The abscisic acid-activated NAC transcription factor SlNAP2 controls leaf senescence and fruit yield in tomato.
Abstract
Leaf senescence is an essential physiological process in plants that supports the recycling of nitrogen and other nutrients to support the growth of developing organs, including young leaves, seeds, and fruits. Thus, the regulation of senescence is crucial for evolutionary success in wild populations and for increasing yield in crops. Here, we describe the influence of a NAC transcription factor, SlNAP2 (Solanum lycopersicum NAC-like, activated by Apetala3/Pistillata), that controls both leaf senescence and fruit yield in tomato (S. lycopersicum). SlNAP2 expression increases during age-dependent and dark-induced leaf senescence. We demonstrate that SlNAP2 activates SlSAG113 (S. lycopersicum SENESCENCE-ASSOCIATED GENE113), a homolog of Arabidopsis (Arabidopsis thaliana) SAG113, chlorophyll degradation genes such as SlSGR1 (S. lycopersicum senescence-inducible chloroplast stay-green protein 1) and SlPAO (S. lycopersicum pheide a oxygenase), and other downstream targets by directly binding to their promoters, thereby promoting leaf senescence. Furthermore, SlNAP2 directly controls the expression of genes important for abscisic acid (ABA) biosynthesis, S. lycopersicum 9-cis-epoxycarotenoid dioxygenase 1 (SlNCED1); transport, S. lycopersicum ABC transporter G family member 40 (SlABCG40); and degradation, S. lycopersicum ABA 8′-hydroxylase (SlCYP707A2), indicating that SlNAP2 has a complex role in establishing ABA homeostasis during leaf senescence. Inhibiting SlNAP2 expression in transgenic tomato plants impedes leaf senescence but enhances fruit yield and sugar content likely due to prolonged leaf photosynthesis in aging tomato plants. Our data indicate that SlNAP2 has a central role in controlling leaf senescence and fruit yield in tomato.
Leaf senescence represents the final stage of leaf development, which is an important part of a deciduous plant’s life cycle. The process is genetically programmed and involves a series of orderly changes that lead to degradation of macromolecules (e.g. proteins) and the mobilization of nutrients to actively growing organs such as young leaves, developing seeds, and fruits. The timing of leaf senescence is a major determinant of crop yield and quality. If senescence occurs early (i.e. premature senescence), the plant’s overall capacity to assimilate CO2 can be reduced (Wingler et al., 2006). Conversely, if senescence is late, then senescence-dependent nutrient recycling is inhibited (Himelblau and Amasino, 2001), which is important for reproductive success. Thus, plasticity in the timing of leaf senescence and the delicate balance between the onset and extent of leaf senescence are essential for ecological success and crop yield.
Leaves undergo massive changes in gene expression throughout senescence (Buchanan-Wollaston et al., 2005; Balazadeh et al., 2008; Breeze et al., 2011). These expression changes are precisely altered to produce a genomic expression program that is customized for the timing, progression, and/or magnitude of leaf senescence in response to different environmental conditions. Therefore, fine-tuning the expression of senescence-related transcriptional regulators is a powerful strategy to manipulate senescence for agronomic purposes, including increased biomass and improved crop yield and production traits.
In the last decade, senescence regulatory transcription factors, particularly those from the NAC family, have been identified. NAC proteins (NAM, ATAF1/2, and CUC2) represent one of the largest plant-specific transcription factor (TF) families with 117 members in Arabidopsis (Arabidopsis thaliana), 151 in rice (Oryza sativa), and 101 in tomato (Solanum lycopersicum; Ooka et al., 2003; Nuruzzaman et al., 2010; Tweneboah and Oh, 2017). NAC proteins harbor a highly conserved N-terminal domain that serves as a DNA binding domain and a variable C-terminal domain that is essential for transcriptional regulation. Several members of the NAC TF family have been reported to be functionally involved in the regulation of leaf senescence in Arabidopsis and other plant species including wheat (Triticum aestivum; Uauy et al., 2006; Zhao et al., 2015), cotton (Gossypium hirsutum; Fan et al., 2015), and rice (Zhou et al., 2013; Mao et al., 2017). For example, Arabidopsis AtNAP (ANAC029; Guo and Gan, 2006) and ORE1 (ANAC092; Kim et al., 2009; Balazadeh et al., 2010) have been identified as central positive regulators of leaf senescence. The microRNA miR164 suppresses accumulation of ORE1 transcripts in young leaves, whereas the transcription factor ETHYLENE-INSENSITIVE3 negatively regulates miR164 expression in an age-dependent manner resulting in decreased expression of miR164 and an increased expression of ORE1 in aging leaves (Kim et al., 2009). It has been demonstrated that ORE1 controls a complex regulatory circuitry that involves direct transcriptional activation of several genes involved in chlorophyll catabolism, ethylene biosynthesis, and senescence activation. Additionally, the ORE1 protein physically interacts with the chloroplast maintenance G2-like transcription factors GLK1 and GLK2, which hinders their transcriptional activity and contributes to the progression of leaf senescence (Rauf et al., 2013; Lira et al., 2017).
The NAC factor AtNAP has been reported to integrate abscisic acid (ABA) signaling and leaf senescence in different plant species (Guo and Gan, 2006; Zhang and Gan, 2012; Liang et al., 2014; Fan et al., 2015). Leaf and silique senescence are delayed in atnap null mutants but promoted in AtNAP inducible overexpression lines of Arabidopsis (Guo and Gan, 2006; Kou et al., 2012). AtNAP binds to the promoter of a Golgi-localized protein phosphatase 2C (PP2C) family gene, SAG113, and activates its expression. Induction of SAG113 inhibits stomatal closure and thereby promotes water loss and accelerates leaf senescence, whereas knocking out the gene delays developmental senescence (Zhang and Gan, 2012). Similarly in rice, OsNAP/PS1 (a functional ortholog of AtNAP) mediates ABA-induced leaf senescence by direct transcriptional activation of several chlorophyll degradation and senescence-associated genes (SAGs) including SGR, NYC1, NYC3, RCCR1, Osh36, OsI57, and Osh69. Overexpression of OsNAP promotes leaf senescence, but knocking down this gene causes a marked delay in senescence. Impeded leaf senescence in OsNAP RNA interference (RNAi) lines occurs concomitantly with a slower decrease in the rate of photosynthesis and ultimately an increased grain yield compared to wild-type plants (Liang et al., 2014). Recently, the cotton putative ortholog of AtNAP, GhNAP, was identified as a positive regulator of ABA-mediated leaf senescence. Reduction in GhNAP expression resulted in delayed senescence and improved cotton yield and fiber quality (Fan et al., 2015).
Tomato is one of the most popular fleshy fruit-bearing crops worldwide. The tomato genome has been sequenced (Tomato Genome Consortium, 2012), and tomato has been used extensively as a model crop for studies of fruit development and physiology. By contrast, very few studies have been conducted on the regulation of leaf senescence and its possible impact on tomato fruit yield and quality. Recently, the closest tomato putative orthologs of Arabidopsis ORE1 (i.e. SlORE1S02, SlORE1S03, and SlORE1S06) were identified and functionally characterized for their roles in regulating tomato leaf senescence (Lira et al., 2017). Like Arabidopsis ORE1, SlORE1s expression is regulated by miR164 in an age-dependent manner, and at the protein level SlORE1s interact with SlGLKs (Lira et al., 2017). Reduced expression of SlORE1s in RNAi lines led to delayed leaf senescence, extended carbon assimilation, and reduced expression of senescence marker genes. Prolonged photosynthetic activity in SlORE1s RNAi lines, compared with wild-type plants, resulted in a significant increase in the supply of photoassimilates to fruits and enhanced fruit yield.
In this study, we report that an ABA-activated NAC transcription factor named SlNAP2 (the tomato putative ortholog of AtNAP from Arabidopsis and OsNAP from rice) plays a central role in regulating leaf senescence. Furthermore, we characterize the influence of SlNAP2 on fruit quality and yield. SlNAP2 is revealed to be a positive regulator of leaf senescence and the senescence regulatory module controlled by SlNAP2 is shown to be highly conserved between tomato and other plant species whose NAP-control mechanisms have been elaborated. SlNAP2 directly controls the expression of the senescence-associated gene SlSAG113 (a homolog of Arabidopsis SAG113) and chlorophyll degradation-related genes SlSGR1 and SlPAO. Intriguingly, we also observed that SlNAP2 directly regulates the expression of both ABA biosynthesis (SlNCED1) and ABA degradation (SlCYP707A2) genes, suggesting the existence of a self-regulation mechanism by which SlNAP2 tunes its dynamic expression in leaves. Transgenic lines with reduced expression of SlNAP2 exhibit a significant delay in leaf senescence, along with an increase in fruit yield and fruit sugar content. Our research further emphasizes the importance of the regulation of leaf senescence for achieving increased fleshy fruit yield and sugar content.
RESULTS
SlNAP2 Is Induced during Senescence
SlNAP2 (Solyc04g005610.2.1) and SlNAP1 (Solyc05g007770.2.1) are closely related (∼72.7% identity at the amino acid level) NAC TFs in tomato (Fig. 1A). Phylogenetic analysis revealed that SlNAP1 and SlNAP2 are homologous to ANAC029/AtNAP (Arabidopsis NAC-like, activated by Apetala3/Pistillata, At1g69490), which is a well-known senescence regulatory NAC TF (Guo and Gan, 2006; Fig. 1B).
Figure 1.
SlNAP1 and SlNAP2 are up-regulated during leaf senescence. A, Protein sequence alignment of AtNAP, SlNAP1, and SlNAP2. Amino acids identical in all three proteins are highlighted with a black background, while conservative substitutions are shown with a gray background. Asterisks indicate the stop codons. B, Phylogenetic analysis of NAC proteins. The phylogenetic tree was constructed by MEGA 5.05 software using the neighbor-joining method with the following parameters: bootstrap analysis of 1,000 replicates, Poisson model, and pairwise deletion. SlNAP1, SlNAP2, NAC-NOR, and SlNAC3 are senescence-induced tomato TFs of the NAP family, while all other TFs are from Arabidopsis. Gene codes are as follows: ATAF1, At1g01720; ATAF2, At5g08790; AtNAC2, At5g39610; AtNAM, At1g52880; AtNAP, At1g69490; CUC2, At5g53950; NAC-NOR, Solyc10g006880; SlNAP1, Solyc05g007770; SlNAP2, Solyc04g005610; and SlNAC3, Solyc07g063420. The numbers at the nodes indicate the bootstrap values. The bar at the bottom indicates the relative divergence of the sequences examined. C, The left panel shows representative images of S. lycopersicum cv Moneymaker leaves at different developmental stages: young leaves (YL), mature leaves (ML), senescent leaves (SL), and late senescent leaves (LS). The right panel denotes the expression levels of SlNAP1 and SlNAP2 in such leaves, determined by RT-qPCR. The y axis indicates expression level (40-dCt). Values are expressed as the difference between an arbitrary value of 40 and dCt, so that high 40-dCt values indicate high gene expression levels. Data are means of three biological replicates ± sd. Asterisks indicate significant difference from young leaves (Student’s t test; **P ≤ 0.01).
To examine the expression patterns of SlNAP1 and SlNAP2 in tomato, we harvested various organs, including roots, stem, flowers, and leaves at different developmental stages (young leaves, mature leaves, early senescent leaves, and late senescent leaves), as well as fruits (immature green, mature green, breaker, and ripe red), and then analyzed transcriptional changes using reverse-transcription quantitative PCR (RT-qPCR). SlNAP1 and SlNAP2 transcripts were detected in all organs examined (Fig. 1C; Supplemental Fig. S1A). However, both genes were significantly induced during leaf senescence and fruit ripening.
Accordingly, histochemical analysis of transgenic SlNAP2pro:GUS tomato plants, expressing the GUS reporter under the control of the 1.5-kb SlNAP2 promoter, revealed elevated GUS activity (indicating enhanced promoter activity) in older parts of the leaves. This was consistent with the results of the RT-qPCR-based expression analyses (Supplemental Fig. S1B). Furthermore, expression analysis by RT-qPCR indicated that both SlNAP1 and SlNAP2 transcript levels increased in leaves during dark-induced senescence (Supplemental Fig. S1C).
SlNAP2 Promotes Leaf Senescence
To elucidate the possible involvement of SlNAP2 in the regulation of leaf senescence, we first generated transgenic tomato lines (cv Moneymaker) constitutively expressing SlNAP2 under the control of the CaMV 35S promoter. Two lines (hereafter called OX-L2 and OX-L10) were selected for further analysis (Supplemental Fig. S2A). Overexpression of SlNAP2 in tomato plants triggered accelerated leaf senescence (Fig. 2A). The two OX lines exhibited a significantly higher number of yellow leaves compared to the wild type 12 weeks after sowing (Fig. 2B). Consequently, the chlorophyll content of the third true leaf decreased faster in OX lines than in the wild type during a period of 6 weeks (Fig. 2C).
Figure 2.
Overexpression of SlNAP2 leads to early developmental leaf senescence. A, Phenotype of wild-type, OX-L2, and OX-L10 plants. Upper panel, 12-week-old plants; lower panel, phenotypes of the third true leaf of 10-week-old plants (leaves were individually photographed and compiled for comparison). B, Ratio of yellow to all leaves in 12-week-old wild-type, OX-L2, and OX-L10 plants. Leaves were counted as yellow if chlorophyll content had declined by more than 50% compared to those leaves in 8-week-old plants. Data are means ± sd (n = 5). C, Chlorophyll loss of the third true leaf (counted from the bottom of the stem) of wild-type, OX-L2, and OX-L10 plants 8, 10, 12, and 14 weeks after sowing (8W–14W). Chlorophyll content was measured using a SPAD analyzer and compared to the content in each genotype at time point 8W (set to 1). Data are means ± sd of three biological replicates. Significant difference from the wild type is denoted by one asterisk (Student’s t test; P ≤ 0.05) or two asterisks (P ≤ 0.01). Red asterisks indicate a significant difference between OX-L2 and the wild type, and blue asterisks indicate a significant difference between OX-L10 and the wild type. D, Expression of senescence marker genes (SlSAG12, SlSAG113, and SlSGR1) in lower positioned leaves of 12-week-old wild-type, OX-L2, and OX-L10 plants, analyzed by RT-qPCR. Data are means ± sd of three biological replicates. E, Expression of senescence marker genes in SlNAP2-IOE plants after 6-h ESTR induction of SlNAP2 expression, compared to expression in mock-treated plants. Data are means ± sd of three biological replicates. Values on the y axes in D and E represent the difference between an arbitrary value of 40 and dCt, so that high 40-dCt values indicate high gene expression level. Asterisks in B, D, and E indicate significant difference from the wild type (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01).
To further analyze the early senescence phenotype of SlNAP2-OX lines at the molecular level, we checked the expression of SAGs. To this end, we selected the following three SAGs: SlSAG12 (Solyc02g076910), a homolog of Arabidopsis SAG12; SlSAG113 (Solyc05g052980), a homolog of Arabidopsis SAG113 and a direct downstream target gene of AtNAP (Zhang and Gan, 2012); and SlSGR1 (Solyc08g080090), which encodes a stay-green protein involved in the degradation of chlorophyll during leaf senescence. We observed that all three genes were transcriptionally induced during natural and dark-induced senescence as well as upon treatment with ABA in wild-type plants (Supplemental Fig. S3, A–C). In accordance with an early senescence phenotype of SlNAP2-OX plants, expression of all three SAGs was significantly higher in the third true leaf of 12-week-old SlNAP2-OX lines compared to the wild type (Fig. 2D).
Next, we generated estradiol-inducible SlNAP2 overexpression lines (SlNAP2-IOE; Supplemental Fig. S2B) and checked the expression by RT-qPCR of SlSAG12, SlSAG113, and SlSGR1 after a 6-h estradiol (ESTR) induction of SlNAP2 expression. The expression of all three SAGs was up-regulated in SlNAP2-IOE plants following ESTR treatment compared to the mock treatment (Fig. 2E). These observations support a role for SlNAP2 in the regulation of developmental leaf senescence by direct or indirect regulation of SAGs.
Knockdown of SlNAP2 Delays Developmental Leaf Senescence
To further investigate the function of SlNAP2 in controlling senescence, transgenic knockdown lines with reduced expression of SlNAP2 were generated using RNAi (Supplemental Fig. S2C). Two lines with reduced expression of SlNAP2 were selected for further analysis (KD-L2 exhibiting an ∼4-fold reduction and KD-L14 with an ∼32-fold reduction). Expression of SlNAP1, the putative ortholog of SlNAP2, was unaltered in these lines, indicating target gene specificity with RNAi (Supplemental Fig. S2D). Considering the high sequence similarity between SlNAP1 and SlNAP2, and the possibility of functional redundancy, we also created SlNAP1 and SlNAP2 double knockdown lines (dKD) using artificial microRNA (amiRNA) technology. Specifically, a 21-bp sequence identical in SlNAP1 and SlNAP2 was selected for generating the amiRNA construct (Supplemental Fig. S2, C and E). In general, the analysis of senescence phenotypes revealed that knocking down SlNAP2 alone (KD) or in combination with SlNAP1 (dKD) resulted in a significant delay of leaf senescence as measured by the number of yellow leaves, the rate of chlorophyll loss, and the expression of senescence marker genes (Fig. 3; see next sections). However, the observed delay in senescence was slightly more pronounced in dKD plants indicating a partial functional redundancy of the two proteins.
Figure 3.
Knocking down SlNAP2 delays developmental leaf senescence. A, Phenotype of SlNAP2 knockdown (KD-L2) and SlNAP2/SlNAP1 double knockdown (dKD-L4) plants. Upper panel, 12-week-old plants; lower panel, phenotypes of the third true leaf of 10-week-old plants. The wild-type plant shown is the same as in Figure 2A (as overexpressor and knockdown lines, together with wild-type plants, were grown side-by-side in the same experiment; plants and leaves were individually photographed and compiled for comparison). B, Ratio of yellow to all leaves in 12-week-old plants. Leaves were counted as yellow if chlorophyll content had declined by more than 50% compared to those leaves in 8-week-old plants. Asterisks indicate significant difference from wild-type plants (Student’s t test; **P ≤ 0.01; n = 5). C, Chlorophyll loss of the third true leaf (counted from the bottom of the stem) of wild-type, KD-L2, and dKD-L4 plants 8, 10, 12, and 14 weeks after sowing (8W–14W). Chlorophyll content was measured using a SPAD analyzer and compared to the content in each genotype at time point 8W (set to 1). Data are means ± sd of three biological replicates. Significant difference from wild type is denoted by one asterisk (Student’s t test; P ≤ 0.05) or two asterisks (P ≤ 0.01). Red asterisks indicate a significant difference between dKD-L4 and the wild type, and blue asterisks indicate a significant difference between KD-L2 and the wild type. D, Metabolite contents of SlNAP2 transgenic lines compared to wild-type plants. The fifth fully expanded leaves were harvested from 8-week-old wild-type, OX-L2, KD-L2, and dKD-L4 plants. Metabolite content was analyzed using gas chromatography-mass spectrometry (n = 4). Log2 fold change (FCh) values of the relative metabolite contents are presented here. Asterisks indicate significant difference from the wild type (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01).
We also analyzed the levels of primary metabolites in fully expanded fifth leaves of 8-week-old SlNAP2 transgenic and wild-type plants. Overall, more significant changes among the genotypes were observed with amino acids (Fig. 3D; Supplemental Table S1). The levels of aromatic amino acids (AAAs; Trp, Tyr, and Phe) were greater in mature leaves of SlNAP2-OX plants but lower in leaves of SlNAP2-KD and dKD plants. An increase in AAA levels was reported previously for dark-induced and nitrate limitation-induced senescence as well as natural senescence (Gibon et al., 2006; Fahnenstich et al., 2007; Araújo et al., 2010, 2011; Watanabe et al., 2013). By contrast, the levels of branched chain amino acids (Ile and Val) were significantly lower in mature leaves of SlNAP2-OX plants. The level of Pro, a stress-induced osmoprotectant, was higher in SlNAP2-KD but significantly lower in SlNAP2-OX compared with wild-type plants. Glu and Asp were lower in mature leaves of SlNAP2-OX plants, and greater in SlNAP2-KD and dKD lines. Conversely, the level of Gln (the major amino acid translocated in the phloem sap during developmental senescence; Guo et al., 20044; Diaz et al., 2005) was higher in SlNAP2-OX but significantly lower in SlNAP2-KD plants. γ-Aminobutyric acid was significantly higher in SlNAP2-OX and lower in SlNAP2-dKD than in the wild type. Accumulation of γ-aminobutyric acid is known to be associated with senescence (Ansari et al., 2005, 2014).
Among tricarboxylic acid cycle intermediates, malic acid and fumaric acid accumulated in SlNAP2 mutants. Accumulation of these metabolites was reported to be associated with older parts of the leaves (Watanabe et al., 2013). These results reveal that SlNAP2-OX and KD plants have varying mature leaf metabolite profiles that reflect their altered senescence phenotypes. Thus, SlNAP2 plays an important role as a positive regulator of leaf senescence in tomato.
SlNAP2 Is Involved in Dark-Induced Senescence
Darkness is widely used as a tool to induce senescence. Several reports indicate that darkening of leaves share many physiological and molecular alterations with developmental senescence, including a decline in photosynthesis and chlorophyll content, leaf yellowing, and enhanced expression of SAGs (Weaver et al., 1998; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Parlitz et al., 2011). To test the involvement of SlNAP2 in dark-induced senescence, young leaves (collected from the upper part of tomato stems) from 10-week-old wild-type and SlNAP2 transgenic lines were subjected to darkness for a period of up to 14 d. SlNAP2-OX plants displayed an early leaf yellowing phenotype under darkness, while the leaves of KD-L2 and dKD-L4 remained greener compared to the wild type under the same condition (Fig. 4A). Dark-induced early leaf yellowing of OX-L2 was accompanied by a dramatic reduction in chlorophyll content (∼70%), while chlorophyll loss was significantly less in the wild type (∼45%), KD-L2 (∼20%), and dKD-L4 (∼14%) leaves (Fig. 4B). Accordingly, the expression of senescence marker genes such as SlSAG12, SlSAG113, and SlAGT1 (Solyc10g076250), as well as of genes involved in chlorophyll degradation, such as SlSGR1, SlPPH (Solyc01g088090), SlPAO (Solyc11g066440), and SlNYC1 (Solyc07g024000), was significantly elevated in OX-L2 but reduced in KD-L2 and dKD-L4 lines compared with the wild type under dark incubation (Fig. 4C). These data suggest that SlNAP2 acts as a positive regulator of both natural and dark-induced leaf senescence by directly or indirectly controlling senescence-associated chlorophyll degradation.
Figure 4.
SlNAP2 accelerates dark-induced senescence. A, Young detached leaves of 10-week-old wild-type and SlNAP2-transgenic lines before (day 0) and after 14 d of dark treatment. Leaves were detached from the top part of the stem. Leaves in each panel were individually photographed and compiled for presentation. B, Chlorophyll content of control and dark-treated leaves, determined using a SPAD analyzer. Data are means of leaves from three plants ± sd. Asterisks indicate significant differences from wild-type plants (Student’s t test; **P ≤ 0.01). C, Expression of SAGs (SlSAG12, SlSAG113, SlAGT1, and SlSAG15) and chlorophyll degradation genes (SlSGR1, SlPPH, SlPAO, and SlNYC1) in control and dark-treated leaves of wild-type and SlNAP2 transgenic lines. The y axis indicates expression level (40-dCt). Data are means ± sd of three biological replicates. Asterisks indicate significant difference from wild-type plant (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01).
Involvement of SlNAP2 in ABA-Mediated Leaf Senescence
ABA is an important hormone involved in the regulation of plant growth and development, including leaf senescence. An increase in endogenous ABA levels during senescence has been reported in several plant species, including oat (Avena sativa; Gepstein and Thimann, 1980), rice (Philosoph-Hadas et al., 1993), maize (Zea mays; He et al., 2005), and Arabidopsis (Balazadeh et al., 2014; Yang et al., 2014). Furthermore, it is known that ABA promotes leaf senescence (Gepstein and Thimann, 1980; Quiles et al., 1995; Yang et al., 2003; Lee et al., 2011; Zhao et al., 2016), the exogenous application of ABA can accelerate chlorophyll degradation (Quiles et al., 1995; Gao et al., 2016), and ABA-deficient mutants display delayed senescence in both rice (Mao et al., 2017) and Arabidopsis (Pourtau et al., 2004).
Transcript levels of SlNAP2 began to increase (∼3-fold change) 2 h after ABA treatment of wild-type seedlings, with additional increases at later time points (e.g. ∼6-fold changes at 8 h; Fig. 5A). Conversely, SlNAP2 expression was significantly reduced in tomato mutants deficient in ABA biosynthesis, such as sitiens (sit) and notabilis (not), further confirming that SlNAP2 is an ABA-activated transcription factor (Fig. 5B). To examine the possible role of SlNAP2 in regulating ABA-induced leaf senescence, we analyzed the phenotype of SlNAP2 transgenic plants upon application of exogenous ABA. Specifically, detached leaves from wild-type and SlNAP2 transgenic plants were exposed to ABA (40 µm) and compared to control (mock) treatments that lacked ABA. When wild-type leaves were treated with ABA, senescence symptoms were induced such as leaf yellowing, chlorophyll loss, and enhanced expression of senescence marker genes (Fig. 5, C–E). Interestingly, ABA-induced early senescence was more pronounced in leaves of SlNAP2 overexpressors than the wild type, while SlNAP2 knockdown lines (KD-L2 and dKD-L4) exhibited stay-green phenotypes upon ABA treatment (Fig. 5, C–E). Activation of SlNAP2 by ABA and the altered senescence phenotype of SlNAP2 transgenic lines upon treatment with ABA implicate SlNAP2 as a regulator of ABA-dependent leaf senescence.
Figure 5.
SlNAP2 regulates ABA-induced leaf senescence. A, Elevated expression of SlNAP2 after ABA treatment. Three-week-old wild-type seedlings were treated with ABA (40 µm) for 2, 4, 8, and 16 h. Data are means ± sd of three biological replicates. Asterisks indicate significant differences from mock-treated plants (Student’s t test; *P ≤ 0.05). B, Reduced expression of SlNAP2 in ABA-deficient mutants, sit and not. Data are means ± sd of three biological replicates. Asterisks indicate significant difference from the wild type (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01). C, Phenotype of detached leaves from 10-week-old wild-type and SlNAP2-transgenic plants before (0 d) and after treatment with 40 µm ABA for 9 d (9 d). Young leaves from the top of the stem were used and individually photographed. D, Chlorophyll content of control and ABA-treated leaves, determined using a SPAD analyzer. Data are means ± sd from six leaves of three different plants. Asterisks indicate significant difference from respective mock-treated leaves (Student’s t test; **P ≤ 0.01). E, RT-qPCR analysis of SlSAG12, SlSAG113, and SlSGR1 expression in control and ABA-treated leaves. The y axis indicates expression level (40-dCt). Asterisks indicate significant difference from the wild type (Student’s t test; **P ≤ 0.01). DAT, days after treatment.
Identification of the Consensus Target Sequence of SlNAP1 and SlNAP2
TFs bind to cis-regulatory elements in promoters of target genes to control their expression. Knowledge of the binding sites favors the identification of TF target genes and helps gain insight into its regulatory functions. Previous work has identified high-affinity binding sequences of TaNAC69 from wheat using an in vitro binding site selection assay employing the CELD fusion method (Xue et al., 2006). SlNAP1 and SlNAP2 are homologs of TaNAC69, suggesting they have overlapping DNA-binding specificities. To identify the target sequences of SlNAP1 and SlNAP2, we analyzed the binding activity of both TFs on 12 randomly selected oligonucleotides with TaNAC69 binding motifs, including SO1, which is considered a high-affinity binding sequence of TaNAC69 (Xue et al., 2006). SlNAP1 and SlNAP2 are capable of binding to TaNAC69-selected motifs containing the YACG (or CGTR) core sequence and share similar binding sequence specificities to that of TaNAC69, with SO1 as the highest-affinity binding motif (Supplemental Fig. S4A).
To assess the specificity of binding, nucleotide mutation (substitution, insertion, or deletion) experiments were performed on the basis of SO1 (SO1m1–SO1m18) and SO48 (SO48m1 and SO48m2) motifs. Our analysis revealed that mutation of nucleotides in the core motifs (oligonucleotides SO1m3, 4, 8, and 9, and SO48m1) led to a dramatic reduction or abolishment of SlNAP1 binding. A significant drop in the binding affinity was noticed upon reducing the distance between the two core motifs from 5 to 4 bp (oligonucleotide SO1m1) or increasing it to 7 bp (oligonucleotide SO1m2; Supplemental Fig. S4, A and B). Based on these results, we conclude that CGT[AG](5N)NACG[ACT][AC][AT][ACG][ACT] and CACG[ACT][AC][AT][AGT][CT] are high-affinity binding sites of SlNAP1. SlNAP2 appears to be more tolerant to the nucleotide mutations of SO1 (which contains two core motifs) than SlNAP1 (Supplemental Fig. S4B). However, the mutation in the single core motif of SO48 (SO48m1) led to almost complete abolishment of its binding activity. To further demonstrate binding of SlNAP2 to the TaNAC69 motif, an electrophoretic mobility shift assay (EMSA) was performed. This experiment revealed that SlNAP2 strongly binds to SO1 (Supplemental Fig. S4C).
Direct Transcriptional Regulation of SAGs and ABA-Related Genes by SlNAP2
To further elucidate the senescence control function of SlNAP2 at the molecular level, we conducted an RT-qPCR analysis to evaluate the expression of 22 senescence-related genes in SlNAP2-IOE plants, including known SAGs, genes involved in chlorophyll degradation, and ABA-associated genes (Supplemental Fig. S2B). Our data revealed that the majority of genes (15 out of 22; SlSAG12, SlSAG15, SlSAG113, SlSGR1, SlAGT1, SlSGR1, SlNYC1, SlPAO, SlNCED1, SlABA3, SITIENS, SlAAO3, SlCYP707A2, SlCYP707A4, and SlABCG40) were induced (log2 fold change > 1) 6 h after estradiol treatment in SlNAP2-IOE plants compared to mock-treated controls (Fig. 6A), suggesting their early and positive regulation by SlNAP2. Expression of all genes examined except one (SlPPH) was down-regulated in KD-L2 and dKD-L4. We then searched for the presence of SlNAP2 binding sites (perfect match) within 1-kb promoters of SlNAP2 early responsive genes to identify potential direct target genes of the NAC TF. Twelve genes (SlSAG12, SlSAG15, SlSAG113, SlSGR1, SlPAO, SlNCED1, SlABA3, SITIENS, SlAAO3, SlCYP707A2, SlCYP707A4, and SlABCG40) harbor SlNAP2 binding sites (perfect match) within their promoters. To test if SlNAP2 interacts with the promoters of early responsive genes in vivo, we generated transgenic lines expressing SlNAP2-GFP fusion protein under the control of the CaMV 35S promoter. As expected for a transcription factor, SlNAP2-GFP fusion protein was located in nuclei, as visualized by fluorescence microscopy of epidermal cells of transgenic tomato leaves (Fig. 6B). Using SlNAP2-GFP lines, we performed chromatin immunoprecipitation coupled withqPCR (ChIP-qPCR) and confirmed direct binding of SlNAP2 to the promoters of SlSAG113, SlSGR1, SlPAO, SlNCED1, SlCYP707A2, and SlABCG40 in vivo (Fig. 6, C and E). Furthermore, employing EMSAs, direct physical interaction of SlNAP2 with the promoters of all six genes was confirmed (Fig. 6, D and F). SlNCED1 encodes 9-cis-epoxycarotenoid dioxygenase, a key enzyme in ABA biosynthesis, and SlCYP707A2 encodes ABA 8′-hydroxylase, a key enzyme in the oxidative catabolism of ABA in tomato (Ji et al., 2014). Positive regulation of both genes by SlNAP2 reveals a complex role for this TF for the regulation of ABA homeostasis.
Figure 6.
Direct regulation of SAGs and ABA-related genes by SlNAP2. A, Heat map showing the fold change (FCh; log2 basis) of the expression ratio of SAGs, chlorophyll degradation as well as ABA biosynthesis and signaling genes in the following samples: 3-week-old SlNAP2-IOE seedlings (line IOE-L5) treated with estradiol (15 µm) for 6 h compared to ethanol (0.15%, v/v) treated seedlings (mock); KD-L2 and dKD-L4 lines, compared to the wild type. Blue, downregulated; red, upregulated (as indicated by the color bar). Data represent means of three biological replicates. Asterisks indicate significant difference from mock-treated and/or wild-type plants (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01). Genes shown in bold are direct targets of SlNAP2 (see C–F). B, Subcellular localization of SlNAP2-GFP fusion protein in epidermal cells of transgenic tomato leaves, visualized by fluorescence microscopy. Top, bright field; bottom, GFP fluorescence (green) under bright field. Bar = 10 µm. C, ChIP-qPCR shows enrichment of SlSAG113, SlSGR1, and SlPAO (but not SlSAG12) promoter regions containing the SlNAP2 binding site. Mature leaves (nos. 3–5) harvested from 8-week-old SlNAP2-GFP plants were used for the ChIP experiment. Values were normalized to the values for Solyc04g009030 (promoter lacking a SlNAP2 binding site). Data are means ± sd of two independent biological replicates, each performed with three technical replicates. D, EMSA showing binding of purified SlNAP2-CELD protein to the 5′-DY682-labeled 40-bp-long promoter fragments of SlSAG113, SlSGR1, and SlPAO, containing the SlNAP2 binding sites. Lane 1, labeled promoter fragment only; lane 2, labeled promoter fragment plus SlNAP2-CELD protein, showing the retardation band (bound probe); lane 3, labeled promoter fragment, SlNAP2-CELD protein plus 100-fold molar access of nonlabeled promoter (competitor). E, ChIP-qPCR. Mature leaves of 8-week-old SlNAP2-GFP plants were harvested for the ChIP experiment. qPCR was performed to quantify the enrichment of SlNCED1, SlCYP70A2, and SlABCG40 promoter regions. Values were normalized to the values for Solyc04g009030 (promoter lacking a SlNAP2 binding site). Data are means ± sd of two biological replicates, each performed in two technical replicates. F, EMSA showing binding of purified SlNAP2-CELD protein to 5′-DY682-labeled 40-bp-long promoter fragments of SlNCED1, SlCYP707A2, and SlABCG40, containing the SlNAP2 binding sites. For description of lanes, see legend to D. G, ABA content. Three-week-old wild-type, KD-L2, and dKD-L4 plants were harvested and ABA content was determined using UPLC-ESI-MS/MS. ABA content is shown as means ± sd of three biological replicates. Asterisks indicate significant difference from the wild type (Student’s t test; *P ≤ 0.05). FW, Fresh weight.
Next, we determined ABA levels in SlNAP2 transgenic lines and wild-type plants. Suppression of SlNAP2 results in higher ABA levels in 3-week-old plants, suggesting its role as an inhibitor of ABA accumulation and subsequently its own accumulation at this stage (Fig. 6G). No change in ABA levels was detected in 8-week-old SlNAP2-KD lines and wild-type plants (Supplemental Fig. S5).
SlNAP2 Knockdown Enhances Fruit Yield and Sugar Content
To test the influence of altered leaf senescence (triggered by manipulating SlNAP2 expression) on fruit yield, we measured various yield-associated parameters. SlNAP2-OX plants started producing flowers around one week earlier than wild-type plants (Supplemental Fig. S6A). The number of fruits per plant significantly increased in SlNAP2-KD plants compared to the wild type, while fruit number decreased in SlNAP2 overexpressors (Supplemental Fig. S6B). Notably, we did not observe a significant difference between genotypes for the mean time span between anthesis and the fruit breaker stage (Supplemental Fig. S6D). Furthermore, fruit size was almost indistinguishable between SlNAP2-KD and wild-type plants but was significantly reduced in SlNAP2 overexpressors (Supplemental Fig. S6C).
Sweetness, which results from enhanced soluble sugar content, is one of the most important traits of tomato fruits. During senescence, sugars are translocated from older leaves (source) to developing fruits (sinks). To test the effect of altered senescence in SlNAP2 transgenics on fruit sweetness, we measured Brix values and soluble sugar content in fruits of different developmental stages. SlNAP2-KD and dKD plants synthesized higher contents of soluble solids in ripe fruits as demonstrated by Brix units (Fig. 7A). The levels of sugars (Fru, Suc, and Glc) were also significantly higher in SlNAP2 KD and dKD lines, particularly at the breaker stage (Fig. 7, B–D). Collectively, our data clearly indicate an association of SlNAP2 deficiency with delayed aging and improved fruit yield and metabolism.
Figure 7.
Fruit Brix value and soluble sugar content. A, The content of total soluble solids of ripe red fruits was determined using a digital refractometer. Values represent the means ± sd of six biological replicates (Student’s t test; *P ≤ 0.05). The contents of Fru (B), Suc (C), and Glc (D) in the pericarps of SlNAP2 transgenic and wild-type fruits at different developmental stages, analyzed by gas chromatography-mass spectrometry (n = 4). Relative metabolite levels were obtained by normalizing the intensity value of each metabolite to the ribitol internal standard. Asterisks indicate significant difference from the wild type (Student’s t test; *P ≤ 0.05 and **P ≤ 0.01). MG, Mature green fruits.
DISCUSSION
The main biological role of senescence is nutrient recycling, which is essential for plant survival, crop yield, and the shelf life of leafy vegetables. Although several studies have demonstrated a connection between leaf senescence and productivity with regard to biomass production and seed/grain yield (for review, see Gregersen et al., 2013), research that has examined the effects of leaf senescence on fleshy fruit production and its nutritional quality is limited. Recently, Lira et al. (2017) demonstrated that a simultaneous suppression of all three homologs of ORE1 in tomato (SlORE1S02, SlORE1S03, and SlORE1S06) resulted in a remarkable delay of leaf senescence, increased fruit yield, and enhanced levels of sugars in the ripe fruits. Furthermore, in apple (Malus sp.), overexpression of the YTH-domain-containing RNA-binding proteins MhYTP1 and MhYTP2 promoted leaf senescence and accelerated fruit ripening (Wang et al., 2017).
In this study, we demonstrate the senescence-regulatory function of SlNAP2, a member of the NAC family of TFs in tomato, and show that the manipulation of leaf senescence by this TF is effective for improving fleshy fruit yield. SlNAP2 is expressed at all stages of leaf and fruit development; however, its expression is significantly induced in senescing leaves and ripe fruits. Similar to its homologs in Arabidopsis, rice and cotton, expression of SlNAP2 is rapidly induced by ABA (one of the main hormones that initiates leaf senescence), indicating conservation of the upstream regulatory pathways that control the ABA-mediated induction of NAP genes across monocot and eudicot plant species (Zhang and Gan, 2012; Liang et al., 2014; Fan et al., 2015).
Transgenic plants with enhanced expression of SlNAP2 exhibit early leaf senescence during typical age-dependent senescence as well as during senescence induced by darkness (Figs. 2 and 4), as measured by reduced chlorophyll content and enhanced expression of senescence marker genes. In contrast, reduced expression of SlNAP2 in knockdown plants results in a substantial delay of natural and dark-induced senescence. The delayed senescence was more pronounced in transgenic lines with reduced expression of both SlNAP2 and its closely related homolog SlNAP1, revealing partially redundant and additive functions of the two genes in the regulation of leaf senescence in tomato.
During senescence, a drastic change occurs in leaf metabolism, which is mainly due to the degradation of metabolites and the subsequent mobilization of nutrients toward developing organs. Metabolomic profiles of fully expanded fifth leaves (from 8-week-old plants) revealed a clear metabolic shift between SlNAP2 overexpression and knockdown lines corresponding to the contrasting senescence phenotypes (Fig. 3). Among amino acids, the levels of aromatic amino acids (particularly Trp and Phe) and Gln were greater in SlNAP2-OX plants but significantly lower in leaves of SlNAP2-KD and dKD plants. Induction of AAAs, and more specifically Trp, during senescence has been shown in several species such as rice (Kang et al., 2009), Arabidopsis (Watanabe et al., 2013; Chrobok et al., 2016), tomato (Araújo et al., 2012), and tobacco (Nicotiana tabacum; Li et al., 2016). Gln is a major source of remobilizable N during senescence and its synthesis is induced by Gln synthetase 1 (GS1) activity in senescent leaves (Tabuchi et al., 2007; Park et al., 2010). Indeed, the transcript level of SlGS1 significantly decreased in SlNAP2-KD and dKD plants in accordance with the dramatic reduction in the level of Gln in those lines (Supplemental Fig. S7). The majority of other amino acids, including branched chain amino acids (Ile and Val), Gly, Ser, Lys, Asp, and Glu, were downregulated in SlNAP2-OX, but upregulated in KD plants. In fact, such a metabolic pattern can be explained in part by enhanced expression of SlGDH (encoding Glu dehydrogenase) in SlNAP2-OX plants and its significant reduction in the KD lines (Supplemental Fig. S7). It has been shown that SlGDH activity increases during senescence, which is essential for deamination of Glu and subsequent catabolism of several amino acids (Masclaux et al., 2000; Masclaux-Daubresse et al., 2006; Miyashita and Good, 2008).
In addition to age- and dark-induced senescence, ABA-induced senescence was also significantly impeded in SlNAP2-suppressed plants, suggesting an important role of SlNAP2 in ABA-induced leaf senescence. Previous reports demonstrated a functionally conserved role for NAP in the regulation of age- and ABA-induced leaf senescence in different eudicot and monocot species such as Arabidopsis (AtNAP), rice (OsNAP), and cotton (GhNAP; Guo and Gan, 2006; Zhang and Gan, 2012; Zhou et al., 2013; Liang et al., 2014; Fan et al., 2015). Reports on downstream regulatory pathways indicate that AtNAP and OsNAP negatively regulate ABA signaling and biosynthesis pathways in Arabidopsis and rice, respectively. In Arabidopsis, AtNAP directly and positively regulates expression of SAG113, a Golgi-localized protein phosphatase 2C (PP2C). Hence, it negatively regulates the ABA-induced promotion of stomatal closure, consequently leading to water loss and triggering leaf senescence (Zhang and Gan, 2012). In rice, OsNAP suppresses ABA biosynthesis genes including OsNCED1, OsNCED3, and OsNCED4, thereby controlling ABA synthesis via a feedback mechanism (Liang et al., 2014).
To unravel the molecular mechanism through which SlNAP2 regulates leaf senescence and gain insight into the level of conservation of regulatory networks controlled by SlNAP2 in tomato and its homologs in other species, we performed RT-qPCR-based expression profiling of senescence-associated genes, chlorophyll degradation genes, and ABA biosynthesis and signaling genes in wild-type plants, SlNAP2 inducible overexpression lines (shortly after SlNAP2 induction by estradiol), and SlNAP2 knockdown lines. Our data revealed that most genes were transcriptionally enhanced after induction of SlNAP2 in estradiol-inducible overexpression plants but reduced in KD and dKD lines. Of the differentially expressed genes (Fig. 6), SlSAG113, SlSGR1, SlPAO, SlNCED1, SlCYP707A2, and SlABCG40 contained bipartite SlNAP2 binding sites within their 1-kb 5′ upstream regulatory regions (promoters), revealing potential direct targets of SlNAP2. The direct interaction between SlNAP2 and the promoters of all six genes was confirmed in vivo by ChIP-qPCR and in vitro by EMSA (Fig. 6). SlSAG113 is a homolog of Arabidopsis SAG113, a direct downstream target gene of AtNAP. SlSGR1 and SlPAO are crucial to chlorophyll degradation in tomato leaves and fruits, and the sgr mutation in tomato results in a stay-green phenotype (Akhtar et al., 1999; Hu et al., 2011; Guyer et al., 2014). Interestingly in rice, OsNAP directly targets the promoters of genes involved in chlorophyll degradation such as SGR, NYC1, NYC3 (PPH), and RCCR, besides other SAGs, and enhances their expression (Liang et al., 2014). SlNAP2 shares high amino acid sequence similarity in the NAC domain with AtNAP (∼92, 2%) and OsNAP (∼79, 2%), explaining the similar target gene profiles.
Intriguingly, SlNAP2 directly promotes the expression of both SlNCED1 and SlCYP707A2, key tomato genes involved in ABA synthesis and catabolism, respectively. It has been shown that the level of ABA in fruits is mainly regulated by SlNCED1 and SlCYP707A2 at the transcriptional levels (Ji et al., 2014). However, functional properties of the enzymes in leaves with respect to regulation of ABA levels and the consequent impact on leaf senescence remain to be investigated. A positive regulation of both genes by SlNAP2 reflects a more complex regulation of ABA homeostasis in tomato leaves. ABA levels were significantly higher in SlNAP2 KD and dKD plants at the early stages of development, while ABA levels were not altered by SlNAP2 at the later stages. This result suggests the existence of a self-regulation mechanism by which SlNAP2 tunes its dynamic expression during leaf development (Fig. 6G). Similarly, a feedback mechanism in ABA-OsNAP1 regulation was previously reported in rice, where ABA amounts were greater in nonsenescent leaves of OsNAP RNAi plants than in those of the wild type (Liang et al., 2014). Together, the direct regulation of SlSAG113 and chlorophyll degradation genes (such as SlSGR1 and SlPAO) by SINAP2, as well as SINAP2’s tight link to ABA signaling demonstrate that the regulation of leaf senescence by SlNAP2 is an evolutionarily conserved pathway for senescence control in eudicots and monocots.
Leaf senescence can significantly impact crop production by remobilization of photoassimilates from older vegetative tissues to sink organs. To examine the influence of altered leaf senescence mediated by a modification of SlNAP2 on tomato productivity, we analyzed different yield-related traits, such as the number and quality of fruits, and the timing of ripening. Importantly, delayed senescence in SlNAP2 knockdown plants was concomitant with an increased number of fruits per plant, while fruit size and speed of fruit ripening were not affected (Supplemental Fig. S6). Furthermore, SlNAP2-KD and dKD plants accumulated higher levels of soluble sugars such as Fru, Glc, and Suc at breaker and ripe fruit stages, and Brix units were significantly higher in dKD than wild-type plants (Fig. 7). Delayed senescence in OsNAP and GhNAP RNAi lines was also reported to be associated with increased yield in rice and cotton, respectively (Liang et al., 2014; Fan et al., 2015). However, it cannot be excluded that the enhanced fruit number and nutritional quality observed in SlNAP2-KD lines may result from a combinatorial effect of SlNAP2 in altering senescence in leaves and modifying the physiology in fruits.
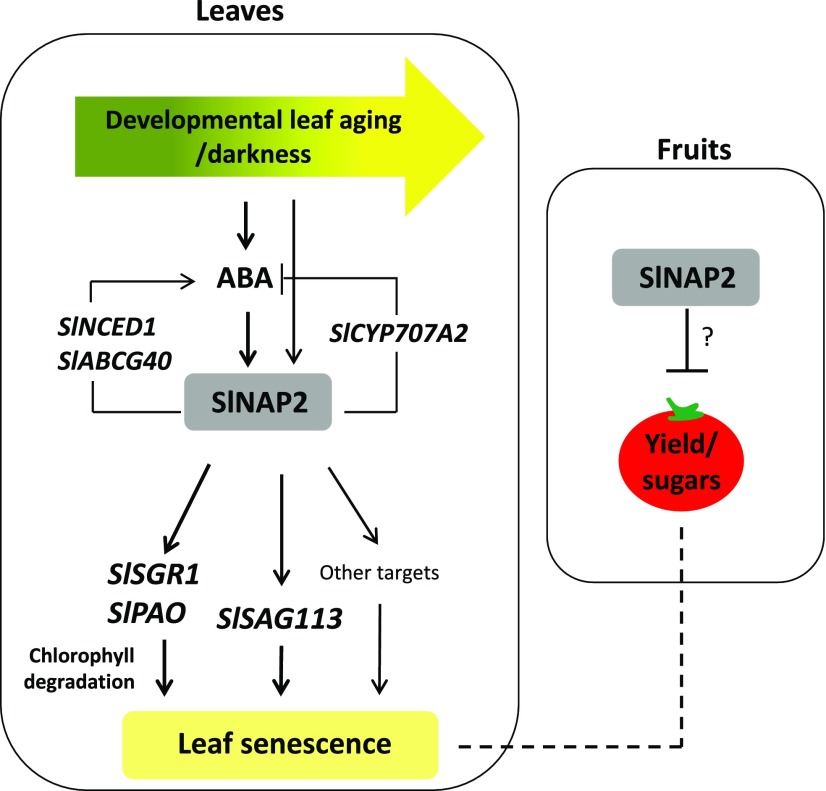
We conclude that SlNAP2 regulates leaf senescence and subsequently fruit yield through a gene regulatory network that combines several target genes including senescence-associated marker genes, chlorophyll degradation genes, and ABA homeostasis-related genes (Fig. 8). The regulation of leaf senescence by SlNAP2 and its positive effect on yield are considerably conserved across species. Manipulation of leaf senescence through a modification of regulatory factors that initiate and control senescence is a powerful strategy to improve agricultural plant productivity, including the production of fleshy fruits.
Figure 8.
Model of SlNAP2 action in tomato. During age-dependent and dark-induced senescence, ABA accumulates in leaves, which leads to an activation of SlNAP2 expression; enhanced expression of SlNAP2 during leaf aging may also be triggered without the involvement of ABA. SlNAP2 activates SlSAG113, chlorophyll degradation genes such as SlSGR1 and SlPAO, and other downstream targets by directly binding to their promoters, thereby promoting leaf senescence. SlNAP2 also directly regulates the expression of ABA biosynthesis (SlNCED1), transport (SlABCG40), and degradation (SlCYP707A2) genes, indicating a complex role in establishing ABA homeostasis. Inhibition of SlNAP2 leads to delayed leaf senescence and enhanced fruit yield and sugar content, likely due to prolonged leaf photosynthesis, although a direct effect of SlNAP2 on fruit development is possible. Arrow-ending lines, positive regulation; T-ending lines, negative regulation. The dashed line indicates a possible but not yet experimentally confirmed interaction between senescing leaves and developing fruits.
MATERIALS AND METHODS
General
Tomato (Solanum lycopersicum) putative orthologs of Arabidopsis (Arabidopsis thaliana) genes were identified using the PLAZA 3.0 database (http://bioinformatics.psb.ugent.be/plaza/; Proost et al., 2015). Genes were annotated using the PLAZA 3.0 and Sol Genomics (https://solgenomics.net/) databases and using information extracted from the literature. Oligonucleotide sequences are listed in Supplemental Table S2. RT-qPCR primers were designed using QuantPrime (www.quantprime.de; Arvidsson et al., 2008), and some primers were designed based on literature data, as indicated. Chemicals and reagents were obtained from Invitrogen, Sigma-Aldrich, and Fluka. Molecular biological kits were obtained from Qiagen and Macherey-Nagel.
Plant Material and Growth Conditions
S. lycopersicum cv Moneymaker wild type was used as control in this study. Seeds were germinated on full-strength Murashige and Skoog medium containing 2% (w/v) Suc, and 3-week-old seedlings were transferred to a mixture of potting soil and quartz sand (2:1, v/v). Plants were grown in a growth chamber at 500 μmol photons m−2 s−1 and 25°C under a 14/10-h light/dark regime in individual pots (18-cm diameter).
DNA Constructs
Primer sequences are listed in Supplemental Table S2. Amplified fragments generated by PCR were sequenced by Eurofins MWG Operon. Constructs were transformed into tomato cv Moneymaker via Agrobacterium tumefaciens GV2260-mediated transformation. For SlNAP2pro:GUS lines, the 1.5-kb SlNAP2 promoter was amplified from wild-type genomic DNA and introduced upstream of the GUS coding sequence in plasmid pKGWFS7,0 (Karimi et al., 2002). For the generation of SlNAP2-KD plants, an 88-bp fragment from the 3′ end of the SlNAP2 coding sequence was amplified from wild-type cDNA and cloned into the Gateway-compatible entry vector pDONR221 (Invitrogen). The fragment was then cloned into the pK7GWIWG2 RNAi vector (Karimi et al., 2002) in sense and antisense orientations, flanking an intervening intron, by Gateway cloning (Invitrogen). To generate SlNAP1/SlNAP2 double knockdown (dKD) plants, an amiRNA construct was engineered by replacing the original miR319a/miR319a* sequence in plasmid pRS300 (Schwab et al., 2006) with a 21-bp sequence (TAATTCCCAGGGATCGAACTT) identical in SlNAP1 and SlNAP2. The amiRNA was designed using Web MicroRNA Designer 3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and subsequently inserted downstream of the CaMV 35S promoter in the pK7WG2 binary vector (Karimi et al., 2002) via Gateway cloning. For 35S:SlNAP2-GFP, the full-length SlNAP2 open reading frame was amplified without its stop codon. The PCR product was cloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning kit (Invitrogen). The sequence-verified entry clone was then transferred to the pK7FWG2 vector (Karimi et al., 2002) by LR recombination (Invitrogen). For SlNAP2-IOE, the SlNAP2 coding sequence was cloned into the pER10-GATEWAY-compatible vector (Zuo et al., 2000). For SlNAP1- and SlNAP2-CELD, the DBP-CELD fusion vector pTacLCELD6XHis was used (Xue, 2005). SlNAP1 and SlNAP2 full coding sequences (without stop codons) were amplified by PCR with a sense primer (including an NheI restriction site) and an antisense primer (including a BamHI restriction site; Supplemental Table S2). The amplified DNA fragments were first inserted into pCR2.1 (Thermo Fisher Scientific) and then inserted N-terminal of CELD using the NheI and BamHI cloning sites of pTacLCELD6XHis to create an in-frame fusion.
Treatments
For estradiol induction, 3-week-old SlNAP2-IOE seedlings were incubated in sterile water containing 15 μm estradiol (control treatment: 0.15% [v/v] ethanol). The seedlings were kept on a rotary shaker for 6 h and then immediately frozen in liquid nitrogen. For ABA treatment, 3-week-old tomato seedlings were incubated in sterile water containing 40 μm ABA. The seedlings were kept on a rotary shaker for 2, 4, 8, or 16 h and then harvested in liquid nitrogen. For dark-induced leaf senescence experiments, detached young leaves from 10-week-old wild-type and SlNAP2 transgenic plants were placed on moist filter papers in petri dishes with the adaxial side facing upwards. The plates were kept in darkness at 22°C for 2 weeks. Filter papers were changed every 5 d. Gene expression levels were determined by RT-qPCR.
Gene Expression Analysis
Total RNA extraction was done using Trizol reagent (Life Technologies). Synthesis of cDNA and RT-qPCR using SYBR Green were performed as described (Balazadeh et al., 2008). PCR was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems). GAPDH (Solyc04g009030) served as reference gene for data analysis. Statistical significance was determined using Student’s t test.
DNA-Binding Site Selection
In vitro binding site selection was performed using the CELD-fusion method with the pTacSlNAP1-LCELD6XHis and pTacSlNAP2-LCELD6XHis constructs, employing biotin-labeled double-stranded oligonucleotides (Xue, 2005). The DNA-binding activity of SlNAP1-CELD and SlNAP2-CELD was measured using methylumbelliferyl β-d-cellobioside as a substrate (Xue, 2002). DNA binding assays with a biotin-labeled single-stranded oligonucleotide or a biotin-labeled double-stranded oligonucleotide without a target binding site were used as controls.
EMSA
SlNAP2-CELD fusion protein was extracted from Escherichia coli Rosetta (DE3) competent cells. 5′-DY682-labeled 40-bp oligonucleotides representing fragments of the SlSAG113 (5′-TCTTCTCATTGGCCCACGTAAATTCAAATCAATAAAATCT-3′), SlSGR1 (5′-ATCGATCGAGCTCCAATACGAATATCGGAATAAGAAAAAA-3′), SlPAO (5′-CATTGTTCATAACTTGCACGCAAATCCTTCTTCTTCTTCT-3′), SlNCED1 (5′-CAATTTCTTTTATATGTCTACGTAATTATTTAAAAAGAAT-3′), SlCYP707A2 (5′-TTGTTGTTTTTTCATTACGTATTTGAAATTCGCGTTAGAG-3′), and SlABCG40 (5′-TTTTTGTGTGTTTGATACGTAATTAAATATAAATAAAAAA-3′) promoters were purchased from Eurofins MWG Operon and annealed with their sequence-complementary oligonucleotides to form DNA probes. Annealing was performed by heating the primers to 99°C, followed by slow cooling to room temperature. The binding reaction was performed at room temperature for 20 min. EMSA reactions were performed using the Odyssey Infrared EMSA kit (LI-COR Biosciences) as described in the manual. DNA-protein complexes were separated on 6% agarose retardation gels, while DY682 signal was detected using the Odyssey Infrared Imaging System (LI-COR Biosciences).
ChIP
ChIP-qPCR was performed from leaves of mature 35S:SlNAP2-GFP plants, and the wild type served as control. ChIP was performed as described (Kaufmann et al., 2010) using anti-GFP antibody to immunoprecipitate protein-DNA complexes. qPCR primers were designed to flank the SlNAP2-binding sites within the promoter regions of SlSAG12, SlSAG113, SlSGR1, SlPAO, SlNCED1, SlCYP707A2, and SlABCG40. Primers annealing to a promoter region of Solyc04g009030 lacking a SlNAP2 binding site were used as a negative control. Primers used for qPCR are listed in Supplemental Table S2.
Chlorophyll Measurements
Chlorophyll content was determined using a SPAD analyzer (N-tester; Hydro Agri).
Metabolite Measurements
Metabolite profiling of tomato mature green leaves and fruits at three developmental stages was carried out by gas chromatography-mass spectrometry (ChromaTOF software, Pegasus driver 1.61; LECO) as described previously (Lisec et al., 2006). The chromatograms and mass spectra were evaluated using TagFinder software (Luedemann et al., 2012). Metabolite identification was manually checked by the mass spectral and retention index collection of the Golm Metabolome Database (Kopka et al., 2005). Peak heights of the mass fragments were normalized on the basis of the fresh weight of the sample and the added amount of an internal standard (ribitol). Statistical differences between groups were analyzed by Student’s t tests on the raw data. Results were determined to be statistically different at a probability level of P < 0.05. Relative metabolite levels were obtained as the ratio between the lines and the mean value of the respective wild type.
ABA Measurement
Plant tissue (∼20 mg fresh weight of each sample) was ground using 3-mm tungsten carbide beads (Retsch) with a MM 301 vibration mill at a frequency of 27.0 Hz for 3 min (Retsch). Internal standard containing deuterium-labeled standard [20 pmol of (+)-3′,5′,5′,7´,7´,7´-2H6-ABA] and 1 mL ice-cold methanol/water/acetic acid (10/89/1, v/v) were added to each of the samples. After 1 h of shaking in the dark at 4°C, the homogenates were centrifuged (20,000g, 10 min, 4°C) after extraction, and the pellets were then reextracted in 0.5 mL extraction solvent for 30 min. The combined extracts were purified by solid-phase extraction on Oasis HLB cartridges (60 mg, 3 mL; Waters), evaporated to dryness in a Speed-Vac (UniEquip), and analyzed by UPLC-ESI (−/+)-MS/MS (Turecková et al., 2009).
Brix Determination
Brix was determined as the concentration of total soluble solids using a digital refractometer (Krüss Optronic) on six ripe red fruit samples per genotype.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SlNAP1 (NP_001316452.1), SlNAP2 (XM_004236996.2), SlSAG12 (XP_004233054), SlSAG113 (XP_004239911.1), SlSGR1 (NP_001234723.1), SlPAO (NP_001234535.2), SlNCED1 (NP_001234455), SlCYP707A2 (XP_004244436.1), SlABCG40 (XP_004247842.1), SlGDH1 (NP_001292722), and SlGS1 (XP_004248690).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of SlNAP1 and SlNAP2.
Supplemental Figure S2. Expression of SlNAP2 in overexpression and knockdown lines.
Supplemental Figure S3. Expression of senescence marker genes.
Supplemental Figure S4. Identification of the binding sequences of SlNAP1 and SlNAP2.
Supplemental Figure S5. ABA content.
Supplemental Figure S6. Effect of SlNAP2 on fruit yield.
Supplemental Figure S7. Expression of SlGDH1 and SlGS1.
Supplemental Table S1. Relative metabolite content of fully expanded leaves of 8-week-old SlNAP2 transgenic and wild-type pants.
Supplemental Table S2. Oligonucleotide sequences.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Karin Koehl and her team (Max Planck Institute of Molecular Plant Physiology) for plant care. We thank the University of Potsdam and the Max Planck Institute of Molecular Plant Physiology for supporting our research.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (FOR 948; BA4769/1-2) and the Max Planck Institute of Molecular Plant Physiology. X.M. received a fellowship from the China Scholarship Council.
Articles can be viewed without a subscription.
References
- Akhtar MS, Goldschmidt EE, John I, Rodoni S, Matile P, Grierson D (1999) Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 50: 1115–1122 [Google Scholar]
- Ansari MI, Lee RH, Chen SCG (2005) A novel senescence-associated gene encoding gamma-aminobutyric acid (GABA): pyruvate transaminase is upregulated during rice leaf senescence. Physiol Plant 123: 1–8 [Google Scholar]
- Ansari MI, Hasan S, Jalil SU (2014) Leaf senescence and GABA shunt. Bioinformation 10: 734–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, Leaver CJ, Fernie AR (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Osorio S, Lohse M, Balbo I, Krahnert I, Sienkiewicz-Porzucek A, Usadel B, Nunes-Nesi A, Fernie AR (2012) Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) 10 (Suppl 1): 63–75 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Schildhauer J, Araújo WL, Munné-Bosch S, Fernie AR, Proost S, Humbeck K, Mueller-Roeber B (2014) Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. J Exp Bot 65: 3975–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. (2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol 172: 2132–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Purdy S, Christ A, Morot-Gaudry JF, Wingler A, Masclaux-Daubresse C (2005) Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. A metabolic profiling approach. Plant Physiol 138: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnenstich H, Saigo M, Niessen M, Zanor MI, Andreo CS, Fernie AR, Drincovich MF, Flügge UI, Maurino VG (2007) Alteration of organic acid metabolism in Arabidopsis overexpressing the maize C4 NADP-malic enzyme causes accelerated senescence during extended darkness. Plant Physiol 145: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, Wang M, Shen H, Wang X (2015) A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J Exp Bot 66: 4669–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Gao J, Zhu X, Song Y, Li Z, Ren G, Zhou X, Kuai B (2016) ABF2, ABF3, and ABF4 promote ABA-Mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol Plant 9: 1272–1285 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Thimann KV (1980) Changes in the abscisic acid content of oat leaves during senescence. Proc Natl Acad Sci USA 77: 2050–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PL, Culetic A, Boschian L, Krupinska K (2013) Plant senescence and crop productivity. Plant Mol Biol 82: 603–622 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Guyer L, Hofstetter SS, Christ B, Lira BS, Rossi M, Hörtensteiner S (2014) Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato. Plant Physiol 166: 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Osaki M, Takebe M, Shinano T, Wasaki J (2005) Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J Exp Bot 56: 1117–1128 [DOI] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Hu ZL, Deng L, Yan B, Pan Y, Luo M, Chen XQ, Hu TZ, Chen GP (2011) Silencing of the LeSGR1 gene in tomato inhibits chlorophyll degradation and exhibits a stay-green phenotype. Biol Plant 55: 27–34 [Google Scholar]
- Ji K, Kai W, Zhao B, Sun Y, Yuan B, Dai S, Li Q, Chen P, Wang Y, Pei Y, et al. (2014) SlNCED1 and SlCYP707A2: key genes involved in ABA metabolism during tomato fruit ripening. J Exp Bot 65: 5243–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Kim YS, Park S, Back K (2009) Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol 150: 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kou X, Watkins CB, Gan SS (2012) Arabidopsis AtNAP regulates fruit senescence. J Exp Bot 63: 6139–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IC, Hong SW, Whang SS, Lim PO, Nam HG, Koo JC (2011) Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol 52: 651–662 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao J, Zhao Y, Lu X, Zhou Z, Zhao C, Xu G (2016) Comprehensive investigation of tobacco leaves during natural early senescence via multi-platform metabolomics analyses. Sci Rep 6: 37976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, Chu C (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira BS, Gramegna G, Trench BA, Alves FRR, Silva EM, Silva GFF, Thirumalaikumar VP, Lupi ACD, Demarco D, Purgatto E, et al. (2017) Manipulation of a senescence-associated gene improves fleshy fruit yield. Plant Physiol 175: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Luedemann A, von Malotky L, Erban A, Kopka J (2012) TagFinder: preprocessing software for the fingerprinting and the profiling of gas chromatography-mass spectrometry based metabolome analyses. Methods Mol Biol 860: 255–286 [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco ( Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211: 510–518 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Good AG (2008) NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot 59: 667–680 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465: 30–44 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Park S, Lee K, Kang K, Kim YS, Lee S, Kweon SJ, Back K (2010) Tryptophan boost caused by senescence occurred independently of cytoplasmic glutamine synthetase. Biosci Biotechnol Biochem 74: 2352–2354 [DOI] [PubMed] [Google Scholar]
- Parlitz S, Kunze R, Mueller-Roeber B, Balazadeh S (2011) Regulation of photosynthesis and transcription factor expression by leaf shading and re-illumination in Arabidopsis thaliana leaves. J Plant Physiol 168: 1311–1319 [DOI] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Hadas E, Aharoni N (1993) Characterization and use in ELISA of a new monoclonal-antibody for quantitation of abscisic acid in senescing rice leaves. Plant Growth Regul 12: 71–78 [Google Scholar]
- Pourtau N, Marès M, Purdy S, Quentin N, Ruël A, Wingler A (2004) Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219: 765–772 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles MJ, Garcia C, Cuello J (1995) Differential effects of abscisic acid and methyl jasmonate on endoproteinases in senescing barley leaves. Plant Growth Regul 16: 197–204 [Google Scholar]
- Rauf M, Arif M, Dortay H, Matallana-Ramírez LP, Waters MT, Gil Nam H, Lim PO, Mueller-Roeber B, Balazadeh S (2013) ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep 14: 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J Exp Bot 58: 2319–2327 [DOI] [PubMed] [Google Scholar]
- Turecková V, Novák O, Strnad M (2009) Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80: 390–399 [DOI] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R (2006) Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Guo TL, Wang P, Sun X, Shao Y, Liang BW, Jia X, Gong XQ, Ma FW (2017) Functional analysis of apple MhYTP1 and MhYTP2 genes in leaf senescence and fruit ripening. Sci Hortic (Amsterdam) 221: 23–32 [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57: 391–399 [DOI] [PubMed] [Google Scholar]
- Xue GP. (2002) Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res 30: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP. (2005) A CELD‐fusion method for rapid determination of the DNA‐binding sequence specificity of novel plant DNA‐binding proteins. Plant J 41: 638–649 [DOI] [PubMed] [Google Scholar]
- Xue GP, Bower NI, McIntyre CL, Riding GA, Kazan K, Shorter R (2006) TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct Plant Biol 33: 43–57 [DOI] [PubMed] [Google Scholar]
- Yang JC, Zhang JH, Wang ZQ, Zhu QS, Liu LJ (2003) Involvement of abscisic acid and cytokinins in the senescence and remobilization of carbon reserves in wheat subjected to water stress during grain filling. Plant Cell Environ 26: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Gan SS (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Derkx AP, Liu DC, Buchner P, Hawkesford MJ (2015) Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol (Stuttg) 17: 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Huang W, Liu L, Chen T, Zhou F, Lin Y (2013) Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol 13: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]