Abstract
Telomere repeats at chromosomal ends, critical to genomic integrity, undergo age-dependent attrition. Telomere length, a polygenic trait, has been associated with risk of several disorders including cancers. In contrast to association of long telomeres with increased risk of several cancers, including melanoma, emerging reports suggest that short telomeres predict poor survival in patients with different cancers. In this study based on 1019 stage I and II cutaneous melanoma patients, we show an association between the patients with short telomeres and poor melanoma-specific survival (HR 2.05, 95% CI 1.33–3.16) compared to patients with long telomeres. Due to inverse correlation between age and telomere length (r -0.19, P < 0.0001), we stratified the patients into quantiles based on age at diagnosis and also carried out age-matched analysis. The effect of short telomeres on survival was determined by using multivariate Cox regression that included composite genetic risk score computed from genotyping of the patients for telomere-length associated polymorphisms. The effect of decreased telomere length on poor melanoma-specific survival was particularly strong in patients within the age quantile below 30 years (HR 3.82, 95% CI 1.10–13.30) and between 30–40 years (HR 2.69, 95% CI 1.03–7.03). Our study shows that in contrast to increased melanoma risk associated with increased telomere length, decreased telomere length predicts poor survival in melanoma subgroups.
Introduction
Cutaneous melanoma with its propensity to metastasize and intrinsic drug resistance remains causal for the majority of skin cancer related deaths1,2. Despite increase in the range of treatments, including targeted and immunotherapies, for eliciting stable response in patients with metastatic melanoma, the long term prospects in terms of treatment remain confined to disease management3. The search for the factors that can identify patients at early stages at risk of poor survival remains crucial in melanoma. The known markers of poor outcome in melanoma include increased Breslow thickness, presence of tumor ulceration, increased tumor mitotic rate, reduced nevus number and the presence of locoregional or distant metastasis at diagnosis4–8. The melanoma genome in general is characterized by one of the highest prevalence of somatic mutations in human cancers and several genetic alterations have been shown to predict outcome in melanoma9–13. In particular, the most frequent somatic mutations in cutaneous melanoma like those in the TERT promoter and BRAF/NRAS reportedly associate with poor disease-free and melanoma-specific survival9,14.
Telomere repeats at chromosomal ends, critical to genomic integrity, are maintained through an elaborate network of proteins and pathways15,16. In humans, TTAGGG repeats account for telomere length that ranges between 10–15 kb17. Inherent limitations of DNA replication and telomerase suppression in most somatic cells through epigenetic reprogramming of the telomerase reverse transcriptase (TERT) gene, cause telomeres to undergo age-dependent incremental attrition16,18. Telomere shortening is further influenced by oxidative damage and replicative stress caused by genetic, epigenetic and environmental factors15,19. However, in most cancers enhanced TERT transcription, due to various mechanisms, and consequent telomerase rejuvenation, imparts tumor cells an infinite capability to surmount proliferative barrier through telomere stabilization16,20,21. Constitutive telomere length, a polygenic trait with high estimated heritability, has hitherto been demonstrated to be associated with nine different loci, with six harboring genes related to telomere homeostasis22–25. Epidemiological data, in general, support an association, with varying magnitudes, between constitutive telomere length and various disorders including cancers26,27. Different investigations over the years, in contrast, have suggested that short telomeres associate with poor patient survival26,28,29.
To test the association between telomere length and melanoma-specific patient survival, we measured telomere length in constitutive DNA from 1019 incident cutaneous melanoma patients with stage I and II disease. We also used telomere length associated polymorphisms to generate composite genetic score for association with the disease outcome. Despite a confounding effect of the age at diagnosis, our results suggest an association between short telomeres and poor patient survival, particularly for melanoma patients in younger age groups.
Results
Relative telomere length and melanoma-specific survival
The study included 1019 patients with AJCC stage I and II cutaneous melanoma. Patients with stage 0, III and IV were excluded from the study (Supplementary Fig. 1). The median age at diagnosis was 52 years [IQR: 39–66] with 463 (45.4%) men and 556 (54.6%) women. Median relative telomere length (represented as the ratio of telomere to single copy gene, T/S) in leukocytes, measured successfully in 995 patients, was 1.20 [inter-quartile range, IQR: 0.96–1.48]. The median follow-up period following the disease diagnosis was 103 months [IQR 60.62–155.11]. Of the 1019 patients, 97 (9.5%) had died due to melanoma. The remaining 922 patients (90.5%) were censored either at their last follow-up date (840 patients) or at death from unrelated causes (82 patients; Supplementary Table S1).
The patients included in the study were further genotyped for rs1317082, rs7726159 and rs6060627 polymorphisms that have been shown to associate with telomere length in genome wide association studies30. Since the risk prediction varied with individual polymorphisms, a weighted genetic score was computed based on genotype data (Supplementary Table S2). The computed weighted genetic risk score due to genotypes with variant alleles for three polymorphisms ranged from −0.34 to 0.39 for rs1317082, −0.61 to 0.00 for rs7726159 and −0.46 to 0.20 for rs6060627 (Supplementary Table S2). The composite genetic risk score, computed for each individual as a sum of weighted scores for all the three polymorphisms, ranged from −1.41 to 0.59 (Median: 0.00; IQR: −0.18 to 0.20).
In univariate Cox regression analysis with telomere length as a continuous variable, an improved melanoma-specific patient survival was observed with increased telomere length (HR 0.65, 95% CI 0.42–1.00, P 0.05). The telomere data was transformed by taking the natural log of telomere length. On this scale, the effect of each positive unit of ln(telomere length) was estimated to have a HR of 0.46 (95% CI 0.26–0.81) on melanoma-specific survival. The data analysis with telomere length as a dichotomous variable based on median distribution also showed that the patients with short telomeres (≤1.20) were at risk of poor survival (log rank P 0.001; HR 2.05, 95% CI 1.33–3.16, Fig. 1) compared to the patients with long telomeres (>1.20). A similar analysis with telomere length as categorical variable based on quartile distribution showed that the patients in the first quartile compared to those in the fourth quartile (longest telomeres) had the worst survival (HR 2.73, 95% CI 1.47–5.07) followed by patients in the second quartile (HR 1.71, 95% CI 0.88–3.32) and the third quartile (HR 1.42, 95% CI 0.72–2.82).
Figure 1.
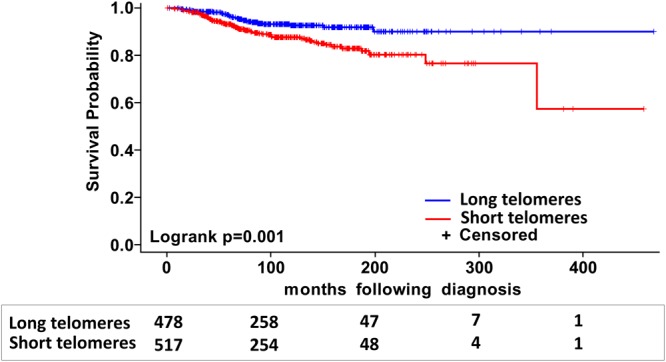
Kaplan-Meier analysis of differences in survival of patients with stage I and II melanoma based on telomere length. Patients were divided into two groups with short (≤1.20) and long (>1.20) telomeres based on median distribution. The numbers of patients at risk in each category, at respective survival intervals, are shown underneath.
Other factors that affected patient survival as determined by Kaplan-Meier and univariate Cox regression analyses included age at diagnosis (P < 0.0001), sex (log rank P 0.0006), nevus count (log rank P 0.03), tumor location (log rank P 0.0005), tumor stage (log rank P < 0.0001), Breslow thickness (log rank P < 0.0001) and tumor ulceration (log rank P < 0.0001) (Supplementary Table S1). Since tumor stage is defined by Breslow thickness, presence/absence of ulceration and tumor mitotic rate, for multivariate analysis only tumor stage was included in the model.
As a continuous variable increased composite genetic risk score showed a statistically significant association with poor melanoma-specific survival (HR for per unit of score: 2.94, 95% CI 1.54–5.62). Similarly, a dichotomous model showed that the patients with higher than median composite genetic risk score (>0) were associated with poor melanoma-specific survival compared to patients with lower than median genetic risk score (log rank P 0.02; HR 1.67, 95% CI 1.12–2.49, Fig. 2a). Segregation of patients based on composite risk score and telomere length into 4 categories, showed that the patient group under high-risk category with short telomeres carried the highest risk for mortality (HR 4.28, 95% CI 1.97–9.28) followed by patients in low-risk category with short telomeres (HR 3.13, 95% CI 1.43–6.88), high-risk category with long telomeres (HR 2.51, 95% CI 1.10–5.74) compared to the low-risk category with long telomeres (Log rank P 0.001, Fig. 2b).
Figure 2.
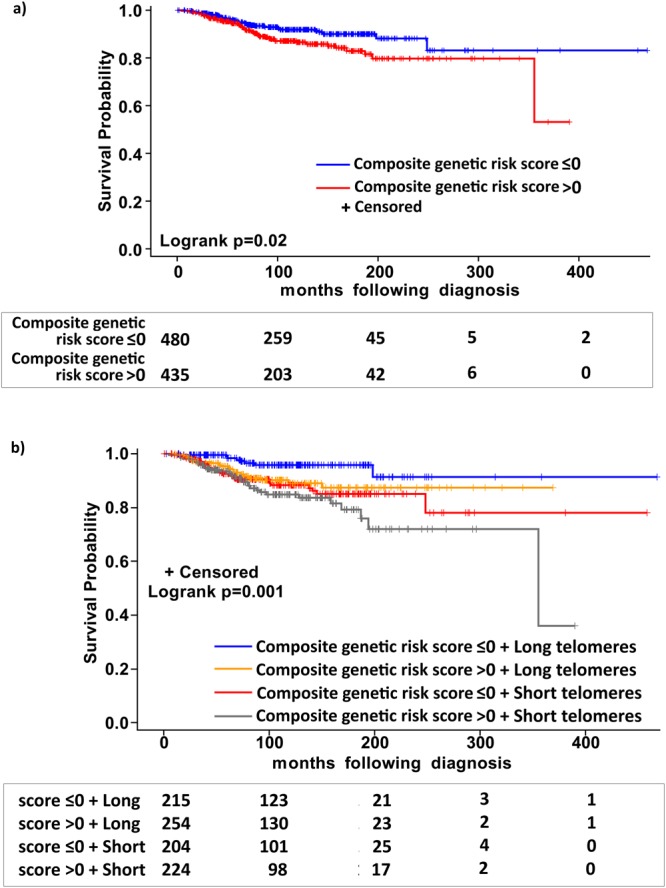
Kaplan-Meier analysis of difference in survival in patients with Stage I and II melanoma; (a) after stratification based on the median distribution of composite weighted genetic score; (b) after stratification into four sub-groups based on median distributions of composite weighted genetic score and telomere length (i) with composite genetic score ≤0 and long telomeres (ii) with composite genetic score >0 and long telomeres (iii) with composite genetic score ≤0 and short telomeres and (iv) with composite genetic score >0 and short telomeres. The numbers of patients at risk in each category are shown underneath at the respective survival intervals.
Telomere-length and patient age at diagnosis
We observed a statistically significant inverse correlation (Pearson correlation r −0.19, P < 0.0001) between telomere length and the patient age at diagnosis. A statistically significant interaction (P interaction 0.05) was also observed in the survival analysis that included age, telomere length and age*telomere length interaction term. With inclusion of age in the model, the association between decreased telomere length and survival was no longer statistically significant (HR 1.50, 95% CI 0.96–2.34). None of the other confounders individually affected the association between telomere length and patient survival (data not shown). Therefore, patients were stratified into quantiles based on the age and the effect of telomere length on melanoma-specific survival was measured using the median telomere length for each subgroup separately. The results showed that the patients below 30 years of age at diagnosis (first quantile) with short telomeres were at risk of poor survival (HR 3.91, 95% CI 1.25–12.29) compared to the patients with long telomeres within that sub-group. The corresponding HR for poor survival for patients with short telomeres compared to patients with long telomeres in the second quantile (30–40 years) was 2.89 (95% CI 1.20–6.91). For the third quantile (40–50 years), the corresponding HR was 2.13 (95% CI 1.13–3.99). The association between patients with short telomeres and poor melanoma-specific survival in the fourth quantile (50–60 years) was also statistically significant (HR 1.57, 95% CI 1.00–2.47). For patients in the fifth (60–70 years) and higher quantiles, the similar associations were no t statistically significant (Table 1).
Table 1.
Effect of telomere-length on melanoma-specific survival in different quantiles based on patients age at diagnosis.
| Univariate analysis* | Multivariate analysisƗ | ||||||
|---|---|---|---|---|---|---|---|
| Age groups | Telomere lengthǂ | Number | Dead | HR (95% CI) | Number | Dead | HR (95% CI)¶ |
| <30 years | Long (T/S > 1.45) | 41 | 0 | ref | 36 | 0 | ref |
| Short (T/S ≤ 1.45) | 48 | 4 | 3.91 (1.25–12.29) | 43 | 4 | 3.82 (1.10–13.30) | |
| 30–40 years | Long (T/S > 1.30) | 87 | 4 | ref | 73 | 4 | ref |
| Short (T/S ≤ 1.30) | 91 | 7 | 2.89 (1.20–6.91) | 76 | 4 | 2.69 (1.03–7.03) | |
| 40–50 years | Long (T/S > 1.27) | 95 | 6 | ref | 80 | 4 | ref |
| Short (T/S ≤ 1.27) | 99 | 9 | 2.13 (1.13–3.99) | 82 | 5 | 1.90 (0.95–3.82) | |
| 50–60 years | Long (T/S > 1.10) | 98 | 3 | ref | 86 | 2 | ref |
| Short (T/S ≤ 1.10) | 100 | 6 | 1.57 (1.00–2.47) | 90 | 6 | 1.34 (0.81–2.23) | |
| 60–70 years | Long (T/S > 1.06) | 95 | 10 | ref | 81 | 9 | ref |
| Short (T/S ≤ 1.06) | 98 | 16 | 1.16 (0.74–1.80) | 89 | 15 | 0.95 (0.58–1.54) | |
| 70–80 years | Long (T/S > 1.02) | 57 | 11 | ref | 45 | 9 | ref |
| Short (T/S ≤ 1.02) | 59 | 11 | 0.85 (0.47–1.56) | 52 | 9 | 0.67 (0.35–1.28) | |
| >80 years | Long (T/S > 0.90) | 11 | 3 | ref | 9 | 3 | ref |
| Short (T/S ≤ 0.90) | 16 | 3 | 0.63 (0.27–1.46) | 15 | 2 | 0.47 (0.19–1.16) | |
*Main effects: Telomere length (P 0.02), Age (P < 0.0001); Interaction effect: telomere length*age (P0.05).
ƗMain effects: Telomere length (P 0.03), Age (P 0.0001); Interaction effect: telomere length*age (P0.03).
ǂTelomere length was dichotomized at median distribution of T/S ratios within corresponding age groups.
¶Hazard ratio (HR) in multivariate model was adjusted for composite genetic risk score, gender, number of nevi, tumor location and tumor stage. HR values in bold indicate statistical significance (P < 0.05).
Multivariate analysis
Multivariate analysis that included age, composite genetic risk score, sex, nevus number, tumor localization, and tumor stage showed that the association of patients with short telomeres (compared to patients with long telomeres) with decreased melanoma-specific survival was not statistically significant (HR 1.17, 95% CI 0.71–1.94, P 0.53; Table 2). A similar result was obtained using telomere length as a continuous variable (HR 0.95, 95% CI 0.67–1.36, P 0.79) and after transformation into log-scale, the HR for the effect of long telomeres on survival was 0.92 (95% CI 0.52–1.62, P 0.76). However, the increased composite genetic risk-score, in the multivariate model, was a statistically significant independent prognostic factor for poor survival (HR 2.75, 95% CI 1.34–5.62). Additionally, the patients with nevus count >50 showed better survival (HR 0.54, 95% CI 0.21–1.38, P 0.20) than those with nevus count <50 (Table 2). Further, multivariate analysis using telomere length and composite genetic risk score as a combined variable showed that the patients under high-risk category with both short and long telomeres showed an increased mortality compared to patients in low-risk category with long telomeres (Table 3). Similarly, a multivariate model that included all confounders showed that the patients with short telomeres in the age group below 30 years (first quantile) were at statistically significant risk of poor survival (HR 3.82, 95% CI 1.10–13.30) compared to patients with long telomeres within the same group. A similar statistically significant association between patients with short telomeres and poor melanoma-specific survival was also observed in the age group 30–40 years (HR 2.69, 95% CI 1.03–7.03). The effect of telomere length on patient survival diminished in subsequent quantiles (Table 1).
Table 2.
Multivariate Cox analysis of the effect of telomere-length on melanoma-specific survival.
| N | Dead | HR (95% CI) | P | ||
|---|---|---|---|---|---|
| Telomere length | Long (ratio >1.20) | 405 | 25 | ref | |
| Short (ratio ≤1.20) | 452 | 51 | 1.17 (0.71–1.94) | 0.53 | |
| Composite genetic risk score | (continuous) | 857 | 76 | 2.75 (1.34–5.62) | 0.006 |
| Age at diagnosis | (continuous) | 857 | 76 | 1.03 (1.02–1.05) | 0.0001 |
| Sex | Males | 389 | 43 | ref | |
| Females | 468 | 33 | 0.75 (0.45–1.23) | 0.25 | |
| Nevus count | <50 | 731 | 71 | ref | |
| >50 | 126 | 5 | 0.54 (0.21–1.38) | 0.20 | |
| Tumor location | Axial | 476 | 50 | ref | |
| Extremities | 322 | 17 | 0.57 (0.32–1.02) | 0.06 | |
| Acral/Mucosal | 59 | 9 | 1.00 (0.47–2.12) | 1.00 | |
| Tumor stage* | 1 (1A, 1B) | 616 | 27 | ref | |
| 2 (2A, 2B, 2C) | 241 | 49 | 4.05 (2.49–6.60) | <0.0001 |
*As tumor stage is defined by Breslow thickness, presence/absence of ulceration and tumor mitotic rate, therefore only tumor stage was included in the analysis.
Table 3.
Multivariate analysis of the effect of telomere length and composite genetic risk score on melanoma-specific survival.
| Composite genetic risk score/Telomere length* | N | Dead | HR (95% CI)Ɨ | P | |
|---|---|---|---|---|---|
| Telomere length | Low risk + long telomeres | 208 | 8 | ref | |
| Low risk + short telomeres | 239 | 24 | 1.74 (0.78–3.99) | 0.17 | |
| High risk + long telomeres | 197 | 17 | 2.41 (1.04–5.61) | 0.04 | |
| High risk + short telomeres | 213 | 27 | 2.19 (0.97–4.91) | 0.06 |
*The composite genetic risk score was dichotomized at median distribution (low risk ≤ 0 and high risk > 0). The telomere length was dichotomized at median distribution of T/S ratios (long > 1.20 and short ≤ 1.20).
ƗHazard ratio (HR) was adjusted for age at diagnosis, gender, number of nevi, tumor location and tumor stage.
Further we also carried age-matched analysis of the patients with “short” and “long” telomeres. Exact matching of the groups (detailed in methods section) resulted in a subset of 676 patients with 338 in each group with a median age of 52 (Supplementary Fig. 2). Cox regression analysis on the age matched group showed that the patients with short telomeres compared to those with long telomere were at statistically significant risk of poor survival (univariate HR 1.89, 95% CI 1.10–3.25, P 0.02). Multivariate data analysis also showed in age-matched groups, the patients with short telomeres were at the risk of poor survival (HR 1.79, 95% CI 1.01–3.17, P 0.05; Table 4) compared to the patients with long telomeres.
Table 4.
Multivariate analysis of effect of telomere length on melanoma-specific survival in age-matched groups.
| N | Dead | HR (95% CI) | P | ||
|---|---|---|---|---|---|
| Telomere length | Long (T/S ratio > 1.20) | 290 | 18 | ref | |
| Short (T/S ratio ≤ 1.20) | 308 | 36 | 1.79 (1.01–3.17) | 0.05 | |
| Composite genetic risk score | (continuous) | 598 | 54 | 2.89 (1.26–6.59) | 0.01 |
| Sex | Males | 254 | 33 | ref | |
| Females | 344 | 21 | 0.49 (0.27–0.87) | 0.02 | |
| Tumor location | Axial | 326 | 34 | ref | |
| Extremities | 228 | 12 | 0.60 (0.30–1.19) | 0.14 | |
| Acral/Mucosal | 44 | 8 | 2.34 (1.03–5.31) | 0.04 | |
| Tumor stage* | 1 (1A, 1B) | 436 | 22 | ref | |
| 2 (2A, 2B, 2C) | 162 | 32 | 3.60 (2.05–6.30) | <0.0001 |
*As tumor stage is defined by Breslow thickness, presence/absence of ulceration and tumor mitotic rate, therefore only tumor stage was included in the analysis.
Discussion
In this study based on stage I and II incident melanoma patients, we observed that short telomeres predispose patients to poor melanoma-specific survival, which contrasts with reported association of long telomere with increased risk of melanoma26,31. Despite a strong inverse correlation between telomere length and age, we observed that the effect of short compared to long telomeres on poor survival was pronounced in patients below 40 years at diagnosis. We also used three polymorphisms located at the TERC, TERT and BCl2L1 loci to generate composite genetic risk score. The polymorphisms that had been previously shown to be associated with telomere length and risk of various cancers through genome wide association studies did not per se show any impact on melanoma-specific survival in this study23,30,32. However, the use of composite genetic risk score together with telomere length strengthened the prediction of melanoma-specific survival in this study. Our results were further corroborated through use of age-matched analysis, which showed that patients with short telomeres were at a statistically significant poorer risk of survival than the patients with long telomeres.
Increased risk of melanoma due to increased telomere length has been postulated as a paradox where sufficient telomere length allows cells to survive until crisis point through continuous division and consequent acquisition of driver mutations33. Multiple alleles that associate with increased telomere length have been shown to increase the risk of various cancers27,31,34–36. Many of the variants associated with constitutive telomere length have been shown to functionally increase telomerase levels37,38. However, it is the critically short telomeres that trigger events leading to genomic instability through end-to-end chromosomal fusion with ultimate emergence of cells from crisis through increased telomerase levels39,40. As shown in this and earlier studies, the shorter telomeres tend to associate with poor survival41. In previous studies on breast cancer, multiple myeloma and renal cell carcinoma, short telomeres were shown to be associated with poor overall survival41–44. Similarly, decreased telomere length was reported to be independently associated with worst survival in patients with idiopathic pulmonary fibrosis; longer donor leukocyte telomere length associated with increase five year survival in patients receiving hematopoietic cell transplantation for severe aplastic anemia but not for patients with acute leukemia45–47. Reduced telomere length in general, in a large prospective study, was shown to be associated with increased risk of infections48.
The observed association of short telomeres with poor melanoma-specific survival seems to be in conformity with emerging data49. Studies have shown that despite ubiquitous telomerase rejuvenation, a continued shortening of telomeres in tumor cells, particularly in presence of the TERT promoter mutations, in the initial stages lead to chromosomal fusions and aneuploidy21,49. Longer telomeres afford ample time for continued cell division until telomerase rejuvenation and telomere stabilization, hence the association with increased risk. In contrast, once a patient has developed the disease, it is the shorter telomeres in tumors that would presumably lead to rapid chromosomal fusions and aneuploidy resulting in observed poor outcome49,50. Evidence suggests that genetic instability drives tumorigenesis51. Distinct types of aneuploidy predict two main hallmarks of cancer, cell proliferation and immune evasion; tumor aneuploidy was reported to correlate inversely with patient survival in clinical trials of immune checkpoint blockade anti-CTLA-4 therapy for metastatic melanoma52.
Previously, we have shown that the TERT promoter mutations associated with poor survival in patients with primary melanoma and our subsequent data showed shorter telomeres in tumors with than without the TERT promoter mutations9,53–55. In this study, telomere length was measured in blood cells and not in tumor tissues; therefore, the association of short telomere length with decreased survival cannot be directly explained by an effect in tumor cells. The telomere attrition in blood leukocytes has been considered as a marker of biological age and also associated with immune dysfunctions56,57. However, we lacked data on immune competency for the patients included in the study to investigate any such correlation.
In this study, however, we show that in contrast to the association between long telomeres and increased melanoma risk, short telomeres predict poor melanoma-specific survival. Other factors that associated with poor survival, besides age at diagnosis, included sex, tumor stage, and reduced nevus count. While increased nevus number is an established a risk factor in melanoma, an earlier report had also shown that high nevus count confers a favorable outcome in melanoma5,58. One report also linked increased nevus number with longer telomeres59. With age as a strong confounder, the risk of poor survival due to short telomeres on survival was in particularly pronounced in younger patients. However, it may be pointed out that the telomere length can be affected a number of other factors including chronic diseases60,61. The lack of relevant data on those factors remains a weakness of the present study and requires cautious interpretation of the results.
Methods
Patient material
The patients included in the present study came from the database maintained at the Instituto Valenciano de Oncologia, a referral skin cancer center for the provinces of Valencia, Alicante and Castellon with a catchment population of approximately 5 million. The database contained a total of 2471 melanoma patients. The number of incident cases recruited between 200 and 2014 was 1712. For this study we included incident cases from which we excluded stage 0 (298 patients), stage III (294 patients), stage IV (31 patients) and 30 patients with melanoma at extra-cutaneous sites. All cases had been staged including lymph node biopsy. Thus, for this study 1059 patients with localized invasive stage I and II melanoma were eligible; however for 40 patients, blood samples were not available. At the end we included 1019 incident cases with histologically confirmed stage I and II melanoma (Supplementary Fig. 1).
Blood samples from the selected patients at the time of diagnosis were collected after informed patient consent between 2000 and 2014 and information remains documented in the departmental database. The patients also provided informed consent to the use of their clinical records in this study. Clinical and pathological data from patients were collected since January 2000 through the review of medical history, personal interview, and clinical examination by expert dermatologists (EN and CR) at the first visit. The available patient information included details about the patient age at diagnosis, sex, nevus count, tumor location, tumor stage, Breslow thickness, tumor ulceration, tumor mitotic rate, histology, lentigines, non-melanoma skin cancer, tumor regression, and smoking (Supplementary Table 1).
The study was approved by the IVO Ethics Committee and all methods were performed in accordance with the relevant guidelines and regulations. DNA from blood was extracted using standard kits (Qiagen).
Relative telomere length measurement
The relative telomere length (TL) was measured in DNA from peripheral blood leucocytes using a monochrome multiplex quantitative real-time PCR (MMqPCR) method that compares telomere repeat copy number (T) to the copy number of a single-copy gene, albumin (S). The relative TL in a given DNA sample was calculated as a corresponding T/S ratio62,63. Each MMqPCR optical 384-well reaction plate contained DNA in triplicates from blood leucocytes, standard DNA and non-template controls. The standard DNA was obtained by pooling the leucocyte genomic DNA from 15 healthy individuals (40–50 years of age). A two-fold serial dilution of standard DNA in seven concentrations (30 ng to 0.47 ng) was used in each plate for quantification. The PCR reaction was performed in a 10 μL reaction volume using 2 μL of 5 × HOT FIREPol Probe qPCR Mix Plus with ROX (Solis BioDyne, Tartu, Estonia), 1.5 μM of Syto 9 (Invitrogen, Carlsbad, CA) and 2–5 ng of genomic DNA. Four primers were used in each reaction to amplify telomere repeats (telg at 200 nM and telc at 400 nM) and albumin gene (albugcr2 at 200 nM and albdgcr2 at 400 nM). The real-time PCR experiments were carried out using the sequence detection system (Applied Biosystems Viia-7) using two simultaneous programs to acquire the respective Ct values for telomere repeats and albumin (control) gene. The conditions for amplification of telomere repeats were 95 °C/15 min, 2 cycles of 95 °C/20 sec and 49 °C/1 min, followed by 25 cycles of 85 °C/20 sec with signal acquisition at 59 °C/30 sec. Thermal conditions for albumin gene were 35 cycles of 95 °C/15 sec, 85 °C/30 sec, with signal acquisition at 84 °C/30 sec. The specificity of all amplifications was determined by melting curve analysis done at default settings (95 °C/15 sec, 60 °C/1 min with continuous signal acquisition at 0.05 °C/sec ramping, 95 °C/15 sec). The efficiency of PCR amplification and T and S values for each sample were determined from the respective standard curves of telomere and albumin reactions. The RTL was expressed as the ratio between T/S by taking mean of the triplicate values for each sample. Inter-assay variation and intra-assay variation was determined by duplicating the reference DNA for all the dilutions in all the assays performed. The PCR efficiencies for telomere and albumin gene assay ranged between 96% and 101%. The interassay coefficients of variation of the telomere and albumin gene assays were 3.5% and 2.67%, respectively. The intra-assay coefficients of variations of the telomere and albumin gene assays were 1.04% and 0.55%, respectively.
Statistical analysis
Differences between the patient groups with short and long telomeres were assessed using chi-square test for categorical variables or two sample t-tests for continuous variables. The differences in telomere length in different sample batches procured at different time points were assessed using one-way ANOVA in generalized linear model. All the statistical tests were two-sided and P < 0.05 was considered significant. The telomere length as T/S ratio was taken as continuous variable (with and without log-transformation) or as categorical variable. The telomere length was dichotomized based on the median distribution, with patients with T/S ratio ≤ 1.20 categorized as the group with ‘short telomeres’ and those with T/S > 1.20 as the group with ‘long’ telomeres. Alternatively, patients were grouped into quartiles based on telomere length with T/S 0.48–0.96 for the first quartile, 0.97–1.19 for the second quartile, 1.20–1.48 for the third quartile and 1.49–4.85 for the fourth quartile (reference). All the other clinical covariates included in the analysis were categorical variables. Patient age was considered as a continuous variable unless otherwise specified.
The Kaplan–Meier method was employed to draw the cumulative survival curves using time period (in months) between the date of diagnosis and the date of cause-specific death (death from melanoma). The patients alive at the end of follow-up period and patients that died from other causes were censored. Death from melanoma was confirmed in most patients by the specialists in charge (E.N. and V.S.), who followed them until the date of death. In a few remaining patients, mainly people living far from the Institute and who spent their last days at home or at the regional hospital, the cause of death was assessed by phone contact with the personnel in charge of the palliative care unit in the area.
Statistical differences between the groups were analyzed by the log-rank test. The association of the independent variables with melanoma specific patient survival was assessed using univariate methods. Hazard ratios (HRs) and 95% confidence intervals (95% CI) were estimated in Cox regression models. For multivariate analysis, the variables that associated statistically significantly (P < 0.05) in univariate analysis, were included in the model. The patient survival time and age were inversely correlated (censored/alive, estimate: −1.08, P > 0.0001; dead, estimate: −1.11, P 0.0004). To measure the telomere length and age interaction on patient survival, a Cox regression model was implemented using telomere length and age as main effects and telomere length*age as interaction effect. For this purpose, patient cohort was divided into 7 quantiles, each representing a 10-year interval based on patient age at diagnosis (1st quantile: <30 years, 2nd quantile: 30–40 years, 3rd quantile: 40–50 years, 4th quantile: 50–60 years, 5th quantile: 60–70 years, 6th quantile: 70–80 years and 7th quantile: >80 years). The telomere length was dichotomized into long and short telomeres based on the median telomere length (T/S ratio) for each quantile. In Cox regression model, telomere length and age were considered as dichotomous and ordinal variable, respectively.
Additionally, to minimize the confounder effect of age in determining the effect of telomere length on patient survival, the two groups of patients with, short (median ≤1.20) and long (>1.20) telomeres were matched for age. Before matching, the number of patients in “short” and “long” telomere group was 517 and 478 with median age 57 years (IQR: 46–68) and 46 years (IQR: 35–61), respectively. To make the groups comparable, an exact matching was carried out using Proc SQL in SAS and the “long” and “short” groups of patients were considered as controls and cases, respectively. The data used for this purpose contained three variables – patient identification, short/long telomeres, and the age at diagnosis. The method of matching was based on (1) for each study case, control cases are matched for age; (2) if a control matched with more than one case, one control was randomly selected; (3) randomly selected desired number of controls for each case. The matching resulted in two identical groups with respect to age, each with 338 patients (Median: 52 years, IQR: 41–65). The method implemented was based on Kawabata et al. (2004) Proceedings of the Twenty-Ninth Annual SAS Users Group International Conference. Cary, NC: SAS Institute Inc.
To determine the effect of polymorphisms on patient survival, the genotype data were analyzed under the additive, dominant and recessive models. The polymorphisms were categorized into carriers and non-carriers based on the association of alleles with short telomeres. Further, a weighted genetic risk score was calculated for the three genotypes of each polymorphism from Cox regression analysis. Using homozygous common genotype as the reference, the beta coefficients for the other two genotypes (heterozygote and homozygous variant genotypes) were calculated from Cox regression analysis and used as the corresponding weight. Only patients with complete genotype data for three polymorphisms were included for computing weighted score. The weighted genetic risk score thus calculated for all three SNPs was summed for each individual based on their genotypes to get composite genetic risk score for that individual. The composite genetic risk score was subsequently implemented in the analyses as a continuous variable in the multivariate analyses. To analyze the combined genetic effect and telomere length on survival, patients were categorized into 4 categories as high risk (composite genetic risk score >0) and long telomeres (median >1.20), high risk (composite genetic risk score >0) and short telomeres (median ≤1.20), low risk (composite genetic risk score ≤0) and long telomeres (median >1.20), low risk (composite genetic risk score ≤0) and short telomeres (median ≤1.20). All statistical analyses were carried out using SAS version 9.4 (SAS Institute Inc., Cary, NC).
Electronic supplementary material
Acknowledgements
The study was supported by a grant from TRANSCAN through the German Ministry of Education and Research (BMBF), Dutch Cancer Society, Norwegian Research Council, Norwegian Cancer Society, Latvian Academy of Sciences and Italian Ministry of Health under the grant number 01KT15511 and German Consortium for Translational Research (DKTK) for the NomCom project. Patient material for the study was retrieved from the Biobank of the Instituto Valenciano de Oncologia.
Author Contributions
R.K., E.N., N.G., A.M., D.P., and M.C. designed the study. S.R. measured telomere length and N.S. performed genotyping. Z.G.-C., C.R., V.T. V.S., and E.N. provided blood samples from patients along with data. V.T. provided pathological confirmation. S.R. with contribution from S.H.M. performed statistical analysis. R.K. together with S.R. drafted the manuscript with input from all for the final version.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eduardo Nagore and Rajiv Kumar contributed equally to this work.
Change history
12/14/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29322-9.
References
- 1.Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383:816–827. doi: 10.1016/S0140-6736(13)60802-8. [DOI] [PubMed] [Google Scholar]
- 2.Braeuer RR, et al. Why is melanoma so metastatic? Pigment cell & melanoma research. 2014;27:19–36. doi: 10.1111/pcmr.12172. [DOI] [PubMed] [Google Scholar]
- 3.Long GV, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nature communications. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 4.In ‘t Hout FE, et al. Prognostic importance of the extent of ulceration in patients with clinically localized cutaneous melanoma. Ann Surg. 2012;255:1165–1170. doi: 10.1097/SLA.0b013e31824c4b0b. [DOI] [PubMed] [Google Scholar]
- 5.Ribero S, et al. High nevus counts confer a favorable prognosis in melanoma patients. International journal of cancer. 2015;137:1691–1698. doi: 10.1002/ijc.29525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balch, C. M. et al. Age as a Prognostic Factor in Patients with Localized Melanoma and Regional Metastases. Ann Surg Oncol, 10.1245/s10434-013-3100-9 (2013). [DOI] [PMC free article] [PubMed]
- 7.Nagore E, et al. Prognostic factors in localized invasive cutaneous melanoma: high value of mitotic rate, vascular invasion and microscopic satellitosis. Melanoma research. 2005;15:169–177. doi: 10.1097/00008390-200506000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Azzola MF, et al. Tumor mitotic rate is a more powerful prognostic indicator than ulceration in patients with primary cutaneous melanoma: an analysis of 3661 patients from a single center. Cancer. 2003;97:1488–1498. doi: 10.1002/cncr.11196. [DOI] [PubMed] [Google Scholar]
- 9.Nagore E, et al. TERT promoter mutations in melanoma survival. International journal of cancer. 2016;139:75–84. doi: 10.1002/ijc.30042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidenreich B, et al. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nature communications. 2014;5:3401. doi: 10.1038/ncomms4401. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomic Classification of Cutaneous Melanoma. Cell161, 1681–1696, 10.1016/j.cell.2015.05.044 (2015). [DOI] [PMC free article] [PubMed]
- 14.Griewank, K. G. et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. Journal of the National Cancer Institute106, 10.1093/jnci/dju246 (2014). [DOI] [PMC free article] [PubMed]
- 15.Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science (New York, N.Y.) 2015;350:1193–1198. doi: 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- 16.Heidenreich B, Kumar R. TERT promoter mutations in telomere biology. Mutat Res. 2017;771:15–31. doi: 10.1016/j.mrrev.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steenstrup T, et al. Telomeres and the natural lifespan limit in humans. Aging. 2017;9:1130–1142. doi: 10.18632/aging.101216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouquerel E, et al. Oxidative guanine base damage regulates human telomerase activity. Nature structural & molecular biology. 2016;23:1092–1100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barthel FP, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nature genetics. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science (New York, N.Y.) 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 22.Hjelmborg JB, et al. The heritability of leucocyte telomere length dynamics. Journal of medical genetics. 2015;52:297–302. doi: 10.1136/jmedgenet-2014-102736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Codd V, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nature genetics. 2013;45(422-427):427e421–422. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broer L, et al. Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur J Hum Genet. 2013;21:1163–1168. doi: 10.1038/ejhg.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codd V, et al. Common variants near TERC are associated with mean telomere length. Nature genetics. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haycock PC, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017;3:636–651. doi: 10.1001/jamaoncol.2016.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh KM, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6:42468–42477. doi: 10.18632/oncotarget.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read J, Wadt KA, Hayward NK. Melanoma genetics. Journal of medical genetics. 2016;53:1–14. doi: 10.1136/jmedgenet-2015-103150. [DOI] [PubMed] [Google Scholar]
- 29.Law MH, et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nature genetics. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pooley KA, et al. A genome-wide association scan (GWAS) for mean telomere length within the COGS project: identified loci show little association with hormone-related cancer risk. Human molecular genetics. 2013;22:5056–5064. doi: 10.1093/hmg/ddt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rode L, Nordestgaard BG, Bojesen SE. Long telomeres and cancer risk among 95 568 individuals from the general population. International journal of epidemiology. 2016;45:1634–1643. doi: 10.1093/ije/dyw179. [DOI] [PubMed] [Google Scholar]
- 32.Renault AL, et al. Telomere length, ATM mutation status and cancer risk in Ataxia-Telangiectasia families. Carcinogenesis. 2017;38:994–1003. doi: 10.1093/carcin/bgx074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aviv A, Anderson JJ, Shay JW. Mutations, Cancer and the Telomere Length Paradox. Trends Cancer. 2017;3:253–258. doi: 10.1016/j.trecan.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh KM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nature genetics. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iles, M. M. et al. The effect on melanoma risk of genes previously associated with telomere length. Journal of the National Cancer Institute106, 10.1093/jnci/dju267 (2014). [DOI] [PMC free article] [PubMed]
- 36.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nature genetics. 2013;45:371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangino M, et al. DCAF4, a novel gene associated with leucocyte telomere length. Journal of medical genetics. 2015;52:157–162. doi: 10.1136/jmedgenet-2014-102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J, et al. Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148. Nature communications. 2017;8:15034. doi: 10.1038/ncomms15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason PJ, Perdigones N. Telomere biology and translational research. Transl Res. 2013;162:333–342. doi: 10.1016/j.trsl.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weischer M, et al. Short telomere length, cancer survival, and cancer risk in 47102 individuals. Journal of the National Cancer Institute. 2013;105:459–468. doi: 10.1093/jnci/djt016. [DOI] [PubMed] [Google Scholar]
- 42.Hyatt S, et al. Telomere length is a critical determinant for survival in multiple myeloma. British journal of haematology. 2017;178:94–98. doi: 10.1111/bjh.14643. [DOI] [PubMed] [Google Scholar]
- 43.Callahan CL, et al. Leukocyte telomere length and renal cell carcinoma survival in two studies. British journal of cancer. 2017;117:752–755. doi: 10.1038/bjc.2017.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duggan C, et al. Change in peripheral blood leukocyte telomere length and mortality in breast cancer survivors. Journal of the National Cancer Institute. 2014;106:dju035. doi: 10.1093/jnci/dju035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai J, et al. Association between telomere length and survival in patients with idiopathic pulmonary fibrosis. Respirology (Carlton, Vic.) 2015;20:947–952. doi: 10.1111/resp.12566. [DOI] [PubMed] [Google Scholar]
- 46.Gadalla, S. M. et al. No association between donor telomere length and outcomes after allogeneic unrelated hematopoietic cell transplant in patients with acute leukemia. Bone marrow transplantation, 10.1038/s41409-017-0029-9 (2017). [DOI] [PMC free article] [PubMed]
- 47.Gadalla SM, et al. Association between donor leukocyte telomere length and survival after unrelated allogeneic hematopoietic cell transplantation for severe aplastic anemia. Jama. 2015;313:594–602. doi: 10.1001/jama.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helby J, Nordestgaard BG, Benfield T, Bojesen SE. Shorter leukocyte telomere length is associated with higher risk of infections: a prospective study of 75,309 individuals from the general population. Haematologica. 2017;102:1457–1465. doi: 10.3324/haematol.2016.161943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiba K, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science (New York, N.Y.) 2017;357:1416–1420. doi: 10.1126/science.aao0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roger L, et al. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. Journal of the National Cancer Institute. 2013;105:1202–1211. doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- 51.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 52.Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science (New York, N.Y.)355, 10.1126/science.aaf8399 (2017). [DOI] [PMC free article] [PubMed]
- 53.Hosen I, et al. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. International journal of cancer. 2015;137:1621–1629. doi: 10.1002/ijc.29526. [DOI] [PubMed] [Google Scholar]
- 54.Heidenreich B, et al. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget. 2015;6:10617–10633. doi: 10.18632/oncotarget.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heidenreich B, Kumar R. Altered TERT promoter and other genomic regulatory elements: occurrence and impact. International journal of cancer. 2017;141:867–876. doi: 10.1002/ijc.30735. [DOI] [PubMed] [Google Scholar]
- 56.Marioni RE, et al. The epigenetic clock and telomere length are independently associated with chronological age and mortality. International journal of epidemiology. 2017 doi: 10.1093/ije/dyx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weng NP. Telomeres and immune competency. Current opinion in immunology. 2012;24:470–475. doi: 10.1016/j.coi.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emery JD, Usher-Smith JA, Walter FM. Predicting the Risk of Melanoma. JAMA dermatology. 2016;152:875–877. doi: 10.1001/jamadermatol.2016.1574. [DOI] [PubMed] [Google Scholar]
- 59.Bataille V, et al. Nevus size and number are associated with telomere length and represent potential markers of a decreased senescence in vivo. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:1499–1502. doi: 10.1158/1055-9965.epi-07-0152. [DOI] [PubMed] [Google Scholar]
- 60.Epel ES, et al. Accelerated telomere shortening in response to life stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rehkopf DH, et al. Leukocyte Telomere Length in Relation to 17 Biomarkers of Cardiovascular Disease Risk: A Cross-Sectional Study of US Adults. PLoS medicine. 2016;13:e1002188. doi: 10.1371/journal.pmed.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen M, et al. A prospective study of telomere length measured by monochrome multiplex quantitative PCR and risk of lung cancer. Lung Cancer. 2011;73:133–137. doi: 10.1016/j.lungcan.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


