Following infection, viruses synthesize nonstructural proteins that mediate viral replication and promote dissemination. Viruses from the family Reoviridae encode nonstructural proteins that are required for the formation of progeny viruses. Although nonstructural proteins of different viruses in the family Reoviridae diverge in primary sequence, they are functionally homologous and appear to facilitate conserved mechanisms of dsRNA virus replication. Using in vitro and cell culture approaches, we found that the mammalian reovirus nonstructural protein σNS binds and stabilizes viral RNA and is required for genome synthesis. This work contributes new knowledge about basic mechanisms of dsRNA virus replication and provides a foundation for future studies to determine how viruses in the family Reoviridae assort and replicate their genomes.
KEYWORDS: RNA-binding proteins, dsRNA replication, reovirus, σNS
ABSTRACT
Viral nonstructural proteins, which are not packaged into virions, are essential for the replication of most viruses. Reovirus, a nonenveloped, double-stranded RNA (dsRNA) virus, encodes three nonstructural proteins that are required for viral replication and dissemination in the host. The reovirus nonstructural protein σNS is a single-stranded RNA (ssRNA)-binding protein that must be expressed in infected cells for production of viral progeny. However, the activities of σNS during individual steps of the reovirus replication cycle are poorly understood. We explored the function of σNS by disrupting its expression during infection using cells expressing a small interfering RNA (siRNA) targeting the σNS-encoding S3 gene and found that σNS is required for viral genome replication. Using complementary biochemical assays, we determined that σNS forms complexes with viral and nonviral RNAs. We also discovered, using in vitro and cell-based RNA degradation experiments, that σNS increases the RNA half-life. Cryo-electron microscopy revealed that σNS and ssRNAs organize into long, filamentous structures. Collectively, our findings indicate that σNS functions as an RNA-binding protein that increases the viral RNA half-life. These results suggest that σNS forms RNA-protein complexes in preparation for genome replication.
IMPORTANCE Following infection, viruses synthesize nonstructural proteins that mediate viral replication and promote dissemination. Viruses from the family Reoviridae encode nonstructural proteins that are required for the formation of progeny viruses. Although nonstructural proteins of different viruses in the family Reoviridae diverge in primary sequence, they are functionally homologous and appear to facilitate conserved mechanisms of dsRNA virus replication. Using in vitro and cell culture approaches, we found that the mammalian reovirus nonstructural protein σNS binds and stabilizes viral RNA and is required for genome synthesis. This work contributes new knowledge about basic mechanisms of dsRNA virus replication and provides a foundation for future studies to determine how viruses in the family Reoviridae assort and replicate their genomes.
INTRODUCTION
Viruses contain the minimal components required to enter host cells and deliver their genetic payloads into the cell interior. For productive infection of a host, viral genomes must be replicated at sites of primary infection and packaged into new virions for dissemination to susceptible cells at local or distant sites. Viruses encode proteins that are not packaged into virions to aid in these postentry replication steps. These proteins, called nonstructural proteins, participate in multiple viral replication steps, including translation of viral mRNAs (1), replication of viral genomes (2), construction of sites for particle assembly (3), and inhibition of innate immune responses (4). Due to constraints on viral genome size (5), most nonstructural proteins play multiple roles during infection (6). Consequently, teasing out individual activities of viral nonstructural proteins is challenging.
Mammalian orthoreoviruses (called reoviruses here) are nonenveloped viruses that infect virtually all mammalian species but cause disease only in the very young (7, 8). The reovirus virion is an ∼80-nm particle composed of two concentric protein shells that encapsidate 10 segments of double-stranded RNA (dsRNA) (9). Reovirus replication begins with recognition of glycan attachment factors (10–12) and cell surface receptors (13, 14). Following internalization, reoviruses traffic within endosomes (15, 16) and undergo acid-dependent proteolytic disassembly (17), which allows core particles to penetrate the endosomes and gain access to the cytoplasm (18). Within the cytoplasm, reovirus cores are transcriptionally active and release viral mRNAs (19, 20). These mRNAs are translated by cellular ribosomes, yielding eight structural and three nonstructural proteins (9). The nonstructural proteins reorganize membranes from the endoplasmic reticulum (21) to form neo-organelles called inclusions or factories (22–24), which serve as sites of viral genome replication and particle assembly (25, 26). By a mechanism that is not understood and is likely aided by nonstructural proteins (27, 28), viral mRNAs are sorted and assembled into a replicase or precore complex (29, 30), followed by dsRNA synthesis and formation of nascent cores (31, 32). These newly synthesized cores are transcriptionally active and release viral mRNAs until transcription is silenced by the coalescence of outer-capsid proteins onto nascent cores (33). Mature reovirus particles are released from cells through an unknown mechanism. Understanding how reoviruses replicate is important, as the viruses have oncolytic potential (34) and may trigger celiac disease (35, 36).
The reovirus nonstructural protein σNS is a 41-kDa protein highly conserved among different reovirus strains (37, 38). Initially incorrectly identified as a poly(C)-dependent RNA polymerase (39), σNS is a single-stranded RNA (ssRNA)-binding protein with a reported specificity for certain regions of reovirus mRNA (40, 41), although it appears to show comparable binding to viral and nonviral RNAs (42). σNS has been defined as a nonclassical helicase, exhibiting strand displacement activity in the absence of ATP hydrolysis and MgCl2 (42). Following reovirus infection of host cells, σNS and μNS, another nonstructural protein that is responsible for nucleation of reovirus inclusions (22), are among the initial translation products (9). σNS and μNS interact with each other in reovirus inclusions (43–45) using a mechanism that partially depends on the presence of RNA (23). Throughout the course of infection, reovirus inclusions mature (46), recruit cellular membranes (21), and attract the host translational machinery (47), probably to localize the synthesis of reovirus proteins to sites of genome replication and particle assembly. These studies give valuable insight into σNS, but its function during reovirus replication remains poorly understood.
To enhance knowledge of the activities of σNS during reovirus replication, we used cells expressing a small interfering RNA (siRNA) targeting the reovirus σNS-encoding S3 gene and quantified the effect of σNS knockdown on various steps of viral replication. We found that σNS is dispensable for primary, but not secondary, rounds of transcription and functions prior to genome replication. We hypothesized that σNS promotes genome replication by stabilizing viral mRNAs. Using complementary in vitro and cell-based assays, we discovered that σNS and RNAs organize into long filamentous structures and that σNS increases the half-life of viral mRNAs. These results define σNS as an RNA stability factor required for genome replication during reovirus infection.
RESULTS
Developing a system to study σNS function.
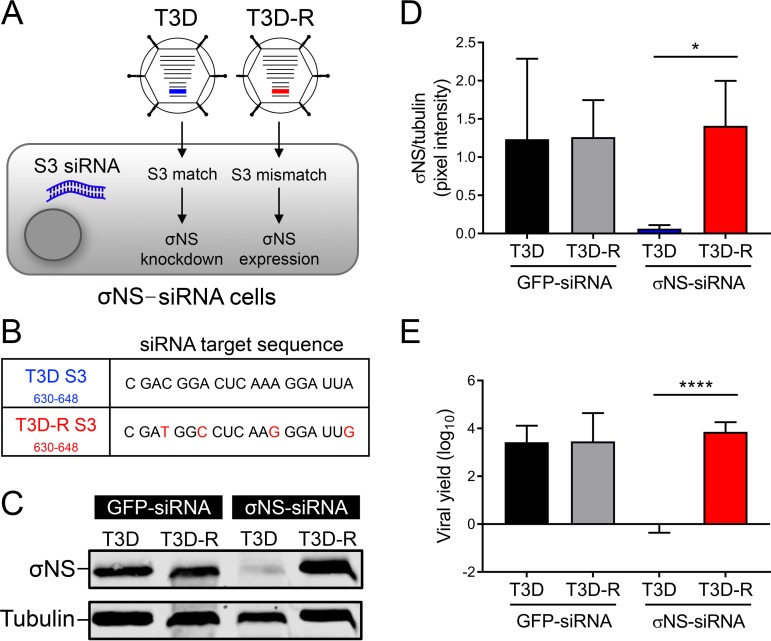
In a previous study (48), we engineered HEK293T cells to stably express an siRNA targeting the reovirus σNS-encoding S3 gene. We sorted the cells by flow cytometry and selected clones that showed the lowest expression of σNS after infection with reovirus strain type 3 Dearing (T3D) (data not shown). These cells were termed σNS-siRNA cells (Fig. 1A). Using the reovirus plasmid-based reverse genetics system (49), we engineered a viral mutant that contained four synonymous mutations within the S3 gene siRNA target site of T3D (Fig. 1B). We termed this virus T3D resistant (T3D-R), as we hypothesized that it would be resistant to the siRNA expressed by σNS-siRNA cells. To determine whether σNS is expressed in T3D-R-infected σNS-siRNA cells, we allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells and quantified σNS protein abundance by immunoblotting (Fig. 1C and D). Infection of σNS-siRNA cells with T3D resulted in a 96% reduction of σNS protein levels relative to infection with T3D-R, indicating that T3D-R effectively escapes RNA interference (RNAi) targeting in σNS-siRNA cells. To confirm that σNS knockdown in T3D-infected σNS-siRNA cells is specific, we allowed either T3D or T3D-R to be adsorbed onto HEK293T cells expressing a nontargeting siRNA (green fluorescent protein [GFP]-siRNA cells) and quantified σNS protein abundance by immunoblotting (Fig. 1C and D). T3D- and T3D-R-infected GFP-siRNA cells expressed comparable amounts of σNS, indicating that the σNS protein of T3D is produced in 293T cells expressing a nontargeting siRNA. To test whether the synonymous mutations introduced in the S3 gene of T3D-R virus alter the production of viral progeny, we allowed either T3D or T3D-R to be adsorbed onto GFP-siRNA and σNS-siRNA cells and quantified viral yields at 24 h postadsorption by plaque assay (Fig. 1E). T3D and T3D-R viruses produced comparable yields at 24 h postadsorption in GFP-siRNA cells, suggesting that the mutations introduced in T3D-R do not alter its replication capacity. Similar results were obtained in replication assays using cells lacking any siRNA (data not shown). In contrast, T3D was incapable of producing progeny in σNS-siRNA cells, indicating that σNS is required for T3D replication. Thus, σNS-siRNA cells effectively and specifically diminish σNS expression when infected with T3D, but not T3D-R.
FIG 1.
(A) Schematic of viruses and cells used to study σNS function. HEK293T cells engineered to stably express an siRNA against the σNS-encoding S3 gene (σNS-siRNA cells) were infected with either T3D or T3D-R virus. Representations of the S3 gene segments for T3D (blue) and T3D-R (red) are highlighted. The S3 gene segment of T3D contains target sequences complementary to the S3 siRNA of σNS-siRNA cells, whereas the S3 gene segment of T3D-R does not. (B) Sequences corresponding to nucleotides 630 to 648 of T3D and T3D-R S3 genes. Synonymous mutations are shown in red. (C) T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells or control cells expressing a nontargeting siRNA (GFP-siRNA cells) at an MOI of 10 PFU per cell and incubated for 24 h. The cell lysates were resolved by SDS-PAGE and immunoblotted using antibodies specific for σNS and α-tubulin. (D) Pixel intensity analysis of the ratio of σNS to α-tubulin in T3D- and T3D-R-infected σNS-siRNA and GFP-siRNA cells for triplicate experiments. (E) T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA and GFP-siRNA cells at an MOI of 1 PFU/cell. Viral yields at 24 h postadsorption were determined by plaque assay. The results are presented as means ± SD. *, P < 0.05, and ****, P < 0.0001, as determined by Student's t test.
Reovirus T3D does not replicate in σNS-siRNA cells.
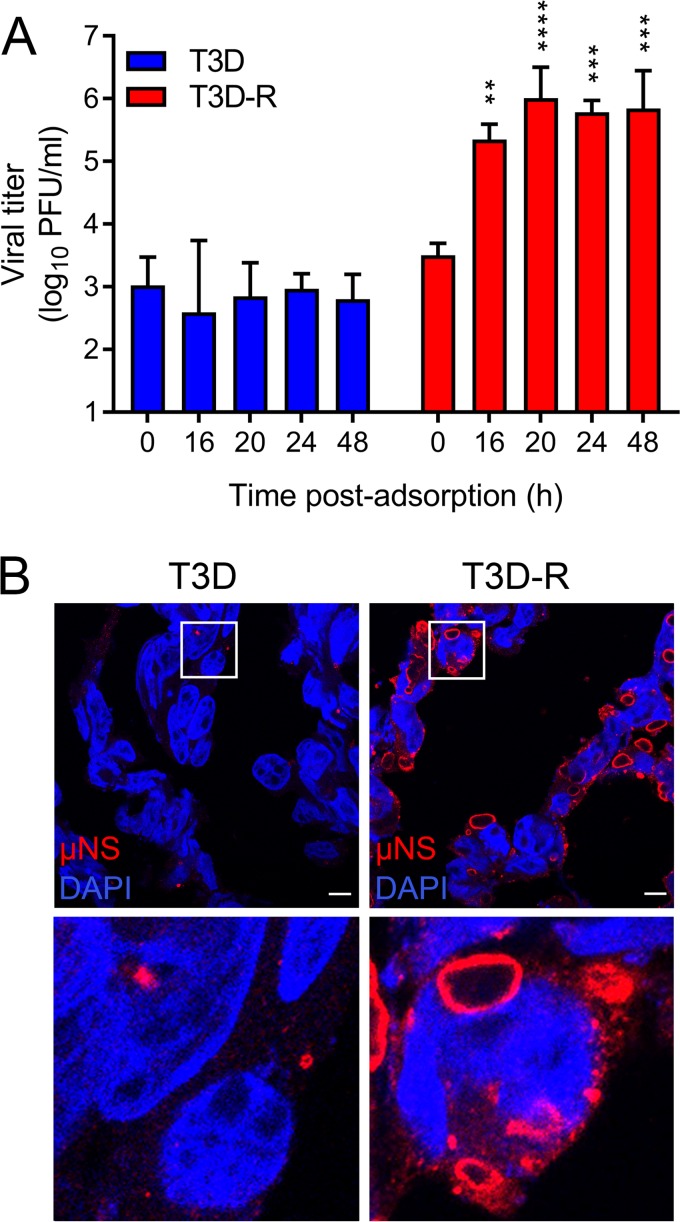
To assess whether σNS is required for formation of progeny virions over a longer interval, we allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells and quantified progeny virus at various times postadsorption by plaque assay (Fig. 2A). T3D was incapable of forming new progeny when infecting σNS-siRNA cells at all intervals tested. In contrast, T3D-R yields increased throughout the time course of infection, reaching ∼70-fold by 16 h and ∼220-fold by 48 h, confirming that σNS is required for progeny particle production.
FIG 2.
Reovirus T3D does not replicate in σNS-siRNA cells. T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells at an MOI of 1 (A) or 100 (B) PFU/cell. (A) Viral titers at 0, 16, 20, 24, and 48 h postadsorption were determined by plaque assay. The results are presented as mean viral titers and SD for three independent experiments. Values that differ significantly from the value at 0 h by one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test are indicated (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (B) Cells were fixed at 24 h postinfection (hpi), stained using μNS-specific antiserum (red) and DAPI (blue) to visualize nuclei, and imaged using confocal microscopy. The boxed regions are shown enlarged below. Scale bars, 10 μm.
To determine whether σNS is required for inclusion formation, we allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells, stained the cells at 24 h with polyclonal serum specific for the reovirus nonstructural protein μNS to visualize inclusions, and imaged the cells by confocal microscopy (Fig. 2B). T3D-infected σNS-siRNA cells contained few small inclusions (<1 μm), similar to those detected early in infection (22). In contrast, T3D-R-infected σNS-siRNA cells were filled with globular inclusions, some of which were more than 10 μm in diameter. These results suggest that σNS is required for replication steps leading to the maturation of reovirus inclusions.
Viral RNA levels are diminished in T3D-infected σNS-siRNA cells.
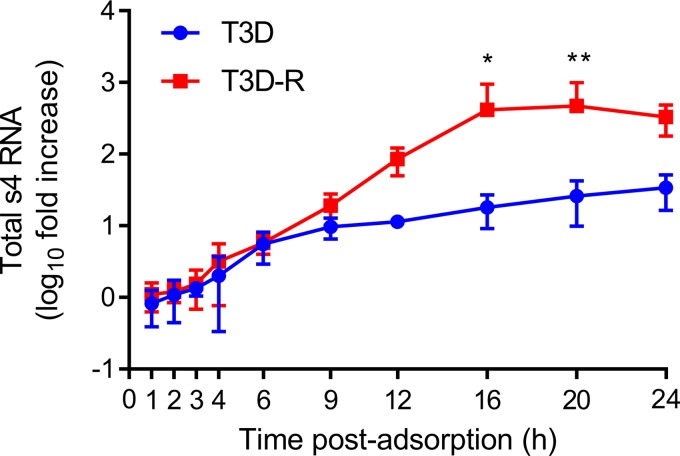
To estimate the timing at which primary and secondary rounds of transcription occur in HEK293T cells, we allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells and quantified s4 RNA by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 3). (A small-case letter preceding a number indicates an ssRNA species by convention.) Levels of s4 RNA in T3D- and T3D-R-infected σNS-siRNA cells increased at similar rates until 6 h postadsorption. At later times, s4 RNA levels increased at higher rates in T3D-R-infected σNS-siRNA cells than in T3D-infected cells. These results suggest that secondary rounds of transcription do not begin until after 6 h postadsorption, as previously observed (50), and that late rounds of gene expression are impaired in T3D-infected σNS-siRNA cells.
FIG 3.
Total s4 RNA levels are diminished in T3D-infected σNS-siRNA cells. T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells at an MOI of 1 PFU/cell. Cells were lysed at the intervals shown, and s4 RNA was quantified by RT-qPCR. The results are presented as mean RNA levels normalized to the RNA levels at 0 h for at least three independent experiments. The error bars indicate SD. Values that differ significantly between T3D- and T3D-R-infected σNS-siRNA cells at each time point by two-way ANOVA followed by Sidak's multiple-comparison test are indicated (*, P < 0.05; **, P < 0.01).
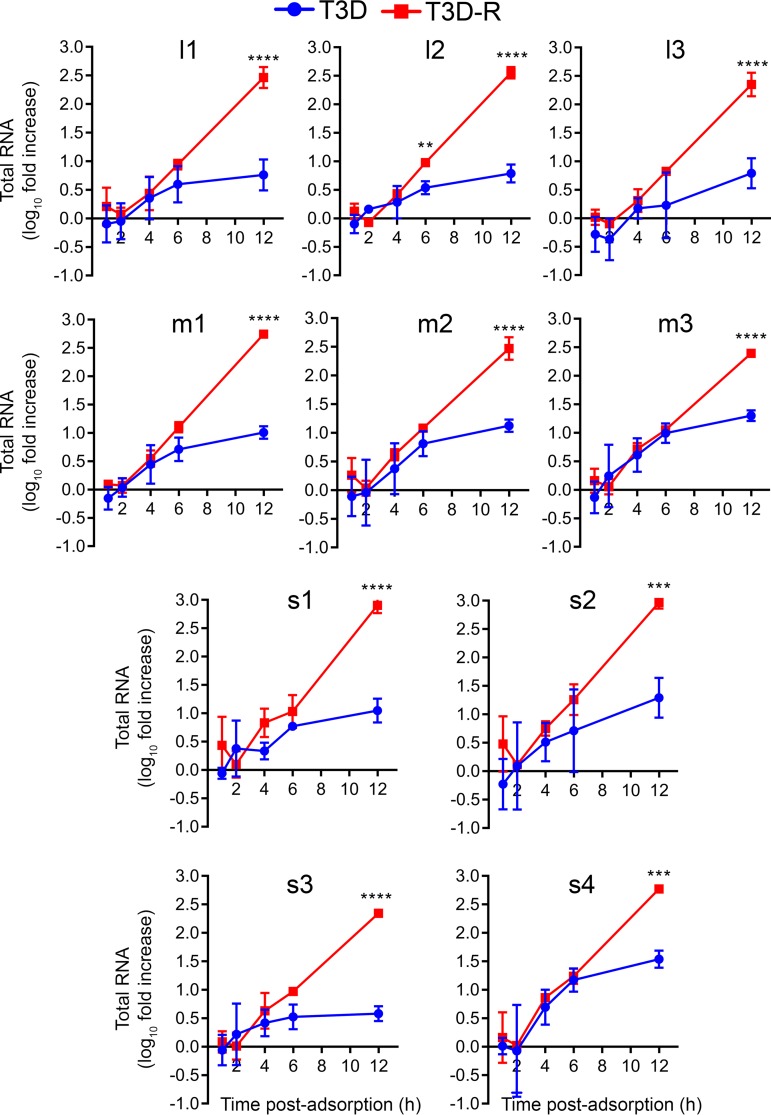
To test whether steady-state levels of viral RNAs are altered by σNS disruption, we quantified RNA levels for the L (large), M (medium), and S (small) gene segments by NanoString (NanoString Technologies, Inc.) analysis during a time course of infection in T3D- and T3D-R-infected σNS-siRNA cells. NanoString technology is based on fluorescent probes that, following hybridization with their target RNAs, allow direct quantification of RNAs in a sample without the requirement for reverse transcription or amplification. We allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells and quantified RNA levels corresponding to the 10 reovirus gene segments at various intervals postadsorption using unique probes targeting each reovirus positive-sense RNA (Table 1 and Fig. 4). The levels of all viral RNAs increased comparably in cells infected with T3D or T3D-R until 4 to 6 h postadsorption, after which time RNA levels in T3D-infected cells increased modestly. In contrast, levels of viral RNAs in cells infected with T3D-R increased exponentially, with levels at 12 h postadsorption elevated ∼250-fold for the L-class RNAs, ∼340-fold for the M-class RNAs, and ∼550-fold for the S-class RNAs relative to the respective levels of these RNAs at the 0-h time point. Because secondary transcription does not occur until after 6 h postadsorption (Fig. 3) (50), these results indicate that σNS is dispensable for primary, but not secondary, rounds of transcription.
TABLE 1.
Custom-designed probes used for NanoStringa
| Gene | Protein | Target sequence (5′→3′) |
|---|---|---|
| L1 | λ3 | GAGCTCAGTCGTCGGTGAGCTTCGTAAACGGACAAAGACGTATGTTAAACATGACTTTGCTTCAGTGAGGTACATTCGTGACGCTATGGCATGTACTAGC |
| L2 | λ2 | TGGACGAAGGCGATCTGATGGTTAGTCGGCTTACGCAACTCCCGTTACGTCCTGATTATGGTAATATCTGGGTCGGCGATGCGCTATCCTATTATGTGGA |
| L3 | λ1 | CCAACGCGCATGGGAACGCCGAATGTATCCAAAATATGTAATTTCGTCGCCTCTTGTGTGCGAAATCGGGTTGGACGGTTTGATCGAGCACAGATGATGA |
| M1 | μ2 | ACGTTTGAGCAGGCGGTTATGGAGATATACAAAGGGATTGCTGGCGTTGACTCGCTGGATGATCTCATCAAGTGGGTGCTGAACTCGGATCTCATTCCGC |
| M2 | μ1 | GCTGGGATCCAAACGGCAAGAAGGTCGGATTCATCGTTTTTCAATCGAAGATACCATTCGAACTTTGGACTGCTGCTTCACAGATCGGTCAAGCCACGGT |
| M3 | μNS | GTGTCGTATGACGTTACGCTCACTCATGAAGACCGGACGCGACGTTGATGCACACAGAGCTTTTCAGCGAGTCCTCTCTCAAGGATACACATCGCTAATG |
| S1 | σ1 | ACTTGCAGAGCTACGCGTTGATCACGACAATCTCGTTGCGAGAGTGGATACTGCAGAACGTAACATTGGATCATTGACCACCGAGCTATCAACTCTGACG |
| S2 | σ2 | GCTTTGCAAGCACAGGCAGATCGAGTGTACGACTGCGATGATTATCCATTTCTAGCGCGTGATCCAAGATTCAAACATCGGGTGTATCAGCAATTGAGTG |
| S3 | σNS | TGGCCACGTCATCTTTAAGTATTTCCCTGGACCGGGGTCGATGGGTGGCGGCTGACGCCAGTGATGCTAGACTGCTGGTTTTTCCGATTCGCGTGTAATG |
| S4 | σ3 | CTGCTCACTGGAAGCGGGGTATGCTGTCCTTCGTTGCGCAGATGCACGAGATGATGAATGACGTGTCGCCAGATGACCTGGATCGTGTGCGTACTGAGGG |
The table shows the target sequence of each reovirus gene segment and protein.
FIG 4.
Levels of all reovirus RNAs are reduced in T3D-infected σNS-siRNA cells. T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells at an MOI of 1 PFU/cell. Cells were lysed at the intervals shown, and RNAs were quantified by NanoString technology using custom-designed probes (Table 1) specific for each reovirus gene segment. The results are presented as mean RNA levels normalized to the RNA levels at 0 h for three independent experiments. The error bars indicate SD. Values that differ significantly between T3D- and T3D-R-infected σNS-siRNA cells at each time point by two-way ANOVA followed by Sidak's multiple-comparison test are indicated (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Reovirus T3D protein synthesis is diminished in infected σNS-siRNA cells.
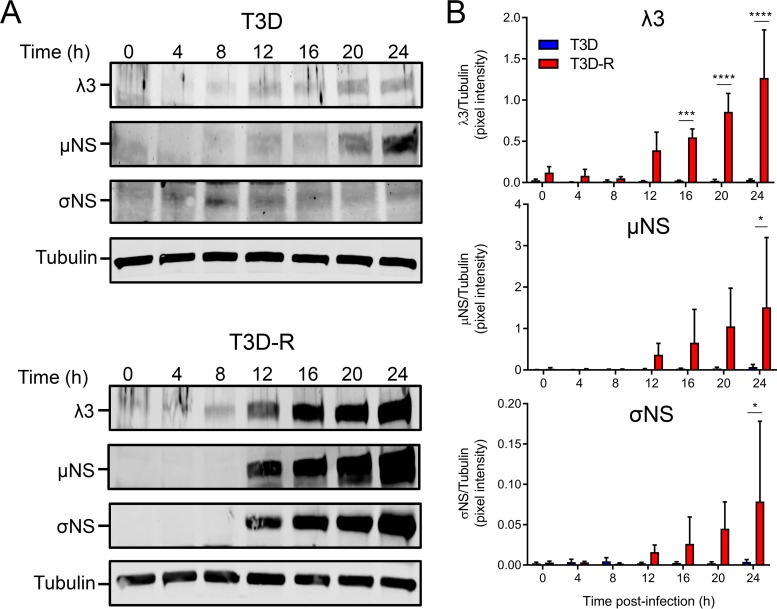
We hypothesized that the reduction in abundance of viral RNAs observed in T3D-infected σNS-siRNA cells would diminish viral protein levels. To test this hypothesis, either T3D or T3D-R was adsorbed onto σNS-siRNA cells, and expression of the RNA-dependent RNA polymerase λ3 and nonstructural proteins μNS and σNS was quantified by SDS-PAGE, followed by immunoblotting at various intervals postadsorption (Fig. 5). Newly synthesized viral proteins were detected at 12 h postadsorption in T3D-R-infected σNS-siRNA cells, and levels of the three immunoblotted viral proteins increased over time. In contrast, viral protein synthesis was impaired in T3D-infected σNS-siRNA cells. These results suggest that σNS is required for replication steps leading to viral protein synthesis.
FIG 5.
T3D protein synthesis is diminished in T3D-infected σNS-siRNA cells. T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells at an MOI of 1 PFU/cell. (A) Cell lysates were collected at the intervals shown and analyzed by immunoblotting using antibodies specific for the λ3, μNS, and σNS proteins. (B) Pixel intensity analysis of the ratios of λ3, μNS, and σNS proteins to α-tubulin. The results are presented as mean pixel intensity and SD for three independent experiments. Values that differ significantly between T3D- and T3D-R-infected σNS-siRNA cells at each time point by two-way ANOVA followed by Sidak's multiple-comparison test are indicated (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001).
Reovirus σNS is required for genome replication.
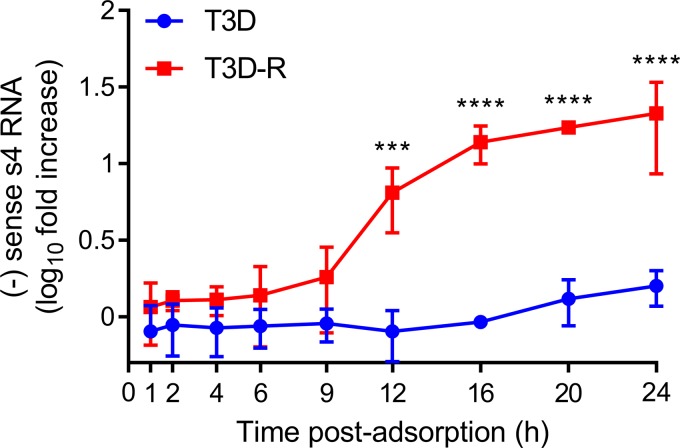
To elucidate whether σNS is required for replication of the reovirus genome, we used a modified qPCR assay to quantify s4 negative-sense ssRNA (50). In this assay, s4 RNA is reverse transcribed using a primer that binds to the negative-sense strand of the S4 dsRNA gene segment. Once the s4 negative-sense cDNA is synthesized, a complementary primer is added, and quantification by qPCR proceeds. As the genome is the only source of negative-sense viral RNA, the assay allows genome copies to be quantified. We allowed either T3D or T3D-R to be adsorbed onto σNS-siRNA cells and quantified s4 negative-sense RNA at various intervals postadsorption (Fig. 6). By this analysis, genome replication appeared to initiate at 9 h postadsorption in T3D-R-infected σNS-siRNA cells, and by 12 h, T3D-R S4 dsRNA levels were ∼10-fold higher than those at the initiation of infection. T3D-R S4 dsRNA levels continued to increase over time, and by 24 h, these levels were ∼20-fold higher than those at 0 h. In contrast, T3D S4 dsRNA was undetectable throughout most of the time course of infection, and by 24 h, T3D S4 dsRNA levels had increased only ∼1.5-fold in σNS-siRNA cells compared with the initiation of infection. These results indicate that σNS functions at a step leading to the replication of the reovirus genome. Thus, by examining the kinetics of individual replication steps, we conclude that σNS mediates a process immediately prior to reovirus genome replication, following primary but before secondary rounds of transcription.
FIG 6.
Reovirus T3D does not synthesize dsRNA in σNS-siRNA cells. T3D or T3D-R virus was allowed to adsorb onto σNS-siRNA cells at an MOI of 1 PFU/cell. Cells were lysed at the intervals shown, and negative (−)-sense s4 RNA was quantified by single-strand RT-qPCR. The results are presented as mean RNA levels at each time point normalized to the RNA levels at 0 h for at least three independent experiments. The error bars indicate SD. Values that differ significantly between T3D- and T3D-R-infected σNS-siRNA cells at each time point by two-way ANOVA followed by Sidak's multiple-comparison test are indicated (***, P < 0.001; ****, P < 0.0001).
The σNS protein protects viral RNA from degradation.
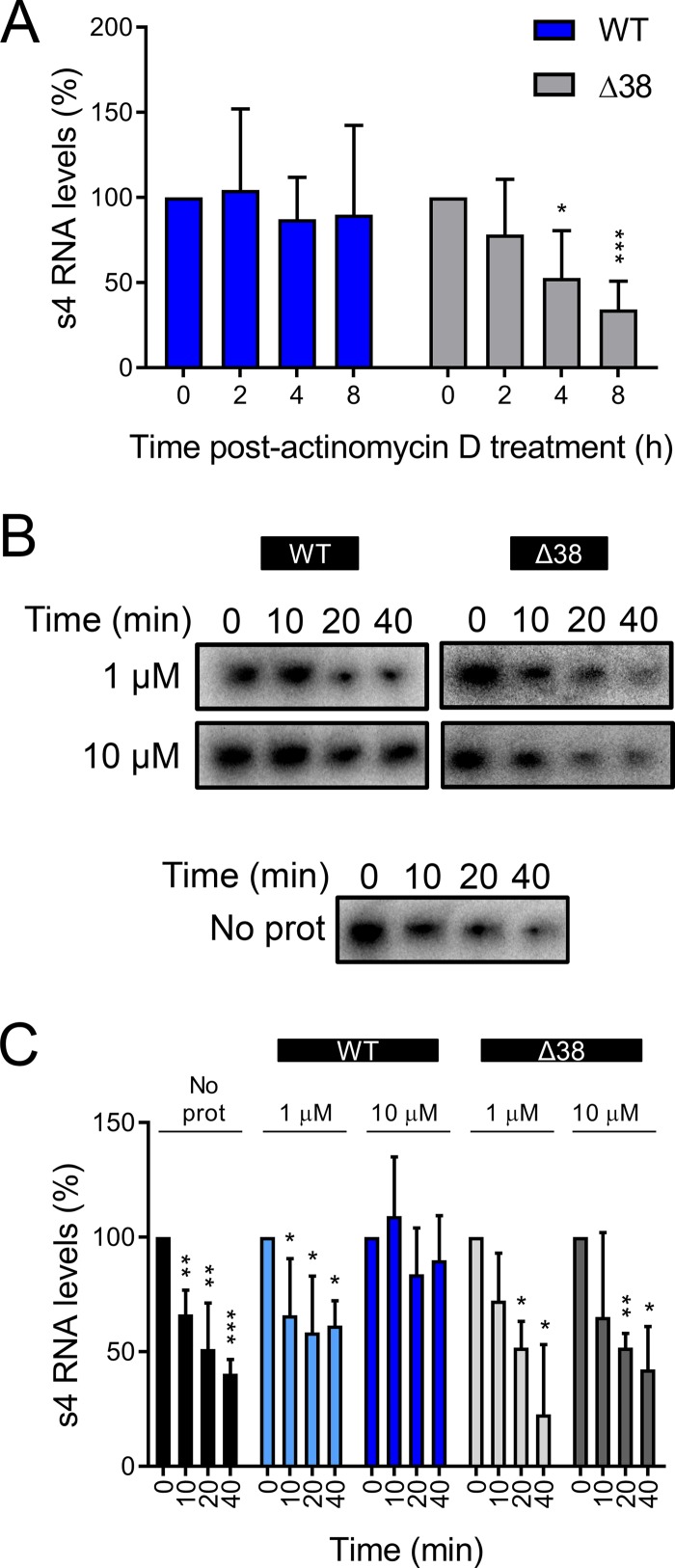
Based on the known affinity of σNS for ssRNA (42, 51) and the requirement for σNS in a step leading to genome replication (Fig. 6), we hypothesized that σNS functions as an RNA stability factor increasing the half-life of viral RNAs. To test this hypothesis, we used two σNS constructs: full-length σNS (wild type [WT]) and a deletion mutant that lacks the first 38 amino acids (Δ38 σNS). Δ38 σNS was identified in a limited proteolysis assay in which purified WT σNS was incubated with trypsin, followed by identification of the digestion products using mass spectrometry (data not shown). The deletion mutant was anticipated to be impaired in RNA binding because it lacks a series of positively charged residues predicted to be required for that activity (51, 52). We first tested whether σNS expression increases RNA half-life in cells. HEK293T cells were transfected with plasmids encoding either WT or Δ38 σNS. At 20 h posttransfection, the cells were transfected again with a plasmid encoding σ3, incubated for 4 h, and treated with actinomycin D to inhibit further transcription. Transcription derived from the σ3-encoding plasmid produces RNAs that are capped and polyadenylated (53). At various times following actinomycin D treatment, we quantified total σ3-encoding s4 RNA by RT-qPCR (Fig. 7A). As actinomycin D inhibits transcription, this approach allowed us to determine the RNA half-life. Transfection of cells with WT σNS led to the maintenance of s4 RNA levels throughout the time course of actinomycin D treatment. At 8 h after treatment initiation, we detected only a 10% reduction in s4 RNA levels. In contrast, transfection of cells with Δ38 σNS failed to protect s4 RNA. At 8 h after treatment initiation, s4 RNA levels had decreased 66% relative to the initiation of actinomycin D treatment. These results suggest that expression of WT σNS increases the half-life of a viral RNA and that this activity requires the amino-terminal 38 amino acids of the protein.
FIG 7.
The σNS protein protects viral RNA from degradation. (A) HEK293T cells were transfected with plasmids encoding either WT or Δ38 σNS and incubated for 20 h, followed by a second transfection with a σ3-encoding plasmid and incubation for 4 h. The cells were treated with 10 μg/ml of actinomycin D and lysed at the intervals shown, and σ3-encoding s4 RNA was quantified by RT-qPCR. The results are presented as mean RNA levels normalized to the RNA levels at 0 h for at least three independent experiments. The error bars indicate SD. (B and C) Radiolabeled uncapped and nonpolyadenylated s4 RNA was incubated without protein (No prot) or with 1 or 10 μM purified recombinant WT or Δ38 σNS at room temperature for 10 min, followed by addition of HeLa S100 lysates. (B) RNA was purified at the intervals shown, resolved by electrophoresis, and visualized by phosphorimaging. (C) Pixel intensity analysis of the s4 RNA bands. The results are presented as mean RNA levels normalized to the RNA levels at 0 min for three independent experiments. The error bars indicate SD. Values that differ significantly from the values at the start of the time course by one-sample t test for each time point are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To more directly test whether σNS protects RNA from degradation, we developed an in vitro cell-free RNA protection assay. We incubated purified, recombinant WT or Δ38 σNS with internally radiolabeled uncapped and nonpolyadenylated s4 RNA. σNS-RNA complexes were incubated with HeLa S100 cytoplasmic lysates for various intervals. RNA was resolved by electrophoresis and quantified by phosphorimaging (Fig. 7B and C). Incubation with WT σNS protected the RNA from degradation in a concentration-dependent manner. At a concentration of 1 μM, σNS did not protect s4 RNA from degradation in HeLa S100 lysates, and similar results were obtained in the presence of Δ38 σNS or in the absence of any additional protein. However, 10 μM σNS protected the s4 RNA, so that by 40 min of incubation, only 10% of the input s4 RNA was lost. Thus, σNS protects viral RNA from degradation.
σNS and RNAs organize in filamentous structures.
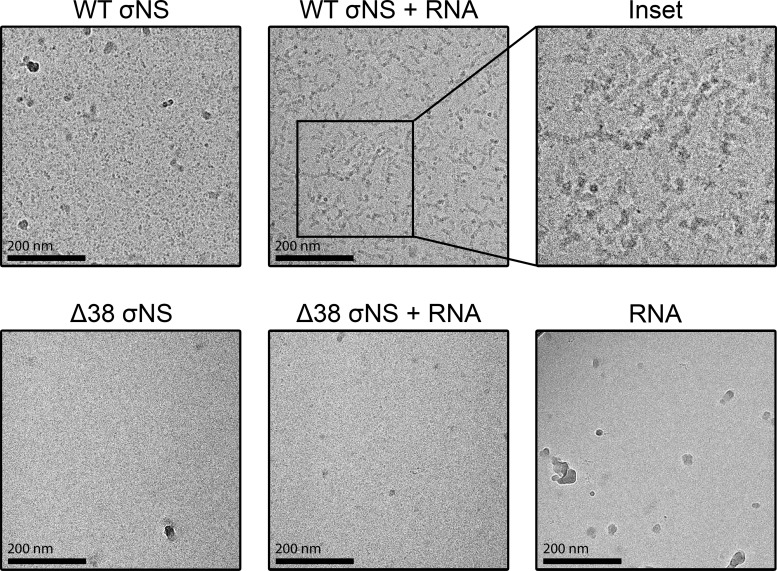
To better understand how σNS interacts with RNA, we used cryo-electron microscopy (cryo-EM) to visualize σNS-RNA complexes. We incubated purified, recombinant WT or Δ38 σNS with uncapped and nonpolyadenylated s4 RNA at a molar ratio of 50:1 on ice for 1 h and imaged σNS-RNA complexes by cryo-EM (Fig. 8). In the absence of RNA, WT σNS appeared to form higher-order structures, as demonstrated by large electron-dense configurations. In contrast, Δ38 σNS appeared smaller and scattered throughout the EM grids. Following incubation with s4 RNA, WT σNS organized into long filamentous structures approaching 200 nm in length. As expected, we did not observe interactions of Δ38 σNS with s4 RNA. These data suggest that WT σNS coats RNA and that these complexes arrange into long filamentous structures.
FIG 8.
The σNS protein forms filamentous structures in the presence of RNA. Uncapped and nonpolyadenylated s4 RNA was incubated with 50× molar excess of purified recombinant WT or Δ38 σNS on ice for 1 h, followed by plunge freezing. Frozen specimens were imaged at ×40,000 magnification with defocus levels ranging from −2.0 μm to −3.5 μm. Representative images for WT σNS, WT σNS complexed with s4 RNA, an enlargement of WT σNS and s4 RNA filaments, Δ38 σNS, Δ38 σNS incubated with s4 RNA, and s4 RNA alone are shown.
The σNS protein forms higher-order complexes with viral and nonviral ssRNAs.
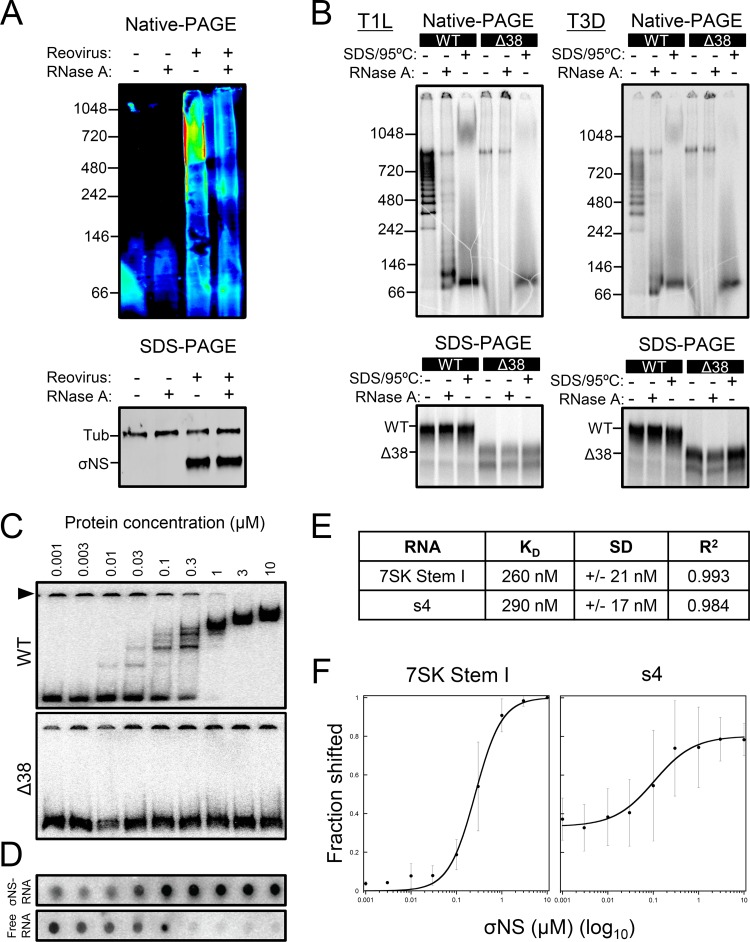
To examine σNS-RNA complexes in detail, we sought to determine whether σNS exists in an RNA-bound complex in infected cells. We allowed reovirus T3D to be adsorbed onto HEK293T cells and, at 24 h postadsorption, we lysed the cells under mild conditions to preserve protein-RNA interactions. Samples were treated with RNase A or left untreated and resolved by native PAGE and SDS-PAGE, followed by immunoblotting using σNS-specific polyclonal serum (Fig. 9A). In the absence of RNase A treatment, σNS migrated predominantly as a high-molecular-mass complex of ∼500 kDa to 1 MDa. Following RNase A treatment, the levels of these high-molecular-mass complexes diminished. Thus, σNS appears to exist as a complex with RNA in infected cells.
FIG 9.
The σNS protein binds viral and nonviral ssRNAs. (A) Reovirus T3D was allowed to adsorb onto HEK293T cells at an MOI of 100 PFU/cell and was incubated for 24 h. The cells were lysed and treated with 0.25 μg/ml of RNase A at 30°C for 1 h. Proteins were resolved by native PAGE or SDS-PAGE, followed by immunoblotting using antibodies specific for σNS and α-tubulin (SDS-PAGE only). Representative immunoblots from triplicate experiments are shown. Pixel intensity is depicted in a rainbow scale (native PAGE only). (B) T1L and T3D WT and Δ38 σNS proteins were translated in vitro in the presence of [35S]methionine and incubated with 0.25 μg/ml of RNase A at 30°C for 1 h after translation, heated at 95°C for 10 min in SDS-PAGE sample buffer, or left untreated. Samples were resolved by native PAGE or SDS-PAGE and visualized by phosphorimaging. (C) Increasing concentrations of purified recombinant WT or Δ38 σNS were incubated with radiolabeled uncapped and nonpolyadenylated 7SK stem I RNA at room temperature for 10 min, followed by native electrophoresis and visualization by phosphorimaging. The arrowhead indicates RNA that failed to enter the gel. (D) Increasing concentrations of purified recombinant WT σNS protein were incubated with radiolabeled uncapped and nonpolyadenylated s4 RNA at room temperature for 10 min. σNS-RNA complexes were spotted onto nitrocellulose membranes, and unbound free RNA was collected on nylon membranes. The membranes were visualized by phosphorimaging. (E) KD, SD, and R2 values for WT σNS and 7SK stem I (panel C, top) or s4 RNA (panel D) were determined using KaleidaGraph. (F) Langmuir isotherm curve fitting for WT σNS and 7SK stem I or s4 RNA. The results are presented as the mean percent shift at each concentration of σNS for at least three independent experiments (7SK stem I RNA, n = 5; s4 RNA, n = 3). The error bars indicate SD.
We next tested the effect of RNase A treatment on the electrophoretic mobility of WT and Δ38 σNS from reovirus strains type 1 Lang (T1L) and T3D produced by coupled in vitro transcription and translation using rabbit reticulocyte lysates in the presence of [35S]methionine. Transcription derived from these σNS-encoding plasmids yielded RNAs that were uncapped and nonpolyadenylated. Following translation, samples were treated with RNase A, denatured with SDS and heat, or left untreated; resolved by native PAGE and SDS-PAGE; and imaged by phosphorimaging (Fig. 9B). In the absence of RNase A, WT σNS migrated as higher-order structures ranging from ∼250 kDa to ∼900 kDa, suggesting that several distinct oligomeric forms of σNS bind RNA. The migration pattern appeared to be regularly spaced, suggesting a specific stoichiometry of σNS-RNA binding. Treatment with RNase A disassembled σNS-RNA complexes. Intermediate-molecular-mass bands became undetectable, and bands with estimated sizes of ∼900 kDa and ∼80 kDa predominated. SDS and heat also disrupted the higher-order structures. As expected, Δ38 σNS did not form higher-order species with RNA. Lastly, we did not observe differences in the migration patterns of T1L and T3D σNS, suggesting that the σNS proteins of both strains interact similarly with RNA. Thus, multiple subunits of σNS bind RNA with an apparently defined stoichiometry.
In a complementary approach, we used electrophoretic mobility shift assays (EMSAs) to examine σNS-RNA complexes. Purified recombinant WT and Δ38 σNS proteins were equilibrated with terminally radiolabeled uncapped and nonpolyadenylated stem I 7SK small nuclear RNA (snRNA). The 7SK snRNA is a highly structured RNA that functions to regulate transcription in metazoans (54). Stem I of the 7SK snRNA encompasses the first 108 nucleotides, which are organized as a GC-rich stem-loop (55). We chose this RNA as a surrogate for a nonviral RNA. Increasing concentrations of WT and Δ38 σNS were incubated with 7SK stem I RNA, and σNS-RNA complexes were resolved by native electrophoresis and quantified by phosphorimaging (Fig. 9C). WT σNS slowed the migration of the 7SK stem I RNA in a concentration-dependent manner, whereas Δ38 σNS was unable to form detectable σNS-RNA complexes. We observed at least four distinct σNS-RNA complexes following incubation of RNA with WT σNS, suggesting that each σNS unit binds ∼27 nucleotides at saturation. Langmuir isotherm curve-fitting yielded an estimated KD (equilibrium dissociation constant) value of 260 nM for the interaction of σNS and 7SK stem I RNA (Fig. 9E and F). Increasing concentrations of WT σNS decreased the amount of free RNA, and at a concentration of 3 μM, no free RNA was detected, suggesting that at this concentration, the 7SK stem I RNA is saturated with σNS. Increasing concentrations of σNS also decreased the amount of aggregated RNA incapable of entering a polyacrylamide gel (Fig. 9C, arrowhead), suggesting that σNS has a function in unwinding RNAs.
To determine the affinity of σNS for viral RNA, we conducted a filter-binding assay using purified recombinant WT σNS and terminally radiolabeled uncapped and nonpolyadenylated s4 RNA. Increasing concentrations of WT σNS were incubated with s4 RNA. σNS-RNA complexes were spotted on a nitrocellulose membrane, and the unbound RNA was collected on a nylon membrane. The radiolabeled RNA on each membrane was quantified by phosphorimaging (Fig. 9D). Increasing concentrations of σNS increased the levels of RNA retained on the nitrocellulose membrane and decreased the free RNA collected on the nylon membrane. Langmuir isotherm curve fitting yielded a KD value of 290 nM for the σNS-s4 RNA interaction, which is comparable to the estimated KD value for the σNS-stem I 7SK RNA interaction (Fig. 9E and F). Collectively, these data indicate that σNS binds a viral RNA with affinity similar to that for a nonviral RNA, suggesting that σNS does not recognize a specific RNA sequence under these conditions.
The σNS protein impairs translation of viral and nonviral RNAs.
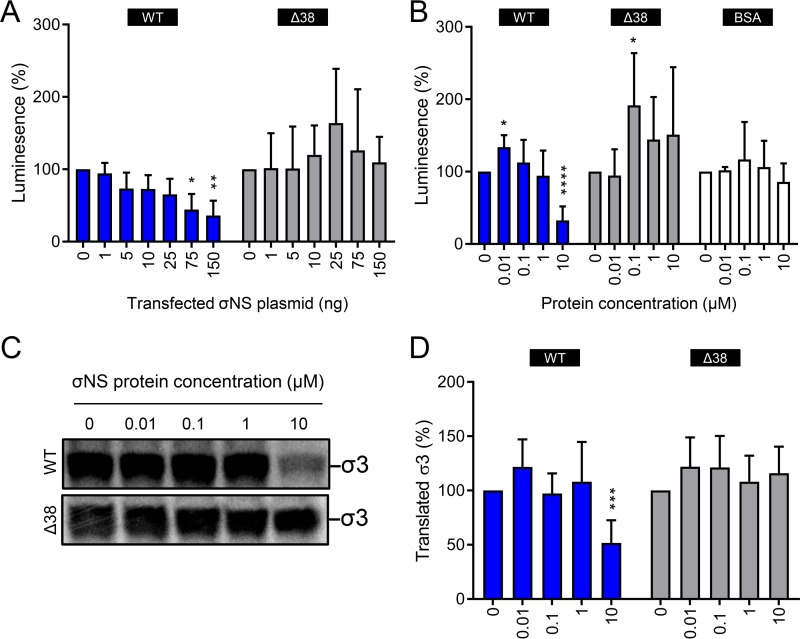
As σNS is an ssRNA-binding protein, we hypothesized that it might alter the translation of RNAs. To test whether σNS affects translation, we cotransfected HEK293T cells with increasing concentrations of plasmids encoding either WT or Δ38 σNS and a fixed concentration of a luciferase-encoding plasmid (Fig. 10A). Transcription derived from these plasmids yields RNAs that are capped and polyadenylated (53). Increasing concentrations of WT σNS plasmid decreased luciferase activity, a marker for luciferase translation. In contrast, transfection of Δ38 σNS did not decrease luciferase activity. These data indicate that σNS impairs translation of a nonviral capped and polyadenylated RNA and that this inhibition depends on the amino-terminal 38 amino acids of σNS.
FIG 10.
The σNS protein diminishes translation of viral and nonviral RNAs. (A) HEK293T cells were transfected with increasing concentrations of plasmids encoding either WT or Δ38 σNS and a fixed concentration of renilla luciferase-encoding plasmid and incubated for 24 h. The cells were lysed, and luciferase levels were quantified. The results are presented as mean luminescence percentages normalized to luciferase levels in the absence of σNS plasmid for at least three independent experiments. The error bars indicate SD. (B) Firefly luciferase uncapped and polyadenylated RNA was incubated with increasing concentrations of purified recombinant WT or Δ38 σNS or BSA at room temperature for 10 min. Protein-RNA complexes were added to wheat germ extracts and incubated at 25°C for 1 h. Luciferase synthesis was quantified by luciferase assay. The results are presented as mean luminescence percentages normalized to luciferase levels in the absence of protein for at least three independent experiments. The error bars indicate SD. (C) Capped, 2′-O-methylated, and nonpolyadenylated s4 RNA was incubated with increasing concentrations of purified recombinant WT or Δ38 σNS at room temperature for 10 min. Protein-RNA complexes were added to wheat germ extracts and incubated at 25°C for 1 h in the presence of [35S]methionine. Samples were resolved by SDS-PAGE and visualized by phosphorimaging. (D) Pixel intensity analysis of the σ3 protein band for at least three independent experiments. The error bars indicate SD. Values that differ significantly from the no-protein condition values by one-sample t test for each time point are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
To test whether σNS displays a broad-spectrum inhibitory effect on translation, we conducted in vitro translation assays using an uncapped and polyadenylated RNA in the presence or absence of σNS. Increasing concentrations of purified recombinant WT or Δ38 σNS or bovine serum albumin (BSA) as a control were incubated with uncapped and polyadenylated luciferase RNA prior to incubation with wheat germ extracts to initiate translation. Wheat germ extracts were chosen for these experiments instead of rabbit reticulocyte lysates because of their higher stringency and dependency on the type of RNA and the reaction conditions for efficient translation (56). After incubation, we quantified luciferase activity (Fig. 10B). WT σNS at a concentration of 10 μM (∼100:1 molar excess) decreased translation of luciferase RNA by 67%, corroborating the observations made in experiments using HEK293T cells. In contrast, Δ38 σNS and the BSA control did not diminish translation efficiency.
Lastly, we examined the effect of σNS on translation of a viral RNA. We incubated increasing concentrations of purified, recombinant WT or Δ38 σNS with capped, 2′-O-methylated and nonpolyadenylated s4 RNA prior to incubation with wheat germ extracts to initiate translation in the presence of [35S]methionine. The protein products were resolved by SDS-PAGE and quantified by phosphorimaging (Fig. 10C and D). Similar to findings made in the luciferase experiments, 10 μM WT σNS (∼80:1 molar excess) significantly impaired translation of the s4 RNA. Thus, when present at high concentrations, σNS inhibits translation of different types of RNAs.
DISCUSSION
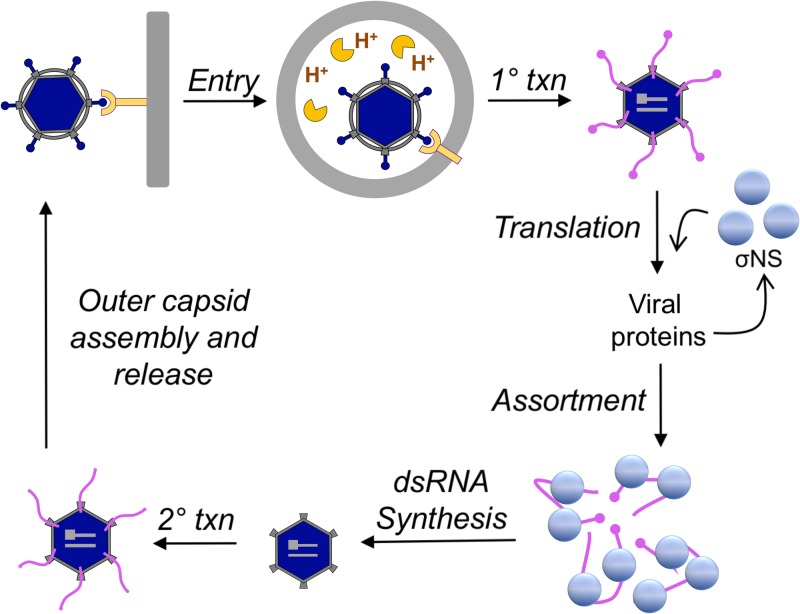
In this study, we found that the mammalian reovirus σNS protein, a nonstructural protein expressed early in reovirus infection (9, 22), is required for a process leading to viral genome replication (Fig. 11). In the absence of σNS, reovirus infection progressed through primary transcription. However, subsequent replication steps, including protein synthesis, genome replication, secondary transcription, formation of reovirus inclusions, and generation of progeny, were impaired. Considering the kinetics of the individual replication steps, we found that the first function of σNS must occur between 6 and 9 h postadsorption (Fig. 3, 4, and 6). Using in vitro and cell-based approaches, we discovered that σNS forms complexes with RNA that organize into long filamentous structures. In addition, we observed that σNS increases the RNA half-life and protects RNA from degradation. These results suggest a function for σNS as an RNA-binding protein that prepares RNA transcripts for replication.
FIG 11.
Model of σNS function during reovirus replication. Reovirus enters cells by receptor-mediated endocytosis, and following acid-dependent proteolytic disassembly, the inner capsid or core is released into the cytoplasm (15, 16). Cores transcribe viral mRNAs corresponding to each gene segment (1° txn), which are subsequently translated to yield each viral protein. We hypothesize that σNS mediates at least two functions. (i) At low concentrations, σNS enhances translation. (ii) At high concentrations, σNS (either alone or with other viral proteins) binds and folds viral mRNAs into a conformation optimal for assortment and genome replication. After synthesis of the negative-sense strand and formation of new cores, secondary rounds of transcription occur (2° txn). The outer capsid assembles, silencing secondary transcription, and particles are released from infected cells by an unknown mechanism.
The σNS protein, originally called essential noncapsid protein σ2A (57), was first identified as a nucleic acid-binding protein in cellular extracts derived from reovirus-infected cells (40). σNS binds nucleic acids with positive cooperativity and covers a length of ∼25 to 27 nucleotides at saturation (42). In experiments using purified σNS and stem I 7SK RNA, a nucleic acid that is 108 nucleotides in length, we observed that at least four units of σNS bound stem I 7SK RNA at saturation (Fig. 9C). These data suggest that σNS covers a length of ∼27 nucleotides, consistent with previous results (42). Residues 2 to 11 of σNS contribute to RNA binding, although Δ2-11 σNS mutants retain the capacity to bind RNA, albeit poorly (51). Using a σNS mutant lacking the amino-terminal 38 amino acids, we found that these residues are absolutely required for RNA binding (Fig. 9C).
By conducting coupled in vitro transcription and translation of WT and Δ38 σNS in the presence of [35S]methionine and resolving the radiolabeled translation products by native PAGE (Fig. 9B), we observed that WT σNS forms higher-order complexes when RNA is present, whereas Δ38 does not. When we treated WT σNS samples with RNase A, most of the high-molecular-mass species disappeared, except one of ∼900 kDa. This ∼900-kDa band was also observed following transcription and translation of Δ38 σNS, further indicating that the existence of the complex does not depend on the presence of RNA. There are at least two possibilities that could account for this ∼900-kDa complex. First, σNS may have the capacity to form large oligomers in the absence of RNA and independently of its amino-terminal 38 amino acids. Second, both WT and Δ38 σNS may interact with a component of rabbit reticulocyte lysates in an RNA-independent manner, forming an ∼900-kDa complex.
Previous competition assays indicated that σNS does not preferentially bind viral over nonviral RNA (42). Our findings corroborate these results, as we calculated similar KD values for σNS binding to viral s4 RNA and nonviral stem I 7SK RNA (290 nM and 260 nM, respectively) (Fig. 9E and F). Studies to quantify the affinity of a protein for RNA are usually conducted in vitro, and it is possible that during infection, other factors contribute to the affinity and specificity of σNS-RNA interactions. For example, sequestration of σNS in discrete cellular environments (22) might account for increased specificity for viral RNAs. In addition, μNS and μ2, each of which interacts with σNS in reovirus inclusions (23, 24, 43, 45), also display affinity for RNA (58, 59). A complex of these proteins and σNS might be responsible for binding viral RNAs. It is also possible that σNS binds cellular RNAs to promote the translation of cellular proteins within these structures (47), as several host proteins are found within inclusions, for example, Hsc70 (60) and the TRiC chaperonin (61).
In the EMSAs conducted in our study, we observed an interesting feature of σNS-RNA interactions (Fig. 9C, arrowhead). Increasing concentrations of σNS resulted in a reduction of aggregated RNA that did not enter the gel. These results suggest that σNS binding to RNA leads to RNA rearrangements allowing the RNA to migrate through the gel matrix. RNA-remodeling proteins that resolve RNA structures are called RNA chaperones (62), which are encoded by some RNA viruses (63). There are three lines of evidence from our study and others suggesting that σNS is an RNA chaperone. First, RNA chaperones differ from other RNA-folding proteins by binding nucleic acids nonspecifically (Fig. 9) and acting in an ATP-independent manner, which has been demonstrated for σNS in strand displacement experiments (42). Second, the avian reovirus σNS protein, which is functionally homologous to mammalian reovirus σNS (64, 65), acts as an RNA chaperone in vitro (27), as does the rotavirus NSP2 nonstructural protein (28, 63). Third, our cryo-EM analysis demonstrated that σNS coats RNAs and forms filamentous structures (Fig. 8). Coating of RNAs by viral proteins has been suggested to mediate the activity of some viral RNA chaperones, and this activity ensures protection and folding of their target RNAs. This is the case for HIV-1 NCp7 (66, 67), poliovirus 3AB (68), and tomato bushy stunt virus p33 (69). Therefore, it is likely that viruses of the family Reoviridae encode RNA chaperones to resolve kinetically trapped RNA conformations and facilitate replication.
Coating of RNAs also might account for the impairment in translation observed when σNS is incubated with RNAs at molar excess, as it might sterically hinder effective ribosome scanning (Fig. 10) (70). In this regard, we envision a bimodal function for σNS in the regulation of translation. At low concentrations of σNS before RNAs become saturated, translation is enhanced, as our in vitro luciferase translation experiments suggested (Fig. 10B). It is possible that σNS directly stimulates translation of viral mRNAs via interactions with a domain that differs from that responsible for RNA binding, as our experiments also showed a tendency for Δ38 σNS to increase translation (Fig. 10). This σNS-mediated translation enhancement effect might be partially responsible for the impairment in protein synthesis observed in T3D-infected σNS-siRNA cells (Fig. 5). At high concentrations of σNS, translation is impaired via a mechanism dependent on the amino-terminal 38 amino acids of the protein (Fig. 10). Concentration-dependent inhibition of translation is a property of other viral proteins, like HIV Gag, which inhibits translation of its own RNA at high concentrations (71), and the coat protein from potato virus A (72). Considering that the same viral RNA is used for translation and replication, a concentration-dependent bimodal function for σNS might be necessary to transition between these replication cycle steps.
There is compelling evidence indicating that σNS is required for genome replication (Fig. 6) (37, 73, 74), and we propose the following model for σNS function during reovirus replication (Fig. 11). Early in infection, σNS concentrations are low, and viral ssRNAs are in excess. Therefore, viral ssRNAs are minimally occupied by σNS. Perhaps at this point, σNS recruits the host translational apparatus to enhance viral translation. Interactions of σNS with the translation initiation machinery have been demonstrated, as σNS colocalizes and coimmunoprecipitates with the ribosomal subunit pS6R and the translation initiation factor eIF3A during infection (47). In addition, stress granules form following infection and disassemble as infection proceeds, and their disruption is required for efficient viral translation (75, 76). σNS is required to disrupt stress granules during infection, as it associates with the stress granule effector protein, G3BP1 (77).
Later in infection, once stress granules are disassembled, inclusions are established, and the translation apparatus is in place, σNS becomes more abundant, and its RNA-binding function serves to prepare ssRNAs for genome replication. Such preparation could involve increasing the RNA half-life (Fig. 7), protecting RNAs from RNases (42), or folding RNAs into a conformation required for assortment, analogous to an activity of rotavirus NSP2 (28). Considering that reovirus RNAs are nonpolyadenylated and uncapped through most of the replication cycle (78, 79) and that capping and polyadenylation increase mRNA stability (80), viral proteins that bind and protect viral RNAs, such as σNS, might be necessary for successful reovirus replication. It is possible that the σNS-mediated RNA protection activity is increased by sequestering RNAs in reovirus inclusions, thus preventing them from being recognized by the cytoplasmic RNA decay machinery (80). σNS also might localize viral RNAs to sites required for interaction with the RNA-dependent RNA polymerase λ3. The σNS-interacting protein μNS has a defined λ3-interacting domain (43) and might be responsible for bridging σNS to the genome replication machinery. σNS also could directly interact with λ3 before the replication of the genome. Finally, an effect of σNS on genome replication could be a consequence of its function to promote translation of λ3 and other viral proteins (Fig. 5 and 11).
We still do not understand the mechanism by which σNS dissociates from viral RNAs before the formation of viral cores. Posttranslational modifications of RNA-binding proteins can neutralize the basic residues responsible for interaction with RNAs. This mechanism has been suggested for reovirus σNS (52) and shown for rubella virus capsid protein (81), tomato bushy stunt virus p33 (82), and hepatitis B virus core protein (83). Rotavirus NSP2 is phosphorylated (84), and similar to the case of σNS, NSP2 temperature-sensitive mutants fail to synthesize dsRNA (85). Whether σNS directly participates in the replication of the dsRNA genome remains a key unanswered question.
Collectively, data gathered in our study provide evidence of a function for σNS in promoting the replication of the reovirus genome and establish a foundation for future studies to determine the mechanism by which σNS acts prior to dsRNA synthesis. This work sets the stage to better understand reovirus RNA assortment and replication.
MATERIALS AND METHODS
Cells and viruses.
HEK293T cells engineered to express an siRNA against the σNS-encoding S3 gene of reovirus strain T3D (48) were single-cell sorted at the Vanderbilt University Medical Center Flow Cytometry Shared Resource. Single-cell sorting was conducted by resuspending ∼2.5 × 106 cells in phosphate-buffered saline (PBS) supplemented to contain 2% fetal bovine serum (FBS) (Gibco) in a volume of 500 μl. Propidium iodide (Sigma) was added at a concentration of 1 μg/ml, and single cells were sorted into wells of a 96-well plate and maintained in 100 μl of Dulbecco's modified Eagle medium (DMEM) (Life Technologies) supplemented to contain 5% FBS, 2 mM l-glutamine (Life Technologies), 100 U/ml of penicillin, 100 μg/ml of streptomycin (Life Technologies), 0.25 mg/ml of amphotericin B (Sigma), and 5 μg/ml of puromycin (Sigma). Cell clones were propagated for approximately 2 weeks, after which time the cells were divided into two plates: one was maintained in supplemented DMEM, and the other was infected with reovirus T3D for 18 h. Infection was scored by fluorescent focus assay using guinea pig σNS polyclonal antiserum (44) as described previously (86). Clones with high knockdown efficiency were selected and passaged. HEK293T cells (ATCC CRL-3216), HEK293T cells expressing an S3-specific siRNA (σNS-siRNA cells), and HEK293T cells expressing a GFP-specific siRNA (GFP-siRNA cells) (48) were maintained in DMEM supplemented to contain 5% FBS, 2 mM l-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, 0.25 mg/ml of amphotericin B, and 5 μg/ml of puromycin (for σNS-siRNA and GFP-siRNA cells). Murine L929 fibroblasts adapted to spinner bottles were maintained in Joklik's modified Eagle's minimal essential medium (JMEM) (Lonza) as described previously (11).
Reoviruses were recovered using plasmid-based reverse genetics (49, 87) and purified as described previously (88, 89). For generation of T3D-R virus, plasmid pT7-S3T3D (Addgene; 33284) was altered by QuikChange (Stratagene) site-directed mutagenesis to contain the synonymous mutations C632T, A635C, A642G, and A649G (Fig. 1B) using T3D_R_QC primers (Table 2). To confirm mutations, viral RNA was extracted from purified virions and subjected to OneStep RT-PCR (Qiagen) using T3D_S3 primers (Table 2). Purified PCR products were analyzed by Sanger sequencing. Viral titers were determined by plaque assay using L929 cells (89).
TABLE 2.
PCR primers
| Primer name | Sequence (5′→3′) |
|
|---|---|---|
| Forward (F) | Reverse (R) | |
| T3D_R_QC | AGATATCCATGCATGCCAATCCCTTGAGGCCATCGTCCAGAAGCTGTTCCG | CGGAACAGCTTCTGGACGATGGCCTCAAGGGATTGGCATGCATGGATATCT |
| T3D_T7S4 | GCGGGTTAATACGACTCACTATAGCTATT | GATGAATGAAGCCTGTCCCACGT |
| T3D_S3 | CCTGATGCGCCAATGTCTAA | CTGTCTCCTCGCAATACAACTC |
| T3D_S4 | GAAGCATTTGCCTCACCATAG | GATCTGTCCAACTTGAGTGTATTG |
| human_GADPH | CCCATCACCATCTTCCAG | ATGACCTTGCCCACAGCC |
| T3D_S4_EagI | AGATGCCATGCCGACGGCCGATGAATGAAGCC | GGCTTCATTCATCGGCCGTCGGCATGGCATCT |
| T1L_S3_RS | CGACGGATCCATGGCTTCCTCACTCAGGGC | ATCACAGGCGGCCGCTTACACGCGAATTGGAAACACCAGC |
| T1L_S3del38 | AACGTTGTTGAGTATCAAATCCGTACAGG | CATGGATCCGAGCTCGGTACCAA |
| T3D_S3_GA | TTGGTACCGAGCTCGGATCCATGGCTTCCTCACTCAGAG | TCTAGACTCGAGCGGCCGCCTTACACGCGAATCGGAAAAAC |
| pcDNA3.1_GA | GGCGGCCGCTCGAGTCTA | GGATCCGAGCTCGGTACC |
| T3D_S3del38_GA | CATGGATCCGAGCTCGGTACC | TTGGTACCGAGCTCGGATCCATGAATGTGGTTGAGTATCAAATTCG |
| T3D_S4_qPCR | CGCTTTTGAAGGTCGTGTATCA | CTGGCTGTGCTGAGATTGTTTT |
Plasmid cloning and DNA transfections.
Plasmids for in vitro translation of the T3D s4 mRNA were engineered by introducing an EagI restriction site at the 3′ end of the S4 cDNA in pT7-S4T3D (Addgene; 33285) using QuikChange site-directed mutagenesis and T3D_S4_EagI primers (Table 2). Plasmids for coupled in vitro transcription and translation of strain T1L σNS were generated by subcloning the open reading frame (ORF) of the T1L S3 cDNA into pcDNA3.1+ using PCR amplification and T1L_S3_RS primers (Table 2). The T1L S3 Δ38 plasmid was engineered by PCR amplification of the S3 cDNA from plasmid pcDNA3.1+ T1L S3 using T1L_S3del38 primers. Plasmids for coupled in vitro transcription and translation and transient expression of strain T3D WT and Δ38 σNS were generated by subcloning the full-length ORF and the Δ38 ORF of the T3D S3 cDNA into pcDNA3.1+ by Gibson assembly (New England BioLabs) using primers T3D_S3_GA, pcDNA3.1_GA, and T3D_S3del38_GA (Table 2). Plasmids for bacterial expression of WT and Δ38 σNS were engineered by Epoch Life Science, Inc. Plasmid T7-FLuc encoding firefly luciferase (FLuc) was purchased from New England BioLabs, and plasmid SV40-RLuc encoding renilla luciferase (RLuc) was provided by Bernardo Mainou (Emory University).
Plasmids for transient protein expression were transfected using FuGene 6 transfection reagent (Promega) according to the manufacturer's instructions. Plasmids for recovering viruses using reverse genetics were transfected using TransIT-LT1 transfection reagent (Mirus) as described previously (49, 87).
Preparation of rabbit λ3-specific polyclonal antiserum.
T3D λ3 was expressed in Sf21 cells using a recombinant baculovirus (90) at the Baculovirus/Monoclonal Antibody Advanced Technology Core Laboratory at Baylor College of Medicine and purified as described previously (90). A rabbit was immunized with 100 μg of λ3 protein in complete Freund's adjuvant at Cocalico Biologicals. Antigen boosts containing 50 μg of λ3 protein in incomplete Freund's adjuvant were administered at days 14, 21, 49, and 134 following the initial inoculation. Antigen boosts containing 100 μg of λ3 protein in incomplete Freund's adjuvant were administered at days 120, 183, and 211 following the initial inoculation. Serum samples were obtained at days 35, 56, 148, and 197 following the initial inoculation and tested for reactivity against purified T3D virions by immunoblotting. Final exsanguination was conducted 225 days after the initial inoculation.
Native PAGE, SDS-PAGE, immunoblotting, and phosphorimaging.
Samples for native PAGE were diluted in 4× native PAGE sample buffer (ThermoFisher) and resolved in 4 to 16% native PAGE Bis-Tris acrylamide gels (ThermoFisher) using the Blue Native PAGE Novex Bis-Tris gel system (ThermoFisher) as described previously (61).
Samples for denaturing SDS-PAGE were diluted in 5× SDS-PAGE sample buffer, incubated at 95°C for 5 min, and resolved in 10% Mini-Protean TGX gels (Bio-Rad).
Native PAGE and SDS-PAGE gels were transferred to polyvinylidene difluoride (PVDF) and nitrocellulose membranes, respectively (61). The following antibodies were used for immunoblotting: guinea pig σNS polyclonal antiserum (44), chicken μNS polyclonal antiserum (47), rabbit λ3 polyclonal antiserum, and mouse α-tubulin monoclonal antibody (Cell Signaling Technology). The following antibodies were used as detection reagents: IRDye800CW donkey anti-guinea pig, IRDye680RD donkey anti-chicken, IRDye800CW goat anti-rabbit, and IRDye680LT goat anti-mouse (Li-Cor).
The membranes were scanned using an Odyssey CLx imaging system (Li-Cor). The pixel intensities of the bands for all gels and membranes were quantified using Image Studio Software (Li-Cor).
Native PAGE and SDS-PAGE gels containing radiolabeled products were incubated in 40% methanol, 10% acetic acid at room temperature for 1 h, washed, and dried. The dried gels were exposed on a phosphorimaging screen and imaged using a PerkinElmer Cyclone phosphor system scanner.
Purification of recombinant σNS protein.
Escherichia coli BL21(DE3)pLysS (Promega) cultures were transformed with T3D WT or Δ38 σNS-encoding plasmids and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma) after reaching an optical density (600 nm) of 0.6 to 0.7. The cells were resuspended in 50 mM Tris-HCl (pH 8), 500 mM NaCl, 10 mM imidazole, and 1 mM dithiothreitol (DTT) and supplemented with protease inhibitor cocktail (Roche). The cells were lysed using a microfluidizer, followed by removal of cell debris by centrifugation. The clarified cell lysates were loaded onto Ni-nitrilotriacetic acid (NTA) columns (Qiagen), and σNS was eluted using a gradient of 50 mM Tris-HCl (pH 8), 0.5 M NaCl, 0.5 M imidazole, and 1 mM DTT. Elution fractions were dialyzed against 20 mM Tris-HCl (pH 8), 100 mM NaCl, 10 mM imidazole, 1 mM EDTA, and 1 mM DTT. The eluted protein was incubated with thrombin (1 U per 100 g of recombinant protein) at 4°C for 12 h to remove the His tag. Protein samples were reloaded onto a Ni-NTA column to remove uncleaved His-tagged σNS. The flowthrough was loaded onto a Superose 6 10/300 GL column (GE Healthcare) in 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 1 mM DTT. The eluted fractions were concentrated using a 30-kDa centrifugal filter unit (Millipore).
Immunofluorescence microscopy of reovirus-infected cells.
Reovirus was allowed to adsorb onto cells at a multiplicity of infection (MOI) of 100 PFU per cell and incubated at 37°C for 24 h. The cells were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in PBS at room temperature for 20 min; permeabilized with 1% Triton X-100 in PBS at room temperature for 5 min; blocked with 0.5% BSA, 0.1% glycine, and 0.05% Tween 20; and stained with chicken μNS polyclonal antiserum, Alexa-Fluor secondary antibody (Invitrogen), and 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) to visualize nuclei. Images were captured using a Zeiss LSM 710 laser scanning confocal microscope and analyzed using a Zeiss LSM5 series image browser.
RNA extraction and purification.
To purify cellular RNA, cells were lysed using TRIzol reagent (ThermoFisher), and RNA was extracted with chloroform. To purify RNA from in vitro transcription reactions, samples were combined with an equal volume of lysis buffer (ThermoFisher) supplemented to contain 1% β-mercaptoethanol (BME). An equal volume of 70% ethanol was added, and the RNA was purified using a PureLink RNA minikit (ThermoFisher) according to the manufacturer's instructions.
S4 quantitative RT-PCR.
Total and single-strand negative-sense s4 RNAs were quantified using qScript XLT one-step RT-qPCR ToughMix, Low ROX (Quanta Bioscience), and T3D_S4_qPCR primers (Table 2) according to the manufacturer's instructions. The following RT-qPCR cycling protocol was used: cDNA synthesis (50°C for 10 min), initial denaturation (95°C for 1 min), and 40 PCR cycles (95°C for 10 s followed by a data collection step at 60°C for 1 min). S4 cDNA was detected using a fluorogenic probe (5′-FAM [fluorescent fluorescein]-AGCGCGCAAGAGGGATGGGA-BHQ [black hole quencher]-1-3′; Biosearch Technologies). For the single-strand RT-qPCR, the following modifications were included: RNA was incubated at 95°C for 3 min and immediately placed on ice; reverse transcription was conducted using only the forward primer T3D_S4_qPCR; and for the quantitative-PCR step, the reverse T3D_S4_qPCR primer was included (50).
NanoString RNA quantification and analysis.
Probes specific for each reovirus gene segment positive-sense RNA were designed by NanoString Technologies using proprietary software (Table 1). RNA was purified from infected cells and incubated with probes in hybridization buffer according to the manufacturer's instructions. Following hybridization, excess probe was removed using an nCounter Prep Station automated liquid handler. Probe-target complexes were transferred to an nCounter cartridge for immobilization and loaded onto an nCounter digital analyzer for imaging and quantification of each target RNA. The quantified expression data were analyzed using nSolver analysis software, including image quality controls and background subtraction. Counts for each probe were normalized using the geometric mean of four cellular transcripts (GUSB, HPRT, SARM1, and RSP6).
In vitro transcription reactions.
Templates for in vitro transcription of the T3D S4 gene segment were generated from pT7-S4T3D by PCR amplification using T3D_T7S4 primers (Table 2). Templates were transcribed in vitro using a HiScribe T7 high-yield RNA synthesis kit (New England BioLabs) according to the manufacturer's instructions with the following modifications. For synthesis of radiolabeled RNA, 125 ng of PCR product was used in the presence of radiolabeled [α-32P]UTP (PerkinElmer) in 20-μl reaction mixtures at room temperature for 2 h. For synthesis of nonradiolabeled RNA, 2 μg of plasmid DNA template was used in 40-μl reaction mixtures at 37°C for 2 h. DNA templates were degraded by on-column incubation with PureLink DNase (ThermoFisher). For some experiments, RNA was capped using vaccinia capping enzyme (New England BioLabs) and 2′-O-methyltransferase (New England BioLabs) in one-step reaction mixtures according to the manufacturer's instructions.
Stem I of 7SK RNA (5′-GGAUGUGAGGGCGAUCUGGCUGCGACAUCUGUCACCCCAUUGAUCGCCAGGGUUGAUUCGGCUGAUCUGGCUGGCUAGGCGGGUGUCCCCUUCCUCCCUCACCGCUCC-3′) was synthesized using T7 RNA polymerase from PCR-generated DNA templates. The RNA was purified from a gel slice following electrophoresis in a 7 M urea–1× Tris-borate-EDTA (TBE)–6% 29:1 polyacrylamide gel.
In vitro cell-free RNA degradation assay and electrophoresis.
s4 RNA transcripts (<10 pM; 5 × 105 cpm) were incubated with various concentrations of σNS in a volume of 10 μl at room temperature for 10 min. σNS-RNA complexes were incubated with 26 μl of a 6-mg/ml stock of HeLa S100 cytoplasmic extracts (Speed Biosystems). At various intervals, RNA was purified from 8-μl aliquots of the treatment mixtures. The RNA was diluted in 2× Novex TBE-urea sample buffer (ThermoFisher), resolved in 6% TBE-urea gels (ThermoFisher), exposed on a phosphorimaging screen, and visualized using a PerkinElmer Cyclone phosphor system scanner.
RNA degradation assay in cells.
HEK293T cells seeded in 12-well plates were transfected using FuGene 6 with 500 ng of plasmid encoding either WT or Δ38 σNS and incubated for 20 h. The cells were transfected a second time using FuGene 6 with 50 ng of pCAG_S4T3D plasmid (49) and incubated for 4 h. The cells were treated with 10 μg/ml of actinomycin D (Sigma) and lysed at various intervals prior to RNA extraction. Expression of s3, s4, and human GADPH RNAs was confirmed by RT-PCR amplification using primers T3D_S3, T3D_S4, and human_GADPH.
In vitro translation reactions.
RNAs were translated in vitro using wheat germ extracts (Promega) according to the manufacturer's instructions with the following modifications. RNAs at a concentration of 10 μg/ml were incubated at 65°C for 3 min, placed immediately on ice, and incubated with various concentrations of WT or Δ38 σNS at room temperature for 10 min. Protein-RNA complexes were incubated with wheat germ extracts supplemented with 80 μM amino acid mixture minus methionine (Promega) and 0.8 U/μl of RNasin (Promega). To synthesize nonradioactive and radioactive proteins, 80 μM amino acid mixture minus leucine (Promega) or 0.7 μCi/μl of [35S]methionine (PerkinElmer) was added, respectively. Translation reaction mixtures were incubated at 25°C for 1 h, followed by freezing at −20°C.
Coupled in vitro transcription and translation reactions.
Coupled in vitro transcription and translation reactions were conducted using the TNT-coupled rabbit reticulocyte lysate system (Promega) according to the manufacturer's instructions. Reaction mixtures were incubated at 30°C for 1 h and terminated by 4-fold dilution in stop buffer (20 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 5 mM magnesium acetate, 5 mM EDTA, 2 mM methionine) supplemented to contain a final concentration of 1 mM DTT and 2 mM puromycin. Protein samples were incubated with RNase A (Qiagen) under variable treatment conditions, depending on the experiment.
5′ labeling of RNA.
RNAs were dephosphorylated using calf intestinal alkaline phosphatase (CIP) (New England BioLabs) according to the manufacturer's instructions with the following modifications. CIP was used at a concentration of 0.5 U per 1 μg of RNA in 20-μl reaction mixtures. Following incubation at 37°C for 30 min, RNA was precipitated using phenol-chloroform (s4 RNAs) or gel purified (7SK stem I RNA) and labeled using T4 polynucleotide kinase (New England BioLabs) and [γ32P]ATP (PerkinElmer). The RNAs were purified using a G25 desalting column (GE Healthcare).
Luciferase assays.
To quantify RLuc activity in transfected cells, HEK293T cells seeded in 96-well plates were transfected with increasing concentrations of WT or Δ38 σNS plasmids (0 to 150 ng), decreasing concentrations of a noncoding plasmid (pT7-S2T3D; Addgene; 33283; 150 to 0 ng), and 50 ng of simian virus 40 (SV40) renilla luciferase plasmid, achieving a final DNA concentration of 200 ng/well, and incubated for 24 h. The cells were lysed with 20 μl of renilla luciferase lysis buffer (Promega) at room temperature for 20 min, followed by addition of 50 μl of renilla luciferase assay reagent (Promega) to each well.
To quantify FLuc activity in in vitro translation reactions, 10 ng of luciferase RNA was incubated with various concentrations of either WT or Δ38 σNS at room temperature for 10 min, followed by incubation with wheat germ extracts at 25°C for 1 h. Two-and-a-half-microliter aliquots from the translation reaction mixtures were added to 50 μl of luciferase assay reagent (Promega) in 96-well plates.
For FLuc and RLuc reactions, luminescence was quantified immediately following addition of the respective luciferase assay reagent with a Synergy H1 BioTek plate reader.
EMSAs.
Snap-cooled 7SK stem I RNA (<10 pM; 500 cpm) was incubated with increasing concentrations of either WT or Δ38 σNS in 100 mM NaCl, 50 mM HEPES (pH 7.2), 10% glycerol, and 1 μg BSA at room temperature for 10 min as described previously (91, 92). The reaction mixtures were loaded into wells of 0.5% native TBE polyacrylamide gels and resolved at 120 V at 4°C for 45 min. The gels were dried, exposed to phosphorimaging screens, visualized using ImageQuant TL (GE), and graphed using KaleidaGraph (Synergy Software).
Filter-binding assay.
s4 RNA (<10 pM; 500 cpm) was incubated with increasing concentrations of WT σNS in 100 mM NaCl, 50 mM HEPES (pH 7.2), 10% glycerol, and 1 μg BSA at room temperature for 10 min. Presoaked nitrocellulose (top) and nylon (bottom) membranes (GE) were used in a 96-well hybrid dot manifold assembly as described previously (93). After filtration, the membranes were air dried, exposed to phosphorimaging screens, and visualized using ImageQuant TL (GE). Data were plotted using KaleidaGraph (Synergy Software).
Electron microscopy of σNS-RNA complexes.
s4 RNA was incubated with either WT or Δ38 σNS at a molar ratio of 50:1 on ice for 1 h before plunge freezing using a Vitribot (MIV). The frozen specimens were imaged at ×40,000 magnification with defocus levels ranging from −2.0 to −3.5 μm using a JEM2010 (200-kV) cryo-electron microscope equipped with a charge-coupled-device (CCD) camera at the CryoEM Center at Baylor College of Medicine.
Statistical methods.
All experiments were conducted independently at least three times. Data are presented as means and standard deviations (SD). All statistical analyses were conducted using GraphPad Prism 7.00 data analysis software.
ACKNOWLEDGMENTS
We thank Christopher Lee, Anthony Lentscher, and Gwen Taylor for critical reviews of the manuscript. We thank Seema Lakdawala and John Parker for helpful discussions. We thank Bernardo Mainou for experimental suggestions and molecular biology expertise. We thank Adams Carroll for helping with illustrations. We thank the Vanderbilt University Medical Center Flow Cytometry Shared Resource for their cell-sorting services. We thank Cocalico Biologicals for their expertise and services in preparing polyclonal antiserum. We thank the Rangos Research Center Cell Imaging Core Laboratory for their assistance with confocal microscopy. We thank the CryoEM Center at Baylor College of Medicine for their electron microscopy support. This project used the University of Pittsburgh HSCRF Genomics Research Core NanoString analysis services.
This work was funded by United States Public Health Service awards R01 AI032539 (B.V.V.P. and T.S.D.), T32 GM007347 (J.J.K.), and F30 AI122563 (J.J.K.). Additional support was provided by the Vanderbilt International Scholar Program (P.F.Z.) and the Vanderbilt Program in Microbial Pathogenesis (P.F.Z.).
REFERENCES
- 1.Phillips MB, Stuart JD, Simon EJ, Boehme KW. 2018. Nonstructural protein sigma1s is required for optimal reovirus protein expression. J Virol 92:e02259-. doi: 10.1128/JVI.02259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu L, Crawford SE, Hyser JM, Estes MK, Prasad BV. 2012. Rotavirus non-structural proteins: structure and function. Curr Opin Virol 2:380–388. doi: 10.1016/j.coviro.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patton JT, Silvestri LS, Tortorici MA, Vasquez-Del Carpio R, Taraporewala ZF. 2006. Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr Top Microbiol Immunol 309:169–187. [DOI] [PubMed] [Google Scholar]
- 4.Stanifer ML, Kischnick C, Rippert A, Albrecht D, Boulant S. 2017. Reovirus inhibits interferon production by sequestering IRF3 into viral factories. Sci Rep 7:10873. doi: 10.1038/s41598-017-11469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes EC. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11:543–546. doi: 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faust TB, Binning JM, Gross JD, Frankel AD. 2017. Making sense of multifunctional proteins: human immunodeficiency virus type 1 accessory and regulatory proteins and connections to transcription. Annu Rev Virol 4:241–260. doi: 10.1146/annurev-virology-101416-041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouattara LA, Barin F, Barthez MA, Bonnaud B, Roingeard P, Goudeau A, Castelnau P, Vernet G, Paranhos-Baccala G, Komurian-Pradel F. 2011. Novel human reovirus isolated from children with acute necrotizing encephalopathy. Emerg Infect Dis 17:1436–1444. doi: 10.3201/eid1708.101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai JH, Williams JV, Edwards KM, Wright PF, Crowe JE Jr, Dermody TS. 2005. Prevalence of reovirus-specific antibodies in young children in Nashville, Tennessee. J Infect Dis 191:1221–1224. doi: 10.1086/428911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dermody TS, Parker JS, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Sutherland DM, Aravamudhan P, Dermody TS. 2018. An orchestra of reovirus receptors: still searching for the conductor. Adv Virus Res 100:223–246. doi: 10.1016/bs.aivir.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Barton ES, Connolly JL, Forrest JC, Chappell JD, Dermody TS. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J Biol Chem 276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- 12.Reiss K, Stencel JE, Liu Y, Blaum BS, Reiter DM, Feizi T, Dermody TS, Stehle T. 2012. The GM2 glycan serves as a functional co-receptor for serotype 1 reovirus. PLoS Pathog 8:e1003078. doi: 10.1371/journal.ppat.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. doi: 10.1016/S0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 14.Konopka-Anstadt JL, Mainou BA, Sutherland DM, Sekine Y, Strittmatter SM, Dermody TS. 2014. The Nogo receptor NgR1 mediates infection by mammalian reovirus. Cell Host Microbe 15:681–691. doi: 10.1016/j.chom.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mainou BA, Dermody TS. 2012. Transport to late endosomes is required for efficient reovirus infection. J Virol 86:8346–8358. doi: 10.1128/JVI.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mainou BA, Zamora PF, Ashbrook AW, Dorset DC, Kim KS, Dermody TS. 2013. Reovirus cell entry requires functional microtubules. mBio 4:e00405-. doi: 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert DH, Deussing J, Peters C, Dermody TS. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J Biol Chem 277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- 18.Sturzenbecker LJ, Nibert ML, Furlong DB, Fields BN. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol 61:2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin AJ. 1976. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence M7 GpppGm pC. J Biol Chem 251:5043–5053. [PubMed] [Google Scholar]
- 20.Skehel JJ, Joklik WK. 1969. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 39:822–831. doi: 10.1016/0042-6822(69)90019-1. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez de Castro I, Zamora PF, Ooms L, Fernandez JJ, Lai CM, Mainou BA, Dermody TS, Risco C. 2014. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio 5:e00931-. doi: 10.1128/mBio.00931-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker MM, Goral MI, Hazelton PR, Baer GS, Rodgers SE, Brown EG, Coombs KM, Dermody TS. 2001. Reovirus sNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J Virol 75:1459–1475. doi: 10.1128/JVI.75.3.1459-1475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller CL, Broering TJ, Parker JS, Arnold MM, Nibert ML. 2003. Reovirus sigma NS protein localizes to inclusions through an association requiring the mu NS amino terminus. J Virol 77:4566–4576. doi: 10.1128/JVI.77.8.4566-4576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broering TJ, Kim J, Miller CL, Piggott CD, Dinoso JB, Nibert ML, Parker JS. 2004. Reovirus nonstructural protein mNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J Virol 78:1882–1892. doi: 10.1128/JVI.78.4.1882-1892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooms LS, Jerome WG, Dermody TS, Chappell JD. 2012. Reovirus replication protein mu2 influences cell tropism by promoting particle assembly within viral inclusions. J Virol 86:10979–10987. doi: 10.1128/JVI.01172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah PNM, Stanifer ML, Hohn K, Engel U, Haselmann U, Bartenschlager R, Krausslich HG, Krijnse-Locker J, Boulant S. 21 July 2017. Genome packaging of reovirus is mediated by the scaffolding property of the microtubule network. Cell Microbiol 19. doi: 10.1111/cmi.12765. [DOI] [PubMed] [Google Scholar]
- 27.Borodavka A, Ault J, Stockley PG, Tuma R. 2015. Evidence that avian reovirus sigmaNS is an RNA chaperone: implications for genome segment assortment. Nucleic Acids Res 43:7044–7057. doi: 10.1093/nar/gkv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borodavka A, Dykeman EC, Schrimpf W, Lamb DC. 2017. Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. Elife 6:e27453. doi: 10.7554/eLife.27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald SM, Patton JT. 2011. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol 19:136–144. doi: 10.1016/j.tim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan EM, Zweerink HJ. 1975. Characterization of transcriptase and replicase particles isolated from reovirus-infected cells. Virology 68:455–466. doi: 10.1016/0042-6822(75)90286-X. [DOI] [PubMed] [Google Scholar]
- 31.Sakuma S, Watanabe Y. 1972. Reovirus replicase-directed synthesis of double-stranded ribonucleic acid. J Virol 10:628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray KE, Nibert ML. 2007. Guanidine hydrochloride inhibits mammalian orthoreovirus growth by reversibly blocking the synthesis of double-stranded RNA. J Virol 81:4572–4584. doi: 10.1128/JVI.02106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farsetta DL, Chandran K, Nibert ML. 2000. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J Biol Chem 275:39693–39701. [DOI] [PubMed] [Google Scholar]
- 34.Simon EJ, Howells MA, Stuart JD, Boehme KW. 2017. Serotype-specific killing of large cell carcinoma cells by reovirus. Viruses 9:E140. doi: 10.3390/v9060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, Meisel M, Kim SM, Discepolo V, Pruijssers AJ, Ernest JD, Iskarpatyoti JA, Costes LM, Lawrence I, Palanski BA, Varma M, Zurenski MA, Khomandiak S, McAllister N, Aravamudhan P, Boehme KW, Hu F, Samsom JN, Reinecker HC, Kupfer SS, Guandalini S, Semrad CE, Abadie V, Khosla C, Barreiro LB, Xavier RJ, Ng A, Dermody TS, Jabri B. 2017. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356:44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown JJ, Short SP, Stencel-Baerenwald J, Urbanek K, Pruijssers AJ, McAllister N, Ikizler M, Taylor G, Aravamudhan P, Khomandiak S, Jabri B, Williams CS, Dermody TS. 2018. Reovirus-induced apoptosis in the intestine limits establishment of enteric infection. J Virol 92:e02062-. doi: 10.1128/JVI.02062-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiener JR, Joklik WK. 1987. Comparison of the reovirus serotype 1, 2, and 3 S3 genome segments encoding the nonstructural protein sNS. Virology 161:332–339. doi: 10.1016/0042-6822(87)90125-5. [DOI] [PubMed] [Google Scholar]
- 38.Goral MI, MochowGrundy M, Dermody TS. 1996. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation. Virology 216:265–271. doi: 10.1006/viro.1996.0059. [DOI] [PubMed] [Google Scholar]
- 39.Gomatos PJ, Stamatos NM, Sarkar NH. 1980. Small reovirus-specific particle with polycytidylate-dependent RNA polymerase activity. J Virol 36:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomatos PJ, Prakash O, Stamatos NM. 1981. Small reovirus particles composed solely of sigma-Ns with specificity for binding different nucleic-acids. J Virol 39:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatos NM, Gomatos PJ. 1982. Binding to selected regions of reovirus mRNAs by a nonstructural reovirus protein. Proc Natl Acad Sci U S A 79:3457–3461. doi: 10.1073/pnas.79.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillian AL, Schmechel SC, Livny J, Schiff LA, Nibert ML. 2000. Reovirus protein sigmaNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J Virol 74:5939–5948. doi: 10.1128/JVI.74.13.5939-5948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller CL, Arnold MM, Broering TJ, Hastings CE, Nibert ML. 2010. Localization of mammalian orthoreovirus proteins to cytoplasmic factory-like structures via nonoverlapping regions of microNS. J Virol 84:867–882. doi: 10.1128/JVI.01571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker MM, Peters TR, Dermody TS. 2003. Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection. J Virol 77:5948–5963. doi: 10.1128/JVI.77.10.5948-5963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broering TJ, Parker JS, Joyce PL, Kim J, Nibert ML. 2002. Mammalian reovirus nonstructural protein microNS forms large inclusions and colocalizes with reovirus microtubule-associated protein micro2 in transfected cells. J Virol 76:8285–8297. doi: 10.1128/JVI.76.16.8285-8297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussiere LD, Choudhury P, Bellaire B, Miller CL. 2017. Characterization of a replicating mammalian orthoreovirus with tetracysteine-tagged muNS for live-cell visualization of viral factories. J Virol 91:e01371-. doi: 10.1128/JVI.01371-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desmet EA, Anguish LJ, Parker JS. 2014. Virus-mediated compartmentalization of the host translational machinery. mBio 5:e01463-. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi T, Chappell JD, Danthi P, Dermody TS. 2006. Gene-specific inhibition of reovirus replication by RNA interference. J Virol 80:9053–9063. doi: 10.1128/JVI.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi T, Antar AA, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ooms LS, Kobayashi T, Dermody TS, Chappell JD. 2010. A post-entry step in the mammalian orthoreovirus replication cycle is a determinant of cell tropism. J Biol Chem 285:41604–41613. doi: 10.1074/jbc.M110.176255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillian AL, Nibert ML. 1998. Amino terminus of reovirus nonstructural protein sNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 52.Richardson MA, Furuichi Y. 1983. Nucleotide sequence of reovirus genome segment S3, encoding non-structural protein sNS. Nucleic Acids Res 11:6399–6408. doi: 10.1093/nar/11.18.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho EJ, Takagi T, Moore CR, Buratowski S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egloff S, Studniarek C, Kiss T. 2018. 7SK small nuclear RNA, a multifunctional transcriptional regulatory RNA with gene-specific features. Transcription 9:95–101. doi: 10.1080/21541264.2017.1344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourbigot S, Dock-Bregeon AC, Eberling P, Coutant J, Kieffer B, Lebars I. 2016. Solution structure of the 5′-terminal hairpin of the 7SK small nuclear RNA. RNA 22:1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDowell MJ, Villakom L, Lodish HF, Joklik WK. 1972. Translation of reovirus messenger RNAs synthesized in-vitro into reovirus polypeptides by several mammalian cell-free extracts. Proc Natl Acad Sci U S A 69:2649–2653. doi: 10.1073/pnas.69.9.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zweerink HJ, McDowell MJ, Joklik WK. 1971. Essential and non-essential non-capsid reovirus proteins. Virology 45:716–723. doi: 10.1016/0042-6822(71)90185-1. [DOI] [PubMed] [Google Scholar]
- 58.Antczak JB, Joklik WK. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal-antibodies directed against reovirus proteins. Virology 187:760–776. doi: 10.1016/0042-6822(92)90478-8. [DOI] [PubMed] [Google Scholar]
- 59.Brentano L, Noah DL, Brown EG, Sherry B. 1998. The reovirus protein mu 2, encoded by the M1 gene, is an RNA-binding protein. J Virol 72:8354–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaufer S, Coffey CM, Parker JS. 2012. The cellular chaperone hsc70 is specifically recruited to reovirus viral factories independently of its chaperone function. J Virol 86:1079–1089. doi: 10.1128/JVI.02662-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knowlton JJ, Fernandez de Castro I, Ashbrook AW, Gestaut DR, Zamora PF, Bauer JA, Forrest JC, Frydman J, Risco C, Dermody TS. 2018. The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat Microbiol 3:481–493. doi: 10.1038/s41564-018-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Xia H, Qian Q, Zhou X. 2015. RNA chaperones encoded by RNA viruses. Virol Sin 30:401–409. doi: 10.1007/s12250-015-3676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Touris-Otero F, Martinez-Costas J, Vakharia VN, Benavente J. 2004. Avian reovirus nonstructural protein microNS forms viroplasm-like inclusions and recruits protein sigmaNS to these structures. Virology 319:94–106. doi: 10.1016/j.virol.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 65.Touris-Otero F, Martinez-Costas J, Vakharia VN, Benavente J. 2005. Characterization of the nucleic acid-binding activity of the avian reovirus non-structural protein sigma NS. J Gen Virol 86:1159–1169. doi: 10.1099/vir.0.80491-0. [DOI] [PubMed] [Google Scholar]
- 66.Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol 410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 67.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. 2005. Disordered RNA chaperone proteins: from functions to disease. Cell Mol Life Sci 62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeStefano JJ, Titilope O. 2006. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J Virol 80:1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stork J, Kovalev N, Sasvari Z, Nagy PD. 2011. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409:338–347. doi: 10.1016/j.virol.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sagar V, Murray KE. 2014. The mammalian orthoreovirus bicistronic M3 mRNA initiates translation using a 5′ end-dependent, scanning mechanism that does not require interaction of 5′-3′ untranslated regions. Virus Res 183:30–40. doi: 10.1016/j.virusres.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson EC, Lever AM. 2006. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J Virol 80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Besong-Ndika J, Ivanov KI, Hafren A, Michon T, Makinen K. 2015. Cotranslational coat protein-mediated inhibition of potyviral RNA translation. J Virol 89:4237–4248. doi: 10.1128/JVI.02915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramig RF, Mustoe TA, Sharpe AH, Fields BN. 1978. A genetic map of reovirus. II. Assignment of the double-stranded RNA-negative mutant groups C, D, and E to genome segments. Virology 85:531–534. [DOI] [PubMed] [Google Scholar]
- 74.Cross RK, Fields BN. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. Virology 50:799–809. doi: 10.1016/0042-6822(72)90434-5. [DOI] [PubMed] [Google Scholar]
- 75.Qin Q, Hastings C, Miller CL. 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J Virol 83:11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qin Q, Carroll K, Hastings C, Miller CL. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2alpha phosphorylation and PKR. J Virol 85:8798–8810. doi: 10.1128/JVI.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choudhury P, Bussiere L, Miller CL. 2017. Mammalian orthoreovirus factories modulate stress granule protein localization by interaction with G3BP1. J Virol 91:e01298-17. doi: 10.1128/JVI.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stoltzfus CM, Shatkin AJ, Banerjee AK. 1973. Absence of polyadenylic acid from reovirus messenger ribonucleic acid. J Biol Chem 248:7993–7998. [PubMed] [Google Scholar]
- 79.Zarbl H, Skup D, Millward S. 1980. Reovirus progeny subviral particles synthesize uncapped mRNA. J Virol 34:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sokoloski KJ, Wilusz CJ, Wilusz J. 2006. Viruses: Overturning RNA turnover. RNA Biol 3:140–144. doi: 10.4161/rna.3.4.4076. [DOI] [PubMed] [Google Scholar]
- 81.Law LM, Everitt JC, Beatch MD, Holmes CF, Hobman TC. 2003. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J Virol 77:1764–1771. doi: 10.1128/JVI.77.3.1764-1771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stork J, Panaviene Z, Nagy PD. 2005. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology 343:79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Gazina EV, Fielding JE, Lin B, Anderson DA. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J Virol 74:4721–4728. doi: 10.1128/JVI.74.10.4721-4728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carpio RV, Gonzalez-Nilo FD, Jayaram H, Spencer E, Prasad BV, Patton JT, Taraporewala ZF. 2004. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J Biol Chem 279:10624–10633. doi: 10.1074/jbc.M311563200. [DOI] [PubMed] [Google Scholar]
- 85.Ramig RF, Petrie BL. 1984. Characterization of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J Virol 49:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ashbrook AW, Lentscher AJ, Zamora PF, Silva LA, May NA, Bauer JA, Morrison TE, Dermody TS. 2016. Antagonism of the sodium-potassium ATPase impairs Chikungunya virus infection. mBio 7:e00693-. doi: 10.1128/mBio.00693-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boehme KW, Ikizler M, Kobayashi T, Dermody TS. 2011. Reverse genetics for mammalian reovirus. Methods 55:109–113. doi: 10.1016/j.ymeth.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furlong DB, Nibert ML, Fields BN. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol 62:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Virgin HWT, Bassel-Duby R, Fields BN, Tyler KL. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J Virol 62:4594–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synt, hesis in a cage—structural studies of reovirus polymerase lambda3. Cell 111:733–745. doi: 10.1016/S0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 91.Lahr RM, Mack SM, Heroux A, Blagden SP, Bousquet-Antonelli C, Deragon JM, Berman AJ. 2015. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5′TOP sequence. Nucleic Acids Res 43:8077–8088. doi: 10.1093/nar/gkv748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lahr RM, Fonseca BD, Ciotti GE, Al-Ashtal HA, Jia JJ, Niklaus MR, Blagden SP, Alain T, Berman AJ. 2017. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife 6:e24146. doi: 10.7554/eLife.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rio DC. 2012. Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harb Protoc 2012:1078–1081. doi: 10.1101/pdb.prot071449. [DOI] [PubMed] [Google Scholar]