The naming of constructs denotes BovHepV nucleotide sequences used in the experiment. Constructs: construct 1-15, BovHepV nt 1 to 15 were included; construct 1-90, BovHepV sequence information starting from nt 1 to the end of domain II was included; construct 1-281, contains virus sequence from nt 1 to the end of domain IIIf; construct 1-297, BovHepV sequence information starting from nt 1 up to translation initiation codon AUG was included; construct 1-301, contains viral sequence from nt 1 to the end of domain IV. Furthermore, two constructs 1-333 and 1-405 harboring partial core coding sequences were analyzed. EMCV, encephalomyocarditis virus IRES sequence; NTR, nontranslated region.
Influence of core protein coding sequences on translation activity.
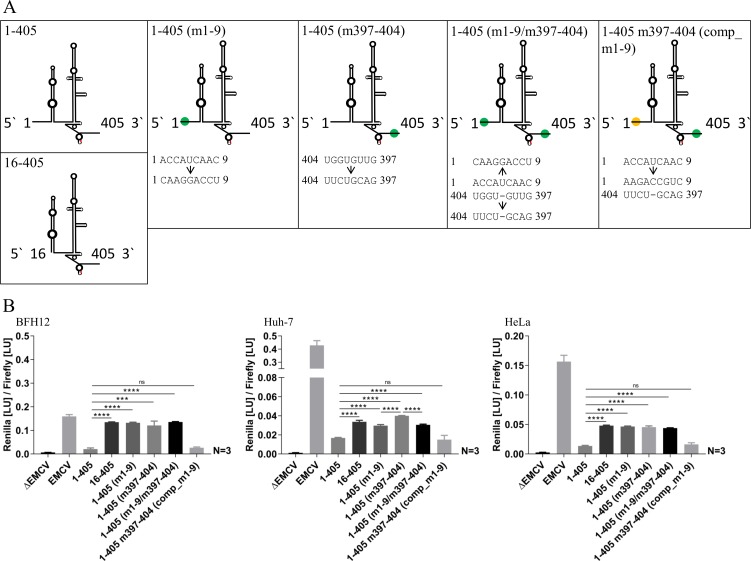
Using construct 16-405, we investigated whether the absence of such interactions with IRES sequence would positively influence translation. Prevention of the putative interaction of nucleotides 1 to 9 with core protein coding sequence restored translation ability in all three cell lines. Furthermore, the nucleotide stretches potentially involved in this process were mutated individually, resulting in mutants 1-405 (m1-9) and 1-405 (m397-404), or in combination, leading to plasmid 1-405 (m1-9/m397-404) (Fig. 4). Analyses of all three mutated constructs revealed significantly enhanced initiation of translation compared to plasmid 1-405 in all investigated cell lines. Subsequently, nucleotides (1 to 9) were mutated to compensate previously introduced mutations in the core coding sequence by using 1-405 (m397-404) as the template. Transfection of the resulting construct 1-405 m397-404 (comp_m1-9) led to a significantly decreased translation initiation in all three cell lines which was comparable to construct 1-405 (Fig. 4).
FIG 4.
(A) Schematic diagrams of investigated BovHepV 5′ NTR secondary structures and mutations on the nucleotide level. Red, translation initiation codon (AUG); green and yellow dots, mutated regions in the 5′ NTR and the core coding sequence. (B) Luciferase activities at 48 h posttransfection in BFH12, Huh-7, and HeLa cells. Plasmid pF/R_EMCV (EMCV) was used as a positive control, and plasmid without the EMCV IRES sequence (ΔEMCV) was used as a negative control. The following plasmids with different partial sequences of BovHepV were transfected: 1-405, virus sequence from nt 1 to 405 (including 111 nt of the core coding sequence); 16-405, BovHepV sequence starting from domain II up to nt 405; 1-405 (m1-9), BovHepV sequence from nt 1 to 405 with mutated IRES sequence (nt 1 to 9); 1-405 (m397-404), BovHepV sequence from nt 1 to 405 with mutated IRES sequence (nt 397 to 404); 1-405 (m1-9/m397-404), BovHepV sequence from nt 1 to 405 containing mutated IRES sequence information at two sites (nt 1 to 9 and nt 397 to 404); 1-405 m397-404 (comp_m1-9), BovHepV sequence from nt 1 to 405 with mutated IRES sequence (nt 397 to 404), which contains additional compensatory mutations of nt 1 to 9. For enhanced illustration of the results, uniform scaling was not used in panel B. EMCV, encephalomyocarditis virus IRES sequence; NTR, nontranslated region.
In the course of this experiment, lower translation efficiencies were observed in Huh-7 when mutants with altered nucleotides 1 to 9 were transfected, illustrated by decreased luciferase activity after transfection of constructs 1-405 (m1-9) and 1-405 (m1-9/m397-404) compared to construct 1-405 (m397-404) (Fig. 4).
miR-122 expression levels in bovine liver tissue and permanent cell lines.
In bovine liver tissue, constantly high expression levels of miR-122 mirrored by cycle threshold (CT) values varying between 25.03 and 25.80 based on 10 ng of total RNA were measured. Significant differences were observed in the three cell lines regarding expression levels of miR-122. In Huh-7 cells, miR-122 was highly abundant (mean CT = 21.40). Extremely small amounts of miR-122 were found in HeLa (mean CT = 32.70), whereas miR-122 was virtually not detected in BFH12 cells (mean CT = 38.68). Normalization of miR-122 to U6 snRNA confirmed very low expression levels of miR-122 in HeLa and BFH12 cells (Table 1).
TABLE 1.
Relative quantification of miR-122 in Huh-7, BFH12, and HeLa cells
| Cell line | Mean CT ± SD/10 ng of total RNA |
Expression fold change compared to HuH-7 cells (2−ΔΔCT) | |
|---|---|---|---|
| miR-122 | U6 snRNA | ||
| Huh-7 | 21.40 ± 0.17 | 20.81 ± 0.02 | 1 |
| BFH12 | 38.68 ± 1.15 | 26.42 ± 0.07 | 3.07 × 10−4 |
| HeLa | 32.70 ± 0.22 | 20.49 ± 0.05 | 3.18 × 10−4 |
Influence of miR-122 abundance on translation activity.
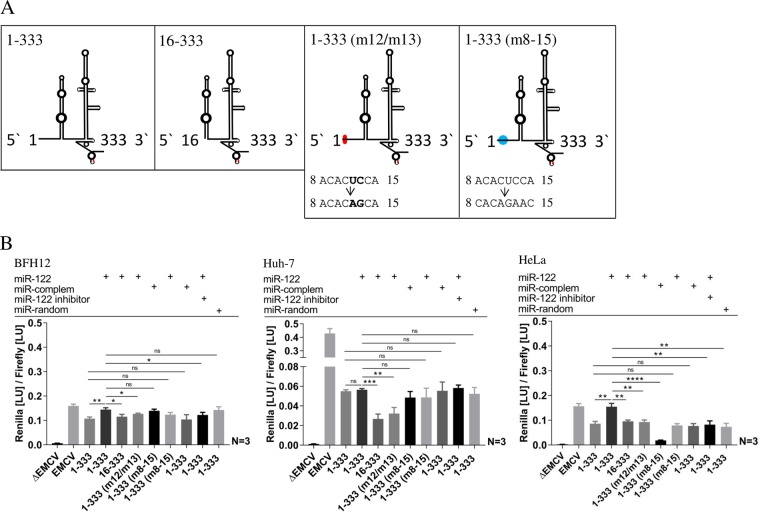
To investigate the impact of the putative BovHepV miR-122 binding site (nt 8 to 15) on translation activity, the effects of complete deletion and the consequences of partial or complete mutation of this sequence in the BovHepV genome were analyzed. Furthermore, cotransfections with miR-122, a complementary miR, miR-122 inhibitor, or a random miR were performed. The translation activity increased slightly in BFH12 cells and was more pronounced in HeLa cells after simultaneous transfection of miR-122 and 1-333 compared to simultaneous transfection of miR-122 and construct 16-333, where the miR-122 binding site was deleted or compared to transfection of 1-333 alone. In contrast, translation could not be enhanced in Huh-7 cells following transfection with plasmid 1-333, together with miR-122. Mutations of nucleotides 12 and 13 within the miR-122 match site of construct 1-333 (m12/m13) cotransfected with miR-122 lead to slightly decreased translation levels in BFH12 and to a marked decrease in the translation in Huh-7 and HeLa cells compared to cotransfection of parental construct 1-333 supplemented with miR-122 (Fig. 5).
FIG 5.
(A) Schematic diagrams of investigated BovHepV 5′ NTR secondary structures and mutations on nucleotide level. Red, translation initiation codon (AUG); blue and red dots, mutated regions in the 5′ NTR. (B) Plasmid pF/R_EMCV (EMCV) was used as a positive control, and plasmid without the EMCV IRES sequence (ΔEMCV) was used as a negative control. Constructs: 1-333, BovHepV sequence information from nt 1 to 333 (including 39 nt of the core coding sequence); 16-333, BovHepV sequence starting from domain II to nt 333; 1-333 (m12/m13), BovHepV sequence from nt 1 to 333 with mutated IRES sequence at positions 12 and 13; 1-333 (m8-15), sequence from nt 1 to 333 with a mutated miR-122 match site from nt 8 to 15. Cotransfections with miR-122, a complementary miR to mutated sequence of 1-333 (m8-15), an miR-122 inhibitor, or a random miR are indicated by plus signs. EMCV, encephalomyocarditis virus IRES sequence; NTR, nontranslated region.
Furthermore, the complete miR-122 match site was mutated, and the resulting construct 1-333 (m8-15) was simultaneously transfected with a microRNA complementary to this mutated sequence replacing the miR-122 match site. Enhanced translation activation capacity similar to translation levels after cotransfection construct 1-333 and miR-122 could be observed in BFH12 and Huh-7 cells. Surprisingly, this effect was not seen in HeLa cells.
Cotransfections of mutant 1-333 (m8-15) with wild-type miR-122 or of the parental plasmid with microRNA duplexes complementary to the mutated nucleotide stretch in mutant 1-333 (m8-15) could not eventually resolve this issue. In BFH12 and HeLa cells, the translation levels did not differ from those where 1-333 was utilized without miR-122, excluding unspecific stimulation of miR-122. However, translation was not impaired in Huh-7 cells following the transfection of construct 1-333 (m8-15) along with miR-122.
Next, 1-333 was transfected simultaneously with miR-122 and miR-122 inhibitor. A pronounced decrease in translation was seen in HeLa cells, whereas a weak decrease could be observed in BFH12 cells. Compared to cotransfection of 1-333 with miR-122, no significant difference was noted in Huh-7 cells. Finally, cotransfection of the same plasmid with a random microRNA did not affect the translation activity in BFH12 or Huh-7 cells but suppressed translation compared to cotransfection with miR-122 in HeLa cells (Fig. 5).
DISCUSSION
Initiation of eukaryotic translation is a protein-mediated procedure and requires canonical eukaryotic initiation factors (eIFs) and 5′-capped mRNA (24, 25). In contrast, several viruses, including HCV, pestiviruses, and picornaviruses, have evolved alternative strategies to initiate translation utilizing cis-acting RNA sequences (26). During the initiation of translation in HCV infection, the 48S complex formation does not require a 5′-cap structure, and the interaction with the 40S ribosomal subunit takes place in the absence of any eukaryotic initiation factors. Instead, translation initiation is mediated by the viral IRES structure comprising domains II to IV and a few 5′-terminal nucleotides of the core protein coding sequence (27).
The bovine hepacivirus exhibits a very similar IRES structure compared to the HCV IRES, which was preliminarily described in an earlier study (28). Accordingly, domains II and III, a pseudoknot formed by IIIf and domain IV, which includes the initiation codon, as well as the first four nucleotides of the core coding sequence, could be identified, while a domain corresponding to HCV IRES domain I is not present in the BovHepV 5′ NTR sequence. In agreement with HCV, BovHepV contains an asymmetric loop in domain II. This loop is important for HCV IRES function because deletion leads to decrease translation efficiency (29). Domain III presents the core of HCV IRES and is part of a pseudoknot structure essential for IRES activity (30). Furthermore, a GGG triplet in stem-loop IIId, which has been reported to be essential for the HCV IRES activity, is also present in the BovHepV IRES (31). Additional highly conserved IRES nucleotides among HCV genotypes are present in the BovHepV IRES structure, underlining a close relationship of the two viruses. HCV stem-loop IV is not essential for IRES activity, but its stability is inversely correlated with translation efficiency (32), implying an equilibrium between folded and unfolded conformation, that is important for controlling interactions of ribosome subunits with viral RNA (33). Regarding the observed structural similarity of the BovHepV and HCV 5′ NTRs, it appears reasonable to assume that comparable mechanistic patterns play a role in the BovHepV life cycle as well.
After analyzing the secondary structure of BovHepV IRES, we studied the function of individual IRES domains and possible interactions between IRES and core coding sequences implicated in translation efficiency, as well as a possible impact of miR-122 on viral translation by using a dual luciferase reporter system. All experiments were performed in three different cell lines of human and bovine origin. Although analyses of individual firefly luciferase light units suggested a slightly enhanced uptake of plasmids by Huh-7 cells compared to BHF12 and HeLa cells (mean values of luciferase units exemplarily given for the data set presented in Fig. 3 were as follows: BFH12, 6.77 × 106; Huh-7, 1.61 × 107; and HeLa, 8.58 × 106), the variability of luciferase ratios observed for the cell lines used correlated with differences in IRES types and structures (EMCV versus BovHepV). While the type II IRES of EMCV initiated translation with highest efficiency in Huh-7 cells, the BovHepV IRES-mediated translation was most effective in the bovine liver cell line BFH12, suggesting adaptation to the bovine host. It was demonstrated that all BovHepV IRES domains are necessary to initiate translation. Interestingly, the construct harboring the IRES sequence, including the initiation codon (1-297), showed significantly higher translation efficiency than the construct which contains the entire domain IV (1-301), indicating mechanistic patterns similar to those previously described for HCV. Although stem-loop IV itself is not required for internal entry of ribosomes, it is able to regulate this process. It has been suggested that stem-loop IV may be stabilized by interactions with viral proteins during viral infection, which constitutes a mechanism for feedback regulation of translation and may be important for viral persistence (32). A similar control mechanism has been described for the translation of bacteriophages (34).
HCV comprises two interaction sites between the IRES and two different short nucleotide sequences situated within the core coding region. The first interaction between domain IV and nt 357 to 372 contributes to the destabilization of domain IV, which enhances translation (22). A second long-range interaction between nt 24 to 38 of HCV IRES overlapping in part with the first miR-122 binding site and nt 428 to 442 of the HCV genome leads to decreased viral translation efficiency (35). We hypothesized that similar interactions might be implicated in BovHepV translation. The first putative interaction sites comprise domain IV of BovHepV and an adenosine-rich part between nt 318 and 331 of the core protein coding sequence corresponding to the interaction of domain IV and nt 357 to 372 of HCV (22). To investigate a possible destabilizing effect of this adenosine-rich part on domain IV, we analyzed translation after the transfection of construct 1-333. In comparison to construct 1-301, which includes the complete IRES structure along with domain IV, a significant increase in translation efficiency was observed, supporting the assumption that destabilization of domain IV is required for effective translation in BovHepV infection as well.
In addition, it was hypothesized that a second long-range interaction between nt 1 to 9 of the BovHepV IRES sequence and nt 397 to 404 corresponding to the interaction between nt 24 to 38 and nt 428 to 442 of HCV might influence translation. Analysis of construct 1-405 mimicking the assumed long-range interactions between the core coding sequence and the 5′ NTR revealed decreased translation capacities. Abolishing this process by introducing mutations at respective interaction sites or by deletion of the anterior nucleotide sequence resulted in restored translation activity, confirming the regulatory mechanisms of virus translation initiation. These results are supported by several previous reports (31, 35). Honda and coworkers showed that a six-nucleotide deletion spanning nt 32 to 37 of the HCV core coding sequence and replacement of the AG dinucleotide at positions 34 and 35 with GA enhanced translational activity. Moreover, it was demonstrated by frameshift mutation that the nucleotide sequence rather than the amino acid sequence of the core protein determined translation efficiency (36). Compensatory mutations introduced at positions 1 to 9 of BovHepV again led to suppressed translation strengthening the concept of an inhibitory downstream core coding sequence. The importance of conserved nucleotides implicated in RNA secondary structures in this region which regulate HCV replication and translation has been discussed earlier (37).
HCV translation is further regulated by host microRNA-122, which is highly expressed in the liver. Binding of miR-122 at seed match sites in the IRES sequence of HCV is supposed to upregulate viral translation (38). According to our results, we hypothesized that the presence of miR-122 might influence the inhibitory long-range interactions between 5′ NTR and core sequences mentioned above. BovHepV constructs with an altered posterior long-range interaction site not involving any changes in the vicinity of the miR-122 seed match sequence were able to initiate translation more efficiently than constructs with mutated nt 1 to 9, which overlap the miR-122 seed match site. These differences were only observed in Huh-7 cells, a liver-derived human cell line which, according to quantitative real-time PCR, also strongly expresses miR-122. This indicates that miR-122 binding enhances translation provided that its annealing site in the BovHepV genome remains unchanged. In BFH12 and HeLa cells, translation could be stimulated by cotransfection of miR-122 duplexes, whereas the addition of miR-122 had no beneficial effect in Huh-7 cells, most probably due to a high endogenous miR-122 expression level.
To further investigate whether translation of BovHepV is dependent on the presence of miR-122 and its seed match sequence in the viral genome, the constructs 16-333 lacking the entire miR-122 binding site and 1-333 (m12/m13) carrying point mutations at positions 12 and 13 of the BovHepV genome were analyzed. A strong decrease in renilla luciferase activity was observed in Huh-7 and HeLa cells and to a lesser extent also in BFH12 cells, confirming previous studies where altering the corresponding nucleotide positions of the second HCV miR-122 seed match site led to reduced translation (20). The assumption that miR-122 is involved in translation was further investigated through mutation of the complete miR-122 binding and cotransfection with an equivalent complementary miR. Thereby, translation could be enhanced in BFH12 and Huh-7 cells, but surprisingly, translation intensity in HeLa cells did not increase. As expected, mutant miR-122 duplexes matching construct 1-333 (m8-15) did not promote the translation of wild-type plasmid 1-333 and wild-type miR-122 did not enhance the translation of mutant 1-333 (m8-15) in BFH12 and HeLa cells. However, translation of this mutant could not be suppressed in Huh-7 cells where miR-122 is highly abundant. Whether cellular components such as other small RNAs might mimic the stimulatory functions of miR-122 and are involved in this process remains to be determined.
Furthermore, very high endogenous miR-122 concentrations in Huh-7 cells seem to outcompete sequestering of miR-122 with antagomir. Accordingly, transfection with random miR also had no effect on translation in this cell line. However, in HeLa cells, translation was strongly impaired in both cases. In BFH12 cells, only minor changes in translation capacities were observed. Obviously, BFH12 cells lost their ability of miR-122 expression during the immortalization process, which is a well-known phenomenon of permanent hepatocytes (15, 39). Mutational analysis of the seed match site revealed only a minor influence of miR-122 abundance and properties of the binding site on translational effectiveness. However, large amounts of miR-122 in bovine liver tissue point at a pivotal role in the BovHepV life cycle. Other functions of miR-122, including protection of the 5′ end against Xrn1 decay and stimulation of RNA replication, will be targeted in future studies. Taken together, our results reveal functional IRES elements in the 5′ NTR of bovine hepacivirus and point to an involvement of liver-specific miR-122 in hepacivirus translation. Future studies will concentrate on the role of miR-122 on RNA stability and virus replication using in vitro-transcribed RNAs and replicons and will clarify the role of BovHepV for liver disease of cattle.
MATERIALS AND METHODS
Determination of BovHepV 5′ NTR sequence and structure.
Complete 5′ NTR sequences of five BovHepV variants from Germany (GenBank accession numbers KP641123 to KP641127) were determined. RNA was isolated from serum samples using the QIAmp viral RNA minikit (Qiagen, Hilden, Germany). The 5′-terminal sequences were resolved by using an RACE DNA purification system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Thereby, two BovHepV-specific primers (GSP1, 5′-AGCCGCACCCACAGAT-3′; GSP2, 5′-CCCTATCAGGCTGTGTGGACTAG-3′) were used in the 5′ RACE procedure. The RNA secondary structure of the BovHepV 5′ NTR “463” (GenBank accession number KP641127) was predicted using mfold bioinformatic software (40). A highly similar RNA secondary structure was predicted by the software program sfold (http://sfold.wadsworth.org).
miR-122 expression levels in permanent cell lines and liver tissue.
Human epithelioid cervix carcinoma cells (HeLa) and human hepatocarcinoma cells (Huh-7) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Bovine fetal hepatocytes (BFH12) (41) were maintained in Williams' medium E (Biochrom GmbH, Berlin, Germany) with 5% fetal bovine serum, 50,000 U of penicillin (Sigma-Aldrich, St. Louis, MO), 50 mg/500 ml streptomycin (Sigma-Aldrich), 2 mM l-alanyl-l-glutamine (Biochrom GmbH), 100 nM dexamethasone (Sigma-Aldrich), and an insulin solution (4.27 mg/500 ml) from bovine pancreas (Sigma-Aldrich). Liver tissue samples were derived from BovHepV-negative tested animals from the Institute of Pathology, University of Veterinary Medicine Hannover. To measure the miR-122 expression level, 106 cells per cell line were lysed with lysis/binding solution (Life Technologies, Carlsbad, CA), and total RNA was isolated with a mirVana miRNA isolation kit (Invitrogen) according to the manufacturer's instructions. Total cellular RNA was extracted from liver tissues from a calf and adult cattle with QIAzol and an RNeasy minikit (Qiagen). The concentration of total RNA was determined spectrophotometrically (NanoDrop 2000; Thermo Scientific, Waltham, MA), and 10 ng of total RNA was applied in each reaction. Reverse transcription and quantitative PCRs were performed in triplicates with a TaqMan MicroRNA reverse transcription kit and TaqMan Universal Master Mix II (no UNG) and TaqMan MicroRNA assays (Applied Biosystems, Foster City, CA). The relative quantification of miR-122 amounts was accomplished by amplification of U6 small nuclear RNA.
Cloning strategy and preparation of constructs.
The plasmid pF/R_wt_EMCV, which includes the sequences of the firefly luciferase genome, encephalomyocarditis virus (EMCV) IRES, and renilla luciferase genome (named “pEMCV”in the following), was used as the template to generate all constructs used in this study (Fig. 2). First, the EMCV IRES sequence downstream of the firefly luciferase coding sequence was replaced by nt 1 to 409 of the BovHepV genome from animal 463 (KP641127). Then, the primers Megaprimer_fwd and Megaprimer_rev were used to generate a 459-bp-spanning amplicon consisting of BovHepV nt 1 to 409 flanked by 22 nucleotides of the 3′ region of the firefly luciferase genome and 28 nucleotides of the 5′ end of the genome encoding the renilla luciferase (Table 2). We used 8 μl of this amplicon and 100 ng of the plasmid pEMCV in a second PCR, followed by DpnI digestion and transfection of Top10 Escherichia coli cells. Ampicillin-resistant clones were selected after colony PCR and were grown in Luria-Bertani medium. Plasmids were prepared from 5-ml cultures, and plasmid integrity was confirmed by Sanger sequencing (LGC Genomics, Berlin, Germany). The resulting plasmid 1-409 included BovHepV nt 1 to 409 flanked by the sequences encoding the firefly and renilla luciferases, respectively. Subsequently, further constructs were obtained by PCR in which the binding primers excluded specific parts of the 1-409 sequence (see Table 2 for primers and resulting plasmids and Fig. 2 to 5 for corresponding BovHepV IRES constructs). As a negative control, the EMCV IRES sequence was completely removed from plasmid pEMCV, resulting in construct ΔEMCV. All PCRs were carried out with Phusion polymerase (Thermo Fisher Scientific, Waltham, MA). PCR products were gel purified (GeneJET gel extraction kit; Thermo Fisher Scientific), followed by phosphorylation with T4 polynucleotide kinase and end-to-end ligation using T4 DNA ligase (New England BioLabs, Ipswich, MA) at 16°C overnight before they were used for transformation of E. coli Top10 cells.
TABLE 2.
Primers used for generation of different BovHepV constructsa
| Primer | Sequence (5′-3′) | Product length (bp) | Resulting plasmid |
|---|---|---|---|
| Megaprimer_fwd | taaacgcgtcgagcatgcatctaACCATCAACACTCCAGGCTCA | 461 | Megaprimer |
| Megaprimer_rev | tgttctggatcataaactttcgaagtcatTTGGAACCACAACGGTGAC | ||
| EMCV_Excl_fwd | CACGATGTCGACATGACTTCG | 8,113 | ΔEMCV |
| EMCV_Excl_rev | GGAATTGGTCGACCTAGATGC | ||
| Renilla_Start_fwd | ATGACTTCGAAAGTTTATGATCCAG | 8,128 | 1-15 |
| 1-15_rev | TGGAGTGTGATGGTTAGATGC | ||
| Renilla_Start_fwd | ATGACTTCGAAAGTTTATGATCCAG | 8,203 | 1-90 |
| 1-90_rev | GGCTCATGGGATAGGCG | ||
| Renilla_Start_fwd | ATGACTTCGAAAGTTTATGATCCAG | 8,393 | 1-281 |
| 1-281_rev | CCGCACCCACAGATCCT | ||
| Renilla_Start_fwd | ATGACTTCGAAAGTTTATGATCCAG | 8,409 | 1-297 |
| 1-297_rev | CATGTTACATGTACAGCCGCA | ||
| Renilla_Start_fwd | ATGACTTCGAAAGTTTATGATCCAG | 8,413 | 1-301 |
| 1-301_rev | CTTCCATGTTACATGTACAGCC | ||
| 1-333_fwd | ATGACTTCGAAAGTTTATGATCC | 8,446 | 1-333 |
| 1-333_rev | GCTTCTGGTTTGAGTTGAAC | ||
| 16-333_fwd | GGCTCATGGATTAAGTTAGGTTCCG | 8,431 | 16-333 |
| 16-333_rev | TAGATGCATGCTCGACGCGTTTATTACAA | ||
| 1-405_fwd | ATGACTTCGAAAGTTTATGATCCAGAACAAAGG | 8,518 | 1-405 |
| 1-405_rev | AACCACAACGGTGACGCCAGGTC | ||
| 16-405_fwd | GGCTCATGGATTAAGTTAGGTTCCG | 8,502 | 16-405 |
| 16-405_rev | TAGATGCATGCTCGACGCGTTTATTACAA | ||
| 1-405_m1-9_fwd | GCATCTAcaaggacctGCTCCAGG | 8,518 | 1-405 (m1-9) |
| 1-405_m1-9_rev | ATGCTCGACGCGTTTATTACAATTTGGACTTTC | ||
| 1-405_m397-404_fwd | GCGTCACgacgtcttTTATGACTTCGA | 8,518 | p1-p405 (m397-404) |
| 1-405_m397-404_rev | CAGGTCTACGCCGCGAGCG | ||
| 1-333_m12/m13_fwd | ATTAAGTTAGGTTCCGCCGAAGCG | 8,446 | p1-p333 (m12/m13) |
| 1-333_m12/m13_rev | CCATGAGCCTGctGTGTTGATGG | ||
| 1-333_m8-15_fwd | CTAACCATCAcacagaacGGCTCATGG | 8,446 | p1-p333 (m8-15) |
| 1-333_m8-15_rev | ATGCATGCTCGACGCGTTTATTACAATTTGG | ||
| 1-405 m397-404_comp_m1-9_fwd | CATCTAAaagaccgtcACTCCAGG | 8,518 | p1-p405 m397-404 (comp_1-9) |
| 1-405 m397-404_comp_m1-9_rev | CATGCTCGACGCGTTTATTACAATTTGG |
Lowercase letters indicate nucleotides of firefly luciferase sequence (Megaprimer_fwd) or Renilla luciferase sequence (Megaprimer_rev), respectively, or mutated nucleotides in the IRES.
Transfection procedure and measurement of renilla and firefly luciferase units.
For transfection, 106 cells were seeded on six-well-plates. Two days later, the cell layers had reached 90% confluence and were transfected with 4 μg of the respective plasmids through lipofection. Simultaneous transfections were carried out with 4 μg of plasmid and 2 μg of miR-122 duplexes, complementary miR duplexes, miR-122 inhibitor duplexes, or random miR duplexes (Dharmacon, Lafayette, CO). The sequences were as follows: miR-122 (mature sequence), 5′-UGGAGUGUGACAAUGGUGUUUG-3′; complementary miR (mature sequence), 5′-CAGAUUGUUCCUGUGCAGCAAC-3′; miR-122 inhibitor (mature sequence), 5′-AACGCCAUUAUCACACUAAAUA-3′; and random miR, 5′-CACGUUAAAACCAUACGCACUACGAAACCCC-3′ (18). The best transfection efficiencies were obtained after transfection of HeLa and Huh-7 cells with Lipofectamine 2000 and of BFH12 cells with Lipofectamine 3000 (Invitrogen). The cells were lysed at 48 h posttransfection with dual-luciferase reporter assay system passive lysis buffer (Promega, Madison, WI), and the luciferase activities were measured with a luminescence reader (GENios Pro; Tecan, Männedorf, Switzerland). Briefly, the firefly luciferase activity was measured using the substrate beetle luciferin; the reaction was stopped with Stop & Glo reagent, and then the renilla luciferase activity was assessed by the addition of coelenterazine substrate. This dual luciferase reporter assay is characterized by direct correlation of the translation initiation capacity to the renilla luciferase activity. The renilla luciferase light units were normalized by dividing the obtained values by the firefly luciferase light units.
Statistical analyses.
Results were tested for statistical differences with an unpaired t test (GraphPad Prism, version 7). The level of significance was set to a P value of <0.05. Each experiment was performed in triplicate.
ACKNOWLEDGMENTS
We thank Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for providing Huh-7 cells and Norbert Tautz (University of Luebeck, Luebeck, Germany) for providing plasmid pF/R_wt.
REFERENCES
- 1.Baechlein C, Fischer N, Grundhoff A, Alawi M, Indenbirken D, Postel A, Baron AL, Offinger J, Becker K, Beineke A, Rehage J, Becher P. 2015. Identification of a novel hepacivirus in domestic cattle from Germany. J Virol 89:7007–7015. doi: 10.1128/JVI.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff AS, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P. 2016. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT. 2017. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter MJ. 2007. Epidemiology of hepatitis C virus infection. World J Gastroenterol 13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart H, Walter C, Jones D, Lyons S, Simmonds P, Harris M. 2013. The non-primate hepacivirus 5′ untranslated region possesses internal ribosomal entry site activity. J Gen Virol 94:2657–2663. doi: 10.1099/vir.0.055764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rijnbrand R, Abell G, Lemon SM. 2000. Mutational analysis of the GB virus B internal ribosome entry site. J Virol 74:773–783. doi: 10.1128/JVI.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspé G, Lemon S, Spaan W. 1995. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett 365:115–119. doi: 10.1016/0014-5793(95)00458-L. [DOI] [PubMed] [Google Scholar]
- 8.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Kieft JS, Zhou K, Jubin R, Doudna JA. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194–206. doi: 10.1017/S1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheel TKH, Kapoor A, Nishiuchi E, Brock KV, Yu Y, Andrus L, Gu M, Renshaw RW, Dubovi EJ, McDonough SP, Van de Walle GR, Lipkin WI, Divers TJ, Tennant BC, Rice CM. 2015. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A 112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr C, Conrad KD, Niepmann M. 2012. Differential stimulation of hepatitis C virus RNA translation by microRNA-122 in different cell cycle phases. Cell Cycle 11:277–285. doi: 10.4161/cc.11.2.18699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson JA, Huys A. 2013. miR-122 Promotion of the hepatitis C virus life cycle: sound in the silence. Wiley Interdiscip Rev RNA 4:665–676. doi: 10.1002/wrna.1186. [DOI] [PubMed] [Google Scholar]
- 13.Machlin ES, Sarnow P, Sagan SM. 2011. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A 108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortimer SA, Doudna JA. 2013. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res 41:4230–4240. doi: 10.1093/nar/gkt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 16.Jangra RK, Yi M, Lemon SM. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goergen D, Niepmann M. 2012. Stimulation of hepatitis C virus RNA translation by microRNA-122 occurs under different conditions in vivo and in vitro. Virus Res 167:343–352. doi: 10.1016/j.virusres.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Jopling CL, Schütz S, Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson JA, Zhang C, Huys A, Richardson CD. 2011. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J Virol 85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts APE, Lewis AP, Jopling CL. 2011. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res 39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jopling C. 2012. Liver-specific microRNA-122. RNA Biol 9:137–142. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rijnbrand R, Bredenbeek PJ, Haasnoot PC, Kieft JS, Spaan WJ, Lemon SM. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585–597. doi: 10.1017/S1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 24.Kapp LD, Lorsch JR. 2004. The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 25.Sachs AB, Sarnow P, Hentze MW. 1997. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831–838. doi: 10.1016/S0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 26.Lozano G, Martínez-Salas E. 2015. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol 12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Fraser CS, Doudna JA. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 28.Corman VM, Grundhoff A, Baechlein C, Fischer N, Gmyl A, Wollny R, Dei D, Ritz D, Binger T, Adankwah E, Marfo KS, Annison L, Annan A, Adu-Sarkodie Y, Oppong S, Becher P, Drosten C, Drexler JF. 2015. Highly divergent hepaciviruses from African cattle. J Virol 89:5876–5882. doi: 10.1128/JVI.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khawaja A, Vopalensky V, Pospisek M. 2015. Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdiscip Rev RNA 6:211–224. doi: 10.1002/wrna.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Le SY, Ali N, Siddiqui A. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 31.Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JYN, Baroudy BM. 2000. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol 74:10430–10437. doi: 10.1128/JVI.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda M, Brown EA, Lemon SM. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T-H, Rijnbrand RCA, Lemon SM. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of Cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J Virol 74:11347–11358. doi: 10.1128/JVI.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernardi A, Spahr P-F. 1972. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci U S A 69:3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. 1999. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J Virol 73:4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A 104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsure Y. 2012. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J Virol 86:1382–1893. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gleich A, Kaiser B, Schumann J, Fuhrmann H. 2016. Establishment and characterisation of a novel bovine SV40 large T-antigen-transduced foetal hepatocyte-derived cell line. Vitro Cell Dev Biol Anim 52:662–672. doi: 10.1007/s11626-016-0018-0. [DOI] [PubMed] [Google Scholar]
- 42.Fraser CS, Hershey JWB, Doudna JA. 2009. The pathway of HCV mRNA recruitment to the human ribosome. Nat Struct Mol Biol 16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]