Abstract
While many introduced invasive species can increase genetic diversity through multiple introductions and/or hybridization to colonize successfully in new environments, others with low genetic diversity have to persist by alternative mechanisms such as epigenetic variation. Given that Phragmites australis is a cosmopolitan reed growing in a wide range of habitats and its invasion history, especially in North America, has been relatively well studied, it provides an ideal system for studying the role and relationship of genetic and epigenetic variation in biological invasions. We used amplified fragment length polymorphism (AFLP) and methylation‐sensitive (MS) AFLP methods to evaluate genetic and epigenetic diversity and structure in groups of the common reed across its range in the world. Evidence from analysis of molecular variance (AMOVA) based on AFLP and MS‐AFLP data supported the previous conclusion that the invasive introduced populations of P. australis in North America were from European and Mediterranean regions. In the Gulf Coast region, the introduced group harbored a high level of genetic variation relative to originating group from its native location, and it showed epigenetic diversity equal to that of the native group, if not higher, while the introduced group held lower genetic diversity than the native. In the Great Lakes region, the native group displayed very low genetic and epigenetic variation, and the introduced one showed slightly lower genetic and epigenetic diversity than the original one. Unexpectedly, AMOVA and principal component analysis did not demonstrate any epigenetic convergence between native and introduced groups before genetic convergence. Our results suggested that intertwined changes in genetic and epigenetic variation were involved in the invasion success in North America. Although our study did not provide strong evidence proving the importance of epigenetic variation prior to genetic, it implied the similar role of stable epigenetic diversity to genetic diversity in the adaptation of P. australis to local environment.
Keywords: DAN methylation, epigenetics, methylation‐sensitive amplification polymorphism, Phragmites australis, plant invasion
1. INTRODUCTION
To survive and spread successfully in a new environment distinct from their original native range, invasive plants need phenotypic variation and plasticity in order to adapt to the new habitat (Davidson, Jennions, & Nicotra, 2011), although a few invaders appear preadapted (Dlugosch & Parker, 2007; Schlaepfer, Glättli, Fischer, & van Kleunen, 2010). Some species could obtain more genetic diversity through multiple introductions and hybridization during the invasion process (Genton, Shykoff, & Giraud, 2005; Kelager, Pedersen, & Bruun, 2013; Rosenthal, Ramakrishnan, & Cruzan, 2008; Suehs, Affre, & Médail, 2004), but other invasive species, especially clonal plants, may maintain low levels of genetic diversity after gene drift accompanied by invasion (Hollingsworth & Bailey, 2000; Lambertini et al., 2010; Lindholm et al., 2005; Loomis & Fishman, 2009). While adaptation is accomplished by genetic changes through mutation, drift and selection are very slow for some introduced species, so epigenetic mechanisms can provide an alternative source of ecologically phenotypic diversity for rapid adjustment (Medrano, Herrera, & Bazaga, 2014). Epigenetic diversity can generate massive heritable variation of ecologically relevant plant traits such as root allocation, drought tolerance and nutrient plasticity (Zhang, Fischer, Colot, & Bossdorf, 2013), and it appears to increase the productivity and stability of plant populations in Arabidopsis thaliana under artificial conditions (Latzel et al., 2013). An increasing number of studies have also demonstrated the common existence and significant role of epigenetic variation in plant populations of herbs (Foust et al., 2016; Herrera, Medrano, & Bazaga, 2014; Medrano et al., 2014; Preite et al., 2015; Schulz, Eckstein, & Durka, 2014), shrubs (Avramidou, Ganopoulos, Doulis, Tsaftaris, & Aravanopoulos, 2015; Herrera & Bazaga, 2013, 2016), and trees (Guarino, Cicatelli, Brundu, Heinze, & Castiglione, 2015; Gugger, Fitz‐Gibbon, PellEgrini, & Sork, 2016; Lira‐Medeiros et al., 2010; Platt, Gugger, Pellegrini, & Sork, 2015; Sáez‐Laguna et al., 2014) under natural conditions. Therefore, epigenetic variation can be a very important mechanism for invasive plant success in a broad range of environments (Douhovnikoff & Dodd, 2014; Richards, Schrey, & Pigliucci, 2012).
Among all identified epigenetic mechanisms, including histone modifications, DNA methylation, and small noncoding RNAs, DNA methylation is relatively stable with transgenerational heritability that can be independent of heritable genes (Bird, 2007; Eichten, Schmitz, & Springer, 2014), with the result that DNA methylation has attracted the most attention in epigenetic studies in ecology and evolution (Alvarez, Schrey, & Richards, 2015). The pattern of DNA methylation can affect ecologically important phenotypes and plasticity (Herrera & Bazaga, 2013; Nicotra et al., 2015; Zhang et al., 2013) and may play a significant role in adaptation to various habitat conditions (Foust et al., 2016; Richards et al., 2012; Schulz et al., 2014). For example, a naturally occurring epiallele named “NMR19‐4” has been discovered in Arabidopsis accessions, and its DNA methylation status is inheritable and independent of genetic variation (He et al., 2018). This epiallele controls leaf senescence and associates with local climates. Moreover, an easy and efficient technique named methylation‐sensitive amplification polymorphism (MS‐AFLP) has been widely used to assess DNA methylation status at a great number of random anonymous loci across the entire genome in nonmodel species without sequenced reference genomes (Alonso, Pérez, Bazaga, Medrano, & Herrera, 2016; Schrey et al., 2013).
Phragmites australis (common reed) has a worldwide distribution and has been considered as a model organism for studying plant invasions (Meyerson, Cronin, & Pyšek, 2016) according to the criteria for identifying model organisms in invasion science adapted by Kueffer, Pyšek, and Richardson (2013). Two main introduced lineages in North America have been detected (Guo, Lambertini, Li, Meyerson, & Brix, 2013; Meyerson, Lambert, & Saltonstall, 2010). One, known as Haplotype M (hereafter “INT”), has spread dramatically across much of the North America, especially in the Great Lakes regions (Saltonstall, 2002). The other, represented by Haplotype M1 and I (hereafter “DELTA” and “LAND”), was native to Mediterranean region, sub‐Saharan Africa, and the Middle East and has expanded along the Gulf Coast of the United States and in the northwest of South America (Lambertini, Sorrell, Riis, Olesen, & Brix, 2012; Lambertini, Mendelssohn et al., 2012). The introduced population of P. australis had a higher level of genetic diversity and heritable phenotypic variation in its invasive range than in parts of its native range, as multiple and uncontrolled immigration events may have occurred from different European regions to North American (Lavergne & Molofsky, 2007). Some heritable traits and ecophysiological differences in the common reed may contribute to invasion success but hitherto cannot be explained by particular genetic lineages (Mozdzer, Brisson, & Hazelton, 2013). However, very few investigations of natural epigenetic variation of P. australis have been reported, and previous studies were conducted just at a small scale such as in midcoast Maine (Spens & Douhovnikoff, 2016) and in the Songnen Prairie of China (Qiu, Jiang, & Yang, 2016). The understanding of potential epigenetic mechanisms in the invasion of P. australis is still very limited.
In this study, we first compared patterns of genetic and epigenetic variation in P. australis grown in a common garden collected from around the globe using AFLP and MS‐AFLP methods. We collected samples from native and introduced groups in North America and samples of P. australis from the native geographic regions from where the introduced groups were believed to have originated, in order to be able to detect the direction and the degree of genetic and epigenetic changes during the invasion of P. australis. We tested the following hypotheses: (1) introduced groups increased epigenetic diversity to compensate for the loss of genetic diversity in response to heterogeneous environments, and (2) introduced groups could acquire epigenetic variation similar to that of the native groups faster than genetic variation. In a word, we would expect rapid epigenetic adaptation of introduced groups to new environments before slow genetic accommodation occurs.
2. MATERIALS AND METHODS
2.1. Plant material
All samples were collected in a common garden at Fanggan Research Station of Shandong University in Shandong Province, China (36°26′N, 117°27′E), where they had been growing for 2 years. This station has a typical warm temperate monsoon climate with a hot rainy summer and a cold dry winter under climate regulation effects of forest (Sun, Li, Guan, Liu, & Zhang, 2017). To avoid developmental epigenetic variation, all leaf samples for molecular analyses were picked at the same position of the plant before flowering phase. A total of 75 specimens of P. australis, representing eight phylogeographic groups, were sampled to investigate genetic and epigenetic variation (Table 1). The invasive P. australis (INT) in the Great Lakes region was likely introduced from the native European counterpart (EU) based on haplotype evidence, and hybridization between the native and invasive groups in nature has not yet been documented in North America (Saltonstall, 2002, 2003; Saltonstall, Lambert, & Rice, 2016). The LAND and DELTA groups of P. australis are two of five identified phenotypes growing sympatrically in the Gulf Coast region. The DELTA group is a typical introduction from the Mediterranean region (MED) and has become one predominant lineage in the Gulf Coast region (Lambertini, Sorrell et al., 2012). The origin of the LAND type is still debated, and we treat this type as a native group in our study due to the following reasons: (1) LAND type has existed for a longer time than DELTA type as the genetic evidence supports an ancient introduction for LAND type and a recent introduction for DELTA type (Lambertini, Sorrell et al., 2012), (2) LAND type is not invasive in this area with only scattered occurrences, and (3) we just used LAND type as a reference for the introduced DELTA group, which coexisted under the homogeneous environment of the Gulf Coast region. Phragmites mauritianus may be a hybrid of LAND type origin (Lambertini, Sorrell et al., 2012). Individuals from Australia (FEAU) were also analyzed as P. mauritianus from Tropical Africa (TA) as an outgroup.
Table 1.
Samples from different regions around the word
| Group | Origin | Country | Sample size | Ploidy level |
|---|---|---|---|---|
| NAT | Great Lakes | United States and Canada | 6 | 4 |
| INT | Great Lakes | United States and Canada | 13 | 4 |
| LAND | Gulf Coast | United States | 13 | 4,6 |
| DELTA | Gulf Coast | United States | 6 | 4,6 |
| EU | Europe | Denmark, Italy, etc. | 11 | 4,8 |
| MED | Mediterranean | Italy, Algeria, etc. | 11 | 4 |
| FEAU | Far East/Australia | Australia | 11 | 8,10 |
| TA | Tropical Africa | Uganda | 4 | — |
2.2. AFLP and MS‐AFLP analysis
Genomic DNA was extracted from fresh leaf tissue according to the cetyltrimethylammonium bromide (CTAB) method (Doyle & Doyle, 1987). The yield and quality of extracted DNA were determined with both 0.8% agarose gels and a microscope spectrophotometer. We investigated all specimens for genetic and epigenetic variation with AFLP and MS‐AFLP methods. The method of MS‐AFLP was adapted from a standard AFLP, replacing the MseI enzyme in two separate runs with the methylation‐sensitive enzymes HpaII and MspI using appropriate adaptors and primers. The AFLP and MS‐AFLP protocols were used referring to Schulz et al.(2014). Fragment analysis was performed on an ABI3730XL DNA capillary sequencer (Applied Biosystems, Foster City, USA) with a Rox‐500 internal size standard (Applied Biosystems) in Shandong Academy of Agricultural Sciences, and then the AFLP and MS‐AFLP fragment profiles were scored with PEAK SCANNER v1.0 (Applied Biosystems). Both AFLP and MS‐AFLP marker systems utilized three selective primer combinations. The digestion and PCR were repeated in part of the samples (16, approximately 21% of all) with all six pairs of primers to verify the reliability of used bands.
2.3. Statistical analysis
To determine the DNA methylation status of every locus from the fragment presence/absence scores of both EcoRI‐MspI and EcoRI‐HpaII reactions, the R‐based statistical package “msap” was carried out (Pérez‐Figueroa, 2013). In the selected scoring strategy, the presence of both EcoRI‐MspI and EcoRI‐HpaII bands is an unmethylated state, and the presence of only one band, either EcoRI–HpaII or EcoRI–MspI, represents a methylated state. However, the absence of both EcoRI–HpaII and EcoRI–MspI bands may reveal either the full methylated state or genetic variation. As these samples were from different ramets with expected high genetic diversity and we cannot ignore the possibility of genetic change in the specific locus, so we adopted the approach considering the absence of both EcoRI‐HpaII and EcoRI‐MspI bands as an uninformative state (i.e., missing data) with the function msap (no.bands = “u”). Then, we screened methylation‐susceptible loci (MSL), for which the observed proportion of methylated status across all samples exceeded the error rate‐based threshold (5% by default).
The “msap” package also provides a report on methylation levels for each group, but it is not forthright to estimate the global level of individual methylation. It is unambiguous that the presence in both bands denotes an unmethylated state, the presence in only EcoRI–HpaII denotes a hemimethylated state in external cytosine, and the presence in only EcoRI‐MspI denotes a full‐ or hemimethylated state in internal cytosine, but no consensus exists on the interpretation of the last pattern where neither band is present. The last pattern caused by full methylation or genetic mutation in the target site should be considered as full methylated data, null data, or missing data. Here, all three strategies were used to compare global cytosine methylation percentage among ploidy levels in common reed. The global methylated cytosine was established using the sum of the internal methylated and the external hemimethylated cytosines.
The genetic and epigenetic diversity of every population was determined using the population genetic software GenAlEx 6.5 (Peakall & Smouse, 2012) in Microsoft Excel to calculate Shannon's Information Index (I), percentage of polymorphic loci (P), and unbiased expected heterozygosity (uHe). Hierarchical AMOVAs were calculated to assess the structure of genetic and epigenetic variation. Pairwise population ΦST values were used to evaluate genetic and epigenetic differentiation among all groups. In addition, principal component analysis (PCA) was conducted with the package “msap” to compare the variance between native and introduced populations in the Great Lakes region and between two introduced populations in the Gulf Coast region.
3. RESULTS
3.1. DNA methylation level
Similar DNA methylation levels among all groups were determined through the MS‐AFLP markers except for the NAT and TA groups, which exhibited a relatively high level of uninformative state (i.e., full methylation or absence of target) (Figure 1). The DNA methylation levels of each group calculated from the MSL data revealed that the unmethylated sites were the most common among all populations of P. australis, followed by internal cytosine methylated and hemimethylated, and uninformative sites. In addition, we did not detect significant differences across ploidy levels in our samples of P. australis when we observed these uninformative loci as full methylated (ANOVA: p = .607), missing (ANOVA: p = .645), or null data (ANOVA: p = .897) (Figure 2).
Figure 1.
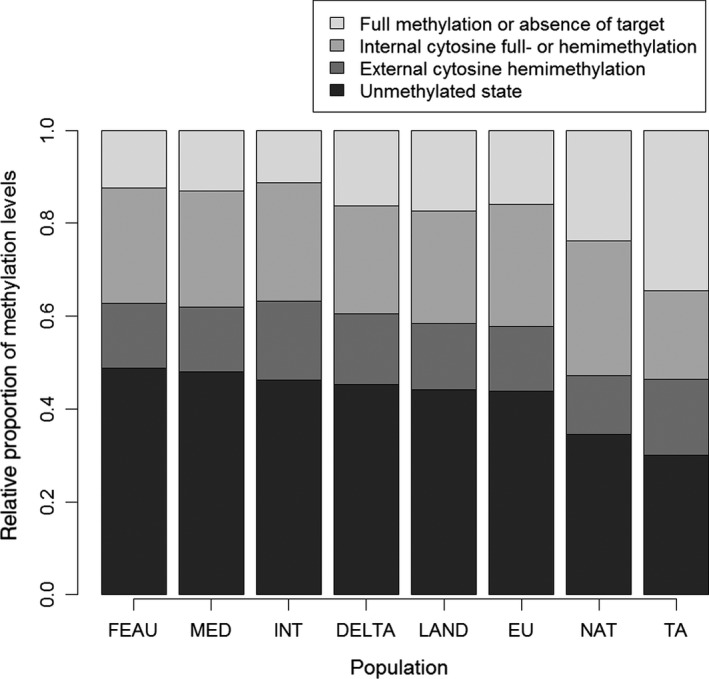
Relative DNA methylation levels in all groups of Phragmites. Among all groups, the percentage of unmethylated states was highest, and internal cytosine methylations were more frequent than external cytosine hemimethylation. The NAT group (native population in the Great Lakes region of Phragmites australis) and the PM group (Phragmites mauritianus) held substantial states of full methylation or absence of target
Figure 2.
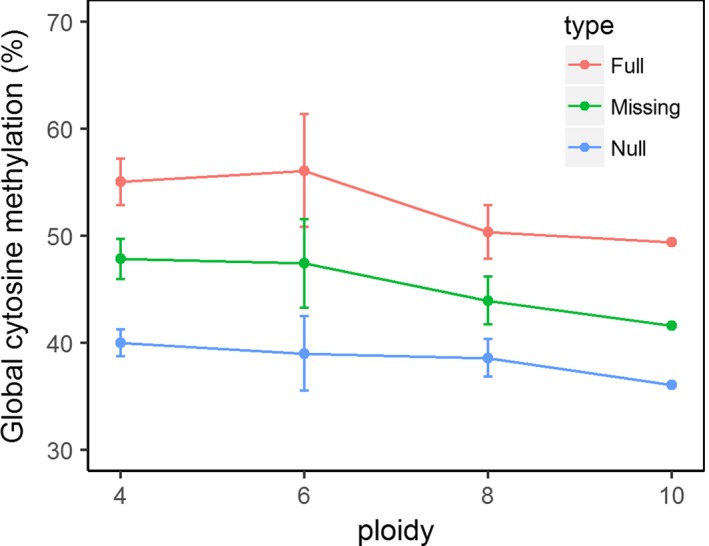
Global cytosine methylation in Phragmites australis leaves from different ploidy levels. Each dot denotes the group mean, bars indicate ± standard error (SE), and colors indicate different scoring methods for treating the last band pattern of MSAP. The global cytosine methylation did not differ significantly across ploidy levels
3.2. Genetic and epigenetic diversity
We obtained 154 polymorphic AFLP loci and 151 MSL loci, and diversity indices showed high levels of both genetic and epigenetic variation (Table 2). Among all groups, LAND had the highest level of genetic diversity while EU had the highest level of epigenetic diversity. Almost all groups of P. australis displayed a higher value of all three diversity indices for epigenetic than genetic variation except for LAND where uHe for genetic variation (uHe = 0.40) was slightly higher than that for epigenetic variation (uHe = 0.39).
Table 2.
Genetic and epigenetic diversity of Phragmites australis
| Pop | I‐gen | %P‐gen | uHe‐gen | I‐epi | %P‐epi | uHe‐epi |
|---|---|---|---|---|---|---|
| DELTA | 0.388 | 0.623 | 0.296 | 0.460 | 0.841 | 0.344 |
| EU | 0.475 | 0.786 | 0.344 | 0.523 | 0.954 | 0.375 |
| FEAU | 0.397 | 0.656 | 0.288 | 0.443 | 0.861 | 0.312 |
| INT | 0.359 | 0.623 | 0.255 | 0.469 | 0.874 | 0.330 |
| LAND | 0.477 | 0.825 | 0.340 | 0.480 | 0.921 | 0.339 |
| MED | 0.385 | 0.630 | 0.280 | 0.487 | 0.940 | 0.343 |
| NAT | 0.241 | 0.429 | 0.178 | 0.312 | 0.530 | 0.240 |
| TA | 0.421 | 0.766 | 0.322 | 0.352 | 0.583 | 0.294 |
| Total | 0.393 | 0.670 | 0.288 | 0.441 | 0.813 | 0.322 |
In the Great Lakes region, the introduced group (INT) exhibited levels of both genetic and epigenetic diversity higher than the native group (NAT) but lower than original groups (EU). Moreover, the native group had the lowest genetic and epigenetic diversity among all groups, and the introduced group followed.
In the Gulf Coast region, the native group (LAND) had a much higher level of genetic diversity than the introduced group (DELTA) but a lower level of epigenetic diversity using the index uHe. The DELTA group had comparable genetic and epigenetic diversity with one possible original group (MED). Compared with another original group, the DELTA group had lower genetic diversity but higher epigenetic diversity.
3.3. Genetic and epigenetic structure
AFLP and MS‐AFLP revealed similar variances among and within groups (see Table 3). Most of the genetic and epigenetic variation existed within rather than among groups, but genetic variance among groups (ΦST = 0.182, p = .001) was more than epigenetic variance (ΦST = 0.072, p = .001).
Table 3.
Pairwise population ΦST values of genetic (above) and epigenetic (below) variation
| DELTA | EU | FEAU | INT | LAND | MED | NAT | TA | |
|---|---|---|---|---|---|---|---|---|
| — | 0.009 | 0.092* | 0.053* | 0.081* | 0.000 | 0.259* | 0.286* | DELTA |
| 0.062* | — | 0.166** | 0.021 | 0.123** | 0.039** | 0.268** | 0.295* | EU |
| 0.034* | 0.067* | — | 0.228** | 0.183** | 0.129** | 0.337** | 0.346* | FEAU |
| 0.008 | 0.029* | 0.084** | — | 0.198* | 0.111** | 0.356** | 0.388** | INT |
| 0.000 | 0.060* | 0.088** | 0.105** | — | 0.130** | 0.204** | 0.230* | LAND |
| 0.103* | 0.018 | 0.046** | 0.052** | 0.039* | — | 0.291** | 0.374* | MED |
| 0.062 | 0.138** | 0.196** | 0.191** | 0.145* | 0.144** | — | 0.334* | NAT |
| 0.062* | 0.079* | 0.113* | 0.167** | 0.116* | 0.114* | 0.163* | — | TA |
9,999 permutations, *p < .05, **p < .001.
In the Great Lakes region, the introduced group INT showed a very low level of genetic (ΦST = 0.021) and epigenetic (ΦST = 0.029) differentiation from the original group EU. The native group NAT had more genetic variance with the introduced group INT (ΦST = 0.356) than the original group EU (ΦST = 0.268), and the NAT group still had more epigenetic variance with the INT group (ΦST = 0.191) than with EU (ΦST = 0.138). The PCA showed less difference in epigenetic variation between the INT and NAT group than in genetic variation, but this tendency was not significant [Figure 3a,b].
Figure 3.
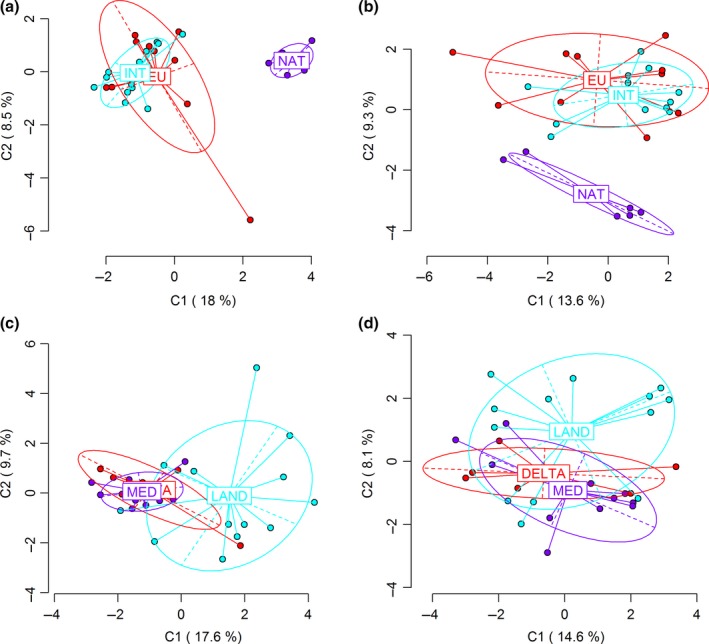
Principal component analysis showing the genetic (a,c) and epigenetic (b,d) variation among several groups in the Great Lakes region (a,b) and the Gulf Coast region (c,d). NAT = the native group in the Great Lakes region; INT = the introduced group in the Great Lakes region; Eu = the noninvasive group in Europe; LAN = the land‐type group in the Gulf Coast region; DELTA = the delta‐type group in the Gulf Coast region; MED = the noninvasive group in the Mediterranean area. Both genetic and epigenetic evidence indicated INT group was similar to EU group (a,b) and DELTA group was similar to MED group (c,d).However, INT group was not closer significantly to NAT group in epigenetic variation (b) relative to genetic variation (a), and DELTA group did not significantly tend toward LAND group in epigenetic variation (c) relative to genetic variation (d)
In the Gulf Coast region, there was low genetic and epigenetic variance (both nearly zero) between the introduced group DELTA and its possible original group MED, and there was a lower level of genetic (ΦST = 0.081) and epigenetic (ΦST = 0.008) differentiation between the LAND group and the DELTA group than that between the LAND group and the MED group (ΦST = 0.130 for genetic variation; ΦST = 0.039 for epigenetic variation). The TA group had the lowest level of genetic differentiation with the LAND group among all groups of P. australis, but not the lowest level of epigenetic differentiation with the LAND group. The PCA displayed a weak decreasing trend of epigenetic differences between the LAND and DELTA group compared with genetic differences [Figure 3c,d].
4. DISCUSSION
This is not an in situ study as all samples were collected from a common garden, which must affect the status of some unstable epigenetic variation. Nevertheless, our results are practical and valuable for the following reasons: (1) it is such a short time in the common garden compared to a hundred years of introduced history that most relatively stable epigenetic variation could remain; (2) the common environment has few chances to cause new epigenetic differences among all individuals. In other words, common garden can separate plastic and heritable components of epigenetic variation (Kilvitis et al., 2014; Richards et al., 2012), and samples in our study still maintain most transgenerational epigenetic variation in which ecologists and evolutionary biologists are interested (Verhoeven, vonHoldt, & Sork, 2016). Moreover, the common garden allowed us to control well the developmental stage, which is the other important factor influencing epigenetic variation besides the environmental one.
4.1. Massive epigenetic variation is correlated with genetic variation
Consistent with the previous study (Spens & Douhovnikoff, 2016), we found a significant difference in epigenetic variation between introduced and native groups. Furthermore, our result revealed that epigenetic structure showed a positive correlation with genetic structure, and epigenetic (as well as genetic) distance also provided coincident evidence for the identified migration of P. australis (Lambertini, Sorrell et al., 2012; Saltonstall, 2002), but epigenetic distance compared with genetic distance was clearly reduced and possibly more related to individual microhabitats. For example, coefficients of genetic differentiation supported the previous haplotype‐based conclusion that P. mauritianus contributed to the hybridization of the LAND type of P. australis (Lambertini, Sorrell et al., 2012), but the epigenetic evidence in our study cannot (Table 3).
The cytosine methylation level in P. australis is relatively steady among geographic and genomic groups, and the detected differences were more due to genetic variation. The reason for the relatively abundant loci with uninformative state in the NAT and TA groups was more likely to be absent from the genetic target rather than full methylation, as the two groups were very different from the other groups in genetic variation (Table 2). Polyploidization is an important evolutionary event in plants and may result in substantial changes in genomewide methylation, but we did not detect the expected increased cytosine methylation with ploidy series (Figure 2). There are very few studies comparing intraspecific DNA methylation levels across wild plants differing in ploidy, and this type of study cannot set apart how much the DNA methylation results were from ploidy versus environmental factors (Alonso, Balao, Bazaga, & Pérez, 2016). MS‐AFLP actually cannot provide a quantitative global DNA methylation forthright (Alonso, Pérez et al., 2016), and the global DNA methylation level in this species should be established by high‐performance liquid chromatography (HPLC) to uncover the role of DNA methylation in natural polyploidizations. Some cytotypes of P. australis can naturally coexist but others cannot (Table 1), so ploidy effects are counteracted by the local environment sometimes. However, the ploidy differences within the samples had little effect on genetic and epigenetic changes during natural invasions in this study.
4.2. The epigenetic diversity may compensate for limited genetic diversity
Many indices have been developed to describe diversity, and all indices perform congruously in common situations; but they may conflict in some cases. In our study, we calculated three indices to compare the genetic and epigenetic diversity among all groups. When divergence occurred, the uHe index was preferred because of the limited number of individuals from each group (Nei, 1978). Consistent with most previous epigenetic studies of natural population differentiation (Choi, Roy, Park, & Kim, 2016; Foust et al., 2016; Kim, Im, & Nkongolo, 2016; Qiu et al., 2016; Schulz et al., 2014), more epigenetic variance existed within groups than genetic variance. One general explanation for this is that a population with limited genetic diversity, especially after genetic drift, can extend its ecological niche through epigenetic variation, which is potentially sensitive to environmental stimulation (Richards, 2006). As some epigenetic variation is affected by developmental factors instead of the environment, common epigenetic variations within populations could be detected. Therefore, numbers of specific loci with ecological effect can and should be determined in rigorous research through generalized linear models or other statistical methods.
Invasive species can increase their genetic diversity through interbreeding with local or other introduced populations in response to diverse habitat environments (Genton et al., 2005; Lavergne & Molofsky, 2007; Rosenthal et al., 2008). Cross‐pollination experiments have demonstrated that that native and introduced populations of Phragmites can hybridize (Meyerson, Viola, & Brown, 2010), but few natural hybridization cases have been detected between the native and introduced lineages (Saltonstall, Castillo, & Blossey, 2014). In the Gulf Coast region, there may be very strong gene flow within and/or among lineages (Meyerson, Lambertini, McCormick, & Whigham, 2012), which caused a relatively high level of genetic diversity. The LAND and DELTA groups were established by independent colonization events (Lambertini, Sorrell et al., 2012), and the LAND group possessed a higher level of genetic diversity than the DELTA group which suggested introduced DELTA group lost some genetic diversity during the invasion process but obtained little genetic variation through interbreeding with other local lineages. This assumption is also supported by the little genetic and epigenetic differentiation between the DELTA and MED groups. Genetic diversity seems to be a key factor explaining its broad ecological adaptation (Kettenring, McCormick, Baron, & Whigham, 2011; Lambertini, Mendelssohn et al., 2012). Compared with LAND group, the DELTA group can adapt to wetter and slightly more saline habitats. Therefore, there is a conflict between lower genetic diversity and the broader ecological amplitude in DELTA group. As the results showed that the epigenetic diversity in the DELTA was as high as in the LAND group, if not higher, the epigenetic diversity partially explained this contradictory. The finding also indicated that epigenetic variation could compensate for decreased genetic variation after an initialized introduction.
4.3. Stable epigenetic differentiation has not occurred
Epigenetic variation in invasive plant populations can contribute to phenotypic variation and plasticity for adaptation in new environments as a fast mechanism during the introduction (Richards et al., 2012; Schulz et al., 2014). In the Great Lakes region, our result predicted a constant threat to the native P. Australis in which the native group exhibited a very low level of epigenetic variation, as well as genetic variation, possibly because of very rare hybridization between the native and introduced groups (Saltonstall, 2011). While the introduced group in the Great Lakes regions also had low genetic diversity relative to those in the Gulf Coast regions, this finding suggested that fewer introductions limited interbreeding there. While the introduced group could not obtain increased genetic variation, it might increase de novo epigenetic variation as a source of phenotypic traits and plasticity in response to the new environment and then could undergo natural selection while spreading through its invasive range, resulting in epigenetic differentiation from the EU group and convergence with the native group. This hypothesis needs more evidences in the further study.
Unlike many invasive clonal species spreading mostly by vegetative reproduction (Gao, Geng, Li, Chen, & Yang, 2010; Lambertini et al., 2010; Richards et al., 2012), the invasion of P. australis in North America contributes to a reproduction strategy combining sexual and vegetative propagation (Albert, Brisson, Belzile, Turgeon, & Lavoie, 2015; McCormick, Kettenring, Baron, & Whigham, 2010). The high genetic diversity in P. australis makes it more difficult to detect the epigenetic changes during the invasion. The epigenetic variation in natural populations can originate from genetic factor (B1), spontaneous epimutations (B2), and environmentally induced epigenetic changes (B3) (Richards et al., 2017). In addition, epigenetic change can create novel genetic variation through regulating transposable elements activity (Richards et al., 2017). In our study, the similarity of genetic and epigenetic structure reflects the drivers B1 and B2. While the structure of B1 and B2 can only be shaped through natural selection, B3 could be direct changed by the environment. The epigenetic differentiation was observed in contrast habitats without variation differentiation (Foust et al., 2016; Gugger et al., 2016; Lira‐Medeiros et al., 2010; Schulz et al., 2014), but it remained unexplored how stable and inheritable the B3 variation is in these studies. Common garden filtered the plastic B3 variation in our study, and epigenetic divergence between populations in the introduced region and in its original region was weakened. Moreover, the heterogeneity of microhabitats within each region can increase epigenetic noise masking the direction of epigenetic changes during the invasion. Finally, it is hard to find the few important adaptive loci using a limited number of anonymous loci provided by AFLP and MS‐AFLP (Schrey et al., 2013). For the above reasons, we failed to detect the significant epigenetic divergence independent of genetic variation.
However, the adaptation of introduced groups cannot be explained entirely by adaptive evolution based on genetic or epigenetic variation in this study. Only a few diverged ecophysiological functional traits of the Mediterranean P. australis M1 lineage were detected between the DELTA and MED groups (Guo et al., 2016), and we determined very little differentiation in genetic and epigenetic variation between them. Given that the climate in the introduced range was more advantageous than that in the original range for the survival of common reed because of abundant precipitation and warm temperature, the introduced environment did not provide enough selection pressures to shape the stable genetic and epigenetic structure of introduced populations. Therefore, we cannot ignore the potential effect of preadaptation and ecological fitting based on the inherently high phenotypic plasticity during the invasion process of common reed (Guo, Lambertini, Nguyen, Li, & Brix, 2014; Guo et al., 2016), which may be correlated with plastic epigenetic mechanisms (Gao et al., 2010).
5. CONCLUSION
In conclusion, epigenetic variation in introduced groups of P. australis is often correlated with genetic variation, suggesting the closely correlated effect of genetic and epigenetic variation in species with high genetic diversity. When the introduced population cannot acquire adequate genetic diversity through hybridization with other introduced populations, the importance of epigenetic variation may rapidly emerge. Faster stable epigenetic convergence between introduced and native groups was not observed maybe due to the strong genetic effect. Further study in situ and in a common garden for several generations is necessary to associate the inheritable genetic and epigenetic loci with invasive traits, and it is better to use a reduced representation bisulfite sequencing approach based on high‐throughput sequencing technology (Robertson & Richards, 2015) for direct comparison with functional genetic context.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
LL, WG, and XG conceived the idea and designed the study. WG and XG transplanted reeds from the field and the cooperation garden to our common garden. SL, ND, and CP contributed to garden management and sample collection. LL and CP performed the molecular laboratory work. LL, ND, and XG analyzed and interpreted the data. LL drafted the manuscript, and all authors participated in manuscript modifications and gave final approval for publication.
ACKNOWLEDGMENTS
We thank Yunfei Cai for helpful guidance on laboratory technology and thank Ting Yu and Song Zhao for looking after these reeds in the common garden. We also want to sincerely thank Hans Brix in Aarhus University of Denmark for his cooperation in the establishment of our Phragmites common garden. This study was funded by National Natural Science Foundation of China (Nos. 31770361 and 31470402), National Key R&D Program of China (No. 2017YFC0505905), and Natural Science Foundation of Shandong Province, China (No. BS2015HZ020).
Liu L, Pei C, Liu S, Guo X, Du N, Guo W. Genetic and epigenetic changes during the invasion of a cosmopolitan species (Phragmites australis). Ecol Evol. 2018;8:6615–6624. 10.1002/ece3.4144
REFERENCES
- Albert, A. , Brisson, J. , Belzile, F. , Turgeon, J. , & Lavoie, C. (2015). Strategies for a successful plant invasion: The reproduction of Phragmites australis in north‐eastern North America. Journal of Ecology, 103, 1529–1537. 10.1111/1365-2745.12473 [DOI] [Google Scholar]
- Alonso, C. , Balao, F. , Bazaga, P. , & Pérez, R. (2016). Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytologist, 212, 571–576. 10.1111/nph.14138 [DOI] [PubMed] [Google Scholar]
- Alonso, C. , Pérez, R. , Bazaga, P. , Medrano, M. , & Herrera, C. M. (2016). MSAP markers and global cytosine methylation in plants: A literature survey and comparative analysis for a wild‐growing species. Molecular Ecology Resources, 16, 80–90. 10.1111/1755-0998.12426 [DOI] [PubMed] [Google Scholar]
- Alvarez, M. , Schrey, A. W. , & Richards, C. L. (2015). Ten years of transcriptomics in wild populations: What have we learned about their ecology and evolution? Molecular Ecology, 24, 710–725. 10.1111/mec.13055 [DOI] [PubMed] [Google Scholar]
- Avramidou, E. V. , Ganopoulos, I. V. , Doulis, A. G. , Tsaftaris, A. S. , & Aravanopoulos, F. A. (2015). Beyond population genetics: Natural epigenetic variation in wild cherry (Prunus avium). Tree Genetics & Genomes, 11, 95 10.1007/s11295-015-0921-7 [DOI] [Google Scholar]
- Bird, A. (2007). Perceptions of epigenetics. Nature, 447, 396–398. 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- Choi, J.‐Y. , Roy, N. S. , Park, K.‐C. , & Kim, N.‐S. (2016). Comparison of molecular genetic utilities of TD, AFLP, and MSAP among the accessions of japonica, indica, and Tongil of Oryza sativa L. Genes & Genomics, 38, 819–830. 10.1007/s13258-016-0426-2 [DOI] [Google Scholar]
- Davidson, A. M. , Jennions, M. , & Nicotra, A. B. (2011). Do invasive species show higher phenotypic plasticity than native species and if so, is it adaptive? A meta‐analysis. Ecology Letters, 14, 419–431. 10.1111/j.1461-0248.2011.01596.x [DOI] [PubMed] [Google Scholar]
- Dlugosch, K. M. , & Parker, I. M. (2007). Molecular and quantitative trait variation across the native range of the invasive species Hypericum canariense: Evidence for ancient patterns of colonization via pre‐adaptation? Molecular Ecology, 16, 4269–4283. 10.1111/j.1365-294X.2007.03508.x [DOI] [PubMed] [Google Scholar]
- Douhovnikoff, V. , & Dodd, R. S. (2014). Epigenetics: A potential mechanism for clonal plant success. Plant Ecology, 216, 227–233. 10.1007/s11258-014-0430-z [DOI] [Google Scholar]
- Doyle, J. J. , & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11–15. 10.2307/4119796 [DOI] [Google Scholar]
- Eichten, S. R. , Schmitz, R. J. , & Springer, N. M. (2014). Epigenetics: Beyond chromatin modifications and complex genetic regulation. Plant Physiology, 165, 933–947. 10.1104/pp.113.234211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust, C. M. , Preite, V. , Schrey, A. W. , Alvarez, M. , Robertson, M. H. , Verhoeven, K. J. F. , & Richards, C. L. (2016). Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Molecular Ecology, 25, 1639–1652. 10.1111/mec.13522 [DOI] [PubMed] [Google Scholar]
- Gao, L. , Geng, Y. , Li, B. , Chen, J. , & Yang, J. (2010). Genome‐wide DNA methylation alterations of Alternanthera philoxeroides in natural and manipulated habitats: Implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant, Cell & Environment, 33, 1820–1827. 10.1111/j.1365-3040.2010.02186.x [DOI] [PubMed] [Google Scholar]
- Genton, B. J. , Shykoff, J. A. , & Giraud, T. (2005). High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Molecular Ecology, 14, 4275–4285. 10.1111/j.1365-294X.2005.02750.x [DOI] [PubMed] [Google Scholar]
- Guarino, F. , Cicatelli, A. , Brundu, G. , Heinze, B. , & Castiglione, S. (2015). Epigenetic diversity of clonal white Poplar (Populus alba L.) populations: Could methylation support the success of vegetative reproduction strategy? PLoS One, 10, e0131480 10.1371/journal.pone.0131480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugger, P. F. , Fitz‐Gibbon, S. , PellEgrini, M. , & Sork, V. L. (2016). Species‐wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Molecular Ecology, 25, 1665–1680. 10.1111/mec.13563 [DOI] [PubMed] [Google Scholar]
- Guo, W.‐Y. , Lambertini, C. , Guo, X. , Li, X.‐Z. , Eller, F. , & Brix, H. (2016). Phenotypic traits of the Mediterranean Phragmites australis M1 lineage: Differences between the native and introduced ranges. Biological Invasions, 18, 2551–2561. 10.1007/s10530-016-1236-9 [DOI] [Google Scholar]
- Guo, W.‐Y. , Lambertini, C. , Li, X.‐Z. , Meyerson, L. A. , & Brix, H. (2013). Invasion of Old World Phragmites australis in the New World: Precipitation and temperature patterns combined with human influences redesign the invasive niche. Global Change Biology, 19, 3406–3422. 10.1111/gcb.12295 [DOI] [PubMed] [Google Scholar]
- Guo, W.‐Y. , Lambertini, C. , Nguyen, L. X. , Li, X.‐Z. , & Brix, H. (2014). Preadaptation and post‐introduction evolution facilitate the invasion of Phragmites australis in North America. Ecology and Evolution, 4, 4567–4577. 10.1002/ece3.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Wu, W. , Zinta, G. , Yang, L. , Wang, D. , Liu, R. , … Zhu, J.‐K. (2018). A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nature Communications, 9, 460 10.1038/s41467-018-02839-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, C. M. , & Bazaga, P. (2013). Epigenetic correlates of plant phenotypic plasticity: DNA methylation differs between prickly and nonprickly leaves in heterophyllous Ilex aquifolium (Aquifoliaceae) trees. Botanical Journal of the Linnean Society, 171, 441–452. 10.1111/boj.12007 [DOI] [Google Scholar]
- Herrera, C. M. , & Bazaga, P. (2016). Genetic and epigenetic divergence between disturbed and undisturbed subpopulations of a Mediterranean shrub: A 20‐year field experiment. Ecology and Evolution, 6, 3832–3847. 10.1002/ece3.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, C. M. , Medrano, M. , & Bazaga, P. (2014). Variation in DNA methylation transmissibility, genetic heterogeneity and fecundity‐related traits in natural populations of the perennial herb Helleborus foetidus . Molecular Ecology, 23, 1085–1095. 10.1111/mec.12679 [DOI] [PubMed] [Google Scholar]
- Hollingsworth, M. L. , & Bailey, J. P. (2000). Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed). Botanical Journal of the Linnean Society, 133, 463–472. 10.1111/j.1095-8339.2000.tb01589.x [DOI] [Google Scholar]
- Kelager, A. , Pedersen, J. S. , & Bruun, H. H. (2013). Multiple introductions and no loss of genetic diversity: Invasion history of Japanese Rose, Rosa rugosa, in Europe. Biological Invasions, 15, 1125–1141. 10.1007/s10530-012-0356-0 [DOI] [Google Scholar]
- Kettenring, K. M. , McCormick, M. K. , Baron, H. M. , & Whigham, D. F. (2011). Mechanisms of Phragmites australis invasion: Feedbacks among genetic diversity, nutrients, and sexual reproduction: Mechanisms of Phragmites invasion. Journal of Applied Ecology, 48, 1305–1313. 10.1111/j.1365-2664.2011.02024.x [DOI] [Google Scholar]
- Kilvitis, H.J. , Alvarez, M. , Foust, C.M. , Schrey, A.W. , Robertson, M. , & Richards, C.L. (2014). Ecological epigenetics In: Landry C.R. & Aubin‐Horth N. (Eds.), Ecological genomics, advances in experimental medicine and biology (pp. 191–210). Dordrecht, the Netherlands: Springer; 10.1007/978-94-007-7347-9_10 [DOI] [PubMed] [Google Scholar]
- Kim, N.‐S. , Im, M.‐J. , & Nkongolo, K. (2016). Determination of DNA methylation associated with Acer rubrum (red maple) adaptation to metals: Analysis of global DNA modifications and methylation‐sensitive amplified polymorphism. Ecology and Evolution, 6, 5749–5760. 10.1002/ece3.2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueffer, C. , Pyšek, P. , & Richardson, D. M. (2013). Integrative invasion science: Model systems, multi‐site studies, focused meta‐analysis and invasion syndromes. New Phytologist, 200, 615–633. 10.1111/nph.12415 [DOI] [PubMed] [Google Scholar]
- Lambertini, C. , Mendelssohn, I. A. , Gustafsson, M. H. G. , Olesen, B. , Riis, T. , Sorrell, B. K. , & Brix, H. (2012). Tracing the origin of Gulf Coast Phragmites (Poaceae): A story of long‐distance dispersal and hybridization. American Journal of Botany, 99, 538–551. 10.3732/ajb.1100396 [DOI] [PubMed] [Google Scholar]
- Lambertini, C. , Riis, T. , Olesen, B. , Clayton, J. S. , Sorrell, B. K. , & Brix, H. (2010). Genetic diversity in three invasive clonal aquatic species in New Zealand. BMC Genetics, 11, 52 10.1186/1471-2156-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini, C. , Sorrell, B. K. , Riis, T. , Olesen, B. , & Brix, H. (2012). Exploring the borders of European Phragmites within a cosmopolitan genus. AoB Plants, 2012, pls020 10.1093/aobpla/pls020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latzel, V. , Allan, E. , Bortolini Silveira, A. , Colot, V. , Fischer, M. , & Bossdorf, O. (2013). Epigenetic diversity increases the productivity and stability of plant populations. Nature Communications, 4, 2875 10.1038/ncomms3875 [DOI] [PubMed] [Google Scholar]
- Lavergne, S. , & Molofsky, J. (2007). Increased genetic variation and evolutionary potential drive the success of an invasive grass. PNAS, 104, 3883–3888. 10.1073/pnas.0607324104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm, A. K. , Breden, F. , Alexander, H. J. , Chan, W.‐K. , Thakurta, S. G. , & Brooks, R. (2005). Invasion success and genetic diversity of introduced populations of guppies Poecilia reticulata in Australia. Molecular Ecology, 14, 3671–3682. 10.1111/j.1365-294X.2005.02697.x [DOI] [PubMed] [Google Scholar]
- Lira‐Medeiros, C. F. , Parisod, C. , Fernandes, R. A. , Mata, C. S. , Cardoso, M. A. , & Ferreira, P. C. G. (2010). Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One, 5, e10326 10.1371/journal.pone.0010326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis, E. S. , & Fishman, L. (2009). A continent‐wide clone: Population genetic variation of the invasive plant Hieracium aurantiacum (Orange Hawkweed; Asteraceae) in North America. International Journal of Plant Sciences, 170, 759–765. 10.1086/599241 [DOI] [Google Scholar]
- McCormick, M. K. , Kettenring, K. M. , Baron, H. M. , & Whigham, D. F. (2010). Spread of invasive Phragmites australis in estuaries with differing degrees of development: Genetic patterns, Allee effects and interpretation. Journal of Ecology, 98, 1369–1378. 10.1111/j.1365-2745.2010.01712.x [DOI] [Google Scholar]
- Medrano, M. , Herrera, C. M. , & Bazaga, P. (2014). Epigenetic variation predicts regional and local intraspecific functional diversity in a perennial herb. Molecular Ecology, 23, 4926–4938. 10.1111/mec.12911 [DOI] [PubMed] [Google Scholar]
- Meyerson, L. A. , Cronin, J. T. , & Pyšek, P. (2016). Phragmites australis as a model organism for studying plant invasions. Biological Invasions, 18, 2421–2431. 10.1007/s10530-016-1132-3 [DOI] [Google Scholar]
- Meyerson, L. A. , Lambert, A. M. , & Saltonstall, K. (2010). A tale of three lineages: Expansion of common reed (Phragmites australis) in the U.S. Southwest and Gulf Coast. Invasive Plant Science & Management, 3, 515–520. 10.1614/IPSM-D-09-00052.1 [DOI] [Google Scholar]
- Meyerson, L. A. , Lambertini, C. , McCormick, M. K. , & Whigham, D. F. (2012). Hybridization of common reed in North America? The answer is blowing in the wind. AoB Plants, 2012, pls022 10.1093/aobpla/pls022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson, L. A. , Viola, D. V. , & Brown, R. N. (2010). Hybridization of invasive Phragmites australis with a native subspecies in North America. Biological Invasions, 12, 103–111. 10.1007/s10530-009-9434-3 [DOI] [Google Scholar]
- Mozdzer, T. J. , Brisson, J. , & Hazelton, E. L. G. (2013). Physiological ecology and functional traits of North American native and Eurasian introduced Phragmites australis lineages. AoB Plants, 5, plt048 10.1093/aobpla/plt048 [DOI] [Google Scholar]
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra, A. B. , Segal, D. L. , Hoyle, G. L. , Schrey, A. W. , Verhoeven, K. J. , & Richards, C. L. (2015). Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecology and Evolution, 5, 634–647. 10.1002/ece3.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research – An update. Bioinformatics, 28, 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Figueroa, A. (2013). msap: A tool for the statistical analysis of methylation‐sensitive amplified polymorphism data. Molecular Ecology Resources, 13, 522–527. 10.1111/1755-0998.12064 [DOI] [PubMed] [Google Scholar]
- Platt, A. , Gugger, P. F. , Pellegrini, M. , & Sork, V. L. (2015). Genome‐wide signature of local adaptation linked to variable CpG methylation in oak populations. Molecular Ecology, 24, 3823–3830. 10.1111/mec.13230 [DOI] [PubMed] [Google Scholar]
- Preite, V. , Snoek, L. B. , Oplaat, C. , Biere, A. , van der Putten, W. H. , & Verhoeven, K. J. F. (2015). The epigenetic footprint of poleward range‐expanding plants in apomictic dandelions. Molecular Ecology, 24, 4406–4418. 10.1111/mec.13329 [DOI] [PubMed] [Google Scholar]
- Qiu, T. , Jiang, L. L. , & Yang, Y. F. (2016). Genetic and epigenetic diversity and structure of Phragmites australis from local habitats of the Songnen Prairie using amplified fragment length polymorphism markers. Genetics and Molecular Research, 15, gmr.15038585. 10.4238/gmr.15038585 [DOI] [PubMed] [Google Scholar]
- Richards, E. J. (2006). Inherited epigenetic variation–revisiting soft inheritance. Nature Reviews Genetics, 7, 395–401. 10.1038/nrg1834 [DOI] [PubMed] [Google Scholar]
- Richards, C. L. , Alonso, C. , Becker, C. , Bossdorf, O. , Bucher, E. , Colomé‐Tatché, M. , … Verhoeven, K. J. F. (2017). Ecological plant epigenetics: Evidence from model and non‐model species, and the way forward. Ecology Letters, 20, 1576–1590. 10.1111/ele.12858 [DOI] [PubMed] [Google Scholar]
- Richards, C. L. , Schrey, A. W. , & Pigliucci, M. (2012). Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecology Letters, 15, 1016–1025. 10.1111/j.1461-0248.2012.01824.x [DOI] [PubMed] [Google Scholar]
- Robertson, M. , & Richards, C. (2015). Opportunities and challenges of next‐generation sequencing applications in ecological epigenetics. Molecular Ecology, 24, 3799–3801. 10.1111/mec.13277 [DOI] [PubMed] [Google Scholar]
- Rosenthal, D. M. , Ramakrishnan, A. P. , & Cruzan, M. B. (2008). Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Molecular Ecology, 17, 4657–4669. 10.1111/j.1365-294X.2008.03844.x [DOI] [PubMed] [Google Scholar]
- Sáez‐Laguna, E. , Guevara, M.‐Á. , Díaz, L.‐M. , Sánchez‐Gómez, D. , Collada, C. , Aranda, I. , & Cervera, M.‐T. (2014). Epigenetic variability in the genetically uniform forest tree species Pinus pinea L. PLoS One, 9, e103145 10.1371/journal.pone.0103145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall, K. (2002). Cryptic invasion by a non‐native genotype of the common reed, Phragmites australis, into North America. PNAS, 99, 2445–2449. 10.1073/pnas.032477999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall, K. (2003). Microsatellite variation within and among North American lineages of Phragmites australis . Molecular Ecology, 12, 1689–1702. 10.1046/j.1365-294X.2003.01849.x [DOI] [PubMed] [Google Scholar]
- Saltonstall, K. (2011). Remnant native Phragmites australis maintains genetic diversity despite multiple threats. Conservation Genetics, 12, 1027–1033. 10.1007/s10592-011-0205-1 [DOI] [Google Scholar]
- Saltonstall, K. , Castillo, H. E. , & Blossey, B. (2014). Confirmed field hybridization of native and introduced Phragmites australis (Poaceae) in North America. American Journal of Botany, 101, 211–215. 10.3732/ajb.1300298 [DOI] [PubMed] [Google Scholar]
- Saltonstall, K. , Lambert, A. M. , & Rice, N. (2016). What happens in Vegas, better stay in Vegas: Phragmites australis hybrids in the Las Vegas Wash. Biological Invasions, 18, 2463–2474. 10.1007/s10530-016-1167-5 [DOI] [Google Scholar]
- Schlaepfer, D. R. , Glättli, M. , Fischer, M. , & van Kleunen, M. (2010). A multi‐species experiment in their native range indicates pre‐adaptation of invasive alien plant species. New Phytologist, 185, 1087–1099. 10.1111/j.1469-8137.2009.03114.x [DOI] [PubMed] [Google Scholar]
- Schrey, A. W. , Alvarez, M. , Foust, C. M. , Kilvitis, H. J. , Lee, J. D. , Liebl, A. L. , … Robertson, M. (2013). Ecological epigenetics: Beyond MS‐AFLP. Integrative and Comparative Biology, 53, 340–350. 10.1093/icb/ict012 [DOI] [PubMed] [Google Scholar]
- Schulz, B. , Eckstein, R. L. , & Durka, W. (2014). Epigenetic variation reflects dynamic habitat conditions in a rare floodplain herb. Molecular Ecology, 23, 3523–3537. 10.1111/mec.12835 [DOI] [PubMed] [Google Scholar]
- Spens, A. E. , & Douhovnikoff, V. (2016). Epigenetic variation within Phragmites australis among lineages, genotypes, and ramets. Biological Invasions, 18, 2457–2462. 10.1007/s10530-016-1223-1 [DOI] [Google Scholar]
- Suehs, C. M. , Affre, L. , & Médail, F. (2004). Invasion dynamics of two alien Carpobrotus (Aizoaceae) taxa on a Mediterranean island: I. Genetic diversity and introgression. Heredity, 92, 31–40. 10.1038/sj.hdy.6800374 [DOI] [PubMed] [Google Scholar]
- Sun, X. , Li, X. , Guan, Z. , Liu, J. , & Zhang, S. (2017). The use of meteorological data to assess the cooling service of forests. Ecosystem Services, 25, 28–34. 10.1016/j.ecoser.2017.03.016 [DOI] [Google Scholar]
- Verhoeven, K. J. F. , vonHoldt, B. M. , & Sork, V. L. (2016). Epigenetics in ecology and evolution: What we know and what we need to know. Molecular Ecology, 25, 1631–1638. 10.1111/mec.13617 [DOI] [PubMed] [Google Scholar]
- Zhang, Y.‐Y. , Fischer, M. , Colot, V. , & Bossdorf, O. (2013). Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytologist, 197, 314–322. 10.1111/nph.12010 [DOI] [PubMed] [Google Scholar]


