Abstract
Background
Twin studies suggest that genetic factors may account for up to 50% increased risk for necrotizing enterocolitis (NEC), but genome-wide association studies (GWAS) for NEC are lacking.
Methods
Genotyping was done on Illumina BeadChip, followed by analysis using PLINK with logistic regression under an additive model.
Results
Among 751 extremely low birth weight (<1000g, >401g) neonates, 30 had surgical NEC. 261 single nucleotide polymorphisms (SNPs) showed association with NEC at P<0.05, of which 35 were significant at P<10−7. Minor allele(s) in a a cluster of SNPs spanning a 43 Kb region of chromosome 8 (8q23.3) conferred an odds ratio of 4.72 (95% CI 2.51-8.88) for elevated risk of NEC. Two smaller clusters on chromosome 14 and chromosome 11 exhibited P values 10−7 – 10−8. The chromosome 8 cluster is in an intergenic region between CUB And Sushi Multiple Domains 3 (−1.43 Mb) and Trichorhinophalangeal Syndrome I (+542 kb). RNA sequencing in this region identified a potential novel open reading frame corresponding to a long interspersed element-1 (LINE-1) retrotransposable element.
Conclusion
Genetic variation in an intergenic region of chromosome 8 is associacted with increased risk for NEC with a mechanismthat is yet to be identified.
INTRODUCTION
Necrotizing enterocolitis (NEC) is one of the most severe life-threatening complications of premature birth. The incidence of NEC in extremely preterm neonates (birth weight 401-1000g or gestational age 22-28 weeks) is approximately 9% (1). The mortality of the disease is approximately 30% in very low birth weight (VLBW) and approximately 50% in ELBW neonates (2). In addition to death in the acute phase, the most devastating long term sequelae of NEC are short bowel syndrome and neurodevelopmental impairment (2, 3).
The potential contribution of genetic predisposition to NEC has been considered in prior studies. Twin studies indicate that genetic factors may account for about 50% of the variance in liability for NEC, although adjusting for covariates negated statistical significance in this relatively small cohort (4). Certain SNPs such as in carbamyl phosphate synthetase (CPS1)(5), IL-12 (p40 promoter CTCTAA/GC)(6), VEGF (C-2578A)(7), and NFKB1(8) have been found to be associated with NEC. Other SNPs in TLR4 (A+896G, C+1196T), CD14, CARD15(9), platelet activating factor acetylhydrolase (PAFAH) (10), macrophage migration inhibitory factor (MIF)(11), mannose-binding lectin (12), angiotensin-converting enzyme (ACE) and ATR1166A/C (13), or other cytokines (14) have not been found to be associated with NEC. However, there are no reports to date of genome-wide association studies (GWAS) for necrotizing enterocolitis. Furthermore, with the exception of the ACE study, the aforementioned negative results were obtained from small cohorts, with low statistical power to detect differences.
There are >10 million SNPs in the human genome of which approximately 70% are in intergenic regions (15). Therefore, attempting to identify disesase-causing genetic variations by hypotheis-driven, targeted analysis of SNPs in specific genes is akin to searching for a needle in a haystack. With the increasing availability of information on variations in the human genome, genome-wide association studies (GWAS) became the most efficacious method to identify relationships between gentic variation and diseases (16).
Our objective was to identify genes and pathways associated with surgical NEC (Bell Stage III), compared to infants surviving without medical or surgical NEC. We found that genetic variations most significantly asoociated with increased risk of surgical NEC were located in a cluster of minor alleles in an intergenic region of chromosome 8 in the 8q23.3 region. Since there was no prior knowledge regarding any potential significance to this intergenic region, our further objectives were to perform in silico analysis to identify potential novel coding sequences or other potentially functional domains that might explain why genetic variations at this location would have physiological or pathological consequences.
METHODS
Cohort
Patients included were a subset of infants enrolled in the Eunice Kennedy Shriver NICHD Neonatal Research Network’s Cytokines study that enrolled infants weighing 401-1000 g at birth, < 72 h age, and free of major congenital anomalies (17). The study was approved by institutional review boards (IRBs) at participating centers, and written informed consent was obtained from parent(s). Additional IRB review was required to allow the federally-funded GWA genotyping results with a limited number of phenotype data to be included in the NHGRI Database of Genotypes and Phenotypes (DbGaP).
Isolation of DNA
DNA was extracted from the earliest age blood spot collected on filter paper. Whole genome amplification was used for samples that did not provide adequate genomic DNA. Genotyping was done on the Illumina HumanOmni1-Quad_v1-0_B BeadChip.
Definitions
NEC was defined as proven NEC (>=Bell Stage II (18, 19)). Surgical NEC was defined as placement of drain or performance of laparotomy (Bell Stage III). Importantly, we needed to exclude infants with Spontaneous Intestinal Perforations (SIP) from analysis and we also needed to exclude neonates that did not live long enough to have a chance to develop NEC. Therefore, we excluded all infants who died or developed perforation (surgical NEC) before postnatal day 7 (to avoid including infants who died from respiratory causes, severe IVH or developed SIP) and evaluated only infants who survived beyond day 7 (at risk of developing NEC)
Ethnicity
Ancestry was classified as African-American, non-Hispanic Caucasians, Hispanic Caucasian, and others including Asian and multi-ancestral using GWASTools (20) to generate eigenvalues for the entire dataset.
Imputation
Imputation was run using beagle 3.3.1. 769,757 SNPs were used for imputation with 7,500,443 SNPs being imputed (21).
GWA analysis
SNPs were analyzed using PLINK(22) using logistic regression under an additive model. Three models were run:
Proven NEC (Stage II or greater) or Death vs. survival without proven NEC,
Surgical NEC or death vs. survival without either Medical or Surgical NEC,
Surgical NEC in survivors vs. survivors without surgical NEC.
The regression models included covariates for GA, small for GA, gender, Apgar at 5 min < 5, antenatal steroids, and the genomic ancestry eigenvalues 1-4. The top 10 SNPs (by lowest p-value) for each of the 3 models were mapped to genes.
Validation cohort details
Replication was attempted in the Gene Targets for Intraventricular Hemorrhage Study (23). This study is of inborn infants with birth weights (BW) 401-1000 g with either IVH or normal cranial ultrasounds enrolled prospectively at 24 universities; additional samples were provided from ELGAN, Iowa Prematurity and Oulu University cohorts. The babies were also evaluated for other birth related endpoints such as NEC.
Pathways
We assigned genes to pathways using the Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/collections.jsp). SNPs were assigned to gene(s) based on being exonic, intronic, untranslated region, or within 20 kb of the ends of the gene model. Pathways were analyzed using Gene Set Enrichment Analysis (GSEA) (24). Reactome pathways significant at FDR<0.15 and p<0.01 were considered relevant.
RNA sequencing (RNAseq)
RNA was isolated from mucosal strips of freshly obtained surgical specimens during bowel resection under IRB-approved protocols. RNA isolation was performed using the RNAeasy Mini kit on the Qiacube platform (Qiagen, Germantown, MD). One sample was collected form a specimen obtained during bowel resection due to NEC and another sample was collected from specimen collected during resection to correct ileal atresia to ensure coverage. RNAseq was performed at Hudson Alpha (Huntsville, AL). Fastq sequences were groomed with FASTQ groomer, and they were aligned to the genome (hg19) by bowtie and tophat (25, 26). Resulting BAM datasets were restricted to a +/− 500 kb region around the NECRISK region using BAM slicer. Clusters of aligned sequences were identified in Galaxy visualizer and the genomic coordinates were used to generate further slices of the BAM file corresponding to individual clusters of alignment. These cluster BAM files were then converted to lists of FASTA files using the convert, merge randomize tool in Galaxy. The resulting FASTA files were converted to contigs using the CAP3 program at the Pôle Rhône-Alpes de Bioinformatique Site Doua (PRABI) (27). The contigs were used with the Blat tool in the UCSC genome browser to align with the genome.
RESULTS
The patient population consisted of all ELBW infants with birth weights ≥ 401 g and ≤ 1000 g for whom blood spots were available in the repository of the Eunice Kennedy Shriver NICHD Neonatal Research Network. We found it important to exclude any overlap between spontaneous intestinal perforation (SIP) and NEC and we also needed to exclude neonates who died in the period when NEC is very rare but SIP is common, i.e., the first postnatal week. Therefore, infants who died or developed intestinal perforation prior to postnatal day 7 were excluded, based on the observations that the vast majority of NEC cases occur past day 7 (28) and the majority of SIPs occur prior to day 7 (29). 751 infants were included in the analysis of which 30 infants were diagnosed with surgical NEC after day 7 (of 40 infants with genomic data, 10 had spontaneous gastrointestinal perforation before day 7, and 30 had surgical NEC diagnosed after day 7).
Major demographic variables describing the cohort analyzed in this study are summarized in Table 1. Table 1 a contains variables at birth and Table 1 b contains variables describing the clinical course. There were no significant differences in patient characteristics at birth including: ancestry, birth weight, gestational age, SGA, gender, Apgar score, Cesarean section delivery. Age at full enteral feeds (p= 0.007), age at first enteral feed (p= 0.02) and days of assisted ventilation (p= 0.001) were significantly different between the surgical NEC group compared with the controls. There was no difference in the need for bag and mask ventilation at birth, PDA, Indocin for PDA or surgery for PDA.
Table 1.
| a: Characteristics of enrolled infants: At birth | |||
|---|---|---|---|
| Variable | Entire population | Surgical NEC vs. Survival without medical or surgical NEC [in infants surviving >7d and without Spontaneous Intestinal Perforation] |
|
| Surgical NEC | Survival without NEC | ||
| Sample size | 751 | 30 | 546 |
| Birth weight in grams (mean, SD) | 758 (140) | 758 (137) | 781.5 (132.5) |
| Gestational age in weeks (mean, SD) | 25.8 (2.0) | 25.5 (1.57) | 26.1 (1.94) |
| Genetic Ancestry (as determined by GWASTools (Gogarten, 2012) | |||
| Non-Hispanic Caucasian | (137) | (4) 13.3 % | (133) 24.4% |
| African American | (303) | (20) 66.7% | (283) 51.8% |
| Hispanic Caucasian | (136) | (6) 20% | (130) 23.8% |
| Multiple gestation (%) | (136) 18.2% | (7) 23.3% | (94) 17.3% |
| Antenatal steroids (%) | (546) 72.7% | (24) 80% | (403) 73.8% |
| SGA (%) | (105) 14% | (3) 10% | (83) 15.2% |
| Male sex | (353) 47.1% | (16) 53.3% | (244) 44.7% |
| Apgar score at 1 minute (SD) | 4.3 (2.3) | 4.0 (2.2) | 4.5 (2.3) |
| Apgar score at 5 minutes (SD) | 6.6 (1.8) | 6.6 (1.5) | 6.7 (1.8) |
| C-section Yes (%) | (430) 57.3% | (19) 63.3% | (330) 60.4% |
| b: Characteristics of enrolled infants at birth: Clinical Course | |||
|---|---|---|---|
| Variable | Entire population | Surgical NEC vs. Survival without medical or surgical NEC [in infants surviving >7d and without Spontaneous Intestinal Perforation] |
|
| Surgical NEC | Survival without NEC | ||
| Any Mechanical ventilation (%) | (689) 91.8% | (30) 100% | (494) 90.5% |
| Age at first enteral feed (Days) | 8.6 | 11.5 | 7.9* |
| Median | 27 | 6 | 6 |
| IQR | (4-10) | (4-14) | (4-9) p=0.016 |
| Age at full enteral feeds (Days) | 31.5 | 41.3 | 29.1* |
| Median | 27 | 30 | 25 |
| IQR | (17-41) | (13-59) | (17-38) P=0.007 |
| Days of assisted ventilation (SD) | 26.9 (27.5) | 42 (29.3) | 24.9* (25.9) P=0.001 |
| Need for bag and mask vent | 697 (92.8%) | 30 (100%) | 494 (90.5%) |
| PDA | 392 (52.2%) | 21 (70.0%) | 279 (51.1%) |
| Indocin for PDA | 332 (44.2%) | 17 (56.7%) | 246 (45.1%) |
| Surgery for PDA | 136 (18.1%) | 7 (23.3%) | 95 (17.4%) |
There were 261 SNPs that exhibited allelic frequencies that differentiated between patients with surgical NEC vs patients without any (medical or surgical) NEC at p<0.05 (Supplemental Table 1), of which 35 were significant at p<10−7 (Table 2). A particularly strong association was found between a cluster of SNPs spanning 43 Kb on chromosome 8 at the location 8q23.3 and the incidence of NEC (Figure 1). Having minor allele(s) in this region conferred an odds ratio of (O.R. 4.72, 2.51 - 8.88) for surgical NEC. We termed this region the NECRISK cluster. Table 3 illustrates allelic frequencies observed in controls vs surgical NEC patients for the SNP (rs7820058) within the NECRISK cluster that showed the highest significance of association with NEC. This increased risk was similar for all three genetic ancestries that were represented in this population (Tables 3A-D). Notably, the minor allele frequencies observed in controls overall and in all three ancestries analyzed corresponded to the minor allele frequency of this SNP in 1000 genomes. Once the diagnostic criteria were loosened, i.e., surgical NEC or detah, or NEC stage II or greater or death were compared to all survivors without NEC, the degree of association with NECRISK SNPs decreased (Supplemental Table S2). Notably, there was no association in minor allele frequency of rs7820058 and stage II NEC (Supplemental Table S3 A) or stage II NEC or death (Supplemental Table S3 B).
Table 2.
SNPs found significantly associated at less than p= 10−6 with surgical NEC by GWAS. (CHR: Chromosome; BP: Base pair location)
| CHR | SNP | Chromosome location (BP) | Any Surgical NEC vs. survivors without any NEC |
|---|---|---|---|
| 8 | rs10505232 | 1.16E+08 | 6.63E−09 |
| 8 | chr8:115870263 | 1.16E+08 | 6.78E−09 |
| 8 | rs62514170 | 1.16E+08 | 7.18E−09 |
| 8 | rs17729017 | 1.16E+08 | 7.26E−09 |
| 8 | rs17729672 | 1.16E+08 | 7.33E−09 |
| 8 | rs17729107 | 1.16E+08 | 7.78E−09 |
| 8 | rs62514171 | 1.16E+08 | 7.91E−09 |
| 8 | rs17667338 | 1.16E+08 | 8.02E−09 |
| 8 | rs62514176 | 1.16E+08 | 8.04E−09 |
| 8 | rs7013533 | 1.16E+08 | 8.04E−09 |
| 8 | rs7820058 | 1.16E+08 | 8.05E−09 |
| 8 | rs17667594 | 1.16E+08 | 8.10E−09 |
| 8 | rs61423644 | 1.16E+08 | 8.11E−09 |
| 8 | rs17729840 | 1.16E+08 | 8.25E−09 |
| 8 | rs62514184 | 1.16E+08 | 2.40E−08 |
| 8 | rs62514166 | 1.16E+08 | 2.44E−08 |
| 8 | rs10955735 | 1.16E+08 | 2.81E−08 |
| 8 | rs62514219 | 1.16E+08 | 2.87E−08 |
| 8 | rs62514192 | 1.16E+08 | 3.08E−08 |
| 8 | rs62514191 | 1.16E+08 | 3.15E−08 |
| 8 | rs6993083 | 1.16E+08 | 3.43E−08 |
| 8 | rs62511960 | 1.16E+08 | 4.09E−08 |
| 8 | rs7015520 | 1.16E+08 | 4.10E−08 |
| 8 | rs62514183 | 1.16E+08 | 5.16E−08 |
| 14 | rs3181382 | 24789310 | 1.18E−07 |
| 14 | rs3742511 | 24784911 | 1.19E−07 |
| 14 | rs2295303 | 24788220 | 1.28E−07 |
| 14 | rs3742510 | 24783681 | 1.86E−07 |
| 16 | rs9927732 | 88076724 | 2.74E−07 |
| 11 | chr11:124744005 | 1.25E+08 | 4.01E−07 |
| 11 | chr11:124745427 | 1.25E+08 | 4.13E−07 |
| 11 | chr11:124739699 | 1.25E+08 | 4.24E−07 |
| 8 | rs10505238 | 1.16E+08 | 6.16E−07 |
| 8 | rs62514182 | 1.16E+08 | 6.41E−07 |
| 11 | chr11:124762541 | 1.25E+08 | 8.37E−07 |
Figure 1.
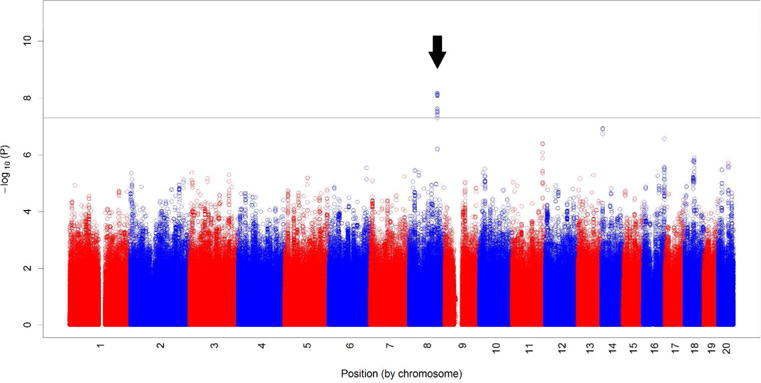
Manhattan Plot of SNPs that exhibited association with surgical NEC vs controls. Data shown are –log P values on the Y axis along vs chromosome locations on the X axis. Arrow points to the NECRISK region.
Table 3.
| A Allelic frequencies of rs7820058 in Surgical NEC in infants surviving >7d vs. survival without medical or surgical NEC across the entire NRN cohort. | ||
|---|---|---|
| P<0.0001 Chi square (O.R. 4.72, 2.51 - 8.88) | NEC (n=60) | NO NEC (n=1092) |
| Major Allele (A) | 45 (75%) | 1020 (93.4%) |
| Minor Allele (G) | 15 (25%) | 72 (6.6%) |
| B Allelic frequencies of rs7820058 in Surgical NEC in infants surviving >7d vs. survival without medical or surgical NEC among non-Hispanic Caucasians | ||
|---|---|---|
| P=0.1135 (OR: 3.52, 95% CI 0.67 - 18.4) | NEC (n=8) | NO NEC (n=266) |
| Major Allele (A) | 6 (75%) | 243 (91.3%) |
| Minor Allele (G) | 2 (25%) | 23 (8.6%) |
| C Allelic frequencies of rs7820058 in Surgical NEC in infants surviving >7d vs. survival without medical or surgical NEC among African-Americans | ||
|---|---|---|
| P<0.01 (OR 3.38: 95% CI 1.46-7.83) Chi square |
NEC (n=40) | NO NEC (n=566) |
| Major Allele (A) | 32 (80%) | 527 (93.1%) |
| Minor Allele (G) | 8 (20%) | 39 (6.9%) |
| D Allelic frequencies of rs7820058 in Surgical NEC in infants surviving >7d vs. survival without medical or surgical NEC among Hispanic Caucasians | ||
|---|---|---|
| P<0.0001 OR (17.86, 95% CI 4.82-66.2) | NEC (n=12) | NO NEC (n=260) |
| Major Allele (A) | 7 (68.3%) | 250 (96.1%) |
| Minor Allele (G) | 5 (41.7%) | 10 (3.9 %) |
A validation cohort consisted of 1018 multi-ancestral extremely preterm neonates from the Gene Targets for Intraventricular Hemorrhage Study (30). This cohort included 52 NEC cases of which 26 had surgical NEC along with 966 controls. Controls were defined as survival > 7 days, but no data was available on spontaneous intestinal perforation and whether the diagnosis of NEC was before or after postnatal day 7. The validation cohort was 50% African American, 42.7% Non-Hispanic Caucasian, and 8.8% Hispanic Caucasian with the balance being other ancestries. The majority of SNPs within the NECRISK region did not differ between cases with NEC and cases without NEC (Supplemental Table S4). However, the minor allele frequency of the only SNP that did exhibit P<0.05 (rs13252246; P=0.02) among the NECRISK SNPs was enriched in NEC cases similar to our discovery cohort. The SNP exhibiting the next lowest P value (P=0.06; rs10755911) was also enriched in NEC.
In addition to the NECRISK region, which showed the highest degree of correlation with the incidence of NEC, in the NRN cohort, a cluster of 4 SNPs on chromosome 14 showed the second highest level of association with surgical NEC with unadjusted P values of 10−7. This chromosome 14 cluster corresponds to the adenylate cyclase 4 (ADCY4) and leukotriene B4 receptor (LTB4R) genes. The next most significant cluster of four SNPs was on chromosome 11 corresponding to the neurogranin gene (P=4*10−7).
In silico analysis of the NECRISK region: As shown in Figure 2A, the nearest two genes to the NECRISK region are CSMD3 (−1.43 Mb) and TRPS1 (+542 kb). The same two genes are located on a syntenic region of murine chromosome 15 with a similar intergenic distance (Figure 2A, B), suggesting a high degree of evolutionary conservation for this region. The chromosomal distances between the NECRISK cluster, CSMD3 and TRPS1 are beyond the typical distances observed in direct regulation of transcription such as the core promoter, proximal promoter, enhancer, silencer, insulator and locus control regions that are typically within 100 kb of the genes they regulate (31). For this reason, we thoroughly analyzed a 200 kb region surrounding the NECRISK SNP cluster in search of known transcripts or potential novel transcripts within a distance that would allow regulation of these transcripts by genetic variation.
Figure 2.
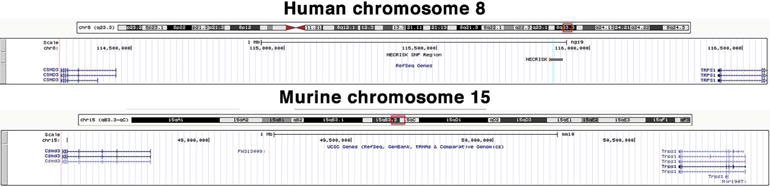
The NECRISK region exhibits a high degree of evolutionary conservation. Image shown is the region between the flanking known genes of TRPS1 and CSMD3 of both human chr 8 and mouse chr 15. The preserved orientation and spacing of genes indicate a high degree of evolutionary conservation.
Based on predicted transcripts annotated in the UCSC genome browser (https://genome.ucsc.edu/), there are three predicted transcripts within the 200 Kb domain surrounding the NECRISK region (Figure 3A). On the other hand, the Ensembl genome browser (http://www.ensembl.org/index.html) shows a single predicted gene in the same region (Figure 3B). Using another gene prediction program (Softberry; http://www.softberry.com/), we identified five additional potential transcripts in the +/− 100 kb domain around the NECRISK region.
Figure 3.

The NECRISK region and contiguous novel transcripts between the known genes of CSMD3 and TRPS1. The NECRISK region is shown in dark blue while potential novel transcripts identified by alignment of RNAseq results with the human genome are shown in yellow.
In order to identify predicted and novel transcripts, we performed RT-PCR and RNAseq on RNA extracted from small intestine samples obtained at the time of bowel resection from two premature neonates. We selected one specimen from an infant who had bowel resection due to NEC and another specimen (control) from an infant with ileal atresia. Exon-spanning RT-PCR attempts failed to verify the existence of any of the predicted transcripts. RNAseq data were aligned to the genome as described in Methods. Alignments between the identified contiguous RNA sequences (contigs) and the human genome was done, along with alignments of human expressed sequence tags (EST). ESTs are sequences derived from cDNA libraries that match the human genome. All the identified RNA contigs matched the human genome and corresponded to ESTs matching at the same position. The RNAseq did not reveal any alignments that would have matched the predicted transcripts in this region. The RNA sequence that aligned closest to the NECRISK region was located on the negative strand approximately 200 kb from the NECRISK region. This RNA contig contains an open reading frame encoding 419 amino acids that exhibited a 96% identity to the long interspersed element-1 (LINE-1) retrotransposable element. The other identified RNA contig sequences all contained potential open reading frames (ORF), including the contig at + 5Kb corresponding to the first exon of TRPS1. This can be viewed as a “positive control” indicating that our strategy of transcript identification can predict known genes.
Pathway analysis performed using SNP results from the initial GWAS results indicated 52 reactome pathways (www.reactome.org) significant at False Discovery Rate (FDR)<0.15 and p<0.01 (Table 4). These pathways include many that are involved in growth factor receptor (IGF1R, FGFR, EGFR) signaling, eicosanoid signaling, T-cell regulation (CD28, CTLA4), as well as signaling mediated via calcium (cytosolic Ca2+, Calmodulin, Ca-permeable Kainate receptor etc) and G-proteins. Interestingly, the phototransduction cascade is also represented, perhaps due to the high representation of G proteins in this pathway.
Table 4.
Pathway analysis listing Reactome pathways significant at False Discovery Rate (FDR) <0.15 and p<0.01.
| PATHWAY_ID | GENESET_LABEL | SIZE | p-value | FDR |
|---|---|---|---|---|
| REACT_150203 | IRS-related events triggered by IGF1R | 82 | 0.001592357 | 0.140394097 |
| REACT_18352 | Eicosanoid ligand-binding receptors | 15 | 0.002004008 | 0.140394097 |
| REACT_163919 | Inactivation, recovery and regulation of the phototransduction cascade | 32 | 0.002094241 | 0.140394097 |
| REACT_19238 | CD28 dependent Vav1 pathway | 11 | 0.002132196 | 0.140394097 |
| REACT_12056 | NGF signalling via TRKA from the plasma membrane | 199 | 0.002314815 | 0.140394097 |
| REACT_21310 | Phospholipase C-mediated cascade | 55 | 0.003120125 | 0.140394097 |
| REACT_19405 | CTLA4 inhibitory signaling | 22 | 0.003243243 | 0.140394097 |
| REACT_22384 | Netrin mediated repulsion signals | 10 | 0.003333333 | 0.140394097 |
| REACT_23765 | Reduction of cytosolic Ca++ levels | 10 | 0.004052685 | 0.140394097 |
| REACT_163722 | The phototransduction cascade | 33 | 0.004188482 | 0.140394097 |
| REACT_12519 | PECAM1 interactions | 11 | 0.004324324 | 0.140394097 |
| REACT_21272 | Downstream signaling of activated FGFR | 142 | 0.004514673 | 0.140394097 |
| REACT_17025 | Downstream signal transduction | 154 | 0.004514673 | 0.140394097 |
| REACT_120736 | Signaling by FGFR in disease | 167 | 0.004524887 | 0.140394097 |
| REACT_9000 | Calmodulin induced events | 28 | 0.004733728 | 0.140394097 |
| REACT_9053 | CaM pathway | 28 | 0.004733728 | 0.140394097 |
| REACT_13477 | Transmission across Chemical Synapses | 195 | 0.004807692 | 0.140394097 |
| REACT_111064 | DAG and IP3 signaling | 33 | 0.004987531 | 0.140394097 |
| REACT_12478 | EGFR interacts with phospholipase C-gamma | 35 | 0.004987531 | 0.140394097 |
| REACT_115720 | PLCG1 events in ERBB2 signaling | 36 | 0.004987531 | 0.140394097 |
| REACT_21346 | Activation of Ca-permeable Kainate Receptor | 12 | 0.004995005 | 0.140394097 |
| REACT_21322 | Ionotropic activity of Kainate Receptors | 12 | 0.004995005 | 0.140394097 |
| REACT_15526 | G-protein mediated events | 45 | 0.005284016 | 0.140394097 |
| REACT_15307 | Ca-dependent events | 30 | 0.00591716 | 0.140394097 |
| REACT_1946 | PKA activation in glucagon signalling | 17 | 0.006012024 | 0.140394097 |
| REACT_15530 | PKA activation | 16 | 0.006012024 | 0.140394097 |
| REACT_1665 | Glucagon signaling in metabolic regulation | 33 | 0.006012024 | 0.140394097 |
| REACT_9470 | Signaling by FGFR | 152 | 0.006772009 | 0.140394097 |
| REACT_147814 | DAP12 signaling | 158 | 0.006802721 | 0.140394097 |
| REACT_147694 | DAP12 interactions | 171 | 0.006802721 | 0.140394097 |
| REACT_115755 | Signaling by ERBB2 | 154 | 0.006880734 | 0.140394097 |
| REACT_15306 | Cytosolic tRNA aminoacylation | 25 | 0.006993007 | 0.140394097 |
| REACT_15497 | PKA-mediated phosphorylation of CREB | 17 | 0.007014028 | 0.140394097 |
| REACT_25149 | Ion transport by P-type ATPases | 40 | 0.007246377 | 0.140394097 |
| REACT_121315 | Glycosaminoglycan metabolism | 122 | 0.007389163 | 0.140394097 |
| REACT_147739 | MPS IX - Natowicz syndrome | 122 | 0.007389163 | 0.140394097 |
| REACT_147853 | Mucopolysaccharidoses | 122 | 0.007389163 | 0.140394097 |
| REACT_147788 | MPS IIIB - Sanfilippo syndrome B | 122 | 0.007389163 | 0.140394097 |
| REACT_147719 | MPS VI - Maroteaux-Lamy syndrome | 122 | 0.007389163 | 0.140394097 |
| REACT_147825 | MPS IV - Morquio syndrome A | 122 | 0.007389163 | 0.140394097 |
| REACT_147860 | MPS IIIC - Sanfilippo syndrome C | 122 | 0.007389163 | 0.140394097 |
| REACT_147759 | MPS VII - Sly syndrome | 122 | 0.007389163 | 0.140394097 |
| REACT_147734 | MPS II - Hunter syndrome | 122 | 0.007389163 | 0.140394097 |
| REACT_147857 | MPS I - Hurler syndrome | 122 | 0.007389163 | 0.140394097 |
| REACT_147749 | MPS IIID - Sanfilippo syndrome D | 122 | 0.007389163 | 0.140394097 |
| REACT_147753 | MPS IIIA - Sanfilippo syndrome A | 122 | 0.007389163 | 0.140394097 |
| REACT_147798 | MPS IV - Morquio syndrome B | 122 | 0.007389163 | 0.140394097 |
| REACT_12079 | PLC-gamma1 signalling | 35 | 0.007604563 | 0.141476557 |
| REACT_1195 | Insulin receptor signalling cascade | 85 | 0.007961783 | 0.142554162 |
| REACT_121206 | Chondroitin sulfate/dermatan sulfate metabolism | 50 | 0.007981756 | 0.142554162 |
| REACT_19183 | CD28 co-stimulation | 29 | 0.008695652 | 0.149889707 |
| REACT_21247 | FRS2-mediated cascade | 37 | 0.00872818 | 0.149889707 |
DISCUSSION
In this study, we used GWAS on a relatively large cohort of ELBW neonates to identify genetic susceptibility to NEC using very stringent inclusion and exclusion criteria. The choice of ELBW patient population was dictated by to the goal of achieving a relatively high case/control ratio; i.e., the population at the highest risk for NEC. This was necessary both for improving statistical power and minimizing cost of genotyping. The stringent inclusion criterion of surgically verified NEC was chosen because it reduces the possibility of diagnostic error. Finally, we excluded all data when death occurred prior to postnatal day seven, because NEC is rare during this time period, whereas the potentially confounding spontaneous intestinal perforation is relatively frequent at this time. As shown in supplemental table 3, broadening the inclusion criteria to NEC or death or inclusion of stage II NEC notably reduced the statistical significance of association. Furthermore, when stage II NEC was analyzed alone, there was no statistically significant association between minor allele frequency in the NECRISK region and stage II NEC (Supplemetal table 4). We reason that the stronger genetic association with surgical NEC is due to a more precise diagnosis, i.e., a direct visual verification of necrotic bowel, as opposed to weaker association with stage II NEC, where the diagnosis is indirect and may be less precise. We identified a very strong genetic association between surgical NEC and a 43 Kb intergenic region of chromosome 8, delineated by 25 SNPs of the Illumina bead chip array that was used. This association was consistent across the three main ancestral groups in the cohort and conferred an odds ratio of 4.3 of risk for surgical NEC.
The same cohort of patients and the same genotyping data used herein was used earlier in GWAS to interrogate potential genetic basis for sepsis (32), intraventricular hemorrhage (23, 30) and bronchopulmonary dysplasia (33). While all these GWAS identified significant associations between SNPs or SNP clusters with the aforementioned neonatal morbidities, none of those were in the same region of chromosome 8 as the NECRISK region, or in regions on chromosomes 11 and 14 that we found to be associated with NEC. These data indicate that specific genetic variations may underlie specific neonatal pathologies in different organs and that the NECRISK region is specific to be associated with NEC.
NEC has been considered to be a multifactorial disease in which prematurity, variations in clinical practice, and altered microbial colonization have been recognized as main contributors to pathogenesis. Genetic susceptibility has been suspected and there have been several studies addressing the role of genetics by targeted analysis of specific SNPs in genes potentially associated with NEC pathogenesis. Although the majority of these efforts were based on reasonable hypotheses, the chance of identifying the most significant contributors of genetic predisposition by such targeted approaches is practically negligible. The human genome has at least 38 million SNPs(15), making genome-wide association studies the only viable strategy to identify the most significant associations with disease susceptibility. Even with the 1 million SNP coverage of the Illumina chip used in the present study with imputation of 7 million other SNPs, there is a high chance of missing potentially highly significant SNPs and this coverage is best suited to identify regions of the genome that associate with disease severity as opposed to identifying single SNPs. Indeed, the three most significant findings in our study pointed to clusters of SNPs as opposed to single SNPs.
Surprisingly, the NECRISK region that showed the most significant correlation with the incidence of surgical NEC in our patient population is located on an intergenic region where the nearest two known genes are ~0.5 and ~1.5 Mb away. These distances are beyond the 100 Kb limit that is generally considered to be the distance that allows direct regulation of genes by common mechanisms. In order to analyze alternate mechanisms that are distinct from regulation of known distant genes we performed in silico analysis as well as RNAseq to identify potential novel transcripts that are within a closer distance to the NECRISK region. RNAseq identified a number of potential transcripts between the CSMD3 and TRPS1 genes. The nearest potential novel transcript to the NECRISK region is 96% homologous to the LINE-1 retrotransposon. The LINE-1 sequence comprises approximately 17% of the human genome, and leads to genomic diversity and alters gene function (34). There are at least three major mechanisms by which LINE-1 affects the genome. The main mechanism that is known to occur in all eukaryotes is that LINE-1 activation results in a coupled reverse-transcription integration event referred to as target-primed reverse-transcription (TPRT), thereby resulting in multiplication of the LINE-1 loci which may result in gene interruptions and or effects of gene expression in the vicinity of integration. A less common, but well-recognized role of LINE-1 is the transposition of non-autonomously mobile sequences. The most common elements transposed this way are Alu (named after the Alu1 restriction endonuclease site that it contains) and SINE-VNTR-Alu-s (SVAs). Additionally, LINE-1 retrotransposon may mediate nonallelic homologous recombination (NAHR), which is a key mechanism in structural variants or copy number variants (CNV) in the human genome. There are an abundance of structural variants that have been reported in the immediate vicinity of the NECRISK region. Therefore we speculate that one of the potential mechanisms that may mediate the effects of the NECRISK region on the incidence of NEC is a de novo structural variation of chromosome 8 involving LINE-1. This hypothesis is supported by the fact that the NECRISK region is a large segment of the genome, as opposed to a single or small group of SNPs. Validation of this hypothesis is beyond the scope of the present study. Murine studies have demonstrated that retrotransposition of LINE-1 is induced by inflammation in colonic mesenchymal cells, and is associated with the severity of colitis (35) although it is not known if similar results will be obtained in murine NEC models.
In addition to the identification of the NECRISK region, it is notable that there are several other SNPs that showed association with NEC. Intriguingly, the next two most significant associations also involve SNP clusters, albeit much smaller clusters than the NECRISK region. While the NECRISK region seemingly does not involve known genes, the next two most significant clusters involve known genes. The second most significant cluster involves the ADCY4 and LTB4R genes. While neither of these two genes have been directly implicated in the pathogenesis of NEC, both are involved in regulating signaling in epithelial cells and in regulating inflammation. Intestinal epithelial cells express various adenylyl cyclase isoforms, including ADCY4, and their expression levels are regulated during cellular differentiation (36). In turn cyclic AMP, the product of adenylyl cyclase, is a key regulator of epithelial functions such as ion transport, proliferation, migration, apoptosis, membrane recycling and macromolecule secretion (37–40). Intriguingly, LTB4 plays a significant role in a toll-like receptor 4 and cyclooxygenase-2 mediated mechanism of intestinal ischemia/reperfusion injury (41). This finding may be related to a potential role for LTB4 in NEC as TLR4 has been well-documented to play a role in animal models of NEC (42, 43). Notably, the LTB4R gene encodes an eicosanoid receptor, and pathway analysis revealed eicosanoid receptor signaling as the second most prominent pathway affected by NEC-associated SNPs. Variation at these two gene loci may be associated with cAMP or eicosanoid signaling and potentially be associated with vascular dysfunction that predispose to NEC. The third most specific cluster of SNPs corresponds to the genomic location of the neurogranin gene. Although neurogranin used to be considered to be exclusive to the brain, it has been shown to play a role in IL-2-dependent survival of T cells, establishing the feasibility of a role in immune/inflammatory signaling (44).
NEC is primarily a disorder of the immature developing intestine, and our pathway analysis confirmed many that are involved in growth factor receptor (IGF1R, FGFR, EGFR) signaling that may be involved in normal gut development. NEC is also characterized by inflammation and necrosis, and pathways related to eicosanoid signaling, T-cell regulation (CD28, CTLA4), signaling mediated via calcium (cytosolic Ca2+, Calmodulin, Ca-permeable Kainate receptor etc) and G-proteins may contribute to predisposition to intestinal injury, apoptosis, or repair pathways.
The strengths of our study are the relatively large cohort (for studies in premature neonates), stringent diagnostic criteria, and the strong genetic association between genotype and phenotype. Weaknesses of the study are our inability to identify a large validation cohort with sufficient information regarding the clinical course that would permit the same stringent criteria that we used in the discovery cohort. However, it is encouraging that despite the low power in the validation cohort one SNP within the NECRISK cluster exhibited a statistical significance and a similar enrichment in the NEC cases vs controls compared to our discoverycohort. Furthermore, due to the intergenic location of the NECRISK cluster and the consequent multiple potential mechanisms for the effect, we were unable to elucidate the mechanism of action that is responsible for disease susceptibility by genetic variation.
In summary, in this study we identified a very strong genetic association between surgical NEC and an intergenic region of chromosome 8, which we labeled the “NECRISK” region, although we were unable to validate this finding in a different GWAS dataset. RNA sequencing identified a RNA sequence similar to the LINE-1 retrotransposable element that aligned to the negative strand approximately 200 kb from the NECRISK region. Pathway analysis identified pathways related to growth factor, calcium, and G-protein signaling, as well as other pathways associated with inflammation and injury that may contribute to NEC.
Supplementary Material
Acknowledgments
The National Institutes of Health (General Clinical Research Center grants M01 RR30, M01 RR32, M01 RR39, M01 RR70, M01 RR80, M01 RR633, M01 RR750, M01 RR997, M01 RR6022, M01 RR7122, M01 RR8084, M01 RR16587, UL1 RR24979) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grants U01 HD36790, U10 HD21364, U10 HD21373, U10 HD21385, U10 HD21397, U10 HD21415, U10 HD27851, U10 HD27853, U10 HD27856, U10 HD27871, U10 HD27880, U10 HD27881, U10 HD27904, U10 HD34216, U10 HD40461, U10 HD40492, U10 HD40498, U10 HD40689, U10 HD53109) provided grant support for the Neonatal Research Network’s Genomics Study. In addition, Dr. Murray received assistance for the GENEVA study from the National Human Genome Research Institute (U01 HG4423).
The funding agencies provided overall oversight for study conduct, but all data analyses and interpretation were independent of the funding agencies. While NICHD staff did have input into the study design, conduct, analysis, and manuscript drafting, the comments and views of the authors do not necessarily represent the views of the NICHD.
We are indebted to our medical and nursing colleagues and the infants and their parents who agreed to take part in this study.
Data collected at participating NRN sites were transmitted to RTI International, the data coordinating center (DCC) for the NRN, which stored, managed, and analyzed the data for this study. On behalf of the network, Drs. Abhik Das (DCC PI) and Grier Page (DCC Statistician) had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis.
The following investigators participated in this study:
NRN Steering Committee Chair: Alan H. Jobe, MD PhD, University of Cincinnati.
Case Western Reserve University, Rainbow Babies & Children’s Hospital (U10 HD21364, M01 RR80) – Michele C. Walsh, MD MS; Avroy A. Fanaroff, MD; Michele C. Walsh, MD MS; Nancy S. Newman, RN; Bonnie S. Siner, RN.
Cincinnati Children’s Hospital Medical Center, and University Hospital (U10 HD27853, M01 RR8084) – Kurt Schibler, MD; Edward F. Donovan, MD; Vivek Narendran, MD MRCP; Barbara Alexander, RN; Cathy Grisby, BSN CCRC; Jody Hessling, RN; Marcia Worley Mersmann, RN CCRC; Holly L. Mincey, RN BSN.
Duke University School of Medicine University Hospital, Alamance Regional Medical Center, and Durham Regional Hospital (M01 RR30, U10 HD40492) – Ronald N. Goldberg, MD; C. Michael Cotten, MD MHS; Kathy J. Auten, MSHS.
Emory University, Children’s Healthcare of Atlanta, Grady Memorial Hospital, and Emory Crawford Long Hospital (U10 HD27851, M01 RR39) – Barbara J. Stoll, MD; Ellen C. Hale, RN BS CCRC.
Eunice Kennedy Shriver National Institute of Child Health and Human Development – Rosemary D. Higgins, MD; Linda L. Wright, MD; Sumner J. Yaffe, MD; Elizabeth M. McClure, Med; Stephanie Wilson Archer, MA.
RTI International (U10 HD36790) – Abhik Das, PhD; W. Kenneth Poole, PhD; Grier Page, PhD; Scott A. McDonald, BS; Betty K. Hastings; Jeanette O’Donnell Auman, BS; Kristin M. Zaterka-Baxter, RN BSN.
Stanford University, Lucile Packard Children’s Hospital (U10 HD27880, M01 RR70) – Krisa P. Van Meurs, MD; David K. Stevenson, MD; M. Bethany Ball, BS CCRC.
University of Alabama at Birmingham Health System and Children’s Hospital of Alabama (U10 HD34216, M01 RR32) – Waldemar A. Carlo MD; Namasivayam Ambalavanan, MD; Monica V. Collins, RN BSN MaEd; Shirley S. Cosby, RN BSN.
University of California – San Diego Medical Center and Sharp Mary Birch Hospital for Women (U10 HD40461) – Neil N. Finer, MD; Maynard R. Rasmussen, MD; David Kaegi, MD; Kathy Arnell, RNC; Clarence Demetrio, RN; Wade Rich, BSHS RRT.
University of Iowa Children’s Hospital and Mercy Medical Center (U10 HD53109, M01 RR59, UL1 TR442) – Edward F. Bell, MD; Jeffrey C. Murray, MD; Karen J. Johnson, RN.
University of Miami Holtz Children’s Hospital (U10 HD21397, M01 RR16587) – Shahnaz Duara, MD; Charles R. Bauer, MD; Ruth Everett-Thomas, RN MSN.
University of Tennessee (U10 HD21415) – Sheldon B. Korones, MD; Henrietta S. Bada, MD; Tina Hudson, RN BSN.
University of Texas Southwestern Medical Center at Dallas, Parkland Health & Hospital System, and Children’s Medical Center Dallas (U10 HD40689, M01 RR633) – Pablo J. Sánchez, MD; Abbot R. Laptook, MD; Walid A. Salhab, MD; Susie Madison, RN; Nancy A. Miller, RN; Gaynelle Hensley, RN; Alicia Guzman.
University of Texas Health Science Center at Houston Medical School, Children’s Memorial Hermann Hospital, and Lyndon B. Johnson General Hospital (U10 HD21373) – Kathleen A. Kennedy, MD MPH; Jon E. Tyson, MD MPH; Kathleen A. Kennedy, MD MPH; Esther G. Akpa, RN BSN; Patty A. Cluff, RN; Claudia I. Franco, RNC MSN; Anna E. Lis, RN BSN; Georgia E. McDavid, RN; Patti Pierce Tate, RCP.
Wayne State University, Hutzel Women’s Hospital, and Children’s Hospital of Michigan (U10 HD21385) – Seetha Shankaran, MD; Beena G. Sood, MD MS; G. Ganesh Konduri, MD; Rebecca Bara, RN BSN; Geraldine Muran, RN BSN.
Funding Source
The National Institutes of Health (General Clinical Research Center grants M01 RR30, M01 RR32, M01 RR39, M01 RR70, M01 RR80, M01 RR633, M01 RR750, M01 RR997, M01 RR6022, M01 RR7122, M01 RR8084, M01 RR16587, UL1 RR24979) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grants U01 HD36790, U10 HD21364, U10 HD21373, U10 HD21385, U10 HD21397, U10 HD21415, U10 HD27851, U10 HD27853, U10 HD27856, U10 HD27871, U10 HD27880, U10 HD27881, U10 HD27904, U10 HD34216, U10 HD40461, U10 HD40492, U10 HD40498, U10 HD40689, U10 HD53109) provided grant support for the Neonatal Research Network’s Genomics Study.
Footnotes
Disclosure of Conflicts of Interest
The authors have no financial relationships relevant to this article to disclose and the study sponsors have not been involved in any of the following: (1) study design; (2) the collection, analysis, and interpretation of data; (3) the writing of the report; and (4) the decision to submit the paper for publication.
Genomics Investigators
C. Michael Cotten, MD MHS1; Grier Page, PhD2; Waldemar A. Carlo MD3; Edward F. Bell, MD4; Ronald N. Goldberg, MD1; Kurt Schibler, MD5; Rosemary D. Higgins, MD6; Beena G. Sood, MD MS7; David K. Stevenson, MD8; Barbara J. Stoll, MD9; Krisa P. Van Meurs, MD7; Karen J. Johnson, RN4; Abhik Das, PhD10; Scott A. McDonald, BS2; Kristin M. Zaterka-Baxter, RN BSN2; Kathleen A. Kennedy, MD MPH11; Pablo J. Sánchez, MD12; Shahnaz Duara, MD13; Michele C. Walsh, MD MS14; Seetha Shankaran, MD15; Sheldon B. Korones, MD16; Neil N. Finer, MD17; Jeffrey C. Murray, MD4; for the Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network
1Department of Pediatrics, Duke University, Durham, NC
2Social, Statistical and Environmental Sciences, RTI International, Research Triangle Park, NC
3Department of Pediatrics, University of Alabama at Birmingham, Birmingham, AL
4University of Iowa, Department of Pediatrics, Iowa City, IA
5Department of Pediatrics, University of Cincinnati, Cincinnati, OH
6Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD.
7Department of Pediatrics, Wayne State University, Detroit, MI
8Department of Pediatrics, Division of Neonatal and Developmental Medicine, Stanford University School of Medicine and Lucile Packard Children’s Hospital, Palo Alto, CA
9Emory University School of Medicine, Department of Pediatrics, Children’s Healthcare of Atlanta, Atlanta, GA
10Social, Statistical and Environmental Sciences, RTI International, Rockville, MD
11Department of Pediatrics, University of Texas Medical School at Houston, Houston, TX
12Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX
13University of Miami Miller School of Medicine, Miami, FL
14Department of Pediatrics, Rainbow Babies & Children’s Hospital, Case Western Reserve University, Cleveland, OH
15Department of Pediatrics, Wayne State University, Detroit, MI
16Department of Pediatrics, University of Tennessee Health Science Center, Memphis, TN
17University of California at San Diego, San Diego, CA
References
- 1.Stoll BJ, Hansen NI, Bell EF, et al. Neonatal outcomes of extremely preterm infants from the NICHD Neonatal Research Network. Pediatrics. 2010;126:443–456. doi: 10.1542/peds.2009-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakely ML, Lally KP, McDonald S, et al. Postoperative outcomes of extremely low birth-weight infants with necrotizing enterocolitis or isolated intestinal perforation: a prospective cohort study by the NICHD Neonatal Research Network. Ann Surg. 2005;241:984–989. doi: 10.1097/01.sla.0000164181.67862.7f. discussion 989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah TA, Meinzen-Derr J, Gratton T, et al. Hospital and neurodevelopmental outcomes of extremely low-birth-weight infants with necrotizing enterocolitis and spontaneous intestinal perforation. J Perinatol. 2012;32:552–558. doi: 10.1038/jp.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhandari V, Bizzarro MJ, Shetty A, et al. Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics. 2006;117:1901–1906. doi: 10.1542/peds.2005-1414. [DOI] [PubMed] [Google Scholar]
- 5.Moonen RM, Paulussen AD, Souren NY, Kessels AG, Rubio-Gozalbo ME, Villamor E. Carbamoyl phosphate synthetase polymorphisms as a risk factor for necrotizing enterocolitis. Pediatr Res. 2007;62:188–190. doi: 10.1203/PDR.0b013e3180a0324e. [DOI] [PubMed] [Google Scholar]
- 6.Bokodi G, Derzbach L, Banyasz I, Tulassay T, Vasarhelyi B. Association of interferon gamma T+874A and interleukin 12 p40 promoter CTCTAA/GC polymorphism with the need for respiratory support and perinatal complications in low birthweight neonates. Arch Dis Child Fetal Neonatal Ed. 2007;92:F25–29. doi: 10.1136/adc.2005.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banyasz I, Bokodi G, Vasarhelyi B, et al. Genetic polymorphisms for vascular endothelial growth factor in perinatal complications. Eur Cytokine Netw. 2006;17:266–270. [PubMed] [Google Scholar]
- 8.Sampath V, Le M, Lane L, et al. The NFKB1 (g.-24519delATTG) variant is associated with necrotizing enterocolitis (NEC) in premature infants. J Surg Res. 2011;169:e51–57. doi: 10.1016/j.jss.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Szebeni B, Szekeres R, Rusai K, et al. Genetic polymorphisms of CD14, toll-like receptor 4, and caspase-recruitment domain 15 are not associated with necrotizing enterocolitis in very low birth weight infants. J Pediatr Gastroenterol Nutr. 2006;42:27–31. doi: 10.1097/01.mpg.0000192246.47959.b2. [DOI] [PubMed] [Google Scholar]
- 10.Sankararaman S, Yanamandra K, Napper D, Caldito G, Dhanireddy R. The prevalence of platelet activating factor acetylhydrolase single nucleotide polymorphisms in relationship to necrotizing enterocolitis in Northwest Louisiana infants. Springerplus. 2013;2:294. doi: 10.1186/2193-1801-2-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prencipe G, Auriti C, Inglese R, Gallusi G, Dotta A, De Benedetti F. The macrophage migration inhibitory factor -173G/C polymorphism is not significantly associated with necrotizing enterocolitis in preterm infants. J Pediatr Surg. 2013;48:1499–1502. doi: 10.1016/j.jpedsurg.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Koroglu OA, Onay H, Erdemir G, et al. Mannose-binding lectin gene polymorphism and early neonatal outcome in preterm infants. Neonatology. 2010;98:305–312. doi: 10.1159/000291487. [DOI] [PubMed] [Google Scholar]
- 13.Spiegler J, Gilhaus A, Konig IR, et al. Polymorphisms in the Renin-Angiotensin system and outcome of very-low-birthweight infants. Neonatology. 2010;97:10–14. doi: 10.1159/000226602. [DOI] [PubMed] [Google Scholar]
- 14.Henderson G, Craig S, Baier RJ, Helps N, Brocklehurst P, McGuire W. Cytokine gene polymorphisms in preterm infants with necrotising enterocolitis: genetic association study. Arch Dis Child Fetal Neonatal Ed. 2009;94:F124–128. doi: 10.1136/adc.2007.119933. [DOI] [PubMed] [Google Scholar]
- 15.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visscher PM, Wray NR, Zhang Q, et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlon DB, Budd AF, Lippe C, Andrew RL. The quantitative genetics of incipient speciation: heritability and genetic correlations of skeletal traits in populations of diverging Favia fragum ecomorphs. Evolution. 2011;65:3428–3447. doi: 10.1111/j.1558-5646.2011.01389.x. [DOI] [PubMed] [Google Scholar]
- 18.Bell MJ, Ternberg JL, Feigin RD, et al. Neonatal necrotizing enterocolitis. Therapeutic decisions based upon clinical staging. Ann Surg. 1978;187:1–7. doi: 10.1097/00000658-197801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh MC, Kliegman RM. Necrotizing enterocolitis: treatment based on staging criteria. Pediatr Clin North Am. 1986;33:179–201. doi: 10.1016/S0031-3955(16)34975-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogarten SM, Bhangale T, Conomos MP, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock DB, Soler Artigas M, Gharib SA, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ment LR, Aden U, Lin A, et al. Gene-environment interactions in severe intraventricular hemorrhage of preterm neonates. Pediatr Res. 2014;75:241–250. doi: 10.1038/pr.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee WH, Soraisham AS, Shah VS, et al. Incidence and timing of presentation of necrotizing enterocolitis in preterm infants. Pediatrics. 2012;129:e298–304. doi: 10.1542/peds.2011-2022. [DOI] [PubMed] [Google Scholar]
- 29.Attridge JT, Clark R, Walker MW, Gordon PV. New insights into spontaneous intestinal perforation using a national data set: (2) two populations of patients with perforations. J Perinatol. 2006;26:185–188. doi: 10.1038/sj.jp.7211439. [DOI] [PubMed] [Google Scholar]
- 30.Ment LR, Aden U, Bauer CR, et al. Genes and environment in neonatal intraventricular hemorrhage. Semin Perinatol. 2015;39:592–603. doi: 10.1053/j.semperi.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan L, Page G, Kirpalani H, et al. Genome-wide association study of sepsis in extremely premature infants. Arch Dis Child Fetal Neonatal Ed. 2017;102:F439–F445. doi: 10.1136/archdischild-2016-311545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambalavanan N, Cotten CM, Page GP, et al. Integrated genomic analyses in bronchopulmonary dysplasia. J Pediatr. 2015;166:531–537 e513. doi: 10.1016/j.jpeds.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otsubo T, Okamura T, Hagiwara T, Ishizaka Y, Dohi T, Kawamura YI. Retrotransposition of long interspersed nucleotide element-1 is associated with colitis but not tumors in a murine colitic cancer model. PLoS One. 2015;10:e0116072. doi: 10.1371/journal.pone.0116072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi LJ, Jenikova G, Hanson E, et al. Coordinate down-regulation of adenylyl cyclase isoforms and the stimulatory G protein (G(s)) in intestinal epithelial cell differentiation. J Biol Chem. 2010;285:12504–12511. doi: 10.1074/jbc.M109.059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradbury NA, Jilling T, Berta G, Sorscher EJ, Bridges RJ, Kirk KL. Regulation of plasma membrane recycling by CFTR [see comments] Science. 1992;256:530–532. doi: 10.1126/science.1373908. [DOI] [PubMed] [Google Scholar]
- 38.Jilling T, Kirk KL. Cyclic AMP and chloride-dependent regulation of the apical constitutive secretory pathway in colonic epithelial cells. J Biol Chem. 1996;271:4381–4387. doi: 10.1074/jbc.271.8.4381. [DOI] [PubMed] [Google Scholar]
- 39.Mandel KG, Dharmsathaphorn K, McRoberts JA. Characterization of a cyclic AMP-activated Cl-transport pathway in the apical membrane of a human colonic epithelial cell line. J Biol Chem. 1986;261:704–712. [PubMed] [Google Scholar]
- 40.Meir M, Flemming S, Burkard N, et al. Glial cell line-derived neurotrophic factor promotes barrier maturation and wound healing in intestinal epithelial cells in vitro. Am J Physiol Gastrointest Liver Physiol. 2015;309:G613–624. doi: 10.1152/ajpgi.00357.2014. [DOI] [PubMed] [Google Scholar]
- 41.Moses T, Wagner L, Fleming SD. TLR4-mediated Cox-2 expression increases intestinal ischemia/reperfusion-induced damage. J Leukoc Biol. 2009;86:971–980. doi: 10.1189/jlb.0708396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jilling T, Simon D, Lu J, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol. 2006;177:3273–3282. doi: 10.4049/jimmunol.177.5.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leaphart CL, Cavallo J, Gribar SC, et al. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. Journal of immunology (Baltimore, Md) 2007;179:4808–4820. doi: 10.4049/jimmunol.179.7.4808. [DOI] [PubMed] [Google Scholar]
- 44.Devireddy LR, Green MR. Transcriptional program of apoptosis induction following interleukin 2 deprivation: identification of RC3, a calcium/calmodulin binding protein, as a novel proapoptotic factor. Mol Cell Biol. 2003;23:4532–4541. doi: 10.1128/MCB.23.13.4532-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


