Carotenoid cleavage dioxygenase 2 and candidate aldehyde dehydrogenase, and UDP-glycosyltransferase enzymes involved in saffron crocin biosynthesis are localized in the chromoplast, the endoplasmic reticulum, and the cytoplasm.
Abstract
Saffron is the dried stigmas of Crocus sativus and is the most expensive spice in the world. Its red color is due to crocins, which are apocarotenoid glycosides that accumulate in the vacuole to a level up to 10% of the stigma dry weight. Previously, we characterized the first dedicated enzyme in the crocin biosynthetic pathway, carotenoid cleavage dioxygenase2 (CsCCD2), which cleaves zeaxanthin to yield crocetin dialdehyde. In this work, we identified six putative aldehyde dehydrogenase (ALDH) genes expressed in C. sativus stigmas. Heterologous expression in Escherichia coli showed that only one of corresponding proteins (CsALDH3I1) was able to convert crocetin dialdehyde into the crocin precursor crocetin. CsALDH3I1 carries a carboxyl-terminal hydrophobic domain, similar to that of the Neurospora crassa membrane-associated apocarotenoid dehydrogenase YLO-1. We also characterized the UDP-glycosyltransferase CsUGT74AD1, which converts crocetin to crocins 1 and 2′. In vitro assays revealed high specificity of CsALDH3I1 for crocetin dialdehyde and long-chain apocarotenals and of CsUGT74AD1 for crocetin. Following extract fractionation, CsCCD2, CsALDH3I1, and CsUGT74AD1 were found in the insoluble fraction, suggesting their association with membranes or large insoluble complexes. Analysis of protein localization in both C. sativus stigmas and following transgene expression in Nicotiana benthamiana leaves revealed that CsCCD2, CsALDH3I, and CsUGT74AD1 were localized to the plastids, the endoplasmic reticulum, and the cytoplasm, respectively, in association with cytoskeleton-like structures. Based on these findings and current literature, we propose that the endoplasmic reticulum and cytoplasm function as transit centers for metabolites whose biosynthesis starts in the plastid and are accumulated in the vacuole.
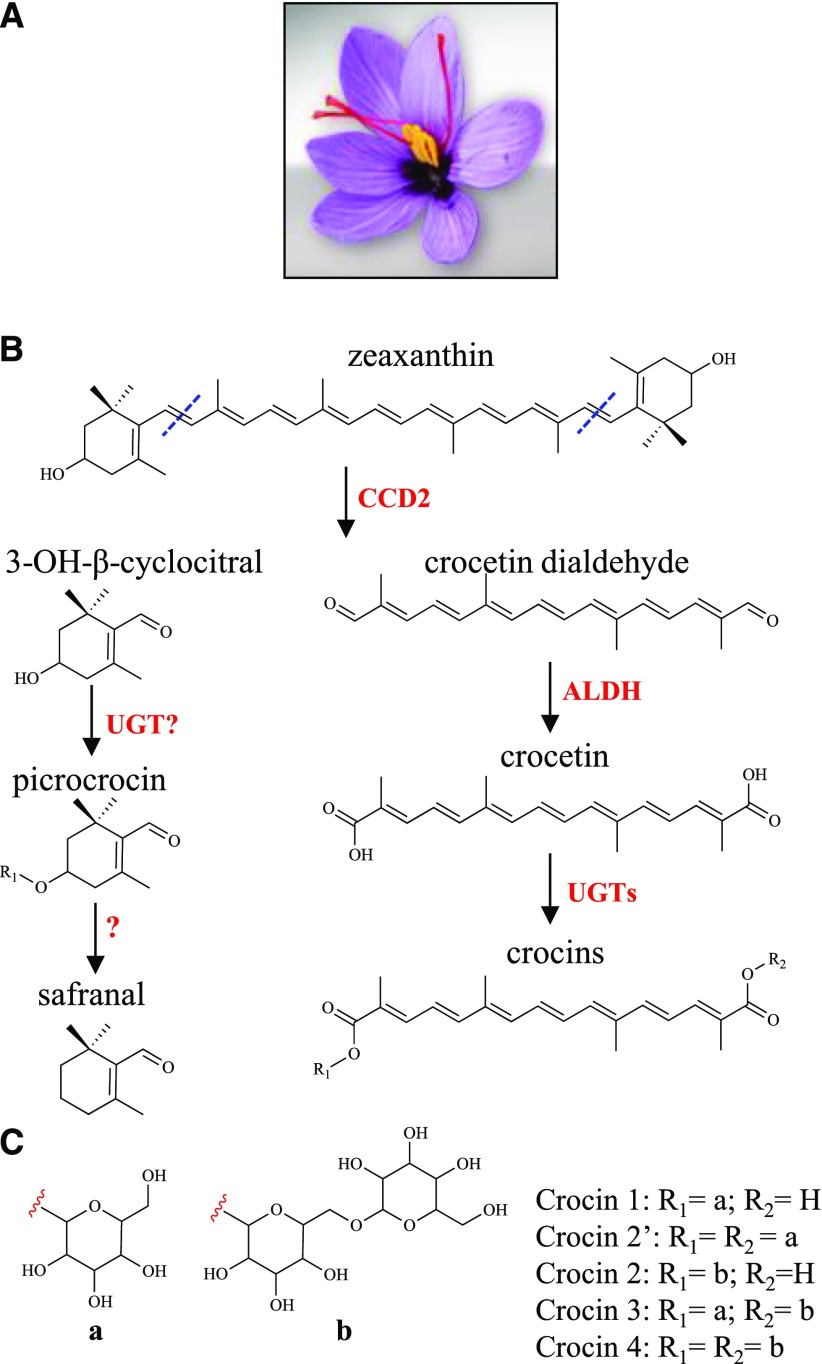
One gram of dried saffron is composed of the stigmas of approximately 150 Crocus sativus flowers, picked by hand. This makes saffron the most expensive spice in the world, with prices ranging from 2 to 10 euros g−1. The main organoleptic properties of the stigmas are due to the accumulation of three apocarotenoid classes: crocins (composed of different glucosyl and gentiobiosyl esters of crocetin), picrocrocin, and safranal (Tarantilis et al., 1995), conferring to saffron its red color, bitter taste, and pungent aroma, respectively. Crocins make up to 10% of stigma dry weight and, by virtue of their glycosylation, are water soluble. They have applications as textile colorants and histochemical stains (Bathaie et al., 2014), as antioxidants (Alavizadeh and Hosseinzadeh, 2014), and in the prevention of age-related macular degeneration (Bisti et al., 2014). Due to their complex structure and abundance of chiral centers, crocins cannot be synthesized chemically, and there is a strong industrial interest in their biotechnological production.
In C. sativus stigmas, crocin biosynthesis starts with symmetric oxidative cleavage of the C7,C8 and C7′,C8′ double bonds in zeaxanthin, which is catalyzed by the enzyme carotenoid cleavage dioxygenase2 (CsCCD2), producing crocetin dialdehyde (Fig. 1; Frusciante et al., 2014). Like many aldehydes, crocetin dialdehyde is highly reactive and is converted into the diacid crocetin by yet unidentified ALDH enzymes. ALDHs are NAD(P)+-dependent oxidoreductases that generally contribute to different processes, such as cytoplasmic male sterility, plant defense, and abiotic stress response (Kirch et al., 2004). The final step of crocin biosynthesis involves the glycosylation of crocetin. This reaction is usually catalyzed by uridine diphosphate glycosyltransferase (UGTs) that mediate the glycosylation of secondary metabolites, xenobiotics, and hormones (Wang, 2009). In Gardenia jasminoides, two crocetin UGTs have been characterized (Nagatoshi et al., 2012): UGT75L6 is responsible for the primary glycosylation of crocetin, producing crocetin monoglucosyl and diglucosyl esters, whereas UGT94E5 is responsible for the secondary glycosylation of the Glc groups, leading to the formation of one or two gentiobiose groups (Fig. 1). Contrasting data have been reported on crocin formation in saffron: a crocetin UGT purified from C. sativus stigmas has been shown to catalyze only primary glycosylation (Cote et al., 2001), whereas a cloned UGT (UGTCs2) has been reported to mediate both types of glycosylation (Moraga et al., 2004).
Figure 1.
Crocin biosynthesis in C. sativus stigmas. A, C. sativus flower at anthesis with red stigmas. B, Proposed scheme of crocin biosynthesis (Frusciante et al., 2014). The CsCCD2 enzyme cleaves zeaxanthin at the 7,8 and 7′,8′ positions, producing 3-OH-β-cyclocitral and crocetin dialdehyde. Crocetin dialdehyde is oxidized to crocetin by an aldehyde dehydrogenase (ALDH), then glycosylated to crocins by UGT enzymes. C, Sugar moieties of the four most abundant C. sativus crocins (crocins 1–4).
The crocin biosynthetic pathway encompasses multiple subcellular compartments: the initial precursor, zeaxanthin, is localized in plastids, whereas the final products, crocins, accumulate in vacuoles (Bouvier et al., 2003b; Gómez-Gómez et al., 2017), like many other plant glycosylated metabolites (Martinoia et al., 2007). We have identified the carotenoid cleavage dioxygenase responsible for the first dedicated step in C. sativus carotenoid biosynthesis, CsCCD2. It has been shown that CsCCD2, the enzyme that catalyzes the cleavage of zeaxanthin in crocin biosynthesis (Frusciante et al., 2014), is localized to the plastid (Ahrazem et al., 2016). In contrast, the subcellular localization of the further intermediates in crocin biosynthesis and the corresponding enzymes remains elusive. There are other examples of glycosylated metabolites, such as steviol (Brandle and Telmer, 2007) and abscisic acid glucosyl ester (ABA-GE) (Nambara and Marion-Poll, 2005), which originate from plastid-localized precursors and accumulate in the vacuole. For these, plastid-derived metabolites are modified and glycosylated in the cytoplasm, forming final glycosylated products that are transported into vacuoles via tonoplast transporters (Martinoia et al., 2007). This compartmentation is consistent with the assumed cytoplasmic localization of the majority of plant UGTs (Ross et al., 2001). A different model for crocin biosynthesis and sequestration was proposed based only on proteomic data and microscopic observation (Gómez-Gómez et al., 2017). This model proposed that crocins are biosynthesized entirely in the plastid, prior to accumulation in plastid-localized vesicles and final transport to the vacuole, where crocins are delivered and stored. A crocetin UGT (UGTCs2) has been identified in C. sativus stigmas (Moraga et al., 2004) and, on the basis of proteomic data, has been proposed to reside in C. sativus stigma chromoplasts (Gómez-Gómez et al., 2017). The ALDH responsible for the second step in crocin biosynthesis has not been identified so far.
To further elucidate the crocin biosynthetic pathway and to determine its compartmentation, we searched a C. sativus stigma transcriptome for putative ALDHs and UGTs and characterized their activity using Escherichia coli as an expression system. We identified an ALDH and a UGT, which together convert crocetin dialdehyde to crocins. In addition, we determined the localization of these two enzymes by immunogold electron microscopy and by analyzing the distribution of GFP fusions in Nicotiana benthamiana cells using confocal microscopy. Based on the data obtained, we propose a model for crocin biosynthesis/compartmentation in C. sativus stigmas.
RESULTS
Identification and Characterization of ALDH Candidate Transcripts in C. sativus Stigmas
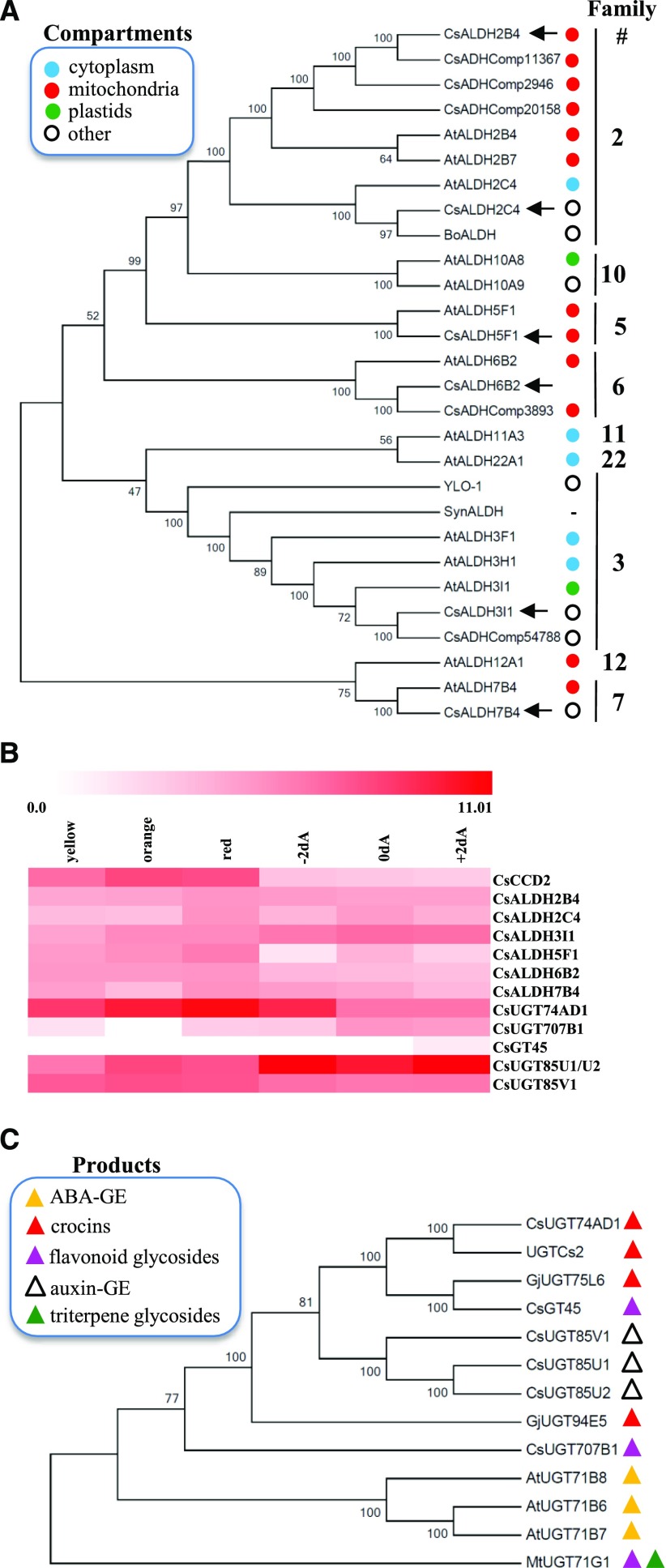
To identify the enzyme(s) involved in the conversion of crocetin dialdehyde into crocetin, an in-house transcriptome assembly obtained from RNA sequencing data from developing C. sativus stigmas (Frusciante et al., 2014) was searched for homologs of transcripts encoding apocarotenoid ALDH enzymes from different organisms: (1) Neurospora crassa YLO-1, a member of family 3 ALDH (EC 1.2.1.5) responsible for the dehydrogenation of β-apo-4′-carotenal into neurosporaxanthin (Estrada et al., 2008) and, together with its Arabidopsis (Arabidopsis thaliana) homologs ALDH3I1 and ALDH3H1 (Stiti et al., 2011a,b), responsible for the oxidation of medium-chain fatty aldehydes; (2) Bixa orellana ALDH (BoALDH), a family 2 ALDH member (EC 1.2.1.3), which has been claimed to convert the apocarotenoid bixin dialdehyde into norbixin (Bouvier et al., 2003a); (3) Synechocystis sp. PCC6803 ALDH (SynALDH), which converts a wide range of apocarotenals and alkanals into the corresponding carboxylic acids (Trautmann et al., 2013); and five ALDHs claimed, based on proteomic data, to reside within stigma chromoplasts (CsADHComp; Gómez-Gómez et al., 2017). Using this approach, we identified six candidate genes expressed in C. sativus stigmas and encoding putative ALDH enzymes (Fig. 2A). CsALDH2C4, a member of family 2, is the closest homolog of BoALDH and AtALDH2C4; a second family 2 member, CsALDH2B4, corresponds to CsADHComp11367 and shows identity of 97.2% and 85.2% to CsADHComp2946 and CsADHComp20158, respectively; CsALDH3I1 (family 3) is a close homolog of AtALDH3I1, AtALDH3H1, YLO-1, SynALDH, and CsADHComp54788; CsALDH5F1, which belongs to family 5, comprising succinic semialdehyde dehydrogenases (EC 1.2.1.24; Brocker et al., 2013), is a close homolog of AtALDH5F1, involved in plant defense against reactive oxygen species (Stiti et al., 2011b); CsALDH6B2 is a family 6 member and a homolog of AtALDH6B2 and CsADHComp3893; and CsALDH7B4 (family 7; EC 1.2.1.31), comprising Δ1-piperideine-6-carboxylate dehydrogenases and α-aminoadipic semialdehyde dehydrogenases (Brocker et al., 2013), is a close homolog of AtALDH7B4 encoded by an osmotic stress-inducible ALDH gene (Kotchoni et al., 2006).
Figure 2.
Candidate genes for crocin biosynthesis in C. sativus stigmas. A, Phylogenetic relationships between ALDHs of C. sativus (Cs), Arabidopsis (At), B. orellana (Bo), Synechocystis spp. (Syn), and N. crassa (YLO-1), inferred using the neighbor-joining method. The subcellular localizations of ALDH enzymes, experimentally demonstrated by Stiti et al. (2011b) or predicted by TargetP, are indicated by colored dots. Accession numbers are as follows: CsALDH2B4 (MG672523), CsALDH2C4 (MF596160), CsALDH3I1 (MF596165), CsALDH5F1 (MF596161), CsALDH6B2 (MG672524), and CsALDH7B4 (MF596162; all described in this article, indicated by black arrows); AtALDHs (Stiti et al., 2011b); BoALDH (AJ548846; Bouvier et al., 2003a); SynALDH (ALJ68758.1; Trautmann et al., 2013); YLO-1 (XP_011394899.1; Estrada et al., 2008); and CsADHComp2946 (AQM36713.1), CsADHComp11367 (AQM36717.1), CsADHComp20158 (AQM36716.1), CsADHComp3893 (AQM36715.1), and CsADHComp54788 (AQM36714.1; Gómez-Gómez et al., 2017). B, Expression of C. sativus CCD2, ALDH, and UGT enzymes in stigma tissue at different developmental stages. Data are expressed as log2 of fragments per kilobase per million. Developmental stages are as follows: yellow, orange, red, 2 d before anthesis (−2dA), day of anthesis (0dA), and 2 d after anthesis (+2dA). C, Phylogenetic relationships between UGTs of C. sativus (Cs), Arabidopsis (At), G. jasminoides (Gj), and M. truncatula (Mt), inferred using the neighbor-joining method. Accession numbers are as follows: CsUGT74AD1 (MF596166; described in this article); UGTCs2 (AAP94878.1; Moraga et al., 2004); CsGT45 (ACM66950.1; Moraga et al., 2009); CsUGT85V1 (AIF76150.1), CsUGT85U1 (AIF76152.1), and CsUGT85U2 (AIF76151.1; Ahrazem et al., 2015; Gómez-Gómez et al., 2017); CsUGT707B1 (CCG85331.1; Trapero et al., 2012); AtUGT71B6 (NP_188815.1), AtUGT71B7 (NP_188816.1), and AtUGT71B8 (NP_188817.1; Dong and Hwang, 2014); GjUGT94E5 (F8WKW8.1) and GjUGT75L6 (F8WKW0.1; Nagatoshi et al., 2012); and MtUGT71G1 (AAW56092.1; Shao et al., 2005). The biosynthetic products of the various UGTs are represented by colored triangles. In A and C, the percentages of replicate trees that clustered together in the bootstrap test are indicated to the left of the branches.
The CsALDH5F1 and CsALDH7B4 amino acid sequences indicate the presence of tetramerization interfaces (Supplemental Fig. S1), whereas CsALDH3I1 and YLO-1 are equipped with a C-terminal hydrophobic domain (Supplemental Fig. S2) flanked by short regions of positively charged amino acids and preceded by an unstructured domain (Supplemental Fig. S1). A similar structure has been found in a rat microsome-localized ALDH and has been shown to mediate its localization to the endoplasmic reticulum (ER; Masaki et al., 1994). Indeed, YLO-1 is a cytosolic, membrane-localized enzyme, although its exact localization is unknown (Estrada et al., 2008). This C-terminal domain has been lost in CsADHComp54788, which represents a truncated form of CsALDH3I1, due to an early stop codon. The subcellular localization of the identified CsALDHs was predicted using TargetP (Supplemental Table S1; Emanuelsson et al., 2000). None of the enzymes contained a plastid transit peptide, excluding localization within this organelle, whereas CsALDH2B4, CsALDH6B2, and CsALDH5F1 are predicted to be mitochondrial. All CsALDH genes were expressed throughout stigma development (Fig. 2B).
CsALDH3I1 Catalyzes the Dehydrogenation of Crocetin Dialdehyde into Crocetin
To check the ability of the different CsALDH enzymes to convert crocetin dialdehyde to crocetin, we expressed them as thioredoxin fusion proteins in E. coli accumulating crocetin dialdehyde (see “Materials and Methods”). Briefly, a strain of E. coli carrying a plasmid for zeaxanthin biosynthesis (Kanr) was cotransformed with the pTHIO-CsCCD2 (Cmr) vector harboring the CsCCD2 enzyme that converts zeaxanthin to crocetin dialdehyde (Frusciante et al., 2014) and a third pTHIO-ALDH vector (Ampr) containing one of the six CsALDH cDNAs or the SynALDH gene (Supplemental Fig. S3A; Trautmann et al., 2013). Immunoblot analysis using an anti-6×His antibody confirmed the expression of CsCCD2-thioredoxin and ALDH-thioredoxin fusion proteins at the expected molecular masses of 81.9 and 69.8 to 76.9 kD, respectively. The intensities of the bands corresponding to ALDH-thioredoxin fusion proteins were similar, indicating that all ALDH proteins were expressed at comparable levels upon induction (Supplemental Fig. S3B).
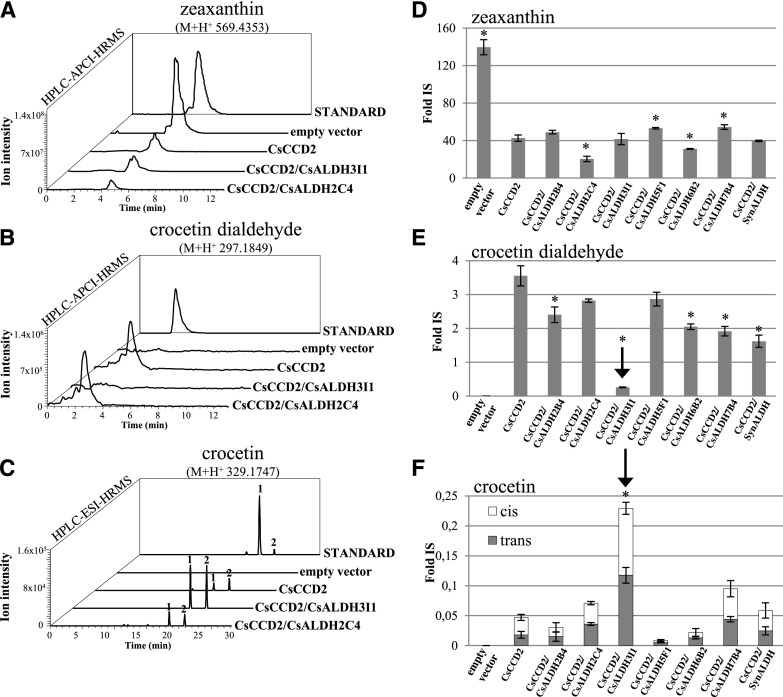
We quantified zeaxanthin, crocetin dialdehyde, and crocetin levels in induced bacterial cells harboring the various constructs. Nonpolar and polar metabolites were analyzed by HPLC-photodiode array detection-high resolution mass spectrometry (HPLC-PDA-HRMS) using a Q-Exactive mass spectrometer. The analysis revealed a decrease of zeaxanthin in all clones expressing CsCCD2 due to the activity of the CsCCD2 enzyme (Fig. 3, A and D). An ion with the mass of crocetin dialdehyde and comigrating with a crocetin dialdehyde authentic standard was detected in all clones expressing CsCCD2, with the exception of the CsCCD2/ALDH3I1 clone, where this metabolite was almost absent (Fig. 3, B and E). Trace levels of an ion with the accurate mass of the ALDH product, crocetin, were observed in all clones expressing CsCCD2, possibly due to the action of an endogenous E. coli ALDH or to the nonenzymatic dehydrogenation of crocetin dialdehyde (Fig. 3, C and F). We observed a different pattern in cells coexpressing CsCCD2 and CsALDH3I1. Compared with all other cells, we detected a much higher reduction in crocetin dialdehyde and, accordingly, a higher increase in crocetin content (Fig. 3, E and F). Thus, since all ALDHs were expressed at similar levels (Supplemental Fig. S3B), we assumed that CsALDH3I1 is the enzyme that converts crocetin dialdehyde into crocetin with high efficiency. The C. sativus ortholog of BoALDH, CsALDH2C4, as well as SynALDH did not show any activity above the E. coli background.
Figure 3.
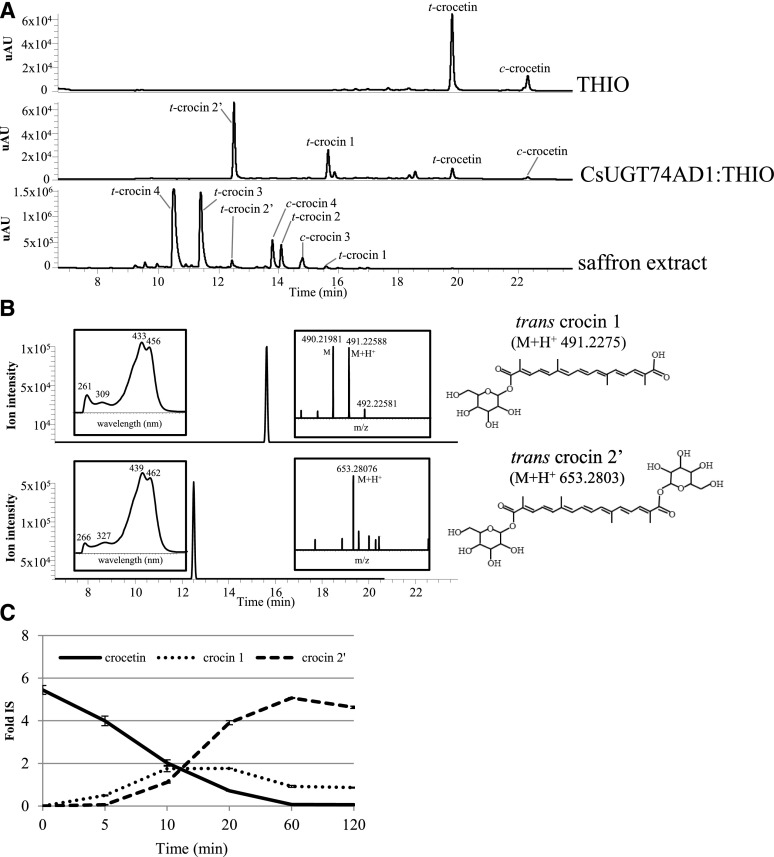
CsALDH3I1 mediates crocetin biosynthesis in E. coli. A to C, Accurate mass chromatograms of zeaxanthin (A), crocetin dialdehyde (B), and crocetin (C) extracted from bacterial clones harboring the pTHIO empty vector or overexpressing CsCCD2 alone or in combination with CsALDH3I1 or CsALDH2C4. Polar and nonpolar fractions were extracted from bacterial cells and run on an HPLC-PDA-HRMS system alongside authentic standards. The metabolites have an accurate mass and a chromatographic mobility identical to those of the authentic standard. Two peaks were detected of crocetin (1, trans-isomer, and 2, cis-isomer; Supplemental Fig. S4). D to F, Relative quantities of zeaxanthin (D), crocetin dialdehyde (E), and crocetin (F) detected by HPLC-atmospheric pressure chemical ionization-HRMS (D and E) and HPLC-electrospray ionization (ESI)-HRMS (F) in recombinant clones. Data are means ± sd of three independent growth batches. Asterisks indicate statistically significant differences (Student’s t test, P < 0.01) compared with CsCCD2 values. Fold IS, Fold internal standard.
Crocetin was resolved by our chromatographic system into two different peaks with identical mass but different chromatographic mobilities (peaks 1 and 2 in Fig. 3C). The commercial standard of crocetin is composed mostly of an all-trans-isomer, which corresponds to the fastest migrating peak (peak 1 in Fig. 3C). Hence, we assume that the two peaks correspond to all-trans- and cis-crocetin with absorption maxima at 426 and 420 nm, respectively, and with an additional absorption peak (cis-peak) at 318 nm (Supplemental Fig. S4; Rubio-Moraga et al., 2010; Chryssanthi et al., 2011). Interestingly, the all-trans- and cis-isomers produced in E. coli were present in approximately equimolar amounts. Because our chromatographic system does not separate the trans- and cis-isomers of crocetin dialdehyde, we are not able to assess whether the cis-configuration is already present in crocetin dialdehyde or is generated during the dehydrogenation reaction.
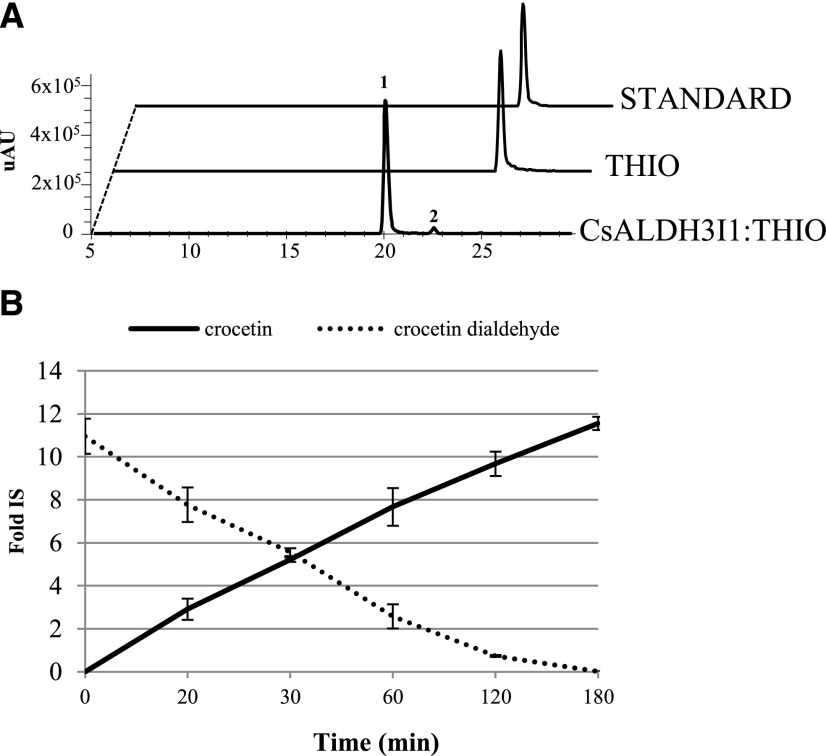
To further characterize the substrate specificity of CsALDH3I1, we introduced the pTHIO-ALDH vector in an E. coli strain harboring the groES-groEL-chaperones system to maximize the amount of correctly folded protein (Nishihara et al., 1998). Proteins were solubilized using Triton X-100 (Supplemental Fig. S5) and utilized in an in vitro assay with different aldehydes according to Trautmann et al. (2013). The control extract from E. coli cell transformed with the void plasmid showed negligible (1.3%) oxidation of crocetin dialdehyde into crocetin (Supplemental Fig. S6), whereas the extract containing CsALDH3I1 converted crocetin dialdehyde into crocetin in a time-dependent manner, with an almost complete conversion after 120 min of incubation (Fig. 4). We also assayed the CsALDH3I1 activity on different apocarotenals and nonapocarotenoid aldehydes. As shown in Supplemental Table S2, CsALDH3I1 displayed a strong preference for long-chain apocarotenals and for diapocarotenoids, showing after 60 min conversion rates of 93.3%, 76.4%, and 3.6% for β-apo-8′-carotenal (C30), crocetin dialdehyde (C20), and retinal (C20), respectively. In contrast, the activity of the enzyme was very low or even not detectable upon incubation with nonapocarotenoid (fatty) aldehydes, such as dodecanal, hexanal, or 4-OH-benzaldehyde.
Figure 4.
CsALDH3I1 mediates crocetin biosynthesis in vitro. A, HPLC chromatograms (absorbance at 440 nm) of in vitro assays performed for 120 min in the absence (THIO) and presence (CsALDH3I1:THIO) of CsALDH3I1. STANDARD = crocetin dialdehyde authentic standard. In the presence of CsALDH3I1, the crocetin dialdehyde is converted into crocetin (1, trans-isomer, and 2, cis-isomer; Supplemental Fig. S4). B, Time course of crocetin dialdehyde conversion in the presence of CsALDH3I1. Data are averages ± sd of three biological replicates and expressed as fold internal standard (Fold IS).
CsUGT74AD1 Catalyzes the Glycosylation of Crocetin into Crocins 1 and 2′
To check whether the previously identified UGT enzyme (UGTCs2; GenBank accession number AY262037.1; Moraga et al., 2004) is involved in crocin biosynthesis and how many sugars it adds, we searched our transcriptome, as well as that of Jain et al. (2016), for the presence of the corresponding transcript. However, we could not find an identical sequence. The most similar UGT transcript present in these transcriptomes was that of CsUGT74AD1, which encodes a version of UGTCs2 (Moraga et al., 2004), showing a four-amino acid extension at the N terminus, lacking seven amino acids at the C terminus, and differing in 44 amino acids (Supplemental Fig. S7). The plant secondary product glycosyltransferase motif, which is responsible for binding the sugar donor UDP-Glc (Gachon et al., 2005), is conserved in CsUGT74AD1, UGTCs2, and G. jasminoides UGTs involved in crocin formation (Nagatoshi et al., 2012), whereas some differences were observed in other catalytic residues. Specifically, both UGTCs2 and GjUGT75L6 lack the Asp-121 residue (Supplemental Fig. S7), which has been shown to be necessary, together with His-22, for the catalytic activity of Medicago truncatula UGT71G1 (Shao et al., 2005). Similar to CsCCD2, CsUGT74AD1 is highly expressed in early stigma development, with transcript levels reaching a maximum at the red stage and declining thereafter (Fig. 2B). A dendrogram that includes CsUGT74AD1, other described C. sativus UGTs (Moraga et al., 2009; Trapero et al., 2012; Ahrazem et al., 2015; Gómez-Gómez et al., 2017), G. jasminoides UGTs involved in crocin biosynthesis (Nagatoshi et al., 2012), and Arabidopsis and M. truncatula UGTs (Shao et al., 2005; Dong and Hwang, 2014) is shown in Figure 2C. Three clusters are observed: one comprising the three C. sativus genes putatively involved in auxin-GE biosynthesis (Ahrazem et al., 2015); a second one comprising CsUGT74AD1, GjUGT75L6, involved in crocetin primary glycosylation (Nagatoshi et al., 2012), and CsGT45, involved in flavonoid glycosylation (Moraga et al., 2009); and a third one comprising Arabidopsis ABA UGTs. GjUGT94E5, involved in crocetin secondary glycosylation (Nagatoshi et al., 2012), and CsUGT707B1, putatively involved in flavonoid glycosylation (Trapero et al., 2012), did not cluster with any other sequence. None of the described CsUGTs contains transmembrane domains (Supplemental Fig. S2) or chloroplast transit peptides (Supplemental Table S1).
To verify the function of CsUGT74AD1, we cloned the corresponding cDNA in the bacterial expression vector pTHIO (Spectr). As expected, coexpression of pTHIO-CsCCD2, pTHIO-CsALDH3I1, and pTHIO-CsUGT74AD1 in a zeaxanthin-accumulating E. coli strain did not result in the accumulation of crocin, probably due to insufficient levels of UDP-Glc in E. coli, which is required for UGT activity (Ross et al., 2001). Thus, we performed an in vitro assay using CsUGT74AD1 produced in the E. coli strain harboring the groES-groEL-chaperones system (Nishihara et al., 1998). Immunoblotting with the anti-His-6 antibody revealed a band corresponding to the CsUGT74AD1-thioredoxin fusion protein (69.7 kD) in soluble extracts of arabinose-induced cells (Supplemental Fig. S8). We performed an in vitro assay using 40 µg of soluble E. coli extract, encapsulated crocetin, and UDP-Glc as substrates (Moraga et al., 2004). The reaction was incubated at 30°C for increasing periods of time, followed by analysis of semipolar and polar metabolites by HPLC-PDA-HRMS. In the assay performed with extracts of cells transformed with the empty pTHIO vector, we only detected two peaks with a maximum A440, corresponding to trans- and cis-crocetin. On the contrary, in the presence of CsUGT74AD1, we detected two additional peaks (Fig. 5A) corresponding to the monoglucosyl and diglucosyl esters of crocetin (crocin 1 and 2′, respectively; Fig. 5B). A time course of the reaction is shown in Figure 5C: the monoglucosyl ester is a reaction intermediate reaching a maximum between 10 and 20 min, whereas an almost complete conversion of crocetin into the diglucosyl ester, crocin 2′, was detected in 60 min. Although we incubated the reaction for up to 120 min and repeated the assay several times, we could not detect the formation of gentiobiosyl esters of crocetin, which are the most abundant forms of crocins in mature stigmas (Fig. 5A). Thus, in our hands, the CsUGT74AD1 enzyme performed only the first step of crocetin glycosylation (the addition of Glc moieties to crocetin), in agreement with previous data obtained in G. jasminoides (Nagatoshi et al., 2012) and C. sativus (Cote et al., 2001).
Figure 5.
CsUGT74AD1 mediates the synthesis of crocins 1 and 2′ in vitro. A, HPLC chromatograms (absorbance at 440 nm) of in vitro assay reactions performed for 60 min in the absence (THIO) and presence (CsUGT74AD1:THIO) of the CsUGT74AD1 enzyme or extracted from mature saffron (saffron extract). t = trans and c = cis. B, HPLC-ESI-HRMS chromatograms (extracted accurate masses) of trans-crocin 1 and trans-crocin 2′ accumulated in the bacterial clone overexpressing the CsUGT74AD1 enzyme. Mass and PDA spectra of peaks are shown in the right and left boxes, respectively. C, Time course of crocetin conversion into trans-crocin 1 and trans-crocin 2′ in the presence of UGT enzyme. Data are averages ± sd of three biological replicates and expressed as fold internal standard (Fold IS).
The activity of CsUGT74AD1 also was tested on a broad range of different substrates known to undergo glycosylation in planta, using UDP-Glc or UDP-Gal as sugar donors. We observed that the enzyme is highly specific, showing a high activity with crocetin and UDP-Glc (81.3% conversion in 30 min; Supplemental Table S3), although we detected a lower activity with UDP-Gal as a sugar donor (35.5% conversion in 30 min). The enzyme also displayed low levels of activity, with UDP-Glc as sugar donor, on flavonoids (i.e. quercetin and naringenin) and on the phenolic acid cinnamic acid, whereas it did not show any activity with ABA or indole 3-acetic acid (Supplemental Table S3). These results suggest that CsUGT74AD1 is the enzyme responsible for the primary glycosylation of crocetin.
CsCCD2, CsALDH3I1, and CsUGT74AD1 Tissue Specificity, Solubility, and Subcellular Localization
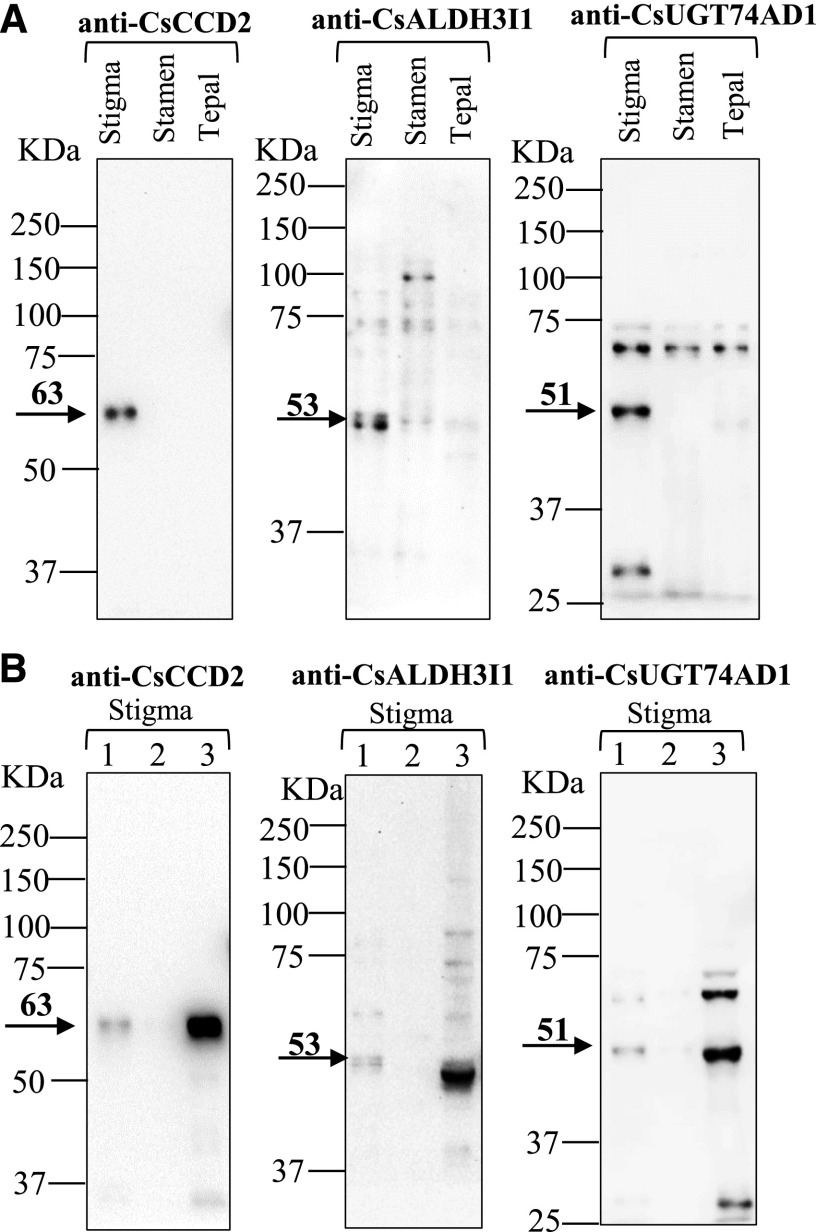
To analyze the tissue-specific expression pattern of the CsCCD2, CsALDH3I1, and CsUGT74AD1 enzymes in flowers of C. sativus, we raised polyclonal antibodies against immunogenic peptides from the three proteins (see “Materials and Methods”) and used them in immunoblot analysis on protein extracts from C. sativus stigmas, stamens, and tepals at anthesis (Fig. 6A). The antibodies raised against CsCCD2 recognized a single band corresponding to the molecular mass of the enzyme (63 kD after the removal of the transit peptide), which was only present in the stigma fraction. The anti-CsALDH3I1 antibodies recognized a major band of 53 kD, corresponding to the molecular mass of the protein, which was highly expressed in stigmas but also present at much lower levels in stamens and tepals. Anti-CsALDH3I1 antibodies also recognized several fainter bands, likely due to cross-reactivity with other ALDH enzymes. The antibodies raised against CsUGT74AD1 recognized a band of 51 kD, which corresponds to the molecular mass of CsUGT74AD1, and two additional bands with molecular masses of 69 and 29 kD. The 29-kD band is too small to represent a functional UGT (Gachon et al., 2005) and corresponds probably to a proteolytic product of CsUGT74AD1. In agreement with this hypothesis, the 51- and 29-kD bands are present together in the stigma fraction and are absent in other tissues. In contrast, we detected the 69-kD band in all three tissues, indicating that it represents an unrelated UGT or a different protein.
Figure 6.
Tissue specificity and solubility of enzymes involved in crocin biosynthesis. A, Immunoblot analysis performed on total proteins (10 µg) extracted from C. sativus stigmas, stamens, and tepals, performed with rabbit polyclonal antibodies raised against CsCCD2, CsALDH3I1, and CsUGT74AD1 peptides. B, Immunoblot analysis performed on total protein extract from C. sativus stigmas. Lane 1, Soluble proteins extracted with PB buffer; lane 2, extrinsic membrane proteins extracted with PB buffer + 200 mm NaCl; lane 3, insoluble proteins extracted with SDS loading buffer (for details, see “Materials and Methods”).
The substrates of CsCCD2 and CsALDH3I1 (zeaxanthin and crocetin dialdehyde, respectively) are hydrophobic molecules that are presumably membrane associated in vivo. Accordingly, both CsCCD2 and CsALDH3I1 contain a transmembrane domain (Supplemental Fig. S2), which in CsALDH3I1 is reminiscent of an ER localization sequence (Masaki et al., 1994). To determine the solubility of the three enzymes involved in crocin biosynthesis, we homogenized C. sativus stigmas in PBS buffer to lyse plastids and then sequentially fractionated the extracts into soluble proteins, extrinsic membrane proteins, and insoluble proteins (lanes 1, 2, and 3, respectively, in Fig. 6B). Interestingly, all three enzymes were found in the insoluble fraction (lane 3 in Fig. 6B). This result was expected for CsCCD2 and CsALDH3I1, which contain a transmembrane domain, but not for CsUGT74AD1, which, like other CsUGTs, does not have any hydrophobic domain (Supplemental Fig. S2).
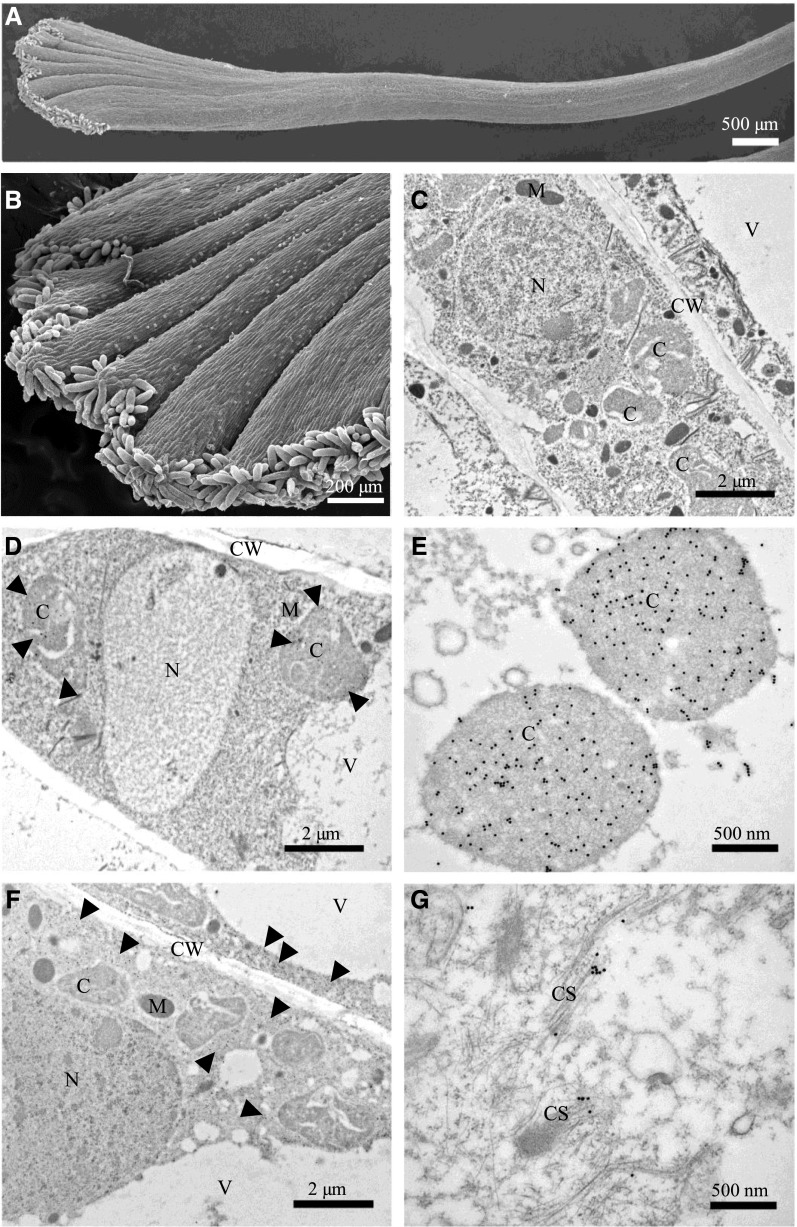
To determine the localization of the three enzymes within stigma cells, we performed immunogold electron microscopy on mature stigmas. Figure 7, A and B, each shows a scanning electron micrograph of a C. sativus stigma. Figure 7C shows an immunoelectron micrograph of a crocin-accumulating cell, decorated with a secondary antibody coupled to colloidal gold. The various organelles (i.e. nucleus, chromoplast, cell wall, and vacuole) are clearly visible without any gold particles. Figure 7, D and E, show low- and high-magnification immunoelectron micrographs, respectively, of cells immunodecorated using the primary anti-CsCCD2 antibodies followed by the secondary antibody coupled to colloidal gold. The gold particles are localized to the chromoplasts, in agreement with the reported plastid localization of a CCD2 from Crocus ancyrensis (Ahrazem et al., 2016) and the predicted presence of a plastid transit peptide for this enzyme (Supplemental Table S1). The CsCCD2 signal was homogenously distributed in the organelles. IEM with anti-CsUGT74AD1 antibodies gave a clear signal in the cytoplasm (Fig. 7, F and G). At the higher magnification, the gold particles showed an association with electron-dense filamentous structures reminiscent of membranes or cytoskeletal elements (Fig. 7G). A cytoskeletal association of UGTs was reported previously for the maize (Zea mays) UDP-Glc starch glycosyltransferase (Azama et al., 2003). The membrane or cytoskeletal association of CsUGT74AD1 could explain its insolubility. Unfortunately, IEM performed using two different anti-CsALDH3I1 antibodies gave no signal, probably due to the fact that the epitopes recognized by these antibodies are not accessible in the folded protein.
Figure 7.
Subcellular localization of CsCCD2 and CsUGT74AD1 in C. sativus stigmas. A and B, Scanning electron microscopy images of C. sativus stigmas. C to G, Immunoelectron microscopy (IEM) of C. sativus stigmas. IEM reactions were performed with an anti-CsCCD2 antibody (D and E) and an anti-CsUGT74AD1 antibody (F and G) followed by an anti-rabbit secondary antibody conjugated to 20-nm gold particles. As a control, labeling was performed omitting primary antibodies (C). For both proteins, a general view (bars = 2 µm) and a 4× detail (bars = 500 nm) are shown. CsCCD2 was detected only in chromoplasts (D and E), while CsUGT74AD1 was detected in the cytoplasm (F), in association with filamentous structures (membranes or cytoskeleton-like structures; G). C, Chromoplast; CS, cytoskeleton; CW, cell wall; M, mitochondrion; N, nucleus; V, vacuole.
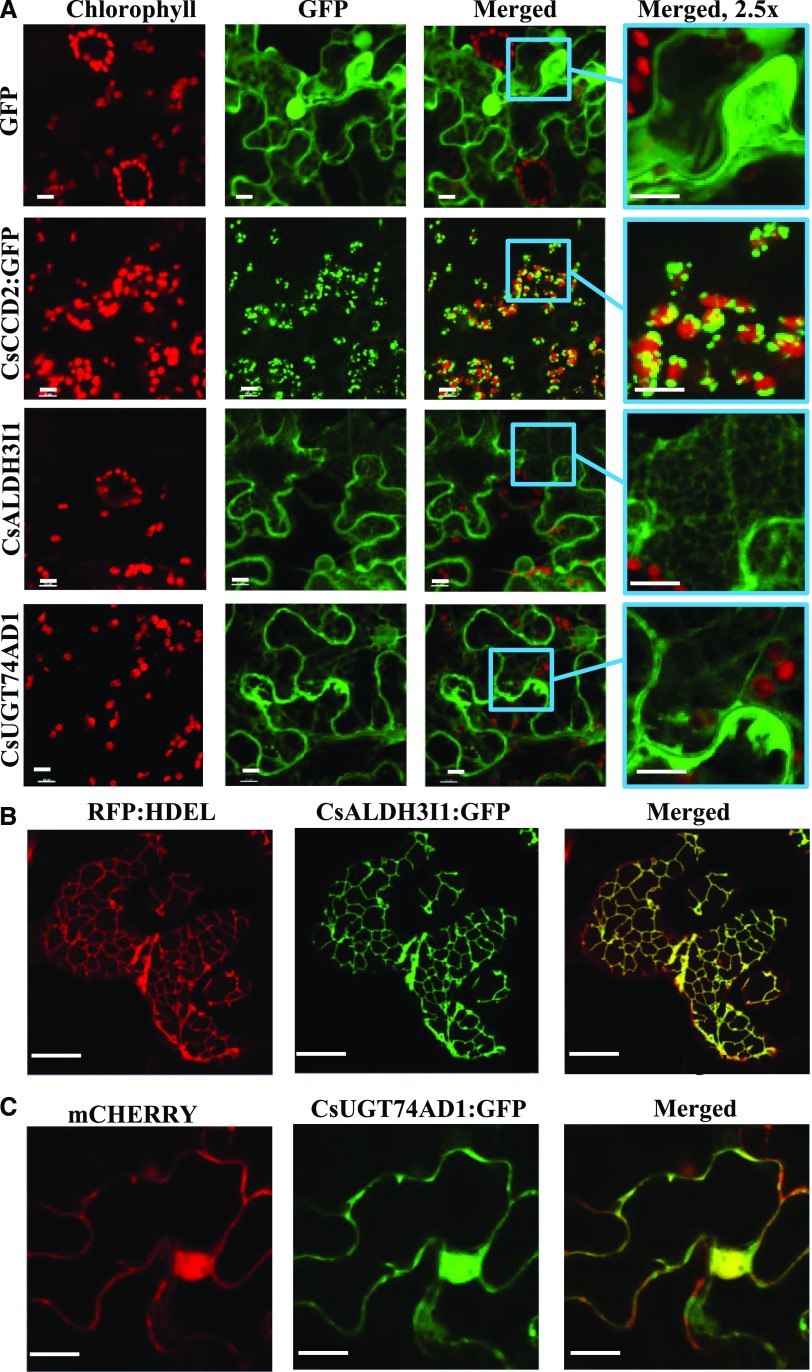
To confirm the data obtained by IEM on CsCCD2 and CsUGT74AD1 and to determine the subcellular localization of CsALDH3I1, we fused the three proteins to enhanced GFP (eGFP; Cinelli et al., 2000) and expressed them in N. benthamiana leaves through infiltration with Agrobacterium tumefaciens (strain C58C1). The confocal microscopy results indicate that CsCCD2:eGFP is localized on chloroplast-associated speckles (Fig. 8A), further confirming the plastid localization demonstrated by IEM. In contrast, CsUGT74AD1 showed a cytosolic localization (Fig. 8A), which was further confirmed by coexpression with cytosolic mCherry (Fig. 8C). Finally, CsALDH3I1 labeled a reticulate membrane network that is likely identical with the ER (Fig. 8A). Indeed, CsALDH3I1 contains a canonical C-terminal retention signal (KKRK; Supplemental Fig. S1; Benghezal et al., 2000). Coexpression of CsALDH3I1 with the ER marker RFP-HDEL showed the colocalization of both proteins, thus confirming that CsALDH3I1 is an ER-associated enzyme (Fig. 8B).
Figure 8.
Subcellular localization of CsCCD2, CsALDH3I1, and CsUGT74AD1 expressed in N. benthamiana leaves. A, For each construct, red (chlorophyll fluorescence), green (GFP fluorescence), merged (overlap of chlorophyll and GFP fluorescence), and a 2.5× zoom of merged are shown. GFP shows atypical cytoplasmic-nuclear localization; CsCCD2 localizes to plastid-associated speckles; CsALDH3I1 shows a cytoplasmic, reticulate localization; CsUGT74AD1 shows a cytoplasmic localization. B, Colocalization of RFP:HDEL (ER marker) and CsALDH3I1:eGFP. C, Colocalization of mCherry (cytoplasmic marker) and CsUGT74AD1:eGFP is cytosol localized. Bars = 10 μm.
All other CsALDH proteins investigated in this study showed a cytosolic localization, as judged by confocal fluorescence microscopy, with the exception of CsALDH5F1, which may be a mitochondrial enzyme (Supplemental Fig. S9; Supplemental Table S1).
DISCUSSION
In this and in a previous work (Frusciante et al., 2014), we have isolated and characterized transcripts expressed in C. sativus stigmas, encoding CCD, ALDH, and UGT enzymes that catalyze the first, second, and third steps, respectively, in C. sativus crocin biosynthesis in E. coli and/or in vitro.
The First Step: Cleavage of Zeaxanthin
The C. sativus enzyme CsCCD2 has been shown previously to catalyze the first dedicated step in crocin biosynthesis (i.e. the cleavage of zeaxanthin to crocetin dialdehyde; Frusciante et al., 2014). A CCD2 from the spring crocus C. ancyrensis, which synthesizes crocins in tepals, is localized to plastids when overexpressed in tobacco leaves (Ahrazem et al., 2016). In this work, both immunoelectron microscopy on C. sativus stigmas and confocal fluorescence microscopy in N. benthamiana leaves confirmed the plastidial localization of CsCCD2, indicating that this localization probably extends to the whole Crocus genus, irrespective of the organ where crocins are synthesized. CsCCD2 is homogenously distributed in C. sativus stigma chromoplasts and, upon extract fractionation, behaves as an insoluble enzyme, consistent with the presence of a large internal transmembrane domain.
The Second Step: Dehydrogenation of Crocetin Dialdehyde
We used an approach similar to that of Frusciante et al. (2014) to identify the ALDH responsible for the second step in crocin biosynthesis. Plant ALDH enzymes have been classified in 13 distinct families (Brocker et al., 2013). In our stigma transcriptome, we identified six expressed ALDHs. In particular, CsALDH3H1 encodes a family 3 member closely related to YLO-1, an N. crassa ALDH that is responsible for the conversion of β-apo-4′-carotenal to neurosporaxanthin (Estrada et al., 2008) and to Synechocystis spp. and Fusarium fujikuroi ALDHs able to convert apocarotenals into the corresponding carboxylic acids (Díaz-Sánchez et al., 2011; Trautmann et al., 2013), whereas CsALDH2C4 is closely related to BoALDH, a B. orellana family 2 ALDH claimed to convert bixin dialdehyde into norbixin (Bouvier et al., 2003a); CsALDH2B4 and CsALDH6B2 are identical or highly similar to CsADH enzymes (Comp11367, Comp2946, Comp20158, and Comp3893) claimed to reside in stigma chromoplasts (Gómez-Gómez et al., 2017); CsALDH5F1 and CsALDH7B4 encode family 5 and 7 enzymes, respectively, reported to oxidize succinic semialdehydes and Δ1-piperideine-6-carboxylate and α-aminoadipic semialdehydes, respectively (Brocker et al., 2013).
By expression in E. coli, we demonstrated that, of the six tested ALDH enzymes whose transcripts are highly expressed in C. sativus stigmas, only CsALDH3I1 is able to catalyze the conversion of crocetin dialdehyde into a mixture of trans- and cis-crocetin, whereas all other ALDHs, including SynALDH, did not show any appreciable activity above the endogenous E. coli background. In vitro assays confirmed the high activity of CsALDH3I1 in converting crocetin dialdehyde into crocetin (Fig. 4) and indicated a strong preference for carotenoid-derived substrates, particularly crocetin aldehyde and long-chain apocarotenals (Supplemental Table S2). The CsALDH3I1 protein is expressed much more in stigmas, the tissue of crocin biosynthesis, than in anthers and tepals. Taken together, these data strongly suggest that CsALDH3I1 is the enzyme catalyzing the dehydrogenation of crocetin dialdehyde in C. sativus stigmas. Interestingly, both CsALDH3I1 and the fungal apocarotenal dehydrogenase YLO-1 contain a C-terminal transmembrane domain, which is flanked by sequences rich in basic amino acids and linked to the ALDH core through an unstructured domain (Supplemental Fig. S1; Estrada et al., 2008). It has been suggested that this domain mediates the YLO-1 association with intracellular membranes (Estrada et al., 2008). Indeed, a rat microsomal ALDH contains very similar C-terminal domain that is responsible for posttranslational targeting to the ER (Masaki et al., 1994).
We were unable to obtain a specific IEM reaction using two distinct anti-CsALDH3I1 antibodies. However, confocal fluorescence in N. benthamiana leaves showed a clear localization in a cytoplasmic reticular structure, similar to that observed for ER-localized proteins harboring a C-terminal KKXX signal (Benghezal et al., 2000), also found in CsALDH3I1. CsALDH3I1 clearly colocalized with an ER marker in N. benthamiana leaves and, upon fractionation of C. sativus stigma extracts, behaved as an insoluble protein, in keeping with its ER localization.
Based on proteomic data, several C. sativus stigma ALDHs have been claimed to reside in the plastid (Gómez-Gómez et al., 2017), including CsADHComp54788, a truncated CsALDH3I1 form that lacks the C-terminal hydrophobic tail likely mediating ER localization. However, neither CsADHComp54788 nor CsALDH3I1 is expected to occur within plastids, as judged by TargetP analysis. Furthermore, none of the additional CsALDH enzymes, many of which are highly similar to those identified by Gómez-Gómez et al. (2017), showed a plastidial localization in N. benthamiana leaves. Of course, it is still possible that a plastid-localized ALDH is expressed at very low levels in C. sativus stigmas and, therefore, is not present in our transcriptome data.
The Third Step: Glycosylation of Crocetin
In spite of several efforts, we were not able to retrieve, in our transcriptome or in that of Jain et al. (2016), a sequence encoding UGTCs2, which has been claimed to perform both primary and secondary glycosylation of crocetin (Moraga et al., 2004) and to reside in stigma chromoplasts (Gómez-Gómez et al., 2017). Instead, we identified the close homolog CsUGT74AD1, which, based on immunoblot analysis estimates, is a very abundant protein in C. sativus stigmas. CsUGT74AD1 possesses all the characteristics of bona fide plant UGTs, including the presence of His-22 and Asp-121, shown to be essential for enzymatic activity (Shao et al., 2005). Using an in vitro assay, we showed that CsUGT74AD1 is able to perform primary, but not secondary, glycosylation of crocetin. CsUGT74AD1 shows a strong preference for crocetin as a substrate and accepts both UDP-Glc and, with lower affinity, UDP-Gal as sugar donors. Similar to CsALDH3I1, the CsUGT74AD1 protein also is much more expressed in stigma than in anthers and tepals. Overall, the data suggest that CsUGT74AD1 mediates the primary glycosylation of crocetin in C. sativus stigmas. Thus, in C. sativus, the situation seems to be similar to that in G. jasminoides, where the biosynthesis of crocins involves two UGTs acting hierarchically: one (GjUGT75L6, a close homolog of CsUGT74AD1) produces crocins 1 and 2′, and a second one (GjUGT94E5) is responsible for the production of crocins 2, 3, and 4 (Nagatoshi et al., 2012). We were unable to find bona fide orthologs of GjUGT94E5 in our transcriptome or in that reported by Jain et al. (2016). The characterization of additional UGTs expressed in C. sativus stigmas will elucidate their possible role in hierarchical crocin glycosylation.
The CsUGT74AD1 protein does not present obvious transmembrane domains, but it is insoluble upon extract fractionation. A similar behavior has been observed for several enzymes in carotenoid biosynthesis, such as phytoene desaturase, which is present in plastids in a soluble complex with HSP70 and in a second, membrane-associated complex containing the active enzyme (Al-Babili et al., 1996). The insolubility of CsUGT74AD1 was consistent with immunogold labeling, showing that the enzyme (or a cross-reacting UGT) localizes to cytoplasmic electron-dense structures reminiscent of membrane or cytoskeletal structures. Confocal microscopy confirmed that the protein colocalizes with mCherry, a cytosolic marker, in N. benthamiana leaves. This localization is in agreement with the cytosolic localization of the majority of plant UGTs (Gachon et al., 2005) but in disagreement with the plastidial localization described, based on proteomic data, for the very similar UGTCs2 (Gómez-Gómez et al., 2017). Given the extremely high abundance of CsUGT74AD1 and its similarity to UGTCs2, we believe that the plastid-associated peptides identified by Gómez-Gómez et al. (2017) and attributed to UGTCs2 belong instead to CsUGT74AD1. One possible explanation of the plastidial localization claimed by Gómez-Gómez et al. (2017) for both ALDH and UGT enzymes is a contamination of the chromoplast membrane preparations with nonplastidial proteins, as suggested by the low-level mitochondrial and peroxisomal contamination observed (Gómez-Gómez et al., 2017).
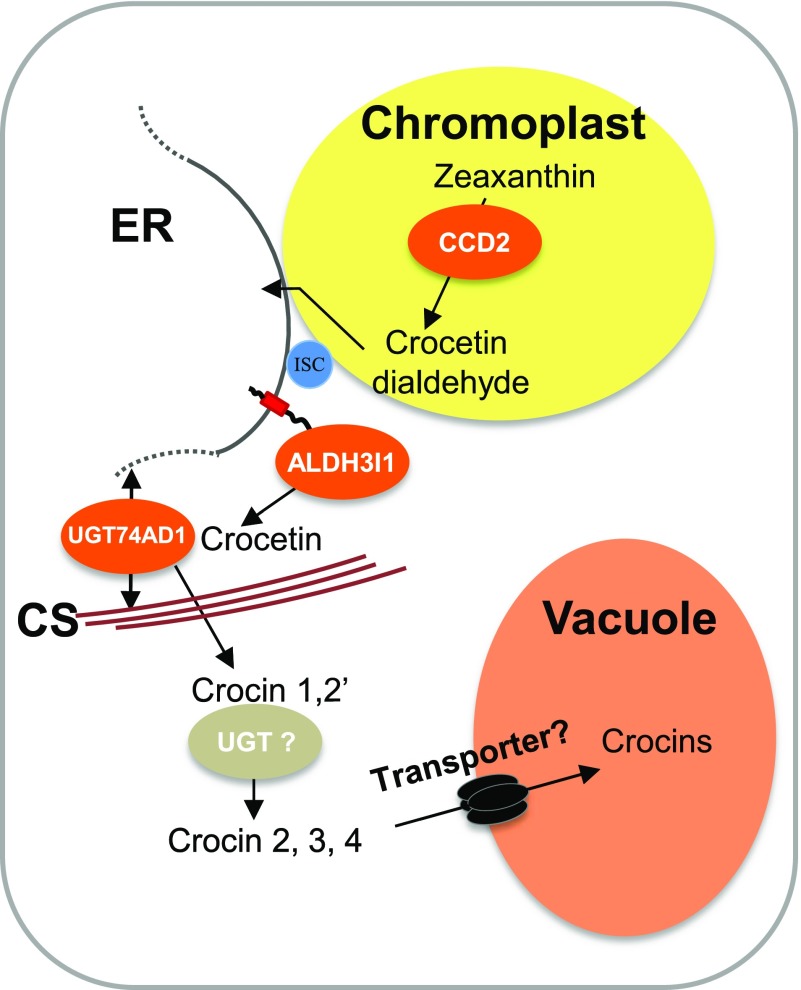
A Model for Crocin Biosynthesis and Trafficking from the Plastid to the Vacuole
Plastids and ER have biochemical continuity, as demonstrated by the fact that mutations in plastid-localized enzymes affecting tocopherol biosynthesis can be complemented by directing the same enzymes to the ER (Mehrshahi et al., 2013). This finding suggests that plastidial and ER membranes are in close contact, allowing hydrophobic molecules like tocopherol biosynthetic intermediates or crocetin dialdehyde to migrate from one compartment to the other (Fig. 9). Our model (Fig. 9) suggests that the ER acts as a transit center for metabolites whose biosynthesis starts in the chloroplast and ends in the vacuole. This type of compartmentation has been described also for the synthesis of steviosides, which are glycosylated diterpenes that accumulate in the vacuoles of Stevia rebaudiana leaves and confer an intensely sweet flavor. Similar to crocins, the first dedicated step in the biosynthesis of steviosides is mediated by a plastid-localized kaurene synthase and the second step by an ER-localized kaurene oxidase (Brandle and Telmer, 2007). The model also predicts that the vacuolar transport of crocins and steviosides, similar to that of other glycosylated secondary metabolites, is mediated by tonoplast-localized transporters (Fig. 9; Martinoia et al., 2007). This model may apply to the subcellular compartmentation of other pathways, such as the biosynthesis of ABA-GE (Nambara and Marion-Poll, 2005; Dong et al., 2015) or of glycosylated apocarotenoids in leaves (Lätari et al., 2015).
Figure 9.
Proposed model for crocin biosynthesis/compartmentation in C. sativus stigmas. CsCCD2 cleaves zeaxanthin in the choromoplast, producing crocetin dialdehyde, which migrates to the ER, where it is converted to crocetin by CsALDH3I1. The ER and plastid membranes are contiguous through the action of an interphase stabilizing complex (ISC; Mehrshahi et al., 2013). Crocetin is converted to crocins 1 and 2′ by CsUGT74AD1, associated with cytoplasmic membranes or with the cytoskeleton (CS). A second, unidentified UGT converts crocins 1 and 2′ into crocins 2, 3, and 4, which are then transported into the vacuole through one or more unidentified tonoplast transporters.
Although the expression pattern, substrate specificity, and tissue specificity of CsALDH3I1 and CsUGT74AD1 strongly suggest that they are involved in the biosynthesis of crocins 1 and 2′, we have no proof yet for their function in planta. Due to the genetic intractability of C. sativus, such proof could be obtained only by heterologous expression in plant tissues containing appropriate levels of the substrates, as already done for CCD2 in maize endosperm (Frusciante et al., 2014). Unfortunately, this tissue contains an endogenous ALDH activity able to dehydrogenate crocetin dialdehyde (Frusciante et al., 2014). Another limitation of the model is that we have not shown that the substrates and products of the three enzymes are indeed localized in the extrapastidial locations (except for crocins, which are localized in the vacuole). This will be the subject of future studies.
Based on microscopic observations, Gómez-Gómez et al. (2017) proposed an alternative model (i.e. that crocin biosynthesis takes place entirely in the plastid and that crocins accumulate in crocinoplast vesicles that move from the plastid to the vacuole). Our model does not exclude the existence of such a pathway. However, none of the ALDH and UGT enzymes we characterized showed a plastidial localization. Similar to the ER-cytoplasmic pathway we describe here, the enzymes involved in the hypothesized plastidial pathway must be isolated and functionally characterized and their subcellular localization must be determined.
MATERIALS AND METHODS
Transcriptomic and Bioinformatic Analyses
Transcript sequences for the putative enzymes were identified by homology with known enzymes in 454 Titanium sequences of Crocus sativus stigmas at different developmental stages (Frusciante et al., 2014) and confirmed in published transcriptomic data (Jain et al., 2016). Expression levels in different stigma developmental stages were obtained with Cufflinks (Trapnell et al., 2012) from in-house data. Heat maps were created using Genesis as described previously (Sturn et al., 2002; Diretto et al., 2010). Evolutionary relationships were inferred using the neighbor-joining method (Saitou and Nei, 1987), and phylogenetic and molecular evolutionary analyses were conducted using MEGA version 7 (Kumar et al., 2016) and the Conserved Domain Database tool of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Subcellular compartmentation was deduced using TargetP 1.1 prediction servers and transmembrane domains using TMHMM 2.0 software (http://www.cbs.dtu.dk/services/TMHMM/).
Cloning of C. sativus Genes
C. sativus ALDH and UGT transcripts were isolated from C. sativus stigma RNA using the Omniscript RT cDNA synthesis kit (Qiagen). Coding sequences were amplified from cDNA using Phusion High Fidelity DNA polymerase (New England Biolabs) with the oligonucleotides listed in Supplemental Table S4. Amplicons were cloned in the pBluescript vector (Stratagene) digested with EcoRV, verified by sequencing, and then reamplified with the oligonucleotides listed in Supplemental Table S4. The PCR products were cloned in the pTHIO vector (Trautmann et al., 2013) fused to an in-frame 5′ thioredoxin gene and to 3′ VP5 epitope and 6×HIS tag. The ALDH genes were cloned in pTHIO (Ampr) vector, obtaining the pTHIO-CsALDHs (Ampr) constructs. CsUGT74AD1 was cloned in a modified pTHIO vector harboring the spectinomycin resistance gene (Spectr) instead of Ampr, producing the pTHIO-CsUGT74AD1 (Spectr) construct. The previously isolated CsCCD2 gene (Frusciante et al., 2014) was cloned in a modified pTHIO vector harboring the chloramphenicol resistance gene (Cmr), obtaining the pTHIO-CsCCD2 (Cmr) construct.
In Bacterio ALDH Assay
The zeaxanthin-accumulating strain of Escherichia coli (Kanr), described previously (Frusciante et al., 2014), was transformed with pTHIO-CsCCD2 (Cmr) and one of the six pTHIO-CsALDHs (Ampr) constructs or with pTHIO-SynALDH (Ampr), which harbors the ALDH1 gene of Synechocystis sp. PCC6803 (Trautmann et al., 2013). As a control, the strain was transformed with pTHIO empty vector or with pTHIO-CsCCD2 (Cmr) alone. Overnight cultures of these clones were inoculated into 50 mL of Luria-Bertani medium containing one-half-strength antibiotics (25 μg mL−1 kanamycin, 12.5 μg mL−1 chloramphenicol, and/or 50 μg mL−1 ampicillin), grown at 37°C to an OD600 of 0.7, and induced with 0.08% (w/v) arabinose at 20°C for 16 h. Cells were pelleted and polar and nonpolar fractions were extracted. Polar and semipolar metabolites were extracted as described previously (Fasano et al., 2016; Rambla et al., 2016) with slight modifications: pellets were resuspended in 10 mL of 75% (v/v) methanol spiked cold (0.5 µg mL−1 formononetin; Sigma-Aldrich), lysed on ice by triplicate sonication at 10 Hz output (10 s each), and centrifuged at 18,000g for 30 min. The supernatant was then dried and redissolved in 200 μL of 75% (v/v) methanol, centrifuged at 18,000g for 20 min to remove particles and aggregates, and subjected to HPLC-PDA-HRMS analysis. Nonpolar metabolites were extracted with the same procedure using 100% acetone spiked cold (0.5 µg mL−1 α-tocopherol acetate; Sigma-Aldrich) for extraction and chloroform for resuspension.
In Vitro ALDH and UGT Assays
The pTHIO-CsALDH3I1 and pTHIO-CsUGT74AD1 plasmids were expressed in the E. coli BL21(pGro7) strain, and protein expression, solubilization with Triton X-100, and preparation of lysates were performed as described (Trautmann et al., 2013; Frusciante et al., 2014). For ALDH assays (Trautmann et al., 2013), 50 μg of protein extract was incubated with different substrates (40 μm final concentration) in a 200-μL volume at 28°C in glass screw-cap vials. In the case of apocarotenals (crocetin dialdehyde, β-apo-8′-carotenal, and retinal), the substrates were micellarized as described (Trautmann et al., 2013) and reactions were stopped by adding 400 µL of acetone, extracted with petroleum/diethyl ether (1:2, v/v), and filtered using a 0.22-μm filter before HPLC-PDA-HRMS analysis. For the other aldehydes (dodecanal, hexanal, and 4-OH-benzaldehyde), the reactions were stopped by derivatization with 2,4-dinitrophenylhydrazine (2,4-DNPH). Briefly, an equal volume of 2 mm 2,4-DNPH in acetonitrile:1 n HCl was added to the reactions and incubated at 40°C for 30 min and then at 4°C overnight. The pH was then adjusted to pH 4, samples were centrifuged at 20,000g for 20 min, and the supernatant was filtered before HPLC-PDA-HRMS analysis. The percentage of substrate conversion was calculated based on the peak area of each substrate in enzyme-containing assays (extract from induced cells transformed with the pTHIO-CsALDH3I1 construct) compared with control incubations (extract from induced cells containing the pTHIO empty vector).
For UGT assays (Moraga et al., 2004), 40 µg of protein extract was incubated in 100 µL of reaction containing 100 µm of the various substrates, either encapsulated crocetin or ABA, naringenin, quercetin, indole 3-acetic acid, and cinnamic acid. UDP-Glc and UDP-Gal (2.5 mm) were included as sugar donors. The reaction was incubated at 30°C and stopped by adding 100 µL of cold ethanol and immediately freezing at −20°C. The HPLC-PDA-HRMS analysis was performed as described below, and the percentage of substrate conversion was calculated as described for CsALDH3I1 assays.
HPLC-PDA-HRMS Analysis
Polar and nonpolar extracts were analyzed with a Q-Exactive mass spectrometer (Thermo Fisher Scientific) coupled to an HPLC system equipped with a photodiode array detector (Dionex). HPLC separation of polar and semipolar metabolites (crocetin, crocins, naringenin, quercetin, cinnamic acid, ABA, and indole 3-acetic acid) and 2,4-DNPH-linked aldehydes was performed as described previously (Alboresi et al., 2016; D’Esposito et al., 2017) with some modifications. One to 5 µL of sample was injected on a C18 Luna reverse-phase column (100 × 2.1 mm, 2.5 µm; Phenomenex) using the mobile phases water + 0.1% (v/v) formic acid (A) and acetonitrile + 0.1% (v/v) formic acid (B) at a total flow rate of 250 µL min−1. The separation of polar and semipolar metabolites was developed using 5% B for 0.5 min, followed by a 24-min linear gradient to 75% B. The separation of the 2,4-DNPH-linked aldehydes was developed using 20% B for 0.5 min, followed by a 24-min linear gradient to 95% B. The ionization of polar and semipolar metabolites was performed using ESI with nitrogen used as sheath and auxiliary gas, set to 45 and 30 units, respectively. The vaporizer temperature was 270°C, the capillary temperature was 30°C, the discharge current was set to 5 μA, and S-lens RF level was set at 50. The acquisition was performed in the mass range 110 to 1,600 mass-to-charge ratio in both positive and negative ion modes with the following parameters: resolution, 70,000; microscan, 1; AGC target, 1e6; maximum injection time, 50. For the ionization of the 2,4-DNPH-linked aldehydes, sheath and auxiliary gas were set to 40 and 10 units, respectively. The vaporizer temperature was 280°C, the capillary temperature was 250°C, the discharge current was set to 3.5 μA, and S-lens RF level was set at 50. The acquisitions were performed as described above.
HPLC separation of nonpolar metabolites (zeaxanthin, crocetin dialdehyde, β-apo-8′-carotenal, and retinal) was performed as described previously (Liu et al., 2014; Su et al., 2015), injecting 1 to 5 µL of sample on a C30 reverse-phase column (100 × 3.0 mm; YMC Europe). The mobile phases used were composed by methanol (A), water:methanol (20:80, v/v) containing 0.2% ammonium acetate (B), and tert-butyl methyl ether (C) at a total flow rate of 800 µL min−1. The separation was developed using 95% A/5% B for 1.3 min, followed by 80% A/5% B/15% C for 2 min, followed by a subsequent 9.2-min linear gradient to 30% A/5% B/65% C. After PDA, the flow was split and 300 µL was sent to the MS source. The ionization of nonpolar metabolites was performed with an atmospheric pressure chemical ionization source. Nitrogen was used as sheath and auxiliary gas, set to 20 and 10 units, respectively. The vaporizer temperature was 300°C, the capillary temperature was 250°C, the discharge current was set to 5.5 μA, and S-lens RF level was set at 50. The acquisition was performed as described for ESI. UV-visible light detection was continuous from 220 to 700 nm. All solvents used were LC-MS grade (Merck Millipore).
Identification was achieved on the basis of accurate masses and by comparison with authentic reference substances. Representative mass chromatograms of the various analytes are shown in Supplemental Figure S10. The ion peak areas were normalized to the ion peak area of the internal standard (formononetin or α-tocopherol acetate for polar/semipolar and nonpolar metabolites, respectively).
Extract Fractionation and Immunoblot Analysis
To analyze protein expression in bacterial clones, total proteins or soluble extracts from 100 μL of induced cultures (OD600 = 0.7) were loaded on a 10% SDS-PAGE gel using a Bio-Rad Mini Protean device. The gel was stained or subjected to immunoblot analysis as described previously (Demurtas et al., 2016), using a mouse monoclonal anti-His6 antibody (Roche, catalog no. 11922416001) at 1:200 dilution, followed by 1 h of incubation with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG antibody (GE Healthcare, catalog no. NA931). To analyze the presence of the CsCCD2, CsALDH3I1, and CsUGT74AD1 enzymes in C. sativus flowers, total proteins were extracted from fresh stigmas, stamens, and tepals (Zafferanami) by grinding the tissues to a fine powder in liquid nitrogen and resuspending in 10 volumes (w/v) of SDS loading buffer. Samples were then lysed on ice by triplicate sonication at 10 Hz output (10 s each) and boiled for 10 min. After centrifugation at 18,000g for 30 min, the supernatant was recovered. Total protein content was measured by colorimetry, loading 10 µL of each sample on an SDS-PAGE gel followed by Coomassie Blue staining and using ImageLab 4.0 software (Bio-Rad) for total protein quantification. An equal amount of total proteins (10 µg) was run on a 10% SDS-PAGE gel, followed by immunoblot analysis (see below).
To analyze the solubility of the CsCCD2, CsALDH3I1, and CsUGT74AD1 enzymes in stigma extracts, the ground material was subjected to sequential extractions. Stigma powder was resuspended and homogenized with an Ultraturrax in 5 volumes of PB buffer (20 mm Na2HPO4 and 2 mm NaH2PO4, pH 7.2) containing protease inhibitors (complete, EDTA-free; Roche), sonicated three times at 10 Hz output (10 s each), and centrifuged at 18,000g for 30 min. The recovered supernatant corresponded to the soluble fraction, whereas the pellet was resuspended in 3 volumes of PB buffer + 200 mm NaCl by vortexing for 5 min at 1,000 rpm, kept in a rotary shaker for 30 min at 4°C, and centrifuged at 18,000g for 30 min. The supernatant recovered after centrifugation contained the extrinsic membrane proteins. Finally, the pellet was resuspended in 3 volumes of SDS loading buffer (insoluble fraction).
For all experiments, PVDF membranes were incubated 2 h at room temperature with 1 µg mL−1 of a polyclonal anti-CsCCD2, anti-CsALDH3I1, or anti-CsUGT74AD1 antibody (affinity-purified sera of rabbits immunized with the synthetic peptides MANKEEAEKRKKKP, YGGKRDEKRLKIAP, or EVMDGERSGKIREN, respectively; GenScript), followed by incubation for 1 h with a secondary anti-rabbit antibody (GE Healthcare) at 1:15,000 dilution. The bound antibody was revealed as described previously (Demurtas et al., 2016).
Electron Microscopy
Fresh stigmas were collected and cut in transverse sections, 2 mm long, and treated as follows. For scanning electron microscopy, samples were fixed with 2% (w/v) paraformaldehyde + 2.5% (w/v) glutaraldehyde in 0.05 m cacodylate buffer, pH 7.2, overnight at 4°C. Samples were washed in the same buffer and then fixed in 1% (w/v) OsO4 in 0.05 m cacodylate buffer, pH 7.2, for 1 h at room temperature and washed again in the same buffer. Samples were then dried according to the critical point method using CO2 in a Balzers Union CPD 020, attached to aluminum stubs using a carbon tape, and sputter coated with gold in a Balzers MED 010 unit. The observation was made by a JEOL JSM 6010LA scanning electron microscope.
For IEM, specimens were fixed with a mixture of 4% (w/v) paraformaldehyde + 0.25% (w/v) glutaraldehyde in 0.05 m phosphate buffer, pH 6.9, for 1 h at room temperature. After rinsing three times in the same buffer for 30 min, samples were dehydrated in a graded ethanol series and embedded in medium-grade LR White resin (Multilab Supplies). The resin was polymerized in tightly capped gelatin capsules for 48 h at 50°C. Ultrathin sections were cut with Reichert Ultracut and LKB Nova ultramicrotomes using a diamond knife and collected on nickel grids. For immunogold staining in postembedding, the incubation protocol suggested by Aurion was used. Residual aldehyde activity was suppressed by using PBS with 0.05 m Gly, pH 7.4, for 20 min. A subsequent block step was made with 5% (w/v) BSA, 5% (v/v) normal goat serum, and 0.1% (w/v) cold-water fish skin gelatin for 30 min. Sections were washed in incubation buffer (PBS and 0.1% BSA-cTM, pH 7.4; 3 × 5 min). Incubation with the primary antibodies was made overnight in a moist chamber at 4°C. Anti-CsCCD2, anti-CsALDH3I1, and anti-CsUGT74AD1 antibodies, described previously, were diluted in incubation buffer and used at concentrations of 5 µg mL−1 (anti-CsCCD2) and 1 µg mL−1 (anti-CsALDH3I1 and anti-CsUGT74AD1). In addition, for CsALDH3I1, another primary antibody (affinity-purified sera of rabbits immunized with the synthetic peptide VKELRESFNKGTTR; GenScript), at 1 µg mL−1 concentration, also was used. Sections were then washed in incubation buffer (6 × 5 min) and incubated for 1 h with a secondary goat anti-rabbit antibody conjugated to 20-nm gold particles (British BioCell International), diluted 1:10 in incubation buffer. After rinsing in incubation buffer (6 × 5 min), the grids were washed in PBS (6 × 5 min). Sections were stained subsequently with uranyl acetate and observed with a JEOL JEM EXII transmission electron microscope at 100 kV. Micrographs were acquired by the Olympus SIS VELETA CCD camera equipped the iTEM software. Control sections in which the primary antibodies were omitted also were analyzed.
Transgene Expression in Nicotiana benthamiana Leaves and Confocal Microscopy
The C termini of CsALDHs and CsUGT74AD1 were fused to the N terminus of eGFP, with gene sequences cloned in the pBI-eGFP vector (Frusciante et al., 2014) using Gibson assembly (Gibson et al., 2009). The vector was digested with XbaI restriction enzyme, whereas CsALDHs and CsUGT74AD1 coding sequences were amplified with Q5 High Fidelity DNA polymerase (New England Biolabs, catalog no. M0491S) from pBluescript-CsALDHs and pBluescript-CsUGT74AD1 constructs. A Pro-Gly-Pro tripeptide was introduced as a linker between the analyzed polypeptides and eGFP. The oligonucleotides used are listed in Supplemental Table S4. The purified PCR fragments were then assembled, generating pBI-CsALDHs:eGFP constructs and pBI-CsUGT74AD1:eGFP constructs, and checked by sequencing before transformation in Agrobacterium tumefaciens strain C58C1. To obtain the pBI-CsCCD2:eGFP construct harboring the full-length CsCCD2 sequence, comprising the chloroplast transit peptide (Ahrazem et al., 2016), the sequence coding for the transit peptide (5′-ATGGAATCTCCTGCTACTAAATTACCTGCACCTCTGCTGATGTTATCTTCTTCTCCATTCCTTCTCCCTTCTCCTAATAAGAGCTCCTCCATCTTCCTTCCACGTAAATTAGGGCCGCTACCTCCAAAATATTATTATTACAATTGCTGCCATCCTAAGAGTAGATCAATCTCAGTAGTATCA-3′) was synthesized by Genscript and inserted by Gibson assembly at the 5′ end of the CsCCD2 gene inserted in the pBI-CsCCD2:eGFP construct, as described by Frusciante et al. (2014). Agroinfiltration of N. benthamiana leaves was performed as described (Hamilton and Baulcombe, 1999). Cultures with OD600 = 0.4 were incubated in a solution containing 10 mm MgCl2, 10 mm MES, and 100 μm acetosyringone for 2 h. Young leaves of 6-week-old N. benthamiana plants, grown under hydroponic conditions, were agroinfiltrated with the C58C1 strain containing one of the following constructs: pBI-eGFP, pBI-CsCCD2:eGFP, pBI-CsALDHs:eGFP, or pBI-CsUGT74AD1:eGFP; at least four independent leaves from three different plants were agroinfiltrated with each construct. After 24 and 48 h, leaves were analyzed by a confocal laser scanning microscope (Olympus FV1000). Lasers 488 nm (argon) and 635 nm (diode) were used to detect the green and red fluorescence of eGFP and chlorophyll signals, respectively. Images of 512 × 512 pixels were acquired in xyz scan mode using a 20× objective (numerical aperture 0.75) with optical zooming 3× or 40× (numerical aperture 1.30) magnification and processed by IMARIS (Bitplane) software. For colocalization studies, leaves were coinfiltrated with C58C1 strains containing pBI121:CsALDH3I1:eGFP and pBI:RFP:HDEL (ER marker; Lee et al., 2013) or pBI121:CsUGT74AD1:eGFP and pBI:mCherry (cytosolic marker). GFP and RFP/mCherry were excited at 488 and 561 nm, respectively, and detected in the 495- to 520-nm and 571- to 638-nm ranges, respectively. Images were acquired on a Zeiss LSM880 confocal microscope using a 63× (numerical aperture 1.4) oil-immersion objective.
Accession Numbers
GenBank accession numbers are as follows: CsALDH2B4 (MG672523), CsALDH2C4 (MF596160), CsALDH3I1 (MF596165), CsALDH5F1 (MF596161), CsALDH6B2 (MG672524), CsALDH7B4 (MF596162), and CsUGT74AD1 (MF596166).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of CsALDH, YLO-1, SynALDH, and BoALDH protein sequences.
Supplemental Figure S2. Hydrophobic profiles of CsCCD2, ALDH, and UGT proteins.
Supplemental Figure S3. Simultaneous expression of CsCCD2 and different ALDHs in E. coli.
Supplemental Figure S4. Identification of trans- and cis-crocetin in E. coli.
Supplemental Figure S5. Expression and solubilization of CsALDH3I1:THIO from E. coli BL21(pGro7).
Supplemental Figure S6. Endogenous E. coli activity on crocetin dialdehyde in vitro.
Supplemental Figure S7. Alignment of plant UGT protein sequences.
Supplemental Figure S8. Expression and solubilization of CsUGT74AD1:THIO from E. coli BL21(pGro7).
Supplemental Figure S9. Subcellular localization of different CsALDH proteins in N. benthamiana leaves.
Supplemental Figure S10. Representative HPLC-HRMS chromatograms of the reactions reported in Supplemental Tables S2 and S3.
Supplemental Table S1. Predicted and experimentally confirmed subcellular localization of CsCCD2, CsALDH, and CsUGT proteins and their characterized Arabidopsis homologs.
Supplemental Table S2. CsALDH3I1 substrate preference.
Supplemental Table S3. CsUGT74AD1 substrate preference.
Supplemental Table S4. Oligonucleotides used to isolate and clone CsALDH and CsUGT74AD1 genes.
Note added in proof
While this manuscript was in press, a paper appeared (Gomez-Gomez et al., 2018, Int. J. Mol. Sci. 2018, 19, 1409) describing the in vitro characterization of several C. sativus ALDHs. A truncated form of CsALDH3I1, lacking the 145 C-terminal amino acids, was found to have only basal levels of activity on crocetin dialdehyde, suggesting that the C-terminal hydrophobic domain is essential for full activity.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Elisabetta Bennici (Energy and Sustainable Economic Development [ENEA]) for the growth of N. benthamiana plants; Salvatore Chiavarini (ENEA) for suggestions on 2,4-DNPH derivatization; Lourdes Gomez-Gomes and Angela Rubio-Moraga (University of Albacete) and Zafferanami for the provision of C. sativus flowers; Gaetano Perrotta, Paolo Facella, and Fabrizio Carbone (ENEA) for 454 sequencing, and Aparna Balakrishna (King Abdullah University of Science and Technology) for technical assistance. Part of the computing resources used for this work were kindly provided by the CRESCO/ENEAGRID High Performance Computing infrastructure and its staff (Ponti et al., 2014).
Footnotes
This work was supported by the European Union (From discovery to products: A next generation pipeline for the sustainable generation of high-value plant products, FP7 Contract 613153) and by baseline funding given to S.A.-B. from King Abdullah University of Science and Technology, and benefited from the networking activities within the European Cooperation in Science and Technology Action CA15136 (EUROCAROTEN).
Articles can be viewed without a subscription.
References
- Ahrazem O, Rubio-Moraga A, Trapero-Mozos A, Climent MF, Gómez-Cadenas A, Gómez-Gómez L (2015) Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci 234: 60–73 [DOI] [PubMed] [Google Scholar]
- Ahrazem O, Rubio-Moraga A, Berman J, Capell T, Christou P, Zhu C, Gómez-Gómez L (2016) The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol 209: 650–663 [DOI] [PubMed] [Google Scholar]
- Alavizadeh SH, Hosseinzadeh H (2014) Bioactivity assessment and toxicity of crocin: a comprehensive review. Food Chem Toxicol 64: 65–80 [DOI] [PubMed] [Google Scholar]
- Al-Babili S, von Lintig J, Haubruck H, Beyer P (1996) A novel, soluble form of phytoene desaturase from Narcissus pseudonarcissus chromoplasts is Hsp70-complexed and competent for flavinylation, membrane association and enzymatic activation. Plant J 9: 601–612 [DOI] [PubMed] [Google Scholar]
- Alboresi A, Perin G, Vitulo N, Diretto G, Block M, Jouhet J, Meneghesso A, Valle G, Giuliano G, Maréchal E, et al. (2016) Light remodels lipid biosynthesis in Nannochloropsis gaditana by modulating carbon partitioning between organelles. Plant Physiol 171: 2468–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azama K, Abe S, Sugimoto H, Davies E (2003) Lysine-containing proteins in maize endosperm: a major contribution from cytoskeleton-associated carbohydrate-metabolizing enzymes. Planta 217: 628–638 [DOI] [PubMed] [Google Scholar]
- Bathaie SZ, Farajzade A, Hoshyar R (2014) A review of the chemistry and uses of crocins and crocetin, the carotenoid natural dyes in saffron, with particular emphasis on applications as colorants including their use as biological stains. Biotech Histochem 89: 401–411 [DOI] [PubMed] [Google Scholar]
- Benghezal M, Wasteneys GO, Jones DA (2000) The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12: 1179–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisti S, Maccarone R, Falsini B (2014) Saffron and retina: neuroprotection and pharmacokinetics. Vis Neurosci 31: 355–361 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Dogbo O, Camara B (2003a) Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300: 2089–2091 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, Mutterer J, Camara B (2003b) Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandle JE, Telmer PG (2007) Steviol glycoside biosynthesis. Phytochemistry 68: 1855–1863 [DOI] [PubMed] [Google Scholar]
- Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang Y, Wang X, Kotchoni SO, Wood AJ, Kirch HH, Kopečný D, et al. (2013) Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237: 189–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chryssanthi DG, Lamari FN, Georgakopoulos CD, Cordopatis P (2011) A new validated SPE-HPLC method for monitoring crocetin in human plasma: application after saffron tea consumption. J Pharm Biomed Anal 55: 563–568 [DOI] [PubMed] [Google Scholar]
- Cinelli RA, Ferrari A, Pellegrini V, Tyagi M, Giacca M, Beltram F (2000) The enhanced green fluorescent protein as a tool for the analysis of protein dynamics and localization: local fluorescence study at the single-molecule level. Photochem Photobiol 71: 771–776 [DOI] [PubMed] [Google Scholar]
- Cote F, Cormier F, Dufresne C, Willemot C (2001) A highly specific glucosyltransferase is involved in the synthesis of crocetin glucosylesters in Crocus sativus cultured cells. J Plant Physiol 158: 553–560 [Google Scholar]
- D’Esposito D, Ferriello F, Molin AD, Diretto G, Sacco A, Minio A, Barone A, Di Monaco R, Cavella S, Tardella L, et al. (2017) Unraveling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol 17: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demurtas OC, Massa S, Illiano E, De Martinis D, Chan PK, Di Bonito P, Franconi R (2016) Antigen production in plant to tackle infectious diseases flare up: the case of SARS. Front Plant Sci 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Sánchez V, Estrada AF, Trautmann D, Al-Babili S, Avalos J (2011) The gene carD encodes the aldehyde dehydrogenase responsible for neurosporaxanthin biosynthesis in Fusarium fujikuroi. FEBS J 278: 3164–3176 [DOI] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, Beyer P, Giuliano G (2010) Transcriptional-metabolic networks in β-carotene-enriched potato tubers: the long and winding road to the Golden phenotype. Plant Physiol 154: 899–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Hwang I (2014) Contribution of ABA UDP-glucosyltransferases in coordination of ABA biosynthesis and catabolism for ABA homeostasis. Plant Signal Behav 9: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Park Y, Hwang I (2015) Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem 58: 29–48 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Estrada AF, Youssar L, Scherzinger D, Al-Babili S, Avalos J (2008) The ylo-1 gene encodes an aldehyde dehydrogenase responsible for the last reaction in the Neurospora carotenoid pathway. Mol Microbiol 69: 1207–1220 [DOI] [PubMed] [Google Scholar]
- Fasano C, Diretto G, Aversano R, D’Agostino N, Di Matteo A, Frusciante L, Giuliano G, Carputo D (2016) Transcriptome and metabolome of synthetic Solanum autotetraploids reveal key genomic stress events following polyploidization. New Phytol 210: 1382–1394 [DOI] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S, et al. (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA 111: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10: 542–549 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Parra-Vega V, Rivas-Sendra A, Seguí-Simarro JM, Molina RV, Pallotti C, Rubio-Moraga Á, Diretto G, Prieto A, Ahrazem O (2017) Unraveling massive crocins transport and accumulation through proteome and microscopy tools during the development of saffron stigma. Int J Mol Sci 18: E76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Jain M, Srivastava PL, Verma M, Ghangal R, Garg R (2016) De novo transcriptome assembly and comprehensive expression profiling in Crocus sativus to gain insights into apocarotenoid biosynthesis. Sci Rep 6: 22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ (2004) The ALDH gene superfamily of Arabidopsis. Trends Plant Sci 9: 371–377 [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lätari K, Wüst F, Hübner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R (2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L (2013) An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol 197: 481–489 [DOI] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan JP, Li Z, Giuliano G, Regad F, Bouzayen M (2014) The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol 203: 206–218 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y (1994) Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J Cell Biol 126: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrshahi P, Stefano G, Andaloro JM, Brandizzi F, Froehlich JE, DellaPenna D (2013) Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc Natl Acad Sci USA 110: 12126–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga AR, Nohales PF, Pérez JA, Gómez-Gómez L (2004) Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta 219: 955–966 [DOI] [PubMed] [Google Scholar]
- Moraga AR, Mozos AT, Ahrazem O, Gómez-Gómez L (2009) Cloning and characterization of a glucosyltransferase from Crocus sativus stigmas involved in flavonoid glucosylation. BMC Plant Biol 9: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatoshi M, Terasaka K, Owaki M, Sota M, Inukai T, Nagatsu A, Mizukami H (2012) UGT75L6 and UGT94E5 mediate sequential glucosylation of crocetin to crocin in Gardenia jasminoides. FEBS Lett 586: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T (1998) Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol 64: 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti G, Palombi F, Abate D, Ambrosino F, Aprea G, Bastianelli T, Beone F, Bertini R, Bracco G, Caporicci M (2014) The role of medium size facilities in the HPC ecosystem: the case of the new CRESCO4 cluster integrated in the ENEAGRID infrastructure. In Bassini S, Nygard M , eds, 2014 International Conference on High Performance Computing & Simulation (HPCS).IEEE, New York, NY, pp 1030–1033 10.1109/HPCSim.2014.6903807 [DOI] [Google Scholar]
- Rambla JL, Trapero-Mozos A, Diretto G, Rubio-Moraga A, Granell A, Gómez-Gómez L, Ahrazem O (2016) Gene-metabolite networks of volatile metabolism in Airen and Tempranillo grape cultivars revealed a distinct mechanism of aroma bouquet production. Front Plant Sci 7: 1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim EK, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Moraga A, Trapero A, Ahrazem O, Gómez-Gómez L (2010) Crocins transport in Crocus sativus: the long road from a senescent stigma to a newborn corm. Phytochemistry 71: 1506–1513 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17: 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiti N, Adewale IO, Petersen J, Bartels D, Kirch HH (2011a) Engineering the nucleotide coenzyme specificity and sulfhydryl redox sensitivity of two stress-responsive aldehyde dehydrogenase isoenzymes of Arabidopsis thaliana. Biochem J 434: 459–471 [DOI] [PubMed] [Google Scholar]
- Stiti N, Missihoun TD, Kotchoni SO, Kirch HH, Bartels D (2011b) Aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Front Plant Sci 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantilis PA, Tsoupras G, Polissiou M (1995) Determination of saffron (Crocus sativus L.) components in crude plant extract using high-performance liquid chromatography-UV-visible photodiode-array detection-mass spectrometry. J Chromatogr A 699: 107–118 [DOI] [PubMed] [Google Scholar]
- Trapero A, Ahrazem O, Rubio-Moraga A, Jimeno ML, Gómez MD, Gómez-Gómez L (2012) Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol 159: 1335–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann D, Beyer P, Al-Babili S (2013) The ORF slr0091 of Synechocystis sp. PCC6803 encodes a high-light induced aldehyde dehydrogenase converting apocarotenals and alkanals. FEBS J 280: 3685–3696 [DOI] [PubMed] [Google Scholar]
- Wang X. (2009) Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett 583: 3303–3309 [DOI] [PubMed] [Google Scholar]