Long noncoding RNAs in rice with major reproductive and Polycomb complex 2-regulated subclasses have conserved members detected in Brachypodium and maize.
Abstract
Long noncoding RNAs (lncRNAs) have been characterized extensively in animals and are involved in several processes, including homeobox gene expression and X-chromosome inactivation. In comparison, there has been much less detailed characterization of plant lncRNAs, and the number of distinct lncRNAs encoded in plant genomes and their regulation by developmental and epigenetic mechanisms remain largely unknown. Here, we analyzed transcriptome data from Asian rice (Oryza sativa) and identified 6,309 long intergenic noncoding RNAs (lincRNAs), focusing on their expression in reproductive tissues and organs. Most O. sativa lincRNAs were expressed in a highly tissue-specific manner, with an unexpectedly high fraction specifically expressed in male gametes. Mutation of a component of the Polycomb Repressive Complex2 (PRC2) resulted in derepression of another large class of lincRNAs, whose expression is correlated with H3K27 trimethylation in developing panicles. Overlap with the sperm cell-specific lincRNAs suggests that epigenetic repression of lincRNAs in the panicles was partially relieved in the male germline. Expression of a subset of lincRNAs also showed modulation by drought in reproductive tissues. Comparison with other cereal genomes showed that the lincRNAs generally have low levels of conservation at both the sequence and structural levels. Use of a novelty detection support vector machine model enabled the detection of nucleotide sequence and structural homology in ∼10% and ∼4% of the lincRNAs in genomes of purple false brome (Brachypodium distachyon) and maize (Zea mays), respectively. This is the first study to report on a large number of lncRNAs that are targets of repression by PRC2 rather than mediating regulation via PRC2. That the vast majority of the lincRNAs reported here do not overlap with those of other rice studies indicates that these are a significant addition to the known lincRNAs in rice.
In eukaryotic genomes, intergenic regions are typically many times larger than those occupied by protein-coding genes, with an estimated 90% of the human genome containing non-protein-coding sequences (Bertone et al., 2004; Cheng et al., 2005; Kapranov et al., 2007). Even in more compact genomes such as Arabidopsis (Arabidopsis thaliana), it has been estimated that noncoding antisense transcription occurs for 70% of protein-coding loci (Wang et al., 2014), and similar levels have been observed in mammals (Katayama et al., 2005). In the past decade, long noncoding RNAs (lncRNAs) have received considerable attention due to the discovery of biological functions associated with several lncRNAs. In animals, these lncRNAs are involved in critical processes such as gene dosage compensation (X-INACTIVE SPECIFIC TRANSCRIPT [Xist] and RNA ON THE X1/2 [roX1 and roX2]), developmental patterning (HOX TRANSCRIPT ANTISENSE RNA [HOTAIR]), the induction and maintenance of pluripotency in embryonic stem cells (long intergenic noncoding RNAs [lincRNA]-RoR and lincRNA-Sox2), immunity (lincRNA-Cox2), cell stress response (lncRNA UCA1), autonomous cell death (lincRNA-p21), and others (for review, see Aune and Spurlock, 2016; Bartonicek et al., 2016; Hu and Shan, 2016; Zhang and Cao, 2016). In plants, the number of lncRNAs that have been functionally characterized are relatively few. Examples are lncRNAs involved in winter dormancy in flowering (COLD ASSISTED INTRONIC NONCODING RNA [COLDAIR] and antisense transcripts [COOLAIR]), regulation of phosphate assimilation (INDUCED BY PHOSPHATE STARVATION1 [IPS1/At4]), nodulation (EARLY NODULIN40), and control of male fertility (LONG-DAY-SPECIFIC MALE-FERTILITY-ASSOCIATED RNA [LDMAR]; Campalans et al., 2004; Franco-Zorrilla et al., 2007; Swiezewski et al., 2009; Heo and Sung, 2011; Ding et al., 2012b,a).
The mechanisms of lncRNA action are thought to arise through interactions with other cellular components, including other RNAs via discrete functional domains that can occur within the same or interacting RNAs (for review, see Mercer and Mattick, 2013). Apart from the functional domains, there are two easily detected domain properties: lncRNA-nucleotide target interactions and lncRNA-protein interactions. The function of lncRNA-nucleotide target interaction is demonstrated by the sequestration of the microRNA miR399 by the plant lincRNA IPS1/At4 involved in phosphate homeostasis (Franco-Zorrilla et al., 2007). The lncRNA-protein interaction group includes lncRNAs that interact with heterogenous nuclear ribonucleoproteins involved in the regulation of transcription and splicing in animals and Polycomb Repressive Complex2 (PRC2), leading to chromatin silencing in plants and animals (Rinn and Chang, 2012; Yu et al., 2015). Of a set of ∼3,300 human lincRNAs, 20% bind to PRC2, suggesting that this is a large and important subset of lncRNAs (Khalil et al., 2009). The process of chromatin silencing via PRC2 plays a critical role in developmental processes. The independent evolutionary origins, for example, X-chromosome inactivation (Xist) and winter-induced flowering control via the Flowering Locus C-associated locus, COLDAIR, suggest that the likely mechanism of action is robust under different regulatory contexts.
High-throughput screening of expressed sequences has been fruitful for the production of lncRNA catalogs in animal systems. In plants, an Arabidopsis transcript-based study identified a set of 6,480 lincRNAs (Liu et al., 2012), two studies in maize (Zea mays) identified 2,492 and 1,704 lncRNAs (Boerner and McGinnis, 2012; Li et al., 2014), and in Asian rice (Oryza sativa), 2,224 lncRNAs and 771 lincRNAs (Komiya et al., 2014; Zhang et al., 2014), and more recently 11,000 lncRNAs (Wang et al., 2015), were identified. For both animal and plant studies, these catalogs of expressed lncRNAs are in the thousands and represent part of a broader phenomenon referred to as pervasive transcription (for review, see Clark et al., 2011). Whether plants have utilized lncRNAs for developmental processes to the same extent as animals has yet to be determined, but efforts at large-scale identification of plant lncRNAs expressed in a diverse set of tissues should lead to the discovery of new lncRNAs that have biological and/or agronomic importance.
Here, we present a set of 6,309 O. sativa lincRNAs detected through analysis of RNA sequencing (RNA-seq) data from predominantly reproductive tissues as well as analysis of their expression and conservation within cereals. Within this set, about one-third of the lincRNAs were dominantly expressed in sperm cells, and another one-third were differentially expressed in developing panicles of a Polycomb Group gene mutant, called embryonic flower2b (emf2b). The wild-type EMF2b gene product is a component of the PRC2 complex that plays an essential role in panicle development in O. sativa and is orthologous to the Arabidopsis EMF2 and FERTILIZATION INDEPENDENT SEED2 components of PRC2 complexes regulating flowering and seed development, respectively (Conrad et al., 2014). To address the issue of biological relevance, a novelty detection support vector machine (SVM) on nucleotide and structure-sensitive alignments of the O. sativa lincRNAs against syntenic regions of Brachypodium distachyon and maize was used to select potentially conserved lincRNAs. The accuracy of the SVM approach in detecting conserved lincRNAs was examined in terms of interspecies nucleotide variation levels detected between O. sativa and African rice (Oryza glaberrima), and the results demonstrated that the SVM approach enabled the detection of conservation not readily detectable by other means.
RESULTS
LincRNA Discovery and Initial Analysis
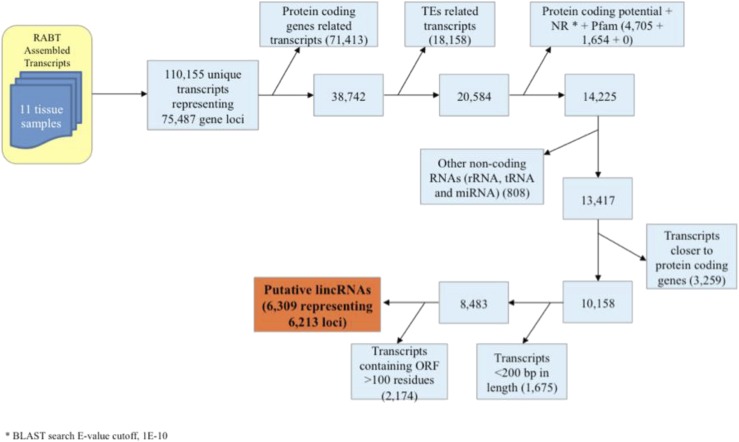
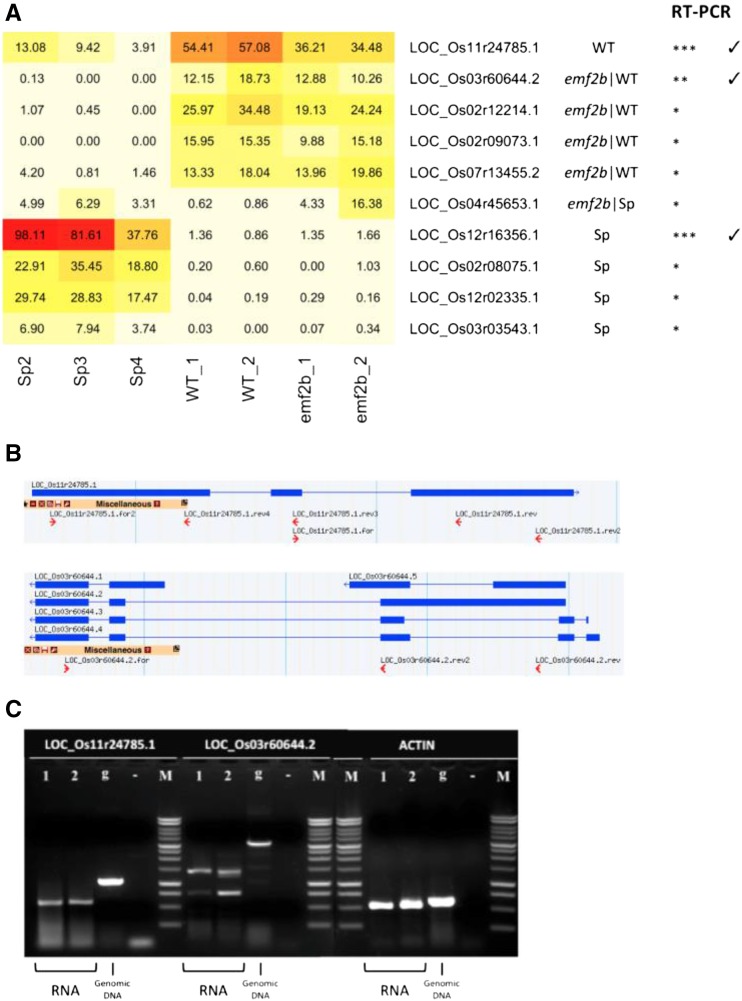
In order to generate an assembled transcriptome that is broadly applicable to plant development, we utilized RNA-seq data from a wide variety of tissues/cell types from japonica rice. These were obtained from prior studies within our laboratory and collaboration as well as from externally available sources (Supplemental Table S1; Chodavarapu et al., 2012; Davidson et al., 2012; Lu et al., 2012; Anderson et al., 2013; Zhang et al., 2013; Conrad et al., 2014; Krishnan et al., 2017). The samples included the three products of meiosis (egg cell, sperm cells, and the vegetative cytoplasm of pollen) as well as other reproduction-related tissues (anther, embryo, endosperm, wild-type panicles, and emf2b panicles) and somatic tissues (leaf, seed, and seedling). These RNA-seq data sets were used to assemble the transcriptome in this study, while expression analysis was performed on data that included additional RNA-seq data sets consisting of drought-related samples and a deeper sequencing of the panicle samples. The 11 assembly data sets were put through a pipeline consisting of transcriptome assembly followed by a series of filtering steps to identify and remove protein-coding RNA transcripts and other RNAs not corresponding to lincRNAs. LincRNAs were defined as lncRNAs if they originated from intergenic regions of the MSU7.0 genome and they met specific criteria (Fig. 1). For an expressed RNA to be regarded as a lincRNA, it must be at least 200 nucleotides long and not code for a protein (Liu et al., 2012). To remove potential protein-coding genes, a set of 110,155 assembled transcripts (75,487 gene loci) were screened by identifying those corresponding to previously annotated loci (nontransposable element protein-coding and transposable element [TE] protein-coding gene models), identifying those with protein-coding potential through homology with known proteins and similarity to protein-coding genes (see “Materials and Methods”), and finally restricting the size of any potential coding sequence to 100 amino acids. Noncoding RNAs that were not considered lncRNAs (rRNA, tRNAs, and miRNAs) also were removed. An additional requirement was that any lincRNA should be at least 500 nucleotides from the nearest annotated gene (Liu et al., 2012). This criterion is more stringent than that usually employed, but it was chosen to avoid the spurious detection of lincRNAs from poor annotation or alternate splicing. The analysis resulted in a final set of 6,309 transcripts representing 6,214 loci that were used as the working set of lincRNAs for the remainder of this study (Fig. 1). Of these, a set of 23 loci, corresponding to 24 lincRNAs, including 10 long lincRNAs (greater than 1 kb; Fig. 2C; Supplemental Fig. S1A) and 14 short lincRNAs (less than 0.5 kb; Supplemental Fig. S1, A and B), was validated via reverse transcription (RT)-PCR (Supplemental Results S1). The lincRNAs were selected for expression detected in sperm cells and wild-type and emf2b panicles (Fig. 2A), with some of these, for example LOC_Os11r24785.1 and LOC_Os03r60644.2, containing predicted introns within the amplified region (Fig. 2, B and C; Supplemental Fig. S1), confirming the validity of the transcript assemblies. These results are described in more detail in Supplemental Results S1. After committing to the set of 6,214 lincRNA loci, 20 of these overlapped with 23 of a previously reported set of 1,349 noncoding RNAs (Supplemental Table S2), as defined by Liu et al. (2013). Of these 23 loci, 10 corresponded to small nucleolar RNAs, while only two of the remaining 13 had been annotated as U2 spliceosomal noncoding RNAs.
Figure 1.
The pipeline used to identify lincRNAs is shown with a reduction in the number of candidate transcripts at each step due to the removal of sequences through the application of a filter as indicated. RABT, Reference Annotation Based Transcript.
Figure 2.
A, Heat map showing the expression of selected lincRNAs as RNA-seq reads normalized to transcripts per million (TPM) . In the columns to the right of this are the dominant expression categories (first), RT-PCR confirmation confidence (second), and evidence of intron splicing (third). WT, Wild type. B, LincRNA loci exon-intron models with RT-PCR primer match locations. C, RT-PCR gel images confirming the expression and splicing of selected lincRNAs. Specifically with products in bp for RNA (DNA) is as follows: LOC_Os11r24785.1.for2 × LOC_Os11r24785.1.rev3 giving 774 (1,029), LOC_Os03r60644.2.rev × LOC_Os03r60644.2.for giving 1,383 (3,339), and OsActin1.for × OsActin1.rev giving 528 (609). The validation of the remaining lincRNAs (in A) is shown in Supplemental Figure S1.
Strand information was obtained for a subset of the 6,309 processed transcript models by the existence of sequence reads that bridge across introns, from direct strand information present for the seedling library data (Lu et al., 2012) and from additional strand-specific RNA-seq data from drought-stressed plants (see below). Due to extremely limited materials from some of the reproductive samples, such as sperm cells and young emf2 mutant panicles, strand-specific RNA-seq data were not available directly for these transcriptomes and were extrapolated using information from other samples. This analysis resulted in strand information for a total of 464 lincRNA loci (562 transcripts). Unlike protein-coding genes, open reading frames (ORFs) are not a general feature of noncoding RNAs. Therefore, detection of a bias of ORFs to the sense strand (i.e. the transcribed strand) within a group of putative lincRNAs may suggest that a significant proportion of these RNAs are actually protein-coding RNAs. With the strand information available, and with the assumption that protein-coding ORFs should be longer than would occur by chance, the longest ORFs for both the sense and antisense sequences were collated. If many of the lincRNAs actually code for proteins, one might expect a bias in favor of larger ORFs to be found on the sense strand as compared with the antisense strand. However, using the Mann-Whitney U test to compare the sense and antisense strands showed no statistically significant difference in the lengths of the longest ORFs (P = 0.19). The lack of apparent bias toward a longer ORF on the sense strand is consistent with the idea that the selected sequences as a group do, in fact, represent noncoding RNAs.
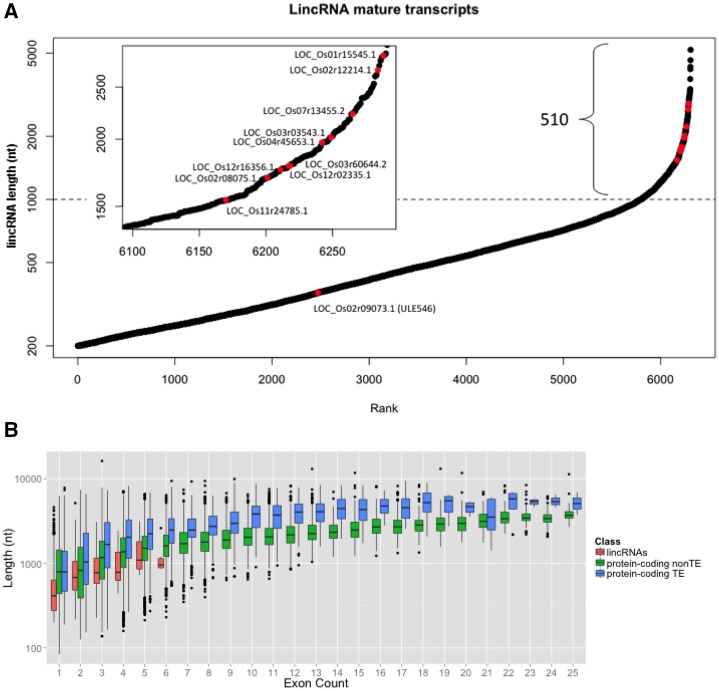
The size distribution of mature lincRNA transcripts ranged from the lower limit of 200 nucleotides to a maximum of 5.2 kb, but about half were less than 500 nucleotides, and only 510 were larger than 1 kb (Fig. 3A). A subset of the lincRNAs larger than 1 kb was selected for molecular confirmation (see below). The dominance of shorter transcripts differs from protein-coding genes that typically have 1- to 2-kb transcripts. The observed transcripts also differ from protein-coding mature transcript models in that a high proportion (94%) had only a single exon, with a maximum of up to six exons. By comparison, 19% of protein-coding genes had a single exon and slightly more had two exons (Supplemental Fig. S2). The existence of fewer exons in lincRNAs may be due to the small size of the transcripts, or it could be a special property of lincRNAs. To determine which of these is most likely, the lengths of the transcripts were plotted as a function of the number of exons, with the data divided into three types: lincRNAs, TE protein coding, and non-TE protein coding. Overall, there was little difference detectable between lincRNA and protein-coding genes, suggesting that there is no special propensity of lincRNA genes to exclude introns (Fig. 3B).
Figure 3.
A, Rank plot shows the size distribution of detected lincRNAs. While many lincRNAs are less than 1 kb, several of those chosen for additional RT-PCR confirmation were mostly larger than 1 kb (inset). B, Box plots show that transcript length roughly correlates with the number of exons within each of the three transcript classes. LincRNAs may have fewer exons as compared with the protein-coding transcripts (TE and non TE). nt, Nucleotides.
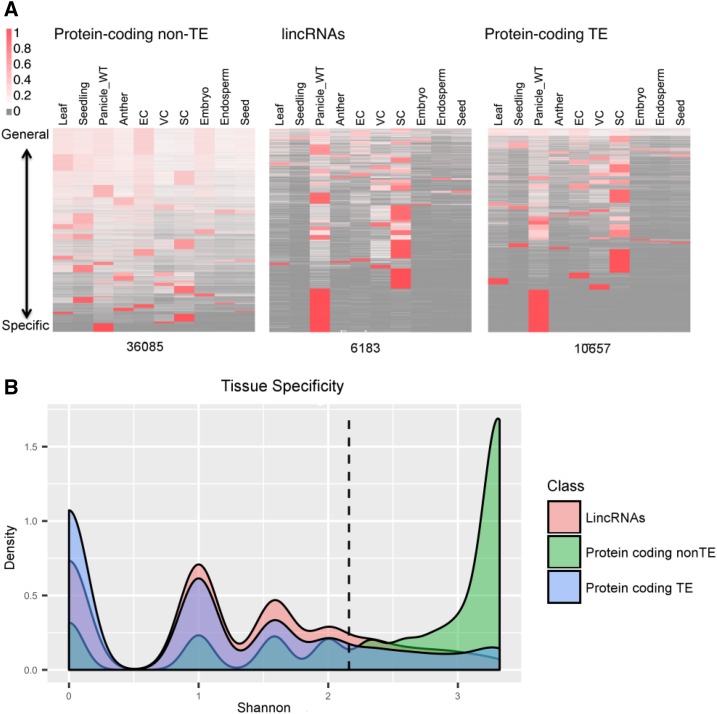
To further characterize the lincRNA loci, the longest isoforms were assessed for their expression levels and patterns within the different RNA-seq source tissues and compared with protein-coding genes and transcripts from TEs. The lincRNAs were expressed at levels lower than non-TE protein-coding genes but at levels higher than TE protein-coding genes, which is in line with what has been found for other studies on lncRNAs (Supplemental Fig. S3; Liu et al., 2012; Li et al., 2014). Despite the average lincRNA being expressed at a level intermediate to non-TE and TE protein-coding genes, the tissue specificity of lincRNAs is similar to that of TE protein-coding genes (Fig. 4). The vast majority of lincRNAs and TE protein-coding loci (82.1% and 82.8%) were expressed in four or fewer tissues/cells, whereas the reverse was true for the non-TE protein-coding loci, which had 71.6% of genes expressed in more than four tissues (Fig. 4B). These results are in agreement with previous reports in rice, Arabidopsis, and maize (Liu et al., 2012; Li et al., 2014; Zhang et al., 2014).
Figure 4.
Tissue specificity is shown for 36,035 non-TE protein-coding genes, 6,183 lincRNAs, and 10,657 TE protein-coding genes via heat map (A) where EC, VC, and SC correspond to Egg Cell, Vegetative Cytoplasm (of mature pollen) and Sperm Cell (of mature pollen), respectively, and more clearly by a density plot of Shannon entropy (log2 of the number of tissues with detected expression; B). For the heat maps, genes were clustered into 33, 33, and 35 groups, respectively, and the expression level in RPKM was row normalized to 1 for each locus. The expression level is shown as a spectrum with high expression (red) to low expression (white), and the absence of reads also is indicated (gray). For the density plots, predominant expression within one, two, and three tissues shows up as peaks at 0, 1, and 1.58. For non-TE protein-coding genes, the peak occurs at a position greater than a value of 3, corresponding to at least eight of the tissues with detectable expression. Most lincRNA and TE loci (82.3% and 82.6%) are expressed in four or fewer tissues/cell types (left of the dashed line), as compared with non-TE protein-coding genes, for which 71.6% of loci are expressed in more than four tissues/cell types (right of the dashed line). WT, Wild type.
LincRNA Expression Defines Overlapping Sperm Cell-Specific and Chromatin-Modulated Classes
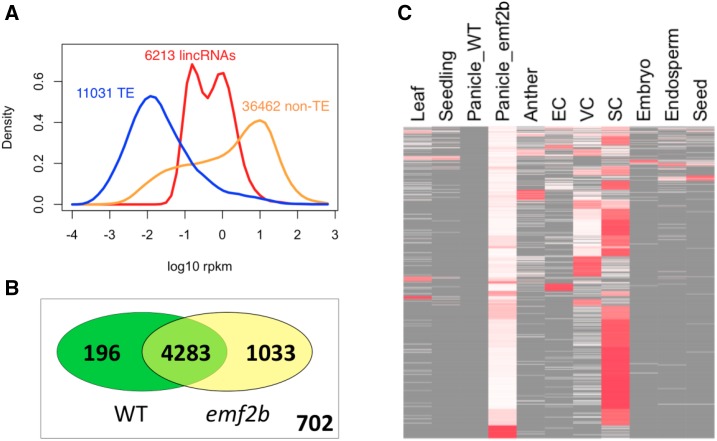
The initial expression analysis of the 10 sample types described earlier was conducted only on wild-type tissues. However, the lincRNAs were assessed additionally in O. sativa panicles that were mutant for EMF2B, a polycomb group protein involved in chromatin silencing through trimethylation of H2K27 via PRC2. The emf2b mutant results in the increased expression of protein-coding genes during panicle development in O. sativa (Conrad et al., 2014), and for some of these genes, this increase is due to a reduction in levels of H3K27 trimethylation (H3K27me3) chromatin marks. Interestingly, when the expression data of lincRNAs and protein-coding genes from the emf2b mutant are included with the wild-type samples, the expression level distribution changes from a single peak (Supplemental Fig. S3) to a double-headed peak (Fig. 5A) in lincRNAs but not in protein-coding genes (TE and non TE). This suggests that a substantial proportion of the lincRNAs become derepressed specifically in the emf2b mutant panicles. The derepression of many lincRNAs is confirmed by the existence of 1,033 lincRNAs with reads in the emf2b mutant panicles that are absent in the RNA-seq data from wild-type panicles (Fig. 5, B and C). Furthermore, 391 of these were not detected in any of the 10 wild-type samples. We define these emf2b-specific lincRNAs as a class of lincRNAs distinct from those expressed predominantly in wild-type panicles. In addition, there also was a set of lincRNAs highly expressed in sperm cells. The sperm-expressed lincRNAs overlap to some extent with the emf2b lincRNAs, as shown for the 1,033 emf2b lincRNAs that are not expressed in wild-type panicles (Fig. 5C).
Figure 5.
A, Expression value distribution plots made using data from the 10 wild-type tissues but also including data from emf2b mutant panicles, which results in a bimodal distribution for the lincRNAs, suggesting that the loss of EMF2B in panicles results in derepression of many lincRNAs. B, Venn diagram showing which of the 6,214 loci correspond to lincRNAs with and without RNA-seq reads in the data for wild-type (WT) panicles and emf2b mutant panicles. The set of 702 lincRNAs refers to those not present in panicles. C, Heat map of 1,033 lincRNAs having reads in emf2b mutant panicles but not in wild-type panicles. Heat colors (white to red) correspond to RPKM values row normalized to 1, with the absence of reads also indicated (gray). The samples EC, VC, and SC correspond to Egg Cell, Vegetative Cytoplasm (of mature pollen), and Sperm Cell (of mature pollen), respectively. The greater amount of red in the column SC shows that many of the lincRNAs expressed in the emf2b mutant panicles are most highly expressed in sperm cells.
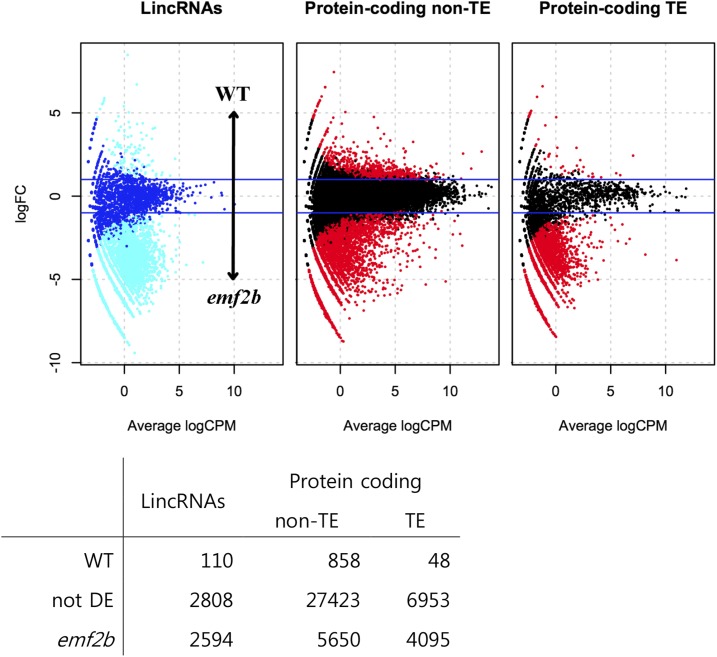
Differential expression analysis between wild-type and emf2b panicles revealed that transcribed loci from all three types of genes (lincRNAs and TE and non-TE protein-coding genes) were derepressed in the mutant panicles as compared with the wild type. However, lincRNAs were most affected, with 47% of the 5,512 loci with panicle reads having a statistically significant increases in expression (5% false discovery rate [FDR]) within the mutant compared with the wild type (Fig. 6). The next most affected were TE protein-coding loci, with about 37% derepressed, followed by non-TE protein-coding loci, with about 17% derepressed in the mutant panicles (Fig. 6). The 2,594 lincRNAs detectably derepressed in the emf2b mutant panicles was considerably larger than the 1,033 lincRNAs that have no reads in wild-type panicles. This suggests that, in the wild-type background, the PRC2 complex might regulate a significant proportion of lincRNAs, potentially up to as much as half of those reported in this study.
Figure 6.
The results of differential expression analysis between wild-type and mutant panicles are shown graphically as minus versus average (MvA) plots (top), with expression level on the x axis (log of counts per million) versus log fold change (logFC), and as a numbers table of differentially expressed (DE) genes (bottom). Transcripts from both coding and noncoding loci have higher expression in emf2b mutant panicles (bottom row of table and negative values on the log fold change scale of plots) as compared with wild-type (WT) panicles. However, the proportions of affected loci in each class differ, with 47.1%, 16.7%, and 36.9% derepressed loci for lincRNAs, non-TE protein-coding, and TE protein-coding loci, respectively.
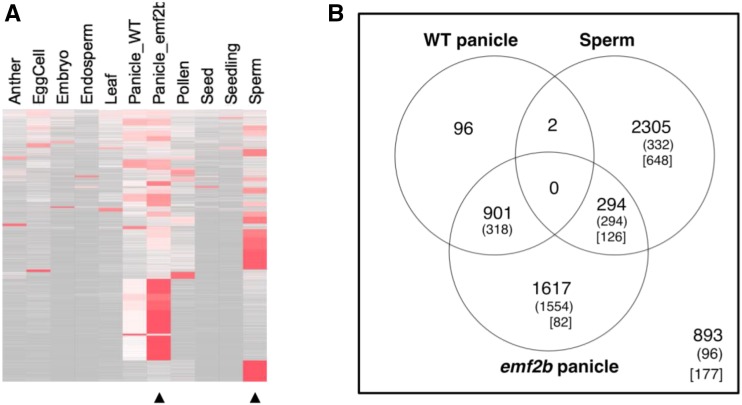
Since many of the 1,033 lincRNAs are more highly expressed in sperm cells than in any of the other tissue/cell types, a comparison of the expression levels of lincRNAs within reproductive tissues was performed. Specifically, this was between wild-type panicles, emf2b panicles, and sperm cells. There were 6,108 lincRNAs with reads in at least one of these three tissue or cell types. Clustering of expression profiles for the full set of 6,213 lincRNA loci across the 11 tissues or cell types showed that many were most highly expressed either in panicles of emf2b mutant plants or in sperm cells (Fig. 7A). Notably, those most highly expressed in mutant panicles also were detectable in wild-type panicles but were expressed at lower levels in sperm cells, while those most highly expressed in sperm cells had comparatively lower expression in panicles of both wild-type and mutant plants. Differential expression analysis confirmed this division of lincRNAs, with 2,305 and 1,617 classified as predominantly from sperm and emf2b panicles, respectively (Fig. 7B). The existence of two distinct lincRNA subclasses within sperm cells, specifically one affected by emf2b and another that is independent of emf2b, supports the existence of a PRC2-regulated class of lincRNAs that is highly developmental in nature.
Figure 7.
A, Heat map for RPKM values row normalized to 1 across 11 tissue/cell types for the 6,213 lincRNAs (original isoform set) grouped into 39 clusters ranked top to bottom by decreasing average Shannon entropy of each cluster. Sperm-specific and emf2b panicle mutant derepressed lincRNA subclasses form a few large blocks of clusters with lower Shannon entropy (higher tissue specificity). B, Dominant expression Venn diagram for 6,108 lincRNAs with reads present within at least wild-type (WT) panicles, emf2b panicles, or sperm cells from mature pollen. LincRNAs are grouped into one of six possible sets for differentially expressed isoforms by using six different contrasts of a negative binomial model of differential expression using the edgeR package (see “Materials and Methods”). Numbers in parentheses refer to those 2,594 lincRNAs derepressed in emf2b panicles relative to wild-type panicles (as determined in the wild-type panicle versus emf2b panicle pairwise analysis) of the lincRNAs belonging to the set of 5,512 expressed in panicles. Numbers in square brackets represent the 1,033 lincRNAs with reads in emf2b panicles but not in wild-type panicles. The set of 893 refers to those not considered differentially expressed in this analysis.
In summary, the lincRNAs were divided into two groups: a set of 2,305 sperm-dominant lincRNAs and a set of 1,617 lincRNAs derepressed in emf2b mutant panicles, which together made up 63% of the lincRNA loci. Within the sperm-dominant set were 648 of the 1,033 lincRNAs with reads from emf2b panicles but not from wild-type panicles (Fig. 7B). These represent a special subclass of lincRNAs normally expressed in sperm cells that are repressed in wild-type panicles in plants with functional EMF2B. These results imply that this subclass of lincRNAs is regulated in a tissue-specific manner by EMF2B, presumptively through PRC2-mediated histone methylation.
A Subset of LincRNA Loci Are Putative Targets of PRC2 Repression
The discovery that a large proportion of the identified lincRNAs (2,595 of 6,309) are derepressed in emf2b panicles as compared with wild-type panicles suggests that lincRNAs may be targets of epigenetic control. Moreover, a number of other developmentally important lncRNAs (Xist, roX1/2, HOTAIR, COLDAIR, and COOLAIR) are known to be involved in epigenetic processes through which they act (Park et al., 2002; Khalil et al., 2009; Swiezewski et al., 2009; Heo and Sung, 2011; Minks et al., 2013). To investigate this further, we performed high-throughput chromatin immunoprecipitation sequencing (ChIP-seq) on wild-type panicles using antibodies that targeted H3K27me3 and analyzed the sites of enrichment both from the point of view of enrichment islands (Sicer) as well as nucleotide position profiles relative to the transcription start site (TSS) and transcription termination site (TTS) for groups of loci.
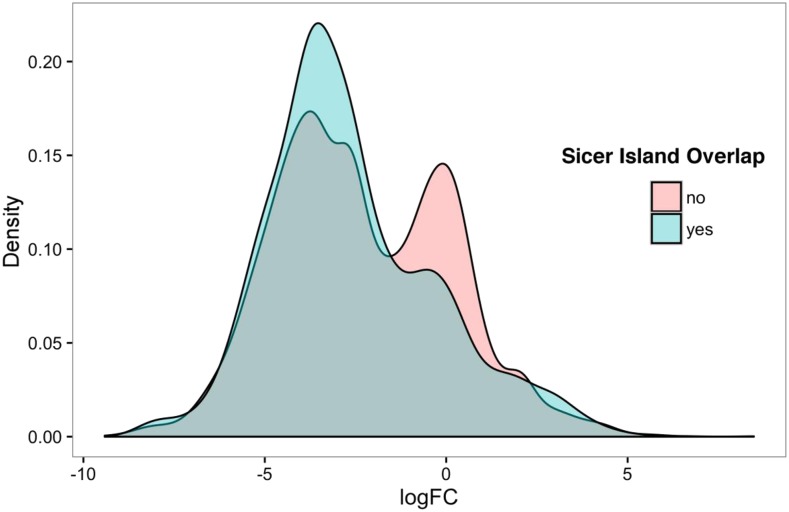
Analysis of the 5,512 lincRNAs expressed in panicles with respect to the locations of H3K27me3-enriched islands revealed a bias (P = 2.96e-10) in proportions consistent with the 2,594 emf2b derepressed lincRNAs having about a 1.45-fold greater tendency to overlap H3K27me3-enriched islands than nondifferentially expressed lincRNAs (Table I). Looking at the relationship from a different perspective, the lincRNAs were classified as overlapping or not overlapping an H3K27me3-enriched island, and the distribution of log fold change expression for wild-type panicles versus emf2b panicles was compared for these two classes (Fig. 8). The distribution of expression values for both groups overlapped, but their median values were not the same (P = 7.7e-06, Mann-Whitney U test). Furthermore, the non-H3K27me3-overlapping class had a clearly bimodal distribution, with a peak near zero (no differential expression) and another peak for negative log fold change values (higher expression in emf2b panicles versus the wild type), whereas the H3K27me3 island-overlapping class had only a small bulge near zero on the side of a larger peak for the negative log fold change values. The bias toward more negative log fold change values for the H3K27me3 island-overlapping class is consistent with this group being more greatly affected by derepression in the emf2b versus wild-type panicles.
Table I. LincRNA derepression in the emf2b mutant shows bias toward those overlapping H3K27me3-enriched islands.
The H3K27me3 peak islands were assessed for overlap (No, Yes) with the location of lincRNAs derepressed in emf2b panicles (−1) versus lincRNAs not differentially expressed (0). Fisher’s exact test gave a P = 2.959e-10 that this would occur by chance alone.
| Overlap with Peak | −1 | 0 | 1 | Total |
|---|---|---|---|---|
| No | 2,076 | 2,426 | 87 | 4,589 |
| Yes | 518 | 381 | 23 | 922 |
| Total | 2,594 | 2,807 | 110 | 5,511 |
Figure 8.
Mann-Whitney U test shows a significant difference in the distribution of edgeR log fold change (logFC) values between lincRNAs that overlap a Sicer peak island (yes) and those that do not (no) for all 5,512 lincRNAs assessed (P = 7.7e-06).
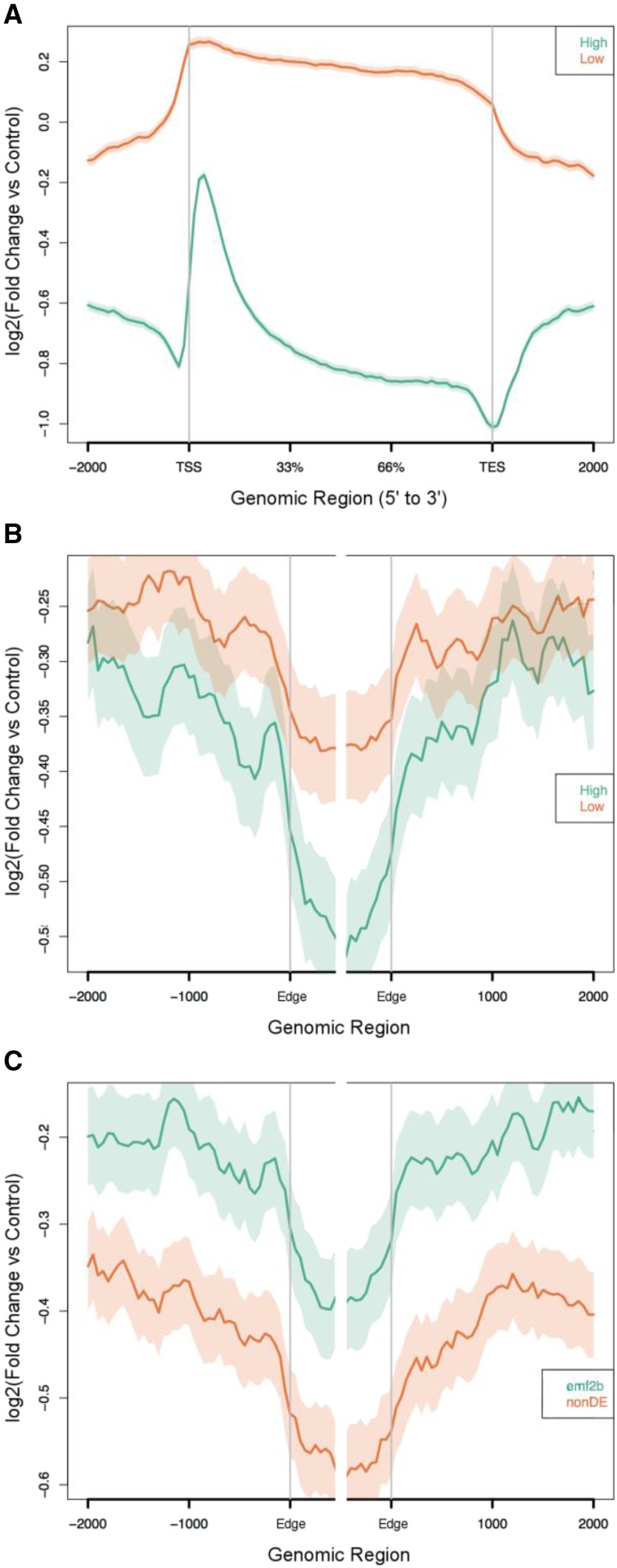
To investigate potential relationships between H3K27me3 marks and TSS and TTS, transcripts from the three different classes (non-TE protein-coding genes, TE protein-coding genes, and lincRNA loci) were divided into two groups (high and low) according to their reads per kilobase of transcript per million mapped reads (RPKM) expression levels in wild-type panicles. Low expression was designated as RPKM values less than 0.5 (log2 value of −1) and high expression as greater than 0.5 (Supplemental Fig. S4). These high- and low-expressing loci were analyzed for H3K27me3 enrichment by comparing the ChIP-seq with input reads and plotting this in relation to the transcribed and flanking regions of each of the three classes of gene (Fig. 9; Supplemental Fig. S5). For protein-coding loci (non TE and TE), this could be done with some precision in relation to TSS and TTS, since these features are reasonably well defined and the strand information is known. However, limited strand information was available for the lincRNAs from immature panicles; therefore, the analysis was performed without incorporating this information. Despite this limitation, clearly higher levels of enrichment for H3K27me3 were seen in the flanking regions than across the lincRNA body for both the low-expressing and the high-expressing groups in panicles (Fig. 9B). This is consistent with these active transcriptional units having lower levels of silencing marks than their surrounding chromatin. Notably, a relatively higher level of H3K27me3 enrichment signal was seen for the low-expressing group, both across the lincRNA body but also the flanking regions, than for the high-expressing group. This pattern also was present for the TE and non-TE protein-coding loci (Fig. 9A; Supplemental Fig. S5). These observations are consistent with H3K27me3 levels modulating the expression of the lincRNA loci in addition to affecting the expression at protein-coding loci.
Figure 9.
Relative H3K27me3 enrichment profiles (ChIP-seq versus input) are shown for non-TE protein-coding genes (A) and lincRNA loci (B and C), where the TSS and TES are shown for the protein-coding loci and, for the lincRNAs, the edges of the transcripts (Edge) are shown where strand information was not available. Within the graphs are plotted two lines, one for each of two classes of loci, where the higher line shows greater enrichment of DNA sequences via ChIP-seq (relative to the input chromatin) and the shaded band indicates the 95% confidence interval (see “Materials and Methods”). For the comparison of high versus low expression classes, both the non-TE protein-coding loci (A) and lincRNAs (B) show greater enrichment of H3K27me3 for the low expression class. For the lincRNA plots (B and C), relative to the lincRNA body, clearer separation is shown for the flanking regions of the emf2b/nondifferentially expressed (nonDE; 2,758/3,456 loci) comparison than for the high/low (2,594/2,808 loci) expression comparison, for which the 95% confidence intervals overlap.
We next examined whether the derepression of lincRNAs observed in the emf2b panicle is likely the direct result of a reduction in H3K27me3 marks or an indirect result via other processes, such as the derepression of PRC2-regulated transcription factors acting on lincRNA loci that are already in an open configuration. To address this, H3K27me3 ChIP-seq enrichment analysis was performed on the 2,594 lincRNAs derepressed in emf2b panicles and the 2,808 lincRNAs that were not differentially expressed in emf2b panicles relative to wild-type panicles (Fig. 9C). This analysis demonstrated a similarly higher ChIP-seq enrichment for reads associated with the emf2b lincRNAs (those derepressed in the mutant) versus those not differentially expressed between wild-type and emf2b panicles, consistent with an association of both H3K27me3 and the wild-type EMF2B with this set of lincRNAs. However, comparison of the ChIP-seq enrichment plots for both the differential expression classes (Fig. 9C) and the expression level classes (Fig. 9B) appears to show a qualitatively clearer separation for the flanking regions for the differential expression classes (derepressed in emf2b versus the nondifferentially expressed class) as compared with the high/low comparison (Fig. 9). Specifically, the 95% confidence intervals for H3K27me3 enrichment overlap in the flanking regions but not the lincRNA bodies for the high/low comparison (Fig. 9B), whereas the H3K27me3 enrichment for the lincRNAs derepressed in emf2b in comparison with the nondifferentially expressed lincRNAs (Fig. 9C) shows a similar difference for the flanking regions and the lincRNA bodies (i.e. similar spacing is seen between the two lines across the whole span of the plot). The two alternative comparisons, high/low versus emf2b/nondifferentially expressed, represent two competing factors potentially associated with H3K27me3 enrichment, with a greater separation of the lines on the plot expected for the factor having a greater association. Therefore, these results are consistent with emf2b/nondifferentially expressed showing greater concordance with H3K27me3 enrichment than the high/low comparison.
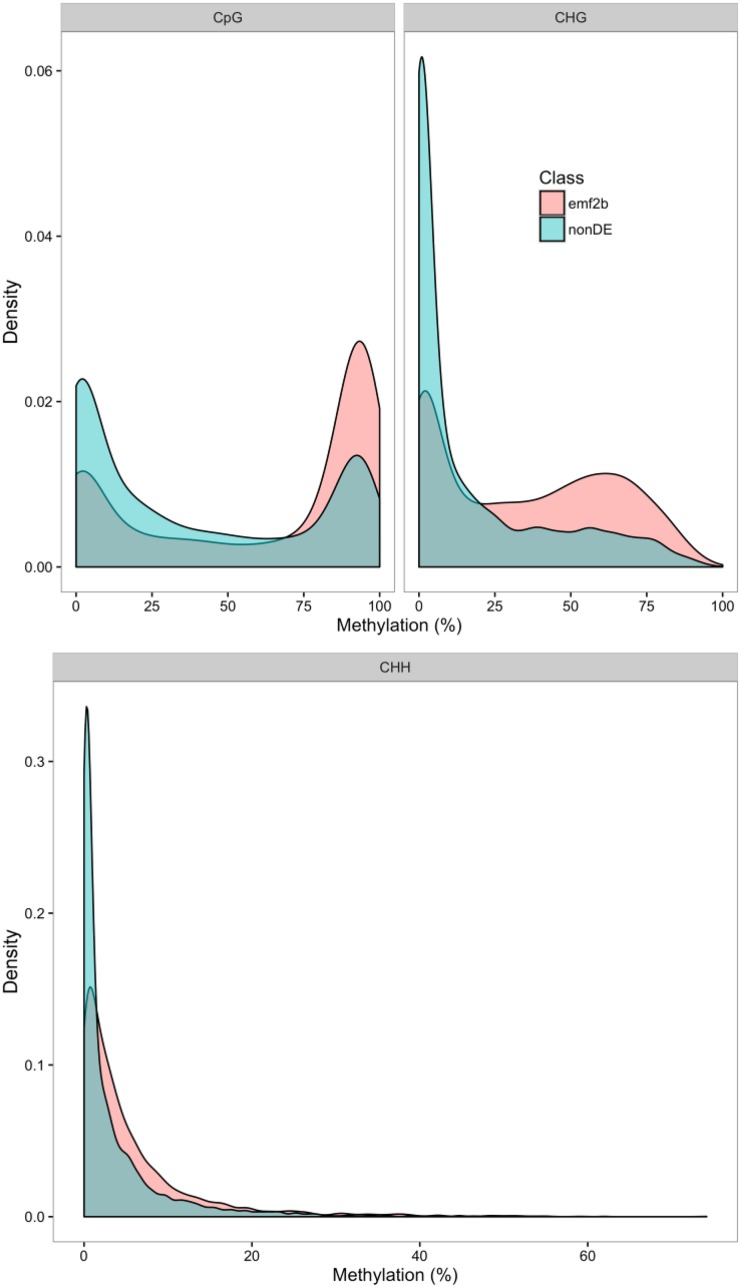
To further investigate epigenetic silencing processes potentially impacting the lincRNAs, the levels of DNA methylation in relation to lincRNA body and flanking regions were assessed using bisulfite sequencing (BS-seq) reads data available for O. sativa leaves (Chodavarapu et al., 2012). The BS-seq reads were remapped to MSU7.0, and the DNA methylation calls were collated for each of the three contexts: CpG, CHG, and CHH (see “Materials and Methods”). The percentage levels of DNA methylation within lincRNA bodies and in flanking regions were assessed across subsets of lincRNAs based on expression levels in panicles and differential expression between wild-type panicles and emf2b panicles as described earlier. The 5-methylcytosine levels of 2,594 lincRNAs derepressed in emf2b panicles and the 2,808 nondifferentially expressed lincRNAs were assessed for (1) the distribution of methylation states within the body of the lincRNAs (Fig. 10) and (2) the position-dependent methylation levels (Supplemental Fig. S6). However, due to the noticeable overdispersed nature of the lincRNA body methylation, the plots of the position-dependent means are not considered reliable. Overall, for the lincRNA bodies in all three contexts, a greater proportion of emf2b derepressed lincRNAs than nondifferentially expressed lincRNAs had high levels of DNA methylation, with CpG and CHG having high peaks at about 90% and 65%, respectively. Thus, relative to the nondifferentially expressed lincRNAs, the emf2b derepressed lincRNAs had a greater association with DNA methylation levels, at least in the leaf.
Figure 10.
Kernel density plots show the distribution of average DNA methylation levels in rice leaf tissue among the 2,594 lincRNAs derepressed in emf2b panicles (emf2b) and the 2,808 nondifferentially expressed lincRNAs (nonDE) as compared with wild-type panicles. The CpG and CHG contexts show clearly bimodal distributions, with a greater proportion of lincRNAs contributing to the high methylation peak for the emf2b class but not for the nondifferentially expressed class. For the CHH context, there is a relative bias toward higher DNA methylation for the emf2b class as compared with the nondifferentially expressed class.
A Subset of Reproductive LincRNAs Is Modulated by Drought
The flowering stage of rice is particularly sensitive to environmental factors, including the availability of water. With a subset of the lincRNAs expressed in panicles and sperm that are likely regulated via epigenetic processes, the available RNA-seq data from a drought experiment at two stages of development (inflorescence and vegetative stages) were assessed for potential modulation by drought within reproductive tissue. To detect any lincRNAs that may be involved in responding to drought, we analyzed the expression of our lincRNA loci within duplicate RNA-seq data sets derived from drought-treated and control plants at the vegetative stage and reproductive (R3) stage of the rice growth cycle.
From a total of 2,303 expressed lincRNAs detected in the samples, lincRNA transcripts were identified as being differentially expressed within various combinations of the watering conditions and growth stages (Table II). Of particular interest was the expression of lincRNAs within the reproductive tissues, for which we detected 208 and 112 lincRNAs up- and down-regulated in response to drought conditions, respectively. Among these were 160 transcripts with higher expression levels in reproductive as compared with vegetative tissues. Of these reproductive-specific lincRNAs, 32 and 25 were detected as up- or down-regulated in response to drought, respectively. Of the 208 lincRNAs up-regulated by drought in inflorescence tissue, five were down-regulated in the vegetative stage. Similarly, for the 112 lincRNAs down-regulated by drought in the inflorescence, six were up-regulated at the vegetative stage. These 11 lincRNAs appear to be responding to an abiotic stress, drought, in a manner that is dependent on the developmental stage.
Table II. LincRNAs are regulated by drought in both vegetative and reproductive stages.
Differential expression analysis categories supported by pairwise comparison using contrasts (inflorescence drought versus inflorescence control and inflorescence versus vegetative) are shown. Also shown are lincRNAs with higher expression in reproductive than vegetative tissues (italics) and a larger set of 208 and 112 lincRNAs (underlined) that respond to drought specifically in reproductive tissues by an increase and decrease in expression, respectively. In total, there are 447 lincRNAs (boldface) that respond to drought.
| Drought versus Control | Inflorescence versus Vegetative | Total | ||||
|---|---|---|---|---|---|---|
| Inflorescence | Vegetative | Down | Up | Not Differentially Expressed | ||
| Down | Down | 4 | 3 | 3 | 10 | 112 |
| Down | Not differentially expressed | 9 | 21 | 66 | 96 | |
| Down | Up | 2 | 1 | 3 | 6 | |
| Not differentially expressed | Down | 27 | 8 | 34 | 69 | 1,983 |
| Not differentially expressed | Not differentially expressed | 192 | 90 | 1,574 | 1,856 | |
| Not differentially expressed | Up | 22 | 5 | 31 | 58 | |
| Up | Down | 3 | 1 | 1 | 5 | 208 |
| Up | Not differentially expressed | 10 | 28 | 147 | 185 | |
| Up | Up | 3 | 3 | 12 | 18 | |
| Total | 272 | 160 | 1,871 | 2,303 | ||
A total of 447 lincRNAs were affected by drought in at least one comparison (Table II). To test whether these lincRNAs might be responding to potential EMF2b-mediated modulation, a contingency table analysis was performed (Supplemental Table S3). If there was a positive association between the emf2b lincRNAs and drought-affected lincRNAs, then the risk ratio (RR) should be greater than 1, which was not the case (RR = 0.366). Also, no positive association could be shown for drought-derepressed lincRNAs within inflorescences (RR = 0.336) or vegetative tissues (RR = 0.636). This meant that we could reject the hypothesis that the 1,617 lincRNAs negatively regulated in panicles by EMF2b also were enriched in the 447 drought-affected lincRNAs, the 208 inflorescence drought-enriched lincRNAs, or the 82 vegetative drought-enriched lincRNAs. However, a negative association was detected for the full set of 447 drought-affected lincRNAs (Fisher’s exact test, P = 5.2e-15) and the subset of 208 lincRNAs within the inflorescence (Fisher’s exact test, P = 3.0e-08). This is consistent with the class of lincRNAs negatively regulated by EMF2b in panicles not being the same class of lincRNAs derepressed by drought in reproductive tissues.
Conservation of LincRNAs across Cereals
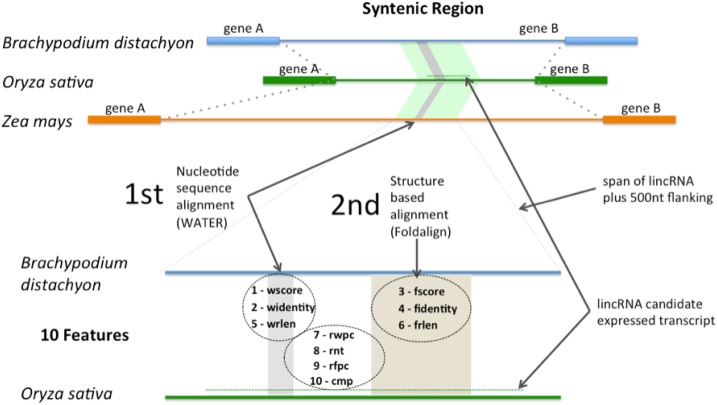
For lncRNAs that typically show low nucleotide sequence conservation, RNA folding has been proposed to be as important as the nucleotide sequence for detecting conservation (Ulitsky et al., 2011). To facilitate the identification of lincRNA orthologs, the existence of synteny was used to greatly reduce the search space to syntenic regions between (1) O. sativa and B. distachyon and (2) O. sativa and maize. A novelty-detection SVM, using features extracted from two different alignments (Fig. 11), was used to identify lincRNAs that may be conserved at the sequence level, the structure level, or both (see “Materials and Methods”; Supplemental Results S2). This approach resulted in the identification of 70 lincRNA isoforms, with 43 uniquely syntenic to B. distachyon, five uniquely syntenic to maize, and 22 with representation in both B. distachyon and maize (Supplemental Fig. S7). Together, this corresponded to 10.2% of the O. sativa lincRNAs having homologs in B. distachyon and 4.2% of the O. sativa lincRNAs having homologs in maize.
Figure 11.
Diagram showing how the syntenic regions between rice, B. distachyon, and maize were processed to extract features used in the homology-detection SVM. To begin, limited nucleotide homology (gray band), detected using the WATER algorithm from EMBOSS, was used to locate a subregion (light green band) for further analysis. The subregion sequence was then scanned for structural (beige band) and sequence homology using Foldalign. Alignment features corresponding to sequence homology (1, 2, and 5) and structure-assisted homology (3, 4, and 6) in addition to other features (7–10) were extracted for modeling.
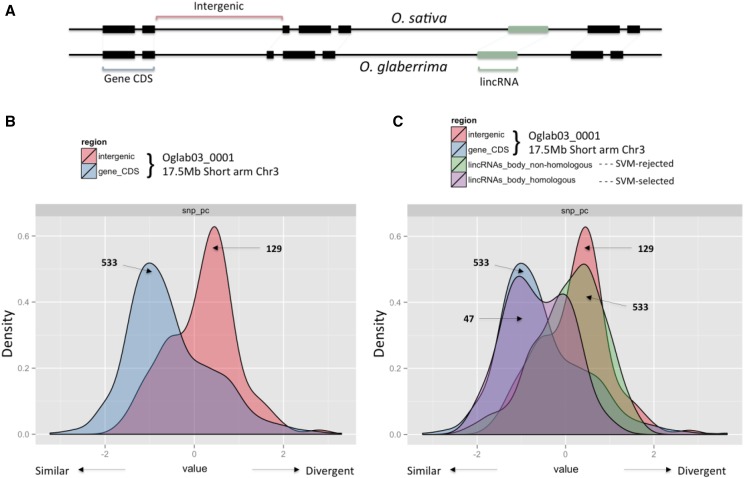
The proportions of O. sativa lincRNAs with SVM-selected alignments for B. distachyon and maize were consistent with O. sativa being more closely related to B. distachyon than to maize (Hedges et al., 2015; http://www.timetree.org/) and are consistent with these lincRNAs being orthologous. However, despite the separation between O. sativa and B. distachyon and O. sativa and maize, medians of ∼47 and ∼48 million years, respectively (Hedges et al., 2015; http://www.timetree.org/ in December 2016), the similarity detected for many could conceivably be due to residual homology rather than to selection. To address this issue, the lincRNAs were aligned with the newly available genome sequence for O. glaberrima (African rice), a species distinct from Asian rice. The frequency of mismatches between O. sativa and O. glaberrima was used as an estimate of single-nucleotide polymorphisms (SNPs) for each lincRNA. This was similarly performed for intergenic regions and those of the coding sequence of protein-coding genes that were present on the relatively complete sequence of the short arm of chromosome 3 of O. glaberrima. From density plots of estimated SNP rate distributions, it can be seen, as expected, that the SNP rate for the protein-coding sequences is lower than that for intergenic regions (Fig. 12, A and B). The SVM-analyzed lincRNAs that had alignments with O. glaberrima sequences were divided into two groups: those that were selected by the SVM as novel and those that were not (i.e. homology and no homology detected; Fig. 12, A and C). Whereas the peak of the non-SVM-selected lincRNAs overlapped the distribution of similarity scores of the intergenic regions, that of the SVM-selected lincRNAs, although bimodal, mostly overlapped the distribution of coding sequence. This indicated that some of those lincRNAs selected by the SVM had an SNP rate distribution that is more similar to protein-coding regions than the intergenic regions, suggesting that these lincRNAs are conserved with their similarity retained due to selective processes.
Figure 12.
A, Schematic of the different region types that were compared. B and C, Distribution mismatch rate (loosely treated as SNP rate) across a log scale for four different classes of sequences (intergenic, protein-coding sequence [CDS], and body of lincRNAs selected and not selected by SVM) that map to the short arm of chromosome 3 of O. glaberrima, where the SVM-selected lincRNAs are considered to be in the homologous class. The 47 homologous lincRNAs (C) are a subset of the 70 SVM-selected lincRNAs that happen to be present in the sequence corresponding to the short arm of chromosome 3 of O. glaberrima. The 47 SVM-selected lincRNAs show a bimodal distribution, with the larger group overlapping the mismatch rate distribution of protein-coding genes, consistent with a similar degree of conservation.
DISCUSSION
Identification and Verification of LincRNAs
This study reports on the identification of 6,214 loci corresponding to 6,309 lincRNAs that were present within 11 different RNA-seq data sets, of which about half were from reproductive cells/tissues. Strand-specific RNA-seq data were not available for the specialized cell types, sperm cells and young emf2 mutant panicles, that provided much of the lincRNAs in this study. Strand information could be extracted for a total of 464 lincRNA loci (562 transcripts) using the strand-specific RNA-seq data from the seedling and drought-stressed samples; therefore, most of the lincRNA loci do not have associated chromosome strand information at this time. We note that previous studies have demonstrated that reliable lincRNA assemblies can be constructed without requiring strand information, as in maize (Li et al., 2014) or tetrapods (Necsulea et al., 2014). However, for those lincRNAs assembled without strand information, it is possible that the delineation of some is not accurate. For example, two distinct but overlapping lincRNAs that are transcribed in antisense to each other would not be resolved and, thus, would appear as a single lincRNA. We confirmed by RT-PCR (Fig. 2C; Supplemental Fig. S1, A and B) the presence of transcripts from lincRNA loci selected for tissue-specific expression, including the presence of predicted introns, providing independent validation for the findings. The broader category of lncRNAs includes the lincRNAs but also transcripts that overlap other transcribed loci in sense/antisense orientations and in exonic/intronic sequences. In this analysis, we restricted the pipeline to intergenic regions at a distance of at least 500 bp from annotated genes to avoid any confusion in the identification of the noncoding RNA loci due to poor annotation or alternate splicing. This necessarily meant that potentially important lncRNAs might have been missed. An example of this is the LDMAR lncRNA (JQ317784.1) that confers male sterility in the cv Nongken of rice 58S (Ding et al., 2012a,b) but that overlaps with the locus LOC_Os12g36030, so was filtered out earlier in the pipeline. However, despite limiting the analysis to intergenic regions, the number of lincRNA loci detected compares favorably with that of other studies in monocotyledonous plants, two of which were in rice and one in maize (Komiya et al., 2014; Li et al., 2014; Zhang et al., 2014).
LincRNAs Identified Lack Overlap with Previous Studies
The earlier rice study (Komiya et al., 2014) detected MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1)-associated short interfering RNAs (siRNAs) generated by loci corresponding to the phased clusters of inflorescence-specific 21-nucleotide small RNAs. In the maize study of the B73 reference genotype (Li et al., 2014), a total of 20,163 putative lncRNAs were detected, of which 18,459 were likely from small RNA-generating loci and another 1,704 were considered high-confidence lncRNAs. A recent RNA-seq-based lncRNA study in rice (Zhang et al., 2014) detected a total of 2,224 lncRNAs from reproductive tissues, of which 1,624 were considered lincRNAs while the remaining 600 were considered long noncoding natural antisense transcripts. This study is most closely related to our study with respect to the types of tissues included, specifically reproductive tissues (i.e. anthers, pistils, and seeds), of which the first should include pollen. Therefore, the detection of overlapping loci between the two studies might be expected. Yet, of their total of 1,624 reproductive lncRNAs, just 333 overlap with 340 of our 6,214 lincRNA loci. This overlap represents only about 5% of the loci, suggesting that the discovery of lncRNAs in rice is far from complete. Lastly, we also looked for overlap of our lincRNAs with 11,229 lincRNAs reported in a more recent study in rice (Wang et al., 2015) that identified lincRNAs associated with agronomic traits. Of the 6,309 lincRNAs from this study, only 1,296 overlapped with the 11,229 lincRNAs. Due to the low frequency of overlap, we checked whether we had excluded potential lincRNAs early in our initial lincRNA pipeline. Interestingly, a check for any overlap of the 110,155 unfiltered transcripts initially obtained in the pipeline (Fig. 1) indicated that 48% to 64% (5,437–7,167 RNAs, depending on the removal of large anomalous transcript models) of the 11,229 lincRNAs from Wang et al. (2015) overlapped with our unfiltered transcripts, but this overlap corresponded to only about 10% of our unfiltered transcripts. In case the relatively large fraction of lincRNAs from Wang et al. (2015) overlapping the unfiltered transcripts was due to the absence of a 500-bp clearance requirement, a minimum overlap of 70% also was assessed. This resulted in a large drop in overlap, from 7,167 to only 1,428 of the 11,229 lincRNAs. Taken together, this suggests that a substantial fraction of the 11,229 lincRNAs from Wang et al. (2015) are lincRNAs closely associated with annotated genes and might potentially include nonannotated extensions of protein-coding genes. Therefore, it is possible that many of the 11,229 lincRNAs are actually alternative transcripts from protein-coding loci. The low proportion of the 6,309 lincRNAs overlapping lincRNAs of other studies might be the result of differences in the source materials used. Of note is the significant diversity of tissues used in this study. Of the 11 tissues/cell types, only about five of these overlapped with the combined diversity of tissues from both the Zhang et al. (2014) and Wang et al. (2015) studies. In addition to this, the emf2b lincRNAs were detected under a different epigenetic state. This suggests that the detection of lincRNAs in rice is perhaps largely incomplete and that sampling of a greater number of tissues under different environmental and epigenetic conditions likely will result in the identification of many more lncRNA loci.
The Tissue Specificity of LincRNAs Provides Insight into Biological Function and Evolution
One of the prevailing questions in the field of lncRNA research is to what extent the transcribed lincRNA loci have biological functions in the organism. One way to address this is to examine the broader properties of the lincRNAs identified. The O. sativa lincRNAs reported in this study are expressed in a highly tissue-specific manner (Shannon entropy < 2), similar to previous studies (Guttman et al., 2010; Liu et al., 2012; Li et al., 2014; Zhang et al., 2014). Interestingly, among the O. sativa lincRNAs, those likely to be more conserved (i.e. selected by the SVM) show more general expression than those not selected (Supplemental Fig. S8). The idea that newly evolved genes would be more tissue specific is supported by the structure of the 35S promoter, the general expression of which is the result of cumulative contributions of its constituent parts (Benfey and Chua, 1990). Hence, the default state of de novo genes may be to be highly tissue specific, and the observed higher tissue specificity of the maize lincRNAs may merely reflect the existence of a greater number of potentially newly expressing RNA loci that one might expect for a genome with a much larger intergenic space than O. sativa. In addition, the results suggested that a large fraction of the lincRNAs are negatively regulated by EMF2B in wild-type panicles but potentially also in sperm cells in a developmental fashion. Interestingly, de novo genes in Drosophila melanogaster tend to be expressed specifically in male reproductive tissues (Zhao et al., 2014). It was proposed that these loci arose recently from sequences in the intergenic regions, putatively from noncoding RNAs. It is interesting that these de novo genes are simpler than typical protein-coding genes, more often consisting of a single exon, and are specific to male reproductive tissues, which is remarkably similar to the sperm-specific O. sativa lincRNAs reported here. Therefore, one possibility is that some of the O. sativa loci are evolving protein-coding genes that are repressed in most tissues except male gametic cells.
Epigenetic Regulation of LincRNAs
The involvement of a PRC2 subunit in the regulation of the O. sativa lincRNAs possibly through H3K27me3 modification stands in contrast to many previous studies that have implicated lncRNAs in the regulation of other genes through the PRC2. It is also interesting that H3K27me3 appears to regulate a larger proportion of the lincRNAs described here, as compared with the protein-coding genes, suggesting that the lincRNAs do not represent typical expressed loci. This is unlike the light-regulated natural antisense lncRNAs reported by Wang et al. (2015), for which histone acetylation rather than H3K27me3 was found to be associated with light responsiveness. In animal systems, H3K27me3 levels across the genome are anticorrelated with DNA 5-methylcytosine (5mc) levels (Reddington et al., 2013), with the methyl groups in DNA suspected as inhibitory to PRC2 interaction with the histones. In plants, there is a similar although less strict relationship (Weinhofer et al., 2010; Zhou et al., 2016), likely due to the availability of three methylation contexts. Hence, it is possible that the large proportion of PRC2-regulated lincRNAs reported here may be associated with lower levels of 5-methylcytosine relative to protein-coding genes, as the latter were less affected by the loss of EMF2b. Despite this possibility, we note that the bodies of emf2b-expressed lincRNAs appeared to be enriched for 5-methylcytosine sites at least in leaf tissue, relative to the other lincRNAs. Similar to the known association with many protein-coding genes, there was a drop in 5-methylcytosine at the loci edges for the emf2b differentially expressed lincRNA genes. This drop was greater than that observed for the nondifferentially expressed lincRNAs, possibly indicative of a role for DNA methylation adjacent to the gene body in PRC2 repression of lincRNA genes. While differences in DNA methylation levels in plants have been associated with differential gene expression, in general, DNA methylation within plants is considered to be relatively consistent between tissues, and this was recently formally confirmed in B. distachyon between leaves and flowers (Roessler et al., 2016). Regardless, it is still possible that the levels of DNA methylation observed in the leaves do not reflect that in the wild-type panicles, and additional experiments would need to be performed within the same tissues in order to confirm a relationship between histone modification and DNA methylation for the O. sativa lincRNA loci reported in this study.
LincRNAs and miRNA Regulation
One of the lincRNAs validated by RT-PCR corresponds to an ultraconserved element that has been published previously (Kritsas et al., 2012), is predicted to have extensive base pairing within an RNA secondary structure, and also is found in B. distachyon (Supplemental Fig. S9). The existence of extensive conservation within ultraconserved elements as a predictor of function was more recently validated for the mammalian lncRNA (Uc.283+A), which was shown to regulate the generation of mature miRNAs via precise interaction with primary transcript miRNA precursor transcripts (Liz et al., 2014), suggesting the potential for lincRNA involvement in miRNA regulation. In addition to the 21-nucleotide phased small RNAs (Supplemental Fig. S10A) first indicated earlier that are associated with MEL1 lncRNA loci, there was also at least one other O. sativa lincRNA locus overlapping with 24-nucleotide phased small RNAs (Supplemental Fig. S10B). Phased small RNAs, typically 21 nucleotides, have been associated previously with second-strand synthesis of RNA molecules during trans-acting siRNA biogenesis, but in this case, the small RNAs were 24 nucleotides, a size class typically associated with de novo DNA methylation via RNA-dependent DNA methylation, suggesting the possibility of a DNA methylation process directed by trans-acting siRNAs. Taken together, this complexity in association with small RNAs indicates that the described O. sativa lincRNAs encompass a diverse set of mechanisms of action with a lot of potential for further research.
In summary, by the application of stringent criteria on transcriptomes assembled from RNA-seq reads from 10 tissue types, a comprehensive set of 6,309 lncRNAs was defined. About two-thirds of these were defined as either dominant within sperm cells or as derepressed within the emf2b mutant. While many previously reported lincRNAs belong to the PRC2 class that direct transcriptional repression through H3K27me3 chromatin marks, this study reported a large number of lincRNAs, as many as 41%, targeted by this process rather than regulated through this process. Apart from these two large classes, there also existed a distinct subset of 447 lincRNAs that were drought responsive, of which 208 were specific to the agronomically important and drought-sensitive reproductive stage of O. sativa. With lincRNAs typically showing very low sequence conservation, we utilized a novel detection SVM model to identify lincRNAs conserved at the nucleotide and structural levels between O. sativa and two other members of the Poales order, B. distachyon and maize, for which 10.2% and 4.2% of the O. sativa lincRNAs were conserved, respectively, and these results are supported by the rates of SNPs detected in the available genomic sequence of O. glaberrima. In conclusion, the novelty of the 6,309 lincRNAs is supported by the fact that about 80% have not yet been reported within other large lincRNA studies in plants. With these covering a large variety of tissues, they represent a baseline for the lincRNAs expressed by the rice genome.
MATERIALS AND METHODS
RNA-Seq Data and Analysis
The short-read sequencing data from 11 different tissues/cell types of Asian rice (Oryza sativa; Supplemental Table S1) were downloaded from various repositories (anther, embryo, endosperm, leaf, seed, and seedling) or were available from internal studies that have been published previously (wild-type panicle, emf2b panicle, egg cell, sperm cell, and vegetative cytoplasm from pollen). Following the identification of lincRNAs (described below), differential expression analysis was performed on a larger number of short-read data sets (Supplemental Table S1) using the R package edgeR. Specifically, for the wild-type panicle, emf2b panicle, and sperm comparison, biological replicate samples of two, two, and three, respectively, were used, and the six differentially expressed classes between these were obtained via contrasts (Supplemental Data File S1), as done previously (Anderson et al., 2013). For the drought experiment differentially expressed analysis, duplicate biological replicates were collected for each of the vegetative drought, vegetative control, reproductive drought, and reproductive control treatments, and differentially expressed classes were obtained in a manner similar to that indicated above.
LincRNA Discovery Pipeline
A total of 568,886,638 RNA-seq reads from 11 different tissue types were aligned separately on the O. sativa genome (cv Nipponbare; MSU7.0) using TopHat (version 2.0.5) [–library-type fr-unstranded–segment-length 18] and, for the seedling data, using the fr-firststrand option. The Reference Annotation Based Transcript assembly was performed for each tissue/mutant type individually using Cufflinks (version 2.0.2) [-u–library-type fr-unstranded-g all.gff3]. Eleven Reference Annotation Based Transcript assemblies (10 wild-type tissues and one emf2b mutant panicle) were merged using Cuffmerge to generate a representative transcript set.
We then categorized the transcripts based on various filters. The transcripts overlapping with known protein-coding genes and TE loci were identified and separated. Furthermore, as informed by Coding Potential Calculator (CPC version 0.9) with UniRef90 as a reference set, the remaining transcripts with coding potential less than zero were considered noncoding. In addition to filtering all transcripts with known coding potential, we performed BLAST searches (E-value cutoff of 1E-10) against the nonredundant protein database from the National Center for Biotechnology Information (NCBI) and an hmmscan against all Pfam domains. The noncoding transcripts were searched (E-value cutoff of 1E-05) for known rRNA, tRNA, and miRNA precursors to separate their O. sativa orthologs. Then, transcripts within 500-bp flanking regions of annotated genes were categorized as gene-associated transcripts (3,259). The other transcripts shorter than 200 bp were defined as other intergenic transcripts. The ORFs were identified using EMBOSS (getorf) for all six reading frames, and transcripts with ORFs greater than 100 residues were classified as transcripts of unknown coding potential. The remaining set of 6,309 transcripts was defined as lincRNAs that represented 6,213 unique lincRNA loci (Supplemental Data Files S2 and S3). The longest isoforms for all lincRNA loci were chosen for further analysis. Bedtools were used extensively whenever two browser extensible data files needed to be compared.
Normalized Expression Values
A perl script was employed to count the uniquely mapped reads on each exon from the TopHat alignment files and to calculate the expression values (RPKM) in each of the tissue/mutant types for all protein-coding genes and lincRNA loci. While using paired-end RNA-seq data, only one of the mapped read pairs was considered for the expression value calculation. The maximal gene expression was calculated using RPKM values for protein-coding genes and lincRNAs. To prepare the heat map depicting tissue specificity, the RPKM values were row-wise normalized (to calculate the fractional abundance of genes across different tissue types). LincRNAs were clustered by X-means clustering using the row-normalized RPKM values.
Molecular Confirmation via RT-PCR
Tissue collection and cDNA synthesis were described previously for sperm cells (Anderson et al., 2013) and wild-type and emf2b mutant panicles (Conrad et al., 2014). RT-PCR was performed with 1 μL of cDNA and the primers listed in Supplemental Table S5 using Bioline MyTaq Red Mix following the manufacturer’s protocol. Cycles were as follows: 95°C for 1 min, followed by 35 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s, and a final extension for 1 min at 72°C. The PCR products were visualized using standard agarose Tris-acetate-EDTA (TAE) gel electrophoresis with ethidium bromide.
Analysis of ChIP-Seq Data
The ChIP-seq data for H3K27me3 modification from seedlings and panicles were obtained from the short-read archive at NCBI and our laboratory, respectively, and our data were stored under accession GSE62550 on NCBI Gene Expression Omnibus. The adaptor contamination was trimmed using the NGS QC Toolkit (version 2.3). These data were aligned to the O. sativa genome using Bowtie (version 0.12.8) [-S -k 15–best–chunkmbs 20000]. The uniquely mapped reads were used for the identification of significant methylation marks using Sicer (version 1.1) [redundancy threshold, window size, fragment size, effective genome fraction, gap size, FDR = > 1 200 250 0.61/0.91 600 0.1] at FDR 0.1. The output WIG file contained a peak value for each 200-bp window where the reads were mapped. These peak values were normalized read counts for that window. The consecutive 200-bp windows were merged to prepare a browser extensible data file containing islands. The peak values of the underlying windows were averaged to represent the peak for the methylation islands. TSS/TTS ChIP-seq enrichment plots were generated using ngsplot (version 2.08; Shen et al., 2014), but the code was modified, so instead of generating shaded margins of se, the shaded margins correspond to 95% confidence intervals (1.96 × se).
DNA Methylation Analysis
BS-seq reads in the series SRR949542 to SRR949552 in fastq files were trimmed with cutadapt 1.5 (Martin, 2011) via trim_galore 0.3.7 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) using default settings. The trimmed reads were aligned to MSU7.0 using Bowtie 2.1.0 (Langmead and Salzberg, 2012) via Bismark 0.12.5 (Krueger and Andrews, 2011) using default settings. Methylation data were extracted via R from merged SAM alignment files using the R package methylKit 0.9.2 (Akalin et al., 2012). The resulting per-base methylation status files were processed using various custom perl scripts to produce data of a suitable format for different purposes. The processed data were plotted using the R package ggplot2 1.0.1 (Wickham, 2009).
Detection of Potentially Conserved LincRNAs
A pipeline was developed to identify potentially conserved lincRNAs between O. sativa and Brachypodium distachyon as well as between O. sativa and maize (Zea mays). Due to potentially poor sequence conservation, an approach was taken to maximize the chance of detection and reduce the compute time. The strategy was to only search within sequence regions of the heterologous genomes that were syntenic to the O. sativa lincRNA being assessed. To reduce processing time and to create features for input into an SVM, the lincRNAs were aligned using two different aligners. First, a nucleotide-only alignment algorithm (Smith-Waterman) was employed to narrow the matching syntenic intergenic regions of the heterologous genome to the approximate size of the O. sativa lincRNA, and then an alignment using a structure-assisted alignment algorithm (Foldalign) was performed. It should be noted that the pipeline was not designed to define the boundaries of the lincRNAs but to aid in the detection of conservation. Consensus structures of conserved lincRNAs were generated using RNAalifold (Lorenz et al., 2011).
Extraction of Syntenic Regions
Syntenic blocks between rice-B. distachyon and rice-maize pairs were obtained from SynMap at CoGe. The protein-coding genes that flanked the intergenic region containing the lincRNA were checked for their orthologs in syntenic blocks, and if both flanking genes were found, the intergenic region was extracted from the respective genome. A total of 978 O. sativa intergenic regions with syntenic B. distachyon regions corresponded to 1,268 loci (1,286 isoforms) of O. sativa lincRNAs, while the 796 O. sativa intergenic regions with syntenic maize regions corresponded to 1,039 loci (1,055 isoforms) of O. sativa lincRNAs. There was a set of syntenic regions encompassing both B. distachyon and maize that consisted of 855 loci (869 isoforms) of O. sativa lincRNAs.
Alignment of LincRNAs to Syntenic Regions
The O. sativa lincRNA sequences were aligned to syntenic regions from B. distachyon and maize using a pipeline that incorporated two alignments. The first was a Smith-Waterman alignment that identified the best local alignment using only nucleotide conservation information. A subsequence of the syntenic region was extracted that included the best local alignment flanked by 500 nucleotides on both sides. The O. sativa lincRNA was then aligned using Foldalign 2.1.1 (Havgaard et al., 2005) to the syntenic subsequence to identify the best local alignment that included nucleotides identified as conserved at the level of base pairing in addition to conservation at the nucleotide level. For a single comparison (e.g. an O. sativa lincRNA versus a B. distachyon subsequence), the sequences encompassing the two alignments (nucleotide only and structure dependent) did not necessarily overlap. However, a lincRNA with a conserved function also might be expected to be conserved with respect to the relative placement of these regions. For example, if the nucleotide-only aligned sequences were 5′ of the structure-dependent aligned sequences in O. sativa, this arrangement also would be expected within the heterologous sequence. Any difference would be consistent with a rearrangement if, indeed, these represented orthologous sequences.
A custom perl script was used to automate alignment of the lincRNAs to the plus and minus strands of syntenic regions using the WATER algorithm from EMBOSS, and sequence pairs were kept for structural alignment if the two alignments (i.e. rice-B. distachyon versus rice-maize) overlapped in the O. sativa nucleotide coordinates. Two more perl scripts were used to perform structure-assisted alignments using the Foldalign algorithm and to summarize and tabulate the interspecies sequence and structure alignment overlap properties for later use in an SVM analysis. A set of sequence and structural alignments to act as a negative control for SVM training was produced by shuffling the O. sativa lincRNAs between the original extracted syntenic regions from B. distachyon and maize. These alignments also were summarized and tabulated for overlap properties.
Novelty-Detection SVM
For each comparison (e.g. one O. sativa lincRNA versus a B. distachyon subsequence), a set of 10 alignment features (Supplemental Table S4) was tabulated. These features included three for the nucleotide-only alignment (wscore, widentity, and wrlen), three for the structure-dependent alignment (fscore, fidentity, and frlen), three for the overlap between these two alignment types (rwpc, rnt, and rfpc), and one for the compatibility status of the placement of these two alignments between O. sativa and the heterologous sequence. These 10 features were then used in a novelty-detection SVM using the svm function of R package e1071 version 1.6-7 (Meyer et al., 2014). The SVM was trained using a negative control data set consisting of similar alignment data produced from nonsyntenic pairs via shuffling the O. sativa lincRNA against the syntenic regions from B. distachyon and likewise for the set of maize syntenic regions. Using a grid to explore the parameter space, parameter selection and training was performed on half the negative control alignments to avoid overfitting with cross-validation using the complete set of negative control and syntenic alignments . Parameters were chosen that maximized novelty detection while minimizing false positives. Of the 1,544 negative control alignments, four were classified as novel (false positives), while 96 of the 2,183 alignments from the real syntenic pairs were classified as novel. The feature data and classification status for the control and real syntenic regions are available . The 96 selected alignments generally showed high values for the alignment scores suggestive of conservation as well as having configurations of nucleotide-only and structure-dependent alignment blocks compatible between O. sativa and the heterologous species.
Accession Numbers
All sequence data will be deposited in the NCBI Gene Expression Omnibus. The accession number for EMF2b is Os09g13630.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-PCR validation of selected lincRNAs.
Supplemental Figure S2. LincRNAs may have fewer introns than protein-coding genes.
Supplemental Figure S3. LincRNAs show expression levels that are intermediate to the TE and non-TE protein-coding genes.
Supplemental Figure S4. Defining high and low expression level classes.
Supplemental Figure S5. The high-expression TE protein-coding class shows greater enrichment for H3K27me3.
Supplemental Figure S6. LincRNAs derepressed in emf2b may have DNA methylation profiles more similar to protein-coding genes.
Supplemental Figure S7. Selection of potentially conserved lincRNAs via SVM.
Supplemental Figure S8. Potentially conserved lincRNAs are less tissue specific.
Supplemental Figure S9. LOC_Os02r09073.1 corresponds to ULE546 of B. distachyon.
Supplemental Figure S10. LincRNAs may be the source of both 21- and 24-nucleotide phased small RNA clusters.
Supplemental Table S1. Summary of short-read data sets.
Supplemental Table S2. Overlap of lincRNAs with Deng noncoding RNAs (as in Liu et al., 2013).
Supplemental Table S3. LincRNAs derepressed in emf2b do not correspond to those derepressed under drought conditions.
Supplemental Table S4. Ten features derived from alignments of syntenic regions used in novelty-detection SVM.
Supplemental Table S5. Primer pairs used for RT-PCR validation of lincRNAs.
Supplemental Data File S1. LincRNA gene models gff.
Supplemental Data File S2. LincRNA predicted transcripts fasta file.
Supplemental Data File S3. LincRNA classes table.
Supplemental Results S1. Validation of selected lincRNAs.
Supplemental Results S2. Synteny analysis of lincRNA regions.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Eugene Lurie for technical assistance.
Footnotes
This research was supported by research grants IOS-1128145 and IOS-1547760 from the National Science Foundation to V.S. and utilized the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by National Institutes of Health S10 Instrumentation Grants S10RR029668, S10RR027303, and S10OD018174. In part, this material is based upon work supported by National Science Foundation Award DBI-0922747 to A.P., for the project titled “Cereal Drought Stress Response and Resistance Networks.”
Articles can be viewed without a subscription.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V (2013) Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76: 729–741 [DOI] [PubMed] [Google Scholar]
- Aune TM, Spurlock CF III (2016) Long non-coding RNAs in innate and adaptive immunity. Virus Res 212: 146–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartonicek N, Maag JLV, Dinger ME (2016) Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer 15: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250: 959–966 [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Boerner S, McGinnis KM (2012) Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 7: e43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Kondorosi A, Crespi M (2004) Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. (2011) The reality of pervasive transcription. PLoS Biol 9: e1000625, discussion e1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad LJ, Khanday I, Johnson C, Guiderdoni E, An G, Vijayraghavan U, Sundaresan V (2014) The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J 80: 883–894 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Buell CR (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J 71: 492–502 [DOI] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012a) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA 109: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Shen J, Mao H, Xie W, Li X, Zhang Q (2012b) RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol Plant 5: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havgaard JH, Lyngsø RB, Stormo GD, Gorodkin J (2005) Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics 21: 1815–1824 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S (2015) Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hu S, Shan G (2016) LncRNAs in stem cells. Stem Cells Int 2016: 2681925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura K (2014) Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J 78: 385–397 [DOI] [PubMed] [Google Scholar]
- Krishnan A, Gupta C, Ambavaram MMR, Pereira A (2017) RECoN: Rice Environment Coexpression Network for systems level analysis of abiotic-stress response. Front Plant Sci 8: 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsas K, Wuest SE, Hupalo D, Kern AD, Wicker T, Grossniklaus U (2012) Computational analysis and characterization of UCE-like elements (ULEs) in plant genomes. Genome Res 22: 2455–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua N-H (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Zhu D, Chen W, Deng W, He H, He G, Bai B, Qi Y, Chen R, Deng XW (2013) A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant 6: 830–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz J, Portela A, Soler M, Gómez A, Ling H, Michlewski G, Calin GA, Guil S, Esteller M (2014) Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell 55: 138–147 [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Zhu C, Lu G, Guo Y, Zhou Y, Zhang Z, Zhao Y, Li W, Lu Y, Tang W, et al. (2012) Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genomics 13: 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20: 300–307 [DOI] [PubMed] [Google Scholar]
- Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F (2014) e1071: Misc Functions of the Department of Statistics. http://cran.rproject.org/web/packages/e1071/index.html
- Minks J, Baldry SE, Yang C, Cotton AM, Brown CJ (2013) XIST-induced silencing of flanking genes is achieved by additive action of repeat A monomers in human somatic cells. Epigenetics Chromatin 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640 [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH (2002) Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623 [DOI] [PubMed] [Google Scholar]
- Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. (2013) Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 14: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler K, Takuno S, Gaut BS (2016) CG methylation covaries with differential gene expression between leaf and floral bud tissues of Brachypodium distachyon. PLoS ONE 11: e0150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E (2014) ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH (2015) Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J 84: 404–416 [DOI] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C (2010) H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) Ggplot2: Elegant Graphics for Data Analysis, Ed 2 Springer Publishing Company, Incorporated [Google Scholar]
- Yu AD, Wang Z, Morris KV (2015) Long noncoding RNAs: a potent source of regulation in immunity and disease. Immunol Cell Biol 93: 277–283 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cao X (2016) Long noncoding RNAs in innate immunity. Cell Mol Immunol 13: 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang J, Marowsky NC, Long M, Wing RA, Fan C (2013) High occurrence of functional new chimeric genes in survey of rice chromosome 3 short arm genome sequences. Genome Biol Evol 5: 1038–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ (2014) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ (2014) Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343: 769–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Liu X, Zhou C, Zhou Q, Zhao Y, Li G, Zhou DX (2016) Cooperation between the H3K27me3 chromatin marker and non-CG methylation in epigenetic regulation. Plant Physiol 172: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]