ABSTRACT
Porcine deltacoronavirus (PDCoV) has recently emerged as an enteric pathogen that can cause serious vomiting and diarrhea in suckling piglets. The first outbreak of PDCoV occurred in the United States in 2014 and was followed by reports of PDCoV in South Korea, China, Thailand, Lao People's Democratic Republic, and Vietnam, leading to economic losses for pig farms and posing a considerable threat to the swine industry worldwide. Our previous studies have shown that PDCoV encodes three accessory proteins, NS6, NS7, and NS7a, but the functions of these proteins in viral replication, pathogenesis, and immune regulation remain unclear. Here, we found that ectopic expression of accessory protein NS6 significantly inhibits Sendai virus-induced interferon beta (IFN-β) production as well as the activation of transcription factors IRF3 and NF-κB. Interestingly, NS6 does not impede the IFN-β promoter activation mediated via key molecules in the RIG-I-like receptor (RLR) signaling pathway, specifically RIG-I, MDA5, and their downstream molecules MAVS, TBK1, IKKε, and IRF3. Further analyses revealed that NS6 is not an RNA-binding protein; however, it interacts with RIG-I/MDA5. This interaction attenuates the binding of double-stranded RNA by RIG-I/MDA5, resulting in the reduction of RLR-mediated IFN-β production. Taken together, our results demonstrate that ectopic expression of NS6 antagonizes IFN-β production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA, revealing a new strategy employed by PDCoV accessory proteins to counteract the host innate antiviral immune response.
IMPORTANCE Coronavirus accessory proteins are species specific, and they perform multiple functions in viral pathogenicity and immunity, such as acting as IFN antagonists and cell death inducers. Our previous studies have shown that PDCoV encodes three accessory proteins. Here, we demonstrated for the first time that PDCoV accessory protein NS6 antagonizes IFN-β production by interacting with RIG-I and MDA5 to impede their association with double-stranded RNA. This is an efficient strategy of antagonizing type I IFN production by disrupting the binding of host pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs). These findings deepen our understanding of the function of accessory protein NS6, and they may direct us toward novel therapeutic targets and lead to the development of more effective vaccines against PDCoV infection.
KEYWORDS: porcine deltacoronavirus, accessory protein, immune evasion, interferon
INTRODUCTION
Porcine deltacoronavirus (PDCoV) is a swine enteropathogenic coronavirus that can lead to acute diarrhea and vomiting in infected nursing piglets (1–3). PDCoV was first detected in Hong Kong in 2012 (4). However, the first outbreak of PDCoV occurred in Ohio in 2014, after which it rapidly spread to other states of the United States (5–9). Subsequently, other countries, including South Korea (10), China (11–13), Thailand (14), Lao People's Democratic Republic (15), and Vietnam (16), have reported the prevalence of PDCoV. Furthermore, a recent report indicated that calves are also susceptible to PDCoV, highlighting the significant threat to animal health posed by this virus and gaining tremendous attention (17, 18).
PDCoV is an enveloped, single-stranded, positive-sense RNA virus belonging to the genus Deltacoronavirus of the family Coronaviridae (4). The full-length genome of PDCoV is approximately 25.4 kb in length, with the essential genes occurring in the order 5′ UTR-ORF1a/1b-S-E-M-NS6-N-NS7-NS7a-3′ UTR and encoding a total of 15 mature nonstructural proteins, four structural proteins, and three accessory proteins (13, 19–21). Coronavirus accessory proteins are species specific, and each coronavirus encodes various amounts of accessory proteins interspaced between viral structural protein genes. For example, feline infectious peritonitis virus (FIPV), which is an alphacoronavirus, and infectious bronchitis virus (IBV), which is a gammacoronavirus, each have four accessory proteins, while another alphacoronavirus, porcine epidemic diarrhea virus (PEDV), has only one accessory protein and the betacoronavirus severe acute respiratory syndrome coronavirus (SARS-CoV) has eight (22). Although coronavirus accessory proteins have been widely considered to be dispensable for viral replication in vitro (23–25), extensive reports have indicated that many accessory proteins are involved in immune regulation, such as SARS-CoV ORF3b, ORF6, and ORF9b (26–28), Middle East respiratory syndrome coronavirus (MERS-CoV) ORF4a and ORF4b (29–31), and mouse hepatitis virus (MHV) ns2 (32, 33). To our knowledge, there is no report on the functions of PDCoV accessory proteins.
In virus-infected cells, certain viral RNA replication intermediates, leader RNAs, or defective interfering RNAs with 5′ triphosphates are generated, and these substances act as pathogen-associated molecular patterns (PAMPs) that are recognized by host pattern recognition receptors (PRRs), such as retinoic acid-induced gene I (RIG-I) and melanoma differentiation gene 5 (MDA5), in the cytoplasm (34–36). Upon PAMP recognition, RIG-I and MDA5 are activated, resulting in the recruitment of mitochondrial antiviral signaling protein (MAVS) (also known as IPS-1, VIAS, or Cardif) to the RIG-I-like receptor (RLR) signalosome; this leads to interferon beta (IFN-β) production via activation of the complex formed by transcription factor IRF3 and NF-κB activator TBK1/IKKε, followed by the subsequent activation of IRF3 and NF-κB (37, 38). However, many viruses, including CoVs, have evolved various mechanisms to antagonize IFN via targeting multiple steps in the IFN signaling pathway (39–44). Previous studies have demonstrated that PDCoV infection suppresses the RIG-I-mediated production of type I IFN (45). However, the details of the molecular mechanism by which PDCoV regulates IFN activity are still largely unknown. Accessory protein NS6 is encoded between the M and N genes in the PDCoV genome; it is expressed in virus-infected cytoplasm and has been detected in purified virions (19). Interestingly, SARS-CoV accessory proteins ORF6 and ORF9b have also been identified as virion-associated proteins as well as IFN antagonists (46–48). Therefore, we aimed to investigate whether or not PDCoV NS6 participates in the regulation of the RLR-mediated IFN signaling pathway.
In this study, our findings clearly reveal that overexpression of PDCoV NS6 inhibits IFN-β production via interacting with RIG-I and MDA5 to disturb their association with PAMP double-stranded RNA (dsRNA), a known initial step of the IFN signaling pathway.
RESULTS
PDCoV NS6 inhibits SeV-induced IFN-β production.
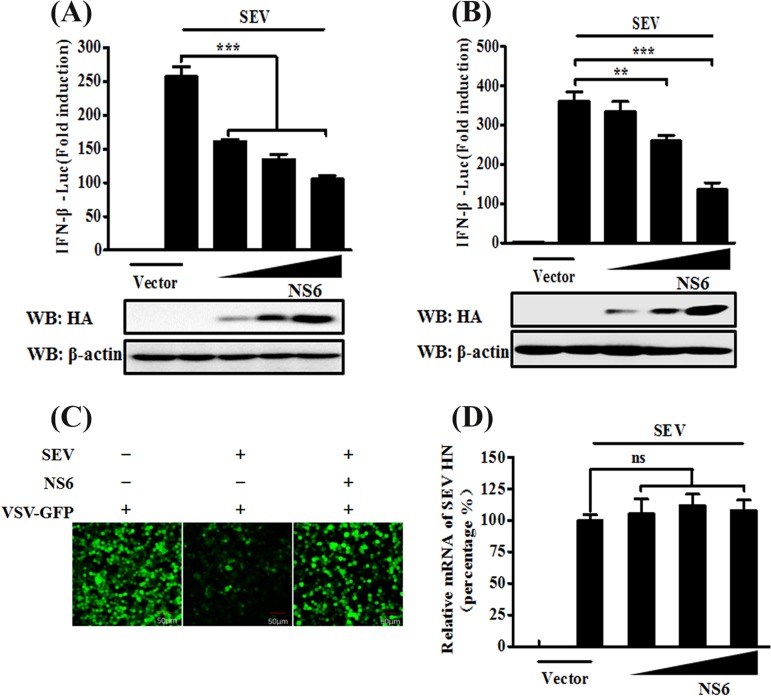
To investigate whether or not PDCoV NS6 is an IFN antagonist, human embryonic kidney (HEK-293T) cells or porcine kidney (LLC-PK1) cells were cotransfected for 24 h with increasing amounts of NS6 expression plasmid (pCAGGS-HA-NS6) or empty vector, together with the firefly luciferase reporter plasmid IFN-β-Luc and Renilla luciferase reporter plasmid pRL-TK (as an internal control), and then infected with Sendai virus (SeV) for 12 h. The cells were lysed, and the resultant lysates were subjected to Dual-Luciferase reporter assays. The results showed that the SeV-induced IFN-β-Luc promoter activation was significantly inhibited by NS6 overexpression in both cell lines (Fig. 1A and B). To further confirm the results from these IFN-β-Luc reporter assays, we performed IFN bioassays by using an IFN-sensitive vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP). The level of VSV-GFP replication is inversely linked to the levels of secreted IFN-α/β from the transfected HEK-293T cells. As seen in Fig. 1C, cellular supernatants from SeV-infected cells significantly inhibited the replication of VSV-GFP in HEK-293T cells. However, the natural replication of VSV-GFP was, to a large extent, restored by the presence of supernatants from cells expressing NS6 compared with that of supernatants from empty vector-transfected cells. To rule out the possibility that the NS6 protein itself affects the replication of SeV, relatively quantitative real-time reverse transcription-PCR (RT-PCR) was performed to detect SeV HN gene expression in pCAGGS-HA–NS6-transfected HEK-293T cells. As shown in Fig. 1D, there was no significant difference in the amount of SeV HN mRNA in pCAGGS-HA-NS6-transfected cells compared with that in empty vector-transfected cells, indicating that the observed NS6-mediated inhibition of IFN expression was not due to a general restriction of SeV replication. These results strongly indicate that PDCoV NS6 antagonizes IFN-β production.
FIG 1.
NS6 inhibits SeV-mediated IFN-β production. (A and B) HEK-293T cells (A) or LLC-PK1 cells (B) cultured in 24-well plates were transfected with IFN-β-Luc plasmid and pRL-TK plasmid, together with increasing amounts (0.2, 0.4, or 0.8 μg) of plasmid pCAGGS-HA-NS6. At 24 h after transfection, cells were left untreated or were infected with SeV (10 hemagglutination units/well). The cells were then subjected to Dual-Luciferase assays at 12 h postinfection. The expression of PDCoV NS6 protein was confirmed by Western blotting with an anti-HA antibody. β-Actin served as a protein loading control. (C) HEK-293T cells were transfected with the indicated amounts of pCAGGS-HA-NS6 or empty vector. At 24 h after transfection, the cells were infected with SeV for 12 h, after which cell supernatants were harvested. Following UV irradiation, the harvested cell supernatants were overlaid onto fresh HEK-293T cells in 24-well plates. At 24 h after treatment, the cells were infected with VSV-GFP, and 12 h postinfection, virus replication was detected via fluorescence microscopy. (D) HEK-293T cells grown in 24-well plates were transfected with increasing quantities of pCAGGS-HA-NS6 or corresponding amounts of empty vector. At 24 h after transfection, cells were infected with SeV for 12 h. The total RNA was then extracted, and the SeV HN gene expression level was analyzed via quantitative real-time RT-PCR with normalization to the GAPDH gene expression level. The results shown are representative of data from three independent experiments: **, P < 0.01; ***, P < 0.001; ns, nonsignificant differences in data.
PDCoV NS6 impairs activation of IRF3 and NF-κB.
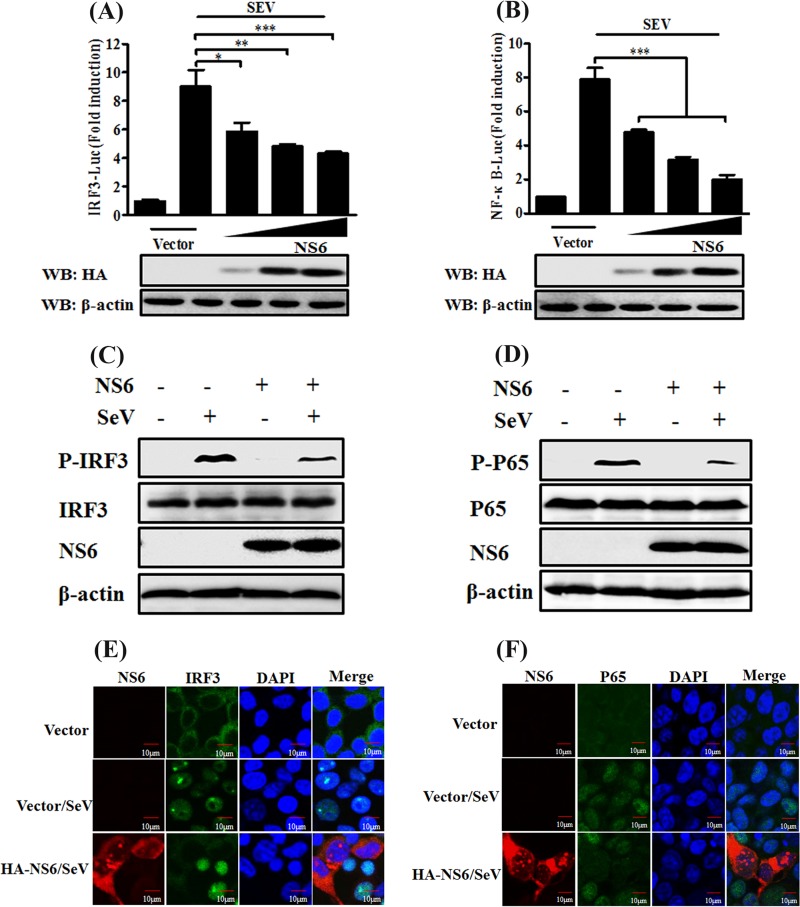
The transcription factors IRF3 and NF-κB are required for the induction of IFN-β production. Since our above-described results indicate that PDCoV NS6 antagonizes IFN-β production, we next explored the effect of NS6 on the activation of IRF3 and NF-κB. To this end, HEK-293T cells were transfected with pCAGGS-HA-NS6 and the luciferase reporter plasmid IRF3-Luc or NF-κB-Luc (each contains four copies of the IRF- or NF-κB-binding motif of the IFN-β promoter upstream of the firefly luciferase reporter gene), along with the internal control plasmid pRL-TK, followed 24 h later by stimulation with SeV for 12 h. As seen in Fig. 2, the SeV-induced activation of both IRF3-dependent (Fig. 2A) and NF-κB-dependent (Fig. 2B) promoters was dose dependently impaired by overexpressing NS6.
FIG 2.
NS6 inhibits activation of IRF3 and NF-κB. (A and B) HEK-293T cells were transfected with the indicated amounts of pCAGGS-HA-NS6 or empty vector, together with IRF3-Luc (A) or NF-κB-Luc (B) and pRL-TK plasmids, followed by stimulation with SeV, and they were analyzed as described in the legend to Fig. 1A. Anti-HA antibody was used to detect the expression of PDCoV NS6, and anti-β-actin antibody was used to detect β-actin protein by Western blotting (WB), which served as a protein loading control. (C and D) HEK-293T cells were transfected with pCAGGS-HA-NS6 or empty vector. After 24 h, the cells were infected with SeV or left untreated for 8 h. Cell lysates were collected for Western blot analysis with primary antibodies against phosphorylated IRF3 (p-IRF3 Ser386) and total IRF3 (C) or phosphorylated p65 (p-p65 Ser536) and total p65 (D), HA, and β-actin. (E and F) HEK-293T cells were transfected with pCAGGS-HA-NS6 or empty vector, followed by mock infection or SeV infection for 8 h as described for panels C and D. The cells were then fixed and subjected to an immunofluorescence assay with rabbit anti-IRF3 and anti-p65 and mouse anti-HA antibodies as primary antibodies, followed by staining with secondary antibodies Alexa Fluor 488-conjugated donkey anti-rabbit IgG (green) or Alexa Fluor 594-conjugated donkey anti-mouse IgG (red). DAPI staining (blue) indicates the locations of the cell nuclei. Fluorescent images were acquired with a confocal laser scanning microscope (FluoView ver. 3.1; Olympus, Japan). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
IRF3 and NF-κB are regarded as critical regulatory factors in the initiation of the innate antiviral response. They are activated via phosphorylation and nuclear translocation upon viral infection, followed by the assembly of coordinately activated transcription factors and the induction of transcription of specific defense genes, such as IFN-β (49, 50). Therefore, we further investigated the impact of NS6 protein on the phosphorylation and nuclear translocation of IRF3 and NF-κB by performing Western blotting and indirect immunofluorescent assays (IFAs). As shown in Fig. 2, the levels of phosphorylated IRF3 and p65 were markedly enhanced in SeV-infected cells compared with those in mock-infected cells. However, the SeV-mediated IRF3 and p65 phosphorylation levels were notably lower in NS6-expressing cells (Fig. 2C and D). In agreement with the Western blot results, the nuclear translocations of IRF3 and p65 were also impeded by NS6 protein (Fig. 2E and F). These results strongly support the idea that PDCoV NS6 acts as an IFN antagonistic protein by blocking the activation of IRF3 and p65.
PDCoV NS6 fails to disrupt IFN-β promoter activation driven by RIG-I, MDA5, MAVS, TBK1, IKKε, or IRF3.
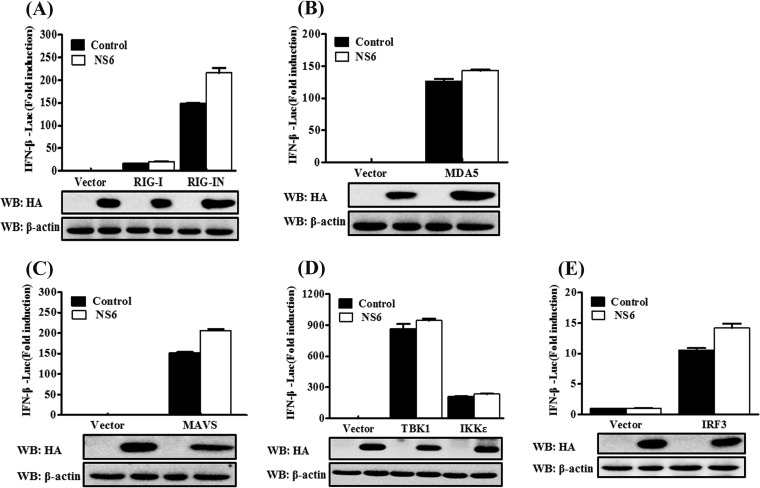
SeV is a strong inducer of the RLR-mediated IFN-β signaling pathway (51). The finding that NS6 protein inhibits the SeV-mediated activation of IRF3 and p65 indicates that NS6 protein blocks the RLR-mediated type I IFN signaling pathway. To investigate this possibility and to determine at which step the NS6 protein displays its activity, we measured the effect of NS6 on the IFN-β production induced by a series of key signaling molecules in the RLR signaling pathway, specifically RIG-I, RIG-IN (a constitutively activated RIG-I mutant), MDA5, MAVS, TBK1, IKKε, and IRF3. Based on a comparison with the corresponding empty vector-transfected cells, NS6 failed to block the activation of the IFN-β promoter in cells overexpressing any of the above-described signaling molecules (Fig. 3). These results suggest that the inhibition of IFN-β production by NS6 occurs via targeting the RLR signaling pathway at the level of RIG-I/MDA5 or the upstream signaling components.
FIG 3.
NS6 fails to inhibit IFN-β production induced by RLR signaling pathway molecules. HEK-293T cells were transfected with IFN-β–Luc, pRL-TK, and pCAGGS-HA-NS6 along with constructs expressing RIG-I/RIG-IN (A), MDA5 (B), MAVS (C), TBK1 and IKKε (D), or IRF3 (E). Dual-Luciferase assays were performed 28 h after transfection. The relative firefly luciferase activity was relative to that of an untreated empty vector control, with normalization to the Renilla reniformis luciferase activity. The presented results represent the means and standard deviations of data from three independent experiments. The expression of NS6 protein was verified by Western blotting with anti-HA antibody. β-Actin served as a protein loading control.
NS6 protein blocks the IFN-β promoter activation induced by the combination of RIG-I/MDA5 and SeV/poly(I·C).
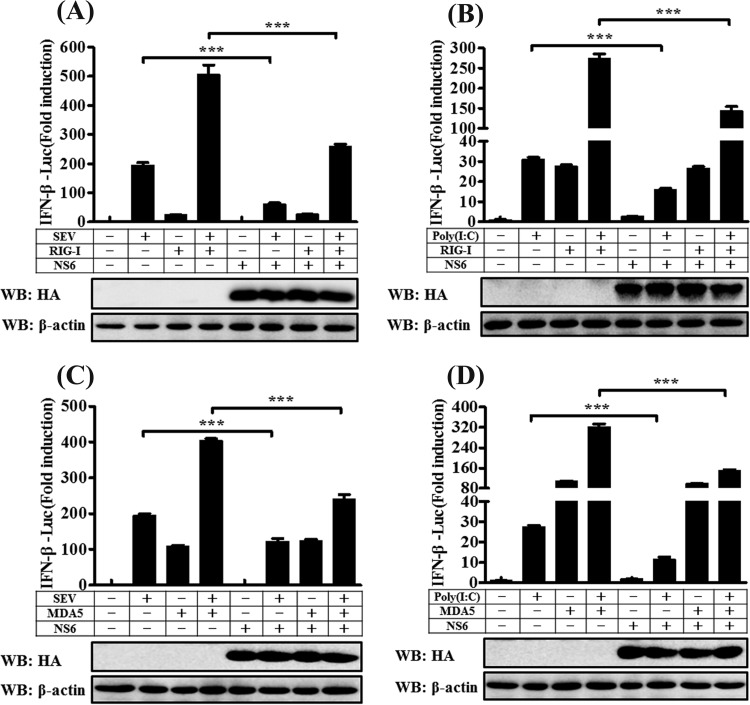
Although NS6 does not inhibit RIG-I/MDA5-mediated IFN-β promoter activation (Fig. 3A and B), the ectopic expression of NS6 significantly inhibits SeV-mediated IFN-β production (Fig. 1A and B). To further investigate the role of NS6, we next examined the effect of NS6 on SeV-induced IFN-β promoter activation in RIG-I- or MDA5-expressing cells. HEK-293T cells were transfected with an expression construct encoding full-length RIG-I or MDA5 or with an empty vector, along with pCAGGS-HA-NS6 or its corresponding empty vector. After 24 h, these cells were stimulated with SeV or poly(I·C) (a synthetic mimic of dsRNA) for 12 h, after which Dual-Luciferase reporter assays were performed. As shown in Fig. 4, SeV/poly(I·C) stimulation notably induced the activation of the IFN-β promoter, but the increased activation was significantly lower in the presence of NS6 protein. Overexpression of either RIG-I or MDA5 resulted in a significant activation of IFN-β promoter, and this activation did not appear to be inhibited by NS6 protein. These results are consistent with those shown in Fig. 1A and 3A and B. RIG-I/MDA5-mediated activation of the IFN-β promoter increased dramatically following stimulation with SeV or poly(I·C). However, the synergistic activation of IFN-β promoter induced by RIG-I (Fig. 4A and B) or MDA5 (Fig. 4C and D) coupled with SeV/poly(I·C) was significantly inhibited by NS6 protein. Based on these findings, we speculate that the inhibition of IFN-β production by NS6 occurs at the RIG-I/MDA5-dsRNA recognition step.
FIG 4.
NS6 disrupts the IFN-β promoter activation induced by RIG-I or MDA5 coupled with SeV or poly(I·C). HEK-293T cells were transfected with IFN-β–Luc, pRL-TK, and the other indicated expression plasmids. At 24 h after transfection, cells were stimulated with SeV/poly(I·C) or were left untreated for 12 h. Dual-Luciferase assays were then performed as described in the legend to Fig. 3. The presented results represent the means and standard deviations of data from three independent experiments. ***, P < 0.001. Western blot analysis with anti-HA antibody shows the expression of NS6 protein, and Western blotting for β-actin served as a protein loading control.
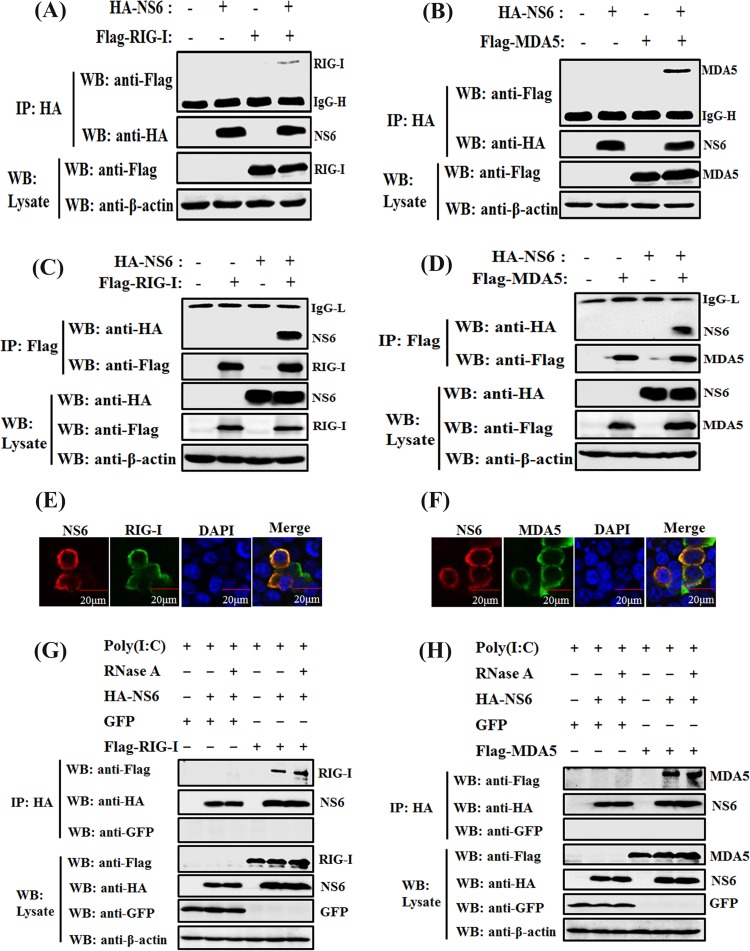
NS6 protein interacts with both RIG-I and MDA5.
To further investigate the hypothesis that NS6 targets the initial RIG-I/MDA5-dsRNA recognition step, we tested if NS6 is able to interact with RIG-I or MDA5, leading to the blockage of their functions. HEK-293T cells were cotransfected with expression plasmids encoding hemagglutinin (HA)-tagged NS6 protein and Flag-tagged RIG-I or MDA5, followed by coimmunoprecipitation (Co-IP) and Western blot analyses with anti-HA and anti-Flag monoclonal antibodies (MAbs), respectively. Both RIG-I and MDA5 were efficiently coimmunoprecipitated with HA-NS6 by anti-HA MAb (Fig. 5A and B). In a reverse Co-IP experiment, NS6 proteins were also efficiently coimmunoprecipitated with RIG-I or MDA5 by anti-Flag MAb (Fig. 5C and D). Furthermore, IFAs also demonstrated that HA-NS6 and Flag-RIG-I or MDA5 were colocalized and were both distributed predominately in the cytoplasm (Fig. 5E and F).
FIG 5.
NS6 interacts with both RIG-I and MDA5. (A to D) HEK-293T cells were cotransfected with pCAGGS-HA-NS6 and Flag-tagged RIG-I (A and C) or Flag-tagged MDA5 (B and D), respectively. At 28 h after transfection, cells were lysed and subjected to immunoprecipitation analysis with anti-HA (IP: HA) or anti-Flag (IP: Flag) antibody. The whole-cell lysates (WCL) and immunoprecipitation (IP) complexes were analyzed via Western blotting using anti-Flag, anti-HA, or anti-β-actin antibodies. (E and F) HEK-293T cells were cotransfected with pCAGGS-HA-NS6 and Flag-tagged RIG-I (E) or MDA5 (F). At 28 h after transfection, the cells were fixed for immunofluorescence assays to detect NS6 protein (red) and RIG-I or MDA5 (green) with anti-HA and anti-Flag antibodies, respectively. (G and H) HEK-293T cells were transfected with pCAGGS-HA-NS6 and pEGFP-C1, Flag-tagged RIG-I (G), or MDA5 (H) expression plasmids for 24 h, followed by the transfection of poly(I·C). Cells were lysed 36 h after transfection, and the clarified supernatants were left untreated or were treated with RNase A (50 μg/ml). The samples were then subjected to immunoprecipitation assays using anti-HA MAb (IP: HA). Cell lysates and immunoprecipitated complexes were subjected to Western blot assays as described for panels A and B.
Previous studies have identified RIG-I and MDA5 as dsRNA-binding proteins (29, 52). Based on this feature, we hypothesized that the interaction between NS6 and RIG-I or MDA5 is mediated by RNA with a tertiary complex form. To test this possibility, HEK-293T cells were cotransfected with NS6 and RIG-I or MDA5 expression plasmids for 24 h, followed by transfection with poly(I·C). To exclude the nonspecific binding of NS6 with RIG-I or MDA5, cells cotransfected with plasmid encoding green fluorescent protein (GFP) were used as a control. The lysates from transfected cells were treated with RNase A (50 μg/ml; TaKaRa) and then subjected to immunoprecipitation with anti-HA. As shown in Fig. 5G and H, both RIG-I and MDA5, but not GFP, could be coimmunoprecipitated with NS6 protein under RNase A treatment, and the Co-IP efficiency did not change with the addition of RNase A. These results indicate that the specific interaction between RIG-I or MDA5 and NS6 is RNA independent.
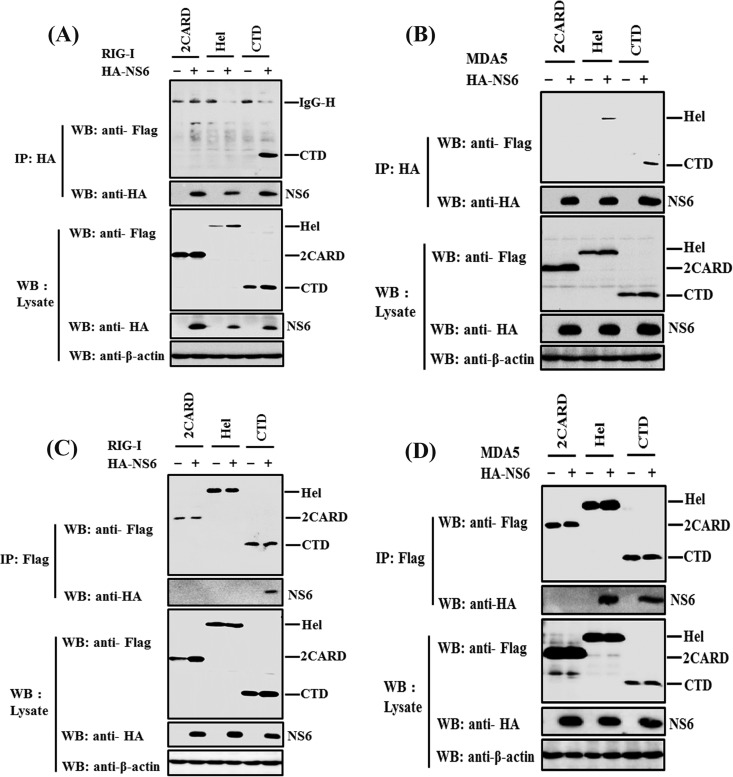
NS6 interacts with the carboxyl terminus domain of RIG-I and the helicase and carboxyl terminus domains of MDA5.
Both RIG-I and MDA5 are RIG-I-like receptors, and they harbor similar functional domains for the activation of type I IFN, including two N-terminal caspase-recruitment domains (CARDs), a central DExD/H-box helicase domain (Hel), and a C-terminal domain (CTD) (53). To explore which domain of RIG-I/MDA5 binds to NS6, various expression plasmids encoding the 2CARD, Hel, or CTD of RIG-I/MDA5 were constructed. HEK-293T cells were cotransfected with HA-NS6 and a Flag-tagged 2CARD, Hel, or CTD expression plasmid of RIG-I/MDA5. At 28 h posttransfection, the cells were harvested and subjected to Co-IP analyses with anti-HA or anti-Flag MAb. When the immunoprecipitation was performed with anti-HA MAb, NS6 coimmunoprecipitated with the CTD of RIG-I, or the Hel and CTD of MDA5, but not with other mutants of RIG-I/MDA5 (Fig. 6A and B). In the reverse Co-IP experiments with anti-Flag MAb, both RIG-I CTD and MDA5 Hel and CTD were able to coimmunoprecipitate with NS6 (Fig. 6C and D). Together, these results indicate that NS6 specifically interacts with the CTD of RIG-I and the Hel and CTD of MDA5.
FIG 6.
NS6 interacts with the carboxyl terminus domain of RIG-I or the helicase and carboxyl terminus domains of MDA5. HEK293T cells were cotransfected with pCAGGS-HA-NS6 and the expression plasmids encoding the 2CARD, Hel, or CTD of RIG-I/MDA5. Immunoprecipitation assays with anti-HA (A and B) or anti-Flag (C and D) antibody (IP: HA or IP: Flag, respectively) and Western blot analysis were performed as described for Fig. 5A and C.
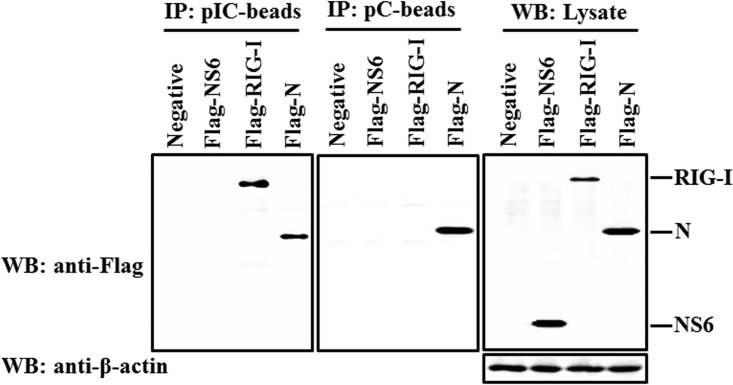
NS6 is not a dsRNA-binding protein.
To investigate whether or not NS6 protein is able to bind RNA molecules, we performed a pulldown experiment with poly(I·C)-coated agarose beads or poly(C)-coated agarose beads (Sigma) (as a negative control). This method has been extensively used to identify viral RNA-binding proteins, such as MERS-CoV 4a and Ebola VP35 protein (29, 54). RIG-I served as a positive control because it has been proven to interact directly with poly(I·C) (52). Previous work demonstrated that the binding of N protein to RNA is a widespread feature for coronaviruses (55), so we investigated whether or not PDCoV N protein has a similar characteristic. As shown in Fig. 7, RIG-I could be detected bound to poly(I·C)-coated agarose beads but not bound to poly(C)-coated agarose beads, further confirming that RIG-I binds dsRNA; PDCoV N protein was found bound for both poly(I·C)-coated agarose beads and poly(C)-coated agarose beads, indicating that PDCoV N protein can bind both double- and single-stranded RNA. However, NS6 protein was not detected bound to either poly(I·C)-coated agarose beads or poly(C)-coated agarose beads, verifying that NS6 is not an RNA-binding protein.
FIG 7.
NS6 is not an RNA binding protein. HEK-293T cells were transfected with plasmids encoding Flag-tagged RIG-I protein, PDCoV NS6 and N protein, or empty vector, respectively. Cells were lysed 28 h after transfection, and the resulting supernatants were incubated with poly(C)- or poly(I·C)-coated agarose beads for 4 h at 4°C. The beads were then washed three times with lysis buffer by centrifugation, followed by Western blotting with mouse anti-Flag antibody. The poly(I·C)-coated agarose beads (pIC-beads) were prepared from poly(C)-coated beads (pC-beads; Sigma) by incubating them with an equal volume of 2 mg of poly(I) (Sigma) per ml for 1 h at 56°C.
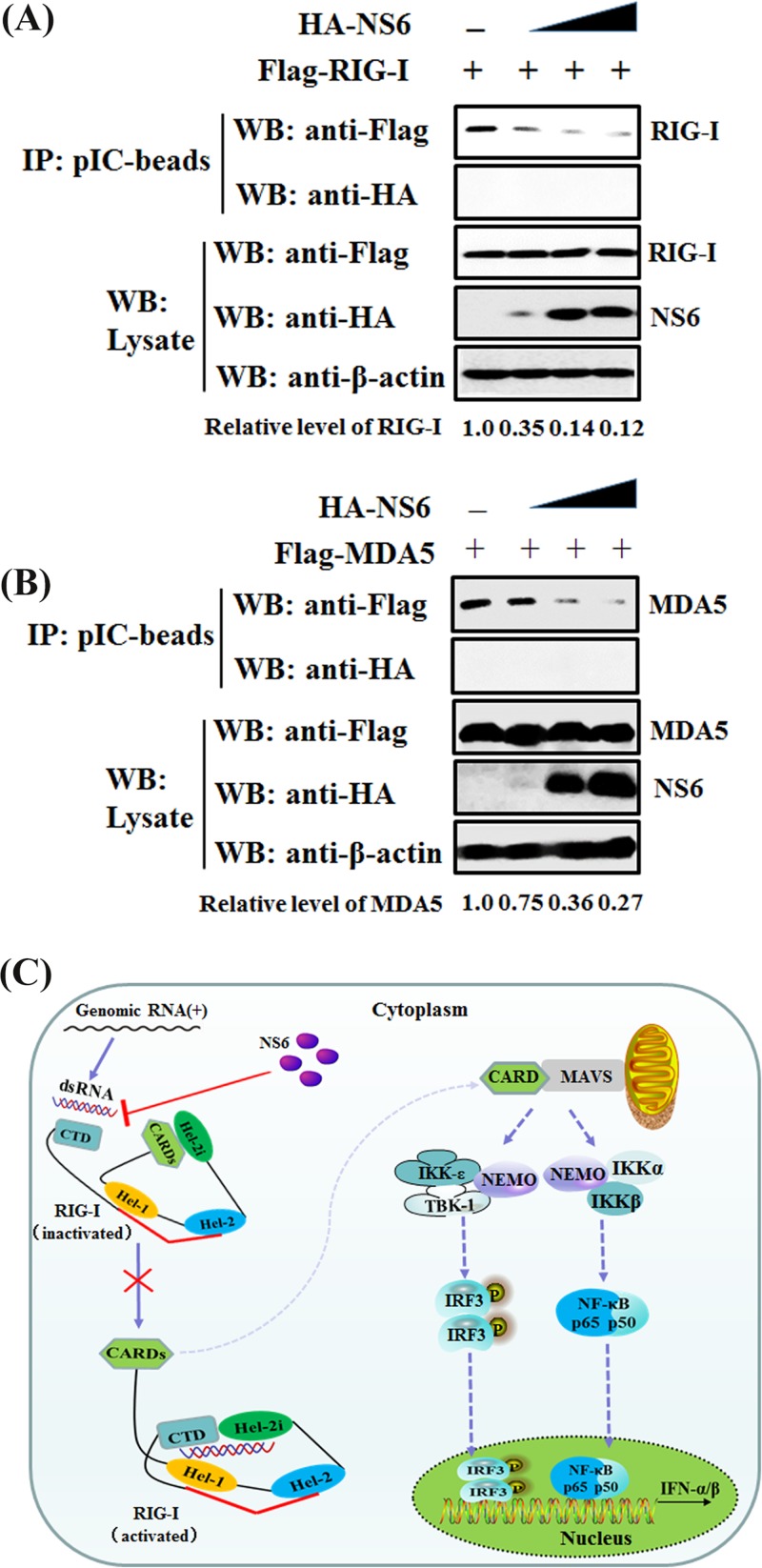
NS6 attenuates the interaction of dsRNA with RIG-I/MDA5.
Given our above-described finding that NS6 is not an RNA-binding protein, the possibility that NS6 inhibits SeV/poly(I·C)-induced IFN-β production by competing with RIG-I/MDA5 for dsRNA binding can be excluded. Thus, we speculated that NS6 protein disrupts or attenuates the binding of dsRNA with RIG-I/MDA5. A competition assay was performed by using a poly(I·C) pulldown assay. HEK-293T cells were transfected with Flag-RIG-I or -MDA5 and increasing amounts of HA-NS6. The clarified lysates from cells transfected with RIG-I or MDA5 expression constructs were incubated with those from cells transfected with increasing concentrations of NS6 expression plasmid, followed by supplementation with prepared poly(I·C)-coated agarose beads for 4 h at 4°C. Bound RIG-I or MDA5 was then detected by Western blotting. As seen in Fig. 8, the expression levels of RIG-I, MDA5, and NS6 proteins were clearly detected in whole-cell lysates; however, significantly smaller amounts of RIG-I (Fig. 8A) and MDA5 (Fig. 8B) coimmunoprecipitated with poly(I·C)-coated agarose beads were detected with increasing concentrations of NS6 protein. These results indicate that NS6 protein at least partially functions to block the recognition or binding of dsRNA by RIG-I or MDA5, leading to the antagonism of IFN-β production.
FIG 8.
NS6 hinders the combination of dsRNA with RIG-I/MDA5. (A and B) HEK-293T cells were individually transfected with plasmids encoding Flag-tagged RIG-I (A), MDA5 (B), and increasing quantities of NS6 expression plasmids for 28 h. Lysates from the cells overexpressing NS6 were incubated with an equal volume of lysates from cells overexpressing RIG-I or MDA5, followed by treatment with poly(I·C)-coated agarose beads for 4 h at 4°C. The beads were then washed three times with lysis buffer by centrifugation and subjected to Western blotting as described in the legend to Fig. 7. The numbers below the images represent the relative level of RIG-I/MDA5 compared to that of the control group via ImageJ software analysis. (C) A schematic diagram, where RIG-I acts as a representative protein, of the mechanism for NS6 protein inhibition of the RLR signaling pathway. In the inactivated state of RIG-I, the CARDs are bound to Hel-2i, which is unavailable for downstream signaling in this autoinhibited state. The CTD, which is tethered to the red bridging helix by a flexible linker, is able to sense RNA PAMPs. Upon virus infection, the CTD-bound dsRNA is preoriented to form a network of interactions with the helicase domains Hel-1 and Hel-2i but not Hel-2, leading to the segregation of interaction of CARDs with Hel-2i and the subsequent availability for interaction with downstream signaling molecules (64).
DISCUSSION
As species-specific proteins of coronaviruses, accessory proteins have received increasingly more attention over the past decade, and novel accessory proteins encoded by coronaviruses have been continually identified in virus-infected cells, such as ORFX of bat SARS-like coronavirus (56) and the NS7a protein of PDCoV (20). In this study, we investigated the function of PDCoV NS6. Our results reveal that NS6 possesses the property of antagonizing IFN-β production, and it does so by interacting with the CTD of RIG-I and the Hel and CTD of MDA5, which attenuates the binding of RIG-I/MDA5-dsRNA.
RIG-I and MDA5 belong to the RIG-I-like helicase group of the SF2 family, and they are important cytoplasmic PRRs that function as viral dsRNA receptors to initiate the type I IFN response against infection with an RNA virus (57). For CoV, MDA5 appears to be more important than RIG-I to recognize CoV replicative intermediates (58, 59). In an effort to evade host immune surveillance, many viral proteins target these two molecules to disrupt IFN signaling. For example, both human respiratory syncytial virus NS2 protein and New World arenavirus Z protein antagonize the activation of IFN-β production via interacting with RIG-I to disturb its association with the downstream signaling molecule MAVS (60, 61). Additionally, the X protein encoded by hepatitis B virus suppresses virus-triggered IFN-β induction via interacting with MDA5 and MAVS to disrupt the formation of the MDA5-MAVS complex (62). Furthermore, influenza A virus nonstructural protein 1 (NS1) interacts with RIG-I and inhibits RIG-I ubiquitination to antagonize RIG-I-mediated IFN-β production (51, 63). In this study, we found that PDCoV NS6 also interacts with RIG-I/MDA5; however, differently from the mechanisms used by the viral proteins mentioned above, PDCoV NS6 does not inhibit IFN-β production by overexpressing RIG-I or MDA5. Moreover, NS6 also does not interact with MAVS and does not disrupt the complex formation of RIG-I and MAVS, TBK1, or IKKε (data not shown), which is not surprising given that NS6 does not inhibit the IFN-β promoter activity induced by RIG-I, MDA5, MAVS, or their downstream molecules (Fig. 3).
Our results also demonstrate that PDCoV NS6 specifically interacts with the CTD of RIG-I; however, it can interact with the Hel and CTD of MDA5. RIG-I and MDA5 have similar domain structures, possessing N-terminal tandem CARDs, a central DExD/H-box type RNA helicase containing two RecA domains (Hel-1 and Hel-2) with a family-specific insertion, named Hel-2i, within Hel-2, and a CTD (53, 64, 65). Based on the different molecular mechanisms for dsRNA recognition by MDA5 and RIG-I, especially the structural mechanism for the divergent RNA recognition by RIG-I and MDA5 (65–68), the different interaction domain between RIG-I and MDA5 with NS6 is reasonable and can be explained. Previous studies demonstrated that the RIG-I CTD caps the dsRNA end and plays a predominant role in high-affinity binding and selectivity for dsRNA (66), while the MDA5 CTD binds to the dsRNA stem (65). Differently from the RIG-I helicase domain, the MDA5 helicase domain also contributes to dsRNA stem recognition, and its role is beyond simply providing additional RNA affinity but likely includes precise positioning of the CTD for efficient recognition of the dsRNA stem (65). Interactions with the CTD of RIG-I and the Hel and CTD of MDA5 make NS6 to block the binding of RIG-I/MDA5 with dsRNA. In the case of RIG-I, for example, the binding of its CTD for viral RNA PAMPs can serve as the first step in initiating the activation of downstream signaling pathways (64). In the ligand-free state, the binding of CARDs to Hel-2i results in the formation of an autorepressed state for this protein by sterically hindering the access of ubiquitination enzymes and of polyubiquitin binding to CARDs. Upon viral infection, the initial binding of viral dsRNA to the CTD results in its functional transformation from an autorepressed state into a signaling-competent configuration and the release of CARDs, subsequently leading to 2CARD oligomerization, followed by its interaction with MAVS as described in the legend to Fig. 8C. However, the interaction between NS6 and the CTD of RIG-I or the Hel and CTD of MDA5 appears to block the dsRNA binding sites (CTDs or Hel), resulting in RIG-I/MDA5 having a reduced dsRNA-binding ability and less subsequent type I IFN production. Because NS6 is not an RNA-binding protein, the possibility that NS6 inhibits SeV/poly(I·C)-induced IFN-β production by competing with RIG-I/MDA5 for dsRNA binding can be excluded (Fig. 7). Thus, it is possible that NS6 competes with dsRNA for binding to RIG-I/MDA5. The competition binding experiment results (Fig. 8) support this hypothesis, which is illustrated in Fig. 8C, using RIG-I as a representative receptor. Indeed, previous work has also shown that the overexpression of the CTD inhibits RIG-I-dependent signaling in response to SeV infection (69), and the possible mechanism for it is mediated through the sequestration of viral RNA produced during SeV infection (52). Overall, our experiments reveal that NS6 utilizes a mechanism that is different from those of other viral proteins previously reported to antagonize RLR-mediated IFN-β production.
The NS6 protein is unique to PDCoV, with no significant homology to other viral proteins of known coronaviruses. Previous work has shown that PDCoV NS6 is expressed during early virus infection and is distributed in the cytoplasm (19). It seems likely that the early expression of NS6 proteins in virus-infected cells is important because their direct interaction with RIG-I or MDA5 functions to prevent the recognition of viral dsRNA by these proteins. It should be noted that the identified function of NS6 to antagonize IFN production in the present study is derived from overexpression and should be further tested in the context of live virus infection. Regrettably, a PDCoV reverse genetics system was not available when this work was conducted, preventing further investigation of the NS6 IFN antagonist activity at the level of virus infection in vivo. We are currently working to establish a PDCoV reverse genetics system, which will allow the generation of mutant or deletion viruses that can be used to further explore the functions of NS6 protein, such as the effects of NS6 deletion on IFN-β production, viral replication, and/or host spectrum.
Interestingly, this study found that PDCoV NS6 appears to interact preferentially with RNA-binding proteins, even though it is not an RNA-binding protein itself. In addition to the well-known RNA-binding proteins RIG-I and MDA5, PDCoV N protein was also found to be an RNA-binding protein (Fig. 7), and NS6 interacts with N protein (data not shown). Our previous study showed that PDCoV NS6 is associated with the purified viral particle, and NS6 is mainly localized in the endoplasmic reticulum (ER) and ER-Golgi intermediate compartments, which are the sites of coronavirus assembly and packaging (19). Whether or not the interaction between NS6 and N protein is associated with viral replication and assembly is under investigation in our laboratory, and the resulting findings will greatly improve our understanding of the role of NS6 protein in viral replication and pathogenicity.
In summary, we report that overexpression of accessory protein NS6 antagonizes IFN-β production via interacting with RIG-I/MDA5 to impede their association with dsRNA, leading to the blockage of the beginning PRR-dsRNA recognition step. To date, only PDCoV nsp5 (70, 71) and NS6 protein (this study) have been identified as IFN antagonists, while at least eight proteins encoded by SARS-CoV have been identified as IFN antagonists (26, 27, 39, 72–74). The further identification and characterization of PDCoV-encoded IFN antagonists will accelerate the elucidation of the association between PDCoV and the IFN signaling pathway, which may lead to the development of novel effective therapeutic strategies and vaccines.
MATERIALS AND METHODS
Viruses and cells.
The PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) used in this study was isolated in China in 2014 from a piglet with severe diarrhea (75). SeV was obtained from the Centre of Virus Resource and Information, Wuhan Institute of Virology, Chinese Academy of Sciences. VSV-GFP was gifted by Zhigao Bu at the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences. HEK-293T cells were obtained from the China Center for Type Culture Collection and maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS). The LLC-PK1 cells used for PDCoV propagation were purchased from the ATCC (ATCC number CL-101) and grown under the same conditions as those described above.
Plasmids and Dual-Luciferase reporter assay.
The NS6 gene from PDCoV strain CHN-HN-2014 was amplified with the primers PDCoV-NS6-F and PDCoV-NS6-R (Table 1) and cloned into pCAGGS-HA-N with an N-terminal HA tag or pCAGGS-Flag-N with an N-terminal Flag tag and named pCAGGS-HA-NS6 and pCAGGS-Flag-NS6, respectively. The PDCoV N gene was also cloned into pCAGGS-Flag-N with an N-terminal Flag tag using the primers PDCoV-N-F and PDCoV-N-R (Table 1), and the resulting plasmid was named pCAGGS-Flag-NP. The luciferase reporter plasmids IFN-β-Luc, NF-κB-Luc, and IRF3-Luc have been described previously (76). The expression plasmids for Flag-tagged RIG-I and its constitutively activated mutant, RIG-IN, as well as MDA5, MAVS, TBK1, and IRF3 and its constitutively activated mutant, IRF3-5D, have also been described previously (77). The GFP expression plasmid (pEGFP-C1) was purchased from TaKaRa (Japan). Three characteristic functional domains of RIG-I or MDA5, including the 2CARD (RIG-I amino acids [aa] 1 to 228; MDA5 aa 1 to 295), the helicase domain (RIG-I aa 229 to 803; MDA5 aa 296 to 827), and the CTD (RIG-I aa 804 to 925; MDA5 aa 828 to 1025), were cloned into the pCAGGS-Flag-N vector using the primers listed in Table 1, and the resulting expression constructs were named pCAGGS-Flag-2CARD(RIG-I), pCAGGS-Flag-2CARD(MDA5), pCAGGS-Flag-Hel(RIG-I), pCAGGS-Flag-Hel(MDA5), pCAGGS-Flag-CTD(RIG-I), and pCAGGS-Flag-CTD(MDA5), respectively. All plasmids were verified by sequencing. For luciferase reporter assays, HEK-293T or LLC-PK1 cells grown in 24-well plates were transfected using Lipofectamine 2000 (Invitrogen) with a luciferase reporter plasmid (IFN-β-Luc, NF-κB-Luc, or IRF3-Luc) and pRL-TK (Promega), together with the indicated expression plasmid or an empty vector. At 24 h after transfection, the cells were stimulated with SeV (10 hemagglutinating activity units/well) or poly(I·C) (InvivoGen, USA) for 12 h. Subsequently, the firefly luciferase and Renilla luciferase activities from lysed cells were evaluated through the Dual-Luciferase reporter assay system according to the instructions from the manufacturer (Promega). Representative data from three independently conducted experiments are expressed as the relative firefly luciferase activities with normalization to the Renilla luciferase activities.
TABLE 1.
Sequences of the primers used for real-time PCR and construction of plasmids
| Primer | Nucleotide sequence (5′–3′) |
|---|---|
| SEV-HN-F | AAAATTACATGGCTAGGAGGGAAAC |
| SEV-HN-R | GTGATTGGAATGGTTGTGACTCTTA |
| h-GAPDH-F | TCATGACCACAGTCCATGCC |
| h-GAPDH-R | GGATGACCTTGCCCACAGCC |
| PDCoV-N-F | ACTGAATTCATGGCTGCACCAGTAGTCCCTAC |
| PDCoV-N-R | CTAATCGATCTACGCTGCTGATTCCTGCTTTAT |
| PDCoV-NS6-F | ACTGAATTCATGTGCAACTGCCATCTGCAGC |
| PDCoV-NS6-R | CTGCTCGAGTTAATTTAATTCATCTTCAAG |
| RIG-I-2CARD-F | TAAATCGATATGACCACCGAGCAGCGA |
| RIG-I-2CARD-R | CAGCTCGAGTCATGGACATGAATTCTC |
| RIG-I-Hel-F | TAAATCGATCCTTCAGAAGTGTCTGATA |
| RIG-I-Hel-R | CAGCTCGAGCTTATCAGGGACAGGTTTTGG |
| RIG-I-CTD-F | AAATCGATGAAAATAAAAAACTGCTCTG |
| RIG-I-CTD-R | CAGCTCGAGTCATTTGGACATTTCTGC |
| MDA5-2CARD-F | GCGATCGATATGTCGAATGGGTATTCCACAGAC |
| MDA5-2CARD-R | GCGCTCGAGTTAATTCTCTTCATCTGAATCACTTC |
| MDA5-Hel-F | TCTATCGATATGGTGGCAGCAAGAGCATCCCCG |
| MDA5-Hel-R | TATCTCGAGTTACTCATCAGCTCTGGCTCGACCA |
| MDA5-CTD-F | TCTATCGATATGAGCACCTACGTCCTGGTTG |
| MDA5-CTD-R | GCGCTCGAGCTAATCCTCATCACTAAATAAAC |
RNA extraction and quantitative real-time RT-PCR.
To confirm the effects of NS6 protein on SeV replication, HEK-293T cells in 24-well plates were transfected with increasing amounts of NS6 expression plasmids. After 24 h, the cells were mock infected or infected with SeV for 12 h. Total RNA was extracted from the treated cells with TRIzol reagent (Invitrogen), followed by first-strand cDNA synthesis using avian myeloblastosis virus (AMV) reverse transcriptase (TaKaRa, Japan) with the indicated primers (Table 1). Each quantitative real-time PCR (qPCR) experiment was performed at least three times and was conducted via the SYBR green PCR assay (Applied Biosystems) using the cDNA described above as the template. The results are expressed as the relative gene expression level with normalization to the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
IFN bioassay.
To measure the effect of NS6 on the amount of IFN production by HEK-293T cells following stimulation by SeV, IFN bioassays were performed as described previously (54).
Western blot analysis.
HEK-293T cells grown in 60-mm dishes were transfected with the indicated plasmids for 24 h. The cells were mock infected or infected with SeV for 8 h. The transfected cells were harvested with lysis buffer (4% SDS, 3% dithiothreitol [DTT], 0.065 mM Tris-HCl [pH 6.8], 30% glycerin) supplemented with a protease inhibitor cocktail and a phosphatase inhibitor cocktail (Sigma). Equal amounts of proteins were subjected to separation by 12% SDS-PAGE and then transferred to a polyvinylidene difluoride membrane, followed by blocking with 5% nonfat milk in PBST with 0.1% polysorbate-20 and subsequent treatment with the indicated primary antibodies, namely, rabbit anti-p-IRF3 and anti-p65 (ABclonal), anti-p-p65 and anti-IRF3 (Cell Signaling Technology), and mouse anti-Flag or -HA (MBL) antibodies at 37°C for 4 h. After washing three times with PBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Beyotime, China) for 45 min at room temperature. After washing three times, the membrane was visualized by enhanced chemiluminescence (ECL) reagents (Bio-Rad). The expression levels of β-actin were detected with a mouse anti-β-actin monoclonal antibody (MBL) and were considered indicative of whether or not the protein sample loading was equal.
Co-IP and Western blot analyses.
Co-IP assays were performed as described previously (77). HEK-293T cells that had been cultured in 60-mm dishes were cotransfected with the indicated expression plasmids containing Flag or HA tags. After 28 h, the cells were harvested and lysed on ice with 0.5 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 10% glycerin, 0.1% SDS, and 2 mM Na2EDTA) for 30 min at 4°C. A portion of each supernatant from the lysed cells was used in the whole-cell extract assays. The remaining portions of the supernatants from the lysed cells were immunoprecipitated with affinity antibodies overnight at 4°C and then treated with protein A+G agarose beads (Beyotime) for 5 h at 4°C. The beads containing immunoprecipitates were washed three times with 1 ml of lysis buffer. Whole-cell extracts and immunoprecipitates were resuspended in SDS-PAGE loading buffer, boiled at 95°C for 5 min, and then subjected to 12% SDS-PAGE and transferred to polyvinylidene difluoride membrane, followed by Western blot analyses with the indicated antibodies.
Poly(I·C) pulldown assay.
HEK-293T cells grown in 60-mm plates were transfected with 4 μg of each of the indicated expression plasmids, including Flag-tagged RIG-I, N and NS6, or empty vector for 24 h. The cells were harvested and lysed on ice with 400 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 10% glycerin, 0.1% SDS, and 2 mM Na2EDTA) supplemented with a cocktail of protease inhibitors (Sigma). The clarified cell lysates were mixed with a prepared suspension of poly(I·C)-coated agarose beads and incubated for 4 h at 4°C. The beads were washed three times with 1 ml of lysis buffer by multiple centrifugations and then subjected to Western blotting by using mouse anti-Flag antibody (MBL) as the primary antibody, followed by treatment with HRP-conjugated goat anti-mouse IgG.
IFA.
Monolayers of HEK-293T cells seeded onto coverslips in 24-well plates were transfected with pCAGGS-HA-NS6 or empty vector for 24 h. The cells were then mock infected or infected with SeV for 8 h. The cells were subsequently fixed with 4% paraformaldehyde for 15 min and then permeated with methyl alcohol for 10 min at room temperature. After three washes with PBST, the cells were blocked with PBST containing 5% bovine serum albumin (BSA) for 1 h, followed by incubation separately with a rabbit polyclonal antibody against IRF3 (1:200) or against p65 (1:200) or a mouse anti-HA antibody (1:200) for 1 h at 37°C. The cells were then stained with secondary antibodies Alexa Fluor 594-conjugated donkey anti-mouse IgG and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Santa Cruz Biotechnology) for 1 h at 37°C, followed by treatment with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime) for 15 min at room temperature. Fluorescent images were visualized with the use of a confocal laser scanning microscope (FluoView 3.1; Olympus, Japan).
Statistical analysis.
Statistical differences were determined by one-way analyses of variance using GraphPad Prism 5.0 software. For all experiments, differences were considered statistically significant when P values were <0.05.
ACKNOWLEDGMENTS
We thank Zhigao Bu for providing VSV-GFP recombinant virus.
This work was supported by the National Natural Science Foundation of China (31730095), the National Key R&D Plan of China (2016YFD0500103), the Key Technology R&D Programme of China (2015BAD12B02), and the Major S&T Project of Hubei Province (2017ABA138).
REFERENCES
- 1.Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ. 2015. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis 21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu H, Jung K, Vlasova AN, Saif LJ. 2016. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch Virol 161:3421–3434. doi: 10.1007/s00705-016-3056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. 2015. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6:e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marthaler D, Jiang Y, Collins J, Rossow K. 2014. Complete genome sequence of strain SDCV/U S A/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc 2:e00218-. doi: 10.1128/genomeA.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. 2014. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis 20:1347–1350. doi: 10.3201/eid2014.140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Byrum B, Zhang Y. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, U S A, 2014. Emerg Infect Dis 20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Byrum B, Zhang Y. 2014. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg Infect Dis 20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J. 2014. Full-length genome sequence of porcine deltacoronavirus strain U S A/IA/2014/8734. Genome Announc 2:e00278-. doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S, Lee C. 2014. Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc 2:e01191-. doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. 2015. Porcine deltacoronavirus in mainland China. Emerg Infect Dis 21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YW, Yue H, Fang W, Huang YW. 2015. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from mainland China. Genome Announc 3:e00945-. doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. 2015. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis 62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S, Amonsin A. 2016. Porcine deltacoronavirus, Thailand, 2015. Emerg Infect Dis 22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorsirigool A, Saeng-Chuto K, Temeeyasen G, Madapong A, Tripipat T, Wegner M, Tuntituvanont A, Intrakamhaeng M, Nilubol D. 2016. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch Virol 161:2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. 2017. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis 64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J. 2016. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res 226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung K, Hu H, Saif LJ. 2017. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol 162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang P, Fang L, Liu X, Hong Y, Wang Y, Dong N, Ma P, Bi J, Wang D, Xiao S. 2016. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang P, Fang L, Hong Y, Liu X, Dong N, Ma P, Bi J, Wang D, Xiao S. 2017. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J Gen Virol 98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, Lee C. 2015. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res 208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. 2014. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res 109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yount B, Roberts RS, Sims AC, Deming D, Frieman MB, Sparks J, Denison MR, Davis N, Baric RS. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol 79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan YJ, Lim SG, Hong W. 2006. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res 72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer F, Peng D, Hingley ST, Weiss SR, Masters PS. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol 71:996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. 2014. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Ferraro D, Zhao J, Hussain S, Shao J, Trujillo J, Netland J, Gallagher T, Perlman S. 2010. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J Virol 84:3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li YZ, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. 2016. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7:e00258. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 2014. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol 95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci U S A 110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 35.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol 81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp R. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 38.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Shi K, Yoo D. 2016. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth-Cross JK, Martinez-Sobrido L, Scott EP, Garcia-Sastre A, Weiss SR. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo J, Fang L, Dong N, Fang P, Ding Z, Wang D, Chen H, Xiao S. 2016. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology 495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Zheng BJ, Zeng R, Lu W, Lin YP, Xue L, Li L, Yang LL, Xu C, Dai J, Wang F, Li Q, Dong QX, Yang RF, Wu JR, Sun B. 2009. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology 388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang C, Peters CJ, Makino S. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J Virol 81:5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett 425:112–116. doi: 10.1016/S0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 50.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell 1:507–518. doi: 10.1016/S1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 51.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 53.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. 2011. Structural insights into RNA recognition by RIG-I. Cell 147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cardenas WB, Loo YM, Gale M Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CK, Hsu YL, Chang YH, Chao FA, Wu MC, Huang YS, Hu CK, Huang TH. 2009. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J Virol 83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng LP, Gao YT, Ge XY, Zhang Q, Peng C, Yang XL, Tan B, Chen J, Chmura AA, Daszak P, Shi ZL. 2016. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J Virol 90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou J, Chang M, Nie P, Secombes CJ. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol 9:85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalinger ZB, Elliott R, Rose KM, Weiss SR. 2015. MDA5 is critical to host defense during infection with murine coronavirus. J Virol 89:12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan L, Briese T, Lipkin WI. 2010. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol 84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ling Z, Tran KC, Teng MN. 2009. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol 83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang XM, Li Y, Mao AP, Li C, Li YK, Tien P. 2010. Hepatitis B virus X protein suppresses virus-triggered IRF3 activation and IFN-beta induction by disrupting the VISA-associated complex. Cell Mol Immunol 7:341–348. doi: 10.1038/cmi.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolakofsky D, Kowalinski E, Cusack S. 2012. A structure-based model of RIG-I activation. RNA 18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 66.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell 29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 67.Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. 2009. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys 488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel DJ. 2010. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu X, Fang L, Wang D, Yang Y, Chen J, Ye X, Foda MF, Xiao S. 2017. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology 502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, Lv M, Ye X, Peng G, Fang L, Xiao S. 2017. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J Virol 91:e00003-. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li SW, Wang CY, Jou YJ, Huang SH, Hsiao LH, Wan L, Lin YJ, Kung SH, Lin CW. 2016. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci 17:E678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. 2013. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One 8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. 2016. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, Fang L, Shi Y, Zhang H, Gao L, Peng G, Chen H, Li K, Xiao S. 2016. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J Virol 90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]