Abstract
The stringent response is characterized by (p)ppGpp synthesis resulting in repression of translation and reprogramming of the transcriptome. In Staphylococcus aureus, (p)ppGpp is synthesized by the long RSH (RelA/SpoT homolog) enzyme, RelSau or by one of the two short synthetases (RelP, RelQ). RSH enzymes are characterized by an N-terminal enzymatic domain bearing distinct motifs for (p)ppGpp synthetase or hydrolase activity and a C-terminal regulatory domain (CTD) containing conserved motifs (TGS, DC and ACT). The intramolecular switch between synthetase and hydrolase activity of RelSau is crucial for the adaption of S. aureus to stress (stringent) or non-stress (relaxed) conditions. We elucidated the role of the CTD in the enzymatic activities of RelSau. Growth pattern, transcriptional analyses and in vitro assays yielded the following results: i) in vivo, under relaxed conditions, as well as in vitro, the CTD inhibits synthetase activity but is not required for hydrolase activity; ii) under stringent conditions, the CTD is essential for (p)ppGpp synthesis; iii) RelSau lacking the CTD exhibits net hydrolase activity when expressed in S. aureus but net (p)ppGpp synthetase activity when expressed in E. coli; iv) the TGS and DC motifs within the CTD are required for correct stringent response, whereas the ACT motif is dispensable, v) Co-immunoprecipitation indicated that the CTD interacts with the ribosome, which is largely dependent on the TGS motif. In conclusion, RelSau primarily exists in a synthetase-OFF/hydrolase-ON state, the TGS motif within the CTD is required to activate (p)ppGpp synthesis under stringent conditions.
Author summary
The stringent response is a general stress response, which allows bacteria to survive nutrient limited conditions and to better tolerate antibiotic treatment. In the human pathogen, Staphylococcus aureus, the stringent response plays an important role for virulence, phagosomal escape and antibiotic tolerance. The response is initiated by the synthesis of the nucleotide derivative (p)ppGpp which in turn leads to growth arrest and reprogramming of gene expression. However, a rapid and controlled inactivation of these growth inhibitory molecules is equally important for the organism. (p)ppGpp synthesis as well as hydrolysis is accomplished by a bi-functional RelA/SpoT homolog, RelSau bearing distinct synthetase, hydrolase and sensory domains. We elucidated how the C-terminal sensory domain of RelSau controls the intermolecular switch between hydrolase and synthetase activities in S. aureus. The switch is crucial for the appropriate response of S. aureus to adapt to changing environment encountered during infection.
Introduction
Bacteria react to nutrient limitation via a stress response that is characterized by the synthesis of pyrophosphorylated GTP (pppGpp) or GDP (ppGpp) (previously reviewed in [1,2,3,4,5,6,7,8,9,10,11]). Synthesis of (p)ppGpp, induced under these stress conditions (stringent conditions), results in many physiological changes, including inhibition of rRNA synthesis, replication and translation but also activation or repression of various genes. In many pathogenic bacteria, (p)ppGpp influences virulence, persistence and host interaction (see reviews [9,10]).
(p)ppGpp is synthesized by cytoplasmic enzymes that contain a conserved synthetase domain. RelA of Escherichia coli was the first such enzyme described and has been shown to synthesize (p)ppGpp under conditions of amino acid limitation [12]. E. coli and many other gram-negative bacteria possess an additional enzyme, SpoT, that possesses (p)ppGpp synthetase and hydrolase activities. The (p)ppGpp synthetase activity of SpoT is stimulated by various conditions, e.g. fatty acid deprivation [13,14]. In Firmicutes, homologous enzymes (Rel) constitute a distinct class of (p)ppGpp synthetases [11,15,16]. RelA, SpoT and Rel enzymes all belong to RSH (for RelA/SpoT homolog) superfamily [15,17]. Similar to SpoT, the Rel enzymes from Firmicutes are bifunctional proteins with (p)ppGpp synthetase and hydrolase activities; however, similar to RelA, the synthetase activity of these enzymes is stimulated upon amino acid starvation [18,19]. RSH enzymes share a multi-domain architecture with a C-terminal regulatory domain (CTD) and an N-terminal enzymatic domain (NTD) containing synthetase and hydrolase motifs. The only available crystal structure of an RSH enzyme is that of the NTD of Rel from Streptococcus equisimilis [20]. The structure indicates two conformations of the enzyme, corresponding to the reciprocal active states of the enzyme: (p)ppGpp-synthetase-ON/hydrolase-OFF (stringent) and synthetase-OFF/hydrolase-ON (relaxed). It has been proposed that the CTD is involved in reciprocal regulation of the enzymatic states. The current model suggests that under non-stringent (relaxed) conditions, the interaction of the CTD with the NTD maintains the enzyme in the synthetase-OFF/hydrolase-ON conformation [21,22,23]The CTD of RelA stimulates (p)ppGpp synthesis in a ribosome-dependent manner when uncharged tRNA, as a consequence of amino acid limitation, is located in the ribosomal A-site [24,25]. Interestingly, Rel from S. equisimilis is responsive to amino acid starvation only within its native genetic background and not when expressed in E. coli [13,21]. Bioinformatic analyses have revealed the presence of three conserved motifs within the CTDs of RSHs: TGS, ACT and DC. The TGS motif (named after the presence in ThrRS, GTPases, and SpoT) was shown to be responsible for the interaction of SpoT with the acyl-carrier protein. The ACT motif (named after three of the allosterically regulated enzymes in which this domain is found: aspartate kinase, chorismate mutase and TyrA) was proposed to be a conserved regulatory ligand-binding fold [26,27]. Recently, major insights into the ribosome-RelA structure were provided by cryo-EM analyses [28,29,30]. The structures revealed that RelA adopts an open conformation in which the CTD is intertwined around an A-site tRNA within the intersubunit cavity of the ribosome, and the NTD extends into the solvent. The structures support a model in which association of monomeric RelA with the ribosome relieves the autoinhibitory effect of the CTD on the NTD. It was hypothesized that autoinhibition in the unbound state is mediated by oligomerization of RelA. Oligomerization was previously demonstrated to occur via a conserved aspartate-cysteine motif (DC) in the CTD [31,32,33]. Interaction of monomeric RelA with the ribosome and putative RelA oligomerization in the unbound state indicate that the switching of enzymatic activities occurs via a complex mechanism that has not yet been elucidated.
In the human pathogen Staphylococcus aureus, the stringent response plays an important role in virulence [18], phagosomal escape [34] and antibiotic tolerance [35]. In S. aureus, in addition to RelSau, two enzymes with (p)ppGpp synthetase activity (RelP and RelQ) are present. These enzymes form homotetramers that lack the CTD and the hydrolase domain [36,37,38] and are transcriptionally induced under conditions of cell-wall stress [35]. The basal (p)ppGpp level produced by these enzymes is controlled by the hydrolase activity of RelSau [35]. The phenotypic consequences of (p)ppGpp accumulation vary among species and can be mediated by different mechanisms. In S. aureus, as in other Firmicutes, (p)ppGpp regulates transcription by an indirect mechanism that strongly relies on the lowering of intracellular GTP levels [39,40,41]. Low GTP levels lead to de-repression of the CodY regulon. CodY, when loaded with GTP and branched-chain amino acids, acts as a repressor of a variety of genes, e.g., genes involved in amino acid synthesis and virulence [41] A decrease in GTP levels could also lead to the repression of sensitive GTP-initiating promoters (e.g., those of stable RNA genes) [42,43]. All these studies illustrate the complex role of (p)ppGpp during the bacterial life cycle. The cellular concentration of (p)ppGpp has to be tightly regulated not only to support survival under stressed conditions but also to avoid toxicity under relaxed conditions. The molecular switch between the synthetase and hydrolase activities of RelSau is crucial for the maintenance of this balance.
Here, we aim to elucidate the role of the CTD in controlling the activity of Rel of the major human pathogen S. aureus (RelSau) in vivo. We show that the (p)ppGpp synthetase activity is restricted in S. aureus and that the synthetase is activated only upon interaction of the CTD with ribosomal partners under stringent conditions. The TGS and DC motifs within the CTD are essential for the enzymatic switch to the synthetase-ON state and play a major role in the interaction between RelSau and the translational apparatus.
Results
CTD-deleted RelSau is in a synthetase-OFF/hydrolase-ON state in its native S. aureus background
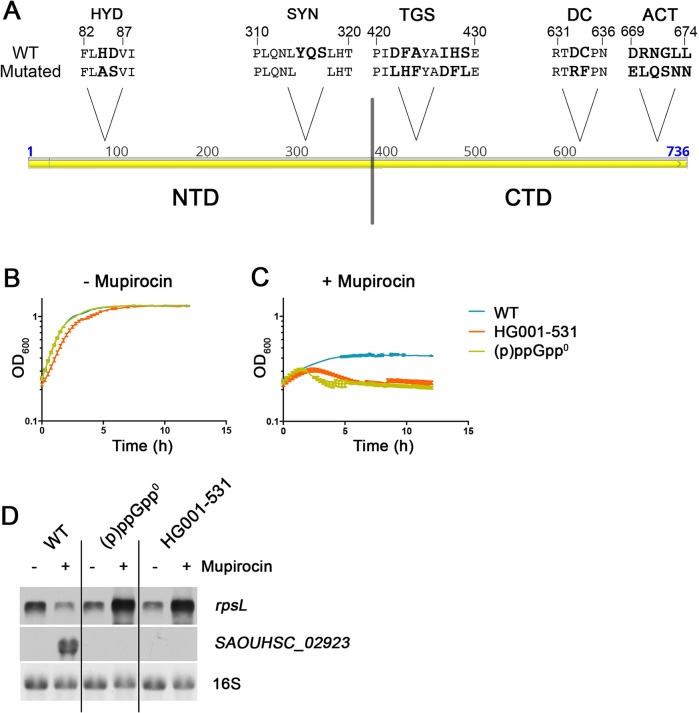
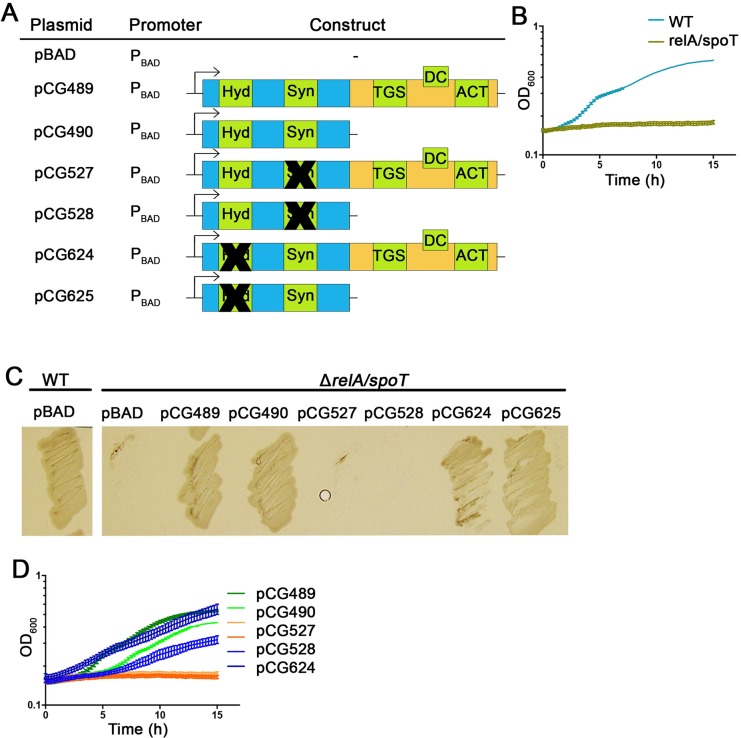
We aimed to analyze the role of the CTD of RelSau in the stringent response in S. aureus. In RelSau the canonical domains and motifs could be identified through alignment with RelA and SpoT from E. coli (Fig 1A). We first established a readout system for (p)ppGpp activity. To this end, we analyzed strain HG001 (wild type) as well as an isogenic mutant of this strain that carries mutations in all three (p)ppGpp enzymes (full deletion of rel, synthetase mutation in relP and relQ) and thus is unable to synthesize pppGpp (designated (p)ppGpp0) [35]. The mutant exhibits no phenotypic difference compared to the wild type when grown in rich medium (Fig 1B). The stringent response can be evoked by mupirocin, an inhibitor of isoleucyl-tRNA synthetase [18,34]. (p)ppGpp synthesis results in higher tolerance towards mupirocin. The (p)ppGpp0 strain exhibited a typical decline in OD600 when treated with mupirocin (Fig 1C). Furthermore, synthesis of (p)ppGpp results in repression of genes coding for ribosomal proteins (e.g., rpsL) and de-repression of the CodY target genes (e.g., SAOUHSC_02923, a putative amino acid transporter) (Fig 1D). Therefore, we used the enhanced mupirocin tolerance and typical transcription pattern (rpsL down, SAOUHSC_02923 up) as a readout for (p)ppGpp synthesis in S. aureus. As a first approach, we deleted the CTD of the wild-type RelSau (SAOUHSC_01742) to generate strain HG001-531. In contrast to a full-length rel deletion (Geiger et al., 2014a), truncation of the CTD had only a slight effect on growth (Fig 1C). It has been previously shown that RelSau is essential due to its hydrolase function [35]. Thus, the CTD does not seem to impede the hydrolase activity in vivo. Northern blot analysis revealed that the CTD mutant was unable to elicit a mupirocin-induced stringent response (Fig 1B). The transcriptional pattern of the marker genes rpsL and SAOUHSC_02923 as well as the mupirocin tolerance (Fig 1D) of the CTD mutant were indistinguishable from those of the (p)ppGpp0 strain. This finding is consistent with the general assumption that the CTD is required for sensing amino acid deprivation. However, hydrolase activity seems to be hardly effected by the CTD.
Fig 1. Experimental set-up for synthetase activity in S. aureus.
(A) The molecular architecture of RelSau including conserved and mutated motifs. S. aureus strains HG001 wild type (WT), (p)ppGpp0 and HG001-531 (rel CTD deleted) were grown in rich medium to OD600 = 0.3 and split in cultures with and without 0.3 μg/ml mupirocin. Growth was monitored after reaching OD600 = 0.3 without (B) or with mupirocin (C).For Northern analysis (D) RNA was isolated from bacteria 30 minutes after reaching OD600 = 0.3 and hybridized with digoxigenin-labelled probes specific for gene encoding ribosomal protein RpsL and the CodY target gene SAOUHSC_02923. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the bottom lane.
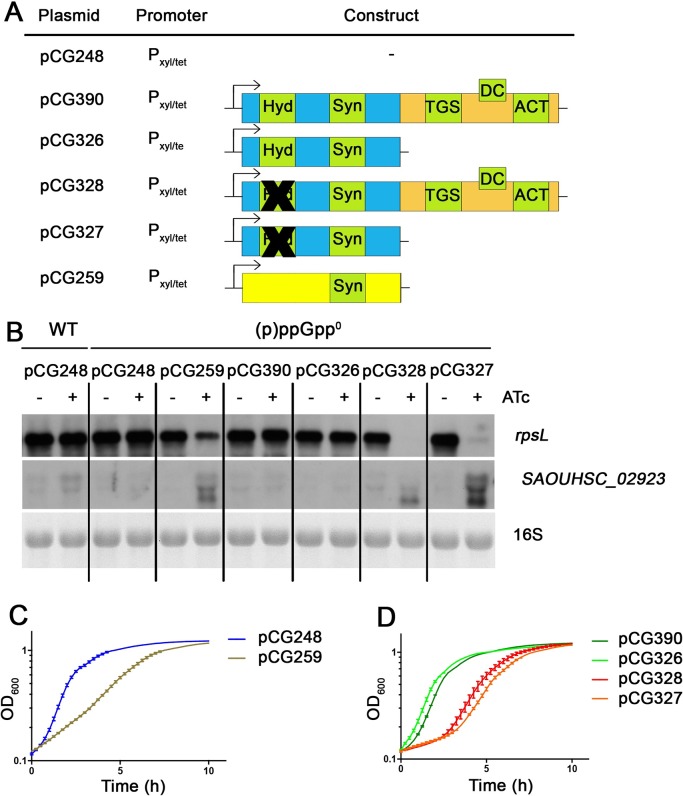
Next, we complemented the (p)ppGpp0 strain with anhydrotetracycline (ATc)-inducible full-length and truncated rel constructs (Fig 2A) and analyzed the effects under relaxed growth conditions (exponential growth phase in nutrient-rich medium). As a positive control, we induced relQ expression. RelQ is a small synthetase without a regulatory CTD and thus can activate the stringent response in a (p)ppGpp0 mutant by transcriptional induction alone. This activation was demonstrated by downregulation of rpsL and upregulation of SAOUHSC_02923 (Fig 2B) and by the immediate growth arrest after relQ induction (Fig 2C).
Fig 2. Influence of the CTD on the synthetase activity in vivo.
ATc inducible rel with and without CTD or hydrolase domain and relQ (A) were expressed in the (p)ppGpp0 mutant and compared to WT HG001. The black crosses (A) represent mutations as indicated in Fig 1A. Strains were grown in rich medium to OD600 = 0.3 and then split in cultures with and without 0.1 μg/ml ATc. For Northern analysis (B) RNA was isolated from bacteria 30 minutes after reaching OD600 = 0.3 and hybridized with digoxigenin-labelled probes. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the bottom lane. rpsL and SAOUHSC_02923 are control markers for the induction by (p)ppGpp Growth was monitored after addition of 0.1 μg/ml ATc. The (p)ppGpp0 was complemented with different plasmids: empty vector (pCG248) and relQ (pCG259), as controls (C), and different rel constructs (D).
In contrast to relQ, transcriptional induction of full-length rel showed no effect on the transcription of marker genes or on growth (Fig 2B and 2D). This finding confirms that additional post-transcriptional activation is required to activate the synthetase activity. Induction of a construct lacking the CTD also failed to induce the stringent response phenotype (Fig 2B and 2D). At first glance, these results may indicate that under relaxed conditions, the enzymatic domain of RelSau, with or without the CTD, is tightly held in a synthetase-OFF conformation. Alternatively, the hydrolase might be hyperactive, so any (p)ppGpp synthesized would be immediately degraded. To test this hypothesis, we mutated the hydrolase domain in full-length and CTD-deleted rel constructs. Indeed, both full-length and truncated rel lacking the hydrolase domain elicited a stringent response pattern similar to that of the wild type, as indicated by transcriptional and growth analyses (Fig 2B and 2D). Thus, we presume that there might be some synthetase activity under relaxed growth conditions. However, due to hydrolase activity, any (p)ppGpp present is efficiently degraded under these conditions.
CTD of RelSau does not impact hydrolase activity in vivo
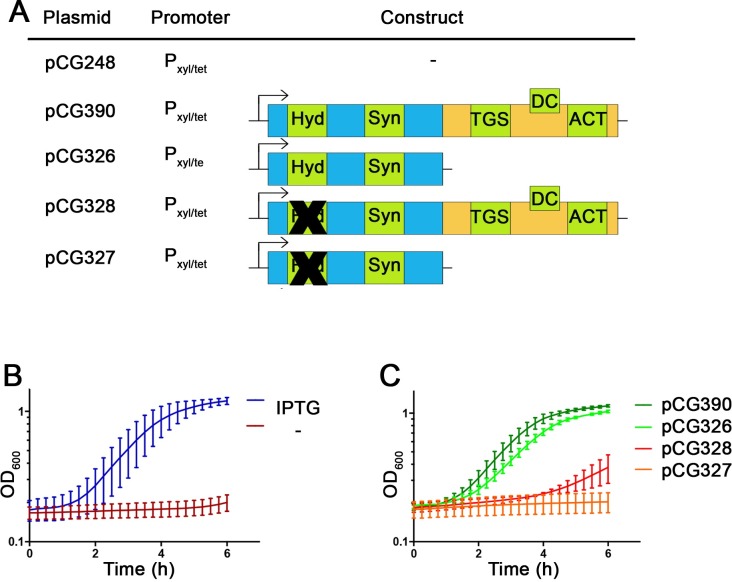
To analyze the hydrolase activity of RelSau in vivo, we used a conditional rel mutant strain (HG001-55) [18] in which genomic rel was placed under an IPTG-inducible promoter complemented with different rel constructs (Fig 3A). Without IPTG, the rel mutant is unable to grow (Fig 3B) because it cannot degrade the (p)ppGpp synthesized by RelP and RelQ [35]. We introduced ATc-inducible full-length or truncated rel into the conditional rel mutant and monitored growth after ATc induction. As expected, constructs with mutated hydrolase could not rescue the growth defect of HG001-55 (Fig 3C). However, full-length and CTD-truncated rel, with intact hydrolase, fully complemented the growth defect of the conditional rel mutant. These results show that the hydrolase was constitutively active, independent of the presence of the CTD. In summary, the data indicate that under relaxed conditions, the wild-type RelSau enzyme, with or without sensory domain, is tightly held in the hydrolase-ON state.
Fig 3. CTD of RelSau has no impact on hydrolase activity in vivo.
The conditional rel mutant HG001-55 (inducible by IPTG) was complemented with ATc inducible rel constructs (A). Strains were grown during preculture in presence of IPTG (0.5 mM) and were diluted to an initial OD600 = 0.1 and growth monitored over time. As control, strain complemented with empty vector was grown with and without IPTG to illustrate the essentially of the Rel hydrolase (B). Rel constructs were grown without IPTG but addition of ATc (0.1 μg/ml) (C).
Role of the CTD in the enzymatic activity of RelSau in vitro
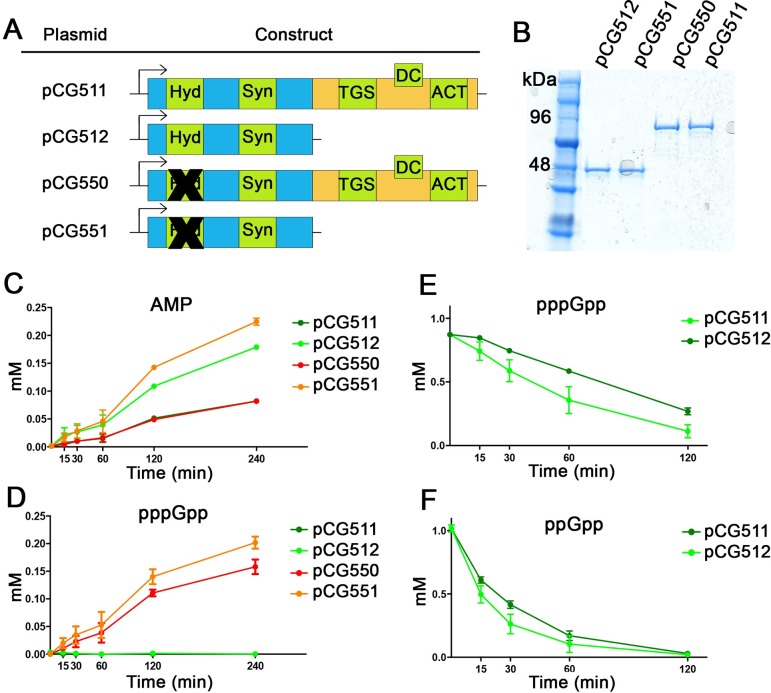
To confirm the data from the in vivo experiments under relaxed conditions, full-length or CTD-truncated RelSau proteins (with or without hydrolase domains) were purified and tested in vitro for enzymatic activities (Fig 4A and 4B). In the synthetase reaction, pyrophosphate is transferred from ATP to GTP, yielding AMP and pppGpp. The presence of both products was measured by HPLC-MS. AMP production was detectable with all constructs (Fig 4C); however, the AMP levels were significantly higher for the constructs that lacked the CTD (Fig 4C), indicating that the CTD negatively interferes with synthetase activity. Interestingly, pppGpp production was not detected for constructs with intact hydrolase (Fig 4D). However, proteins with mutated hydrolases synthesized detectable amounts of (p)ppGpp. The enzyme lacking the CTD showed slightly higher pppGpp synthetase activity than the full-length RelSau supporting the inhibitory effect of the CTD on the synthetase domain. Thus, RelSau exhibits strong hydrolase activity, which prevents pppGpp accumulation. This finding was confirmed by the rapid degradation of pppGpp (Fig 4E) and ppGpp (Fig 4F) by full-length and CTD deleted RelSau. Notably, RelSau preferentially degraded ppGpp over pppGpp. The CTD apparently has a minor impact on hydrolase activity.
Fig 4. Role of the CTD on the enzymatic activity in vitro.
2 μM of purified proteins with and without CTD or hydrolase domain (A) were loaded on SDS-PAGE, RelSau (85 KDa) and CTD-deleted (48KDa) (B) and used to assay synthetase (C, D) and hydrolase activity (E, F). The black crosses (A) represent mutations as indicated in Fig 1A. Synthetase activity was determined in the presence of the two substrates ATP and GTP and 2 μM of enzymes. The reaction products AMP (C) and pppGpp (D) were monitored over time at 37°C by HPLC-MS. Hydrolase activity was assayed in the presence of pppGpp (E) or ppGpp (F) and 0.1 μM of enzymes. Decrease of 1 mM of pppGpp or ppGpp was monitored for 120 minutes at 37°C.
Role of the CTD of RelSau in E. coli
Our analysis of RelSau in its native background seemed to be inconsistent with the results of previous studies, in which different CTD-deleted enzymes from other organisms were expressed in E. coli [20,21,44,45]. These studies indicated that RSH enzymes that lack CTDs are in a synthetase-ON/hydrolase-OFF state. Thus, based on these studies, we also expressed full-length and CTD-deleted rel in E. coli using an arabinose-inducible promoter. We tested the capacity of different rel constructs (Fig 5A) to complement the defective phenotype of MG1655, a relA/spoT mutant, under stringent conditions (Fig 5B and 5C). Full-length and CTD-deleted RelSau were able to complement the relA/spoT mutation. The complementation could be attributed to (p)ppGpp synthetase activity: mutation within the synthetase domain abolished complementation, whereas mutation within the hydrolase domain did not affect the complementation assay. Thus, in E. coli, CTD-deleted RelSau, similar to other RSH enzymes, is predominantly in a synthetase-ON/hydrolase-OFF state, whereas in S. aureus, this enzyme is primarily in a hydrolase-ON state.
Fig 5.
Role of CTD of RelSau in E. coli. rel from S. aureus with and without CTD, hydrolase or synthetase domain (A) were expressed in relA/spoT mutant of E. coli MG1655. The black crosses (A) represent mutations as indicated in Fig 1A. As controls WT and relA/spoT mutant were transformed with empty vector (B and C). Strains were grown under stringent conditions streaked on M9 agar plates (C) or M9 medium for growth analysis (D). Under these conditions the relA/spoT mutant was unable to grow.
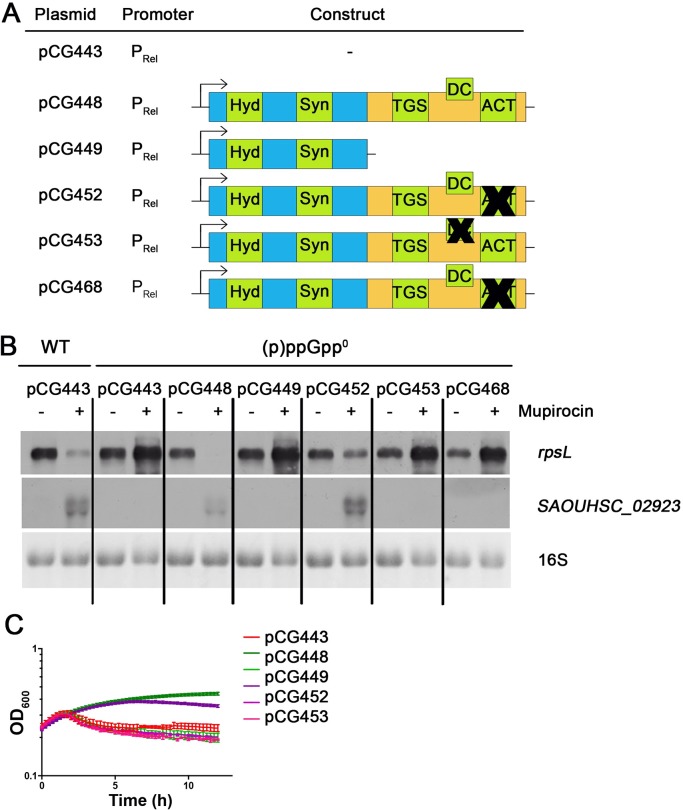
Role of CTD motifs for sensing in vivo
Within the CTDs of RSHs, several conserved motifs can be identified. The conserved TGS, DC and ACT motifs of RelSau were predicted based on sequence alignments, and the critical residues of these motifs were mutated (Fig 1A). Wild-type and CTD-mutated rel were cloned to be under the control of the native rel promoter and introduced into the (p)ppGpp0 strain (Fig 6A). The stringent response upon mupirocin treatment was analyzed by Northern blotting and growth analysis (Fig 6B and 6C). A (p)ppGpp0 strain containing the empty vector showed the typical decrease in OD600 after mupirocin treatment. Induction of full-length rel in the (p)ppGpp0 mutant fully complemented the mutant phenotype, whereas the CTD-deleted rel was unable to do so. Mutation of the ACT motif resulted in slightly impaired complementation. However, mutation of the TGS or DC motif resulted in complete inactivation of the stringent response. Expression of these mutated rel genes resulted in a phenotype that was not distinguishable from the phenotype of the (p)ppGpp0 strain in terms of growth and gene expression pattern. Thus, the TGS and DC motifs are required for stringent response, while the ACT motif plays only a minor role.
Fig 6. Role of CTD motifs for sensing of RelSau in vivo.
(p)ppGpp0 mutant was complemented with different rel constructs expressed from the native promoter (A). The black crosses (A) represent mutations as indicated in Fig 1A. Strains were grown in rich medium to OD600 = 0.3 and then split in cultures with and without 0.3 μg/ml mupirocin. For Northern analysis (B) RNA was isolated from cells 30 minutes after reaching OD600 = 0.3 and hybridized with digoxigenin-labeled probes. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the bottom lane. Growth was monitored after reaching OD600 = 0.3 after which 0.3. μg/ml mupirocin was added (C).
Co-immunoprecipitation (Co-IP) of RelSau
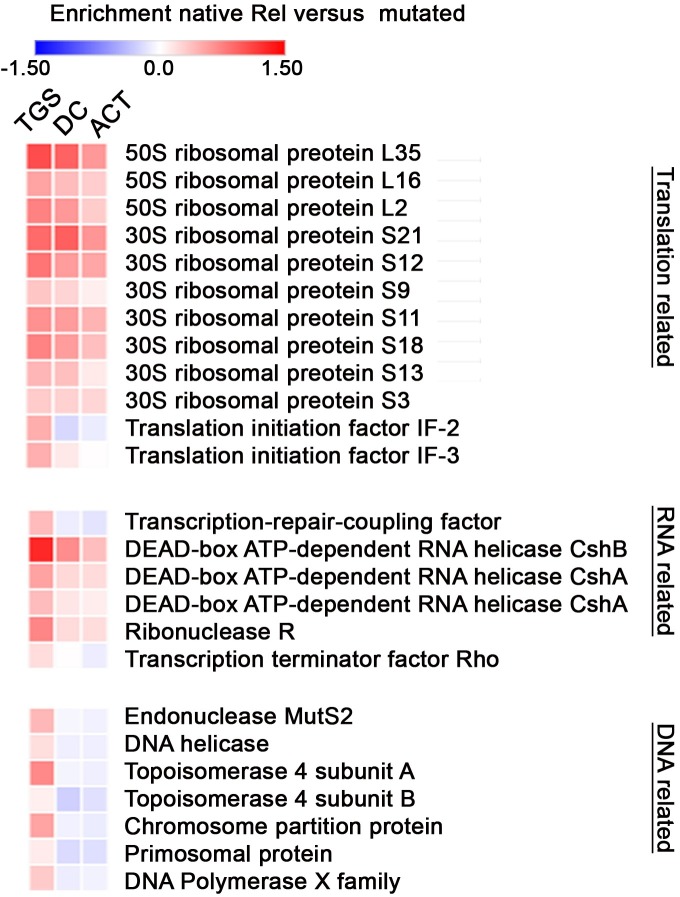
We aimed to analyze the role of the conserved motifs within the CTD for interaction with cytosolic proteins. Therefore, we performed Co-IP experiments using whole-cell lysates of (p)ppGpp0 mutants expressing wild-type or mutated (ACT, DC and TGS see Fig 1A) versions of RelSau. For each pull-down experiment, the wild-type or mutant RelSau was the most abundant protein detected, with no significant difference observed between wild type and mutant proteins (Data S1 Dataset) and the expression of all proteins was similar as shown by Western blot analysis (Blot in S1 Fig). Mainly ribosomal proteins were co-immuno-precipitated with native RelSau. When RelSau with mutated TGS motif was used as bait significant less proteins were enriched (Fig 7 first column). Most of these putative TGS interacting proteins were also found to be effected when RelSau harboring mutations in ACT or DC motifs were used, although to a lesser extent. Immuno-precipitated proteins that were strongly influenced by the TGS mutation are ribosomal proteins, proteins associated with RNA degradation and proteins involved in DNA-related pathways. In summary, the results indicate that all three motifs within the CTD work together to dock RelSau onto the translational apparatus. The strongest interaction is mediated by TGS, whereas the ACT motif seems to have a low impact. The TGS motif seems to mediate also interaction with non-ribosomal proteins.
Fig 7. Influence of the CTD motifs on RelSau interactions.
S. aureus (p)ppGpp0 was complemented with native and mutated RelSau. Cell lysates were mixed with magnetic beads, coated with RelSau antibodies and enriched proteins identified by MS. Proteins significantly (t-test difference > 0.05) less abundant using TGS mutated versus native RelSau as bait are shown in the first column. The effects of the DC or ACT mutation on interaction with these proteins are shown in the second and third column, respectively.
Discussion
RSH enzymes are major players in the synthesis and hydrolysis of the second messenger (p)ppGpp. There is still limited information about the molecular switch that regulates the two activities, both present in long RSH enzymes. Here, we analyzed how the CTD of RelSau influences the enzymatic activities in vivo. We showed that RelSau exists primarily in a synthetase-OFF/hydrolase-ON conformation. Only under stringent growth conditions was the switching to the synthetase-ON conformation detectable, and this switching occurred only when the CTD possessed intact TGS and DC motifs.
In S. aureus and probably in other Firmicutes, Rel combines the functions of the two prototypic RSH enzymes, RelA and SpoT, from Proteobacteria. The synthetase activity is needed to elicit a stringent response phenotype, presumably via interaction with ribosomes and uncharged tRNA, as previously shown for RelA [24,25]. However, similar to SpoT, RelSau also possesses strong hydrolase activity, which is necessary to counteract the (p)ppGpp production by the small synthetases RelP and RelQ present in Firmicutes. The equilibrium between these two activities needs to be tightly regulated in order to attain an appropriate level of (p)ppGpp based on the growth conditions. So far, potential differences between RelA, SpoT and Rel associated with the molecular switch could not be inferred from the sequence or in vitro analyses. In vivo activities of different RSH enzymes were mainly analyzed by heterologous expression in E. coli. These analyses indicated that without CTDs, RSH enzymes possess strong synthetase activity [21,44,45]. Similarly, RelSau, with or without the CTD, can complement an E. coli relA/spoT mutant, also indicating that (p)ppGpp synthesis can occur with or without the CTD. However, analysis of the same construct in the native background clearly showed that RelSau lacking the CTD is tightly held in the hydrolase-ON state, and synthetase activity is detectable only in constructs that lack the hydrolase. Thus, RelSau, with or without the CTD, exhibits net (p)ppGpp synthetase activity when expressed in E. coli but net hydrolase activity when expressed in S. aureus. It would be interesting to see whether enzymes from other organisms show a similar discrepancy between E. coli and native backgrounds. Our results indicated that the enzymatic activity of RelSau is influenced by species-specific interactions of the enzymatic NTD with unknown factors. To date, there is no evidence that the NTD alone interacts with the ribosome. Thus, other interaction partners or intracellular properties of the NTD should be elucidated in the future. An alternative possibility is that less (p)ppGpp is needed to complement the phenotype of a pppGpp mutant in E. coli allowing growth even if RelSau has a weak synthetase. However, this is not supported by our in vitro results, showing that synthetase activity is only detectable when the hydrolase is mutated in constructs with or without CTD. Analysis of full-length or truncated RelSau in vitro largely confirmed the results obtained with the in vivo data obtained in S. aureus. The presence of an intact hydrolase abrogates the synthetase activity. Synthesis of pppGpp was detectable only in hydrolase-deficient constructs. Moreover, we show that the CTD has an inhibitory effect on synthetase activity since truncated versions of RelSau showed higher accumulation of the reaction products AMP and pppGpp compared to the full-length enzyme. However, the CTD had only a minor impact on the strong hydrolase activity of the purified enzymes. Interestingly, RelSau preferentially hydrolyzes ppGpp over pppGpp. In vivo, it was shown that RelP and RelQ mainly produce ppGpp [35], which is toxic at high concentrations and requires efficient hydrolysis. This observation could explain the preference of RelSau for ppGpp hydrolysis.
The Co-IP results indicate that RelSau interacts with the translation machinery and that the TGS strongly influence this interaction. This is largely consistent with previous data obtained for RelA [24,25,28,29,30]. Among the 10 detected interacting ribosomal proteins, L16, S13, and S12 are homologous to E. coli proteins that have been previously identified to interact with RelA [28,29,30]. Of note, the TGS motif also seems to hamper the putative interaction of RelSau with other proteins of the RNA and DNA pathways. Whether such interactions are specific and involved in the molecular function of RelSau remains to be investigated.
The in vivo analyses combined with the Co-IP results provided some clues regarding the roles of the different motifs of the CTD of RelSau. Of the three motifs, the TGS motif showed the strongest effect, and the ACT motif showed the weakest effect, on (p)ppGpp activation and ribosomal interaction. Thus, the role of the ACT motif remains to be elucidated but seems to be minor. The TGS motif is clearly required for synthetase activation, most likely interacts with the ribosome to sense whether or not the tRNA in the A-site is aminoacetylated as shown for RelA [28,29,30,46]. Similar to the TGS motif, the DC motif was also found to be required for synthetase activity and to influence interactions with ribosomal proteins. DC has also been reported to interact with 23S rRNA ASF [28] which is critical to RelA activation in E. coli [46], presumably through stabilizing ribosome interaction.
This finding contradicts the simple model in which the DC motif causes oligomerization and thereby autoinhibition [31,32,33]. This would imply that DC mutation alleviates autoinhibition leading to increased synthase activity. In contrast, our data showed that the DC-mutated RelSau is held in a synthetase-OFF state. Thus, our data support a model in which the DC motif participates in specific activation upon ribosomal contact, and that this interaction is involved in the intramolecular switch.
Materials and methods
Strains and growth conditions
Strains and plasmids are listed in the table in S1 Table. For strains carrying resistance genes, antibiotics (10 μg/ml erythromycin, 5 μg/ml tetracycline, 10 μg/ml chloramphenicol, and 100 μg/ml ampicillin) were used only in precultures. For the conditional mutant HG001-55, IPTG (final concentration of 0.5 mM) was added only in the preculture. S. aureus strains were grown in CYPG (10 g/l casamino acids, 10 g/l yeast extract, 5 g/l NaCl, 0.5% glucose and 0.06 M phosphoglycerate) medium [47]. Bacteria from an overnight culture were diluted to an initial optical density (OD600) of 0.05 in fresh medium and grown with shaking (220 rpm) at 37°C to the desired growth phase. Expression of cloned proteins was induced in exponential phase (OD600 = 0.3) with 0.1 μg/ml anhydrotetracycline (ATc) and in stringent conditions by addition of 0.3 μg/ml mupirocin. E. coli strains were grown in an overnight preculture in LB medium. Stringent conditions were applied by growing cells in modified M9 medium (33.7 mM NaHPO4, 22 mM KH2PO4, 8.55 mM NaCl, 9.35 mM NH4Cl, 1 mM MgSO4, 0.3 mM CaCl2, 1 μg/ml thiamine hydrochloride, 0.4% glycerol, 1 mM serine, 1 mM methionine and 1 mM glycine) [48]. For growth on solid media, single colonies grown on LB agar were streaked on M9 agar plates. For growth curve analyses, bacteria were inoculated to the desired OD600 (S. aureus initial OD600 = 0.05; E. coli initial OD600 = 0.1) in a 96-well plate, and growth was monitored in an Infinite M200 Pro microplate reader (Tecan).
Construction of vectors for expression of full-length and truncated rel
All oligonucleotides are listed in S2 Table. ATc-inducible plasmids, derived from pCG248, were generated with a restriction enzyme cloning strategy. Amplicons and vector were digested with EcoRI restriction enzyme. Substitution of the hydrolase domain and the ACT, TGS and DC mutations were achieved by overlapping PCR. For expression of the rel constructs under the native promoter, the shuttle vector pCG443 was designed based on pJL77 [49]. pJL77 was digested with AscI and SphI to remove the previous insert, including the promoter. The rel promoter was amplified from genomic DNA, digested using the same restriction enzymes, and ligated to generate pCG443. Full-length, truncated and mutated versions of rel were subcloned from the pCG248 plasmids into AscI-digested pCG443 by Gibson assembly [50].
All inserts were verified by sequencing (4base lab AG advanced molecular analysis), electroporated into the restriction-deficient S. aureus strain RN4220, and then transduced into the final S. aureus strains. All S. aureus strains were tested by PCR for the presence of the correct plasmid.
For expression in E. coli, different derivatives of rel were cloned into the EcoRI site of pBAD30 via Gibson assembly using the oligonucleotides listed in Table S2. Resulting vectors were verified by sequencing and moved to MG1655 E. coli strains (wild type and relA/spoT mutant) [51,52].
For protein purification, different rel derivatives were subcloned from the pCG248-based plasmids into BamHI-digested pET15b using Gibson assembly.
Generation of S. aureus mutant strains
The markerless rel CTD-deletion mutant was obtained using the ATc-inducible suicide vector pBASE6 [34]. Deletion was introduced by overlapping PCR with the primers listed in S2 Table, and the amplicon was cloned into BglII- and SalI-digested pBASE6 by Gibson assembly. The resulting plasmid was verified by sequencing and electroporated into RN4220, from which the plasmid was transduced into HG001. Mutagenesis was performed as described previously [34]. Mutation was verified by PCR.
RNA isolation and Northern blot analysis
RNA isolation and Northern blot analysis were performed as described previously [53]. Briefly, 5 ml of bacteria were collected at the desired time point (30 minutes after induction) and centrifuged. The pellet was resuspended in 1 ml of TRIzol reagent (Thermo Fisher Scientific) with 0.5 ml of zirconia/silica beads (0.1-mm diameter) and lysed using a high-speed homogenizer (Thermo Fisher Scientific). RNA was isolated following the instructions provided by the TRIzol manufacturer. For the detection of specific transcripts on the Northern blot, digoxigenin-labeled probes were generated using the DIG-labeling PCR Kit as described by the manufacturer (Roche Life Science).
Purification of different RelSau constructs
E. coli BL21 (DE3) (New England Biolabs) cells that were freshly transformed with plasmids carrying full-length rel constructs were grown for 16 hours at room temperature under constant shaking (150 rpm) in LB medium supplemented with D(+)-lactose-monohydrate (12.5 g/l) and ampicillin (100 μg/ml). Cells were harvested (20 minutes, 3000 x g, 4°C) and resuspended in ice-cold high-KCl buffer A (20 mM HEPES (pH 7.4), 20 mM NaCl, 20 mM MgCl2, 1 M KCl, 30% (v/v) glycerol, and 40 mM imidazole) supplemented with 10 μg/ml DNAse and cOmplete protease inhibitor cocktail (Roche). Cells were lysed by a French press at 1000 psi. The lysate was centrifuged (50,000 x g, 45 minutes, 4°C), and the clear supernatant was filtered (0.22-μm pore size) before being loaded onto a 1-ml HisTrap HP column (GE Healthcare Life Sciences) equilibrated with high-KCl buffer A. Purification was performed with an ÄKTA purification system (GE Healthcare Life Sciences), and elution was carried out with an imidazole gradient to a final concentration of 500 mM. Fractions were analyzed by SDS-PAGE, and the fractions containing the protein of interest were collected and concentrated to 5 ml with an Amicon Ultracel-50K ultracentrifugal device, with a cut-off of 50 kDa (Merck Millipore). Protein was further purified by size-exclusion chromatography (HiLoad 16/600 Superdex 200 pg, GE Healthcare Life Sciences). The size-exclusion column was previously equilibrated with ice-cold high-KCl SEC buffer (20 mM HEPES (pH 7.0), 20 mM NaCl, 20 mM MgCl2, 1 M KCl, and 30% (v/v) glycerol). Protein-containing fractions were pooled, concentrated by ultra-filtration with a 50-kDa cut-off, aliquoted and stored at -80°C. For purification of CTD-truncated constructs, the same procedure was followed using different buffers: low-KCl buffer A (20 mM HEPES (pH 7.4), 200 mM NaCl, 20 mM MgCl2, 20 mM KCl, 30% (v/v) glycerol, and 40 mM imidazole) for affinity purification and low-KCl SEC buffer (20 mM HEPES (pH 7.0), 200 mM NaCl, 20 mM MgCl2, 20 mM KCl, and 30% (v/v) glycerol) for size exclusion. For concentration of truncated RelSau, Amicon Ultracel-30K (Merck Millipore) was used.
In vitro assay for enzymatic activity and HPLC-MS analysis
Synthetase assays were performed in reaction buffer (20 mM HEPES (pH 7.0), 200 mM NaCl, 20 mM MgCl2, and 20 mM KCl) with 1 mM ATP, 1 mM GTP and 2 μM purified enzyme. Hydrolase assays were performed in the same reaction buffer with 1 mM ppGpp or 1 mM pppGpp (both from Jena Biosciences) and 0.1 μM purified enzyme. Assays were performed at 37°C; aliquots were taken at the indicated times; and the enzyme reactions were stopped by addition of an equal volume of chloroform. The mixtures were briefly vortexed and centrifuged (3 minutes, 11,000 × g). The aqueous phase containing the nucleotides was collected and stored at -20°C prior to analysis. Nucleotide analysis was performed using an ESI-TOF mass spectrometer (micrO-TOF II, Bruker) operated in negative-ion mode and connected to an UltiMate 3000 high-performance liquid chromatography (HPLC) system (Dionex). 5 μl of each sample at 10°C was injected onto the SeQuant ZIC-pHILIC column (Merck, PEEK 150 × 2.1 mm, 5 μm), and the system was run at 30°C as previously described [43]. The following 40-minute gradient program was used at a flow rate of 0.2 ml/min: 5 minutes of 82% buffer A (CH3CN) and 18% buffer B (100 mM (NH4)2CO3, pH 9.2); 25 minutes of a linear gradient to 42% buffer A; and finally, 10 minutes of 82% buffer A. The DataAnalysis program (Bruker) was used to present the nucleotide masses as extracted-ion chromatograms, and the peak areas were calculated and quantified with Prism 5 (GraphPad). Dilution series of commercially available nucleotides ppGpp (m/z, 601.95), pppGpp (m/z, 681.92) and AMP (m/z, 346.06) were used for calibration to quantify the amounts of nucleotides in the reactions.
Co-IP of RelSau with native S. aureus proteins
To generate RelSau-specific antibodies, 0.5 mg of purified full-length protein was sent to Davids Biotechnologie GmbH to generate antiserum and affinity-purified IgG. The specificity of the IgG was verified by Western blot analysis (Blot in S1 Fig). For Co-IP, bacteria were grown in 100 ml of CYPG medium to an OD600 of 1 and centrifuged (5,000 μ g, 5 minutes). The pellet was washed 2 times with PBS and resuspended in 500 μl of cold Co-IP buffer (20 mM HEPES (pH 7.0), 200 mM NaCl, 20 mM MgCl, 20 mM KCl, 0.5 mM DTT, 0.2% (v/v) Tween 20 and cOmplete protease inhibitor cocktail). The resuspended pellet was lysed with 0.5 ml of zirconia-silica beads (0.1 mm diameter) using a high-speed homogenizer (two times, 6,500 rpm, 20 s). Lysed cells were centrifuged for 1 hour at 14,000 x g at 4°C, and the supernatant was aliquoted (100 μl) and frozen at -80°C. Co-IP was performed with Dynabeads (Thermo Fisher Scientific) following the manufacturer’s instructions with some minor modifications. Briefly, 50 μl of Dynabeads slurry was used for each sample. The storage solution was removed, and the beads were incubated with 30 μg of Anti-RelSau IgG resuspended in PBS (pH 7.4) with 0.02% Tween 20 for 30 minutes at room temperature under constant rotation. Coated beads were pelleted using a magnetic rack; the supernatant was removed; and 100 μl of the cell lysates were added and incubated for 30 minutes at room temperature under constant rotation. After incubation, the beads were gently washed 3 times with Co-IP buffer using a magnetic rack. Washing solution was removed, and the beads were resuspended in SDS sample buffer, boiled at 95°C for 5 minutes, and run approximately 1 cm into an SDS-PAGE gel. The gel slice was subsequently analyzed by mass spectrometry.
Quantitative label-free proteomics
Three biological replicates of (p)ppGpp0 complemented with WT and mutant RelSau were analyzed. Gel slices were digested as described previously [54]. Peptide mixtures were then separated on an EasyLC nano-HPLC (Proxeon Biosystems) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) as described elsewhere [55] with the following modifications: peptides were eluted with an 87-min segmented gradient of 5–33–90% HPLC solvent B (80% acetonitrile in 0.5% acetic acid). Each sample was run in triplicate. The acquired MS spectra were processed with the MaxQuant software package, version 1.5.2.8 [56] with the integrated Andromeda search engine [57] as described previously [55]. Database searches were performed against a target-decoy S. aureus all-strains database obtained from UniProt, containing 126,225 protein entries and 248 commonly observed contaminants. The label-free algorithm was enabled, as was the “match between runs” option [58]. Label-free quantification (LFQ) protein intensities from the MaxQuant data output were used for relative protein quantification. Downstream bioinformatic analysis (ANOVA and two-sample t-tests) was performed using the Perseus software package, version 1.5.0.15. P < 0.05 was considered to be statistically significant. For the heatmap, among the 4 different proteins, those that showed significant differences according to ANOVA were selected (Data in S1 Dataset). For these selected candidates, the t-test differences, indicating changes in the amount, were calculated between the protein immunoprecipitated with WT or mutant RelSau and plotted on the heatmap.
Statistical analysis
The results for the growth and in vitro analyses represent the mean ± SD of at least three biological replicates. Significance was calculated using Prism 5 by one-way ANOVA with Bonferroni correction.
Supporting information
S. aureus (p)ppGpp0 complemented with RelSau, wild type and different domain mutants, under the native promoter. Strains were grown in rich medium to OD600 = 1 and then harvested. For Worthern analysis, lysate was obtained and the different RelSau constructs were detected with anti-Rel specific antibody.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Acknowledgments
We thank Natalya Korn, Isabell Samp and Vittoria Bisanzio for excellent technical assistance. We thank Elsa Germain–Maisonneuve for providing E. coli strains and vectors.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), www.dfg.de: TR34/B1 to CW; DFG SFB766/A15, and GRK1708/B2 to CM; SFB766/Z02 to BM; SPP1879 to GB, SFB766 to SW. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1.Cashel M (1975) Regulation of bacterial ppGpp and pppGpp. Annual review of microbiology 29: 301–318. 10.1146/annurev.mi.29.100175.001505 [DOI] [PubMed] [Google Scholar]
- 2.Chatterji D, Ojha AK (2001) Revisiting the stringent response, ppGpp and starvation signaling. Current opinion in microbiology 4: 160–165. [DOI] [PubMed] [Google Scholar]
- 3.Wagner R (2002) Regulation of ribosomal RNA synthesis in E. coli: effects of the global regulator guanosine tetraphosphate (ppGpp). J Mol Microbiol Biotechnol 4: 331–340. [PubMed] [Google Scholar]
- 4.Gralla JD (2005) Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Molecular microbiology 55: 973–977. 10.1111/j.1365-2958.2004.04455.x [DOI] [PubMed] [Google Scholar]
- 5.Magnusson LU, Farewell A, Nystrom T (2005) ppGpp: a global regulator in Escherichia coli. Trends in microbiology 13: 236–242. 10.1016/j.tim.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 6.Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J (2006) New horizons for (p)ppGpp in bacterial and plant physiology. Trends in microbiology 14: 45–54. 10.1016/j.tim.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 7.Jain V, Kumar M, Chatterji D (2006) ppGpp: stringent response and survival. J Microbiol 44: 1–10. [PubMed] [Google Scholar]
- 8.Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annual review of microbiology 62: 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 9.Srivatsan A, Wang JD (2008) Control of bacterial transcription, translation and replication by (p)ppGpp. Current opinion in microbiology 11: 100–105. 10.1016/j.mib.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Xie J (2009) Magic spot: (p) ppGpp. J Cell Physiol 220: 297–302. 10.1002/jcp.21797 [DOI] [PubMed] [Google Scholar]
- 11.Wolz C, Geiger T, Goerke C (2010) The synthesis and function of the alarmone (p)ppGpp in firmicutes. International journal of medical microbiology: IJMM 300: 142–147. 10.1016/j.ijmm.2009.08.017 [DOI] [PubMed] [Google Scholar]
- 12.Cashel M (1969) The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. The Journal of biological chemistry 244: 3133–3141. [PubMed] [Google Scholar]
- 13.Battesti A, Bouveret E (2009) Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. Journal of bacteriology 191: 616–624. 10.1128/JB.01195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battesti A, Bouveret E (2006) Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Molecular microbiology 62: 1048–1063. 10.1111/j.1365-2958.2006.05442.x [DOI] [PubMed] [Google Scholar]
- 15.Atkinson GC, Tenson T, Hauryliuk V (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PloS one 6: e23479 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittenhuber G (2001) Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotechnol 3: 585–600. [PubMed] [Google Scholar]
- 17.Steinchen W, Bange G (2016) The magic dance of the alarmones (p)ppGpp. Molecular microbiology 101: 531–544. 10.1111/mmi.13412 [DOI] [PubMed] [Google Scholar]
- 18.Geiger T, Goerke C, Fritz M, Schafer T, Ohlsen K, et al. (2010) Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infection and immunity 78: 1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendrich TM, Marahiel MA (1997) Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Molecular microbiology 26: 65–79. [DOI] [PubMed] [Google Scholar]
- 20.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R (2004) Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117: 57–68. [DOI] [PubMed] [Google Scholar]
- 21.Mechold U, Murphy H, Brown L, Cashel M (2002) Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. Journal of bacteriology 184: 2878–2888. 10.1128/JB.184.11.2878-2888.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, et al. (1991) Overexpression of the relA gene in Escherichia coli. The Journal of biological chemistry 266: 3760–3767. [PubMed] [Google Scholar]
- 23.Jain V, Saleem-Batcha R, China A, Chatterji D (2006) Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci 15: 1449–1464. 10.1110/ps.062117006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haseltine WA, Block R (1973) Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proceedings of the National Academy of Sciences of the United States of America 70: 1564–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH (2002) Dissection of the mechanism for the stringent factor RelA. Mol Cell 10: 779–788. [DOI] [PubMed] [Google Scholar]
- 26.Chipman DM, Shaanan B (2001) The ACT domain family. Current Opinion in Structural Biology 11: 694–700. [DOI] [PubMed] [Google Scholar]
- 27.Grant GA (2006) The ACT domain: a small molecule binding domain and its role as a common regulatory element. J Biol Chem 281: 33825–33829. 10.1074/jbc.R600024200 [DOI] [PubMed] [Google Scholar]
- 28.Loveland AB, Bah E, Madireddy R, Zhang Y, Brilot AF, et al. (2016) Ribosome*RelA structures reveal the mechanism of stringent response activation. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown A, Fernandez IS, Gordiyenko Y, Ramakrishnan V (2016) Ribosome-dependent activation of stringent control. Nature 534: 277–280. 10.1038/nature17675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arenz S, Abdelshahid M, Sohmen D, Payoe R, Starosta AL, et al. (2016) The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic acids research 44: 6471–6481. 10.1093/nar/gkw470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gropp M, Strausz Y, Gross M, Glaser G (2001) Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. Journal of bacteriology 183: 570–579. 10.1128/JB.183.2.570-579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, et al. (2005) Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44: 9913–9923. 10.1021/bi0505316 [DOI] [PubMed] [Google Scholar]
- 33.Yang X, Ishiguro EE (2001) Dimerization of the RelA protein of Escherichia coli. Biochem Cell Biol 79: 729–736. [DOI] [PubMed] [Google Scholar]
- 34.Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, et al. (2012) The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS pathogens 8: e1003016 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiger T, Kastle B, Gratani FL, Goerke C, Wolz C (2014) Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. Journal of bacteriology 196: 894–902. 10.1128/JB.01201-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinchen W, Vogt MS, Altegoer F, Giammarinaro PI, Horvatek P, et al. (2018) Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ. Scientific reports 8: 2195 10.1038/s41598-018-20634-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinchen W, Schuhmacher JS, Altegoer F, Fage CD, Srinivasan V, et al. (2015) Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proceedings of the National Academy of Sciences of the United States of America 112: 13348–13353. 10.1073/pnas.1505271112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manav MC, Beljantseva J, Bojer MS, Tenson T, Ingmer H, et al. (2018) Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP. The Journal of biological chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu K, Bittner AN, Wang JD (2015) Diversity in (p)ppGpp metabolism and effectors. Current opinion in microbiology 24: 72–79. 10.1016/j.mib.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaca AO, Colomer-Winter C, Lemos JA (2015) Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. Journal of bacteriology 197: 1146–1156. 10.1128/JB.02577-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiger T, Wolz C (2014) Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. International journal of medical microbiology: IJMM 304: 150–155. 10.1016/j.ijmm.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 42.Krasny L, Gourse RL (2004) An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. The EMBO journal 23: 4473–4483. 10.1038/sj.emboj.7600423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastle B, Geiger T, Gratani FL, Reisinger R, Goerke C, et al. (2015) rRNA regulation during growth and under stringent conditions in Staphylococcus aureus. Environmental microbiology 17: 4394–4405. 10.1111/1462-2920.12867 [DOI] [PubMed] [Google Scholar]
- 44.Bag S, Das B, Dasgupta S, Bhadra RK (2014) Mutational analysis of the (p)ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. Archives of Microbiology 196: 575–588. 10.1007/s00203-014-0996-9 [DOI] [PubMed] [Google Scholar]
- 45.He P, Deng C, Liu B, Zeng L, Zhao W, et al. (2013) Characterization of a bifunctional enzyme with (p)ppGpp-hydrolase/synthase activity in Leptospira interrogans. FEMS microbiology letters 348: 133–142. 10.1111/1574-6968.12279 [DOI] [PubMed] [Google Scholar]
- 46.Kudrin P, Dzhygyr I, Ishiguro K, Beljantseva J, Maksimova E, et al. (2018) The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic acids research 46: 1973–1983. 10.1093/nar/gky023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novick RP (1991) Genetic systems in staphylococci. Methods Enzymol 204: 587–636. [DOI] [PubMed] [Google Scholar]
- 48.Neidhardt FC, Bloch PL, Smith DF (1974) Culture Medium for Enterobacteria. Journal of Bacteriology 119: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liese J, Rooijakkers SH, van Strijp JA, Novick RP, Dustin ML (2013) Intravital two-photon microscopy of host-pathogen interactions in a mouse model of Staphylococcus aureus skin abscess formation. Cellular microbiology 15: 891–909. 10.1111/cmi.12085 [DOI] [PubMed] [Google Scholar]
- 50.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 51.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, et al. (1991) Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. The Journal of biological chemistry 266: 5980–5990. [PubMed] [Google Scholar]
- 52.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- 53.Goerke C, Campana S, Bayer MG, Doring G, Botzenhart K, et al. (2000) Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infection and immunity 68: 1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burian M, Velic A, Matic K, Gunther S, Kraft B, et al. (2015) Quantitative proteomics of the human skin secretome reveal a reduction in immune defense mediators in ectodermal dysplasia patients. J Invest Dermatol 135: 759–767. 10.1038/jid.2014.462 [DOI] [PubMed] [Google Scholar]
- 55.Carpy A, Krug K, Graf S, Koch A, Popic S, et al. (2014) Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast). Mol Cell Proteomics 13: 1925–1936. 10.1074/mcp.M113.035824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26: 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 57.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, et al. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research 10: 1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 58.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, et al. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32: 279–289. 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. aureus (p)ppGpp0 complemented with RelSau, wild type and different domain mutants, under the native promoter. Strains were grown in rich medium to OD600 = 1 and then harvested. For Worthern analysis, lysate was obtained and the different RelSau constructs were detected with anti-Rel specific antibody.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.