Abstract
Background
Lignocellulosic biomass is seen as an abundant renewable source of liquid fuels and chemicals that are currently derived from petroleum. When lignocellulosic biomass is used for ethanol production, the resulting liquid residue (stillage) contains large amounts of organic material that could be further transformed into recoverable bioproducts, thus enhancing the economics of the biorefinery.
Results
Here we test the hypothesis that a bacterial community could transform the organics in stillage into valuable bioproducts. We demonstrate the ability of this microbiome to convert stillage organics into medium-chain fatty acids (MCFAs), identify the predominant community members, and perform a technoeconomic analysis of recovering MCFAs as co-products of ethanol production. Steady-state operation of a stillage-fed bioreactor showed that 18% of the organic matter in stillage was converted to MCFAs. Xylose and complex carbohydrates were the primary substrates transformed. During the MCFA production period, the five major genera represented more than 95% of the community, including Lactobacillus, Roseburia, Atopobium, Olsenella, and Pseudoramibacter. To assess the potential benefits of producing MCFAs from stillage, we modeled the economics of ethanol and MCFA co-production, at MCFA productivities observed during reactor operation.
Conclusions
The analysis predicts that production of MCFAs, ethanol, and electricity could reduce the minimum ethanol selling price from $2.15 to $1.76 gal−1 ($2.68 gal−1 gasoline equivalents) when compared to a lignocellulosic biorefinery that produces only ethanol and electricity.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1193-x) contains supplementary material, which is available to authorized users.
Keywords: Medium-chain fatty acids, Mixed culture fermentation, Carboxylate platform, Chain elongation, Lignocellulose, Stillage
Background
The production of food, fuels, pharmaceuticals and many chemicals depends on microbial fermentations. When one considers the sum of microbial biomass, excreted metabolic end-products, and non-metabolized nutrients, there is considerable residual organic matter in the liquid residue (stillage) remaining after distillation. One common co-product of ethanol production is biogas, which is generated by anaerobic digestion of stillage. Combusting lignin and biogas creates heat and power used to operate the biorefinery, and any excess electricity can be sold as a co-product [1]. In a techno-economic analysis (TEA) conducted by the National Renewable Energy Laboratory (NREL), a 61 million gallon per year lignocellulosic ethanol biorefinery produced fuel at a price of $2.15 gal−1 ($3.27 gal−1 gasoline-equivalents) and electricity worth $6.57 million year−1 [2].
The Renewable Fuel Standards (RFS), created by the Energy Policy Act of 2005 and expanded by the Energy Independence and Security Act of 2007, set production goals for many renewable energy sources, including lignocellulosic-derived ethanol [3, 4]. While several lignocellulosic biorefineries have been opened, total lignocellulosic ethanol production in the United States remains short of original targets. The high costs of obtaining biomass and producing enzymes to hydrolyze biomass are cited as barriers to achieving an acceptable level of profitability for lignocellulosic biorefineries [2].
One way to potentially improve the economics of lignocellulosic fuel production is to produce valuable co-products, such as medium-chain fatty acid (MCFA), from stillage. MCFAs are monocarboxylic acids containing six to twelve carbon atoms and are utilized for the production of rubbers, dyes, pharmaceuticals, and antimicrobials [5]. They can also be used as precursors for chemicals currently derived from fossil fuels [6]. In addition to being valuable, MCFAs also have decreased solubility compared to short-chain fatty acids (SCFA), which should allow for easier extraction from an aqueous medium.
In this study, we investigated the valorization of switchgrass-derived stillage to MCFAs. Switchgrass has been identified as a promising feedstock for biofuel production that can be cultivated on marginal lands [7]. In this study, we tested the ability of using mixed culture anaerobic fermentation, as in the so-called carboxylate platform [8, 9], to valorize stillage to MCFAs. Here MCFA is the sum of hexanoate and octanoate since it is still largely unknown how to direct metabolism to production of only one MCFA. In several past studies, ethanol has been utilized as an electron donor to drive MCFA production from either added acetate or acetate produced by the community as a fermentation intermediate [10–13]. Conversion of lactic acid to MCFA has also been investigated [14, 15]. Recently, a pure culture of Megasphaera elsdenii was used to convert glucose in lignocellulosic hydrolysate to MCFAs [16]. Stillage from corn-derived ethanol has also been used to produce MCFAs [17]. Andersen et al. utilized a mixture of lignocellulosic stillage and dilute ethanol to produce MCFAs at titers greater than their solubility concentrations [18]. However, MCFA production from industrial streams having minimal amounts of glucose or ethanol remains largely unexplored. In addition, there is no published TEA investigating production of MCFAs from stillage.
While past studies have investigated MCFA production from lignocellulosic materials, none have evaluated production of ethanol followed by MCFA production from the resulting stillage in a biorefinery. Thus, the objectives of this study are to (1) test the hypothesis that a stillage-fed microbial community can sustain production of MCFAs; (2) investigate the stability of the microbiome and potential roles of abundant community members in the MCFA-producing reactor; and (3) evaluate the technoeconomics of producing MCFAs from ethanol stillage. To achieve the third objective, we modeled a modified lignocellulosic biorefinery producing MCFA as a co-product of ethanol and electricity (Fig. 1). After accounting for the amount of organic matter in stillage that is directed to MCFA production, the reduction in overall biogas and electricity production, and the increased capital and operational costs associated with MCFA production, our data predict that the potential revenue from producing MCFAs at levels observed in this study would have a positive impact on the economics of lignocellulosic biorefining.
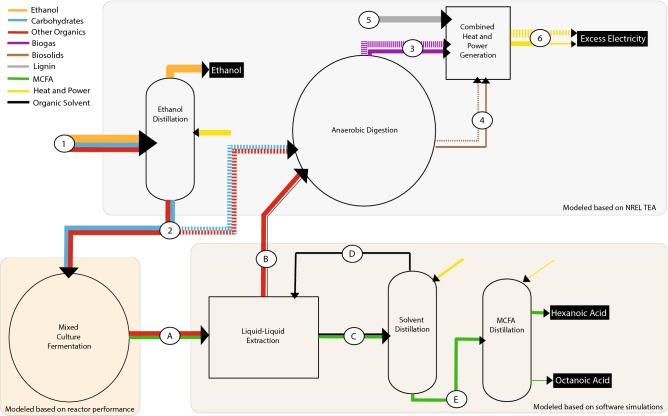
Fig. 1.
Energy balance through a proposed lignocellulosic biorefinery that simultaneously produces ethanol, electricity, and MCFAs. For this study, we modeled anaerobic digestion and combined heat and power generation using criteria used in a 2011 technoeconomic analysis performed by NREL. We modeled a MCFA-producing mixed culture fermentation process based on productivities observed in this study. Commercially available software was utilized for the modeling of downstream processes to extract and separate hexanoic and octanoic acids. In the proposed biorefinery, a portion of organic matter remaining in stillage is converted to MCFAs, which can be recovered via liquid–liquid extraction and distillation. Remaining organic matter undergoes anaerobic digestion to produce biogas that is combusted, along with lignin and biosolids, to produce electricity. Dashed lines represent current process flows for converting organic materials in stillage directly to biogas. Solid lines represent proposed processes for producing MCFA as a co-product. Line weights are proportional to approximate energy content represented as chemical oxygen demand (COD) for liquid and gas streams and as megawatts (MW) for heat and power. Process streams include (1) fermented hydrolysate; (2) stillage; (3) biogas; (4) biosolids; (5) lignin; (6) heat and power; (A) fermented stillage; (B) aqueous phase; (C) organic phase; (D) organic solvent; and (E) MCFAs
Methods
Switchgrass stillage production
Shawnee switchgrass, grown in 2010 at the Arlington Agricultural Research Center in Wisconsin, USA, was used as the biomass source for this study. Switchgrass was treated using ammonia-fiber expansion (AFEX), enzymatically hydrolyzed, and fermented, as described previously [19]. During processing, hydrolysate is filtered to remove insoluble components, including insoluble lignin. Past work has demonstrated that switchgrass hydrolysates generated with this process contain sufficient nutrients and trace elements to sustain microbial growth [19]. Ethanol fermentations of switchgrass hydrolysate were performed with Saccharomyces cerevisiae Y128, an engineered yeast strain with improved xylose utilization and lignotoxin tolerance [20]. Ethanol was removed post-fermentation using a glass distillation apparatus consisting of a 1-L boiling flask, heating mantle, distillation column, and condenser. During distillation, the fermented hydrolysate was heated to approximately 100 °C to maintain a distillation neck temperature of 78 °C. Therefore, the distillation process not only removed ethanol but also sterilized the stillage. The stillage remaining after distillation was stored at 4 °C until fed to the bioreactor.
Mixed culture fermentation bioreactor
A mixed culture fermentation bioreactor was inoculated with sludge from an acid-phase digester at the Nine Springs Wastewater Treatment Plant in Madison, Wisconsin. The bench-scale reactor consisted of a vessel with a 150-mL working volume that was continuously stirred at 150 rpm with a magnetic stir bar and maintained at 35 °C using a water bath. The reactor was sealed with a rubber stopper and vented so that any gas produced was released to the atmosphere. For all experiments, the solid retention time (SRT) is equal to the hydraulic retention time.
Initially, we conducted short-term (6 day) experiments to assess if microbial growth could be sustained in stillage and to determine the primary fermentation end products under different pH conditions. For these initial experiments, the pH was either uncontrolled or controlled at set points of 5.0, 5.5, 6.0, or 6.5 with 5 M KOH. A hydraulic retention time of 2 days was utilized for these initial experiments by pumping 75 mL day−1 (3.13 mL h−1) both into and out of the reactor. A shorter SRT was utilized to allow for fast turnover and stabilization of the microbial community. While this short SRT resulted in production of MCFAs, we elected to increase the SRT for a long-term experiment in an attempt to improve overall MCFA titers. For the long-term (252 day) sustained experiment, the pH was controlled at a set point of 5.50 with 5 M KOH and the SRT was controlled at 6 days by pumping 25 mL day−1 (1.04 mL h−1) into and out of the reactor.
Chemical analyses
We collected samples from the reactor and stillage for chemical analyses. All samples were filtered using 0.22-μm syringe filters (ThermoFisher Scientific SLGP033RS, Waltham, MA, USA). Soluble chemical oxygen demand (COD) analysis was performed using High-Range COD Digestion Vials (Hach 2125915, Loveland, CO, USA) as per standard methods [21]. Soluble carbohydrates were measured with the anthrone method [22]. Total soluble proteins were measured with the bicinchoninic acid assay using the Pierce™ BCA Assay Kit (ThermoFisher Scientific 23225, Waltham, MA, USA) and the Compat-Able™ Protein Assay Preparation Reagent Set (ThermoFisher Scientific 23215, Waltham, MA, USA) [23].
Glucose, xylose, acetic acid, formic acid, lactic acid, succinic acid, pyruvic acid, glycerol and xylitol were analyzed with high-performance liquid chromatography (HPLC) and quantified with an Agilent 1260 Infinity refractive index detector (Agilent Technologies, Inc. Palo Alto, CA) using a 300 × 7.8 mm Bio-Rad Aminex HPX-87H column with Cation-H guard (BioRad, Inc., Hercules, CA). A column temperature of 50 °C was used and 0.02 N H2SO4 was used for the mobile phase with a flow rate of 0.50 mL min−1.
Acetamide, ethanol, n-propionic acid, n-butyric acid, iso-butyric acid, n-pentanoic acid, iso-pentanoic acid, n-hexanoic acid, iso-hexanoic acid, n-heptanoic acid, and n-octanoic acid were analyzed with tandem gas chromatography–mass spectrometry (GC–MS). An Agilent 7890A GC system (Agilent Technologies, Inc. Palo Alto, CA) with a 0.25-mm Restek Stabilwax DA 30 column (Restek 11008, Belefonte, PA) was used. The GC–MS system was equipped with a Gerstel MPS2 (Gerstel, Inc. Baltimore, MD) autosampler and a solid-phase micro-extraction gray hub fiber assembly (Supelco, Bellefonte, PA). The MS detector was a Pegasus 4D TOF–MS (Leco Corp., Saint Joseph, MI). Stable isotope-labeled internal standards were used for each of the analytes measured with GC–MS.
Aromatic compounds were analyzed with liquid chromatography–tandem mass spectrophotometry (LC–MS/MS). For LC–MS/MS analyses, an Ultimate HPG-3400RS pump and WPS-3000RS autosampler (ThermoFisher) were mated to an ACQUITY UPLC HSS T3 reversed-phase column (2.1 × 150 mm, 1.8 μm particle diameter, Waters Corporation) with a guard cartridge. Gradient elution was performed at 0.400 mL min−1. The LC system was coupled to a TSQ Quantiva Triple Quadrupole mass spectrometer (Thermo Scientific). The Ion Transfer Tub Temp was kept at 350 °C as was the vaporizer temperature. Analytes measured with LC–MS/MS included vanillamide, 4-hydroxybenzyl alcohol, syringamide, coumaryl amide, 4-hydroxybenzoic acid, feruloyl amide, vanillic acid, p-coumaric acid, ferulic acid, and benzoic acid. Detailed chemical analysis data are provided in Additional file 1.
Microbial community analysis
Amplification and sequencing of the V3–V4 region of the 16S rRNA gene were performed to classify and determine the relative abundance of bacteria in the reactor. For the initial short-term (6 day) experiments, biomass samples were collected from the inoculum acid digester sludge and from the reactor every 2 days for 6 days. For the long-term (252 day) experiment, biomass samples were collected from the inoculum acid digester sludge and from the reactor at days 2, 4, and 6, and then every 6 days for the duration of the experiment. Biomass was harvested by centrifuging samples at a relative centrifugal force of 10,000g for 10 min and decanting supernatant. Biomass was then stored at − 80 °C until DNA extraction was performed.
DNA was extracted using a Power Soil® DNA Isolation Kit (MoBIO Laboratories 12888, Carlsbad, CA). The purity of extracted DNA was analyzed using a NanoDrop spectrophotometer (Thermo Fisher Scientific ND-2000, Waltham, MA), and DNA was quantified using a Qubit 3.0 (Thermo Fisher Scientific Q33126, Waltham, MA). The V3 and V4 regions of the 16S rRNA gene were amplified using the primer set S-D-Bact-0341-b-S-17/S-D-Bact-1061-a-A-17 as described by Klindworth et al. [24]. Amplicons were sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA) using pair-end 250 base pair kits at the University of Wisconsin-Madison Biotechnology Center.
Paired-end reads were merged with Fast Length Adjustment of Short Reads (FLASH) using default parameters [25]. The merged reads were analyzed with the Qiime pipeline, utilizing the split libraries command to remove low-quality sequences [26]. Sequences were clustered into operational taxonomic units (OTUs) using uclust [27]. Sequences were aligned with PyNast, and chimera detection was performed with ChimeraSlayer [28, 29]. Singleton OTUs were removed, and the samples were rarefied to an equal depth, with 130,000 sequences retained for the long-term (252 day) reactor experiment and 45,000 sequences retained for the short-term (6 day) reactor experiments. A representative sequence for each OTU was taxonomically classified using the SILVA database [30]. Tables of OTUs with taxonomic assignments are provided in Additional file 2. The Phyloseq package version 1.14.0 was used for data visualization and heat maps were generated with the superheat package [31, 32]. To construct phylogenetic trees, multiple sequence alignments were performed using MUSCLE, and maximum-likelihood phylogenetic trees were constructed with RAxML using the GTRGAMMA method with 1000 bootstraps [33, 34].
Statistical analysis of microbial community data was performed using multivariate repeated measures ANOVA with the nlme package in R to generate generalized least square models in which time was correlated to all predictor variables using the corAR1 structure [35]. Redundancy analysis was also performed using the rda command in the vegan package [36]. Environmental factors were iteratively selected until all were statistically significant (p < 0.1) based on 999 model permutations.
Technoeconomic analysis
To estimate the economic impact of producing MCFAs from ethanol stillage, a TEA was performed based on information provided in the National Renewable Energy Laboratory (NREL) TEA for a 61 million gallon per year lignocellulosic ethanol facility [2]. We assumed that switchgrass has a similar feedstock cost to corn stover ($58.50 U.S. dry ton−1), which is within the range of costs assumed for switchgrass feedstock in other studies [37, 38]. Instead of assuming all stillage undergoes anaerobic digestion, we assumed that a portion of the organic matter was converted to organic acids using data obtained in this study and then simulated the extraction of hexanoic and octanoic acids with ASPEN (AspenTech, Bedford, MA) to select an organic solvent and determine process separation efficiencies, heating demands, and sizes for reactors and equipment. We selected 2-octanol as the solvent for liquid–liquid extraction due to the high extraction efficiencies predicted with ASPEN. We assumed that the organic matter in the aqueous phase that remains after extracting the MCFAs was fed to the anaerobic digester to produce biogas. The specific methane yield (g methane produced per g COD consumed) and biosolid yield (g biomass produced per g COD consumed) were assumed to be the same as in the NREL TEA [2]. The efficiency of combined heat and power generation by combusting biogas, lignin, and biosolids was also assumed to have the same efficiency as the NREL TEA, with a total of 21% of the energy in the combusted material converted to usable heat and power [2].
The costs for additional reactors and distillation columns were estimated by scaling-related costs presented in the NREL TEA [2]. Costs for the liquid–liquid extraction were determined based on the volumetric flow rate and equations available in Seider et al. [39]. The KOH usage was calculated based on experimental reactor data. The 2-octanol demand (2-octanol lost to the aqueous phase) was based on modeling the liquid–liquid extraction with ASPEN. Prices for hexanoic acid, octanoic acid and 2-octanol were obtained from Zauba for imported quantities greater than 1000 kg in 2016 (Additional file 3) [40]. For consistency with past reporting, all costs and profits are reported in 2007 United States Dollars (USD). To convert from 2016 to 2007 USD, cost indices from the St. Louis Federal Reserve were used [41]. Electricity prices from the NREL TEA were used [2]. A 30-year cash flow was calculated using the cash-flow calculation tool available with the NREL TEA [42], and the minimum ethanol selling price (MESP) was determined by setting the net present value to zero based on a target 10% internal rate of return, consistent with the NREL TEA [2]. Detailed information related to the TEA is provided in Additional file 3.
COD calculations
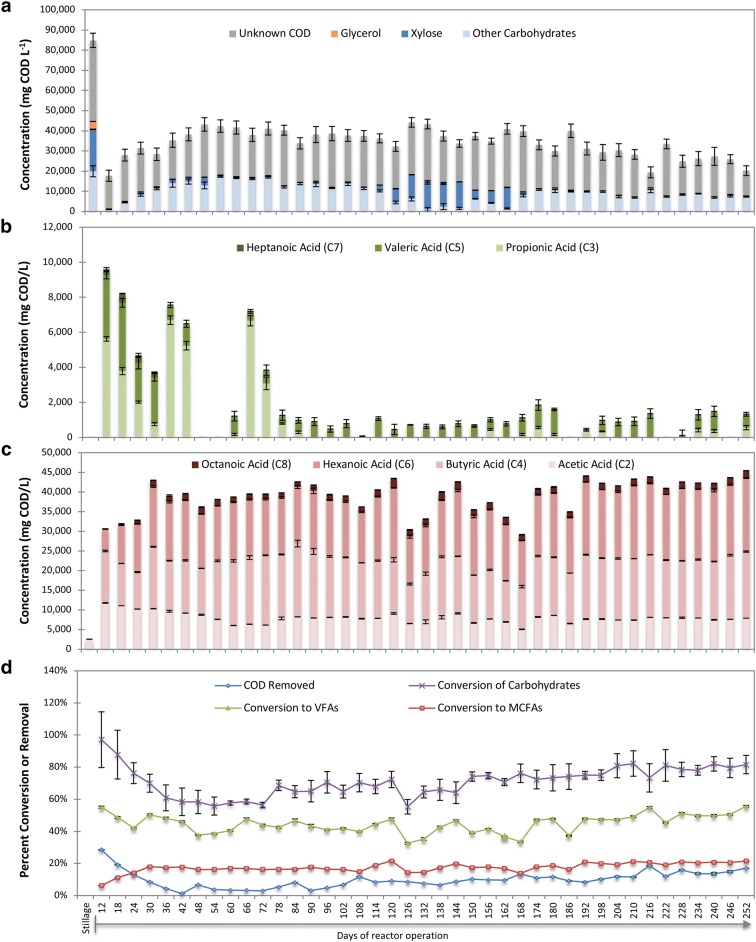
Unless otherwise noted, we report concentrations as mass of COD per unit volume. This allows for the direct comparison of relative reducing equivalents contained within each of the compounds consumed and created. The theoretical COD of each compound, or the theoretical amount of oxygen needed to fully oxidize the compound, was used to convert the measured mass units to COD. Protein was assumed to have 1.5 g COD per g of protein, which is consistent with the COD of albumin. A COD of 1.06 g COD per g carbohydrate was used to convert total carbohydrates measured with the anthrone method to COD. This value is consistent with the COD of glucose and xylose. The “Unknown COD” represents the measured COD minus the COD of known components. Where provided, error bars represent standard deviation of technical replicates. The “COD Removed” is calculated as the percentage of COD removed at each time point. “Conversion of Carbohydrates” is calculated based on the difference between total carbohydrates in the switchgrass stillage and the reactor sample for each time point. “Conversion to SCFA” is based on the amount of COD converted to carboxylic acids containing two to five carbons, and “Conversion to MCFA” is based on the COD converted to monocarboxylic acids containing six to eight carbons.
Results
Chemical analyses of switchgrass stillage
In a lignocellulosic biorefinery, an ethanologenic microorganism ferments biomass sugars to ethanol and the ethanol is removed via distillation, producing an organic-rich stillage fraction. The concentrations of compounds remaining in stillage are, therefore, dependent on the efficiency of the upstream fermentation. For this study, two batches of stillage (Table 1) were produced from switchgrass hydrolysate fermented with S. cerevisiae Y128, a strain with improved utilization of xylose [20]. The starting glucose and xylose concentrations in the hydrolysate prior to fermentation were 56,000 ± 300 mg COD L−1 and 36,000 ± 200 mg COD L−1, respectively. After the fermentation, the ethanol concentration was 51,000 ± 2900 mg COD L−1 with nearly 100% of the glucose and 47% of the xylose consumed. Glycerol, a common byproduct of yeast fermentation [43], reached a final concentration of 2500 ± 100 mg COD L−1. Acetic and formic acids decreased slightly during the ethanologenic fermentation, and only a small amount of lactic acid (30 ± 1 mg COD L−1) was detected (Additional file 1: Table S1). The total COD of the two batches of fermented hydrolysate was 160,000 ± 1500 mg COD L−1 (Additional file 1: Table S2).
Table 1.
Major chemical components contained within hydrolysate and fermented hydrolysate after fermentation with Saccharomyces cerevisiae Y128
| Hydrolysate | Fermented hydrolysate | |
|---|---|---|
| Glucose | 56,000 ± 300 | 44 ± 1.7 |
| Xylose | 36,000 ± 230 | 19,000 ± 4500 |
| Glycerol | 310 ± 0.86 | 2500 ± 130 |
| Acetic acid | 2065 ± 30 | 1600 ± 68 |
| Ethanol | < 100 | 51,000 ± 2900 |
The COD remaining in stillage, after distilling ethanol from the fermented hydrolysate, was approximately 60% of the COD in the fermented hydrolysate. The major chemical energy components in the stillage included xylose, acetamide (derived from acetate during ammonia-based pretreatment of switchgrass), glycerol, and acetic acid (Table 2). Residual glucose was minimal (Table 1), and the ethanol that was not removed in distillation (Table 2) represented less than 3% of the ethanol present in the original fermentation broth (Table 1). Carbohydrates, excluding xylose, accounted for 18% of the COD, while proteins accounted for only 2.2% of the COD in the stillage. In addition, a large portion of the COD is comprised of components with undetermined chemical identity. This “Unknown COD” likely contains a variety of compounds that are either produced during biomass deconstruction, originate from the switchgrass, or are produced during the yeast ethanol fermentation.
Table 2.
Composition of major organic matter components and aromatic compounds in the two batches of stillage fed to the mixed culture fermentation bioreactor
| Stillage batch 1 | Stillage batch 2 | |
|---|---|---|
| Major stillage components (mg COD L−1) | ||
| Soluble COD | 95,400 ± 432 | 95,800 ± 982 |
| Unknown COD | 38,300 ± 3250 | 42,100 ± 3190 |
| Xylose | 20,800 ± 148 | 20,900 ± 168 |
| Other carbohydrates | 19,300 ± 2310 | 15,500 ± 2230 |
| Acetamide | 4030 ± 270 | 4200 ± 340 |
| Glycerol | 3900 ± 32.1 | 3920 ± 36.3 |
| Acetic acid | 2550 ± 21.1 | 2580 ± 20.5 |
| Proteins | 2200 ± 145 | 1910 ± 162 |
| Ethanol | 1220 ± 305 | 1590 ± 161 |
| Aromatic compounds (μg COD L−1) | ||
| Coumaroyl amide | 13,000 ± 250 | 5400 ± 200 |
| Feruloyl amide | 12,000 ± 130 | 3200 ± 83 |
| p-Coumaric acid | 3500 ± 43 | 1100 ± 34 |
| Benzoic acid | 1700 ± 102 | 2000 ± 22 |
| Vanillamide | 290 ± 0.95 | 230 ± 0.50 |
| 4-Hydroxybenzoic acid | 380 ± 15 | 320 ± 0.46 |
| Vanillic acid | 320 ± 0.09 | 370 ± 4.6 |
| Ferulic acid | 250 ± 13 | 90 ± 3.2 |
| 4-Hydroxybenzyl alcohol | 240 ± 3.7 | 110 ± 1.9 |
| Syringamide | 230 ± 0.06 | 138 ± 2.3 |
Major stillage components are reported in mg COD L−1 whereas aromatic compounds are reported in μg COD L−1
While major COD components between the two batches of stillage were similar, the aromatic compounds, including known lignotoxins [19, 44], varied between the stillage batches (Table 2). Feruloyl amide, p-coumaroyl amide, and coumaric acid were higher in batch 1 than in batch 2. Only benzoic acid and vanillic acid were higher in batch 2. From a reducing-equivalent standpoint, these aromatic compounds account for less than 0.05% of the COD in stillage, but these concentrations are within the range of lignotoxins shown to inhibit fermentation activity by pure cultures of ethanologenic organisms [20].
Stillage fermentation under different pH conditions
Due to the relatively low concentration of six-carbon sugars, the complexity of remaining organic materials, and the potential toxicity of aromatic compounds, bacterial growth on stillage derived from AFEX-treated hydrolysate was expected to be challenging. We, therefore, conducted short-term experiments to determine if a microbial community could metabolize organic materials remaining in stillage. Using inoculum from an acid-phase anaerobic digester, we fermented stillage at different pH conditions (uncontrolled, 5.0, 5.5, 6.0, and 6.5) for 6 days utilizing a SRT of 2 days and analyzed both the extracellular end products and the microbial community. Acid-phase digester sludge was used as inoculum because the microbial consortia were expected to contain a variety of fermenting organisms and not expected to contain high levels of methanogens [45, 46].
Conditions in which the pH was uncontrolled led to the pH stabilizing at 3.6 and accumulation of lactic and acetic acids (Additional file 4). SCFA accumulated in the reactor when the pH was maintained between 5.0 and 6.5. Maintaining a pH of 5.5 resulted in the highest accumulation of MCFAs (Additional file 4). Analysis of the microbial community by 16S rRNA gene sequencing showed variations in composition with pH (Additional file 5), with OTUs associated with the genera Lactobacillus (89.9%) and Acetobacter (9.9%) becoming the most abundant when the pH was uncontrolled. Lactobacillus was present in the reactors at all pH conditions. At pH 5.0, Megasphaera was enriched (46.3%), while at pH 5.5, OTUs related to Pseudoramibacter (14.3%) and Olsenella (14.1%) were abundant. At pH 6.0, Mitsuokella (20.8%), Acetitomaculum (17.0%), and Megasphaera (14.2%) were all abundant. When the reactor was maintained at pH 6.5, more OTUs related to the Bacteroidetes phylum were abundant, including OTUs related to the genera Prevotella (12.3%) and Bacteroides (40.8%).
These results demonstrated that a community derived from an acid digester sludge inoculum could ferment stillage to carboxylic acids, including MCFAs, under a variety of pH conditions. Further, organisms identified in the stillage-fed reactors included members of the Clostridia (Megasphaera, Pseudoramibacter) that have previously been associated with MCFA production [5, 10, 13, 15, 18, 47]. Members of Clostridia have been enriched in other MCFA-producing bioreactors under similar pH conditions [12, 18, 48]. In agreement with our observation of Lactobacillus at all pH conditions, Lactobacillus is a common genus in MCFA-producing microbiomes [10, 15, 17, 18, 47]. In total, the fermentation product (Additional file 4) and community (Additional file 5) data confirmed that materials in stillage could be converted to MCFAs by a microbial community originating from a full-scale wastewater treatment plant acid digester.
Sustained MCFA production from switchgrass stillage
Based on these results, we chose to control the reactor pH at 5.5 for a long-term experiment to demonstrate sustained production of MCFAs. Initially, xylose and other carbohydrates were consumed, and a mixture of odd- and even-chain linear fatty acids was produced (Fig. 2a–c). The maximum utilization of carbohydrates was achieved at Day 12, with 97 ± 17% of the measured initial carbohydrates consumed (Fig. 2d). During the first 30 days of operation, accumulation of monocarboxylic acids steadily increased, reaching nearly 50% conversion of COD in stillage to monocarboxylic acids (Fig. 2d). As reactor operation continued, the concentration of odd-chain monocarboxylic acids (C3, C5 and C7) decreased (Fig. 2b) while that of even-chain acids increased (Fig. 2c). From Day 30 through Day 252, the average conversion of COD in stillage to MCFAs was 18 ± 2.1%, and MCFAs accounted for 41 ± 7.0% of the total monocarboxylic acids produced.
Fig. 2.
Mixed culture fermentation reactor performance for 252 days. a Compounds removed from stillage; b production of odd-chain propionic (C3), valeric (C5), and heptanoic (C7) acids; c production of even-chain acetic (C2), butyric (C4), hexanoic (C6), and octanoic (C8) acids; d removal of COD, percent conversion of carbohydrates, and percent conversions of COD to SCFA (C2–C5) and MCFAs (C6–C8)
Microbial community analysis
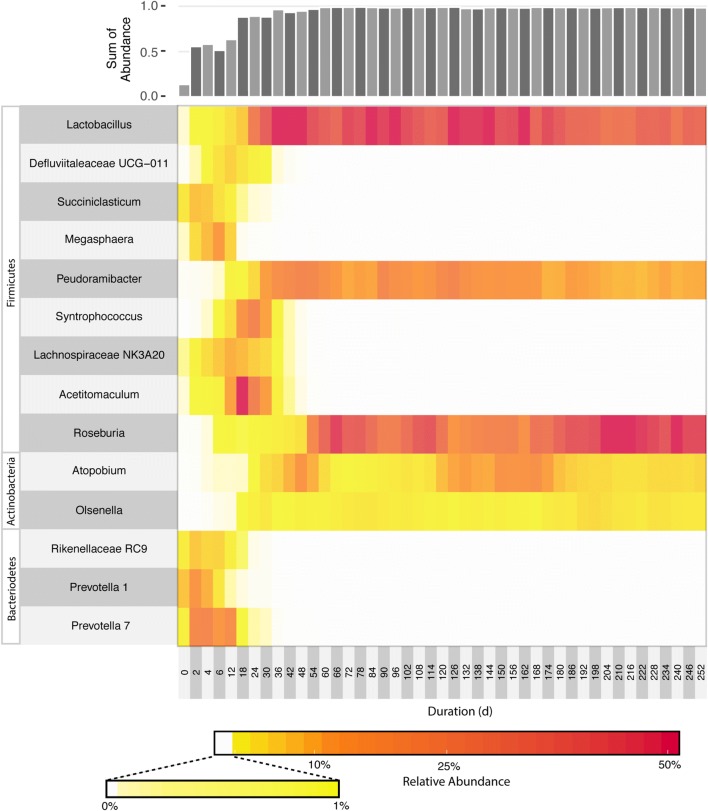
We used 16S rRNA gene amplicon sequencing to assess the members of the microbial community in this bioreactor and any changes that occurred in its composition as a function of time (Fig. 3; Additional file 6). The initial microbial community contained many Proteobacteria, Firmicutes, and Bacteroidetes (Additional file 6). Early on in reactor operations, Bacteroidetes and Firmicutes became the most abundant organisms, with Prevotella species accounting for most of the Bacteroidetes. The increase in abundance of Prevotella 7 (Fig. 3) corresponds with the time of increased carbohydrate conversion (p < 0.001), in agreement with Prevotella’s described ability to degrade polysaccharides and other complex substrates [49]. Megasphaera, an organism known to produce odd-chain fatty acids (OCFA) [50], was present in the inoculum and increased in abundance during the early phase of reactor operation. The high abundance of Megasphaera (p = 0.0023) and Prevotella 7 (p = 0.0016) at early stages of reactor operation corresponded with a period of higher OCFA production.
Fig. 3.
Relative abundance of bacteria in the mixed culture fermentation reactor for 252 days. Day 0 corresponds to the acid digester sludge inoculum. Bacterial abundance is summarized based on the genera assigned by annotating representative sequences with the SILVA database. The sum of abundance represents the percentage of OTUs contained within the indicated genera. A heat map of the top 100 OTUs is provided in Additional file 3: Figure S3 and a table of all OTUs are provided in Additional file 4: Table S1
After extended operation, we found that the community composition stabilized and was dominated by organisms from five genera, including three Firmicutes (Lactobacillus, Pseudoramibacter and Roseburia) and two Actinobacteria (Olsenella and Atopobium). At later time points (Day 30–Day 252), the OTUs corresponding to these five genera accounted for greater than 95% of the total 16S rRNA gene sequences (Fig. 3). The relative abundance of Pseudoramibacter (p = 0.0045), Lactobacillus (p = 0.0022), and Olsenella (p = 0.014) all correlated with the period of increased MCFA production. Neither Roseburia (p = 0.147) nor Atopobium (p = 0.546) are significantly correlated to increased MCFA production.
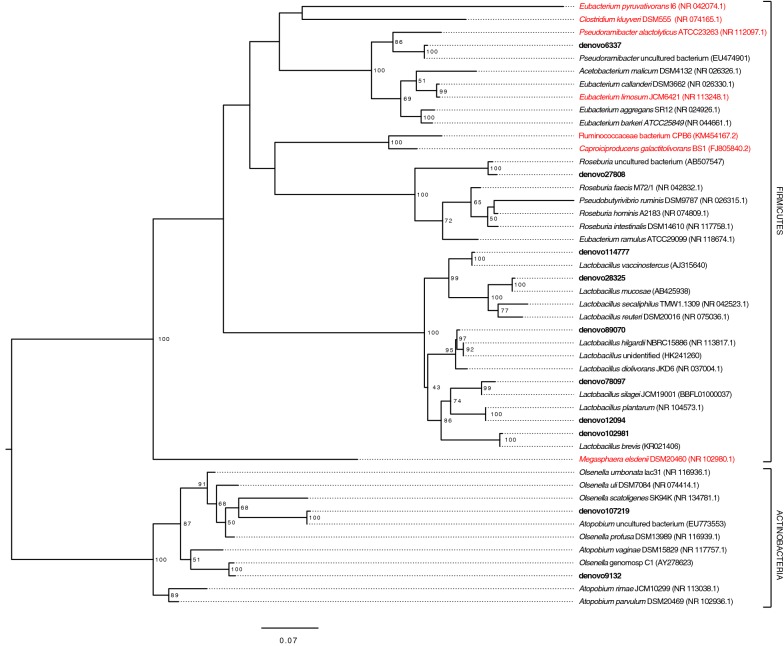
Representative sequences for the most abundant OTUs were used to construct a maximum-likelihood phylogenetic tree (Fig. 4). The six high-abundance Lactobacillus OTUs (denovo114777, denovo28325, denovo102981, denovo12094, denovo78097, and denovo89070) clustered with known xylose-consuming, heterofermentative Lactobacilli (L. mucosae, L. plantarum, L. silagei, L. brevis, L. vaccinostercus and L. diolivorans) [51–58]. As lactic acid has previously been demonstrated as a substrate for MCFA production [14, 15], it may be a key intermediate for MCFA production in a microbial community [18]. While significant lactic acid accumulation was not observed during steady-state sampling, when we monitored time-dependent changes in the reactor after stillage was spike-fed, lactic acid transiently accumulated to detectable levels in the medium (Fig. 5) suggesting that lactic acid is produced but consumed by other community members.
Fig. 4.
Phylogenetic tree including the top ten most abundant OTUs at Day 252. OTUs from this study are shown in bold text. Known chain-elongating bacteria are shown in red text. Bootstrap values greater than 50 are shown, and the phylogenetic tree is rooted to the Actinobacteria phylum. The horizontal branch distance corresponds to the mean nucleotide substitutions per sequence site. The accession numbers for 16S rRNA gene sequences for the indicated bacteria are provided in parentheses
Fig. 5.
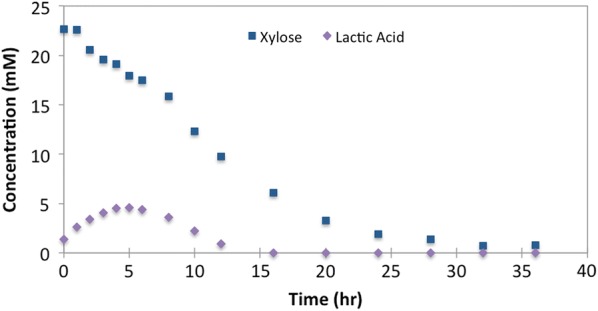
Time-dependent changes of xylose and lactic acid concentrations. Lactic and xylose were measured after adding a spike feed of 25 mL stillage to the reactor at 252 days of operation. As xylose is removed from the media, lactic acid accumulates at approximately 1 mol of lactic acid per mol of xylose consumed. Extracellular lactic acid begins to decrease 6 h after the addition of stillage
The two OTUs within the Actinobacteria phylum, denovo9132 and denovo107219, clustered with members of the Atopobium and Olsenella genera, respectively (Fig. 4), in the Coriobacteriaceae family. Several Atopobium and Olsenella species have been shown to consume carbohydrates and produce lactic acid [59–62]. The most abundant OTU at 252 days of reactor operation (denovo27808) clustered with Roseburia, which are known to utilize carbohydrates and acetic acid and produce butyric and lactic acids [63–65]. Another high-abundance OTU identified in the microbial community (denovo6337) clustered with Pseudoramibacter alactolyticus (previously Eubacterium alactolyticum), a bacterium that has been described as producing hexanoate and octanoate from lactic acid and glucose [66].
Starting at Day 120, the feed changed from stillage batch 1 to stillage batch 2, which contained lower concentrations of aromatic compounds (Table 2). While initial changes in community compositions occurred (Fig. 3), with an increase in Atopobium and decrease in Roseburia, the major genera remained consistent and the community eventually re-stabilized. This initial change in stillage feed source coincided with a reduction in xylose utilization (Fig. 2); however, xylose utilization eventually increased and overall MCFA production was not impacted by this change in the stillage source (p = 0.415).
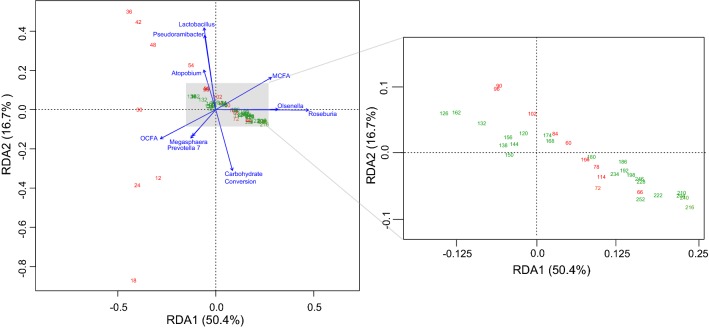
We also performed redundancy analysis to relate the community composition with MCFA production, odd-chain fatty acid production (OCFA), and carbohydrate conversion and to investigate co-occurrence of abundant bacteria in the reactor (Fig. 6). For early time points (Days 12–24), the abundance of Prevotella and Megasphaera correlates with OCFA production. The analysis also showed that higher relative abundance of Lactobacillus is associated with higher relative abundance of Pseudoramibacter and higher relative abundance of Roseburia correlates with higher relative abundance of Olsenella (Fig. 6). These correlations suggest that these organisms may work in tandem during stillage metabolism. In the case of Lactobacillus and Pseudoramibacter, the Lactobacillus may be producing lactate that Pseudoramibacter converts to MCFAs. This relationship is analogous to that suggested by Andersen et al. in which Megasphaera utilized lactate generated by Lactobacillus [17]. Similarly, Olsenella may be producing intermediates, such as acetate, that are known to be utilized by Roseburia.
Fig. 6.
Redundancy analysis of microbial community and fermentation reactor performance. The percent variance explained by each axis is indicated in parentheses. The numbers within the plot indicate the day of sampling. Red numbers indicate time points in which stillage batch 1 was fed to the reactor, and green numbers indicate time points in which stillage batch 2 was fed to the reactor. Arrows indicate environmental factors related to community composition and the sum of individual genera within the bacterial community. These include the sum of propionic, valeric, and heptanoic acids (OCFA; p = 0.001), the sum of hexanoic, heptanoic, and octanoic acids (MCFAs; p = 0.001), and the total percentage of carbohydrates converted to fermentation products (carbohydrate conversion; p = 0.001)
Overall, the bacterial community results indicate that a stable fermenting community containing only five genera was enriched from the acid-digester sludge inoculum during growth on stillage. We suspect that Clostridia-related organisms (Pseudoramibacter and/or Roseburia) are responsible for MCFA production and the remaining community members ferment sugars to intermediate compounds (acetate, lactate, and/or ethanol) that provide substrates for MCFA production.
Economic analysis of MCFA production from stillage
Based on the sustained production of MCFAs in this study, we evaluated the potential value of this process. We did this by modifying the NREL TEA for a lignocellulosic ethanol biorefinery to include a process in which stillage is used to produce MCFAs. Using average percent conversions in the bioreactor between Day 30 and Day 252, we estimated that the COD remaining in stillage was converted to end products at the following percentages: 5.4% acetic acid, 15% butyric acid, 16% hexanoic acid, and 1.7% octanoic acid. Further, based on reactor operations during the same time period, 9.1% of the COD is removed from the system as off-gas.
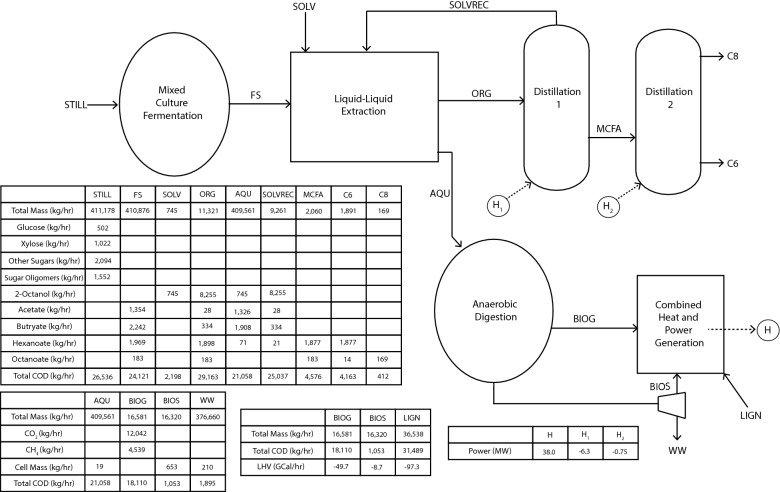
Based on these conversions, a new mass and energy balance for the biorefinery was determined (Fig. 7). The MCFA-producing fermentation reactor was sized for a SRT of 6 days, yielding an estimated total reactor volume of 16 million gallons. Software simulations predicted that a solvent flow rate of 9000 kg h−1 was needed to recover 99.9% of the octanoic acid and 96.4% of the hexanoic acid, respectively. Software simulations further predicted that of the 9000 kg h−1 2-octanol feed, 745 kg h−1 separates into the aqueous phase and needs to be replenished. In our TEA, the organic phase undergoes a column distillation to remove 2-octanol that has a volume of 630 ft3 and requires a total heating duty of 6.3 MW. After distilling the solvent, the model assumes that hexanoic and octanoic acids are separated in a second distillation column with a volume of 240 ft3 that requires a total heating duty of 0.75 MW.
Fig. 7.
Proposed mass and energy balance for co-production of ethanol and MCFAs from lignocellulosic ethanol stillage. The following process flow indicators are used: STILL stillage, FS fermented stillage, SOLV organic solvent, AQU aqueous phase, SOLVREC recovered solvent, BIOG biogas, BIOS biosolids, WW wastewater, LIGN lignin, H generated heat and power, H1 heating duty for distillation column 1, H2 heating duty for distillation column 2
After the liquid–liquid extraction, the aqueous phase is fed to biogas-producing anaerobic digesters (Fig. 7). Anaerobic digestion of lignocellulosic stillage [67] and acid-digested stillage [68] for biogas production has been demonstrated by others. The mass flow rate of COD to the anaerobic digesters, including stillage, lignin, and biosolids, is 21,000 kg h−1, resulting in biogas production of 16,600 kg h−1 (compared to 21,900 kg h−1 if the stillage is used directly as in the NREL TEA) [2]. The overall power generation from the remaining organics after MCFA removal is reduced from 41.0 MW to 38.0 MW. The reduction in overall power generation is small because lignin contributes the majority of COD to the anaerobic digesters.
As a result of the additional heating demands for the MCFA distillation columns, the net electricity that can be sold decreased from 13.7 to 3.8 MW (Table 3). In addition, the capital costs associated with the stillage fermentation reactor, liquid–liquid extraction, and distillation columns increased the total capital investment from $423 million to $441 million. The additional chemical costs for KOH and 2-octanol added annual operating costs of $14 and $8.3 million, respectively. However, the MCFA products increased revenue by $57 million ($7.5 million from octanoic acid, and $47.5 million from hexanoic acid). Based on a 30-year cash flow with a 10% internal rate of return, the minimum ethanol selling price was determined to be $1.76 per gallon ($2.68 per gallon gasoline-equivalent) (Additional file 7). This is 18% lower than the $2.15 per gallon for when electricity is generated as the only co-product to ethanol [2].
Table 3.
Summary of technoeconomic analysis for co-production of ethanol, electricity, and MCFAs in a lignocellulosic biorefinery
| Description | Size | No. | Installation factor | Total capital and installation cost |
|---|---|---|---|---|
| Additional capital expenses for MCFA production | ||||
| Mixed culture fermentation reactors | 4 MG | 4 | 1.0a | $7,540,000 |
| Mixed culture fermentation agitators | 30 hp | 4 | 1.5 | $317,000 |
| Mixed culture fermentation feed pumps | 2500 gpm | 4 | 1.0a | $227,000 |
| Caustic feed system | 300 gph | 4 | 1.0a | $29,800 |
| Liquid–liquid extraction | 4700 ft3 | 2 | 2.4 | $860,000 |
| Solvent feed system | 50 gpm | 1 | 2.4 | $29,200 |
| Distillation column 1 (solvent recovery) | 630 ft3 | 1 | 2.4 | $1,420,000 |
| Distillation column 2 (MCFA separation) | 240 ft3 | 1 | 2.4 | $797,000 |
| Description | Cost per ton | Annual operating expense | ||
|---|---|---|---|---|
| Additional operating expenses for MCFA production | ||||
| KOH | $866 | $14,000,000 | ||
| 2-Octanol | $1402 | $9,660,000 | ||
| Description | Production | Annual revenue | ||
|---|---|---|---|---|
| Additional revenue for MCFA production | ||||
| Electricity | 3759 kW | $1,810,000 | ||
| HA | 1877 kg h−1 | $47,450,000 | ||
| OA | 169 kg h−1 | $7,490,000 | ||
| Description | Ethanol–electricity co-productionb | Ethanol–electricity–MCFA co-production | ||
|---|---|---|---|---|
| Comparison of ethanol–electricity co-production to ethanol–electricity–MCFA co-production | ||||
| Total co-product revenue | $6,600,000 | $56,800,000 | ||
| Total capital investment | $422,900,000 | $441,200,000 | ||
| MESP | $2.15 | $1.76 | ||
| MESP (gasoline equivalents) | $3.27 | $2.68 | ||
Discussion
Our work illustrates the potential of using microbial communities to convert stillage into valuable co-products. In the stillage-fed bioreactor, productivities of hexanoic (2.6 ± 0.3 g L−1 day−1) and octanoic (0.27 ± 0.04 g L−1 day−1) acids were sustained for 214 days with titers at 66 ± 8.2 and 97 ± 15% of their solubility in water, respectively. These productivities are consistent with other studies investigating the conversion of organic substrates derived from lignocellulosic materials or ethanol production wastes to MCFAs (Additional file 8: Table S17). Our system is unique, however, in that the primary carbohydrate consumed is xylose and the stillage has already been depleted of a large portion of fermentable sugars and the ethanol that others have used to produce MFCA. While we are proposing the co-production of ethanol and MCFAs in this study, recent work has also explored production of MCFA as the main product of a lignocellulosic biorefinery. In work performed by Nelson et al., Megasphaera consumed glucose in lignocellulosic hydrolysate to generate hexanoic acid, but xylose was not consumed [16]. The microbial community like the one presented in this study could be utilized to convert the remaining xylose to MCFAs.
The simplicity of the microbial community enriched in this study positions it well as a model community for MCFA production. Others have shown enrichments containing OTUs related to primary sugar fermenters, such as Lactobacillus, and OTUs related to Clostridia that may be involved in converting intermediate fermentation products to MCFA [13–15, 17, 18, 47, 69]. In our microbial community, at Day 252, only 10 OTUs are present at greater than 1% relative abundance, and these OTUs make up 89.3% of the total OTUs (Additional file 5: Figure S3). The statistical analyses indicate that Pseudoramibacter and Lactobacillus are co-enriched, and their abundance correlates with higher MCFA production. We, therefore, propose that Lactobacillus converts xylose to lactate and acetate by heterofermentation, and the lactate is elongated to MCFAs by Pseudoramibacter. While 16S rRNA gene sequencing allows for the phylogeny of abundant organisms to be estimated, the function of community members should be investigated further utilizing metagenomic approaches. Due to the simplicity of the microbial community obtained in this study, this microbiome is well positioned for further investigation with metagenomic tools. Furthermore, its simplicity makes this a candidate microbiome for simulation with synthetic communities in the future. Of the OTUs that became enriched in the reactor, only Roseburia (denovo27808) and Pseudoramibacter (denovo6337) emerged as likely MCFA-producing bacteria. While Pseudoramibacter have been shown to produce MCFAs [66], to our knowledge, the ability of Roseburia to produce MCFAs has not been studied.
The TEA shows that even at the modest productivities of hexanoic and octanoic acids obtained in this study, MCFAs produced from ethanol stillage could improve the economic feasibility of lignocellulosic biorefining if the productivity can be maintained at industrial scale. Improvements in the overall conversion of stillage COD to MCFAs and production of a higher proportion of octanoic acid would further increase the revenue that can be generated by this strategy. Increasing MCFA product specificity towards octanoic acid is an ongoing area of research. One strategy to increase octanoic acid production is to utilize pertractive extraction of MCFAs to reduce product inhibition, as has been performed in past studies [13, 14]. Recent work has also shown that increasing the ratio of ethanol to acetate increases selectivity of octanoic acid production [13]. The model of increasing the ratio of reduced electron donors to acetate suggests that, in the absence of ethanol, increasing the production of lactate as a fermentation intermediate (rather than acetate) could further drive octanoic acid production.
The economy of co-producing MCFAs may also be affected by upstream biomass processing (i.e., the conversion of plant polymers to their constituent monomer units) and ethanol fermentation. For example, utilization of xylose by industrial yeast strains, such as S. cerevisiae, is limited [20], although attempts to improve pentose utilization by ethanol producers is an area of intense research activity [70]. Even though the S. cerevisiae Y128 strain used in this study was engineered for improved xylose utilization, it only consumed 47% of the xylose available in the switchgrass hydrolysate. Future ethanologenic organisms used in a lignocellulosic biorefinery may leave less xylose available for MCFA production. However, given the higher price of MCFAs compared to ethanol, decreasing xylose consumption by the ethanologenic organism may actually result in an improved economy of the lignocellulosic biorefinery.
Another simple opportunity for improving the economic potential of co-producing MCFAs is utilizing sodium hydroxide for pH control, instead of KOH, as sodium hydroxide is roughly one-sixth the cost of KOH. In our current model, the cost of KOH (Table 3) is a major expense. Alternatives to controlling pH with chemicals, such as electrolytic extraction which both controls the pH and extracts the acid products [17], should also be explored further.
Conclusion
In this study, we tested the hypothesis that microbial communities could be used to produce valuable compounds from lignocellulosic stillage. We developed conditions for sustained MCFA production by an anaerobic microbiome that uses stillage produced during lignocellulosic biorefining. By fermenting switchgrass stillage, we maintained productivities of hexanoic and octanoic acids of 2.6 ± 0.3 and 0.27 ± 0.04 g L−1 day−1, respectively. To our knowledge, this is the first demonstration of MCFA production with xylose and other organics in lignocellulosic ethanol stillage as the primary substrates. The MCFA-producing microbial community was derived from a diverse wastewater treatment ecosystem, but over time it became enriched with OTUs representing only five genera, including members of the Firmicutes phylum (Lactobacillus, Roseburia, and Pseudoramibacter) and of the Actinobacteria phylum (Olsenella and Atopobium). Pseudoramibacter are Clostridia related to known MCFA-producing organisms, some of which have been shown to produce hexanoic and octanoic acids [66].
A TEA, based on an update to an industry-accepted model, shows that, at the productivity of MCFAs achieved in this study, valorizing lignocellulosic ethanol stillage to MCFAs could improve the economic sustainability of a biorefinery. For example, using the MCFA production experimentally observed, if 16% of the COD remaining in stillage is converted to hexanoic acid and 1.7% is converted to octanoic acid, the minimum ethanol selling price could be reduced by 18%, from $2.15 to $1.76 gal−1. Optimization of microbiome MCFA productivities, MCFA extraction, solvent recovery and selection of the ethanologenic organism may contribute further to improving the economy of the lignocellulosic biorefinery.
Additional files
Additional file 1. Chemical analyses of stillage production and mixed culture fermentation reactor. Table S1. Results from two fermentations of switchgrass hydrolysate with Saccharomyces cerevisiae Y128. Table S2. Soluble COD results for fermented switchgrass hydrolysate. Table S3. Summary of organic components in switchgrass stillage and the mixed culture fermentation reactor. Table S4. Total soluble COD in switchgrass stillage and mixed culture fermentation reactor. Table S5. Components in stillage and mixed culture fermentation reactor samples measured by GC-MS and HPLC analyses. Table S6. Total soluble carbohydrates measured in stillage and mixed culture fermentation reactor with the anthrone method. Table S7. Total soluble proteins in stillage and mixed culture fermentation reactor measured with the BCA assay. Table S8. Summary of aromatic compounds in stillage.
Additional file 2. Rarefied tables of operational taxonomic units (OTUs) and classification with Silva. Table S9. OTU table for 6d experiments testing different pH conditions. Table S10. OTU table for 252 days of reactor operation at pH 5.5.
Additional file 3. Data used for development of the technoeconomic analysis (TEA). Table S11. Summary of process modeling for liquid-liquid extraction, solvent distillation, and MCFA separation with ASPEN. Table S12. Summary of capitol expenses added to the NREL TEA. Table S13. Summary of operating expenses and additional product revenues added to NREL TEA. Table S14. 30 year cash flow from NREL TEA updated to include MCFA production expenses and revenues. Table S15. Calculations for additional capital expenses. Table S16. Chemical costs for octanoic acid, hexanoic acid and 2-octanol obtained from Zauba.
Additional file 4. Chemical analysis for mixed culture fermentations after 6 days under different pH conditions.
Additional file 5. Heat map of most abundant OTUs in the initial mixed culture fermentation experiments under different pH conditions.
Additional file 6. Heat map of 100 most abundant OTUs from 252 days of mixed culture fermentation reactor operations.
Additional file 7. Summary of Technoecomoic Analysis (Output from NREL TEA Model).
Additional file 8: Table S17. Comparison of hexanoic (C6) and octanoic (C8) acid productivities and titers for this study and other studies utilizing cellulosic or ethanol-production derived substrates.
Authors’ contributions
MJS designed and performed the experiments, analyzed the data, and wrote the manuscript. GL and MD assisted with data collection, technoeconomic analyses, and manuscript preparation. MM developed GC–MS methods and performed chemical analyses of samples. TJD and DRN assisted with study design and were major contributors to writing the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Yaoping Zhang, Ed Pohlmann, and Dan Xie for providing the fermented switchgrass hydrolysate used in this study and Tate Hattervig, Chantel Roberts, and Jesus Santiago for performing testing related to reactor monitoring. We further thank Matt Seib and the Madison Metropolitan Sewerage District for providing the acid digester sludge inoculum and Duygu Celik for assistance with initial reactor setup. We also thank Jason Russell and the Coon Laboratory for LC–MS/MS analyses. We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing Illumina sequencing services.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The raw sequencing reads from 16S rRNA gene amplicon sequencing are available under NCBI BioProject ID PRJNA393345 (252-day experiment) and NCBI BioProject ID PRJNA432807 (6-day experiments). Stillage and reactor data are available in Additional file 1 and data used for the TEA are provided in Additional file 3.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science Grants DE-FC02-07ER64494 and DE-SC0018409). MJS is supported by the National Science Foundation Graduate Research Fellowship Program under Grant no. DGE-1256259.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1193-x) contains supplementary material, which is available to authorized users.
Contributor Information
Matthew J. Scarborough, Email: mscarborough@wisc.edu
Griffin Lynch, Email: gtlynch@wisc.edu.
Mitch Dickson, Email: mwdickson@uwalumni.com.
Mick McGee, Email: mmcgee2@wisc.edu.
Timothy J. Donohue, Email: tdonohue@bact.wisc.edu
Daniel R. Noguera, Email: dnoguera@wisc.edu
References
- 1.Gerbrandt K, Chu PL, Simmonds A, Mullins KA, MacLean HL, Griffins WM, Saville BA. Life cycle assessment of lignocellulosic ethanol: a review of key factors and methods affecting calculated GHG emissions and energy use. Curr Opin Biotechnol. 2016;38:63–70. doi: 10.1016/j.copbio.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Humbird D, Davis R, Tao L, Kinchin C, Hsu D, Aden A, Schoen P, Lukas J, Olthof B, Worley M et al. Process design and economics for biochemical conversion of lignocellulosic biomass to ethanol. NREL Technical Report: NREL; 2011.
- 3.Senate and house of representatives of the United States of America. Energy Policy Act of 2005. In: Public Law 109-58. Washington, D.C.: US Government Publishing Office; 2005.
- 4.Senate and house of representatives of the United States of America. Energy Independence and Security Act of 2007. In: America SaHoRotUSo (eds). Public Law 110–140. Washington, D.C.: US Government Publishing Office; 2007.
- 5.Angenent LT, Richter H, Buckel W, Spirito CM, Steinbusch KJJ, Plugge CM, Strik D, Grootscholten TIM, Buisman CJN, Hamelers HVM. Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ Sci Technol. 2016;50:2796–2810. doi: 10.1021/acs.est.5b04847. [DOI] [PubMed] [Google Scholar]
- 6.Sarria S, Kruyer NS, Peralta-Yahya P. Microbial synthesis of medium-chain chemicals from renewables. Nat Biotechnol. 2017;35:1158–1166. doi: 10.1038/nbt.4022. [DOI] [PubMed] [Google Scholar]
- 7.Gelfand I, Sahajpal R, Zhang X, Izaurralde RC, Gross KL, Robertson GP. Sustainable bioenergy production from marginal lands in the US Midwest. Nature. 2013;493:514–517. doi: 10.1038/nature11811. [DOI] [PubMed] [Google Scholar]
- 8.Agler MT, Wrenn BA, Zinder SH, Angenent LT. Waste to bioproduct conversion with undefined mixed cultures: the carboxylate platform. Trends Biotechnol. 2011;29:70–78. doi: 10.1016/j.tibtech.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Holtzapple MT, Granda CB. Carboxylate platform: the MixAlco process part 1: comparison of three biomass conversion platforms. Appl Biochem Biotechnol. 2009;156:95–106. doi: 10.1007/s12010-008-8466-y. [DOI] [PubMed] [Google Scholar]
- 10.Agler MT, Spirito CM, Usack JG, Werner JJ, Angenent LT. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates. Energy Environ Sci. 2012;5:8189. doi: 10.1039/c2ee22101b. [DOI] [Google Scholar]
- 11.Urban C, Xu J, Sträuber H, dos Santos Dantas TR, Mühlenberg J, Härtig C, Angenent LT, Harnisch F. Production of drop-in fuels from biomass at high selectivity by combined microbial and electrochemical conversion. Energy Environ Sci. 2017;10:2231–2244. doi: 10.1039/C7EE01303E. [DOI] [Google Scholar]
- 12.Vasudevan D, Richter H, Angenent LT. Upgrading dilute ethanol from syngas fermentation to n-caproate with reactor microbiomes. Bioresour Technol. 2014;151:378–382. doi: 10.1016/j.biortech.2013.09.105. [DOI] [PubMed] [Google Scholar]
- 13.Kucek LA, Spirito CM, Angenent LT. High n-caprylate productivities and specificities from dilute ethanol and acetate: chain elongation with microbiomes to upgrade products from syngas fermentation. Energy Environ Sci. 2016;9:3482–3494. doi: 10.1039/C6EE01487A. [DOI] [Google Scholar]
- 14.Kucek LA, Nguyen M, Angenent LT. Conversion of l-lactate into n-caproate by a continuously fed reactor microbiome. Water Res. 2016;93:163–171. doi: 10.1016/j.watres.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Tao Y, Liang C, Li X, Wei N, Zhang W, Zhou Y, Yang Y, Bo T. The synthesis of n-caproate from lactate: a new efficient process for medium-chain carboxylates production. Sci Rep. 2015;5:14360. doi: 10.1038/srep14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R, Peterson D, Karp E, Beckham G, Salvachúa D. Mixed carboxylic acid production by megasphaera elsdenii from glucose and lignocellulosic hydrolysate. Fermentation. 2017;3:10. doi: 10.3390/fermentation3010010. [DOI] [Google Scholar]
- 17.Andersen SJ, Candry P, Basadre T, Khor WC, Roume H, Hernandez-Sanabria E, Coma M, Rabaey K. Electrolytic extraction drives volatile fatty acid chain elongation through lactic acid and replaces chemical pH control in thin stillage fermentation. Biotechnol Biofuels. 2015;8:221. doi: 10.1186/s13068-015-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen SJ, De Groof V, Khor WC, Roume H, Props R, Coma M, Rabaey K. A Clostridium group IV species dominates and suppresses a mixed culture fermentation by tolerance to medium chain fatty acids products. Front Bioeng Biotechnol. 2017;5:8. doi: 10.3389/fbioe.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong RG, Higbee A, Bottoms S, Dickinson Q, Xie D, Smith SA, Serate J, Pohlmann E, Jones AD, Coon JJ, et al. Inhibition of microbial biofuel production in drought-stressed switchgrass hydrolysate. Biotechnol Biofuels. 2016;9:237. doi: 10.1186/s13068-016-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parreiras LS, Breuer RJ, Avanasi Narasimhan R, Higbee AJ, La Reau A, Tremaine M, Qin L, Willis LB, Bice BD, Bonfert BL, et al. Engineering and two-stage evolution of a lignocellulosic hydrolysate-tolerant Saccharomyces cerevisiae strain for anaerobic fermentation of xylose from AFEX pretreated corn stover. PLoS ONE. 2014;9:e107499. doi: 10.1371/journal.pone.0107499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Public Health Association WEF, and American Water Works association standard methods for the examination of water and wastewater, 21st edition. American Public Health Association; 2005.
- 22.Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochem J. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 24.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barter RL, Yu B. Superheat: An R package for creating beautiful and extendable heatmaps for visualizing complex data. LIbrary CU (ed). Statistics applications; 2017. [DOI] [PMC free article] [PubMed]
- 33.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinheiro J. Package ‘nlme’. In. 2017.
- 36.vegan: Community Ecology Package. R package version 2.4-1. https://CRAN.R-project.org/package=vegan. Accessed 1 Dec 2015
- 37.Kumar A, Sokhansanj S. Switchgrass (Panicum vigratum L.) delivery to a biorefinery using integrated biomass supply analysis and logistics (IBSAL) model. Bioresour Technol. 2007;98:1033–1044. doi: 10.1016/j.biortech.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 38.Sokhansanj S, Mani S, Turhollow A, Kumar A, Bransby D, Lynd L, Laser M. Large-scale production, harvest and logistics of switchgrass (Panicum virgatum L.)—current technology and envisioning a mature technology. Biofuels Bioprod Bioref. 2009;3:124–141. doi: 10.1002/bbb.129. [DOI] [Google Scholar]
- 39.Seider WD, Seader JD, Lewin DR, Widagdo S. Product and process design principles: synthesis, analysis, and evaluation, 3rd edn. New Jersey: Wiley; 2009.
- 40.Search Import Export Data. https://www.zauba.com/shipment_search. Accessed 15 Apr 2017
- 41.Producer price index by commodity for chemicals and allied products: industrial chemicals (WPU061).
- 42.Process design for biochemical conversion of biomass to ethanol (2002 and 2011 design reports). http://www.nrel.gov/extranet/biorefinery/aspen_models. Accessed 6 Sep 2016
- 43.Wang ZX, Zhuge J, Fang HY, Prior BA. Glycerol production by microbial fermentation: a review. Biotechnol Adv. 2001;19:201–223. doi: 10.1016/S0734-9750(01)00060-X. [DOI] [PubMed] [Google Scholar]
- 44.Austin S, Kontur WS, Ulbrich A, Oshlag JZ, Zhang W, Higbee A, Zhang Y, Coon JJ, Hodge DB, Donohue TJ, et al. Metabolism of multiple aromatic compounds in corn stover hydrolysate by Rhodopseudomonas palustris. Environ Sci Technol. 2015;49:8914–8922. doi: 10.1021/acs.est.5b02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh S, Conrad JR, Klass DL. Anaerobic acidogenesis of wastewater sludge. J Water Pollut Control Fed. 1975;47:30–45. [PubMed] [Google Scholar]
- 46.Strauber H, Lucas R, Kleinsteuber S. Metabolic and microbial community dynamics during the anaerobic digestion of maize silage in a two-phase process. Appl Microbiol Biotechnol. 2016;100:479–491. doi: 10.1007/s00253-015-6996-0. [DOI] [PubMed] [Google Scholar]
- 47.Kucek LA, Xu JJ, Nguyen M, Angenent LT. Waste Conversion into n-Caprylate and n-Caproate: resource recovery from wine lees Using anaerobic reactor microbiomes and in-line extraction. Front Microbiol. 1892;2016:7. doi: 10.3389/fmicb.2016.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge S, Usack JG, Spirito CM, Angenent LT. Long-term n-Caproic acid production from yeast-fermentation beer in an anaerobic bioreactor with continuous product extraction. Environ Sci Technol. 2015;49:8012–8021. doi: 10.1021/acs.est.5b00238. [DOI] [PubMed] [Google Scholar]
- 49.Dodd D, Kocherginskaya SA, Spies MA, Beery KE, Abbas CA, Mackie RI, Cann IKO. Biochemical Analysis of a beta-d-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in prevotella ruminicola 23. J Bacteriol. 2009;191:3328–3338. doi: 10.1128/JB.01628-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon BS, Choi O, Um Y, Sang BI. Production of medium-chain carboxylic acids by Megasphaera sp. MH with supplemental electron acceptors. Biotechnol Biofuels. 2016;9:129. doi: 10.1186/s13068-016-0549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleckwedel J, Teran LC, Bonacina J, Saavedra L, Mozzi F, Raya RR. Draft genome sequence of the mannitol-producing strain Lactobacillus mucosae CRL573. Genome Announc. 2014;2:01292. doi: 10.1128/genomeA.01292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roos S, Karner F, Axelsson L, Jonsson H. Lactobacillus mucosae sp. nov., a new species with in vitro mucus-binding activity isolated from pig intestine. Int J Syst Evol Microbiol. 2000;50(Pt 1):251–258. doi: 10.1099/00207713-50-1-251. [DOI] [PubMed] [Google Scholar]
- 53.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chao SH, Sasamoto M, Kudo Y, Fujimoto J, Tsai YC, Watanabe K. Lactobacillus odoratitofui sp. nov., isolated from stinky tofu brine. Int J Syst Evol Microbiol. 2010;60:2903–2907. doi: 10.1099/ijs.0.019307-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Vadlani PV. Lactic acid production from biomass-derived sugars via co-fermentation of Lactobacillus brevis and Lactobacillus plantarum. J Biosci Bioeng. 2015;119:694–699. doi: 10.1016/j.jbiosc.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 56.Garde A, Jonsson G, Schmidt AS, Ahring BK. Lactic acid production from wheat straw hemicellulose hydrolysate by Lactobacillus pentosus and Lactobacillus brevis. Bioresour Technol. 2002;81:217–223. doi: 10.1016/S0960-8524(01)00135-3. [DOI] [PubMed] [Google Scholar]
- 57.Dellaglio F, Vancanneyt M, Endo A, Vandamme P, Felis GE, Castioni A, Fujimoto J, Watanabe K, Okada S, et al. Lactobacillus durianis Leisner, is a later heterotypic synonym of Lactobacillus vaccinostercus Kozaki and Okada 1983. Int J Syst Evol Microbiol. 2002;2006(56):1721–1724. doi: 10.1099/ijs.0.64316-0. [DOI] [PubMed] [Google Scholar]
- 58.Krooneman J, Faber F, Alderkamp AC, Elferink SJ, Driehuis F, Cleenwerck I, Swings J, Gottschal JC, Vancanneyt M. Lactobacillus diolivorans sp. nov., a 1,2-propanediol-degrading bacterium isolated from aerobically stable maize silage. Int J Syst Evol Microbiol. 2002;52:639–646. doi: 10.1099/00207713-52-2-639. [DOI] [PubMed] [Google Scholar]
- 59.Collins MD, Wallbanks S. Comparative sequence analyses of the 16 s ribosomal-RNA genes of Lactobacillus-Minutus, Lactobacillus-Rimae and Streptococcus-Parvulus—proposal for the creation of a new genus Atopobium. FEMS Microbiol Lett. 1992;95:235–240. doi: 10.1111/j.1574-6968.1992.tb05372.x. [DOI] [PubMed] [Google Scholar]
- 60.Jovita MR, Collins MD, Sjoden B, Falsen E. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. Int J Syst Bacteriol. 1999;49:1573–1576. doi: 10.1099/00207713-49-4-1573. [DOI] [PubMed] [Google Scholar]
- 61.Dewhirst FE, Paster BJ, Tzellas N, Coleman B, Downes J, Spratt DA, Wade WG. Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae: description of olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella profusa sp. nov. Int J Syst Evol Microbiol. 2001;51:1797–1804. doi: 10.1099/00207713-51-5-1797. [DOI] [PubMed] [Google Scholar]
- 62.Li X, Jensen RL, Hojberg O, Canibe N, Jensen BB. Olsenella scatoligenes sp. nov., a 3-methylindole-(skatole) and 4-methylphenol-(p-cresol) producing bacterium isolated from pig faeces. Int J Syst Evol Microbiol. 2015;65:1227–1233. doi: 10.1099/ijs.0.000083. [DOI] [PubMed] [Google Scholar]
- 63.Stanton TB, Savage DC. Roseburia cecicola gen. nov., sp. nov., a Motile, obligately anaerobic bacterium from a mouse cecum. Int J Syst Bacteriol. 1983;33:618–627. doi: 10.1099/00207713-33-3-618. [DOI] [Google Scholar]
- 64.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 65.Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 66.Holdeman LV, Elizabeth CP, Moore WEC. Amended description of Ramibacterium alactolyticum Prevot and Taffanel with propoosal of a neotype strain. Int J Syst Bacteriol. 1967;17:323–341. doi: 10.1099/00207713-17-4-323. [DOI] [Google Scholar]
- 67.Tian ZL, Mohan GR, Ingram L, Pullammanappallil P. Anaerobic digestion for treatment of stillage from cellulosic bioethanol production. Bioresour Technol. 2013;144:387–395. doi: 10.1016/j.biortech.2013.06.119. [DOI] [PubMed] [Google Scholar]
- 68.Nasr N, Elbeshbishy E, Hafez H, Nakhla G, El Naggar MH. Comparative assessment of single-stage and two-stage anaerobic digestion for the treatment of thin stillage. Bioresour Technol. 2012;111:122–126. doi: 10.1016/j.biortech.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 69.Hegner R, Koch C, Riechert V, Harnisch F. Microbiome-based carboxylic acids production: from serum bottles to bioreactors. RSC Adv. 2017;7:15362–15371. doi: 10.1039/C6RA28259H. [DOI] [Google Scholar]
- 70.Devarapalli P, Deshpande N, Hirwani RR. Xylose utilization in ethanol production: a patent landscape. Biofuel Bioprod Bioresour. 2016;10:534–541. doi: 10.1002/bbb.1664. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Chemical analyses of stillage production and mixed culture fermentation reactor. Table S1. Results from two fermentations of switchgrass hydrolysate with Saccharomyces cerevisiae Y128. Table S2. Soluble COD results for fermented switchgrass hydrolysate. Table S3. Summary of organic components in switchgrass stillage and the mixed culture fermentation reactor. Table S4. Total soluble COD in switchgrass stillage and mixed culture fermentation reactor. Table S5. Components in stillage and mixed culture fermentation reactor samples measured by GC-MS and HPLC analyses. Table S6. Total soluble carbohydrates measured in stillage and mixed culture fermentation reactor with the anthrone method. Table S7. Total soluble proteins in stillage and mixed culture fermentation reactor measured with the BCA assay. Table S8. Summary of aromatic compounds in stillage.
Additional file 2. Rarefied tables of operational taxonomic units (OTUs) and classification with Silva. Table S9. OTU table for 6d experiments testing different pH conditions. Table S10. OTU table for 252 days of reactor operation at pH 5.5.
Additional file 3. Data used for development of the technoeconomic analysis (TEA). Table S11. Summary of process modeling for liquid-liquid extraction, solvent distillation, and MCFA separation with ASPEN. Table S12. Summary of capitol expenses added to the NREL TEA. Table S13. Summary of operating expenses and additional product revenues added to NREL TEA. Table S14. 30 year cash flow from NREL TEA updated to include MCFA production expenses and revenues. Table S15. Calculations for additional capital expenses. Table S16. Chemical costs for octanoic acid, hexanoic acid and 2-octanol obtained from Zauba.
Additional file 4. Chemical analysis for mixed culture fermentations after 6 days under different pH conditions.
Additional file 5. Heat map of most abundant OTUs in the initial mixed culture fermentation experiments under different pH conditions.
Additional file 6. Heat map of 100 most abundant OTUs from 252 days of mixed culture fermentation reactor operations.
Additional file 7. Summary of Technoecomoic Analysis (Output from NREL TEA Model).
Additional file 8: Table S17. Comparison of hexanoic (C6) and octanoic (C8) acid productivities and titers for this study and other studies utilizing cellulosic or ethanol-production derived substrates.
Data Availability Statement
The raw sequencing reads from 16S rRNA gene amplicon sequencing are available under NCBI BioProject ID PRJNA393345 (252-day experiment) and NCBI BioProject ID PRJNA432807 (6-day experiments). Stillage and reactor data are available in Additional file 1 and data used for the TEA are provided in Additional file 3.