The induction of ER stress is a common outcome in cells infected with coronaviruses. The UPR initiated by ER stress is actively involved in viral replication and modulates the host innate responses to the invading viruses, but these underlying mechanisms remain incompletely understood. We show here that infection with the alphacoronavirus TGEV elicited ER stress in vitro and in vivo, and the UPR PERK-eIF2α branch was predominantly responsible for the suppression of TGEV replication by ER stress. Furthermore, the PERK-eIF2α axis inhibited TGEV replication through direct inhibition of viral proteins due to global translation inhibition and type I IFN induction. These findings highlight a critical role of the UPR PERK-eIF2α pathway in modulating host innate immunity and coronavirus replication.
KEYWORDS: endoplasmic reticulum stress, unfolded protein response, protein kinase R-like ER kinase, transmissible gastroenteritis virus, translation attenuation, interferon
ABSTRACT
Coronavirus replication is closely associated with the endoplasmic reticulum (ER), the primary cellular organelle for protein synthesis, folding, and modification. ER stress is a common consequence in coronavirus-infected cells. However, how the virus-induced ER stress influences coronavirus replication and pathogenesis remains controversial. Here, we demonstrated that infection with the alphacoronavirus transmissible gastroenteritis virus (TGEV) induced ER stress and triggered the unfolded protein response (UPR) in vitro and in vivo, and ER stress negatively regulated TGEV replication in vitro. Although TGEV infection activated all three UPR pathways (activating transcription factor 6 [ATF6], inositol-requiring enzyme 1 [IRE1], and protein kinase R-like ER kinase [PERK]), the virus-triggered UPR suppressed TGEV replication in both swine testicular (ST) and IPEC-J2 cells primarily through activation of the PERK-eukaryotic initiation factor 2α (eIF2α) axis, as shown by functional studies with overexpression, small interfering RNA (siRNA), or specific chemical inhibitors. Moreover, we demonstrated that PERK-eIF2α axis-mediated inhibition of TGEV replication occurs through phosphorylated eIF2α-induced overall attenuation of protein translation. In addition to direct inhibition of viral production, the PERK-eIF2α pathway activated NF-κB and then facilitated type I IFN production, resulting in TGEV suppression. Taken together, our results suggest that the TGEV-triggered PERK-eIF2α pathway negatively regulates TGEV replication and represents a vital aspect of host innate responses to invading pathogens.
IMPORTANCE The induction of ER stress is a common outcome in cells infected with coronaviruses. The UPR initiated by ER stress is actively involved in viral replication and modulates the host innate responses to the invading viruses, but these underlying mechanisms remain incompletely understood. We show here that infection with the alphacoronavirus TGEV elicited ER stress in vitro and in vivo, and the UPR PERK-eIF2α branch was predominantly responsible for the suppression of TGEV replication by ER stress. Furthermore, the PERK-eIF2α axis inhibited TGEV replication through direct inhibition of viral proteins due to global translation inhibition and type I IFN induction. These findings highlight a critical role of the UPR PERK-eIF2α pathway in modulating host innate immunity and coronavirus replication.
INTRODUCTION
The endoplasmic reticulum (ER) is the major cellular organelle for protein synthesis, folding, modification, and trafficking (1–3). When proteins entering the ER overwhelm its folding capacity, misfolded/unfolded proteins accumulate in the ER and cause ER stress. To restore ER homeostasis, the three primary ER stress sensors (protein kinase R-like ER kinase [PERK], activating transcription factor 6 [ATF6], and inositol-requiring enzyme 1 [IRE1]) are activated by accumulated misfolded/unfolded proteins in the ER and initiate the unfolded protein response (UPR) (1, 3–6). The activated PERK phosphorylates eukaryotic initiation factor 2α (eIF2α), attenuating global translation and selectively enhancing translation of activating transcription factor 4 (ATF4) (7, 8). Phosphorylation of eIF2α decreases translation of most mRNAs by impeding the recycling of eIF2α to its active GTP-bound form and inhibiting the delivery of the initiator Met-tRNAi to the initiation complex, allowing cells to conserve resources and to effectively restore homeostasis in the ER (9). The replication of coronaviruses, a family of important animal and human pathogens, is structurally and functionally associated with the ER (2, 10, 11). During coronavirus infection, several viral proteins are synthesized in the ER (12, 13). Moreover, double-membrane vesicles, the site of coronavirus RNA synthesis, and viral envelopes are derived from the ER membrane (14, 15). Upon completion of the replication and assembly cycle, virions bud from the ER-Golgi intermediate compartment (15, 16). Coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), mouse hepatitis virus (MHV), and infectious bronchitis virus (IBV), can all induce significant ER stress following infection (17–20). Several lines of evidence have indicated a link between viral replication and the PERK pathway (8, 21, 22). IBV infection suppresses phosphorylation of eIF2α by upregulating GADD34, a component of the PP1 complex responsible for eIF2α dephosphorylation, to enhance viral replication (22). Viral replication is dependent on the host cell protein-synthetic machinery for producing viral proteins and viral particles. The UPR PERK axis is involved in modulating cellular protein translation under ER stress. However, how the PERK signaling cascades specifically manipulate coronavirus replication is still unclear.
Transmissible gastroenteritis virus (TGEV), a member of the alphacoronavirus family (16), is the cause of an economically important swine disease. TGEV infects and destroys villous epithelial cells of the jejunum and ileum, which results in lethal watery diarrhea and dehydration in piglets (23, 24). A greater understanding of its pathogenesis is critical for the development of a new strategy to treat TGEV infection. In contrast to most CoVs, such as SARS-CoV, which inhibit type I interferon (IFN-I) production, TGEV infection activates nuclear factor-kappa B (NF-κB) and induces significant IFN-I production both in vivo and in vitro (25–28). IFN-Is are well-known innate cytokines that play a major role in host defenses against viral infection. NF-κB is a master regulator of the proinflammatory response and is a key transcription factor for the regulation of IFN-I production (29). However, the underlying mechanism(s) exploited by TGEV to activate NF-κB and induce IFN-I remain unclear. Several recent studies have demonstrated the critical roles of UPR in modulating the innate immune responses (9, 30). In an inert cellular state, NF-κB is sequestered in the cytoplasm by inhibitors of NF-κB (IκBs), the most prominent and well-studied being IκBα (29). The basal level of IκBα is maintained by constitutive synthesis and degradation of the protein. Under ER stress induced by amino acid starvation or UV irradiation, reduced general protein synthesis by phosphorylation of eIF2α results in a net decrease of the inhibitory protein IκBα, thereby activating NF-κB (30, 31). Nonetheless, the actual contributions of the UPR to NF-κB-mediated cytokine induction during coronavirus infection remain unclear.
Given the pivotal role of the ER in coronavirus replication, we explored the ability of TGEV to induce ER stress and investigated how the TGEV-triggered UPR affects viral replication. We found that TGEV upregulated GRP78 and triggered the UPR in vitro and in TGEV-infected ileum tissues. ER stress triggered by a chemical inducer or TGEV infection decreased TGEV replication. Furthermore, we demonstrated that the TGEV-induced UPR negatively regulated viral replication, primarily by activating the PERK-eIF2α pathway, although all three UPR pathways are activated by TGEV infection. The PERK-dependent UPR branch emerges as a cellular antiviral response that antagonizes TGEV replication by reducing global protein synthesis and inducing IFN-I production. Our findings highlight the role of the PERK-eIF2α pathway in inhibiting TGEV replication and suggest a possible therapeutic target for the treatment of TGEV.
RESULTS
TGEV infection induces ER stress in vitro.
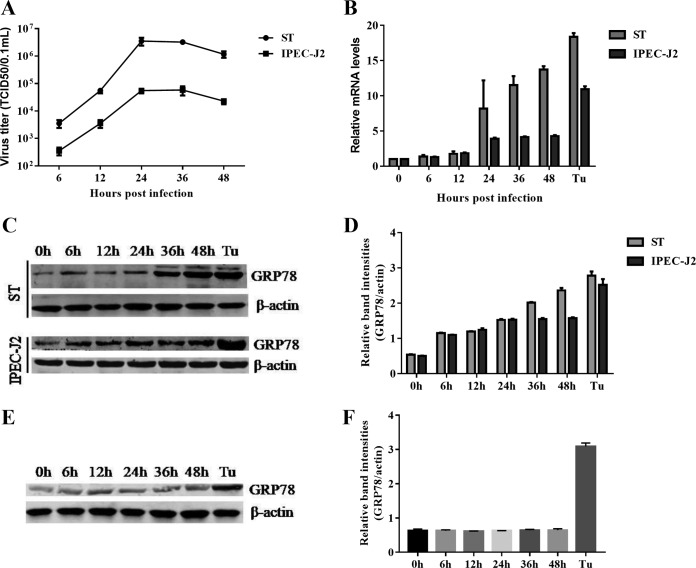
To determine whether TGEV infection induces cellular UPR in vitro, we monitored the expression of the major marker (GRP78) of the UPR following TGEV infection in swine testicular (ST) cells, and the ER stress inducer tunicamycin (Tu) was used as a positive control. The growth curve of the virus showed an initial exponential rise and reached a maximal constant value at 24 to 36 h postinfection (hpi) (Fig. 1A). The GRP78 mRNA levels were upregulated starting at 6 hpi and then gradually increased up to 48 hpi (Fig. 1B). The upregulated GRP78 expression was confirmed by Western blotting of TGEV-infected cells compared with the mock-infected control cells (Fig. 1C and D). These results clearly demonstrated that TGEV infection triggered ER stress in ST cells.
FIG 1.
TGEV infection induces ER stress in ST and IPEC-J2 cells. ST cells and IPEC-J2 cells were infected with TGEV H87 at an MOI of 1; samples were collected at 0, 6, 12, 24, 36, and 48 hpi. (A) One-step growth curve of TGEV in ST cells and IPEC-J2 cells. (B) A time-dependent increase of GRP78 expression was revealed by qPCR in ST cells and IPEC-J2 cells. Total RNA was isolated, and the transcriptional levels of GRP78 were measured by qPCR at different time points (0 to 48 h) after infection. (C) Elevated protein expression of GRP78 was confirmed by Western blotting in ST cells and IPEC-J2 cells. Tu (2 μg/ml) was used as a positive control for UPR activation; β-actin was used as a loading control. (D) Relative GRP78 levels. (E) TGEV-induced UPR was dependent on active viral replication. ST cells were infected with UV-inactivated TGEV or treated with Tu (2 μg/ml). The GRP78 expression in ST cells at different time points was determined by Western blotting using anti-GRP78 antibody. (F) Relative GRP78 levels. Means and SD of the results from three independent experiments are shown.
TGEV primarily infects and replicates in porcine small intestinal epithelial cells in vivo (32). To further verify the induction of ER stress by TGEV infection in primary target cells, we monitored the ER stress in IPEC-J2 cells following TGEV infection. IPEC-J2 is a nontransformed cell line originating from jejunum epithelium isolated from a neonatal unsuckled piglet and is widely used as an in vitro model system for studying porcine intestinal pathogen-host interactions and porcine-specific pathogenesis (33, 34). The growth curve of TGEV in IPEC-J2 cells was similar to that in ST cells (Fig. 1A). Upregulation of GRP78 expression was determined by monitoring transcripts (Fig. 1B) and by blotting GRP78 protein in TGEV-infected IPEC-J2 cells (Fig. 1C and D). In contrast to actively replicating TGEV, UV-inactivated TGEV did not trigger the upregulation of GRP78 protein in ST cells, as shown by measurement of GRP78 protein (Fig. 1E and F). These findings indicated that TGEV-induced ER stress depends on active viral replication. Together, these data showed that TGEV infection triggers ER stress in both ST and IPEC-J2 cells.
TGEV infection activates all three UPR pathways in vitro.
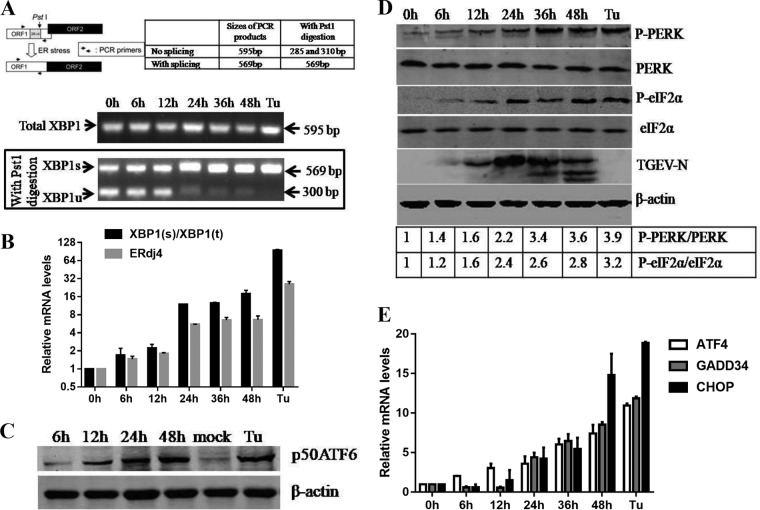
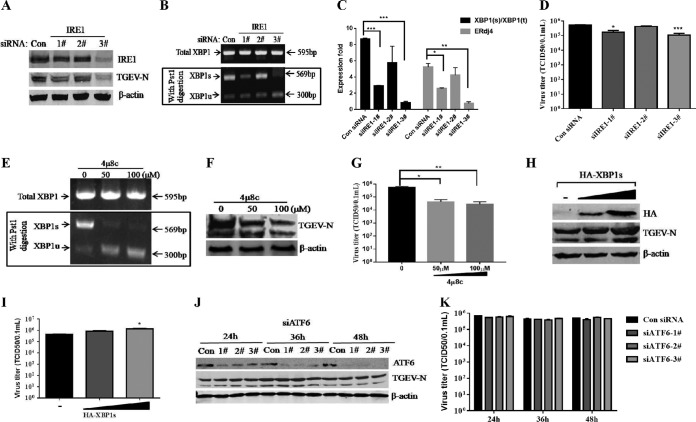
ER stress potentially triggers the three UPR signaling pathways (PERK, IRE1, and ATF6). Thus, we initially analyzed the UPR signaling pathways following TGEV infection. IRE1 activation promotes endoribonuclease activity of IRE1 that selectively cleaves a 26-nucleotide (nt) segment from XBP1 mRNA and creates the active spliced X-box binding protein 1 (XBP1s) transcription factor, whose target genes enhance the ER protein-folding capacity. The XBP1 cDNA was amplified by reverse transcriptase (RT)-PCR and digested by PstI, which has a recognition site located within the 26-nt region of XBP1 cDNA that is removed by IRE1-mediated splicing, as previously described (35, 36) (Fig. 2A). The spliced XBP1s form increased after TGEV infection, consistent with a decrease in the unspliced XBP1 (XBP1u), indicating that IRE1 was activated in the later phase of infection (Fig. 2A). Consistent with these findings, the XBP1 splicing ratio increased from approximately 2-fold to 19-fold in TGEV-infected cells (Fig. 2B). The activation of the IRE1-XBP1 axis by TGEV infection was further confirmed by measuring the transcripts of the XBP1s downstream target ER-localized DnaJ homologue 4 (ERdj4) gene (37) (Fig. 2B). In response to ER stress, ATF6 translocates from the ER to the Golgi apparatus, and then, the 90-kDa full-length ATF6 (p90 ATF6) is processed to its active 50-kDa form (38). The results of Western blotting showed that generation of cleaved ATF6 (50-kDa protein) was gradually increased and peaked at 48 hpi (Fig. 2C). These results indicated that TGEV infection activates the IRE1 and ATF6 UPR pathways.
FIG 2.
TGEV infection activates all three UPR signaling pathways in vitro. (A) TGEV H87 induces XBP1 mRNA splicing in ST cells. Shown is the analysis scheme for XBP1 mRNA splicing. The sizes of PCR-amplified fragments from spliced and unspliced XBP1 with or without PstI cleavage are shown. ST cells were treated with Tu (2 μg/ml) and mock infected or infected with TGEV at an MOI of 1. Cells were harvested at 6, 12, 24, or 48 hpi for RT-PCR analysis. The PCR product of XBP1 was further digested with PstI. The digested PCR products were separated by 2% agarose gel electrophoresis and photographed with a gel documentation system (Vilber Lourmat, Marne-la-Vallée, France). (B) XBP1 mRNA splicing and mRNA expression of the ERdj4 gene. The expression of selected UPR IRE1 downstream genes was quantified by real-time relative qPCR. (C) Increase in cleaved ATF6 (p50 ATF6) in TGEV-infected ST cells. ST cells were treated with 2 μg/ml Tu for 8 h or infected with TGEV at an MOI of 1 at 6, 12, 24, or 48 hpi before the cell lysates were harvested for immunoblotting with anti-ATF6 (Abcam) and anti-actin antibodies. (D) Phosphorylation of PERK and eIF2α in TGEV-infected cells. ST cells were infected with H87 at an MOI of 1 and harvested at 0, 6, 12, 24, 36, and 48 hpi. Cell lysates were subjected to Western blot analysis with antibodies against p-PERK, total PERK, p-eIF2α, total eIF2α, and the TGEV-N protein. β-Actin was used as a loading control. Band intensities for p-PERK and p-eIF2α were normalized to those for total PERK and total eIF2α, respectively. The fold increases in phosphorylation are indicated below the blots, with the phosphorylation at 0 hpi given a value of 1. (E) PERK pathway downstream ATF4, CHOP, and GADD34 gene expression. Cells were lysed to extract total RNA, which was used to determine the expression of ATF4, CHOP, GADD34, and GAPDH genes by qPCR. Means and SD of the results from three independent experiments are shown.
Among the three UPR signaling pathways, PERK is a key molecule that alleviates the accumulation of misfolded proteins in the ER by phosphorylating eIF2α, which attenuates mRNA translation under ER stress via inhibition of the recycling of eIF2α to its active GTP-bound form and blocking the initiation phase of polypeptide chain synthesis (2). We detected a gradual induction of PERK phosphorylation starting at 6 h following TGEV infection or in cells treated with Tu compared to mock-infected cells (Fig. 2D). The induction of phosphorylation and activation of PERK upon TGEV infection correlated with the eIF2α phosphorylation pattern (Fig. 2D). The profiles of phosphorylated PERK (p-PERK) and p-eIF2α were similar to the kinetics of the viral nucleocapsid (N) protein expression (Fig. 2D). As expected, PERK-eIF2α activation following TGEV infection resulted in elevated mRNA levels of the eIF2α downstream ATF4, CHOP, and GADD34 target genes (Fig. 2E). These results suggest that TGEV infection activates the PERK-eIF2α-ATF4-CHOP pathway. Collectively, TGEV infection triggers all three UPR signaling branches, although not simultaneous activation, to variable degrees in vitro.
TGEV infection activates all three UPR pathways in vivo.
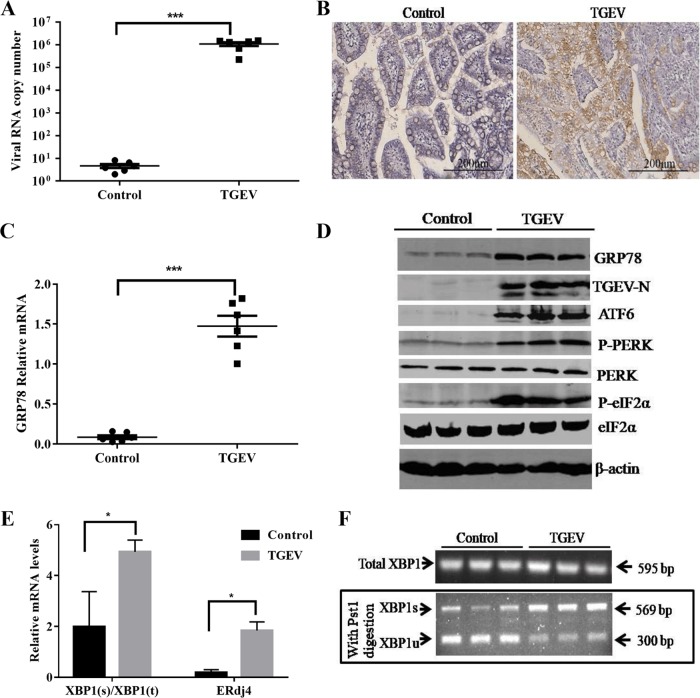
To explore whether TGEV infection induces ER stress in vivo, ER stress was monitored in the primary target of TGEV infection in vivo, ileum tissues from TGEV-infected piglets at 48 hpi. TGEV infection was confirmed by the measurement of viral loads (Fig. 3A) and immunohistochemistry with a specific monoclonal antibody (MAb) to the TGEV N protein (Fig. 3B). TGEV infection resulted in a more than 15-fold increase in GRP78 expression in ileum tissues compared to that of the control (GRP78 mRNA, 1.47 ± 0.35 for TGEV infection versus 0.08 ± 0.05 for mock infection; P < 0.0001) (Fig. 3C). The increased GRP78 expression was further verified by Western blotting of GRP78 protein in TGEV-infected ileum tissues, which showed elevated GRP78 protein levels compared with those of the control (Fig. 3D). These results demonstrated that TGEV infection induces cellular ER stress in vivo.
FIG 3.
TGEV infection activates all three UPR signaling pathways in vivo. Twelve 2-day-old SPF piglets were orally inoculated with TGEV strain H87 or with DMEM as uninfected controls. All the piglets were euthanized by the end of the study, which was terminated at 48 hpi. (A) Virus replication in the ileum was tested by qPCR. (B) Representative microphotographs of viral antigen immunochemical staining in TGEV-noninfected and -infected ileum tissues (magnification, ×200). (C) GRP78 expression in ileum tissues was detected by qPCR. (D) Protein levels of GRP78, ATF6, p-PERK, PERK, p-eIF2α, and eIF2α in ileum samples from TGEV-noninfected and -infected piglets. (E) XBP1s/XBP1t (XBP1-spliced/XBP1-total) ratio and mRNA expression of the ERdj4 gene in ileum samples. (F) XBP1 mRNA splicing in ileum samples from TGEV-noninfected and -infected piglets. XBP1s, spliced XBP1; XBP1u, unspliced XBP1. Means and SD of the results from three independent experiments are shown. *, P < 0.05; ***, P < 0.001.
To further determine if all three UPR branches were activated in vivo as they were in vitro, we analyzed the UPR signaling pathways in TGEV-infected piglets. The phosphorylation of PERK and eIF2α and of cleaved ATF6 (50-kDa protein) all increased in TGEV-infected ileum tissues compared with control samples (Fig. 3D), indicating the activation of PERK and ATF6. TGEV-infected ileum tissues exhibited an increased ratio of XBP1s to the total XBP1 and elevated mRNA levels of ERdj4 compared with those of the mock control (Fig. 3E), indicating that IRE1 is slightly activated in vivo. Consistent with these findings, the XBP1 spliced form increased along with a decrease in the unspliced form in the ileum samples from TGEV-infected piglets (Fig. 3F). Altogether, these data demonstrated that TGEV infection in vivo activates all three UPR pathways.
ER stress is detrimental to TGEV replication.
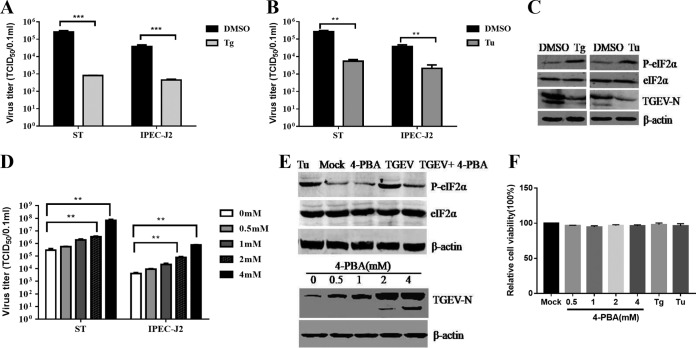
Growing evidence shows that the virus-induced UPR modulates viral replication (39–43). To explore the role of ER stress in TGEV replication, we initially investigated the effects of Tu and thapsigargin (Tg) treatments on TGEV replication; these two chemical ER stress inducers are widely used as positive controls for the UPR (44). Addition of the ER stress inducer Tg (1 μM) or Tu (2 μg/ml) substantially inhibited TGEV infection in both ST and IPEC-J2 cells (Fig. 4A and B). TGEV suppression by Tg or Tu was further confirmed by monitoring TGEV N protein expression (Fig. 4C). The viral suppression by Tg or Tu was not due to cellular cytotoxicity, as no significant cytotoxicity was observed by measuring cell viability with a Cell Counting Kit 8 (CCK-8) assay (Beyotime, Hangzhou, China) (Fig. 4F).
FIG 4.
UPR suppresses TGEV replication in both ST and IPEC-J2 cells. ST cells and IPEC-J2 cells were pretreated with Tg (1 μM), Tu (2 μg/ml), 4-PBA, or DMSO carrier control 2 h before infection and maintained at that concentration after infection. (A, B, and D) TGEV titers on ST cells and IPEC-J2 cells treated with Tg, Tu, 4-PBA, or control. (C and E) Western blotting was performed to test p-eIF2α, eIF2α, and TGEV N expression. β-Actin was used as a sample loading control. (F) Cell viability was detected with a CCK-8 assay after treatment with Tg, Tu, and 4-PBA. Means and SD of the results from three independent experiments are shown. **, P < 0.01; ***, P < 0.001.
To further clarify the role of ER stress in TGEV replication, we pretreated ST cells and IPEC-J2 cells with the chemical chaperone 4-phenylbutyric acid (4-PBA), which was reported to alleviate ER stress by inhibiting the phosphorylation of eIF2α (45, 46). The 4-PBA pretreatment decreased eIF2α phosphorylation (64% decrease) in TGEV-infected cells (Fig. 4E), suggesting that 4-PBA alleviates TGEV-induced ER stress. The 4-PBA treatment potently enhanced TGEV replication in both cell lines, as shown by TGEV N protein levels and viral titers, in a dose-dependent manner (Fig. 4D and E). Enhancement of TGEV replication by 4-PBA treatment indicated that alleviation of ER stress by inhibiting eIF2α phosphorylation promotes TGEV propagation. The enhanced TGEV replication following 4-PBA treatment was not due to an increase in cell numbers because the 4-PBA concentrations in this study did not alter cell viability, as shown by CCK-8 assays (Fig. 4F). Collectively, these results demonstrated that the elicited UPR negatively regulates TGEV replication.
The virus-activated ATF6 and IRE1 pathways do not suppress TGEV replication.
TGEV infection triggered three UPR signaling pathways (PERK, IRE1, and ATF6) (Fig. 2 and 3). We next determined which UPR pathway primarily accounts for the reduction in TGEV replication. To clarify the role of IRE1 in TGEV infection, we examined TGEV infection in the presence of IRE1 knockdown or overexpression by using small interfering RNA (siRNA) duplexes that target IRE1 and by transfecting cells with the tandem construct hemagglutinin (HA)-tagged XBP1s, respectively. The knockdown efficiency of IRE1 siRNAs was confirmed by IRE1 protein blotting, XBP1 splicing, and ERdj4 induction, and IRE1-specifc siRNA duplex number 3 exhibited the greatest efficiency (Fig. 5A, B, and C). IRE1 knockdown decreased TGEV N protein and virus production (Fig. 5A and D), indicating that the IRE1 branch of the UPR facilitates TGEV replication. These findings are consistent with the effects of an IRE1-specific inhibitor (4μ8C) (47) on viral replication. The inhibition of IRE1 activity by 4μ8C decreased the TGEV titers by up to 4-fold (Fig. 5E, F, and G). To further confirm the role of the IRE1 pathway in TGEV infection, ST cells were transiently transfected with the HA-XBP1s-expressing plasmid and the empty vector pCAGGS-HA for 24 h and then infected with TGEV H87 at a multiplicity of infection (MOI) of 1. TGEV N protein and viral titers increased in a dose-dependent manner when XBP1s was overexpressed (Fig. 5H and I). These data demonstrated that the IRE1 signaling pathway promotes TGEV infection instead of suppressing TGEV replication.
FIG 5.
The IRE1 and ATF6 pathways did not suppress TGEV infection. (A, B, C, and D) ST cells were transfected for 24 h with siRNA duplexes that target IRE1 and then challenged with TGEV. (A) At 24 hpi, Western blotting was performed to detect IRE1 and TGEV N. (B and C) XBP1 mRNA splicing and mRNA expression of the ERdj4 gene in IRE1 knockdown cells were determined. (D) Virus titers. (E, F, and G) ST cells were treated with different concentrations of 4μ8c, followed by infection with TGEV H87. Cells were harvested at 24 hpi and subjected to XBP1 splicing assay and Western blotting using antibodies against TGEV N protein, and the virus titers were calculated. (H and I) ST cells were transiently transfected with the HA-XBP1s-expressing plasmid and the empty vector pCAGGS-HA for 24 h and then infected with TGEV H87 at an MOI of 1. At 24 hpi, Western blotting was performed to detect HA and TGEV N, and the virus titers were calculated. (J and K) ST cells were transfected with siRNA of ATF6 for 24 h, 36 h, and 48 h and then infected with TGEV H87. At 24 hpi, Western blotting was performed using antibodies against ATF6 and TGEV N protein, and the virus titers were calculated. Means and SD of the results from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To monitor the effect of the ATF6 pathway on TGEV replication, we performed a time course knockdown by ATF6-specifc siRNA duplexes. The efficiency of ATF6 knockdown was confirmed by Western blotting, which showed that the best knockdown efficiency was observed at 48 h after transfection (Fig. 5J). However, no significant difference in TGEV replication was observed after ATF6 knockdown by siRNA compared with that of control siRNA (Fig. 5J and K). Taken together, these results indicate that activation of the IRE1 and ATF6 branches does not account for the suppression of TGEV replication by ER stress.
The PERK-eIF2α pathway inhibits TGEV replication.
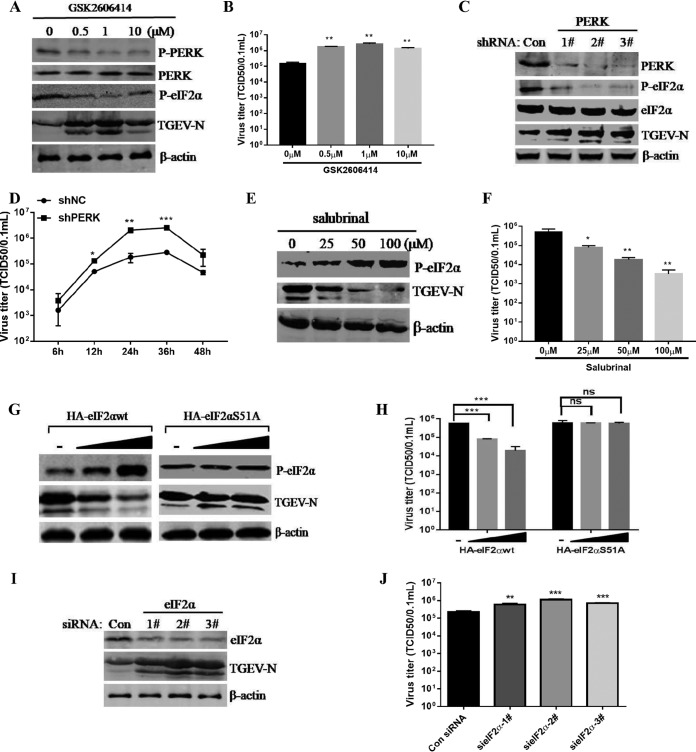
Given that suppressing eIF2α phosphorylation by 4-PBA treatment enhanced TGEV replication (Fig. 4E), we hypothesized that the PERK-eIF2α pathway of the UPR primarily accounts for the viral inhibition of the TGEV-induced UPR. To verify the role of the eIF2α kinase PERK in eIF2α phosphorylation and TGEV replication, we initially disrupted the PERK signaling pathway with the PERK-specific inhibitor GSK2606414 (PERKi) or by short hairpin RNA (shRNA) knockdown of PERK. PERK inhibition by PERKi was confirmed by Western blotting p-PERK and p-eIF2α (Fig. 6A). We found that PERKi treatment substantially elevated TGEV replication in ST cells (Fig. 6A and B). In addition, the increase in TGEV replication was correlated with the inhibition efficacy of p-PERK and p-eIF2α; the most significant upregulation of the TGEV N protein was observed at 1 μM PERKi compared to 0.5 μM and 10 μM. Transfection of shPERK significantly reduced the expression of PERK (71.3% decrease for number 1, 89.1% for number 2, and 81.2% for number 3 shRNAs) and p-eIF2α (68.8% decrease for number 1, 87.9% for number 2, and 79.1% for number 3 shRNAs) at the protein levels compared to those in the cells transfected with the control shRNA (Fig. 6C). The corresponding reduction of p-eIF2α by PERK knockdown and PERKi implied that p-eIF2α is primarily mediated through activation of PERK instead of other eIF2α kinases in TGEV infection. Consistent with these results of PERKi treatment, PERK knockdown promoted TGEV replication (Fig. 6C). Moreover, we performed a time course experiment to assess the replication of TGEV in PERK knockdown cells and found that TGEV showed significantly increased titers in PERK knockdown cells at 12, 24, and 36 hpi (Fig. 6D). These results demonstrated that PERK activation suppresses TGEV infection.
FIG 6.
The activated PERK-eIF2α pathway of the UPR inhibits TGEV replication. (A and B) Inhibition of PERK by GSK2606414 promoted TGEV replication in ST cells as measured by Western blotting using antibodies against the TGEV N protein (A) and viral titers (B). (C) ST cells were transfected with shRNA targeting PERK or control shRNA for 24 h, and then the cells were challenged with TGEV. At 24 hpi, Western blot analyses for PERK, eIF2α, p-eIF2α, TGEV N, or β-actin were performed. (D) ST cells were transfected with PERK shRNA number 2 or control shRNA for 24 h and then challenged with TGEV for 6, 12, 24, 36, and 48 h. Viral titers were calculated at different time points. (E and F) ST cells were treated with the indicated concentrations of salubrinal for 24 h and then infected with TGEV H87 (MOI = 1) in the continued presence of salubrinal. (E) After the virus adsorbed to the cells, medium containing the indicated concentrations of salubrinal was added. Cell lysates of ST cells were analyzed by Western blotting using antibody against p-eIF2α and the TGEV N protein. (F) The TGEV TCID50 in the supernatants was titrated on ST cells. (G and H) ST cells were transfected with HA-eIF2αwt or HA-eIF2αS51A for 24 h and then infected with TGEV H87. At 24 hpi, Western blot analysis was performed to detect p-eIF2α, TGEV N, or β-actin, and the viral titers were calculated. (I and J) ST cells were transfected with siRNA of eIF2α for 24 h and then infected with TGEV H87. After 24 hpi, Western blot analysis was performed to detect eIF2α, TGEV N, or β-actin, and the viral titers were calculated. Means and SD of the results from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further clarify the impact of the PERK pathway on TGEV replication, we quantified TGEV replication in TGEV-infected ST cells in the presence of different concentrations of salubrinal (a selective inhibitor of eIF2α dephosphorylation) for 24 h. Salubrinal treatment significantly increased the phosphorylated eIF2α in TGEV-infected cells compared with dimethyl sulfoxide (DMSO)-treated control cells (Fig. 6E). In contrast to the inhibition of p-eIF2α, the increase in phosphorylated eIF2α by salubrinal treatment caused a decrease in TGEV replication ranging from 30- to 551-fold for TGEV in ST cells; the reduction of TGEV replication displayed a dose-dependent reaction to different salubrinal concentrations (Fig. 6F). These results further demonstrated that eIF2α phosphorylation negatively regulates TGEV replication. The decreased replication of TGEV by salubrinal treatment was not due to cellular cytotoxicity, since we and others did not observe cytotoxicity at the concentrations used in the study (reference 48 and data not shown).
To further elucidate the role of eIF2α Ser-51 phosphorylation in the replication of TGEV, we examined TGEV infection in ST cells following transfection with wild-type eIF2α (HA-eIF2αwt) or an unphosphorylatable mutant form of eIF2α containing Ala instead of Ser at residue 51 (HA-eIF2αS51A). While inhibition of TGEV replication occurred in the eIF2αwt cells, no decrease in TGEV replication was detected in the eIF2αS51A cells (Fig. 6G and H). Furthermore, we examined the importance of eIF2α phosphorylation in the inhibition of TGEV replication in response to eIF2α siRNA knockdown, which decreased eIF2α phosphorylation. In contrast to the overexpression of eIF2α, eIF2α knockdown by siRNA in ST cells enhanced TGEV replication (Fig. 6I and J). Thus, these data showed that activation of the PERK-eIF2α pathway substantially suppresses TGEV replication.
The PERK-eIF2α pathway attenuates global protein synthesis, including viral proteins.
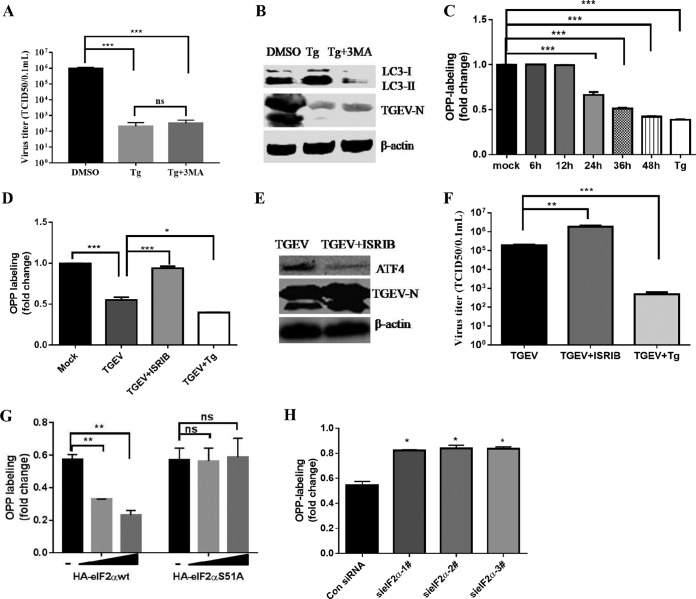
Next, we sought to elucidate how the UPR PERK-eIF2α pathway inhibits TGEV replication. The activation of PERK-eIF2α potentially causes multiple cellular changes, including autophagy and reversible stalled protein translation. A previous study demonstrated that autophagy negatively regulates TGEV infection (49). However, we did not observe rescue of Tg-mediated reduction of TGEV replication after combining the Tg treatment with the autophagy inhibitor 3-methyladenine (3MA) compared to the Tg treatment alone (Fig. 7A and B), indicating that autophagy is not involved in PERK-eIF2α-mediated suppression of TGEV infection. Phosphorylation of eIF2α briefly halts the initiation of mRNA translation and reduces global protein synthesis (8). As TGEV infection induced eIF2α phosphorylation, to explore whether p-eIF2α-mediated translation attenuation accounts for the TGEV suppression by the PERK-eIF2α axis, we monitored nascent protein synthesis in TGEV-infected ST cells using cotranslational labeling with O-propargyl-puromycin (OPP) as described previously (50). TGEV infection significantly reduced OPP labeling from 24 hpi to 48 hpi (40% to 60% reduction) (Fig. 7C). These findings are consistent with the kinetics of TGEV replication and the expression of TGEV N protein, which peaked at 24 hpi and gradually decreased from 24 hpi to 48 hpi (Fig. 1A and 2D).
FIG 7.
Analysis of mRNA translation by OPP labeling. (A and B) ST cells were pretreated with Tg (1 μM) or a combination with 3MA (5 mM) 2 h before infection and maintained at that concentration after infection. TGEV titers were calculated (A), and Western blotting was performed to test LC3 and TGEV-N expression, and β-actin was used as sample loading control (B). (C) TGEV infection decreased global protein synthesis. ST cells were infected with TGEV H87 (MOI = 1) or mock infected for 6, 12, 24, 36, or 48 h before OPP labeling. The graph shows the fold increase in OPP labeling (means and SD) in ST cells, with values for mock-infected cells set to 1.0. Statistical comparisons between control and TGEV-infected cells are shown (Student's t test). Cells treated with Tg (1 μM) were used as a positive control. (D, E, and F) ST cells were treated or not with 200 nM ISRIB or Tg (1 μM) for 2 h, followed by infection with TGEV H87 for 24 h before OPP labeling. (D) Graph showing fold increase in OPP labeling, with values for untreated cells set to 1.0. Statistical comparisons between groups are shown (Student's t test). (E) Immunoblot analysis of ATF4 and TGEV-N in TGEV-infected ST cells in the presence or absence of 200 nM ISRIB. (F) Virus titers were measured in ISRIB-treated cells. (G) ST cells were transfected with HA-eIF2αwt or HA-eIF2αS51A and analyzed for ongoing translation. (H) ST cells were transfected with eIF2α siRNA or control siRNA for 24 h and then infected with TGEV H87. At 24 hpi, global protein synthesis was measured by OPP labeling. Means and SD of the results from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
To confirm that the decreased mRNA translation following TGEV infection is related to eIF2α phosphorylation, we used ISRIB, a PERK branch-specific inhibitor, to investigate the mRNA translation (51). ISRIB was used at 200 nM based on previous publications (52) and our own pilot studies. Addition of ISRIB rescued the mRNA translation attenuation of TGEV (Fig. 7D) and significantly increased the expression of the TGEV N protein and viral titers (Fig. 7E and F). These findings were further confirmed by overexpression and knockdown of eIF2α. By using OPP labeling of nascent proteins, we demonstrated that overexpression of wild-type eIF2α (HA-eIF2αwt) inhibited the synthesis of proteins in transfected cells, while overexpression of the unphosphorylatable mutant form of eIF2α (HA-eIF2αS51A) showed no difference in nascent protein synthesis (Fig. 7G). In contrast, knockdown of eIF2α rescued nascent protein synthesis (Fig. 7H). These results suggest that the suppression of TGEV replication by the PERK-eIF2α axis occurs at least partially through eIF2α phosphorylation-mediated overall attenuation of protein translation.
PERK phosphorylation of eIF2α suppresses TGEV replication due to IFN-I induction via NF-κB activation.
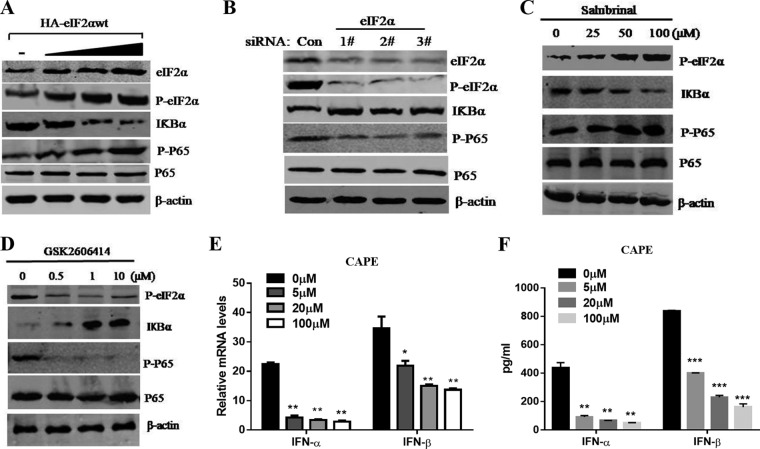
Given previous studies showing that PERK phosphorylation of eIF2α activates NF-κB in response to noninfectious stress conditions (31) and that IFN-Is have critical roles in antiviral innate immunity, we explored whether eIF2α phosphorylation in response to TGEV is linked to induction of IFN-I production and suppression of viral replication via NF-κB activation. Following transfection with HA-tagged eIF2α and increased eIF2α phosphorylation, there was a significant increase in activation of NF-κB, which was determined by immunoblotting using specific antibodies against phospho-p65, with a marked reduction in IκBα levels (Fig. 8A). In comparison, this activation of NF-κB was largely diminished in ST cells following siRNA knockdown of eIF2α (Fig. 8B). Knockdown of eIF2α by siRNA resulted in increased IκBα protein levels and decreased NF-κB activation in TGEV-infected cells (Fig. 8B). Consistent with these results, salubrinal enhanced eIF2α phosphorylation in TGEV-infected ST cells (Fig. 8C), which caused substantial reduction of IκBα levels and enhanced NF-κB activation (Fig. 8C). In striking contrast to salubrinal, GSK2606414 resulted in increased IκBα protein levels and decreased NF-κB activation in TGEV-infected cells (Fig. 8D). These results indicate that phosphorylation of eIF2α by PERK contributes to the activation of NF-κB in response to TGEV infection.
FIG 8.
Activation of NF-κB by PERK phosphorylation of eIF2α in TGEV-infected ST cells. (A) ST cells were transfected with different concentrations of HA-eIF2αwt for 24 h and then infected with TGEV H87 (MOI = 1). Cell lysates of ST cells were analyzed by Western blotting using specific antibodies against p-eIF2α, eIF2α, IκBα, phospho-p65, P65, and β-actin. (B) ST cells were transfected with eIF2α siRNA or control siRNA for 24 h prior to TGEV infection and harvested at 24 hpi for Western blot analyses with the indicated antibodies. (C) ST cells were treated or not (exposed to equal amounts of DMSO) with different doses of salubrinal for 24 h and then infected with TGEV H87 (MOI = 1) in the continued presence of salubrinal. Cell lysates of ST cells were analyzed by Western blotting using specific antibodies against p-eIF2a, IκBα, phospho-p65, P65, and β-actin. (D) ST cells were treated with DMSO or different concentrations of GSK2606414 for 2 h prior to TGEV infection and harvested at 24 hpi for Western blot analyses with the indicated antibodies. (E and F) The effect of TGEV-induced IFN-α/β was reduced in a dose-dependent manner after treatment with the NF-κB inhibitor CAPE. The mRNA and protein levels of IFN-α/β were analyzed after ST cells were pretreated with CAPE. Means and SD of the results from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
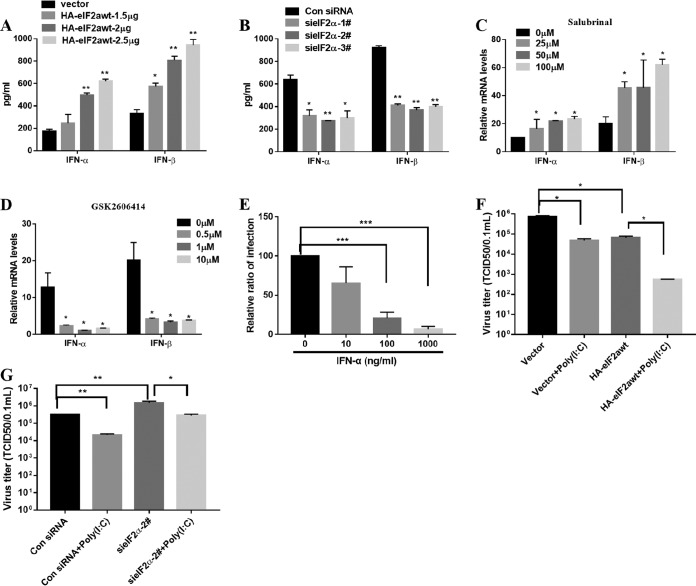
Previous studies have demonstrated that TGEV infection potently induces IFN-α (25–27), as well as IFN-β, in IPEC-J2, ST, and PK-15 cells (28, 53–55) and that the NF-κB signaling pathway has a fundamental role in TGEV-induced IFN-I production in PK-15 cells (28, 56). To investigate the role of activated NF-κB in IFN-I production of ST cells, we analyzed the protein levels of IFN-α/β after ST cells were pretreated with caffeic acid phenethyl ester (CAPE), an NF-κB-specific inhibitor. As shown in Fig. 8E and F, the production of TGEV-induced IFN-α/β was reduced in a dose-dependent manner after treatment with CAPE, indicating that NF-κB is highly involved in IFN-I production by TGEV infection in ST cells.
Because PERK phosphorylation of eIF2α is involved in TGEV-induced NF-κB activation, theoretically, it should have an impact on TGEV-induced IFN-α/β expression. The expression levels of IFN-α/β in eIF2α overexpression and knockdown cells were analyzed after TGEV infection. As expected, the enhancement of eIF2α phosphorylation significantly promoted IFN-α/β expression, while eIF2α knockdown substantially inhibited IFN-α/β expression (Fig. 9A and B). Meanwhile, the enhancement of eIF2α phosphorylation by salubrinal significantly promoted IFN-α/β expression, while PERK disruption by PERKi substantially inhibited IFN-α/β expression compared with that in cells treated with the DMSO control (Fig. 9C and D). These results demonstrate that the UPR PERK-eIF2α axis has a vital role in IFN-I induction in response to TGEV infection. To confirm the role of IFN-I in controlling TGEV replication, we measured the viral titers after treatment with porcine IFN-α and found that IFN-α suppressed TGEV replication in a dose-dependent manner, as previously reported (57) (Fig. 9E). Poly(I·C), a ligand of TLR3, is well known to activate NF-κB and induce IFN-I (58). To determine that the reduction of TGEV replication in these cells is a direct consequence of the increased NF-κB activation and IFN production, we monitored TGEV replication in ST cells with eIF2α overexpression or knockdown in the presence or absence of poly(I·C), which activates NF-κB and induces IFN-I. Consistent with a previous report (53), poly(I·C) treatment directly inhibited TGEV infection (Fig. 9F) and eIF2α overexpression enhanced the poly(I·C)-mediated TGEV inhibition (Fig. 9F), whereas eIF2α silence undermined poly(I·C)-mediated TGEV inhibition (Fig. 9G). This further shows that the eIF2α pathway is involved in NF-κB activation and IFN-I production. Thus, these results suggest that PERK-eIF2α phosphorylation in response to TGEV infection promotes IFN-I production and then suppresses viral replication.
FIG 9.
Phosphorylation of eIF2α in response to TGEV is linked to induction of IFN-I production and suppression of viral replication. (A) The expression levels of supernatant IFN-α/β in HA-eIF2αwt-transfected ST cells were analyzed by ELISA after TGEV infection. (B) The expression levels of supernatant IFN-α/β in eIF2α knockdown cells were analyzed by ELISA after TGEV infection. (C) The expression levels of IFN-α/β in salubrinal-treated ST cells were analyzed after TGEV infection. (D) The expression levels of IFN-α/β in PERK inhibitor-treated ST cells were analyzed after TGEV infection. (E) IFN-α suppressed TGEV replication in a dose-dependent manner. Viral replication was measured after treatment with porcine IFN-α. The relative quantification of TGEV infection is presented. (F and G) ST cells were transfected with 2.5 μg/ml HA-eIF2αwt (F) and eIF2α siRNA number 2 or control siRNA (G) for 24 h, stimulated or not with poly(I·C) for 12 h, and then infected with TGEV H87 (MOI = 1). The virus titers were measured. Means and SD of the results from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
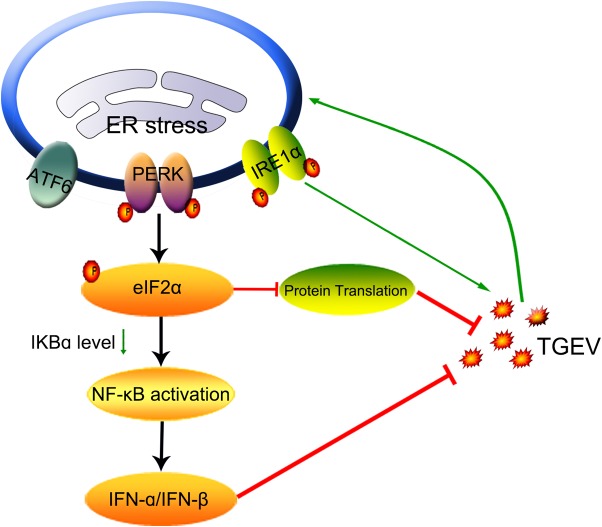
Coronavirus infection generally induces ER stress and initiates the UPR, which is actively involved in viral replication and modulates the host innate responses to the invading viruses. In this study, we found that infection with the alphacoronavirus TGEV elicited ER stress in vitro and in vivo and that ER stress had a detrimental effect on TGEV replication. Although TGEV infection triggered all three UPR signaling pathways, the suppression of TGEV replication by the UPR was largely mediated by activation of the PERK-eIF2α branch. Moreover, the mechanisms of the PERK-eIF2α axis to curtail TGEV replication are through eIF2α phosphorylation, which mediates the overall attenuation of protein translation and IFN-I production (Fig. 10).
FIG 10.
Model depicting viral suppression by PERK-eIF2α pathway activation during TGEV infection. TGEV infection induces ER stress by activating all three UPR signaling pathways (ATF6, IRE1, and PERK). Phosphorylation of eIF2α by PERK activation attenuates global protein synthesis, which decreases the levels of cytoplasm IκBα and leads to reduced inhibition of IκBα on NF-κB, promoting IFN-α/β production. The stalled translation and IFN-α/β production negatively regulate TGEV replication. The arrows indicate activation, and the blunt-ended lines indicate inhibition.
ER stress has been observed in cells infected by coronaviruses, such as MHV, IBV, human coronavirus (HCoV)-OC43, and SARS-CoV (2, 17, 18, 59, 60). Here, TGEV infection upregulated GRP78 and activated all three UPR pathways following TGEV infection in vitro and in vivo (Fig. 1 to 3). These findings are consistent with those of a previous study showing that TGEV induces ER stress, but Cruz et al. did not observe the induction of the GRP78-PERK UPR instead of the PKR-eIF2α pathway following TGEV infection (24). The discrepancy in activation of the TGEV-induced UPR pathway might result from the amino acid sequence variation of the spike proteins of different TGEV strains used. In our study, TGEV H87 shows enteric tropism instead of the respiratory tropism of the strain used in the previous study. In addition, TGEV activated the other two UPR signaling pathways, ATF6 and IRE1. The activation of multiple UPR pathways by a viral infection has been reported in other coronavirus infections, such as IBV, MHV, HCoV-OC43, and SARS-CoV (2, 13, 18, 60). Importantly, we also observed significant ER stress in TGEV-infected ileum tissues, the primary tissue targets in vivo (Fig. 3C to F), indicating that ER stress is potentially involved in the pathogenesis of TGEV in vivo.
The specific roles of the three UPR branches in viral replication are not the same, although TGEV infection induced all three UPR signaling branches. The ATF6 pathway does not modulate TGEV replication, based on the results of siRNA knockdown (Fig. 5J and K). Blockade of the IRE1 pathway by specific inhibitors or siRNA knockdown decreased TGEV replication, whereas overexpression of the spliced XBP1s, the major effector of the IRE1 signaling pathway, enhanced TGEV replication (Fig. 5A to I), indicating that IRE1 activation facilitates viral replication. The UPR PERK branch is highly associated with viral replication and pathogenesis (8, 21, 22, 61). Here, we demonstrated that the TGEV-induced UPR PERK activation primarily accounted for the increased eIF2α phosphorylation and negatively regulated TGEV replication. Induction of the UPR PERK has also been reported to suppress another alphacoronavirus, porcine epidemic diarrhea virus (PEDV) (61). This finding is different from the results in IBV, whose replication is not significantly affected by the suppression of eIF2α dephosphorylation by salubrinal (18, 20). Thus, the UPR PERK pathway during coronavirus infection exerts different effects on different coronaviruses. While deciphering the roles of the three UPR pathways in viral replication, one should keep in mind that there is complex cross-talk among the three UPR pathways, as more than one UPR pathway is commonly activated in response to stimuli, such as TGEV infection or Tg or Tu treatment, which induced all three UPR pathways. For TGEV, the inhibitory effect of PERK-eIF2α (more than 30-fold decrease by eIF2α overexpression in ST cells [Fig. 6H]) overwhelms the enhancement of IRE1 (only 4-fold increase by XBP1s overexpression) in viral replication.
For alleviation of ER stress, phosphorylation of eIF2α reduces translation of most mRNAs by inhibiting delivery of the initiator Met-tRNAi to the initiation complex, allowing cells to conserve resources (9). Consistent with the results of a previous study (24), TGEV infection triggered phosphorylation of eIF2α, which led to a significant reduction in global protein synthesis (Fig. 7). Although the accumulation of phosphorylated eIF2α began at 6 hpi, significant global translation inhibition was observed after 24 hpi (Fig. 2D and 7C), suggesting that one or more TGEV gene products act to delay this inhibitory event. These findings are consistent with the results of a previous study showing that TGEV protein 7 promotes eIF2α dephosphorylation by physically interacting with protein phosphatase 1 (PP1), the major enzyme responsible for eIF2α dephosphorylation (24). The later inhibition of protein synthesis coincides with the decrease in viral replication, indicating that the global translation attenuation does not discriminate between host and viral mRNAs and is responsible for the eIF2α-mediated viral suppression. Translational attenuation is also reported in other coronavirus infections, such as MHV, HCoV-OC43, and SARS-CoV (60, 62, 63). Generally, the global translation attenuation mediated by eIF2α phosphorylation is detrimental to viral replication and represents a host defense. This is why many viruses have evolved different strategies to counteract the negative regulation of eIF2α phosphorylation in viral replication, such as TGEV protein 7 (24) and vesicular stomatitis virus (VSV) M protein (64). Reoviruses compartmentalize the viral translational machinery within viral factories to coerce the host translational machinery into synthesizing viral proteins in the face of ongoing global protein synthesis under ER stress (65). Thus, the global inhibition of translation largely mediated by eIF2α phosphorylation represents a critical part of the cellular antiviral response that blocks viral protein synthesis, effectively dampening virus production.
NF-κB, as a master transcription regulator, regulates the genes involved in immune and inflammatory responses, stress remediation, and cell growth (29). Previous studies have shown that PERK and eIF2α phosphorylation is required for NF-κB in the response to diverse noninfectious stress conditions (9, 30). Consistent with these findings, PERK-eIF2α phosphorylation induced by TGEV infection was shown to actively contribute to the reduced basal level of IκBα protein in the cytoplasm caused by the global translation shutdown, which resulted in significant NF-κB activation and then massive production of IFN-I in vitro (Fig. 8). Thus, we identified another mechanism of coronaviruses to activate NF-κB via the PERK-eIF2α pathway (Fig. 10). Given the pivotal roles of NF-κB in the host inflammatory and immune responses, further characterizing the role of the PERK-eIF2α pathway in NF-κB activation during other viral infections is worthwhile and necessary.
Our and other in vitro studies have demonstrated that NF-κB activation accounts for most of the IFN-I production induced by TGEV infection (56). Consistent with previous studies showing that IFN-I production elicited by TGEV infection is mediated through multiple mechanisms, including NF-κB and IRF3 (56), blockade of NF-κB by CAPE significantly inhibited IFN-I but did not completely block IFN-α/β production in ST cells (Fig. 8), indicating that another existing pathway(s) alternatively activates IFN-I production independent of the NF-κB-mediated pathway. Given that IFN-I proteins potently inhibit the TGEV infection observed in this study (Fig. 9E) and in previously published studies (57), the PERK-eIF2α pathway may inhibit TGEV replication, at least partially, by enhancing IFN production, in addition to the direct stalled translation of viral proteins. More importantly, we observed the significant activation of PERK-eIF2α in TGEV-infected ileum in vivo, indicating that PERK-eIF2α might actively contribute to virus resolution in vivo.
Collectively, our data demonstrated that TGEV infection induced ER stress and activated all three UPR pathways and that the activated PERK pathway negatively regulated TGEV replication by attenuating protein translation and promoting IFN-I production (Fig. 10). Moreover, substantial ER stress was induced in the ileal tissues of TGEV-infected pigs. This work reveals a previously unrecognized mechanism of coronavirus-activated PERK-eIF2α to suppress viral growth in the host and highlights the critical role of the virus-induced UPR in viral replication. Further characterization of the role of the UPR during TGEV infection could provide novel insights into coronavirus biology and new antiviral drug targets in coronavirus infection.
MATERIALS AND METHODS
Cells and viruses.
ST cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and antibiotics (100 μg/ml streptomycin and 100 U/ml penicillin) in a humidified incubator at 37°C and 5% CO2. The porcine small intestinal epithelial cell line IPEC-J2 was kindly provided by Anthony Blikslager (North Carolina State University, Raleigh, NC) and was grown in DMEM–nutrient mixture F-12 (Ham) (1:1) (DMEM-F12) (Gibco), supplemented with 5% FBS, 5 μg/ml insulin-transferring selenium supplements (Gibco), 5 ng/ml epidermal growth factor (Gibco), and 1% penicillin-streptomycin. TGEV strain H87, derived from the virulent strain H16 (GenBank accession no. FJ755618), was propagated and titrated in ST cells.
Inactivation of TGEV was performed by subjecting the above-mentioned virus stock to 254-nm short-wave UV radiation overnight on ice in a 60-mm tissue culture dish. Following exposure, the samples were harvested and stored at −80°C; 100% inactivation by UV overnight was ascertained by infection assay in ST cells.
Cloning and construction of plasmids.
pHA-eIF2αwt and pHA-XBP1s were generated by PCR amplification of the corresponding cDNAs from ST cells. Synthesis and amplification of cDNA (total mRNA from ST cells) was performed using the SuperScript one-step RT-PCR system kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Primers P1 and P2 of porcine eIF2α and XBP1s were each added at a final concentration of 0.2 μM. The PCR cycling conditions were 94°C for 2 min (to inactivate the reverse transcriptase), 94°C for 30 s (DNA denaturation), 57°C for 30 s (annealing), and 72°C for 10 min (extension) for a total of 35 to 40 cycles. The RT-PCR primers eIF2αwt P1 (5′-GGGGTACCATGCCGGGTCTGAGTTGTAGA-3′ [the KpnI cutting site is underlined]) and eIF2αwt P2 (5′-CCGCTCGAGTTAATCTTCAGCTTTGGCTTCC-3′ [the XhoI cutting site is underlined]) were used to amplify a fragment covering the coding sequence of porcine eIF2α. The RT-PCR primers XBP1s P1 (5′-GGGGTACCATGGTGGTGGTGGCAGCTGCGCAGAG-3′ [the KpnI cutting site is underlined]) and XBP1s P2 (5′-CCGCTCGAGTCACTTCATTAATGGCTTCCAGCTTGGC-3′ [the XhoI cutting site is underlined]) were used to amplify a fragment covering the coding sequence of porcine XBP1s. The purity and size of the amplified product were verified by 1.2% agarose gel electrophoresis. The cloned PCR products were sequenced by Comate Bioscience Co., Ltd. (Changchun, China), and analyzed with the GenBank database BLAST (www.ncbi.nlm.nih.gov/) program. Then, the PCR products were inserted into a pCAGGS-HA vector (Clontech) to obtain pHA-eIF2αwt and pHA-XBP1s using KpnI and XhoI. The recombinant plasmids were confirmed by sequencing. pHA-eIF2αS51A was generated using PCR-mediated site-directed mutagenesis to introduce the ser51Ala substitution (primers, 5′-TCTTCTTAGTGAGCTCTCCAGAAGGCGTATCC-3′ and 5′-GGATACGCCTTCTGGAGAGCTCACTAAGAAGA-3′). DNA sequence analysis confirmed the presence of the mutation.
Cell culture, virus infection, and treatments with chemicals.
ST cells and IPEC-J2 cells were infected with TGEV H87 (MOI = 1) or mock infected with phosphate-buffered saline (PBS). After 2 h of incubation at 37°C, unbound viruses were removed by washing the cells three times with PBS, and the cells were cultured in DMEM supplemented with 1% DMSO and 0.3% trypsin (0.25%; Gibco) at 37°C for different times.
Tu, Tg, 4-PBA, salubrinal (an eIF2α dephosphorylation inhibitor), ISRIB (an experimental drug that reverses the effects of eIF2α phosphorylation), 4μ8C (a potent specific inhibitor of IRE1), and poly(I·C) were purchased from Sigma. GSK2606414 (a selective inhibitor of PERK; Selleckchem) and CAPE (a potent and specific inhibitor of NF-κB activation; Selleckchem) were dissolved in DMSO. ST cells and IPEC-J2 cells were pretreated with different concentrations of chemicals or the same volume of DMSO, followed by inoculation with TGEV H87 (MOI = 1). Poly(I·C) was transfected at a final concentration of 1 μg/ml. After incubation for 2 h, the supernatant was removed and replaced with cell culture media containing different doses of the chemicals. Cells were harvested at 24 hpi and then subjected to SDS-PAGE and Western blotting using antibodies against p-eIF2α, eIF2α, or TGEV N protein. Supernatants were harvested at 24 hpi for viral titration.
Experimental infection of piglets.
Twelve 2-day-old specific pathogen-free (SPF) piglets were randomly divided into two groups. The SPF piglets in group 1 were orally inoculated with 5 ml of 1 × 105 50% tissue culture infective doses (TCID50) TGEV strain H87. SPF piglets in group 2 were inoculated with DMEM, serving as uninfected controls. After virus infection, clinical signs were recorded on a daily basis. All the piglets were euthanized by the end of the study, which was terminated at 48 hpi (Animal Ethics Committee approval number Heilongjiang-SYXK-2006-032). Small-intestine samples from the piglets were collected for quantitative PCR (qPCR) and immunohistochemistry staining. The immunohistochemistry staining was performed as previously described (66). The slides were visualized by ×200 magnification microscope photographs.
SDS-PAGE and Western blot analysis.
ST cells were infected with TGEV and harvested at the indicated time points. An equal number of cells were lysed with the cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 0.1% SDS, 5 mM sodium orthovanadate) containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF) protease inhibitor cocktail (Roche Molecular Biochemicals) for 30 min. Equal amounts of total cell lysates were separated by SDS-PAGE. The proteins in the gel were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA), which were then blocked with 5% nonfat dry milk in TBST (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20) at 4°C overnight and incubated for 2 h with different primary antibodies. Antibodies against GRP78 (ab21685), p-PERK (ab192591), p-eIF2α (Ser51) (ab32157), ATF6 (ab122897), ATF4 (ab1371), and β-actin (ab6276) were purchased from Abcam (Cambridge, MA). Antibodies against total eIF2α (sc-11386) and total PERK (sc-13073) were from Santa Cruz Biotechnology (Dallas, TX). Antibodies against IκBα (L35A5), phospho-NF-κB P65 (P-P65) (93H1), and NF-κB P65 (L8F6) were purchased from Cell Signaling Technology (Beverly, MA). Monoclonal antibodies against TGEV N protein were prepared and stocked by our team. After washing three times with TBST, the membrane was incubated with 1:10,000-diluted IRDye800-conjugated anti-mouse IgG, Alexa Fluor 680 goat anti-rabbit IgG, or Alexa Fluor 680 rabbit anti-goat IgG (Invitrogen, Carlsbad, CA) in blocking buffer for 1 h at room temperature. The membrane was scanned in an Odyssey infrared imaging system (Li-Cor Biosciences) after washing with TBST. The fluorescence intensity of each band was measured using Odyssey 2.1 software (Li-Cor Biosciences).
RNA interference.
siRNAs against ATF6, IRE1, and eIF2α; shRNAs against PERK; and control scrambled shRNA were purchased from Genepharma (Shanghai, China), and their sequences are listed in Table 1. ST cells were seeded in a 6-well plate and grown to 70 to 80% confluence. siATF6, siIRE1, sieIF2α, shPERK, and nontarget control shRNA were transfected using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. At 24 h posttransfection, cells were infected with TGEV and harvested at the indicated time points for protein and virus titration analyses.
TABLE 1.
Sequences of sense strands of siRNA used to ablate ATF6, eIF2ɑ, IRE1, and PERK protein expression in ST cells
| Target | siRNA sequence | Sequence (5′→3′) |
|---|---|---|
| ATF6 | 1st | GGGUUAGAAGCAAGGUUAATT |
| 2nd | GGUAUAUUGGAACAGGAUUTT | |
| 3rd | CCAGAAGUUAUCAAGACUUTT | |
| eIF2ɑ | 1st | GGAAUACAACAACAUCGAAGG |
| 2nd | GCAGAUAUUGAAGTGGCUUGU | |
| 3rd | GCCCAAAGUGGUUACAGAUAC | |
| IRE1 | 1st | GCACAGACCUGAAGUUCAATT |
| 2nd | GGAGGUUAUCGACCUGGUUTT | |
| 3rd | CCAUCAUCCUGAGCACCUUTT | |
| PERK | 1st | CGACAACCCGAAUUACAACAA |
| 2nd | AGGUCUAGGGAGCGAACCUCC | |
| 3rd | CUGCAGAUUGUGGAGGCGGUA | |
| Control | Nontargeting control | UUCUCCGAACGUGUCACGUTT |
RNA isolation and real-time quantitative RT-PCR.
To detect UPR induction, ST cells and IPEC-J2 cells were infected with TGEV H87 at an MOI of 1; the cells were collected at 0, 6, 12, 24, 36, and 48 hpi. The cellular mRNA was purified according to the manufacturer's instructions for the RNeasy minikit (Qiagen Sciences, Hilden, Germany). RNA was reverse transcribed using a PrimeScript II 1st-strand cDNA synthesis kit (TaKaRa, Dalian, China). qPCR was performed in triplicate using Power SYBR green PCR master mix (TaKaRa, Dalian, China). The primers were designed using Oligo 6 software and are shown in Table 2. All the data were acquired with LightCycler 480 real-time PCR machines (Roche) and analyzed with LightCycler 480 software 1.5 based on the cycle threshold (ΔΔCT) method. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as the internal control.
TABLE 2.
Real-time qPCR primers used for UPR detection
| Target | Orientation | Sequence |
|---|---|---|
| GRP78 | Forward | 5′-ATGGCCGTGTGGAGATCATC-3′ |
| Reverse | 5′-GAGCTGGTTCTTGGCTGCAT-3′ | |
| ATF4 | Forward | 5′-CCCTTTACGTTCTTGCAAACTC-3′ |
| Reverse | 5′-GCTTCCTATCTCCTTCCGAGA-3′ | |
| GADD34 | Forward | 5′-AAGAGCCTGGAGAGAGGAGAG-3′ |
| Reverse | 5′-GTCCCCAGGTTTCCAAAAGCA-3′ | |
| CHOP | Forward | 5′-CTCAGGAGGAAGAGGAGGAAG-3′ |
| Reverse | 5′-GCTAGCTGTGCCACTTTCCTT-3′ | |
| ERdj4 | Forward | 5′-CAGAGAGATTGCAGAAGCATATGA-3′ |
| Reverse | 5′-GCTTCTTGGATCGAGTGTTTTG-3′ | |
| XBP1s | Forward | 5′-GAGTCCGCAGCAGGTG-3′ |
| Reverse | 5′-CCGTCAGAATCCATGGGG-3′ | |
| XBP1t | Forward | 5′-TCCGCAGCACTCAGACTACGT-3′ |
| Reverse | 5′-ATGCCCAAGAGGATATCAGACTC-3′ | |
| GAPDH | Forward | 5′-CCTTCCGTGTCCCTACTGCCAAC-3′ |
To assess TGEV replication, the S gene of TGEV was used as a standard for the TGEV genome. Primers based on the TGEV S gene were synthesized for quantification of the TGEV genome in real-time quantitative RT-PCR: forward, 5′-GCTTGATGAATTGAGTGCTGATG-3′, and reverse, 5′-CCTAACCTCGGCTTGTCTGG-3′. Total viral RNA was isolated as described above, and qPCR was conducted using Power SYBR green PCR master mix (TaKaRa, Dalian, China) according to the manufacturer's instructions.
Virus titration.
ST cells and IPEC-J2 cells grown in 6-well culture plates (Costar, Cambridge, MA) were treated with chemicals (4-PBA, Tg, Tu, ISRIB, 4μ8C, GSK2606414, salubrinal, or DMSO control) and infected with TGEV for 24 h or transfected with siATF6, siIRE1, sieIF2α, or shPERK for 24 h and then infected with TGEV at an MOI of 1. At 24 hpi, the culture supernatants were collected, and the cells were subjected to three freeze-thaw cycles. The cultures were serially 10-fold diluted from 10−1 to 10−10 and added to confluent ST cells or IPEC-J2 cells in 96-well plates (Costar, Cambridge, MA). After 72 h of incubation, the supernatant was removed, the cells were fixed with 4% polyformaldehyde, and viral antigen was detected using immunofluorescence. Viral titers were calculated using the Reed-Muench method and expressed as TCID50/0.1 ml.
Protein synthesis assay.
Click-iT assays were performed using 1 × 105 cells per assay. A commercial kit was employed according to the manufacturer's recommendations (Click-iT Plus OPP Alexa Fluor 647 protein synthesis assay kit; Molecular Probes; no. C10458). Simply, OPP (20 μM; Life Technologies) was added to the cells and incubated for 30 min. The cells were washed in ice-cold PBS and then fixed with 3.7% formaldehyde and permeabilized using 0.5% Triton X-100 in PBS. Alexa Fluor 647 (Life Technologies) was conjugated to OPP as described in the manufacturer's instructions. The plate was analyzed with the Operetta high-content screening system (PerkinElmer).
Immunohistochemistry.
Representative sections of ileum tissues were fixed with 4% paraformaldehyde and stored in 70% ethanol at 4°C. Paraffin embedding, sectioning, and hematoxylin-and-eosin (H&E) staining were performed by the histology service of the Harbin Veterinary Research Institute. Four-micrometer sections were immunostained for TGEV N protein. Briefly, samples were deparaffined at 60°C and rehydrated by successive incubations in 100% xylol, 100% ethanol, and 96% ethanol. Endogenous peroxidase was blocked at 37°C in darkness with 1% H2O2 diluted in methanol. Samples were incubated with a MAb specific for TGEV N protein (1:50). Bound primary antibodies were detected with biotinylated antibodies specific for mice, using an ABC peroxidase staining kit and a metal-enhanced DAB substrate kit (Pierce), following the manufacturer's recommendations.
ELISA.
An IFN-α enzyme-linked immunosorbent assay (ELISA) was performed on supernatants using a porcine IFN-α ELISA kit obtained from Sigma (RAB1131) according to the instruction manual. IFN-β sandwich ELISA was performed using an ELISA kit purchased from Sangon Biotech (Shanghai, China) (D740048). A plate reader was used to measure the optical density at 450 nm (OD450) to calculate IFN-α/β concentrations.
Cell viability measurement.
Cell viability was determined using the CCK-8 assay (Beyotime, Hangzhou, China) according to the manufacturer's instructions.
Statistical analysis.
All the results shown in the figures are presented, where appropriate, as means and standard deviations (SD) from the results of three independent experiments and were analyzed with GraphPad Prism (GraphPad Software, Inc.). Differences were considered significant if the P value was <0.05.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China (2016YFD0500100 and 2017YFD0502200), the China Postdoctoral Science Foundation (2015M570185), and the Heilongjiang Province Science Foundation for Youths (QC2017031).
REFERENCES
- 1.Hetz C, Papa FR. 2018. The unfolded protein response and cell fate control. Mol Cell 69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Fung TS, Huang M, Liu DX. 2014. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res 194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 4.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A 102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karagoz GE, Acosta-Alvear D, Nguyen HT, Lee CP, Chu F, Walter P. 2017. An unfolded protein-induced conformational switch activates mammalian IRE1. Elife 6:e30700. doi: 10.7554/eLife.30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol 18:7499–7509. doi: 10.1128/MCB.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J Virol 77:3578–3585. doi: 10.1128/JVI.77.6.3578-3585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34:7–11. doi: 10.1042/BST0340007. [DOI] [PubMed] [Google Scholar]
- 10.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher T, Perlman S. 2013. Public health: broad reception for coronavirus. Nature 495:176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung TS, Liu DX. 2014. Coronavirus infection, ER stress, apoptosis and innate immunity. Front Microbiol 5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan CP, Siu KL, Chin KT, Yuen KY, Zheng B, Jin DY. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol 80:9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol 80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masters PS. 2006. The molecular biology of coronaviruses. Adv Virus Res 66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Versteeg GA, van de Nes PS, Bredenbeek PJ, Spaan WJ. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J Virol 81:10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y, Fung TS, Huang M, Fang SG, Zhong Y, Liu DX. 2013. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J Virol 87:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Regla-Nava JA, Alvarez E, Oliveros JC, Zhao J, Fett C, Perlman S, Enjuanes L. 2011. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog 7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krahling V, Stein DA, Spiegel M, Weber F, Muhlberger E. 2009. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol 83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datan E, Roy SG, Germain G, Zali N, McLean JE, Golshan G, Harbajan S, Lockshin RA, Zakeri Z. 2016. Dengue-induced autophagy, virus replication and protection from cell death require ER stress (PERK) pathway activation. Cell Death Dis 7:e2127. doi: 10.1038/cddis.2015.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J Virol 83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Nieuwstadt AP, Zetstra T, Boonstra J. 1989. Infection with porcine respiratory coronavirus does not fully protect pigs against intestinal transmissible gastroenteritis virus. Vet Rec 125:58–60. doi: 10.1136/vr.125.3.58. [DOI] [PubMed] [Google Scholar]
- 24.Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol 72:8636–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Bonnardiere C, Laude H. 1981. High interferon titer in newborn pig intestine during experimentally induced viral enteritis. Infect Immun 32:28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riffault S, Carrat C, van Reeth K, Pensaert M, Charley B. 2001. Interferon-alpha-producing cells are localized in gut-associated lymphoid tissues in transmissible gastroenteritis virus (TGEV) infected piglets. Vet Res 32:71–79. doi: 10.1051/vetres:2001111. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Wu W, Xie L, Wang D, Ke Q, Hou Z, Wu X, Fang Y, Chen H, Xiao S, Fang L. 2017. Cellular RNA helicase DDX1 is involved in transmissible gastroenteritis virus nsp14-induced interferon-beta production. Front Immunol 8:940. doi: 10.3389/fimmu.2017.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 30.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, Wek RC. 2003. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol 23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang HY, Wek RC. 2005. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J 385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F, Li G, Wen K, Bui T, Cao D, Zhang Y, Yuan L. 2010. Porcine small intestinal epithelial cell line (IPEC-J2) of rotavirus infection as a new model for the study of innate immune responses to rotaviruses and probiotics. Viral Immunol 23:135–149. doi: 10.1089/vim.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geens MM, Niewold TA. 2011. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnology 63:415–423. doi: 10.1007/s10616-011-9362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 36.Yu CY, Hsu YW, Liao CL, Lin YL. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol 80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galindo I, Hernaez B, Munoz-Moreno R, Cuesta-Geijo MA, Dalmau-Mena I, Alonso C. 2012. The ATF6 branch of unfolded protein response and apoptosis are activated to promote African swine fever virus infection. Cell Death Dis 3:e341. doi: 10.1038/cddis.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, Legge K, Monick MM. 2012. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem 287:4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, Shalimar Nayak B, Ranjith Kumar CT, Surjit M. 2016. Endoplasmic reticulum stress induced synthesis of a novel viral factor mediates efficient replication of genotype-1 hepatitis e virus. PLoS Pathog 12:e1005521. doi: 10.1371/journal.ppat.1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid SP, Shurtleff AC, Costantino JA, Tritsch SR, Retterer C, Spurgers KB, Bavari S. 2014. HSPA5 is an essential host factor for Ebola virus infection. Antiviral Res 109:171–174. doi: 10.1016/j.antiviral.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Su YC, Wu JL, Hong JR. 2011. Betanodavirus up-regulates chaperone GRP78 via ER stress: roles of GRP78 in viral replication and host mitochondria-mediated cell death. Apoptosis 16:272–287. doi: 10.1007/s10495-010-0565-x. [DOI] [PubMed] [Google Scholar]
- 44.Prostko CR, Brostrom MA, Malara EM, Brostrom CO. 1992. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J Biol Chem 267:16751–16754. [PubMed] [Google Scholar]
- 45.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. 2008. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A 109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 49.Guo L, Yu H, Gu W, Luo X, Li R, Zhang J, Xu Y, Yang L, Shen N, Feng L, Wang Y. 2016. Autophagy negatively regulates transmissible gastroenteritis virus replication. Sci Rep 6:23864. doi: 10.1038/srep23864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeomans A, Thirdborough SM, Valle-Argos B, Linley A, Krysov S, Hidalgo MS, Leonard E, Ishfaq M, Wagner SD, Willis AE, Steele AJ, Stevenson FK, Forconi F, Coldwell MJ, Packham G. 2016. Engagement of the B-cell receptor of chronic lymphocytic leukemia cells drives global and MYC-specific mRNA translation. Blood 127:449–457. doi: 10.1182/blood-2015-07-660969. [DOI] [PubMed] [Google Scholar]
- 51.Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, Okreglak V, Ashkenazi A, Hann B, Nader K, Arkin MR, Renslo AR, Sonenberg N, Walter P. 2013. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 26 February 2015. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 4. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L, Yang X, Mou C, Yang Q. 2017. Transmissible gastroenteritis virus does not suppress IFN-beta induction but is sensitive to IFN in IPEC-J2 cells. Vet Microbiol 199:128–134. doi: 10.1016/j.vetmic.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cruz JL, Becares M, Sola I, Oliveros JC, Enjuanes L, Zuniga S. 2013. Alphacoronavirus protein 7 modulates host innate immune response. J Virol 87:9754–9767. doi: 10.1128/JVI.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becares M, Pascual-Iglesias A, Nogales A, Sola I, Enjuanes L, Zuniga S. 2016. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol 90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Z, An K, Xie L, Wu W, Zhang R, Wang D, Fang Y, Chen H, Xiao S, Fang L. 2017. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology 507:170–178. doi: 10.1016/j.virol.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jordan LT, Derbyshire JB. 1995. Antiviral action of interferon-alpha against porcine transmissible gastroenteritis virus. Vet Microbiol 45:59–70. doi: 10.1016/0378-1135(94)00118-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 59.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. 2009. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Favreau DJ, Desforges M, St-Jean JR, Talbot PJ. 2009. A human coronavirus OC43 variant harboring persistence-associated mutations in the S glycoprotein differentially induces the unfolded protein response in human neurons as compared to wild-type virus. Virology 395:255–267. doi: 10.1016/j.virol.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Li JR, Sun MX, Ni B, Huan C, Huang L, Li C, Fan HJ, Ren XF, Mao X. 2014. Triggering unfolded protein response by 2-deoxy-d-glucose inhibits porcine epidemic diarrhea virus propagation. Antiviral Res 106:33–41. doi: 10.1016/j.antiviral.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol 9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Connor JH, Lyles DS. 2005. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J Biol Chem 280:13512–13519. doi: 10.1074/jbc.M501156200. [DOI] [PubMed] [Google Scholar]
- 65.Desmet EA, Anguish LJ, Parker JS. 2014. Virus-mediated compartmentalization of the host translational machinery. mBio 5:e01463-14. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Guo L, Jia X, Wang T, Wang J, Sun Z, Wang L, Li X, Tan F, Tian K. 2015. Construction of a triple gene-deleted Chinese pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine 33:2432–2437. doi: 10.1016/j.vaccine.2015.03.094. [DOI] [PubMed] [Google Scholar]