Abstract
The development of myeloid and lymphoid neoplasms related to overexpression of FGFR1 kinases as a result of chromosome translocations depends on the promotion of a stem cell phenotype, suppression of terminal differentiation and resistance to apoptosis. These phenotypes are related to the stem cell leukemia/lymphoma syndrome (SCLL), which arises through the effects of the activated FGFR1 kinase on gene transcription which includes microRNA dysregulation. In a screen for miRNAs that are directly regulated by FGFR1, and which stimulate cell proliferation and survival, we identified miR-339-5p, which is highly upregulated in cells carrying various different chimeric kinases. Overexpression of miR-339-5p in SCLL cell types enhances cell survival and inhibition of its function leads to reduced cell viability. miR-339-5p overexpression protects cells from the consequences of FGFR1 inactivation, promoting cell cycle progression and reduced apoptosis. Transient luciferase reporter assays and qRT-PCR detection of endogenous miR-339-5p expression in stably transduced cell lines demonstrated that BCR-FGFR1 can directly regulate miR-339-5p expression. This correlation between miR-339-5p and FGFR1 expression is also seen in primary human B-cell precursor acute lymphoblastic leukemia. In a screen to identify targets of miR-339-5p, we identified and verified the BCL2L11 and BAX genes, which can promote apoptosis. In vivo, SCLL cells forced to overexpress miR-339-5p show a more rapid onset of disease and poorer survival compared with parental cells expressing endogenous levels of miR-339-5p. Analysis of human primary B-cell precursor ALL shows a significant higher expression of miR339-5p compared with the two cohorts of CLL patient samples, suggesting direct roles in disease progression and supporting the evidence generated in mouse models of SCLL.
Keywords: MicroRNA 339-5p, AML, SCLL, BAX, BCL2L11
Introduction
MicroRNAs (miR) provide a mechanism of post-translational regulation of gene expression either by targeting specific mRNAs for degradation or interfering with their translation into proteins [1]. The net effect is a suppression of function for the target genes. miR action is DNA sequence directed, and so individual miRs have the capacity to simultaneously target many genes, providing extensive regulation of the cellular gene expression profile. It is now clear that cancer development is intimately associated with miR regulation of genes involved with proliferation, invasion and other hallmarks of cancer [2]. Analysis of acute myelogenous leukemia (AML), for example, has identified a broad involvement of MiRs which can be specific to the particular molecular subtype [3]. In our recent studies involving analysis of a stem cell leukemia/lymphoma syndrome (SCLL), expression analysis identified a core group of MiRs that affected proliferation and survival of the leukemias and lymphomas that have been generated in mouse models of this disease [4].
SCLL develops as a result of the ligand independent, constitutive activation of FGFR1 kinase as a result of chromosome translocations that lead to chimeric FGFR1 kinases [5]. There are a variety of different chromosome rearrangements described in SCLL but FGFR1 is always involved and in all cases the partner genes provide the dimerization capability needed for constitutive FGFR1 activation [6]. FGFR1 is a transcription factor that regulates specific genes but the fusion kinases also activate cytoplasmic proteins through tyrosine phosphorylation [7]. During our screen for miRs potentially activated by FGFR1, we identified a series that were downregulated when FGFR1 function was pharmacologically suppressed [4]. Multiple members of a closely related family, miR-17/92, were shown to have a significant effect on growth and survival of SCLL cells but, during this analysis, several other individual miRs also had profound effects on growth, such as miR-339-5p. Here we establish that miR-339-5p promotes proliferation and survival of SCLL cells through downregulation of the BCL2L11 and BAX pro-apoptotic genes.
Materials and Methods
Cells and cell culture
BBC2, KG1 and BaF3 cells were cultured in RPMI 1640 medium containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin. NIH3T3 and HEK293T cells were cultured in DMEM medium containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin. For acid-induced apoptosis, cell lines were grown in standard culture medium, adjusted to pH 5.6 with HCl and then filtered as described previously [27]. Cells were then collected for analysis at the indicated time points. Retroviral transduction was performed as described previously [4, 10]. In brief, phoenix-ampho packaging cells were transfected with DNA plasmids using lipofectamine 2000. The viral supernatant was collected after 48 and 72 h and was used for RetroNectin-mediated infection, following the manufacturer’s protocol. Selection with puromycin was performed at a concentration of 1 µg/ml for 4 days, followed by culture in the absence of the selection agent. SCLL cell lines were generated in-house and verified through the expression of the fusion kinases that define them. BaF3 cells are confirmed by virtue of their IL3 dependence, 3T3 cells were obtained from ATCC and used within 5 passages from recovery from frozen. Identity of these cells was confirmed by constant morphology analysis as recommended by ATCC. Mycoplasma testing was not routinely performed.
Plasmid constructs
For overexpression studies, the ~500bp fragment encompassing miR-339-5p, including the primary miRNA and flanking sequence, was cloned into pMSCV-PIG (Addgene Plasmid #21654). MicroRNA sponge constructs, which serve as competitive inhibitors of the target miR, were generated by multiple insertion of oligoduplexes (Fisher Scientific, Hampton, NH) containing 3 bulged miRNA binding sites (MBS) against miR-339-5p into the pMSCV-PIG-sp vector [28]. The promoter region of miR-339-5p was PCR amplified from genomic DNA and inserted into pGL3 Luciferase Reporter Vectors. Oligonucleotides (~90 bp) containing the original or mutated target sites were cloned into the pMIR-REPORT miRNA expression reporter vector (Fisher Scientific, Hampton, NH).
RT-PCR and western blot analysis
Quantitative reverse transcription polymerase chain reaction (RT-PCR) and western blotting assays were carried out as described previously [4].
Flow cytometry
For the GFP competition assay, cells containing exogenous constructs or empty vector (GFP+) were mixed 50:50 with parental cells (GFP-) and the GFP+/GFP− ratio was determined at different time points using FACSCanto II flow cytometry (BD Bioscience, San Jose, CA). Cell cycle profiles were determined with the LSR II flow cytometer (BD Bioscience, San Jose, CA) following Hoechst 33342 (Thermo Fisher Scientific, Waltham, MA) labeling. To measure apoptosis, cells were stained with APC Annexin V DNA binding dye (Biolegend, San Diego, CA) according to the manufacturer’s protocol and analyzed using FACSCanto II flow cytometry (BD Bioscience, San Jose, CA).
Luciferase reporter assay
Promoter assays were performed using HEK293T cells following co-transfection of a promoter reporter plasmid derived from pGL3 with or without pMSCV-BCR-FGFR1 containing plasmids as described previously [4]. For miRNA target assays, the luciferase target site-containing plasmids were cotransfected with the miR-339-5p overexpression plasmid into HEK293T cells. The transfected cells were then harvested 48 h after transfection and analyzed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Renilla luciferase was used to normalize for transfection efficiency, and the ratio of firefly/ Renilla luciferase activities defined the relative promoter activity. The pMIR-REPORTER system was used to verify miR target sites where the predicted target sites were introduced into the multiple cloning site in the 3’UTR of the luciferase gene (see results).
In vivo studies
Approximately 1 × 106 cells were injected into the tail veins of 6–8 week old, female, sub-lethally (600 Rad) irradiated Balb/C mice [10]. Disease progression was monitored for the expression of GFP through weekly analysis of peripheral blood derived from the tail vein. At the end of the experiment, mice were sacrificed and organs harvested for evaluation of tumor burden using flow cytometry and morphological examination. Standard histopathological analysis of organs was performed as described previously [8,10]. Animal protocols followed guidelines and procedures approved by the IACUC of Augusta University.
Results
miR-339-5p is overexpressed and promotes cell growth in SCLL
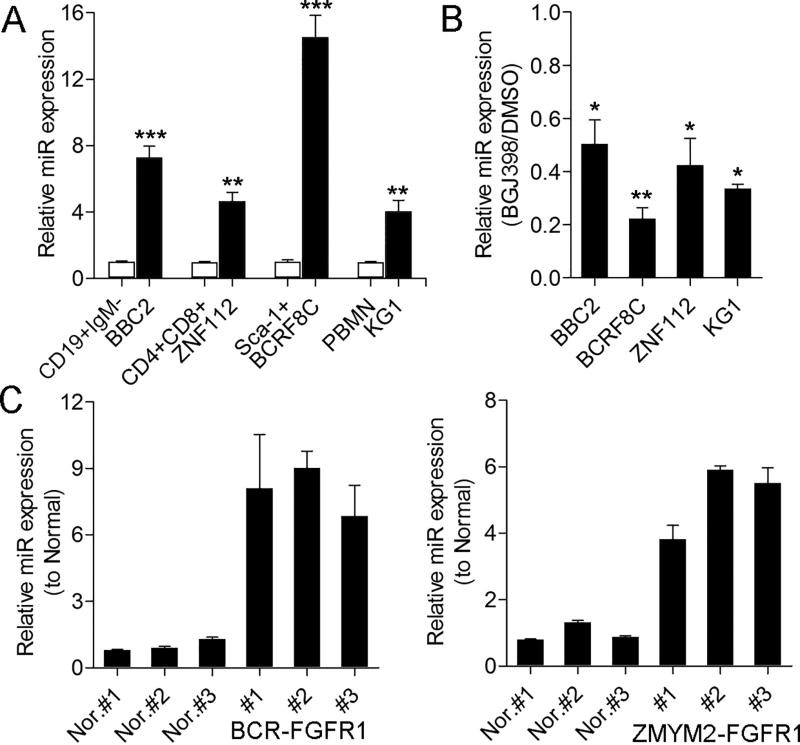
Suppression of FGFR1 activity using the BGJ398 kinase inhibitor in SCLL cell lines showed significant downregulation of miR-339-5p [4]. To analyze the relative expression levels of miR-339-5p in SCLL cells, we confirmed the reported immunophenotypes of three murine cell lines (Supplemental Figure 1); BBC2, which expresses BCR-FGFR1, was CD19+ IgM−, ZNF112, which expresses ZMYM2-FGFR1, was CD4+CD8+ and BCRF8C, which also expresses BCR-FGFR1, was Sca1+. When miR-339-5p levels were measured in the SCLL cells, compared with flow sorted normal counterparts (Supplemental Figure 1), there was a highly significant upregulation in all three cell lines (Figure 1A). In addition, miR-339-5p was also upregulated in FGFR1OP2-FGFR1 expressing human KG1 AML cells. When the same four cell lines are treated with BGJ398 (Figure 1B), which has been shown to suppress chimeric FGFR1 kinase activation in SCLL [12], there is a 50–80% reduction in miR-339-5p expression levels (depending on the cell line). The same increase in miR-339-5p expression (Figure 1C) was seen in tumor cells from BCR-FGFR1 and ZMYM2-FGFR1 expressing primary leukemias from murine models [8, 10].
Figure 1. miR-339-5p expression levels are elevated in cells expressing chimeric FGFR1 kinases.
Analysis of miR-339-5p expression levels in SCLL cell lines, relative to their normal cell counterparts, shows highly significant upregulation (A). When the same four cell lines were treated with the BGJ398 FGFR1 inhibitor (B) there were significant reductions in miR-339-5p levels compared with DMSO-treated cells. Analysis of miR-339-5p expression levels in primary lymphomas from the BCR-FGFR1 (C, left) and ZMYM2-FGFR1 (C, right) mouse models compared with normal splenic cells, shows significant upregulation. * p = <0.05, ** p = <0.01, *** p = <0.001, **** p = <0.0001.
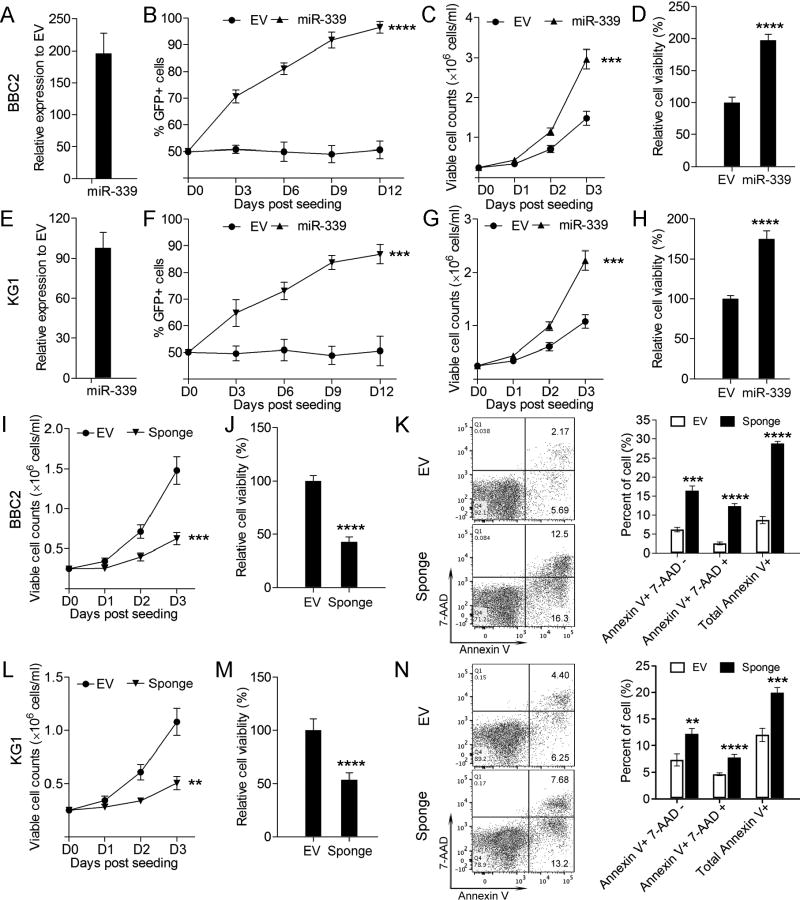
Using the GFP competition assay (see methods), cell growth was significantly increased when miR-339-5p was overexpressed in BBC2 cells (Figure 2A, B). Trypan blue exclusion analysis (Figure 2C) and luminescence cell viability assays (Figure 2D) further support this increased growth rate/survival. The same proportional increase in proliferation and viability was seen in the human SCLL KG1 cell line (Figure 2E–H). To inhibit miR-339-5p, we developed dominant-negative, bulged sponge constructs which are complementary to miR-339-5p and so, when expressed at high levels, can specifically and efficiently inhibit the miR-339-5p function. When miR-339-5p function was suppressed using these microRNA sponges in BBC2 (Figure 2I–J) cell proliferation and viability was significantly reduced. Flow cytometric analysis, comparing the control cells with those treated with the sponge constructs, shows significantly increased levels of Annexin V positive cells, demonstrating increased apoptosis in BBC2 (Figure 2K). The same reduced proliferation and cell viability was seen in human KG1 cells treated with the miR sponges (Figure 2L–M) and apoptosis was also increased (Figure 2N) These observations confirm the potential tumor promotion role of miR-339-5p in SCLL.
Figure 2. Effects of miR-339-5p overexpression on cell growth and viability.
Q-PCR analysis demonstrates high-level miR-339-5p overexpression in transduced BBC2 cells relative to endogenous levels (A). BBC2 cells overexpressing miR-339-5p show a highly significant increase in cell growth in GFP competition assays (B) as well as cell viability using trypan blue exclusion assays (C) and luminescence viability assays (D), compared with the empty vector transduced cells. In the analysis using human KG1 cells, the same effects of miR-339-5p overexpression were seen (E–H). When BBC2 cells were treated with miR-339-5p sponges (SP), cell viability demonstrated by trypan blue exclusion (I) and luminescence assays (J) was significantly reduced. Flow cytometric analysis (K) demonstrates the sponge-treated cells show a significant increase in apoptosis. The same effects were seen for KG1 cells treated with the miR-339-5p sponges (L–N). *** p = <0.001, **** p = <0.0001.
Overexpression of miR-339-5p mediates resistance to BGJ398 induced cell cycle inhibition and apoptosis
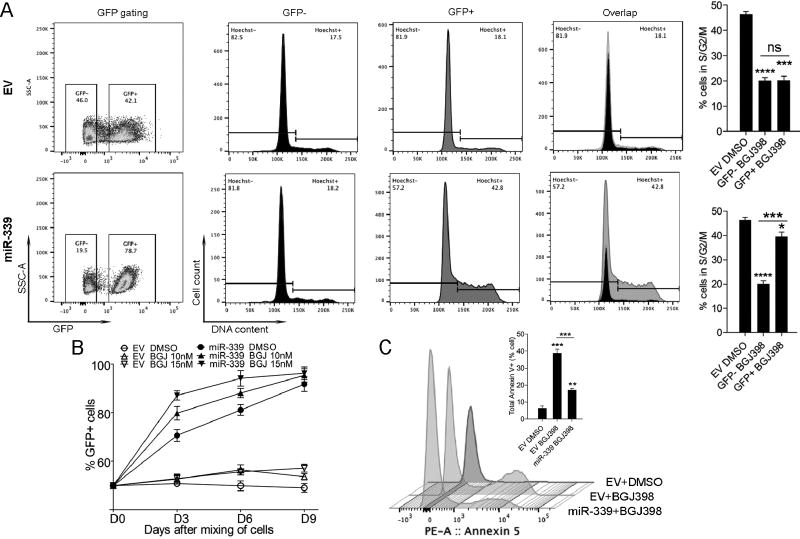
The BGJ398 FGFR1 inhibitor leads to growth suppression and increased apoptosis in SCLL cells [12]. To investigate how miR-339-5p responds to FGFR1 inhibition, BBC2 cells overexpressing either the empty vector (GFP+) or miR-339-5p (GFP+) were mixed 50:50 with the parental cell line (GFP−) and treated with BGJ398 for 72 hours. Cells transduced with the empty vector showed a growth rate approximately equal to the parental BBC2 cells in the presence of BGJ398, whereas cells overexpressing miR-339-5p showed a growth advantage over the parental cells, with increased GFP+ ratios from 50% to 78.7% (Figure 3A). Flow cytometric analysis of the cell cycle in these cells showed a significant reduction in the proportion of cells in S/G2/M in both the EV expressing cells and parental BBC2 cells. In contrast, overexpression of miR-339-5p, led to a disproportionate increase in the number of cells in S/G2/M compared with the parental cells (Figure 3A). Thus, overexpression of miR-339-5p can mitigate the cellular effect caused by inhibition of FGFR1 signaling and in a dose dependent manner (Figure 3B). In addition to its effect on the cell cycle, miR-339-5p overexpression also mitigated sensitivity to apoptosis induced by BGJ398 (Figure 3C). Together, these observations suggested a direct involvement of miR-339-5p in FGFR1 fusion kinase-driven disease progression.
Figure 3. Suppression of FGFR1 leads to reduced cell cycle progression and increased apoptosis.
Flow cytometry analysis of BBC2 cells expressing either the empty vector (EV, GFP+, above) or exogenous miR-339-5p (GFP+, below) following a 72 hour treatment with BGJ398 shows an increase in GFP+ cells only when miR-339-5p is overexpressed (A, left). Cell cycle analysis in the EV group shows BGJ398 treatment equally affects the GFP+ (EV transduced) and GFP− (parental BBC2 cell) population of cells, reducing the proportion in S/G2/M. In contrast, overexpression of exogenous miR-339-5p leads to a relative increase in the proportion of cells in S/G2/M, partially overcoming the cell cycle inhibition caused by BGJ398 treatment (A). This selective enrichment of miR-339 expressing cells following BGJ398 treatment is dose dependent (B). Analysis of the Annexin-V population of cells shows that BGJ398 treatment of cells transduced by EV leads to a highly significant increase in apoptosis (C), but these effects are significantly counteracted by overexpression of miR-339-5p. ** p = <0.01, *** p = <0.001, **** p = <0.0001.
miR-339-5p expression is regulated by chimeric FGFR1 kinases
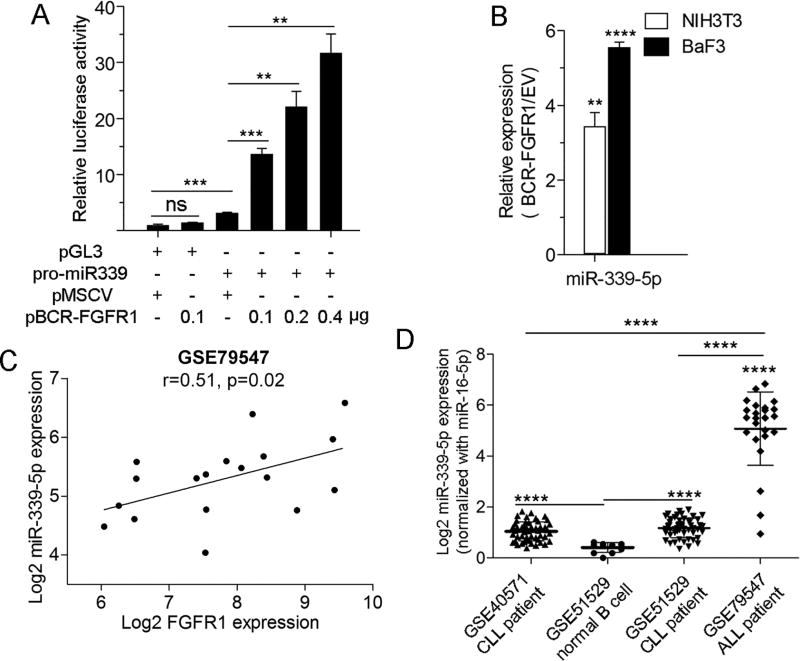
The correlation between expression levels of FGFR1 and miR-339-5p suggested that its expression is regulated by FGFR1. We therefore cloned the miR-339 promoter region (−1626 to −7 bp) into the pGL3 luciferase reporter vector and co-transfected into 293T cells with varying amounts of the BCR-FGFR1 construct. As shown in Figure 4A, luciferase expression was activated in a dose dependent manner in the presence of BCR-FGFR1, providing direct evidence that miR-339-5p is activated as a consequence of FGFR1 expression. This suggestion was further supported by the demonstration that NIH3T3 and BaF3 cell lines, stably transduced with BCR-FGFR1, also showed increased expression of miR-339-5p (Figure 4B). The relationship between FGFR1 and miR-339-5p expression was further supported by an analysis of miRNA and mRNA expression profiles in a cohort of patients with B-cell precursor acute lymphoblastic leukemia (ALL), which revealed a significant, positive correlation between the expression of FGFR1 and miR-339-5p (Figure 4C). There was no correlation in a similar analysis of two cohorts of patients with relatively benign CLL, possibly due to relative low expression levels of miR-339-5p in these leukemias (Supplementary Figure S2A–C). A similar comparison between B-cell precursor ALL and data sets for either normal donor B cell or CLL samples, showed a significantly higher expression of miR-339-5p (Figure 4D and Supplementary Figure S2D) compared with the CLL patients when normalized either with housekeeping miR-16 or Let-7a. In contrast, no differential expression for control housekeeping miRNA Let-7a and miR-16-5p was seen in these cohorts (Supplementary Figure S2E–F). Together, these data support the importance of miR-339-5p in chimeric FGRF1-induced SCLL.
Figure 4. Relationship between FGFR1 and miR-339-5p expression.
In luciferase reporter assays (A) activation of the miR-339-5p promoter is proportional to the expression levels of BCR-FGFR1 compared with expression of the empty vector (MCSV). Forced expression of BCR-FGFR1 in NIH3T3 or BaF3 cells leads to an increase in miR-339-5p expression levels (B). In an analysis of the GEO database, miR expression set GSE79547 for B-cell precursor ALL, there is a proportional relationship between expression of FGFR1 and miR-339-5p (C). miR-339-5p expression levels are relatively higher in GSE40571 and GSE51529 reporting data from low malignant CLL cells compared with normal B-cells from GSE51529. More dramatically increased expression levels of miR-339-5p are seen in tumors from ALL patients (D). ** p = <0.01, *** p = <0.001, **** p = <0.0001.
miR-339-5p regulates activity of BCL2L11 and BAX
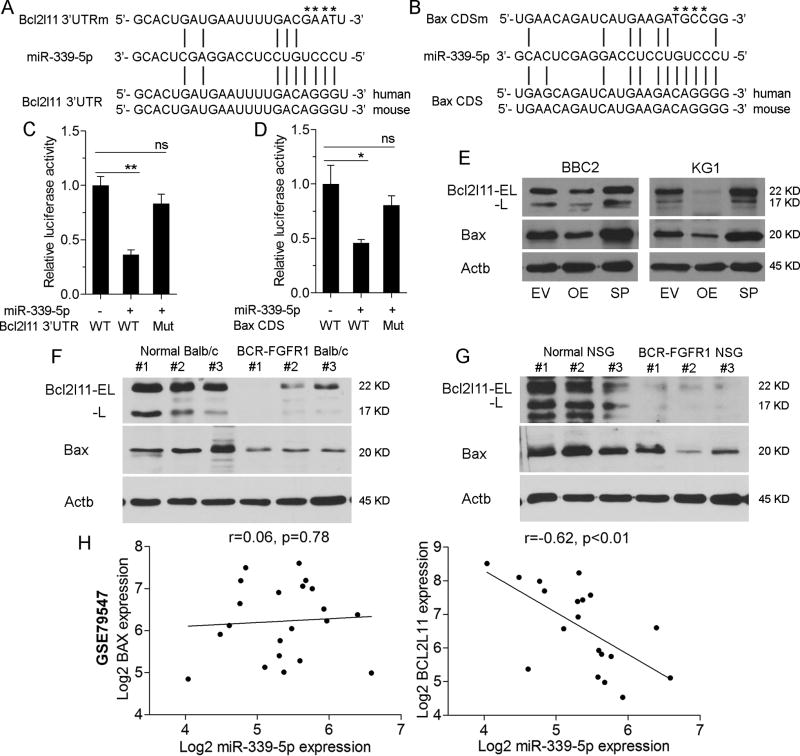
The demonstration that miR-339-5p promotes cell viability in SCLL cells, suggested that it targets transcripts related to this phenotype. Bioinformatics analysis, using sequence-based prediction algorithms, including TargetScan and miRWalker2.0 [13–14], were used to identify potential miR-339 mRNA targets (Supplemental Figure S3, Supplemental Table T1). Since miR-339-5p appears to affect apoptosis (Figure 3), we filtered genes that affected this phenotype and identified BCL2L11 and BAX as candidate targets (Figure 5A, B; Supplemental Figure S3). Target sites in these genes were identified in the 3’UTR of BCL2L11 and in the coding region of BAX. To determine whether miR-339-5p directly regulates BCL2L11 and BAX expression, we performed luciferase reporter assays where the target sites, with ~90bp flanking sequences, were cloned into the pMIR-REPORT luciferase reporter vector. When 293FT cells were co-transfected with miR-339-5p and luciferase constructs, there was a 50% reduction in luciferase activity of both BCL2L11 and BAX in the presence of miR-339-5p (Figure 5C, D). Mutation of the target sites (Figure 5A–B) in each case abrogated the suppressive effect of miR-339-5p. Western blot analysis showed that miR-339-5p overexpression reduced BCL2L11 and BAX proteins levels, and miRNA sponges targeting miR-339-5p led to an increase in BCL2L11 and BAX protein levels in both BBC2 and KG1 cells, further confirming that BCL2L11 and BAX were direct targets of miR-339-5p (Figure 5E). Analysis of primary SCLL spleen cells (Figure 5F) from both the BCR-FGFR1 murine model and the BCR-FGFR1 human cell model demonstrated downregulation of BCL2L11 and BAX compared with normal splenic cells (Figure 5G) [8, 15]. These data indicate that miR-339-5p negatively regulates BCL2L11 and BAX in SCLL.
Figure 5. miR339-5p targets BCL2L11 and BAX.
miR339-5p target sites within the 3’UTR of BCL2L11 (A) and the coding region of BAX (B) were identified using prediction algorithms. In each case mutations of specific bases (shown by * above the sequence) in the binding regions were generated. In luciferase reporter assays (C) co-expression of miR-339-5p in cells carrying the wild type (WT) miR-339-5p target site in BCL2L11 shows a significant reduction in activation compared with a construct in which BCL2L11 sites have been mutated. The same relationship is observed in luciferase expression assays where the miR-339-5p target and mutated sequences for BAX are expressed (D). In BBC2 and KG1 cells, overexpression (OE) of miR-339-5p led to reduced expression levels of BCL2L11 (isoforms; EL = extra long and L = long) and BAX but co-expression of sponges (SP) targeting miR-339-5p prevents the suppressive effects on these two genes (E). In the murine SCLL model of BCF-FGFR (F), expression levels of BCL2L11 and BAX are reduced compared with splenic cells from normal mice. The same reduction in BCL2L11 and BAX expression levels is seen in primary AML cells derived from human cell SCLL that have been developed in immunocompromised (NSG) mice (G). Analysis of the pre-B-precursor ALL (GSE79547) data set, shows no change in relative levels of BAX expression compared to miR-339-5p expression (H) but a highly significant proportional decrease in BCL2L11 expression with increasing miR-339-5p expression. * p = <0.05, ** p = <0.01, ns = not significant.
To further validate the specificity of miR-339-5p for the predicted target site in the 3’UTR of BCL2L11, we used the pMIR-REPORT Luciferase assay and introduced these BCL2L11 target sites into the 3’UTR (Supplemental Figure S4A). In this assay miRNA targeting of these sites leads to destabilization of the luciferase mRNA resulting in reduced activity. When the Luciferase reporter was co-transfected into 293T cells with the miR339-5p expression vector, luciferase activity was highly significantly reduced (Supplemental Figure S4B). Next, we prepared oligonucleotides with 3–9 repeats of the miR339-5p target sequence in the pMCSV-PIG-Sp vector (Supplemental figure S4A) and co-transfected these into 293T cells together with the miR339-5p expression vector and the luciferase gene harboring the BCL2L11 target sites. It was anticipated that the introduced miR339-5p target sites would compete for miR339-5p binding and so suppress the effect on luciferase mRNA degradation. As shown in Supplemental Figure S4B, luciferase activity in the presence of the multiple binding site (MBS) constructs increased proportionally to the number of tandem binding sites, further supporting the specificity of miR339-5p for the BCL2L11 target sites. To further support the specific effects of the sponge constructs in suppressing miR339-5p, we again used the luciferase-BCL2L11 reporter assay where the miR339-5p expression construct was co-transfected with different ratios of the sponge constructs (Supplemental Figure S4C) where the higher the concentration of sponges correlated with increased luciferase activity. These data provide further evidence that the sponge constructs can efficiently target and suppress miR339-5p activity.
To broaden the context of the effects of miR339-5p, we analyzed the relative mRNA expression levels of miR-339-5p, BAX and BCL2L11 in a data set (GSE79547) available in GEO for B-cell precursor ALL. No significant correlation (Figure 5H) was observed in the expression levels of BAX and miR-339-5p, which might have been expected since miR-339-5p interferes with protein translation rather than mRNA stability. In contrast, there is a highly significant correlation between miR-339-5p and BCL2L11 expression in this analysis, where higher levels of miR-339-5p led to reduced levels of BCL2L11, which is expected since miR-339-5p targets sequences in the 3’UTR affecting mRNA stability as described above. This observation supports the idea that miR-339-5p can promote leukemogenesis and SCLL progression through targeting BAX and BCL2L11.
miR-339-5p protects SCLL cells from induced apoptosis
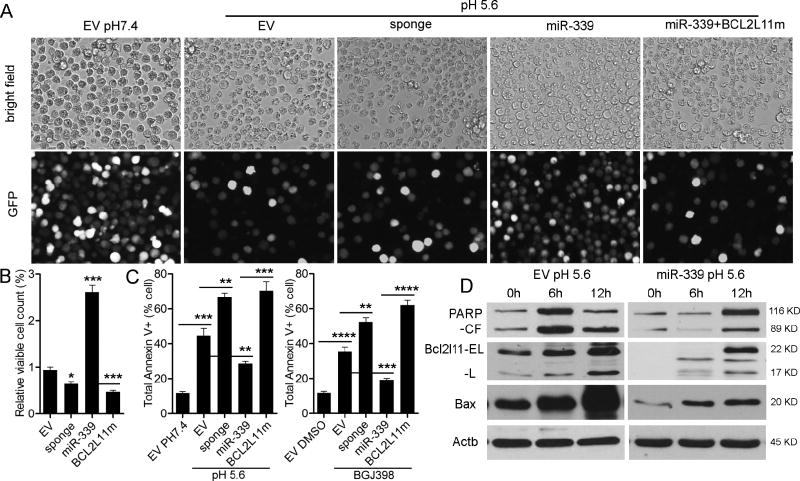
Reduced apoptosis as a result of miR-339-5p overexpression in BGJ398 treated cells, as well as miRNA target analysis identifying BCL2L11 and BAX, suggested that miR-339-5p could promote SCLL through protection against apoptosis. To investigate this hypothesis, we induced apoptosis using reduced acidic conditions. When miR-339-5p overexpressing cells were placed in pH5.6 culture conditions, there was a significant increase in cell viability, compared with cells transfected with the empty vector alone (Figure 6A, B). Cell viability was again reduced when miR-339-5p sponges were introduced into the cells. When the BCL2L11 overexpression construct that lacked miR-339-5p target sites in the 3’UTR was co-expressed in cells overexpressing miR-339-5p, cell survival was also reduced in the low pH environment (Figure 6A, B). Analysis of the Annexin V levels in these different cell lines demonstrated that reduction in cell viability was consistent with increased levels of apoptosis (Figure 6C, Supplemental Figure S5A). These observations support the conclusion that miR-339-5p can directly target pro-apoptotic BCL2L11 and affect apoptosis of SCLL cells. This conclusion is further supported by the fact that the same effects on apoptosis were seen when cells were treated with BGJ398 (Figure 6C; Supplemental Figure S5B).
Figure 6. Expression of miR-339-5p suppresses apoptosis.
Fluorescence microscopy of GFP+ BBC2 cells, transduced with either the empty vector or miR-339-5p, when subjected to low pH culture conditions (A), shows a significant reduction in viable cell numbers transduced with the empty vector compared with overexpression of miR-339-5p. Cells treated with the miR-339-5p sponges show reduced viability, as do miR-339-5p expressing cells transduced with BCL2L11 in which the miR-339-5p target sites have been mutated (A–B). This reduced sensitivity to low pH-induced apoptosis is confirmed by analysis of Annexin V expression (C, left), which is significantly reduced in miR-339-5p overexpressing cells, but significantly increased in cells expressing the miR-339-5p sponge construct or the mutant BCL2L11. The same response is observed following treatment of the same cells with the BGJ398 FGFR1 inhibitor (C, right). Comparison of BBC2 cells expressing either the empty vector or miR-339-5p (D), shows PARP cleavage (CF = cleaved form) after 12 hours is impaired in miR-339-5p overexpressing cells compared with EV expressing cells and is accompanied by reduced levels of BCL2L11 (isoforms; EL = extra-long and L = long) and BAX following exposure to acidic conditions. *** p = <0.001, **** p = <0.0001.
At the protein level, the reduced apoptosis phenotype was accompanied by an impaired activation of BCL2L11 and BAX in the miR-339-5p overexpressing cells, compared with the empty vector control (Figure 6D). The resistance of miR-339-5p expressing cells to acid induced apoptosis was also confirmed by the delay in the activation and cleavage of PARP (Figure 6D), a hallmark of apoptosis [16]. These data demonstrate miR-339-5p can protect cells from apoptosis by targeting BCL2L11 and BAX..
miR-339-5p enhances SCLL development in vivo
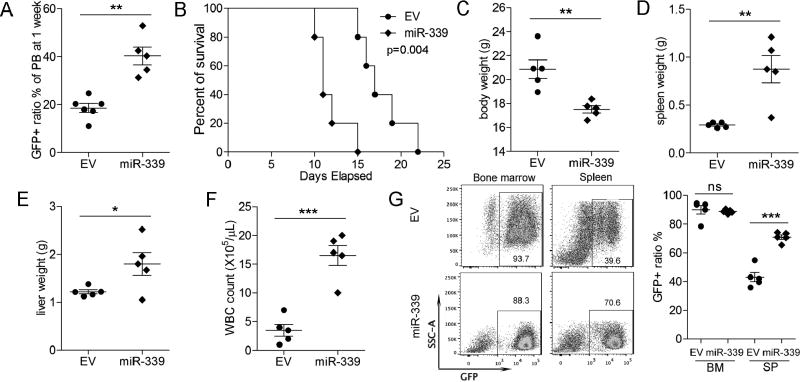
To evaluate the oncogenic effect of miR-339-5p in FGFR1 fusion kinase driven leukemogenesis in vivo, 1×106 BBC2 cells, either stably overexpressing miR-339-5p or the vector control, were injected into the tail veins of 6- to 8-week old (n=5), sub-lethally irradiated, syngeneic BALB/c mice as described previously [10]. Consistent with the in vitro cell proliferation effects, overexpression of miR-339-5p increased the proportion of GFP+ BBC2 cells in the peripheral blood from 20.29% to 40.33% (p = 1.75e-03) one week after transplantation (Figure 7A). Aggressive disease subsequently developed in miR-339-5p overexpressing cells within 10–25 days (Supplemental Figure S6A–B) and Kaplan-Meyer comparison between mice injected with the miR-339-5p overexpressing BBC2 cells and parental BBC2 cells, shows a highly significant (p = 0.004) reduction in survival (Figure 7B). In addition, the mice transplanted with the BBC2 cells with forced expression of miR-339-5p, displayed reduced body weight due to disease burden (Figure 7C), as well as significantly enlarged spleens (Figure 7D) and livers (Figure 7E), compared with empty vector control cells. Consistently, the white blood cell count in the miR-339-5p group on termination of the experiment was significantly higher than for the empty vector control group (Figure 7F). Flow cytometry analysis of cells in the bone marrow and spleens from mice engrafted with BBC2 that express either the empty vector or miR-339-5p showed that the vast majority were GFP+, and the more enlarged spleens from the miR-339-5p overexpressing group had an even higher ratio of GFP+ cells (Figure 7G). Histopathology analysis confirmed these findings, with increased cellularity in the peripheral blood of mice overexpressing miR-339-5p, which was associated with hypercellularity in the bone marrow and increased infiltration into the spleen and liver (Supplemental Figure S6C).
Figure 7. miR-339-5p promotes more aggressive development of SCLL in vivo.
When BBC2 cells overexpressing miR-339-5p are injected into the tail veins of sub-lethally irradiated Balb/C mice, the ratio of GFP+ cells to GFP− cells in the peripheral blood is significantly increased after only 7 days (A). Survival of mice is significantly reduced when transplanted with miR-339-5p overexpressing cells (B) compared with parental (EV) cells. This rapid disease progression is reflected in the reduced body weight (C) and increased spleen size (D) in miR-339-5p over expressing cells. Similarly, liver weight (E) and total white cell counts in peripheral blood (F) are also increased. Flow cytometry analysis (G) shows almost complete replacement of cells in the BM and spleens from mice transplanted with GFP+ BBC2 leukemia cells. * p = <0.05, ** p = <0.01, *** p = <0.001, **** p = <0.0001.
Discussion
FGFR1 activation has been suggested to lead to extensive gene regulation, either directly though promoter binding or indirectly through the activation of other co-factors such as CBP, that activate a diverse series of transcription factors7. Here we show that FGFR1 can also regulate specific miR expression profiles, which adds to the complexity of the transformation process through posttranslational modification of gene function. Some miRs detected in our screen appear to have more profound effects on growth and survival, presumably as a result of the genes they regulate. Maintenance of an undifferentiated state is observed in SCLL development [17], as well as suppression of apoptosis. In this respect, the ability of miR-339-5p to suppress the function of genes that promote apoptosis directly affects survival of the tumor cells. miR-339-5p has previously been shown to affect expression of genes such as PRL1 [18], and many others through prediction algorithms that have not been functionally confirmed.
Here we show for the first time that miR-339 can affect production of BAX and BCL2L11. BAX interacts with the BCL2 apoptosis suppressor to promote metastasis through destabilization of the mitochondrial membrane promoting release of cytochrome C. BCL2L11, also known as BIM or BimEL, which is prominently expressed in cells of the hematopoietic linage [19], is an essential pro-apoptotic protein involved in the initiation of apoptotic pathways by binding to anti-apoptotic proteins of the BCL2 family [20]. Growth factors such as IL-3 promote survival of hematopoietic progenitors through downregulation of BCL2L11, and this IL-3-dependent downregulation is mediated through activation of Raf/MAPK and PI3K/mTOR pathways [21]. BCL2L11 is also important for determining the lifespan of myeloid and lymphoid cells, as these cells show increased numbers in mice lacking BCL2L11 [22]. BCL2L11 can also be targeted by several other oncomiRs, such as miR-17~92, miR-106~25, miR-181, miR-148 and miR-221/222, which promote tumor progression [20]. Here, we identified a new oncomiR, miR-339-5p, that promotes SCLL through double targeting pro-apoptotic genes BCL2L11 and BAX. Interestingly, BCL2L11 is also regulated by miR17–92, which we have shown is also upregulated in FGFR1 driven SCLL [4], suggesting FGFR1 fusion kinases simultaneously regulate multiple miRNAs to inhibit key apoptotic genes to promote leukemogenesis.
Although data from SCLL suggests that miR-339-5p strongly promotes cell survival, other studies using MCF7 breast cancer cells [23], for example, showed miR-339-5p could influence levels of p53. This effect was mediated through its targeting of MDM2, thus having an anti-proliferation effect while apparently not affecting apoptosis. Correlative studies in other breast cancer models also suggested a role for miR-339-5p in the suppression of invasion and metastasis [24]. Similar conclusions were reached in colon cancer [25], apparently though its regulation of PRL1. Different miRs can be generated from the same pre-miR but often have different effects and different targets. Thus, the alternatively derived miR-339-3p has also been reported as a suppressor of invasion in melanoma cells with no effect on cell proliferation [26]. In this latter case, MCL1 appears to be a target which also has a role in promoting apoptosis. The clear ability of miR-339-5p to promote survival in FGFR1 expressing myeloid and lymphoid malignancies, in contrast to the opposite effect seen in cells from solid tumors, suggests a cell context-specific action.
Supplementary Material
Acknowledgments
This work was supported by grant CA076167 from the National Institutes of Health (J. Cowell)
Footnotes
Conflict of Interest statement: The authors declare no conflicts of interest associated with this work.
References
- 1.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–33. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 2.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130:1290–301. doi: 10.1182/blood-2016-10-697698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu T, Chong Y, Qin H, Kitamura E, Chang CS, Silva J, et al. The miR-17/92 cluster is involved in the molecular etiology of the SCLL syndrome driven by the BCR-FGFR1 chimeric kinase. Oncogene. 2017 doi: 10.1038/s41388-017-0091-1. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann H, Kunapuli P, Tracy E, Cowell JK. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. J Biol Chem. 2003;278:16198–208. doi: 10.1074/jbc.M300018200. [DOI] [PubMed] [Google Scholar]
- 6.Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol. 2010;41:461–76. doi: 10.1016/j.humpath.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–66. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren M, Tidwell JA, Sharma S, Cowell JK. Acute progression of BCR-FGFR1 induced murine B-lympho/myeloproliferative disorder suggests involvement of lineages at the pro-B cell stage. PLoS One. 2012;7:e38265. doi: 10.1371/journal.pone.0038265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva J, Chang CS, Hu T, Qin H, Kitamura E, Hawthorn L, et al. Gene expression profiling during progression of FGFR1 driven AML in a mouse model of Stem Cell Leukemia Lymphoma syndrome. BMC Cancer. 2017 doi: 10.1016/j.ygeno.2018.10.015. submitted. [DOI] [PubMed] [Google Scholar]
- 10.Ren M, Li X, Cowell JK. Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood. 2009;114:1576–84. doi: 10.1182/blood-2009-03-212704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furley AJ, Reeves BR, Mizutani S, Altass LJ, Watt SM, Jacob MC, et al. Divergent molecular phenotypes of KG1 and KG1a myeloid cell lines. Blood. 1986;68:1101–07. [PubMed] [Google Scholar]
- 12.Wu Q, Bhole A, Qin H, Karp J, Malek S, Cowell JK, et al. SCLLTargeting FGFR1 to suppress leukemogenesis in syndromic and de novo AML in murine models. Oncotarget. 2016;7:10. doi: 10.18632/oncotarget.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowell JK, Qin H, Chang CS, Kitamura E, Ren M. A model of BCR-FGFR1 driven human AML in immunocompromised mice. Br J Haematol. 2016;175:542–45. doi: 10.1111/bjh.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gobeil S, Boucher CC, Nadeau D, Poirier GG. Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): implication of lysosomal proteases. Cell Death Differ. 2001;8:588–94. doi: 10.1038/sj.cdd.4400851. [DOI] [PubMed] [Google Scholar]
- 17.Ren M, Cowell JK. Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease. Blood. 2011;117:6837–47. doi: 10.1182/blood-2010-07-295725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Lu Y, Li X. miR-339-3p inhibits proliferation and metastasis of colorectal cancer. Oncol Lett. 2015;10:2842–48. doi: 10.3892/ol.2015.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Reilly LA, Cullen L, Visvader J, Lindeman GJ, Print C, Bath ML, et al. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. American Journal of Pathology. 2000;157:449–61. doi: 10.1016/S0002-9440(10)64557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sionov RV, Vlahopoulos SA, Granot Z. Regulation of Bim in Health and Disease. Oncotarget. 2015;6:23058–134. doi: 10.18632/oncotarget.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, et al. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol. 2001;21:854–64. doi: 10.1128/MCB.21.3.854-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–38. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 23.Jansson MD, Damas ND, Lees M, Jacobsen A, Lund AH. miR-339-5p regulates the p53 tumor-suppressor pathway by targeting MDM2. Oncogene. 2015;34:1908–18. doi: 10.1038/onc.2014.130. [DOI] [PubMed] [Google Scholar]
- 24.Wu ZS, Wu QA, Wang CQ, Wang XN, Wang Y, Zhao JJ, et al. MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer. 2010;10:542. doi: 10.1186/1471-2407-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou C, Liu G, Wang L, Lu Y, Yuan L, Zheng L, et al. MiR-339-5p regulates the growth, colony formation and metastasis of colorectal cancer cells by targeting PRL-1. PLoS One. 2013;8:e63142. doi: 10.1371/journal.pone.0063142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber CE, Luo C, Hotz-Wagenblatt A, Gardyan A, Kordass T, Holland-Letz T, et al. miR-339-3p is a tumor suppressor in melanoma. Cancer Res. 2016;76:3562–71. doi: 10.1158/0008-5472.CAN-15-2932. [DOI] [PubMed] [Google Scholar]
- 27.Sharma V, Kaur R, Bhatnagar A, Kaur J. Low-pH-induced apoptosis: role of endoplasmic reticulum stress-induced calcium permeability and mitochondria-dependent signaling. Cell Stress Chaperones. 2015;20:431–40. doi: 10.1007/s12192-014-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, et al. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.