Herpes simplex virus (HSV) can be genetically engineered to endow cancer-selective replication and oncolytic activity. γ34.5, a key neurovirulence gene, has been deleted in all oncolytic HSVs in clinical trial for glioma. Glioblastoma stem-like cells (GSCs) are a subpopulation of tumor cells thought to drive tumor heterogeneity and therapeutic resistance. GSCs are nonpermissive for γ34.5− HSV, while non-stem-like cancer cells from the same patient tumors are permissive. GSCs restrict true late protein synthesis, despite normal viral DNA replication and transcription of all kinetic classes. This is specific for true late translation as early and leaky late transcripts are translated late in infection, notwithstanding shutoff of cellular protein synthesis. Expression of Us11 in GSCs rescues the replication of γ34.5− HSV. We have identified a cell type-specific innate response to HSV-1 that limits oncolytic activity in glioblastoma.
KEYWORDS: oncolytic virus, cancer stem cell, HSV, translation, glioma, Us11, herpes simplex virus
ABSTRACT
Oncolytic viruses, including herpes simplex viruses (HSVs), are a new class of cancer therapeutic engineered to infect and kill cancer cells while sparing normal tissue. To ensure that oncolytic HSV (oHSV) is safe in the brain, all oHSVs in clinical trial for glioma lack the γ34.5 genes responsible for neurovirulence. However, loss of γ34.5 attenuates growth in cancer cells. Glioblastoma (GBM) is a lethal brain tumor that is heterogeneous and contains a subpopulation of cancer stem cells, termed GBM stem-like cells (GSCs), that likely promote tumor progression and recurrence. GSCs and matched serum-cultured GBM cells (ScGCs), representative of bulk or differentiated tumor cells, were isolated from the same patient tumor specimens. ScGCs are permissive to replication and cell killing by oHSV with deletion of the γ34.5 genes (γ34.5− oHSV), while patient-matched GSCs were not, implying an underlying biological difference between stem and bulk cancer cells. GSCs specifically restrict the synthesis of HSV-1 true late (TL) proteins, without affecting viral DNA replication or transcription of TL genes. A global shutoff of cellular protein synthesis also occurs late after γ34.5− oHSV infection of GSCs but does not affect the synthesis of early and leaky late viral proteins. Levels of phosphorylated eIF2α and eIF4E do not correlate with cell permissivity. Expression of Us11 in GSCs rescues replication of γ34.5− oHSV. The difference in degrees of permissivity between GSCs and ScGCs to γ34.5− oHSV illustrates a selective translational regulatory pathway in GSCs that may be operative in other stem-like cells and has implications for creating oHSVs.
IMPORTANCE Herpes simplex virus (HSV) can be genetically engineered to endow cancer-selective replication and oncolytic activity. γ34.5, a key neurovirulence gene, has been deleted in all oncolytic HSVs in clinical trial for glioma. Glioblastoma stem-like cells (GSCs) are a subpopulation of tumor cells thought to drive tumor heterogeneity and therapeutic resistance. GSCs are nonpermissive for γ34.5− HSV, while non-stem-like cancer cells from the same patient tumors are permissive. GSCs restrict true late protein synthesis, despite normal viral DNA replication and transcription of all kinetic classes. This is specific for true late translation as early and leaky late transcripts are translated late in infection, notwithstanding shutoff of cellular protein synthesis. Expression of Us11 in GSCs rescues the replication of γ34.5− HSV. We have identified a cell type-specific innate response to HSV-1 that limits oncolytic activity in glioblastoma.
INTRODUCTION
Glioblastoma (GBM), a fatal brain tumor, affects thousands of patients each year (1). Conventional treatment of GBM involves surgical removal of accessible tumor followed by radiation and chemotherapy with temozolomide, extending survival by several months, but ultimately the tumor recurs resistant to therapy (2). GBM tumors are heterogeneous genetically, cellularly, and morphologically, both within and between patients (3–6). The leading theory for GBM tumor heterogeneity and recurrence hypothesizes the presence of a cancer stem cell population which can self-renew, populate the tumor with differentiated cells, and resist therapy, leading to inevitable recurrence (3, 7). GBM stem-like cells (GSCs) were first isolated by culturing patient tumor specimens under neural stem cell (NSC) conditions in serum-free, growth factor-supplemented neural basal medium as nonadherent spheres (7). Single-cell transcriptome analysis (RNA-seq) indicates that GSCs are present in situ, as well as more differentiated tumor cells derived from GSCs and expressing markers of more mature multilineage cell types (4, 5). Culturing tumor specimens in serum generates primary adherent serum-cultured GBM cells (ScGCs), representative of differentiated or bulk tumor cells (8). GSCs are highly tumorigenic in immunodeficient mice, adopting similar patterns of brain invasion, cellular heterogeneity, and vascularity as observed in the original patient tumor, while ScGCs are poorly tumorigenic, epigenetically distinct from GSCs, and do not faithfully recapitulate the patient's tumor (9–12).
Due to the high mortality of GBM, researchers have sought novel therapies to improve patient prognosis, such as oncolytic herpes simplex virus 1 [HSV-1] (oHSV). oHSVs are genetically engineered to selectively replicate in and kill cancer cells and not normal cells by targeting cells with cancer hallmarks such as unregulated cell division or lack of normal antiviral cell responses (13, 14). HSV-1 infects and deposits its DNA genome into the nucleus, where an ordered transcriptional cascade begins. Immediate early (α, IE) gene products are expressed first, which facilitate expression of early (β, E) genes, encoding proteins involved in HSV-1 DNA replication, and leaky late (γ1, LL) genes (15). Only after de novo viral DNA replication are true late (γ2, TL) genes expressed. Once synthesized, the late proteins (structural and tegument) assemble capsids, package newly synthesized HSV-1 DNA, and generate infectious virions (15).
G207, the first oHSV to enter clinical trial in the Unites States (16), has the ICP6 gene (UL39; ribonucleotide reductase large subunit) inactivated by insertion of the Escherichia coli LacZ gene, and both copies of the γ34.5 gene are deleted (17). The γ34.5 protein directs protein phosphatase 1α (PP1α) to dephosphorylate eIF2α, which maintains protein synthesis despite stress signaling from eIF2 kinases, like PKR (18, 19). Loss of γ34.5 greatly reduces neurovirulence (20), which is further decreased by ICP6 inactivation (17), and contributes to selective replication in cancer cells (17, 21). Thus, all oHSVs that have been in clinical trial for GBM have deletions of γ34.5 (13). However, HSV-1s with deletions of γ34.5 (γ34.5− viruses) are somewhat attenuated for replication in many cancer cells (22, 23). Deletion of ICP47 (Us12) complements γ34.5 loss, likely due to placement of TL Us11 under the ICP47 IE promoter (24–26). Us11 binds double-stranded RNA and antagonizes PKR, inhibiting eIF2α phosphorylation and overcoming loss of γ34.5 activation of PP1α (25, 26). In order to create a more efficacious oHSV, ICP47 was removed from G207 to generate G47Δ, which grows in many of the cancer cell lines and GSCs which restrict γ34.5− HSV-1 (9, 22). The ability of Us11 expression in trans in nonpermissive cancer cells, such as GSCs, to rescue γ34.5− HSV-1 has not been tested.
We found that every GSC line tested was nonpermissive for G207, while the matched ScGC lines were all permissive. In contrast, all GSC and ScGC lines tested were permissive for G47Δ. This held true regardless of the primary or recurrent status of the patient's tumor. In addition, the genetic heterogeneity between patient tumors had no noticeable effect on oHSV replication. Here, we show that γ34.5− oHSV G207 is prevented from producing new infectious virus in GSCs due to a translational block that occurs late in virus infection. Viral DNA replication and transcription, including TL gene transcription, occur normally. Despite shutoff of cellular protein synthesis late in infection, E and LL viral proteins continue to be translated. We demonstrate that expression of full-length Us11 protein in GSCs is sufficient to complement the loss of γ34.5 and rescue G207 replication.
RESULTS
ScGCs, but not GSCs, are permissive to γ34.5− oHSV replication.
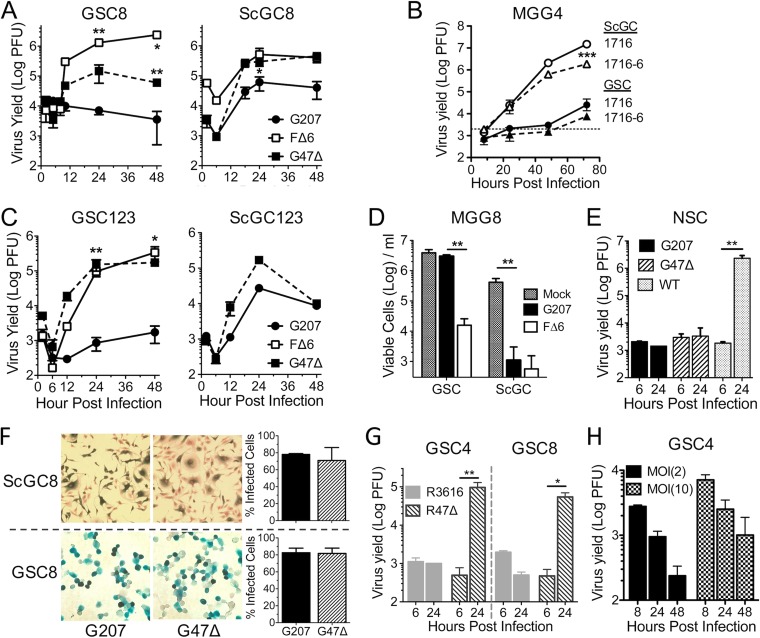
We have isolated matched GSCs and ScGCs from the same patients' tumor specimens (identified by number, e.g., GSC8 and ScGC8 are from specimen MGG8) and shown that they have different phenotypes (i.e., tumorigenicity and gene expression) (9, 11, 12). When we first isolated GSCs, we found that γ34.5− oHSVs, except for G47Δ, replicated poorly or not at all in these cells (9, 23), in contrast to the previously shown replication of G207 and other γ34.5− oHSVs in most established glioma cell lines (17, 21, 23, 27). Here, we compared the permissivity of patient-matched GSCs and ScGCs to G207 replication as well as to that with G47Δ and FΔ6 (γ34.5+). We performed single-step growth experiments at a multiplicity of infection (MOI) of 2 to determine virus yield during a single cycle of virus replication. ScGC8 cells were permissive for G207, G47Δ, and FΔ6 viral growth (Fig. 1A), while GSC8 cells were nonpermissive for G207 (Fig. 1A), as previously reported (9). A similar result was seen with 1716-6 (strain 17 with a deletion of γ34.5 and ICP6), in a multistep growth experiment after infection at a low MOI of 0.1 (Fig. 1B). G47Δ replicates better than G207 and reached similar titers as FΔ6 in ScGCs (Table 1).
FIG 1.
Glioblastoma stem-like cells are not permissive to γ34.5− HSV-1 without ICP47 deletion. (A) Virus growth in GSC8s and ScGC8s after infection at an MOI of 2. Cells and supernatant were collected at indicated times, and titers were determined on Vero cells. The number of PFU was measured (n = 2). Significance was determined versus results with G207 (**) and for results with FΔ6 versus those with G47Δ (*) by a t test. (B) Virus growth of 1716 and 1716-6 after infection at an MOI of 0.1 in MGG4 cells (dashed line, input virus). Significance was determined for results with 1716 and 1716-6 with ScGC versus those with GSC by (n = 3; t test and ANOVA). (C) Virus growth on recurrent GSC123s (n = 3) and ScGC123s (n = 2 or 1) after infection at an MOI of 2. Significance was determined for results with FΔ6 and G47Δ versus those with G207 (t test). (D) Cell viability after G207 or FΔ6 infection of MGG8 cells at an MOI of 0.1 measured 8 days p.i. counting trypan blue-excluding cells. (n = 3; t test and ANOVA). (E) Virus growth in normal human neural stem cells (NSCs) after infection of wild-type F strain, G207, or G47Δ at an MOI of 2 (n = 3; t test). (F) Infectivity assay of MGG8 cells infected with G207 or G47Δ at an MOI of 2. Infected X-Gal+ cells at 6 hpi (left) were counted to determine percent infected cells (right; n = 3). (G) Virus growth of R3616 and R47Δ after infection at an MOI of 2 of GSC4s and GSC8s (n = 2; ANOVA). (H) Virus growth of G207 after infection at an MOI of 2 or 10 of GSC4s (n = 3). Graphs are replicate samples from single experiments. Values are means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 1.
oHSV replication in GSC and ScGC lines
| Tumor type and patient specimen | Virus | Titer ratioa in: |
|
|---|---|---|---|
| GSCs | ScGCs | ||
| Primary | |||
| MGG4 | G207 | 0.7 | 9.7 |
| FΔ6 | 13.7 | 136.6 | |
| G47Δ | 32.7 | 415.7 | |
| MGG8 | G207 | 0.5 | 63.2 |
| FΔ6 | 154.9 | 34.5 | |
| G47Δ | 42.3 | 327.6 | |
| MGG18 | G207 | 3.5 | |
| FΔ6 | NT | NT | |
| G47Δ | 343 | ||
| MGG29 | G207 | 1.3 | 83.3 |
| FΔ6 | 25.8 | 83.3 | |
| G47Δ | 21.4 | 68.5 | |
| MGG75 | G207 | 0.5 | 8.2 |
| FΔ6 | 6.3 | 27.3 | |
| G47Δ | NT | NT | |
| Recurrent | |||
| MGG31 | G207 | 0.97 | 69.7 |
| FΔ6 | N/T | 612 | |
| G47Δ | 192 | 207 | |
| MGG70R | G207 | 0.4 | |
| FΔ6 | 29.7 | None | |
| G47Δ | 8.1 | ||
| MGG70RRb | G207 | 0.7 | |
| FΔ6 | 39.2 | None | |
| G47Δ | 7.0 | ||
| MGG123 | G207 | 0.7 | 99 |
| FΔ6 | 198.5 | NT | |
| G47Δ | 20.7 | 614 | |
The titer at 24 hpi was divided by the titer measured at 5 to 8 hpi. Cells were considered permissive if the ratio was at least 5 or represented half a log increase from the value at eclipse. Values were calculated from single experiments with technical triplicates. None, cells do not exist; NT, not tested.
MGG70RR was a re-recurrent tumor from MGG70R.
GBM inevitably recurs after treatment, so it was important to determine whether GSCs isolated from recurrent GBMs retained their permissivity to oHSV. The growth kinetics of G207 and G47Δ in recurrent MGG123 GSCs and ScGCs were similar to those in primary GBM cells (Fig. 1C). Titers from GSCs and ScGCs infected at an MOI of 2 reached their eclipse around 5 to 8 h postinfection (hpi) and plateaued after 24 hpi (Fig. 1A and C). To examine whether nonpermissiveness was a hallmark of GSCs in general or related to specific genetic abnormalities, we extended these studies to nine other GSC lines and six matched ScGC lines (Table 1). All GSCs tested were nonpermissive to γ34.5− G207 regardless of the primary or recurrent status of the patient's tumor, while all ScGCs were permissive (Table 1). Similarly, GSCs cultured adherently on laminin were not permissive to G207 (data not shown). Because the ScGCs were cultured from the same tumor specimens as their matched GSCs, any differences in replication are likely due to the stem-like phenotype of GSCs. Our GSCs contain a wide variety of genetic alterations (10), suggesting that no common genetic mutation is responsible for the restriction of G207. In line with replication permissivity, G207 was cytotoxic to ScGCs in vitro but not to GSCs, while FΔ6 was cytotoxic to both (Fig. 1D). Normal human neural stem cells (huNSCs) were nonpermissive for both G207 and G47Δ replication but permissive for wild-type (wt) F strain HSV-1, demonstrating cancer selectivity (Fig. 1E).
The difference in permissivities was not due to an inability of G207 to infect GSCs as measurements of infectivity displayed no significant differences between GSCs and ScGCs or between G207 and G47Δ in GSCs (Fig. 1F). Inactivation of ICP6 did not contribute to cell permissivity in the context of γ34.5− HSV-1, as demonstrated with R47Δ (ICP6+ G47Δ) and its parent virus R3616 (γ34.5−) (Fig. 1G) and 1716 (Fig. 1B). Growth of some HSV-1 mutants, i.e., ICP0-null HSVs (28), can be rescued by infection at a high MOI. However, GSCs infected at an MOI of 10 were still nonpermissive to G207 virus growth (Fig. 1H).
Viral capsids are not produced after G207 infection of GSCs.
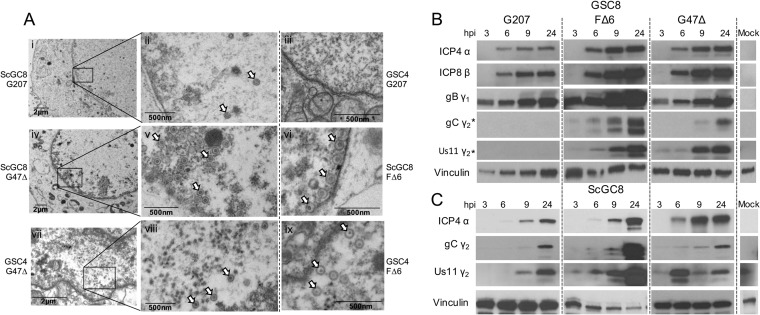
We used transmission electron microscopy (EM) to evaluate capsid assembly in oHSV-infected GSCs and ScGCs late in infection to further ascertain the temporal block. Viral capsids were present in the nucleus of G207-infected ScGCs but not in G207-infected GSCs, while they were present in both G47Δ- and FΔ6-infected GSCs and ScGCs (Fig. 2A). Forty-eight percent and 68% of GSC4s examined (n = 25) during G47Δ and FΔ6 infection, respectively, contained at least one capsid, while none were seen in GSCs infected with G207.
FIG 2.
Capsids and true late (TL) proteins do not accumulate during G207 infection of GSCs. (A) Electron micrographs of ScGC8 and GSC4 cells infected with G207, G47Δ, and FΔ6 at an MOI of 2. Micrographs were taken at 24 hpi in ScGC8s (frames i, ii, and iv to vi) or 16 hpi in GSC4s (iii and vii to ix). Boxed areas (periphery of nucleus) are magnified in panels on the right. Arrowheads indicate virus capsids. Whole-cell extracts from GSC8 (B) and ScGC8 (C) cells infected with G207, FΔ6, or G47Δ at an MOI of 2 for 3, 6, 9, and 24 hpi were separated by SDS-PAGE and immunoblotted using antibodies against viral IE (α), E (β), LL (γ1), and TL (γ2) proteins or vinculin (loading control). Blotting performed on different membranes is denoted with an asterisk (*).
True late protein synthesis is blocked in GSCs after G207 infection.
To assess why capsids were not being produced during G207 infection, we evaluated viral protein synthesis using Western blot analysis of different kinetic classes of viral proteins after infection of GSCs and ScGCs. All viruses (G207, G47Δ, and FΔ6) synthesized IE, E, and LL proteins in GSC8 and ScGC8 (Fig. 2B and C). However, G207-infected GSCs did not synthesize TL proteins (gC and Us11), while G47Δ- and FΔ6-infected GSCs did (Fig. 2B).
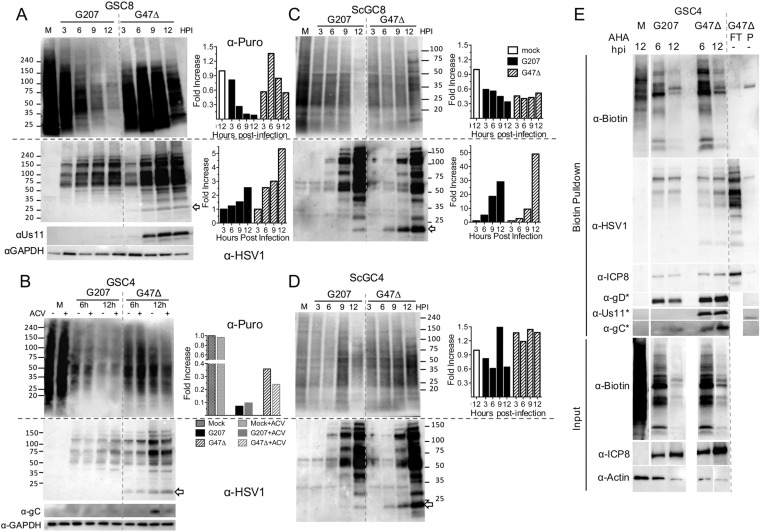
Since viral TL protein synthesis was inhibited in G207-infected GSCs, we examined whether this was selective to HSV proteins or also affected cellular proteins by monitoring global protein synthesis with puromycin. Puromycin incorporates into nascent peptides, terminating their elongation while also labeling de novo protein synthesis (29). G207-infected GSCs underwent a marked translational shutoff around 6 hpi that was not seen with G47Δ-infected GSCs (Fig. 3A). Addition of the HSV-1 DNA synthesis inhibitor acyclovir (ACV) did not influence protein synthesis shutoff during infection of GSCs (Fig. 3B), in contrast to what was seen during γ34.5− HSV-1 infection of neuroblastoma cells (30). ACV treatment did prevent TL protein synthesis of gC in G47Δ-infected GSCs (Fig. 3B, α-gC). A pan-HSV-1 antibody revealed that viral proteins accumulate to a lesser extent in G207-infected GSCs but were not noticeably impacted by host protein synthesis shutoff or ACV (Fig. 3A and B). Us11 was absent from G207-infected GSCs while present in G47Δ-infected GSCs by 6 hpi and after ACV treatment, demonstrating its IE kinetics after ICP47 deletion (Fig. 3A and B). Puromycin labeling was less efficient in ScGCs (compare M lane GSCs and ScGCs), while protein synthesis shutoff in G207-infected ScGCs was much less than that in GSCs (Fig. 3C and D, graphs). Viral protein accumulation occurred similarly in either G207- or G47Δ-infected ScGCs (Fig. 3C and D). Interestingly, the increased accumulation of proteins detected with anti-HSV-1 was much greater with time after infection with G207 and G47Δ in ScGCs than in GSCs (Fig. 3C and D versus A and B; note 10-fold-increased scale in ScGCs). These data suggest that differing abilities to respond to HSV-1-induced protein shutoff contribute to different degrees of permissivity to G207 and G47Δ.
FIG 3.
G207 infection of GSCs inhibits protein synthesis. (A) Western blot of GSC8s infected at an MOI of 2 for indicated times (lane M is mock infected) and pulsed with 10 μg/ml puromycin 15 min before lysates were harvested and probed with anti-puromycin antibody (α-Puro) and stripped and probed with an anti-HSV-1 (α-HSV1) and anti-Us11 and anti-GAPDH (loading control) antibodies. Numbers at the side represent kilodaltons of markers. The arrow indicates Us11. Quantitation of the Western blot is shown at right. Anti-puromycin intensity was normalized to that of M, and anti-HSV-1 intensity was normalized to that of 3 hpi. (B) GSC4s were infected with G207 and G47Δ at an MOI of 2 for 6 or 12 hpi in the presence (+) or absence (−) of 10 μM acyclovir (ACV) and pulsed with puromycin before lysates were harvested and probed as described for panel A, except with anti-gC (α-gC). Quantitation of anti-puromycin blot lanes at 12 hpi with or without ACV is shown on the right (intensity normalized to that of lane M as 1). Western blots of ScGC8s (C) and ScGC4s (D) infected at an MOI of 2 for indicated times, pulsed with puromycin, and immunoblotted as described for panel A. Numbers at the side represent kilodaltons of markers. The arrow indicates Us11. Quantitation of Western blots is shown on the right. Anti-puromycin intensity was normalized to that of lane M, and anti-HSV-1 intensity was normalized to that of 3 hpi. (E) Western blot analysis for AHA-labeled proteins. Cell lysates from G207- or G47Δ-infected GSC4s at 6 and 12 hpi or mock infected (M) after AHA treatment were subjected to Click-iT biotin conjugation and pulled down with streptavidin-coated beads. Lane P, biotin pulldown (control); lane FT, flowthrough for non-AHA-treated (−) lysates. Membranes were analyzed by immunoblotting using anti-biotin (streptavidin), anti-HSV-1, and anti-ICP8 (E), -gD (LL), -Us11, and -gC (TL) antibodies. Blotting performed on different membranes is denoted with an asterisk (*). Input samples (prior to biotin pulldown; bottom three panels) were probed with anti-biotin (streptavidin), anti-ICP8, and anti-actin (loading control).
We could not ascertain whether protein synthesis shutoff was specifically affecting TL proteins or all kinetic classes of HSV-1 proteins late in infection because puromycin truncates polypeptides, and so the products are not detectable with antibody. Therefore, we used amino acid analog azidohomoalanine (AHA)-coupled click chemistry to label nascent peptides with biotin (31). AHA incorporation in GSCs was reduced late after both G207 and G47Δ infection (Fig. 3E), supporting a generalized protein shutoff, as with puromycin. Despite the shutoff, HSV-1 E and LL proteins ICP8 and gD were still synthesized at 12 hpi with both viruses, as were proteins recognized by the pan-HSV-1 antibody (Fig. 3E). In contrast, TL Us11 and gC were not synthesized during G207 infection (Fig. 3E), demonstrating a specific shutoff of TL proteins. GSCs infected with G47Δ maintained TL protein synthesis despite the protein shutoff (Fig. 3E).
Viral DNA replication is not inhibited during G207 infection of GSCs.
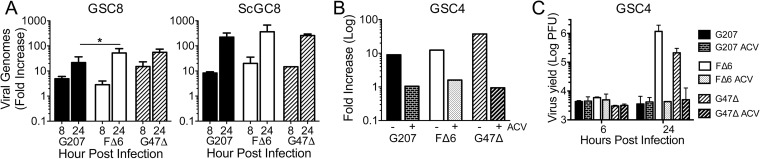
HSV-1 TL proteins require de novo viral DNA replication for synthesis (32). Therefore, we examined whether G207 DNA replication was occurring in infected GSCs. Quantitative PCR (qPCR) was performed on DNA harvested from oHSV-infected GSCs to determine the fold increase in HSV-1 genomes from an initial 2-hpi time point. Viral DNA amplification occurs in G207-infected GSCs and ScGCs at levels similar to those of G47Δ and FΔ6 although approximately 10-fold more viral genomes are made in infected ScGCs (Fig. 4A). Addition of ACV prevented viral DNA replication and viral growth (Fig. 4B and C). To confirm that viral infection and nuclear transit occur similarly, we performed qPCR on nuclei from GSCs infected at an MOI of 2 and observed about 1.8 HSV-1 genomes per GSC nucleus (data not shown), with nearly equal amounts of G207, FΔ6, and G47Δ genomes in infected nuclei (1.0:1.1:0.9 ratio).
FIG 4.
GSCs support G207 DNA replication. (A) Quantitative PCR using HSV-1 ICP4 and cellular GAPDH primers on DNA isolated from MGG8 cells infected with G207, FΔ6, and G47Δ at an MOI of 2 at 8 and 24 hpi. Fold increase was calculated using ΔΔCT of ICP4 and GAPDH in relation to a 2-hpi time point (n = 2 to 9). There is no significant difference between replication levels of viruses in ScGC8s. *, P < 0.05 (ANOVA). (B) qPCR of GSC4s infected with G207, FΔ6, and G47Δ at an MOI of 2 for 24 h in the presence of 10 μM acyclovir (ACV; +) or vehicle (−) (n = 1). (C) Corresponding plaque assay of GSC4s infected in the presence or absence of ACV (n = 2).
Viral true late gene transcription is not inhibited during G207 infection of GSCs.
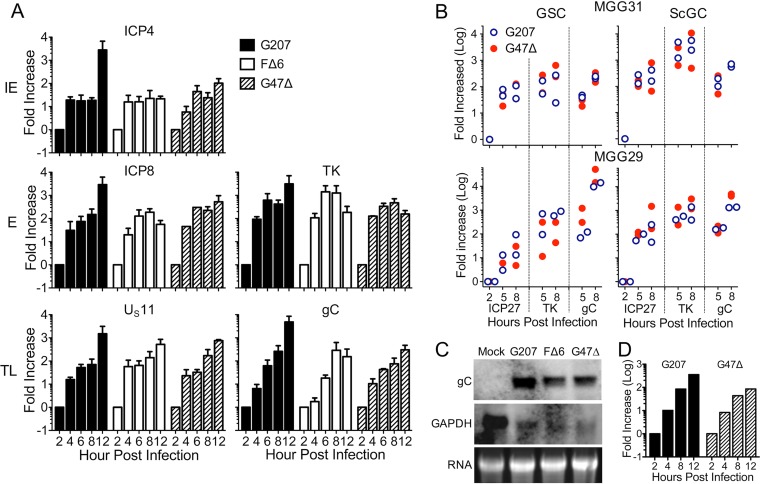
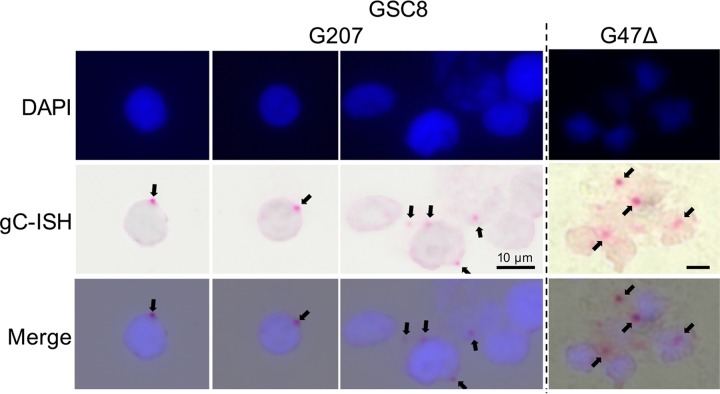
We next examined virus transcription, which is kinetically regulated. Quantitative reverse transcription-PCR (qRT-PCR) showed that G207-infected GSCs produced amounts of IE and E gene transcripts similar to those of FΔ6- and G47Δ-infected GSCs (Fig. 5A), as expected from protein synthesis. Unexpectedly, TL gC and Us11 transcripts were expressed in G207-infected GSCs at levels similar to those with FΔ6 or G47Δ infection (Fig. 5A). This pattern was reproduced in MGG31 and MGG29 cells using TaqMan qRT-PCR, with no large differences between GSCs and ScGCs (Fig. 5B). To demonstrate that the gC transcripts were full-length and not truncations or qRT-PCR artifacts, we performed Northern blotting on total RNA. Full-length gC RNA (∼2.3 kb) was detected with all viruses in infected GSC4 cells (Fig. 5C). We observed a decrease in spliced glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts in infected GSCs (Fig. 5C), as reported previously (33). Therefore, we repeated our qRT-PCR experiments using 18S rRNA as a calibrator gene and observed the same increase in gC transcription (Fig. 5D). We used RNAScope immunohistochemistry to determine the cellular localization of TL gC transcripts, which were similarly clustered adjacent to the nucleus after both G207 and G47Δ infection of GSC8s (Fig. 6). These observations demonstrate that in GSCs, G207 replicates its DNA and synthesizes TL RNAs but is unable to translate them, suggesting that GSCs restrict TL translation that γ34.5 surmounts.
FIG 5.
GSCs support G207 true late RNA transcription. (A) Fold increase (log) of ICP4, ICP8, TK, Us11, and gC viral RNAs in relation to GAPDH (calibrator) as measured by qPCR of cDNA from infected GSC8s (n = 2 to 5). (B) Fold increase of ICP27 (IE), TK (E), and gC (TL) RNAs during infection of MGG29 and MGG31 in relation to 2-hpi values measured by qRT-PCR using TaqMan probes (n = 2). (C) Northern blot of whole-cell RNA from GSC4 cells infected with oHSV at an MOI of 2 for 20 h. Glycoprotein C (gC; 2.3 kb) and control (intronless GAPDH; 1.4 kb) were probed using digoxigenin-labeled DNA probes. (D) Fold increase of gC using the ΔΔCT method during G207 or G47Δ infection of GSC8s in relation to the level of the 18S rRNA calibrator gene (n = 1).
FIG 6.
RNAScope visualization of HSV-1 gC transcripts. HSV-1 TL gC mRNA was visualized using RNAScope immunohistochemistry. GSC8s were infected with G207 or G47Δ and fixed at 12 hpi. Slides were lightly counterstained with hematoxylin and mounted in a DAPI-containing medium. Arrows indicate gC transcripts. ISH, in situ hybridization.
Translation initiation factor phosphorylation is not a direct contributor to G207 replication.
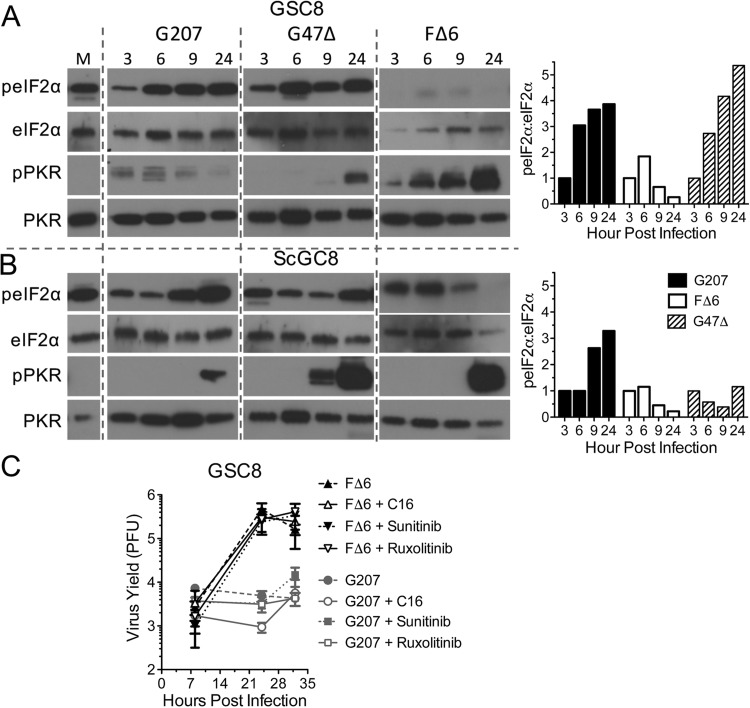
Us11 and γ34.5 prevent shutoff of protein translation by reducing phosphorylation of eIF2α (18, 26). Therefore, we examined whether G47Δ was better able to suppress eIF2α phosphorylation due to PKR inhibition by IE Us11 expression. G207 induced phosphorylated eIF2α (p-eIF2α) in GSC8s although PKR was only minimally activated (Fig. 7A). Perplexingly, G47Δ also elicited similarly high levels of p-eIF2α (Fig. 7A). In contrast, FΔ6 completely inhibited p-eIF2α accumulation in GSC8s, despite broad activation of PKR (Fig. 7A). In ScGCs, p-eIF2α levels continued to rise at late times after infection with G207 but not with G47Δ (Fig. 7B) even though ScGCs support G207 growth. Similar results were observed with GSC4s and ScGC4s (data not shown). This suggests (i) that an eIF2α kinase other than PKR may be phosphorylating eIF2α, (ii) that PKR might not play a key role in restricting G207, (iii) that PKR is acting through other pathways, and/or (iv) that eIF2α phosphorylation may not be the dominant regulator of TL translation in HSV-1-infected GSCs. GSCs expressing a short hairpin RNA (shRNA) to PKR (about 70% inhibition of PKR) remained nonpermissive to G207 (data not shown). We further tested whether a number of inhibitors affecting antiviral responses and oncolytic virus replication might rescue G207 replication in GSCs. We treated GSCs with C16 (PKR inhibitor) (34), ruxolitinib (JAK1/2 inhibitor) (35, 36), and sunitinib (multikinase inhibitor) (37, 38). None of these inhibitors rescued G207 replication or affected replication of FΔ6 in GSCs (Fig. 7C).
FIG 7.
Eukaryotic initiation factor 2α and PKR phosphorylation do not correlate with cell permissivity. Western blotting was performed of GSC8 (A) and ScGSC8 (B) cells infected with G207, G47Δ, or FΔ6 at an MOI of 2 for the time (hours postinfection) indicated at the top. Immunoblotting was performed using antibodies for total and phosphorylated eIF2α and PKR. Quantitation of the Western blot (right) is shown as the ratio of anti-p-eIF2α/eIF2α intensity normalized to the level at 3 hpi (set as 1). (C) GSC8 cells infected with G207 or FΔ6 at an MOI of 2 in the presence of PKR inhibitor (C16), JAK1/2 inhibitor (ruxolitinib), or pan-kinase inhibitor (sunitinib). The cells and supernatant were collected between 8 and 36 hpi, and titers were determined on Vero cells. Values are means ± standard deviations (n = 3).
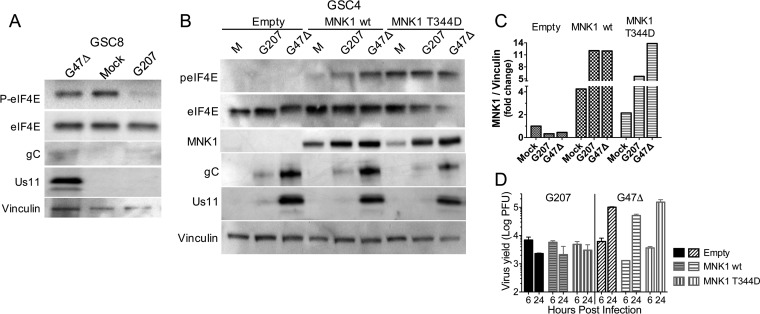
The eIF4F complex, regulated by mTOR, facilitates cap-dependent ribosome association with mRNAs to promote protein synthesis during normal homeostasis and shutoff during endoplasmic reticulum (ER) stress (39). In HSV-1-infected cells, ICP6 promotes eIF4F assembly (40). The eIF4E subunit of the complex, which binds cap structures, has been implicated in tumorigenesis (41). It is phosphorylated by MNK1/2, in response to extracellular signal-regulated kinase (ERK) or p38 mitogen-activated protein kinase (MAPK), which promotes transformation and alters translation of selective mRNAs (42–46). Inhibition of MNK1 reduces HSV-1 replication and protein synthesis in fibroblasts (47, 48). Therefore, we examined whether MNK1/p-eIF4E affect GSC permissivity. p-eIF4E was lost during infection of GSC8s by G207 but not by G47Δ (Fig. 8A). In contrast, while p-eIF4E was lost in G207-infected GSC4s, it was elevated in ScGC4 cells (data not shown). To test whether p-eIF4E could rescue TL translation and/or whether loss of p-eIF4E contributes to the selective shutdown of protein synthesis after G207 infection, we overexpressed MNK1 and a constitutively active MNK1 mutant, T344D (49). Overexpression of wt MNK1 increased p-eIF4E after oHSV infection, and mutant MNK1 (T344D) further elevated p-eIF4E (Fig. 8B and C). Despite sustained induction of p-eIF4E, neither G207 TL protein synthesis nor virus replication was rescued in MNK1-overexpressing GSC4s (Fig. 8B and D). This indicates that elevated p-eIF4E is not sufficient for TL protein synthesis in GSCs.
FIG 8.
Phosphorylation of eIF4E does not restore TL protein translation in G207-infected GSCs. (A) Western blot for eIF4E protein from whole-cell lysates from GSC8 cells infected with G47Δ and G207 (24 hpi) at an MOI of 2 and immunoblotted with antibodies against p-eIF4E, eIF4E, gC, Us11, and vinculin (loading control). (B) Whole-cell extracts from GSC4s expressing MNK1 wild-type or the constitutively active mutant T344D and infected with G207 or G47Δ (24 hpi) at an MOI of 2, or mock (M). Immunoblotting was performed with antibodies against eIF4E, p-eIF4E, MNK1, gC, Us11, and vinculin (loading control). (C) Quantification of Western blot for MNK1 expression as fold change in MNK1/vinculin normalized to the level of mock infection with an empty vector. (D) Virus growth of G207 and G47Δ on MNK1-transduced GSC4s from the experiment shown in panel C. Values are means ± standard deviations (n = 3). G47Δ yield at 24 hpi is significantly different from that at 6 hpi (P < 0.01; ANOVA).
Us11 expression in GSCs complements γ34.5 loss.
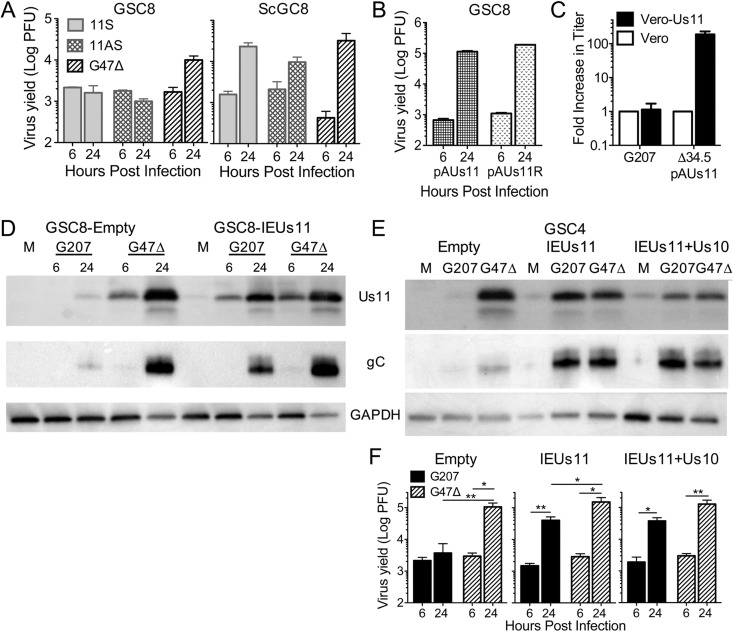
We wanted to directly examine whether IE Us11 expression in GSCs could complement γ34.5 loss. A γ34.5− oHSV with IE Us11 expression (11S) was previously shown to replicate in U373 glioma cells, which are nonpermissive to γ34.5− HSV-1 lacking IE Us11 expression (24, 25). 11S grew in ScGC8s to similar titers as G47Δ but could not replicate in GSC8s (Fig. 9A), indicating that GSCs are more restrictive than U373 cells. We next tested the requirement for Us11 in the context of wild-type γ34.5. Us11 is not necessary for growth in GSCs when γ34.5 is present, as demonstrated by the growth of pAUs11 (γ34.5+ Us11−) in GSC8s (Fig. 9B). However, loss of Us11 in a γ34.5− version of pAUs11 (Δ34.5pAUs11) attenuates the virus even in Vero cells, reducing viral protein synthesis (19). Transduction of Vero cells with a lentivirus expressing Us11 under an HSV IE4/5 promoter increased Δ34.5pAUs11 virus yield by 180-fold but did not alter G207 growth (Fig. 9C), demonstrating the functionality of the Us11 construct.
FIG 9.
Strong Us11 expression is required to complement γ34.5 deletion. (A) Virus growth of 11S, 11AS, or G47Δ on GSC8 (left) and ScGC8 (right) cells, as measured by plaque assay (n = 3). (B) Virus growth of pAUs11 (γ34.5+ Us11−) or pAUs11R (γ34.5+ Us11+) on GSC8 cells. (C) Virus plaque assay of G207 or Δ34.5 pAUs11 on Vero cells transduced with IEUs11 lentivirus. Fold increase in titer was compared to that on Vero cells (n = 3 for IEUs11; n = 1 for Vero cells). (D) Western blot of GSC8s transduced with empty or IEUs11 lentivirus vector and infected with G207 or G47Δ at an MOI of 2 for 6 and 24 hpi. Immunoblotting was performed using anti-Us11, -gC, and -GAPDH (loading controls) antibodies. (E) Western blot of GSC4s transduced with empty, IEUs11, or IEUs11+Us10 lentivirus vector and infected with G207 or G47Δ at an MOI of 2 for 24 hpi. Immunoblotting was performed as described for panel D. (F) Virus growth of G207 or G47Δ at an MOI of 2 for 6 and 24 hpi from transduced GSC4s (as described for panel E) (n = 3). Values are means ± standard deviations. *, P < 0.05; **<0.01 (t test).
We next examined whether G207 replication could be rescued in nonpermissive GSCs by ectopic expression of Us11. GSCs were transduced with lentivirus vectors expressing the Us11 open reading frame (ORF) or full-length Us11 mRNA (also containing the Us10 ORF) under an HSV-1 IE4/5 promoter (IEUs11 or IEUs11+Us10, respectively). G207 infection of transduced GSC4s and GSC8s induced Us11 expression, as well as TL gC (Fig. 9D and E). These Us11-expressing GSC4s became permissive to G207 infection, increasing G207 growth by 27- and 36-fold over the 6-hpi time point with IEUs11 and IEUs11+Us10, respectively, with G47Δ replicating somewhat better (3- to 6-fold) (Fig. 9F). There was no significant difference observed between IEUs11 and IEUs11+Us10 expressing GSC4s (Fig. 9F), suggesting that full-length Us11 is sufficient to complement γ34.5− HSV-1 when expressed in trans.
DISCUSSION
In these studies, we used matched GSC and ScGC lines isolated from the same patient's tumor specimens, allowing us to study two phenotypically distinct cell populations from the same tumor. GSCs differ from ScGCs epigenetically, with altered transcriptional circuits and gene expression profiles; GSCs are more related to neural stem cells (NSCs), whereas ScGCs are more similar to astrocytes (11, 12). While GBM has been classified by expression subtype, different cells within an individual tumor can fall into different subtypes, illustrating the large heterogeneity present within a single GBM (5, 50). Between patients there is also large variability. Here, we identify a unique feature of GSC biology not present in ScGCs: repression of HSV-1 TL translation that is overcome by γ34.5 or Us11 expression. Despite the large genetic differences between tumor specimens and GSCs from different patients (10), there is a consistent cellular physiology that renders all tested GSCs nonpermissive and ScGCs permissive for γ34.5− oHSV. Importantly, G47Δ did not replicate in NSCs, illustrating a further level of restriction in normal cells and validating the oncolytic selectivity of G47Δ.
The inability of GSCs to translate TL transcripts after γ34.5− oHSV infection suggests an issue with translation initiation. Elevated protein synthesis and dysregulation of translation initiation are common attributes of cancer, with the eIF4F complex being a central control node (39). How translation initiation is regulated/altered in cancer stem cells is not well understood, nor are the translational regulatory differences between GSCs and ScGCs. Viruses are dependent on host translational machinery and go to great lengths to co-opt it (51). HSV-1 targets initiation through modifying eIF2α phosphorylation (with γ34.5, Us11, and gB) and eIF4F complex formation (with ICP6 and ICP27), as well as degrading excess mRNAs (virion host shutoff, Vhs) (51). Only a few of the multitude of cancer cell lines tested restrict γ34.5− HSV-1, including U373 (24, 25) and SK-N-SH (30), and these lines exhibit a similar shutoff of protein synthesis during infection (25, 30). It is not understood why U373 or SK-N-SH cells are nonpermissive. However, both γ34.5 and Us11 can prevent translational shutoff by antagonizing the phosphorylation/repression of eIF2α (18, 19, 25, 26). In both GSCs and ScGCs infected with either G207 or G47Δ, levels of p-eIF2α were similarly increased, while activated p-PKR was elevated in ScGCs at 24 hpi by both viruses. Similarly, p-eIF2α levels were previously shown not to correlate with reduced TL expression during Vhs− HSV-1 infection (52). In the absence of Vhs, loss of TL translation was proposed to be due to mRNA overload unless transcripts were present before overload (53). We did not directly test this, as all oHSVs were Vhs+, but the similar transcription kinetics between G207, G47Δ, and FΔ6 in GSCs does not support this hypothesis. Elevation of p-eIF2α, despite low p-PKR, could be due to other eIF2α kinases: PKR-like endoplasmic reticulum kinase (PERK), inhibited by HSV-1 (54), GCN2, or HRI (51).
The regulation of translational initiation is complex and perturbed in many cancers (55), while the significance of p-eIF4E in regulating translation is unclear. In contrast to other viruses, which impair eIF4F activity, HSV-1 usually stimulates it, with ICP6 (51) and/or ICP27 binding to poly(A)-binding protein (PABP) to promote initiation (56). G207, G47Δ, and FΔ6 all have LacZ insertions inactivating the relevant ICP6 domain; however, ICP6+ γ34.5− HSV-1 R3616 and 1716 (23) were also restricted in GSCs, suggesting that ICP6 is not essential for viral translation. We observed that p-eIF4E was present at late times postinfection in GSCs infected with G47Δ but not with G207. This was the only distinct molecular difference we observed between G207- and G47Δ-infected GSCs in relation to protein synthesis shutoff. Activation of eIF4E by transduction with constitutively active MNK1 did not rescue G207 replication or TL protein synthesis, indicating that p-eIF4E is not sufficient to restore HSV-1 TL protein synthesis or replication.
Despite our observations that eIF2α and eIF4E do not appear to play major roles in restricting γ34.5− HSV-1 in GSCs, early ectopic Us11 expression rescues viral growth. The relative unimportance of p-eIF2α levels raises questions as to whether Us11 rescue is due to inhibition of PKR phosphorylation (19, 57, 58). In previous studies, early expression of Us11 from recombinant γ34.5− HSV-1 rescued replication in nonpermissive cancer cells due to purported inhibition of PKR (24, 25, 59). 11S, a γ34.5− HSV-1 with an IE ICP27 promoter-driven Us11, was reported to replicate in U373 cells (25). Despite this IE promoter, this virus expresses very low levels of Us11 (25), which may have contributed to its inability to replicate in GSCs. Both Us11 and γ34.5 are present in the tegument of wild-type HSV-1 but are not expressed until late in infection. Even then, Us11 transcripts are expressed at higher levels than those of γ34.5 (60). We hypothesize that high levels of Us11 are necessary in some cells, like GSCs, for rescuing γ34.5− oHSV replication, which may be independent of eIF2α phosphorylation or PKR. Both γ34.5 and Us11 have multiple diverse functions that could contribute, possibly in unknown ways. Deletion of the beclin-binding domain of γ34.5, which inhibits autophagy, does not restrict replication (23). Additional activities of Us11 include the following: preventing apoptosis; binding PACT, nucleolin, HIPK2, and PAT1; and inhibiting activation of 2′,5′-oligoadenylate synthase (OAS), RIGI, and MDA5 (13), which could be involved in overcoming γ34.5 loss.
A complex issue facing the oncolytic virus field is how to balance viral cytotoxicity and efficacy with safety. As oncolytic viruses are replication competent, there is potential for normal tissue toxicity. This is why γ34.5, the major viral neurovirulence gene, was deleted in many oHSVs (13). This includes a number of γ34.5− oHSVs, in addition to G207 and G47Δ, that are in or entering clinical trial: 1716 (Seprevhir) (61), C134 (62), M032 (63), rQNestin34.5 (64), and talimogene laherparepvec (T-Vec) (14). T-Vec, approved for use in advanced melanoma, is similar to G47Δ, except that it has an intact ICP6 and expresses granulocyte-macrophage colony-stimulating factor (GM-CSF) (65), but may retain some neurovirulence (66). Expressing PKR inhibitors (human cytomegalovirus [HCMV] IRS1 in C134 and HSV-1 Us11 in C122) from the CMV IE promoter can overcome the growth defects of γ34.5− HSV-1; however, C122 (ICP47+ ICP6+) exhibited some neurovirulence in mice (62). The extreme negative effect of γ34.5 loss on virus replication became fully apparent only when GSCs were isolated and tested. GSCs and ScGCs are closely related to primary tumor cells, as opposed to cancer cell lines extensively passaged on plastic. This highlights the importance of examining virus-host interactions in representative cell types. In GSCs, LL γ34.5 functions to overcome a block to TL translation. This restriction lies in an active antiviral system in GSCs, antagonized by γ34.5 and Us11 and not active in ScGCs. This difference between GSCs and ScGCs may also be operative with other oncolytic viruses modulating translation and relevant to stem-like cells from other cancers. These differences provide insights for the development of cancer stem-like cell-targeted therapeutics and understanding host defenses to HSV-1.
MATERIALS AND METHODS
Cells.
GSCs and ScGCs were isolated from GBM patient specimens at Massachusetts General Hospital (MGH) with approval of the Institutional Review Board (9, 10). GSCs were cultured in neural basal medium supplemented with epidermal growth factor (EGF) and fibroblast growth factor (FGF2) at a final concentration of 20 ng/ml and passaged using trypsin LE or mechanical separation. ScGCs and 293T cells (obtained from the Massachusetts Institute of Technology [MIT]/ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) (HyClone; GE Healthcare). Vero cells (ATCC) used for plaque assay were cultured in DMEM with 10% calf serum (HyClone; GE Healthcare). All cells were confirmed to be mycoplasma free (LookOut mycoplasma kit; Sigma).
Viruses.
FΔ6 was constructed from HSV-1 strain F by insertion of LacZ into the ICP6 (UL49) gene (9). G207 was constructed from R3616 (gift from B. Roizman, University of Chicago), with deletion of both copies of γ34.5 (20), by insertion of LacZ into ICP6 (17). G47Δ and R47Δ were constructed by deleting ICP47 from G207 and R3616, respectively (22). 1716 (γ34.5− strain 17) was provided by N. Fraser (University of Pennsylvania). 1716-6 (γ34.5Δ, ICP6−) was described previously (23). Patton strains HSV-1 pAUs11, Δ34.5pAUs11, pAUs11R, 11S, and 11AS were gifts of Ian Mohr (New York University) (19, 25). Briefly, pAUs11 has an insertion of a polyadenylation sequence immediately downstream of the Us11 start codon, causing termination of Us11; Δ34.5-pAUs11 was built from pAUs11 by deleting both copies of the γ34.5 gene; pAUs11R is the rescue virus; 11S has the sequence of Us11 through Us10, and 11AS has the sequence in the antisense orientation inserted into the thymidine kinase (TK) gene under an ICP27 promoter, with both copies of γ34.5 deleted (25). All viruses were purified as described previously (67).
Virus yield assay.
GSCs were dissociated mechanically in phosphate-buffered saline (PBS)-glucose (Sigma) and infected for 1.5 h. ScGCs were plated the day before infection and infected in 1% heat-inactivated fetal calf serum (IFCS)-PBS. After infection cells were washed twice with 1× PBS and incubated in appropriate medium until harvest. Infected whole-cell lysates (supernatant and cells) were frozen, thawed, and sonicated before being used to determine virus yield by plaque assay on Vero cells. Samples were incubated on Vero cells in 1% IFCS, 0.1% pooled human IgG (McKesson Plasma Biologics), and DMEM for 4 days. Wells were fixed with 0.2% glutaraldehyde–2% paraformaldehyde (PFA), stained for β-galactosidase with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) stain (0.2 mg/ml X-Gal, 1 mM ferricyanide, 1 mM ferrocyanide, 2 mM MgCl2 in 1× PBS), and counterstained with neutral red dye.
Infectivity assay.
Cells were infected with virus and fixed in 0.2% glutaraldehyde–2% PFA at 6 to 8 hpi. Infected fixed GSCs were stained with X-Gal (1 mg/ml in 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM magnesium chloride in PBS) for 2 h at 37°C, washed, and then applied to coverslips and dried. ScGCs in 24-well plates were fixed, X-Gal stained, and counterstained with neutral red. LacZ+ blue cells were counted to determine infectivity.
Viability assay.
Cells were seeded in 48-well plates, infected at an MOI of 2, and incubated at 37°C for 8 days. GSCs and ScGCs were dissociated by pipetting and trypsin-EDTA, respectively, and viable (trypan blue-excluding) cells were counted on a hemocytometer.
Western blotting.
Cells were harvested in radioimmunoprecipitation assay (RIPA) buffer (Boston Bioproducts) supplemented with complete protease inhibitor (Roche) and phoSTOP phosphatase inhibitor (Roche), sonicated, and centrifuged. Protein in Laemmli buffer (Boston Bioproducts) was separated on SDS–4 to 15% polyacrylamide gels (Bio-Rad) and transferred semidry onto polyvinylidene difluoride (PVDF) membranes (Turbo-blot; Bio-Rad). Membranes were blocked in 5% skim milk in Tris-buffered saline with Tween 20 (TBS-T; Boston BioProducts) and incubated with the following primary antibodies diluted in milk against HSV-1 proteins: glycoprotein C (3G9, 1:3,000; Abcam), ICP4 (H2033-21B, 1:1,000; US Biological), ICP8 (1:10,000; gift from D. Knipe, Harvard Medical School), glycoprotein D (1:1,000; EastCoast Bio), ICP5 (1:1,000; EastCoast Bio), Us11 (monoclonal, 1:2,000; gift from B. Roizman University of Chicago), Us11 (polyclonal, 1:5,000; gift from M. Mulvey, BeneVir), and Us11 (polyclonal, 1:2,000; gift from D. Coen, Harvard University). Additional antibodies against the following were used: pThr446 PKR, pThr451 PKR, PKR, and pSer251eIF2α (all, 1:1,000; Abcam), eIF2α (1:1,000; Cell Signaling Technologies [CST]), p-eIF4E (1:1,000 in TBST; CST), eIF4E (1:1,000; CST), GAPDH (1:10,000; ACRIS), β-actin (1:10,000; Sigma), streptavidin-horseradish peroxidase (HRP) (1:10,000; CST), vinculin (1:1,000; CST), and anti-puromycin C terminus polyclonal antibody (1:1,000; gift from P. Walter, University of California, San Francisco [UCSF]). Puromycin pulse experiments were performed by adding 10 μg/ml of puromycin to cells for 15 min before washing. Primary antibody was detected with goat anti-rabbit-HRP and rabbit anti-mouse-HRP (Bio-Rad) and developed using Clarity Western ECL substrate (Bio-Rad). Membranes were imaged on a Gel Doc XR system with software (Bio-Rad).
Western blot quantification.
Blot signals were quantified in arbitrary units using the Bio-Rad ImageLab or ImageJ software. For total protein, lanes showing HSV infection were compared to mock infection; for anti-HSV-1, comparison was made to the lanes representing the earliest infection; for MNK1, comparison was made to lanes representing mock infection or empty vector. To correct for different exposure lengths, a correction factor was determined by the change in signal intensity for unsaturated bands at both exposures.
Electron microscopy.
Cells were infected at an MOI of 2 for 16 to 24 h before being fixed in 1.25% formaldehyde, 2.5% glutaraldehyde, and 0.03% picric acid or 2% glutaraldehyde in 0.1 M sodium cacodylate buffer. Fixed cells were stained with 1% osmium tetroxide–1.5% potassium ferrocyanide and incubated in a 1% uranyl acetate solution in maleate buffer or 1% osmium tetroxide in cacodylate buffer. Samples were dehydrated in ascending 70%, 90%, and 100% ethanol solutions, followed by a final dehydration in propylene oxide; they were embedded in 1:1 Epon-propylene oxide and polymerized at 60°C. Cut (100-μm) sections were mounted onto electro-sprayed grids or Formvar-coated grids, stained with uranyl acetate and lead citrate, and imaged using a Tecnai G2 Spirit BioTWIN or JEOL 1011 transmission electron microscope with an AMT 2k charge-coupled-device (CCD) camera.
RNA/DNA isolation.
DNA was purified using a Qiagen core kit B for use in qPCR (100 ng per run). Total RNA was harvested using TRIzol, followed by DNase treatment with RQ1 (Promega), and 1 μg was used for cDNA synthesis with a high-capacity cDNA reverse transcription kit (Applied Biosystems). cDNA from about 1,000 cells was used per qPCR run. qPCR was performed using SYBR Select master mix (Applied Biosystems) or TaqMan master mix and probes (Applied Biosystems). Fold increase was calculated using the ΔΔCT (where CT is threshold cycle) method. GAPDH or 18S RNA was used as a calibrator gene, while ICP4 was used for viral quantification in qPCR. A 2-h postinfection sample was used as the baseline for measuring fold increase. qPCR was carried out on a StepOne Plus PCR machine (Applied Biosystems) using an initial 10-min denaturation at 98°C, followed by 40 cycles of 60°C for 30 s and denaturation at 98°C for 15 s, after which a melting curve was performed up to 98°C. Primers were the following: GAPDH, 5′-CCCACTCCTCCACCTTTGAC (forward [Fwd]) and 5′-TTTTTCTGAGCCAGCCACCA (reverse [Rev]); GAPDH spanning intron, 5′-GTCAGCCGCATCTTCTTT (Fwd) and 5′-CGCCCAATACGACCAAAT (Rev); 18S RNA, 5′-GTAACCCGTTGAACCCCATT (Fwd) and 5′-CCATCCAATCGGTAGTAGCG (Rev); ICP4, 5′-TGATGAAGGAGCTGCTGTTG (Fwd) and 5′-GTACGCCCTGATCACGC (Rev); glycoprotein C (gC), 5′-CCCCAACAATGTCACACAAA (Fwd) and 5′-GGTGTTGTTCTTGGGTTTGG (Rev); Us11, 5′-GGCGACCCAGATGTTTACTTA (Fwd) and 5′-ACCCGAATCTCCACATTGC (Rev); TK, 5′-AAAAACGGAAGCGGGTAGGT (Fwd) and 5′-CATGCCGGATTTTAGCCGTG (Rev).
Plasmid lentivirus construction.
Lentiviral backbone plasmid CD513B-1 (CMV promoter) was purchased from Systems Biosciences (SBI). Us11 cDNA was amplified from FΔ6 viral DNA using primers to insert BmtI and NotI sites as well as a Kozak sequence (5′-GCTAGCGCCACCATGAGCCAGACCCAACC and 5′-GCGGCCGCCTATACAGACCCGCGAGCCGTACGTG; restriction sites are underlined). CMV-Us11 lentivirus plasmid was created by digesting plasmid and Us11 PCR product with BmtI and NotI sequences and ligation using T4-ligase (NEB). HSV IE4/5 promoter from pFLS-HSVIE4/5, created by cloning the pHSV-LAC IE promoter (68) into the FLS shuttle vector (69), was excised by SpeI and AvrII digest and ligated into the CMV-Us11 plasmid at SpeI and XbaI sites to create the IE4/5-Us11 lentivirus plasmid. Hemagglutinin (HA)-MNK1 cDNAs in pCDNA3.1, provided by M. Gromeier (Duke University) (49), were excised and inserted into pCD513B-1. Plasmid sequencing was performed at the MGH Center for Computational and Integrative Biology plasmid sequencing core. Infectious lentivirus was generated from 293T cells transfected with pVSVg, psPAX2, and lentivirus plasmids using FuGene (Promega), harvested 48 h posttransfection, and filtered through a 0.45-μm-pore-size polyethersulfone filter. Cells were transduced by incubation of cells with lentivirus and selection with puromycin (0.5 μg/ml for GSCs and 1 to 10 μg/ml for all other cell lines).
Northern blotting.
Total RNA (20 μg) was denatured at 95°C for 5 min in RNA loading buffer (50% [vol/vol] glycerol, 1 mM EDTA, pH 8.0, 0.25% bromophenol blue, 0.25% xylene cyanol) and electrophoresed on a denaturing 1.0% agarose formaldehyde gel in running buffer (10 mM morpholinepropanesulfonic acid [MOPS], pH 7.0, 8 mM sodium acetate, 1 mM EDTA, and 2.2 M formaldehyde). After electrophoresis, the gel was rinsed in water and washed twice in 20× SSC (3 M NaCl, 300 mM sodium citrate; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Transfer onto a positive nylon membrane (BrightStar) was performed in 20× SSC using an upward capillary transfer method. RNA was UV cross-linked to the membrane, prehybridized in Perfect HyB hybridization buffer (Sigma) for 3 h, and hybridized with digoxigenin (DIG)-labeled probes at 68°C overnight. Probes were created using DIG incorporation and primers amplifying the spliced GAPDH gene (Fwd, 5′-TCTAGAGTCAGCCGCATCTT; Rev, 5′-AAGCTTTCATTGATGGCAACAATA) from GSC cDNA and the gC (Fwd, 5′-TCTAGAGAGGAGGTCCTGACGAA; Rev, 5′-AAGCTTAACCAGACAAACTCCACG) and Us11 (Fwd, 5′-TCTAGAATGAGCCAGACCCAACC; Rev, 5′-AAGCTTCTATACAGACCCGCGAG) genes from HSV-1 DNA. Detection of probe was carried out using an anti-DIG antibody fused to HRP (CST).
RNAScope.
RNA immunohistochemistry was performed using an HSV-1 UL44 RNAScope probe (Advanced Cell Diagnostics [ACD]) and RNAScope, version 2.5, HD Duplex Assay (ACD) as instructed. GSCs were infected at an MOI of 0.1 to 1 on laminin-coated coverslips and fixed with 4% PFA. After RNAScope was completed, slides were briefly incubated in hematoxylin counterstain and mounted with 4′,6′-diamidino-2-phenylindole (DAPI) medium (Vectashield).
Azidohomoalanine labeling.
A Click-iT protein reaction buffer kit, containing azidohomoalanine (AHA), copper sulfate, biotin label, and Click-iT buffers, was from Thermo Fisher. Infected cells were washed three times with warm 1× PBS before being incubated in methionine-free DMEM (Thermo Fisher). During incubation in methionine-free medium, AHA was added to a final concentration of 50 μM for 2 h before cells were harvested. AHA-labeled cells were lysed in RIPA buffer as described above in the paragraph “Western blotting.” Click chemistry was performed on lysates, and biotin-labeled proteins were pulled down on streptavidin-coated agarose beads (Pierce). Biotin pulldown was performed for 1 h at room temperature in the Click-iT reaction mixture. After biotin pulldown, beads were centrifuged and washed with immunoprecipitation buffer (25 mM Tris-HCl, pH 7.4, 130 mM NaCl, 2.7 mM KCl, 1% EDTA pH 8.0, 1% IGEPAL) four times before being boiled in Laemmli SDS buffer for 10 min.
Statistics.
All statistical analysis was performed using the GraphPad Prism analysis tool's unpaired two-tailed student t test and one-way analysis of variance (ANOVA) (followed by Tukey's multiple-comparison test).
ACKNOWLEDGMENTS
We thank the following: Ian Mohr for providing 11S, 11AS, pAUs11, and pAUs11Δ34.5 viruses; Matt Mulvey, Bernard Roizman, and Don Coen for providing anti-Us11 antibodies; David Knipe for providing anti-ICP8 antibody; Lior and Adina Nissim and Erez Pery for lentivirus plasmids and assistance; Joy Jarnagin and Anthony Wong for assistance with lentivirus construction; Mario Suva for assistance with RNAScope; staff of the Harvard Medical School EM Facility for technical assistance; and members of the Molecular Neurosurgery Laboratory, especially Fares Nigim, Hammad Talat, and Andranik Kahramanian for discussion and Melissa Humphrey for production of cell and virus stocks.
Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology (Massachusetts General Hospital), which is partially supported by Inflammatory Bowel Disease grant DK43351 and Boston Area Diabetes and Endocrinology Research Center (BADERC) Award DK57521, and the Harvard Medical School EM Facility. These studies were supported in part by National Institutes of Health grants F31CA192453 (to C.P.), R01NS032677 (to R.L.M.), and RO1CA160762 (to S.D.R.) and by the Thomas A. Pappas Chair in Neurosciences (to S.D.R.).
S.D.R. and R.L.M. are inventors on patents relating to oHSV that are owned by Georgetown University and Massachusetts General Hospital, for which royalties have been received.
REFERENCES
- 1.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. 2017. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19(Supple 5):v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, Singh SK. 2017. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol 28:1448–1456. doi: 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- 4.Lan X, Jorg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB. 2017. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JK, Wang J, Sa JK, Ladewig E, Lee HO, Lee IH, Kang HJ, Rosenbloom DS, Camara PG, Liu Z, van Nieuwenhuizen P, Jung SW, Choi SW, Kim J, Chen A, Kim KT, Shin S, Seo YJ, Oh JM, Shin YJ, Park CK, Kong DS, Seol HJ, Blumberg A, Lee JI, Iavarone A, Park WY, Rabadan R, Nam DH. 2017. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 49:594–599. doi: 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. 2015. Cancer stem cells in glioblastoma. Genes Dev 29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, Jeyaretna DS, Debasitis J, Pruszak J, Martuza RL, Rabkin SD. 2009. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res 69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr, Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, Martuza RL, Rabkin SD. 2012. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol 14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheinbay E, Suva ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, Louis DN, Kasif S, Chi AS, Bernstein BE. 2013. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep 3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. 2014. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters C, Rabkin SD. 2015. Designing herpes viruses as oncolytics. Mol Ther Oncolytics 2:15010. doi: 10.1038/mto.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bommareddy PK, Peters C, Saha D, Rabkin SD, Kaufman HL. 2018. Oncolytic herpes simplex viruses as a paradigm for the treatment of cancer. Annu Rev Cancer Biol 2:155–173. doi: 10.1146/annurev-cancerbio-030617-050254. [DOI] [Google Scholar]
- 15.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Rancaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott, Williams &Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Markert JM, Medlock MD, Rabkin SD, Gillespie GY, Todo T, Hunter WD, Palmer CA, Feigenbaum F, Tornatore C, Tufaro F, Martuza RL. 2000. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther 7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 17.Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL. 1995. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med 1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 18.He B, Gross M, Roizman B. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A 94:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77:10917–10928. doi: 10.1128/JVI.77.20.10917-10928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 34.5, a gene nonessential for growth in culture. Science 250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 21.McKie EA, MacLean AR, Lewis AD, Cruickshank G, Rampling R, Barnett SC, Kennedy PG, Brown SM. 1996. Selective in vitro replication of herpes simplex virus type 1 (HSV-1) ICP34.5 null mutants in primary human CNS tumours—evaluation of a potentially effective clinical therapy. Br J Cancer 74:745–752. doi: 10.1038/bjc.1996.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todo T, Martuza RL, Rabkin SD, Johnson PA. 2001. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc Natl Acad Sci U S A 98:6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai R, Zaupa C, Sgubin D, Antoszczyk SJ, Martuza RL, Wakimoto H, Rabkin SD. 2012. Effect of γ34.5 deletions on oncolytic herpes simplex virus activity in brain tumors. J Virol 86:4420–4431. doi: 10.1128/JVI.00017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr I, Gluzman Y. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J 15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey M, Poppers J, Ladd A, Mohr I. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J Virol 73:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassady KA, Gross M, Roizman B. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J Virol 72:8620–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreansky S, Soroceanu L, Flotte ER, Chou J, Markert JM, Gillespie GY, Roizman B, Whitley RJ. 1997. Evaluation of genetically engineered herpes simplex viruses as oncolytic agents for human malignant brain tumors. Cancer Res 57:1502–1509. [PubMed] [Google Scholar]
- 28.Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol 67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 30.Chou J, Roizman B. 1992. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci U S A 89:3266–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A 103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavromara-Nazos P, Roizman B. 1987. Activation of herpes simplex virus 1 γ2 genes by viral DNA replication. Virology 161:593–598. doi: 10.1016/0042-6822(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 33.Schang LM, Phillips J, Schaffer PA. 1998. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J Virol 72:5626–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delwar ZM, Kuo Y, Wen YH, Rennie PS, Jia W. 2018. Oncolytic virotherapy blockade by microglia and macrophages requires STAT1/3. Cancer Res 78:718–730. doi: 10.1158/0008-5472.CAN-17-0599. [DOI] [PubMed] [Google Scholar]
- 35.Jackson JD, Markert JM, Li L, Carroll SL, Cassady KA. 2016. STAT1 and NF-κB inhibitors diminish basal interferon-stimulated gene expression and improve the productive infection of oncolytic HSV in MPNST Cells. Mol Cancer Res 14:482–492. doi: 10.1158/1541-7786.MCR-15-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allagui F, Achard C, Panterne C, Combredet C, Labarriere N, Dreno B, Elgaaied AB, Pouliquen D, Tangy F, Fonteneau JF, Gregoire M, Boisgerault N. 2017. Modulation of the type I interferon response defines the sensitivity of human melanoma cells to oncolytic measles virus. Curr Gene Ther 16:419–428. doi: 10.2174/1566523217666170102110502. [DOI] [PubMed] [Google Scholar]
- 37.Jha BK, Dong B, Nguyen CT, Polyakova I, Silverman RH. 2013. Suppression of antiviral innate immunity by sunitinib enhances oncolytic virotherapy. Mol Ther 21:1749–1757. doi: 10.1038/mt.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawson KA, Mostafa AA, Shi ZQ, Spurrell J, Chen W, Kawakami J, Gratton K, Thakur S, Morris DG. 2016. Repurposing sunitinib with oncolytic reovirus as a novel immunotherapeutic strategy for renal cell carcinoma. Clin Cancer Res 22:5839–5850. doi: 10.1158/1078-0432.CCR-16-0143. [DOI] [PubMed] [Google Scholar]
- 39.Chu J, Cargnello M, Topisirovic I, Pelletier J. 2016. Translation initiation factors: reprogramming protein synthesis in cancer. Trends Cell Biol 26:918–933. doi: 10.1016/j.tcb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20:461–472. doi: 10.1101/gad.1375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proud CG. 2015. Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta 1849:766–773. doi: 10.1016/j.bbagrm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, Pandolfi PP, Saad F, Sonenberg N. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A 107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, Larsson O, Muller WJ, Miller WH, Sonenberg N. 2015. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korneeva NL, Song A, Gram H, Edens MA, Rhoads RE. 2016. Inhibition of mitogen-activated protein kinase (MAPK)-interacting kinase (MNK) preferentially affects translation of mRNAs containing both a 5′-terminal cap and hairpin. J Biol Chem 291:3455–3467. doi: 10.1074/jbc.M115.694190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geter PA, Ernlund AW, Bakogianni S, Alard A, Arju R, Giashuddin S, Gadi A, Bromberg J, Schneider RJ. 2017. Hyperactive mTOR and MNK1 phosphorylation of eIF4E confer tamoxifen resistance and estrogen independence through selective mRNA translation reprogramming. Genes Dev 31:2235–2249. doi: 10.1101/gad.305631.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bramham CR, Jensen KB, Proud CG. 2016. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E Axis. Trends Biochem Sci 41:847–858. doi: 10.1016/j.tibs.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18:660–672. doi: 10.1101/gad.1185304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, Furic L, Dowling RJO, Sylvestre A, Rong L, Colina R, Costa-Mattioli M, Fritz JH, Olivier M, Brown E, Mohr I, Sonenberg N. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13:543–550. doi: 10.1038/ni.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V, Bigner DD, Gromeier M. 2014. Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol 88:13135–13148. doi: 10.1128/JVI.01883-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh D, Mathews MB, Mohr I. 2013. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5:a012351. doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dauber B, Pelletier J, Smiley JR. 2011. The herpes simplex virus 1 Vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J Virol 85:5363–5373. doi: 10.1128/JVI.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dauber B, Saffran HA, Smiley JR. 2014. The herpes simplex virus 1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload. J Virol 88:9624–9632. doi: 10.1128/JVI.01350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruggero D. 2013. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 5:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith RWP, Anderson RC, Larralde O, Smith JWS, Gorgoni B, Richardson WA, Malik P, Graham SV, Gray NK. 2017. Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding. Proc Natl Acad Sci U S A 114:6310–6315. doi: 10.1073/pnas.1610417114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryant KF, Macari ER, Malik N, Boyce M, Yuan J, Coen DM. 2008. ICP34.5-dependent and -independent activities of salubrinal in herpes simplex virus-1 infected cells. Virology 379:197–204. doi: 10.1016/j.virol.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulvey M, Camarena V, Mohr I. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and γ134.5 gene products. J Virol 78:10193–10196. doi: 10.1128/JVI.78.18.10193-10196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. 1997. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J Virol 71:6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harkness JM, Kader M, DeLuca NA. 2014. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J Virol 88:6847–6861. doi: 10.1128/JVI.00516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Streby KA, Geller JI, Currier MA, Warren PS, Racadio JM, Towbin AJ, Vaughan MR, Triplet M, Ott-Napier K, Dishman DJ, Backus LR, Stockman B, Brunner M, Simpson K, Spavin R, Conner J, Cripe TP. 2017. Intratumoral injection of HSV1716, an oncolytic herpes virus, is safe and shows evidence of immune response and viral replication in young cancer patients. Clin Cancer Res 23:3566–3574. doi: 10.1158/1078-0432.CCR-16-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cassady KA, Bauer DF, Roth J, Chambers MR, Shoeb T, Coleman J, Prichard M, Gillespie GY, Markert JM. 2017. Pre-clinical assessment of C134, a chimeric oncolytic herpes simplex virus, in mice and non-human primates. Mol Ther Oncolytics 5:1–10. doi: 10.1016/j.omto.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel DM, Foreman PM, Nabors LB, Riley KO, Gillespie GY, Markert JM. 2016. Design of a phase I clinical trial to evaluate M032, a genetically engineered HSV-1 expressing IL-12, in patients with recurrent/progressive glioblastoma multiforme, anaplastic astrocytoma, or gliosarcoma. Hum Gene Ther Clin Dev 27:69–78. doi: 10.1089/humc.2016.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kambara H, Okano H, Chiocca EA, Saeki Y. 2005. An oncolytic HSV-1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res 65:2832–2839. doi: 10.1158/0008-5472.CAN-04-3227. [DOI] [PubMed] [Google Scholar]
- 65.Liu BL, Robinson M, Han ZQ, Branston RH, English C, Reay P, McGrath Y, Thomas SK, Thornton M, Bullock P, Love CA, Coffin RS. 2003. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther 10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 66.Hughes T, Coffin RS, Lilley CE, Ponce R, Kaufman HL. 2014. Critical analysis of an oncolytic herpesvirus encoding granulocyte-macrophage colony stimulating factor for the treatment of malignant melanoma. Oncolytic Virother 3:11–20. doi: 10.2147/OV.S36701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ning J, Wakimoto H, Peters C, Martuza RL, Rabkin SD. 2017. Rad51 degradation: role in oncolytic virus-poly(ADP-ribose) polymerase inhibitor combination therapy in glioblastoma. J Natl Cancer Inst 109:1–13. doi: 10.1093/jnci/djw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geller AI, Breakefield XO. 1988. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science 241:1667–1669. doi: 10.1126/science.2843986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuroda T, Martuza RL, Todo T, Rabkin SD. 2006. Flip-Flop HSV-BAC: bacterial artificial chromosome based system for rapid generation of recombinant herpes simplex virus vectors using two independent site-specific recombinases. BMC Biotechnol 6:40. doi: 10.1186/1472-6750-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]