ABSTRACT
Transcription factors of the Sox protein family contain a DNA-binding HMG box and are key regulators of progenitor cell fate. Here, we report that expression of Sox30 is restricted to meiotic spermatocytes and postmeiotic haploids. Sox30 mutant males are sterile owing to spermiogenic arrest at the early round spermatid stage. Specifically, in the absence of Sox30, proacrosomic vesicles fail to form a single acrosomal organelle, and spermatids arrest at step 2-3. Although most Sox30 mutant spermatocytes progress through meiosis, accumulation of diplotene spermatocytes indicates a delayed or impaired transition from meiotic to postmeiotic stages. Transcriptome analysis of isolated stage-specific spermatogenic cells reveals that Sox30 controls a core postmeiotic gene expression program that initiates as early as the late meiotic cell stage. ChIP-seq analysis shows that Sox30 binds to specific DNA sequences in mouse testes, and its genomic occupancy correlates positively with expression of many postmeiotic genes including Tnp1, Hils1, Ccdc54 and Tsks. These results define Sox30 as a crucial transcription factor that controls the transition from a late meiotic to a postmeiotic gene expression program and subsequent round spermatid development.
KEY WORDS: Sox30, Male germ cell, Spermiogenesis, Gene regulation, Mouse
Summary: Analysis of Sox30 mutants using ChIP-seq, meiotic analysis and RNA-seq identifies the direct targets of Sox30 and reveals that Sox30 is a transcription factor of postmeiotic genes in mouse testes.
INTRODUCTION
The developmental progression of round spermatids into mature spermatozoa, termed spermiogenesis, requires multiple molecular events, including profound nuclear restructuring and condensation, formation of an acrosome and assembly of the flagellum. Accordingly, a large number of proteins required for these events have to be synthesized at the onset of and throughout spermiogenesis. For example, spermatid-specific histone variants, transition proteins (TPs), and protamines (Prms) progressively replace nucleosomal histones during chromatin condensation (Meistrich et al., 2003; Oliva, 2006; Barral et al., 2017). Histone variants are first incorporated into the chromatin to loosen histone-DNA interactions, followed by efficient loading of TPs and finally histone removal by Prms. Owing to a relatively inaccessible chromatin state during the late steps of spermiogenesis, pachytene spermatocytes and round spermatids are transcriptionally active (Hecht, 1998; Paronetto and Sette, 2010), and specific mRNAs produced during these stages are stored and stabilized in cytoplasmic ribonucleoprotein (RNP) particles until their translation is required in later stage spermatids (Iguchi et al., 2006).
Studies have focused on the intricate mechanisms for precise activation of the haploid-stage expression program, which involves both transcriptional and post-transcriptional regulation (Bettegowda and Wilkinson, 2010). In mammals, one factor central to round spermatid differentiation is cyclic AMP-responsive element modulator (CREM), a transcriptional activator that drives transcription of a set of post-meiotic genes involved in the structuring of the spermatozoon (Nantel et al., 1996; Martianov et al., 2010; Kosir et al., 2012). Crem-deficient mice exhibit complete arrest of spermiogenesis at step 5, with increased apoptosis in arrested spermatids (Blendy et al., 1996; Nantel et al., 1996; Zhou et al., 1996). Transcription factor II D (TFIID) complex components encompass another group of essential transcriptional factors during spermiogenesis. TFIID is a large complex that consists of the TATA box-binding protein (TBP) and a group of TBP-associated factors (TAFs) (Hernandez, 1993). Mice deficient in TBP-like factor (TRF2; TBPL1) or TAFs (TAF7L) display male infertility due to arrested spermiogenesis (Martianov et al., 2001; Cheng et al., 2007; Zhou et al., 2013), at a later stage than that of Crem mutants. Interestingly, spermiogenesis also requires components of RNP granules, which are structures involved in RNA storage and processing for spermatogenesis. Mice that lack granule components, such as Miwi (Piwil1), display spermiogenic arrest at the early round spermatid stage (Deng and Lin, 2002; Reuter et al., 2011). As a consequence of disrupted nuage architecture, post-meiotic gene expression or small RNA processing is dysregulated in the respective mutant mice.
SRY box (Sox) proteins are a family of transcriptional factors that contain more than 50% homology to the conserved high mobility group (HMG)-box domain that mediates DNA binding and interaction with co-factors (Wegner, 1999). Numerous genetically modified mouse models have established Sox proteins as crucial regulators of cell fate decisions (Sarkar and Hochedlinger, 2013), and some of them have been shown to participate in spermatogenesis. For example, the SoxB group member Sox3 is expressed in a small population of undifferentiated spermatogonia at the basement membrane of the seminiferous tubules. Spermatogonia lacking Sox3 expression lose the competence to differentiate, accompanied by a downregulation of the spermatogonial differentiation-associated genes and an upregulation of the pluripotency genes, such as Pou5f1 (also known as Oct4) (Raverot et al., 2005; Laronda and Jameson, 2011). In contrast with Sox3, which is expressed in perinatal germ cells, expression of the sole member of the SoxH group, Sox30, is largely restricted to postnatal germ cells (Osaki et al., 1999). While this manuscript was under preparation, the spermiogenic-arrest phenotype of a germ cell-specific (Vasa-Cre) Sox30-deficient mouse model was reported (Feng et al., 2017). However, the molecular events by which Sox30 deficiency in mouse testes leads to spermiogenic arrest and the identity of its genome-wide target genes remain unexplored.
In this study, we generated mice with targeted disruption of Sox30 in exon 2, which encodes the HMG DNA-binding domain. Male mice lacking functional Sox30 displayed spermatogenic defects and uniformly arrested at the early stage of round spermatid development (step 2-3), similar to Crem and Miwi mutants. Sox30 mutant germ cells progressed through meiosis but exhibited defects in spermatocyte development. By subsequent RNA sequencing (RNA-seq) of total testicular germ cells and purified stage-specific spermatogenic cells from normal and Sox30 mutant testis and genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) studies, we have uncovered the molecular function of Sox30 and demonstrate that Sox30 is a testis-specific transcription factor that is essential for the activation of haploid differentiation programs during later stages of meiotic cell differentiation into haploid post-meiotic cells.
RESULTS
Sox30 is largely restricted to the nuclei of late meiotic and post-meiotic germ cells
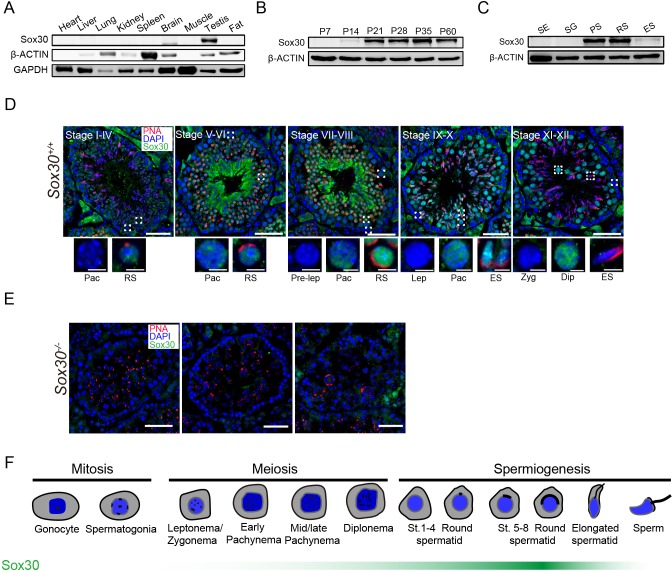
To explore the biological function of Sox30 in the mouse, we first characterized tissue- and developmental stage-specific distribution of Sox30 protein. Western blot analysis confirmed that in adult male mice, Sox30 protein is expressed highly and exclusively in the testis (Fig. 1A), consistent with mRNA expression patterns detected by quantitative RT-PCR (qRT-PCR) (Fig. S1A) (Feng et al., 2017). Analysis of the developmental-specific expression of Sox30 transcript by qRT-PCR predicted a significant increase in expression during spermatogenesis [∼135-fold increase in testis mRNA levels between postnatal day (P) 7 to 35; Fig. S1B]. Consistently, Sox30 protein was undetectable in P7 mouse testis, detectable at P14, with substantially increased expression by P21 when round spermatids first appear (Fig. 1B). We next characterized expression of Sox30 transcript and protein in testicular cell populations isolated by STA-PUT velocity sedimentation, including Sertoli cells, spermatogonia, pachytene spermatocytes, round spermatids and elongating spermatids. The purity of each isolated cell fractions was assessed based on their distinct morphological characteristics and RT-PCR analysis with specific marker genes (Fig. S2A,B). Western blot analysis of protein lysates from stage-specific germ cell populations indicated that Sox30 protein was predominantly expressed in pachytene spermatocytes and round spermatids (Fig. 1C). Moreover, Sox30 protein level was dramatically reduced in elongated spermatids (Fig. 1C).
Fig. 1.
Sox30 is highly enriched in mouse testes. (A-C) Western blot analysis of Sox30 protein in lysates from adult mouse tissues (A), from mouse testis tissue collected at different time points during postnatal development (B) and from isolated spermatogenic cell populations, including Sertoli cells (SE), spermatogonia (SG), pachytene spermatocytes (PS), round spermatids (RS) and elongating spermatids (ES). β-actin serves as an internal loading control. (D,E) Immunostaining of testis sections from 8-week-old wild-type (D) and Sox30−/− mutant (E) mice for Sox30 (green) and PNA (red). PNA is an acrosome marker. DNA was counterstained with DAPI. Lower panels show magnifications of the boxed area in the upper panels. Dip, diplotene; ES, elongating spermatids; Pac, pachytene; pre-lep, pre-leptotene; RS, round spermatids; Zyg, zygotene. Scale bars: 50 μm. (F) Graphic representation of Sox30 expression (green bar) during spermatogenesis.
Immunostaining of adult testis sections revealed that Sox30 protein was absent in early pachytene spermatocytes at stage I-IV, but was detectable in the nuclei of later pachytene spermatocytes in seminiferous tubules at stage V-VI (Fig. 1D). This finding is consistent with the presence of low levels of Sox30 in P14 testis (Fig. 1B), which is enriched for early pachytene spermatocytes. As seminiferous tubules progressed to stage VII-VIII, Sox30 signal progressively increased in pachytene spermatocytes, and intense staining was visible throughout the entire nucleus of round spermatids (Fig. 1D). At stage IX-X, Sox30 expression remained detectable in the nuclei of spermatids at step 9-10, when round spermatids transition into elongating spermatids (Fig. 1D). Strikingly, at stage XI-XII, Sox30 expression disappeared and was undetectable in spermatids at step 11-12 and thereafter (Fig. 1D). Notably, Sox30 signal was robust in the nucleus of diplotene spermatocytes at this stage. The observed nuclear signal of Sox30 is specific, as evident from the absence of signal in corresponding cell types in testis tubules from mice homozygous for a defective Sox30 allele (Sox30−/−; see detail in next paragraph) (Fig. 1E). Collectively, these data indicate that Sox30 is a nuclear protein that is expressed beginning in late pachytene stage and diplotene spermatocytes, peaks in round spermatids at step 7-8, and remains expressed in early elongated spermatids but becomes downregulated at later stages of elongated spermatids (Fig. 1F). This distinct expression pattern of Sox30 during spermatogenesis suggests that it may function during the transition from late meiotic to post-meiotic stages and subsequent round spermatid development.
Sox30 mutants are sterile and arrest at the round spermatid stage
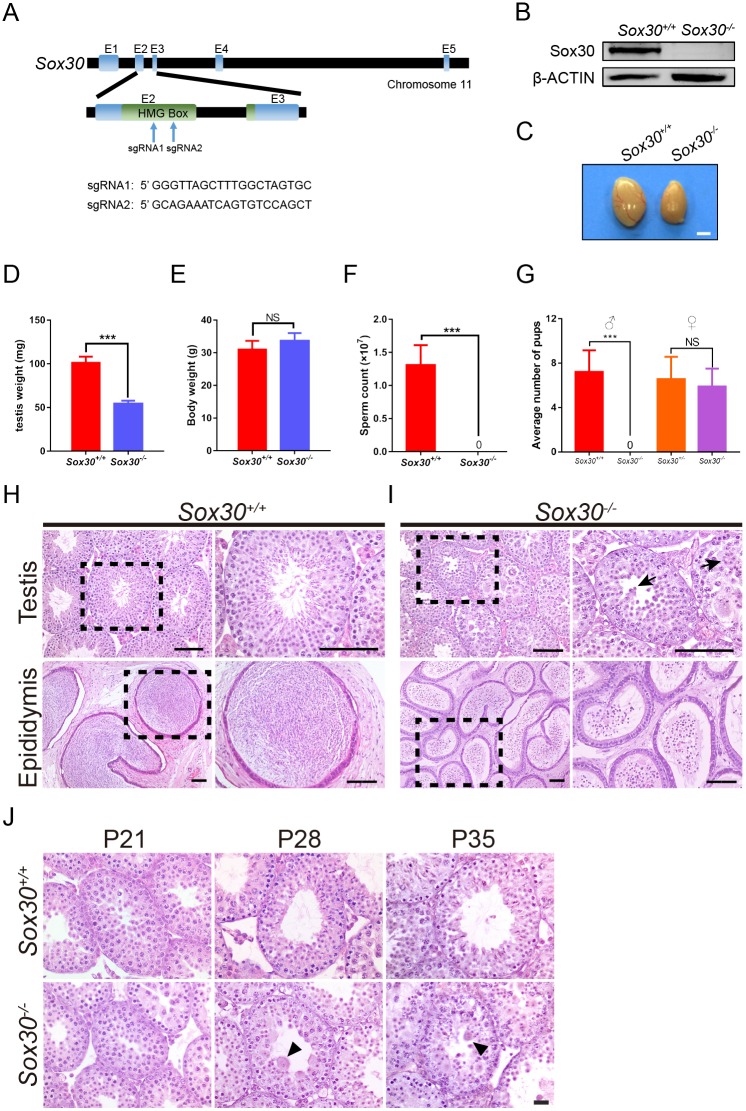
To elucidate the in vivo function of Sox30, we generated Sox30 knockout (KO) mice using CRISPR-Cas9 gene editing technology. Two small guide RNAs (sgRNAs) were designed to target exon 2, which encodes the conserved HMG-box domain (Fig. 2A). Cas9 mRNAs and two sgRNAs were electroporated into zygotes, which were subsequently transferred to pseudopregnant females. To validate nucleotide changes at the targeted loci, the corresponding genomic regions of Sox30 were evaluated by cloning and sequencing (Fig. S3A). The first generation of Sox30 mutants contained seven founders with indels or deletions on Sox30 alleles. In founders #1, #2 and #3, both Sox30 alleles were mutant, and founder #3 was homozygous for a mutant allele with a single bp insertion and 9-bp deletion (Fig. S3A,B). These three founders grew normally into adulthood but were sterile. Histological analysis revealed arrest of spermatogenesis at the round spermatid stage in each of the three founders (Fig. S4A-C). The remaining four founders were heterozygous for a mutant and the wild-type Sox30 allele and were fertile as expected (Fig. S3A). We next established a mutant mouse line with a Sox30 allele containing a 47-bp deletion identified in male founder #4 (Fig. S3B). This deletion produces a premature termination codon, and genotyping PCR and RT-PCR confirmed the absence of full-length transcripts in animals homozygous for the mutant allele (hereafter referred to as Sox30−/−) indicating successful targeting of Sox30 (Fig. S3C-E). Western blot analysis demonstrated absence of Sox30 protein in testis lysates from adult Sox30−/− mice (Fig. 2B). Sox30 mutants grew normally into adulthood without obvious developmental defects. Intercrossing of Sox30+/− mice produced Sox30+/+, Sox30+/− and Sox30−/− offspring at the predicted Mendelian ratio. Sox30−/− females were fertile. However, Sox30−/− males were sterile. The testis size of Sox30 mutant mice was dramatically reduced (Fig. 2C). The weight of Sox30−/− testes (54.38±3.40 mg/pair) from 8-week-old mice was 53.52% that of age matched wild-type testes (101.60±6.57 mg) (Fig. 2D), whereas the body weight was similar (Fig. 2E). Mature spermatozoa were absent in the caudal epididymis of Sox30−/− mice (Fig. 2F). Consistent with this, no pups were obtained from adult Sox30−/− males in mating tests with normal fertile females (Fig. 2G).
Fig. 2.
Sox30 mutant mice arrest at the round spermatid stage. (A) Diagram illustrating the CRISPR/Cas9 targeting strategy, including position and sequence of guide RNAs (sgRNAs). E, exon. (B) Western blot analysis confirms absence of Sox30 protein in lysates from 8-week-old Sox30−/− mutant mice. β-actin serves as an internal loading control. (C) Morphological appearance of testis from Sox30−/−mice versus Sox30+/+ (wild-type) mice revealing smaller size. (D-F) Comparison of testis weight (D), body weight (E) and epididymal sperm counts (F) of 8-week-old Sox30+/+ and Sox30−/− mice. Data are presented as mean±s.d. (D,E: n=6 per group; F: n=5) ***P<0.001; NS, not significant (Student's t-test). (G) Fertility test for adult Sox30−/− males and Sox30−/− females. Each genotype shown was coupled with fertile wild-type mates. n=3 for each genotype. ***P<0.001; NS, not significant (Student's t-test). (H,I) Hematoxylin and Eosin staining of testis tubules from 8-week-old wild-type (H) and Sox30−/− (I) mice. Sox30−/− tubules were devoid of elongating spermatids. Black arrows mark multinucleated cells. Right-hand panels show magnifications of the boxed areas on the left. (J) Sox30+/+ (wild-type) epididymal tubules were full of spermatozoa whereas epididymal tubules from Sox30−/− mice were devoid of mature sperm but contained degenerating cells with round-shaped nuclei (black arrowheads). Scale bars: 100 μm.
Histological examination of adult wild-type testes sections revealed seminiferous tubules full of spermatogenic cells at various developmental stages, including spermatogonia, spermatocytes and spermatids (Fig. 2H). In contrast, Sox30−/− seminiferous tubules lacked any elongating spermatids and were uniformly arrested at the round spermatid stage (Fig. 2I). Spermatids undergoing cellular elongation or nuclear condensation were not detectable; instead, round spermatids formed large multinucleated cell clusters (Fig. 2I, arrows). Epididymides from Sox30−/− mice did not contain mature spermatozoa but were filled with large numbers of degenerating germ cells (Fig. 2I). These findings indicate that Sox30−/− round spermatids failed to proceed through spermiogenesis and were prematurely released to the epididymis.
To determine the onset of the spermatogenic arrest in Sox30−/− males, we examined the first wave of spermatogenesis. Histological analysis of P21 testes showed no discernible abnormality in Sox30−/− testes, and both wild-type and mutant testes contained germ cells that had differentiated into round spermatids (Fig. 2J, left). We next examined P28 testes, when the first wave of germ cells has progressed to the elongating spermatid stage. Strikingly, elongating spermatids were completely absent in the Sox30−/− tubules (Fig. 2J, middle) but giant multinucleated cell clusters were first present in mutant testes at P28, and accumulated in P35 Sox30−/− testes (Fig. 2J, right). These data revealed that round spermatid arrest during the first wave of spermatogenesis in Sox30−/− testes.
Fusion failure of proacrosomic vesicles and germ cell apoptosis in Sox30−/− mice
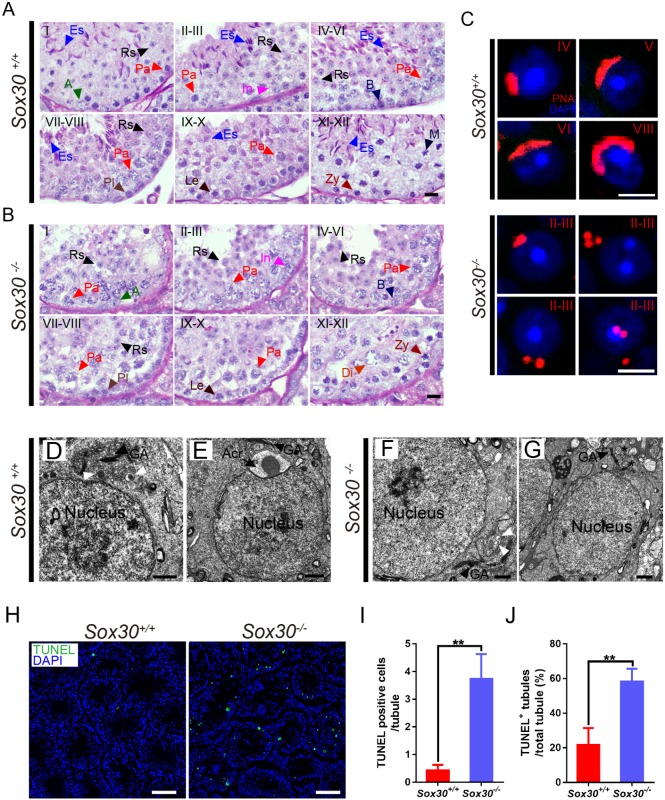
The differentiation of round spermatids into mature sperm encompasses 16 steps. To determine at which step Sox30−/− round spermatids arrested, we stained testis sections with periodic acid-Schiff (PAS), which specifically labels acrosomal glycoproteins. The acrosome is a spermatid-unique organelle derived from the Golgi apparatus. Normally, with the differentiation of spermatids, proacrosomic vesicles are formed (step 2-3), fused into a single acrosomal vesicle that attaches to the nuclear surface of round spermatids (step 4), and spread further over the nuclear surface (step 5-16). As shown in Fig. 3A, testis sections from 8-week-old wild-type males contained spermatids at all steps of differentiation (Fig. 3A). Round spermatids in adult Sox30−/− testes contained PAS-positive proacrosomic vesicles (step 2-3) (Fig. 3B); however, round spermatids with flattened acrosomal vesicles were not detected (Fig. 3B), indicating that round spermatids in Sox30−/− mice were arrested at step 2-3. Immunostaining with the acrosome marker peanut agglutinin (PNA) confirmed the complete absence of spermatids beyond step 2-3 in the mutant as observed in the histological images (Fig. 3C). Instead of flattening to form a cup-shaped acrosome, as seen in controls, Sox30−/− spermatids frequently contained proacrosomic vesicles that had sloughed off the nuclear surface (Fig. 3C).
Fig. 3.
Sox30-deficient round spermatids exhibit impaired fusion of proacrosomic vesicles and arrest at step 2-3. (A,B) PAS- and Hematoxylin-stained testis sections from 8-week-old Sox30+/+ (A) and Sox30−/− (B) mice. The seminiferous tubule stage (top left) was identified based on the PAS staining pattern and arrangement of spermatogenic cells. A, type A spermatogonia; B, type B spermatogonia; Es, elongating spermatids; In, intermediate spermatogonia; Le, leptotene spermatocytes; M, meiotic division; Pa, pachytene spermatocytes; Pl, preleptotene spermatocytes; Rs, round spermatids; Zy, zygotene spermatocytes. Scale bars: 50 μm. (C) Fluorescently tagged PNA-staining (red) of testis sections from 8-week-old mice shows acrosome formation from proacrosomic vesicles in step 4-8 wild-type spermatids but failure of vesicle fusion in cells from Sox30−/− mice. DNA was counterstained with DAPI. Scale bars: 10 μm. (D-G) Electron microscopic analysis of testis tissue from 8-week-old Sox30+/+ (D,E) and Sox30−/− (F,G) mice. Step 2-3 wild-type round spermatids contain proacrosomal vesicles (white arrowheads) located between the Golgi apparatus and the nucleus (D), and a newly assembled acrosome is visible in a step 4 round spermatid (E). (F,G) Sox30−/− spermatids at step 2-3 contain proacrosomic vesicles, but a large acrosome was never observed. A subset of proacrosomic vesicles were not located between the Golgi apparatus and the nucleus (G). White arrowheads mark proacrosomal vesicles; asterisks identify proacrosomal vesicles with abnormal localization. Acr, acrosome; GA, Golgi apparatus. Scale bars: 1 μm. (H-J) Increased apoptosis in Sox30−/− testis. (H) TUNEL assay in testis sections from 8-week-old Sox30+/+ and Sox30−/− males. Scale bars: 100 μm. (I) Quantification of TUNEL-positive cells per tubule. n=3 for each genotype. (J) Percentage of TUNEL-positive tubules in testis from 8-week-old Sox30+/+ and Sox30−/− mice. Tubules examined: Sox30+/+, n=96, Sox30−/−, n=156. Data presented are mean±s.d. **P<0.01 (Student's t-test).
To further characterize the defects in acrosome formation in Sox30−/− spermatids, we performed transmission electron microscopy (TEM) to visualize acrosome biogenesis, which normally includes the formation of one or two proacrosomic vesicles from the Golgi apparatus during step 2-3 located between the Golgi apparatus and the nucleus (Fig. 3D), and fusion into a giant acrosomal vesicle docked to the nuclear envelope during step 4 (Fig. 3E). In Sox30−/− mice, proacrosomic vesicles were observed (step 2-3), but a large acrosome was never observed (Fig. 3F,G). Moreover, some proacrosomic vesicles did not locate between the Golgi apparatus and the nucleus (Fig. 3G).
The multinucleated giant cells observed in the testes from 8- and 4-week-old Sox30−/− males (Fig. 2I,J) may correspond to degenerating or apoptotic germ cells. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) revealed increased numbers of apoptotic cells in tubules of Sox30−/− testes (Fig. 3H-J), with approximately 60% of mutant tubules containing TUNEL-positive cells (Fig. 3J). Moreover, large numbers of apoptotic cells were found in the epididymal tubules from Sox30−/− mice (Fig. S5A). Such uniform arrest during the early stage of round spermatid development leading to apoptosis was also observed in Crem and Miwi mutants (Nantel et al., 1996; Deng and Lin, 2002), suggesting that Sox30 is an essential gene for the progression of early spermiogenesis.
Accumulation of diplotene spermatocytes and enhanced spermatocytes apoptosis in Sox30-deficient testes
Although TUNEL signal was prevalent in multinucleated cells, TUNEL-positive cells were also observed in germ cells from the basal lamina to the luminal layer of the seminiferous epithelium (Fig. 3H). Based on nuclear characteristics, as revealed by DAPI staining, these apoptotic cells likely correspond to meiotic cells. Immunostaining for the Sertoli cell marker Sox9 showed that wild-type and Sox30−/− tubules contained similar numbers of morphologically normal Sertoli cells (Fig. S5B). Sox30−/− testes contained similar numbers of PLZF (ZBTB16)-positive undifferentiated spermatogonia compared with controls (Fig. S5C), suggesting no discernible defects in Sertoli and spermatogonia. These observations indicate that, prior to arrest at the round spermatid stage, Sox30−/− mice might have defects during meiotic stages, consistent with Sox30 expression in late pachytene spermatocytes (Fig. 1D).
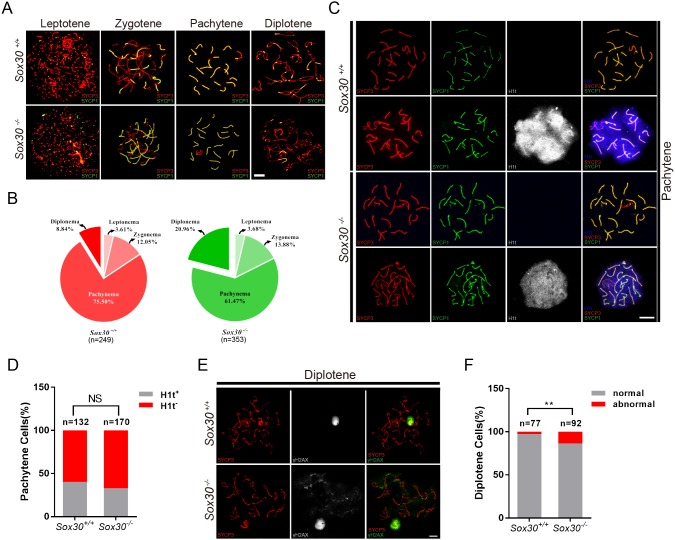
To define potential defects of meiotic progression in Sox30−/− mice, we first examined chromosomal synapsis by double staining spread spermatocyte nuclei with synaptonemal complex protein 1 (SYCP1) and SYCP3. SYCP1 and SYCP3 are the central and lateral element of the synaptonemal complex, respectively (Schalk et al., 1998; Yuan et al., 2000; de Vries et al., 2005; Yang et al., 2006), and their distribution pattern is unique for each developmental stage of the meiotic prophase, identifying leptotene, zygotene, pachytene, diplotene and diakinesis spermatocytes. Although the testes from 8-week-old Sox30−/− mice contained spermatocytes at all meiotic stages (Fig. 4A), the proportion of diplotene spermatocytes in age-matched Sox30−/− testes was significantly increased compared with control (Fig. 4B, Sox30+/+ 8.84%, Sox30−/− 20.96%). This increase of diplotene cells suggested that Sox30−/− spermatocytes successfully progressed to the pachytene stage. We therefore immunostained wild-type and Sox30−/− spread nuclei for testicular H1 histone (H1t; HIST1H1T) (Fig. 4C), a histone variant that specifically accumulates after the mid-pachytene stage (Drabent et al., 1996), and found no difference in the ratio of H1t-positive cells to H1t-negative cells between Sox30+/+ (0.67) and Sox30−/− mice (0.49) (Fig. 4D). Thus, the accumulation of diplotene spermatocytes in Sox30−/− testes appears to be largely due to the defects in the progression of diplotene spermatocytes into later stages, such as diakinesis and metaphase stage. Immunostaining of spread nuclei for γH2AX (phosphorylated form of the histone variant H2AX) revealed that γH2AX was abnormally retained on autosomal chromatin in a small proportion of Sox30−/− diplotene spermatocytes (Fig. 4E,F). Normally, γH2AX disappears from autosomes with progression of meiosis and becomes restricted to the unpaired X and Y chromosomes of the sex body at the pachytene and diplotene stage (Fig. 4E). These data showed that spermatocyte defects are present in Sox30−/− mice, although it is possible that the presence of γH2AX-positive cells may correlate with increased apoptosis in Sox30−/− testes.
Fig. 4.
Increased proportion of diplotene spermatocytes in Sox30−/− testis. (A,B) Immunolabeling of spermatocyte spread nuclei from 8-week-old wild-type and Sox30−/− mice was performed using antibodies against SYCP1 (green) and SYCP3 (red), which are the transverse and lateral elements of synaptonemal complex that forms on the chromosome axis. Differential stages of meiotic I prophase I (leptotene, zygotene, pachytene and diplotene) were identified based on the staining pattern of SYCP1 and SYCP3. At the leptotene stage, axis elements are not completely formed; SYCP3 staining appears as short and discrete lines in the nucleus and SYCP1 signal is absent (Yuan et al., 2000; Yang et al., 2006). During the subsequent zygotene stage, homologous chromosomes start to pair and are associated via the transverse element SYCP1. Axis elements become thicker and SYCP1 is detected at the regions where chromosomes pair. At pachytene, autosomes fully synapse except for the X and Y chromosomes, which only synapse at a short pseudoautosomal region (PAR). SYCP1 and SYCP3 co-localized at autosomes, and the SYCP1 signal is detected in the short region where XY chromosomes synapse (Zickler and Kleckner, 1999; Perry et al., 2001). At diplotene, synapsed chromosomes start to separate from each other and the lateral elements become disassociated in certain regions, where SYCP1 signal disappears. (B) Percentage of spermatocytes at each stage of meiotic prophase I (leptonema, zygonema, pachynema and diplonema) in 8-week-old Sox30+/+ and Sox30−/− mice (spermatocytes analyzed: Sox30+/+, n=249; Sox30−/−, n=353). (C,D) Triple immunostaining for SYCP3 (red), SYCP1 (green), and H1t (white) reveals a normal proportion of early (H1t−) and mid-to-late (H1t+) pachytene spermatocytes in 8-week-old Sox30−/− mice. NS, not significant (χ2 test). (E) Double immunostaining for γH2AX (green) and SYCP3 (red) identifies abnormal localization of γH2AX in diplotene spermatocytes from 8-week-old Sox30−/− mice. (F) Quantification analysis for diplotene spermatocytes with abnormal γH2AX distribution. Sox30+/+, n=77; Sox30−/−, n=92. **P<0.01 (χ2 test). Scale bars: 10 μm.
Sox30 controls a set of haploid genes expressed in pachytene spermatocytes and round spermatids
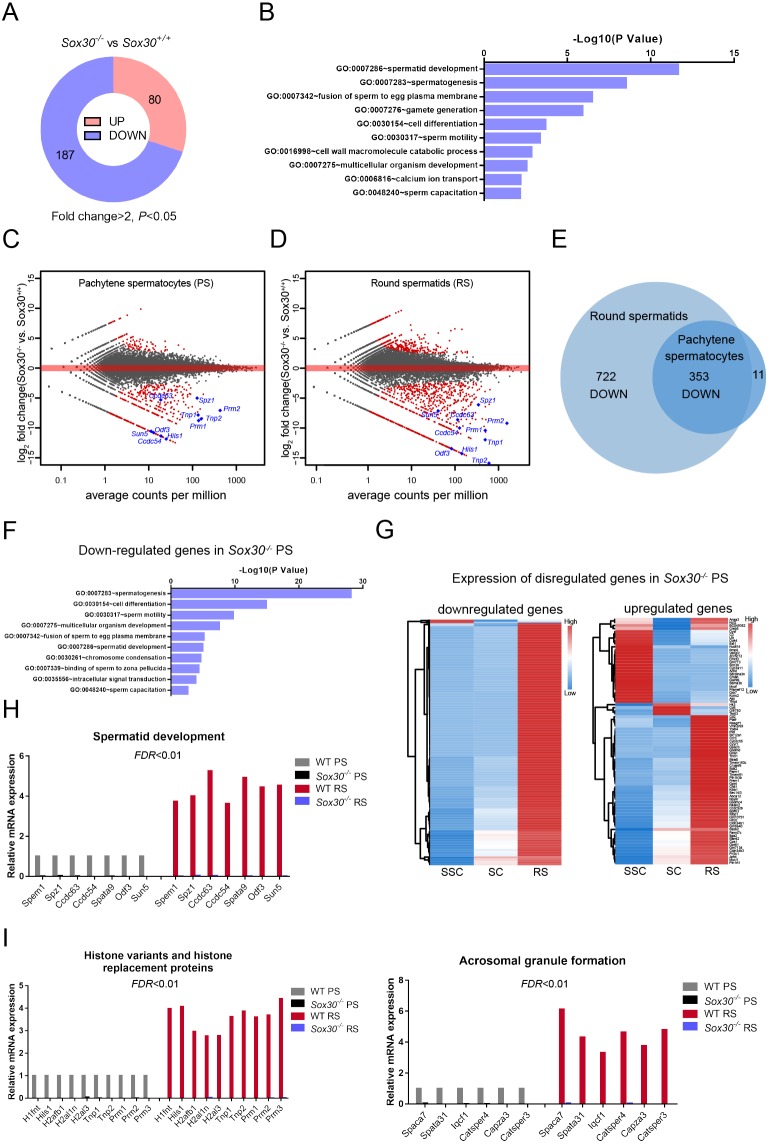
To investigate the molecular events underlying the early spermiogenic arrest in Sox30-deficient males, we first performed RNA-seq of P21 wild-type and Sox30−/− testes (three per genotype). P21 represents a developmental stage concomitant with the first appearance of round spermatid in the first wave of spermatogenesis, and no significant change in the distribution of germ cell populations was observed at this time point between Sox30−/− and control mice (Fig. 2J). We found that, relative to control, 187 genes were downregulated and 80 genes were upregulated in the mutant testes (P<0.05, fold change >2) (Fig. 5A, Table S1). Gene ontology (GO) analysis revealed that downregulated transcripts were strongly associated with the differentiation progression of round spermatids, including spermatid development, spermatogenesis and fusion of sperm to egg plasma membrane (Fig. 5B).
Fig. 5.
Sox30−/− deficiency produces global and cell type-specific gene expression changes in the testis. (A) Number of genes exhibiting significant (fold change >2, P<0.05) up- or downregulation in Sox30−/− versus Sox30+/+ testis at P21. n=3 for each genotype, each of which were deep sequenced separately. (B) Gene ontology associations of downregulated genes in Sox30−/− in testis identified by RNA-seq. (C,D) Scatter plot of differentially expressed transcripts in Sox30−/− pachytene spermatocytes (C) and round spermatids (D) compared with Sox30+/+ cells. Pachytene spermatocytes and round spermatids were isolated from 8-week-old wild-type and Sox30−/− mice by the STA-PUT method. Wild-type, n=6; Sox30−/−, n=8. Each red dot represents a gene that was significantly changed (fold change >2, FDR<0.01). (E) Analysis of differential gene expression in Sox30−/− pachytene spermatocytes and round spermatids identifies 353 transcripts that are downregulated in both stages. (F) GO term enrichment analysis for downregulated transcripts in pachytene spermatocytes of Sox30−/− mice. (G) Genes upregulated or downregulated in Sox30−/− pachytene spermatocytes were used to produce a heat map exhibiting their expression pattern in stage-specific spermatogenic cells. RS, round spermatids; SC, pachytene spermatocytes; SSC, spermatogonial stem cells. (H,I) Expression changes in genes encoding factors essential for spermatid development (H), histone variants and replacement associated proteins (I) and factors involved in acrosomal granule formation (I).
We next performed RNA-seq on purified pachytene spermatocytes and round spermatids from adult Sox30 wild-type and mutant males. Comparison of mRNA expression profiles revealed that 85 and 364 genes were significantly up- and downregulated, respectively, in Sox30−/− versus wild-type pachytene spermatocytes (Fig. 5C, Table S2). In Sox30−/− round spermatids, expression of a larger number of genes was significantly different from control, including 314 upregulated and 722 downregulated genes (Fig. 5D, Table S3). Notably, genes that were downregulated in both Sox30−/− pachytene spermatocytes and round spermatids exhibited a large overlap (pachytene spermatocytes: 353/364, 97%) (Fig. 5E), indicating that these belong to a core or common set of genes subject to regulation by Sox30. The larger number of affected genes in Sox30−/− round spermatids may be due to secondary effects resulting from developmental arrest of round spermatids. GO analysis of downregulated genes in Sox30−/− pachytene spermatocytes and round spermatids found enrichment of genes involved in spermatogenesis and cell differentiation (Fig. 5F, Fig. S6).
To determine whether Sox30 is required for gene expression in haploid male germ cells, we assigned the genes deregulated by Sox30 loss to the previously reported transcriptomes of isolated spermatogenic cell types, specifically using datasets from three spermatogenic cell types (spermatogonial stem cells, spermatocytes and round spermatids) (Davis et al., 2017). Strikingly, this comparison revealed that the majority of downregulated genes in Sox30−/− pachytene spermatocytes and round spermatids was preferentially expressed at the haploid round spermatid stage (Fig. 5G, Fig. S7), whereas genes upregulated in Sox30−/− cells were generally expressed at higher levels in spermatogonial stem cells or round spermatids (Fig. 5G, Fig. S7). Furthermore, many genes downregulated in Sox30−/− pachytene spermatocytes shared a similar developmental expression pattern during spermatogenesis, with upregulation of expression in pachytene spermatocytes followed by high expression level in haploid round spermatids. We detected decreased expression of genes essential for spermatid development (Spem1, Spz1, Ccdc63, Ccdc54, Spata9, Odf3 and Sun5; Fig. 5H). Concomitantly, a group of genes that encode testis-specific histone variants and histone replacement-associated proteins (H1fnt, Hils1, H2afb1, H2al1n, H2al3, Tnp2, Prm1, Prm2 and Prm3; Fig. 5I) as well as genes necessary for acrosomal granule formation (Spaca7, Spata31, Iqcf1, Catsper4, Capza3 and Catsper3; Fig. 5I) were drastically reduced in Sox30−/− cells. These differentially expressed genes between wild-type and Sox30−/− cells were further validated by qRT-PCR assays (Fig. S8A). These results show that Sox30 is specifically required for the expression of haploid round spermatid-specific genes involved in spermiogenesis.
Direct genomic binding sites of Sox30 in mouse testes
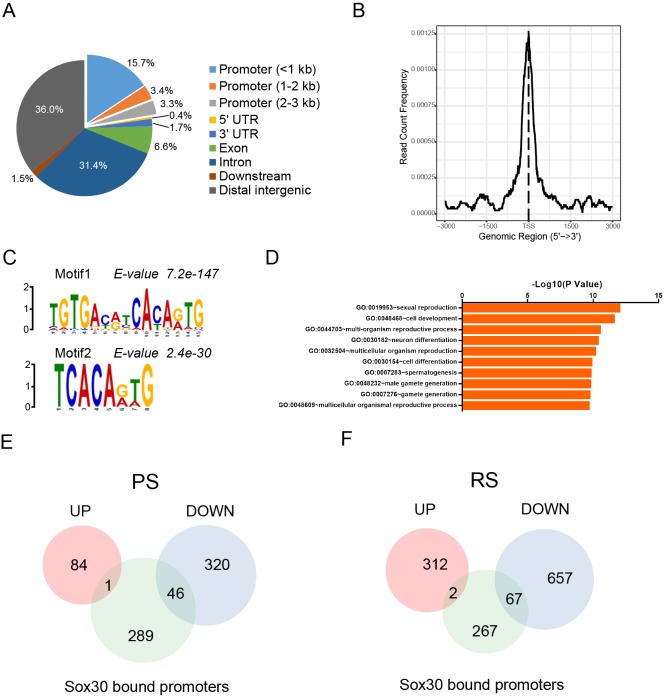
To determine whether Sox30 specifically binds to genomic DNA and to identify genes directly regulated by Sox30, we carried out ChIP-seq for Sox30, using P28 testes, where Sox30 is abundantly expressed. Model-based analysis of ChIP-seq (MACS2) identified 1600 peaks with significant Sox30 enrichment (Table S4). Mapping of ChIP DNA from deep sequencing to genomic regions revealed that 22.4% of Sox30 peaks located within promoter regions, and the remaining peaks mainly resided in intergenic regions (36.0%) and introns (31.4%) (Fig. 6A, Table S4). Consistent with this, the majority of Sox30 binding sites was close to the center of transcription start sites (TSS) (Fig. 6B). De novo motif analysis of Sox30 ChIP peaks using MEME identified a consensus sequence, ‘CACA(G/A)TG’, as the most enriched motif prevalent among Sox30 ChIP peaks (Fig. 6, Table S5). An early in vitro study using an electrophoretic mobility shift assay showed that ectopically expressed recombinant Sox30 bound DNA sequence enriched in ‘ACAAT’ (Osaki et al., 1999). This predicted sequence matches to part of the motif sequence identified in our study. Gene ontology analysis revealed that genes with significant Sox30 peaks correlated with reproduction and cell development (Fig. 6D).
Fig. 6.
ChIP-seq analysis revealed Sox30-binding sites in mouse testis. ChIP-seq experiments with anti-Sox30 antibody were performed using wild-type mouse testes at P28, when Sox30 is abundantly expressed. (A) Genomic locations of Sox30 binding sites. (B) Distribution of Sox30 ChIP reads on gene bodies is plotted. Sox30 was enriched at the regions from −1 kb to +1 kb relative to the TSS. The y-axis represents the frequency of Sox30 ChIP-seq counts at a specific site, normalized by total read counts. (C) De novo motif analysis of Sox30-binding sites using MEME. The most enriched de novo motifs are shown (Motif 1: E value=7.2e−147; Motif 2: E value=2.4e−30). (D) The gene ontology terms analyzed from the top 50% of Sox30-bound genes. (E,F) Identification of direct target genes of Sox30 by ChIP-seq and RNA-seq analysis. The total number of genes with Sox30 ChIP-seq peaks at promoters was compared with disregulated genes upon Sox30 deletion in pachytene spermatocytes (E) and round spermatids (F).
Sox30 preferentially binds to a unique set of postmeiotic genes and regulates their expression
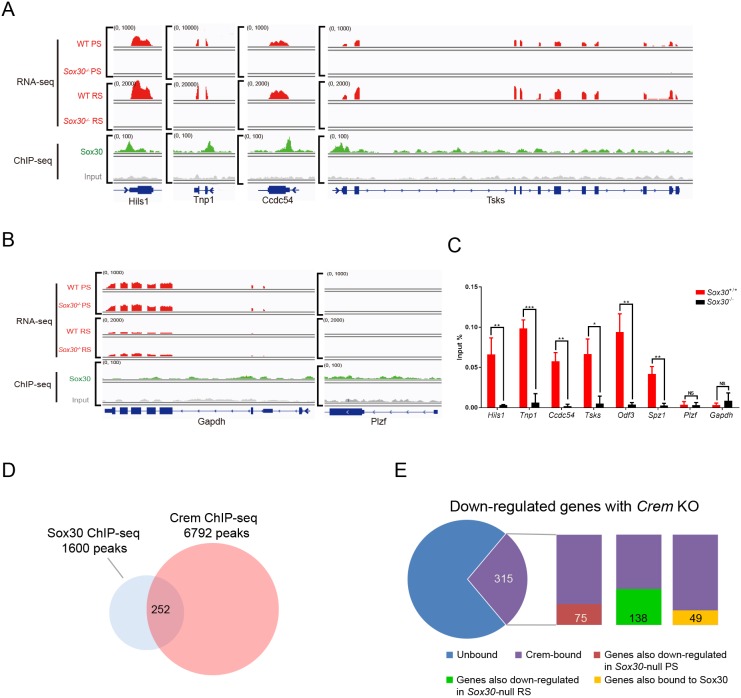
We next investigated whether the recruitment of Sox30 subsequently affects expression of the respective target gene. Of 336 genes with Sox30 ChIP-seq peaks at their promoters, 46 and 67 genes were downregulated upon Sox30 KO in pachytene spermatocytes and round spermatids, respectively (Fig. 6E,F, Table S6). On the contrary, only one or two genes bearing Sox30-binding sites at their promoters were upregulated in Sox30−/− pachytene spermatocytes and Sox30−/− round spermatids (Fig. 6E,F, Table S6), indicating that Sox30 primarily activates the transcription of a set of postmeiotic genes. Notably, Sox30 bound to the promoter regions of key genes involved in haploid cell development including Tnp1, Hils1, Ccdc54 and Tsks (Fig. 7A). Indeed, RNA-seq showed that the absence of Sox30 caused a pronounced reduction in the transcription of these haploid genes in isolated cells (Fig. 7A). Interestingly, strong ChIP-seq binding peaks were observed at the Sox30 promoter, indicating that Sox30 autoregulates its expression by targeting its own mRNA (Fig. S8B). Autoregulation is one of the most efficient regulatory loops for maintaining gene expression levels, and has also been reported for the transcription factor CREM, which occupies its own promoter (Martianov et al., 2010). Conversely, Sox30 did not bind to promoters/gene bodies of genes associated with spermatogonial self-renewal (Plzf) or the housekeeping gene Gapdh (Fig. 7B). ChIP-qPCR analysis further confirmed Sox30 enrichment at promoter regions of representative target genes in wild-type but not Sox30−/− P28 testes (Fig. 7C). Significant reduction in the expression of these target genes in pachytene spermatocytes and round spermatids was validated by q-PCR (Fig. S8A).
Fig. 7.
Sox30 specifically bound and directly regulated expression of postmeiotic genes. (A,B) Genome browser view of Sox30 ChIP-seq and RNA-seq reads on representative gene loci in testes or isolated cells from wild-type and Sox30−/− mice. (C) ChIP-qPCR analysis of Sox30 enrichment on promoter regions of Hils1, Tnp1, Ccdc54, Tsks, Odf3, Spz1, Plzf and Gapdh in P28 testes of wild-type and Sox30−/− mice. The chromatin occupancy at Plzf and the housekeeping gene Gapdh was low and was similar in wild type and mutant. Each sample represents a pool of three individual mice. Data are presented as mean±s.d. *P<0.05, **P<0.01, ***P<0.001 (Student's t-test). (D) Overlapping binding sites of Sox30 and CREM ChIP-seq peaks in testis. (E) Out of a total of 820 genes downregulated in Crem−/− testis, 315 genes contained CREM binding peaks. Of those, 75 and 138 genes were also reduced in Sox30−/− pachytene spermatocytes and Sox30−/− pachytene spermatocytes, respectively. Of the 315 direct targets of CREM, 49 genes were bound by Sox30 at promoters or intragenic regions.
CREM is the master transcription factor that functions at the postmeiotic stage, and the phenotype of Sox30 mutants resembles that of Crem mutants (Blendy et al., 1996; Nantel et al., 1996; Zhou et al., 1996). Comparison of our Sox30 ChIP-seq with the CREM ChIP-seq data set showed that 3.71% CREM-binding sites at genomic regions contained Sox30 ChIP peaks (Fig. 7D). We next performed microarray analysis and found that 820 genes were downregulated greater than 2-fold in Crem−/− testis, and 315 genes were bound by CREM (Fig. 7E) (Martianov et al., 2010; Kosir et al., 2012), suggesting that these are direct targets of CREM. Among these direct targets identified by ChIP-seq and microarray analysis, 75 genes (23.8%) showed similarly reduced expression in Sox30 KO pachytene spermatocytes (Fig. 7E), and 138 genes (43.8%) were similarly reduced in Sox30 KO round spermatids (Fig. 7E). Furthermore, 49 genes (15.6%) of downregulated genes bound by CREM were also bound by Sox30 at promoters and/or gene bodies (Fig. 7E). This observation indicates that Sox30 controls a core set of postmeiotic genes, and that the direct targets of Sox30 and CREM partially overlap during spermiogenesis.
Transcription and processing of piRNA precursors is not affected in Sox30−/− mutant testes
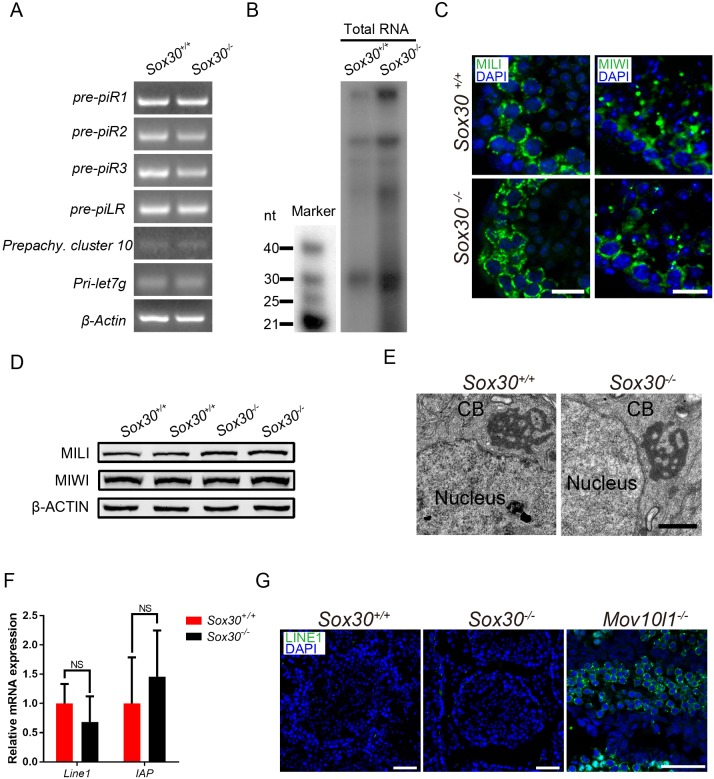
The results described above establish that Sox30 is a transcription factor required for initiating a core set of postmeiotic genes and that mice lacking this protein display impaired spermiogenesis arresting at the early round spermatid stage. A similar phenotype is observed in mouse mutants deficient in piRNA biogenesis or components of the piRNA pathway such as Miwi−/− (Deng and Lin, 2002; Grivna et al., 2006; Reuter et al., 2011) or Mael−/− (Soper et al., 2008; Castaneda et al., 2014) mice. Primary piRNA biogenesis initiates with the transcription of piRNA precursors, and A-MYB (MYBL1) has also been identified as a transcription factor promoting the transcription of both pachytene piRNA precursors from intergenic loci and mRNAs for piRNA biogenesis factors (Bolcun-Filas et al., 2011; Li et al., 2013). Therefore, Sox30 may regulate pachytene piRNA production by affecting piRNA precursor transcription, and spermiogenic arrest of Sox30 mutants could result from altered piRNA abundance. To explore this possibility, we first performed examined expression levels of precursors of four pachytene piRNAs (piR1, piR2, piR3 and piLR) and the precursor of miRNA (Pri-let7g) and they were similar in adult Sox30−/− mutant testes versus control (Fig. 8A). As expected, the expression level of pre-pachytene piRNA precursors (cluster 10) was low at this stage (Fig. 8A). Thus, Sox30 deficiency did not affect the primary transcription of piRNA precursors. Analysis of total RNA isolated from adult testes revealed comparable abundance of piRNAs (24-31 nt in length) in wild type and mutant (Fig. 8B). Furthermore, the piRNA pathway components Mili (Piwil2) and Miwi normally localized to the intermitochondrial cement (IMC) of spermatocytes and aggregated in chromatoid bodies (CBs) of round spermatids in adult mutant testis (Fig. 8C), and their protein abundance remained similar (Fig. 8D). The intact architecture of CBs in Sox30−/− round spermatids was further confirmed by electron microscopy analysis (Fig. 8E). We observed neither upregulation of long interspersed nuclear element-1 (LINE1, L1) and intracisternal A-particle (IAP) transcripts in mutant testes nor detectable levels of the LINE1 retrotransposon-derived open reading frame 1 (ORF1) protein (Fig. 8F,G). In contrast, protein abundance of ORF1 was substantially increased in mouse testis lacking Mov10l1, a gene known to participate in piRNA processing (Fig. 8G) (Zheng et al., 2010; Zheng and Wang, 2012). Thus, Sox30 does not appear to be involved in piRNA-mediated transposon silencing.
Fig. 8.
Sox30−/− deficiency does not affect transcription and processing of piRNA precursors in the testis. (A) Expression of piRNA precursors by semi-quantitative RT-PCR analysis in adult wild-type and mutant testes. (B) The level of piRNA abundance was not reduced in Sox30 mutant testes. 5′ labeling of total small RNAs in 8-week-old Sox30+/+ and Sox30−/− testis. (C) Immunostaining of the piRNA pathway components MILI and MIWI on frozen sections of adult Sox30+/+ and Sox30−/− testes. Scale bars: 10 μm. (D) Western blot analysis of MILI and MIWI in P21 wild-type and mutant testes. (E) Electron micrographs of wild-type and Sox30-deficient round spermatids. Scale bar: 1 μm. (F) Q-PCR analysis of LINE1 and IAP transcripts in P21 wild-type and mutant testes. NS, not significant (Student’s t-test). n=3. (G) Immunofluorescence detection of LINE1 ORF1p from adult wild-type, Sox30−/− and Mov10l1-deficent testes. Scale bars: 50 μm.
Upon initiation of meiosis, pachytene piRNAs account for 95% of piRNAs in the mouse testis and are predominantly derived from non-repetitive pachytene piRNA clusters (Vourekas et al., 2012; Li et al., 2013). Sox30 ChIP-seq reads were mapped to the 119 annotated pachytene piRNA clusters (Vourekas et al., 2012), and only three clusters contained Sox30 ChIP-seq reads (Fig. S9A). We next examined the top ten piRNA clusters that produce the largest amounts of piRNAs. Genome browser track analysis showed that Sox30 does not bind to these piRNA clusters, as illustrated for representative examples in Fig. S9B. Sox30 ChIP-seq peaks at genomic regions were compared with the A-MYB ChIP-seq data set (Bolcun-Filas et al., 2011; Li et al., 2013), and only 1.07% A-MYB binding sites contained Sox30 ChIP peaks (Fig. S9C). Moreover, the binding motif identified by Sox30 ChIP-seq is distinct from that of A-MYB ChIP-seq, suggesting they do not overlap. These results show that Sox30 does not have a regulatory role in transcribing pachytene piRNA precursors.
DISCUSSION
The present study reveals the physiological role of Sox30 in the male germline. Sox30 functions as a testis-specific transcription factor essential for the expression of a core set of haploid cell-specific genes and required for spermiogenesis. In Sox30-deficient testes, proacrosomic vesicles fail to form a single acrosomal organelle and spermatids become uniformly arrested at the early round spermatid stage (step 2-3) (Fig. 3A-C). Our transcriptome analyses of stage-specific fractionated germ cells show that loss of Sox30 results in downregulation of a set of genes in meiotic pachytene spermatocytes and postmeiotic round spermatids (Fig. 5C,D). Subsequent GO analysis revealed that these downregulated transcripts are associated with spermatid nucleus reorganization, acrosome formation and flagella development, all of which represent important molecular events during spermiogenesis. Similarly to the Sox30 mutant, failed fusion of proacrosomic vesicles into a single large acrosome has also been observed in mice lacking the acrosome formation-associated gene Hrb (Agfg1) (Kang-Decker et al., 2001). However, spermiogenesis of Hrb-deficient mice still progressed to develop spermatozoa with a round sperm head and a tail. Thus, the early and clean spermiogenic arrest at step 2-3 in Sox30 mutants is likely a combined effect of defects in acrosome formation, nuclear condensation and flagella development due to the perturbed expression program of spermiogenesis. Further genome-wide ChIP-seq analysis confirmed that Sox30 binds to specific DNA sequences and directly regulates expression of a core set of postmeiotic genes. Through genetic and molecular biological approaches, we demonstrate that Sox30 is one of the few genes that participate in late meiotic stage and spermiogenesis by directly regulating specific haploid genes expression.
During mouse spermatogenesis, transcription of a subgroup of haploid genes begins in late meiotic cells and is elevated dramatically at the round spermatid stage (Iguchi et al., 2006). Although the major defects in Sox30−/− mutants occur at the spermiogenic stage, sporadic spermatocyte defects are present (Fig. 3H, Fig. 4E,F). It is interesting to note that transcription of a set of core haploid genes was reduced at the pachytene stage in Sox30−/− mutants (Fig. 5C), concomitant with increased expression of Sox30 at this stage. These results suggest that Sox30 may function during the transition from the meiotic to the post-meiotic phase by facilitating late meiotic cells to enter a post-meiotic cell differentiation program. Indeed, we observed increased accumulation of diplotene cells in the mutant, despite the fact that the majority of Sox30−/− germ cells underwent meiosis and formed round spermatids. This defect could be, in part, a result of an altered transcriptome in late meiotic cells.
Our data demonstrate that the Sox30 mutation phenocopies the Crem mutation in mice. Sox30 mutant exhibits a complete spermiogenic arrest at step 2-3 (Fig. 3A-C). Similarly, the spermatogenesis of Crem-deficient mice is arrested at the early round spermatid stage (by step 5), accompanied by the absence of postmeiotic cell-specific gene expression (Blendy et al., 1996; Nantel et al., 1996; Zhou et al., 1996). The similarity in spermatogenic phenotypes suggests that Sox30 and CREM may have overlapping functions, despite the normal expression of Crem in Sox30−/− mutant (Fig. S8C). A comparison of CREM-regulated genes with those identified in the Sox30−/− round spermatids reveals a large overlapping set of genes (Table S6, 49.3%) that are downregulated in both mutants. Of the 315 genes that are bound by CREM and display reduced expression in Crem-deficient testis (Martianov et al., 2010; Kosir et al., 2012), 49 genes (15.6%) were bound by Sox30 with similar reduced expression in Sox30−/− pachytene spermatocytes and round spermatids (Fig. 7E). Regulatory factor X 2 (Rfx2) is also an essential transcription factor during spermiogenesis. Rfx2-deficient mice display a complete block of spermiogenesis at the transition from round to elongating spermatids (Kistler et al., 2015; Wu et al., 2016). However, only a few Rfx2-regulated genes (21, 5.8%; data not shown) are downregulated in the Sox30−/− mutant. These analyses indicate that Sox30 and CREM may co-regulate a small cohort of genes, whereas Rfx2 regulates a distinct set of target genes responsible for flagella development.
IMCs and CBs serve as cytoplasmic granules to store RNA and RNA-binding proteins that are involved in the maintenance or processing of mRNAs and small RNAs. Sox30 mutant testis tissue exhibits normal architecture of CBs in round spermatids as well as IMCs in spermatocytes (Fig. 8C,E). Normal function of these granules was also supported by normal piRNA expression in Sox30−/− testes (Fig. 8B). In contrast, these RNA processing-associated granules are significantly impaired in the Miwi−/− mutant, which exhibits a clear-cut arrest of spermatogenesis at the beginning of spermiogenesis (Deng and Lin, 2002). The dysregulated gene expression in the Miwi−/− mutant could result from their instability or it may be a consequence of altered piRNAs, which can target and regulate endogenous mRNAs (Goh et al., 2015; Zhang et al., 2015). Indeed, RNA immunoprecipitation assay has demonstrated that MIWI is an RNA-binding protein that forms complexes with some CREM-target genes (Deng and Lin, 2002). Remarkably, a large cohort of genes found to be downregulated in pachytene cells from Miwi-deficient testes are also dramatically reduced in Sox30-deficient pachytene spermatocytes (240, 68%; data not shown). Thus, the similar spermiogenic arrest in Miwi and Sox30 mutants could be a common consequence of dysregulated haploid genes expression, although through distinct mechanisms.
Although in vitro meiosis and gametogenesis from mouse embryonic stem cell-derived primordial germ cells has been successful, a low frequency of apparent haploid cells and haploid cell-derived offspring is obtained (Zhou et al., 2016). Identifying essential factors or compounds to induce regulatory networks that promote transition of meiosis to postmeiosis and spermatid differentiation may further improve protocols for in vitro spermatogenesis. Many Sox family members are crucial regulators of cell fate decisions. Sox2 has been shown to function as a core transcription factor for the generation of induced pluripotent stem cells (Takahashi and Yamanaka, 2006). Forced expression of Sox2 in somatic cells, in combination with other synergistic factors, enhance embryonic stem cell marker genes. Here, our studies demonstrate an essential role for Sox30 in differentiation of late meiotic cells and initiation of the postmeiotic gene expression program and postmeiotic haploid differentiation. Future studies will be needed to elucidate the endogenous mechanism as well as the signaling molecules that establish precise activation and simultaneous repression of different Sox factors at distinct developmental stages.
MATERIALS AND METHODS
Generation of Sox30 knockout mice by CRISPR/Cas9
Exon 2 of the Sox30 gene, which contains the HMG-box domain, was targeted by two sgRNAs. Oligos for sgRNA expression plasmids were annealed and cloned into the BsaI sites of pUC57-sgRNA (Addgene 51132). Oligo sequences were as follows: sgRNA1_up: taggGGGTTAGCTTTGGCTAGTGC; sgRNA1_down: aaacGCACTAGCCAAAGCTAACCC; sgRNA2_up: taggGCAGAAATCAGTGTCCAGCT; sgRNA2_down: aaacAGCTGGACACTGATTTCTGC. The Cas9 expression construct pST1374-Cas9-N-NLS-Flag-linker (Addgene plasmid #44758, deposited by Xingxu Huang) was linearized with AgeI, transcribed using the T7 Ultra Kit (Ambion), and further purified using RNeasy Mini Kit (Qiagen). pUC57-sgRNA expression vectors were linearized by DraI and transcribed in vitro using the MEGA shortscript Kit (Ambion) as described previously (Shen et al., 2014). Cas9 mRNA (20 ng/μl) and two sgRNAs (5 ng/μl each) were introduced into the cytoplasm and male pronucleus of C57BL/6J mouse zygotes by electroporation. Embryos were implanted into pseudo-pregnant C57BL/6J females according to standard procedures. Founder mice were backcrossed to C57BL/6J. Mice were maintained under specific pathogen-free conditions according to the guidelines of the Institutional Animal Care and Use Committee of Nanjing Medical University.
Isolation of spermatogenic cells
We isolated spermatogonia (primitive type A, mature type A and type B spermatogonia), pachytene spermatocytes, round spermatids and elongated spermatids by the STA-PUT method described by Bellvé with minor modifications (Bellvé, 1993). Sertoli and spermatogonia cells were isolated from mice at P6-8, and pachytene spermatocytes and round spermatids were isolated from adult mice. Briefly, mice testes were harvested after euthanasia and digested with collagenase IV (1 mg/ml). The dispersed seminiferous tubules was washed with DMEM and centrifuged at 500 g. The pellet was further digested with 0.25% Trypsin containing DNase I (1 mg/ml) and filtered to prepare a single-cell suspension. The single-cell suspension was loaded into a cell separation apparatus (ProScience, Canada), followed by a 2-4% bovine serum albumin (BSA) gradient (2% BSA and 4% BSA in DMEM were loaded into the separation apparatus chamber). After 1.5-3 h of sedimentation, cell fractions were harvested. We determined different cell types according to morphological characteristics, cell diameter and DAPI staining under a light microscope. The purity of spermatogonia, pachytene spermatocytes and round spermatids reached about 90%.
Generation of anti-Sox30 polyclonal antibodies
The Sox30 antibody was custom-generated by ABclonal and is now commercially available (ABclonal, A11759). The Sox30 cDNA fragment corresponding to amino acids 1-150 was cloned into the pGEX-4T-1 vector. The GST-Sox30 (1-150) fusion protein was expressed in Rosetta bacteria, purified with glutathione-sepharose beads. Two rabbits were immunized with recombinant Sox30 protein. The anti-Sox30 antiserum was collected, followed by affinity purification with antigens. The purified Sox30 antibody was used for western blot (1:1000), immunofluorescence (1:100) and ChIP-seq assay (6 µg).
Quantitative and semi-quantitative RT-PCR assay
Total RNA was extracted from testes, isolated spermatogenic cells and other tissues with Trizol reagent (Thermo Fisher Scientific), and was treated with DNaseI (Amp grade 1.5 µ/μl, Invitrogen RNase free). Total RNA (1 µg) was reverse transcribed into cDNA with PrimeScriptRT Master Mix (RR036A, Takara, Japan). For quantitative RT-PCR, 2 µl diluted cDNA was used for each real-time PCR containing SYBR Green Premix Ex Taq II (RR820A, Takara). Gene expression was normalized to Rplp0 (also known as 36b4) within the log phase of the amplification curve. For semi-quantitative RT-PCR, we used a typical reaction containing 500 nM forward and reverse primers for individual target genes. For cDNA input, 20 µl cDNA product from the reverse transcription reaction above was diluted 5-fold and 2 µl diluted cDNA was used as the template for each reaction. The PCR reaction products were analyzed by gel electrophoresis after appropriate amplification cycles, identified through a series of different cycles. β-actin was used as a loading control. The primer sequences are listed in Table S7.
Histology and transmission electron microscopy analysis
Testes and epididymides were fixed in Bouin's solution (Sigma, SLBJ3855V) overnight. Then the testes were dehydrated with increasing concentration of ethanol (70%, 80%, 90%, 100%), cleared in xylene, embedded in paraffin and cut into 5-μm-thick sections. The sections were deparaffinized, rehydrated, stained with Hematoxylin and Eosin (H&E; Sigma-Aldrich) or stained with PAS reagent. For transmission electron analysis, samples were fixed with 5% glutaraldehyde in 0.2 M cacodylate buffer at 4°C overnight. They were washed with 0.2 M cacodylate buffer, dehydrated in a graded series of ethanol, and embedded and polymerized by automated microwave tissue processor (Leica EMAMW). After polymerization, the specimens were cut into small pieces (1 mm3) using a LEICA Ultracut UCT ultramicrotome (Leica Microsystems). Ultrathin sections were stained with uranyl acetate and lead citrate, and were examined by TEM (JEOL, JEM-1010).
Immunofluorescence, TUNEL assay and chromosome spread
To obtain cryosections, testes were fixed in 4% paraformaldehyde at 4°C, dehydrated in sucrose, embedded in OCT as previously described (Zheng et al., 2010) then cut at 5 μm thickness using a cryotome (Thermo Scientific Cryotome FSE). Immunostaining was performed on testis cryosections with the following primary antibodies: anti-PLZF (1:200; R&D Systems, AF2944), anti-SOX9 (1:500; Millipore, AB5535), anti-MILI (1:100; Abcam, ab36764), anti-MIWI (1:100; Abcam, ab12337), anti-LINE1 ORF1p (1:200; a gift from Prof. Ramesh Pillai, University of Geneva, Switzerland) and anti-Sox30. For acrosome staining, cryosections were incubated for 1 h at 37°C with Rhodamine-conjugated PNA (Vector Laboratories, RL-1072). Nuclear DNA was stained with DAPI (Sigma, F6057). TUNEL staining was also performed using the ApopTag Fluorescein In Situ Apoptosis Detection kit (Millipore, S7110). Chromosome spreads of prophase I spermatocytes were performed as previously described (Kolas et al., 2005). Primary antibodies were as follows: anti-SYCP1 (1:200; Abcam, ab15090), anti-SYCP2 (1:200; a gift from Jeremy Wang, University of Pennsylvania, USA), anti-SYCP3 (1:200; Abcam, ab97672), anti-γH2AX (1:800; Millipore, 16-202A) and anti-H1t (1:50; Santa Cruz Biotechnology, sc-247171). All the staining of testis sections and spread confocal images were visualized on a confocal microscope (Carl Zeiss, LSM700).
Immunoblotting
Tissues were rinsed with PBS and homogenized in cold RIPA buffer containing protease inhibitor cocktail tablets (Roche, 4693124001). Tissue lysates were rotated at 4°C for 1 h and centrifuged at 11,000 g. The supernatant was collected and the protein concentration was determined by a BCA assay. Aliquots of 20 μg protein were separated by SDS-PAGE.
RNA-seq
For whole testes, wild-type and Sox30 mutant mice testes were collected at P21. Total RNA was extracted from the sample using Trizol reagent (Thermo Fisher Scientific). Three biological replicates were carried out and used for sequencing in both wild-type and Sox30 mutant mice. For isolated spermatogenic cells, total RNA was isolated from pachytene cells and round spermatids from wild-type and Sox30−/− mice (wild type, n=6; Sox30−/−, n=8). Strand-specific libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation kit (Illumina) according to the manufacturer's instructions before submitting to the Illumina Hiseq X ten system. RNA-seq library preparation and sequencing were performed at BGI Tech (Shenzhen, China). Clean data were obtained by trimming the adaptor and filtering rRNAs. Clean reads were then mapped to the mouse genome (mm10) with TopHat (v.2.0.9; Johns Hopkins University, Baltimore, MD, USA) with a GTF file download from the Ensemble database (The European Bioinformatics Institute, Cambridgeshire, UK). The uniquely mapped reads of genes were counted using HTSeq followed by DESeq2 normalization to evaluate gene expression as normalized counts per million. Significant differentially expressed genes were identified as those with a P-value or false discovery rate (FDR) value above the threshold (P21 testes: P<0.05; pachytene spermatocytes and round spermatids: FDR<0.01) and fold-change >2 using DESeq2 software.
ChIP-seq and data analysis
Testes tissue from wild-type and Sox30 mutant was collected at P28 and cross-linked with 1% formaldehyde, followed by quenching with glycine solution. Chromatin fragmentation was performed by sonication in ChIP SDS lysis buffer (50 mM Tris-HCl pH 8, 10 mM EDTA pH 8, 1% SDS) using the Covaris-S220 sonicator. Cross-linked chromatin was incubated with anti-Sox30 antibodies in ChIP dilution buffer (50 mM HEPES pH 7.5, 155 mM NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate) with protease inhibitors overnight. Cross-linking was reversed overnight at 65°C, and DNA extracted using phenol/chloroform/isoamyl alcohol. Precipitated DNA was amplified for deep sequencing or analyzed by quantitative PCR with SYBR Green Premix Ex Taq II (RR820A, Takara). Primer sequences used and ChIP-qPCR analysis can be found in Table S7. For ChIP-seq, DNA was amplified according to the ChIP Sequencing Sample Preparation Guide provided by Illumina using adaptors and primers. Deep sequencing was performed by the Computational Biology Omics Core of the CAS-MPG Partner Institute using Illumina Hiseq X-10 (2X150). Sequenced reads were mapped to the mouse genome (mm10) using BOWTIE software (bowtie2). Peak calling for Sox30 was carried out by using MACS2 (version 2.1.1.20160309, options-mfold 05, 50, -Pvalue 1e−4), on Sox30 ChIP file against the input file. Genome-wide signal coverage tracks at every base pair were also computed by MACS2 and visualized in the Integrative Genome Browser (IGV). Peaks were annotated to the nearest genes within 3 kb of the TSS using Bioconductor package ChIPSeeker 1.12.1. Peaks overlapping by at least one nucleotide with unique gene model promoters (±3 kb of each unique gene model TSS) were considered to be ‘promoter located’. De novo motif searches of ChIP-seq peaks were performed using MEME.
Statistical analysis
All data are reported as mean±s.d. unless otherwise noted in the figure legends. Significance was tested using two-tailed unpaired Student's t-tests (*P<0.05; **P<0.01; ***P<0.001) using Prism 7.0 (GraphPad Software). For meiotic analyses, χ2 test (Prism 7.0) was used to reveal the statistical differences between Sox30+/+ and Sox30−/− mutant cells.
Supplementary Material
Acknowledgements
We thank Sigrid Eckardt for help with manuscript preparation and Mingshuang Li (Bio-Med Big Data Center, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for ChIP-seq DNA library preparation.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.Z., L.Y., X.G.; Methodology: L.Y., S.B., K.Z.; Software: S.B., J.Z., H.F., C.X., M.L.;Validation: K.F., H.Y.; Formal analysis: S.B., J.Z., H.Y.; Investigation: S.B., K.F., H.Y., Q.Y., Y.C., B.S., L.C., H.T., X.L., Y.G., Y.Z., J.X., W.H., W.L., Y.W.; Resources: H.F., C.X.; Data curation: S.B., X.G., K.F.; Writing - original draft: L.Y., S.B., K.Z.; Writing - review & editing: K.Z., Z.S., L.Y.; Visualization: L.Y., S.B., K.Z.; Supervision: K.Z., X.G., L.Y.; Project administration: K.Z., S.B., L.Y.; Funding acquisition: L.Y., K.Z., X.G., Z.S.
Funding
This work was funded by the Ministry of Science and Technology of the People's Republic of China (2016YFA0500902 to K.Z.; 2016YFA0503300 to X.J.G.), the National Natural Science Foundation of China (31471228 and 31771653 to K.Z.; 81471502 to L.Y.; 31471403 and 81771641 to X.J.G.), Jiangsu Science and Technology Department (BK20150047 to K.Z.), the Natural Science Foundation of Jiangsu Province (BK20140897 and 14KJA180005 to K.Z.; 15KJA180006 to L.Y.), the Innovative and Entrepreneurial Program of Jiangsu Province (L.Y.), Jiangsu Provincial Department of Education (BRA2016386 to X.J.G.), Six Talent Peaks Project in Jiangsu Province (YY-019 to X.J.G.), the Fok Ying Tung Education Foundation (161037 to X.J.G.), the National Institutes of Health (ES027544, DK111436 and CA215591 to Z.S.) and the American Heart Association (30970064 to Z.S.). Z.S. was supported by a Taishan scholar Foundation of Shandong Province. Deposited in PMC for release after 12 months.
Data availability
RNA-seq and ChIP-seq data are available from the NCBI SRA database (BioProject No: PRJNA433934; Study accession No: SRP143508) and the National Omics Data Encyclopedia (http://www.biosino.org/node/project/detail/OEP000012; accession No. OEP000012).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.164855.supplemental
Reference
- Barral S., Morozumi Y., Tanaka H., Montellier E., Govin J., de Dieuleveult M., Charbonnier G., Coute Y., Puthier D., Buchou T. et al. (2017). Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Mol. Cell 66, 89-101 e8. 10.1016/j.molcel.2017.02.025. [DOI] [PubMed] [Google Scholar]
- Bellvé A. R. (1993). Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225, 84-113. 10.1016/0076-6879(93)25009-Q [DOI] [PubMed] [Google Scholar]
- Bettegowda A. and Wilkinson M. F. (2010). Transcription and post-transcriptional regulation of spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1637-1651. 10.1098/rstb.2009.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy J. A., Kaestner K. H., Weinbauer G. F., Nieschlag E. and Schütz G. (1996). Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380, 162-165. 10.1038/380162a0 [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E., Bannister L. A., Barash A., Schimenti K. J., Hartford S. A., Eppig J. J., Handel M. A., Shen L. and Schimenti J. C. (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319-3330. 10.1242/dev.067645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda J., Genzor P., van der Heijden G. W., Sarkeshik A., Yates J. R. III, Ingolia N. T. and Bortvin A. (2014). Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 33, 1999-2019. 10.15252/embj.201386855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Buffone M. G., Kouadio M., Goodheart M., Page D. C., Gerton G. L., Davidson I. and Wang P. J. (2007). Abnormal sperm in mice lacking the Taf7l gene. Mol. Cell. Biol. 27, 2582-2589. 10.1128/MCB.01722-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. P., Carrieri C., Saini H. K., van Dongen S., Leonardi T., Bussotti G., Monahan J. M., Auchynnikava T., Bitetti A., Rappsilber J. et al. (2017). Transposon-driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Rep. 18, 1231-1247. 10.15252/embr.201744059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. and Lin H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819-830. 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- de Vries F. A., de Boer E., van den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J. G., van Zeeland A. A., Heyting C. and Pastink A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376-1389. 10.1101/gad.329705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabent B., Bode C., Bramlage B. and Doenecke D. (1996). Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 106, 247-251. 10.1007/BF02484408 [DOI] [PubMed] [Google Scholar]
- Feng C.-W. A., Spiller C., Merriner D. J., O'Bryan M. K., Bowles J. and Koopman P. (2017). SOX30 is required for male fertility in mice. Sci. Rep. 7, 17619 10.1038/s41598-017-17854-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh W. S. S., Falciatori I., Tam O. H., Burgess R., Meikar O., Kotaja N., Hammell M. and Hannon G. J. (2015). piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 29, 1032-1044. 10.1101/gad.260455.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna S. T., Pyhtila B. and Lin H. (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 103, 13415-13420. 10.1073/pnas.0605506103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht N. B. (1998). Molecular mechanisms of male germ cell differentiation. BioEssays 20, 555-561. [DOI] [PubMed] [Google Scholar]
- Iguchi N., Tobias J. W. and Hecht N. B. (2006). Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc. Natl. Acad. Sci. USA 103, 7712-7717. 10.1073/pnas.0510999103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang-Decker N., Mantchev G. T., Juneja S. C., McNiven M. A. and van Deursen J. M. (2001). Lack of acrosome formation in Hrb-deficient mice. Science 294, 1531-1533. 10.1126/science.1063665 [DOI] [PubMed] [Google Scholar]
- Kistler W. S., Baas D., Lemeille S., Paschaki M., Seguin-Estevez Q., Barras E., Ma W., Duteyrat J.-L., Morlé L., Durand B. et al. (2015). RFX2 is a major transcriptional regulator of spermiogenesis. PLoS Genet. 11, e1005368 10.1371/journal.pgen.1005368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Marcon E., Crackower M. A., Höög C., Penninger J. M., Spyropoulos B. and Moens P. B. (2005). Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma 114, 92-102. 10.1007/s00412-005-0334-8 [DOI] [PubMed] [Google Scholar]
- Kosir R., Juvan P., Perse M., Budefeld T., Majdic G., Fink M., Sassone-Corsi P. and Rozman D. (2012). Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis. PLoS ONE 7, e31798 10.1371/journal.pone.0031798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laronda M. M. and Jameson J. L. (2011). Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology 152, 1606-1615. 10.1210/en.2010-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W., Xu J., Moore M. J., Schimenti J. C., Weng Z. et al. (2013). An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67-81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Fimia G.-M., Dierich A., Parvinen M., Sassone-Corsi P. and Davidson I. (2001). Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7, 509-515. 10.1016/S1097-2765(01)00198-8 [DOI] [PubMed] [Google Scholar]
- Martianov I., Choukrallah M.-A., Krebs A., Ye T., Legras S., Rijkers E., Van Ijcken W., Jost B., Sassone-Corsi P. and Davidson I. (2010). Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics 11: 530 10.1186/1471-2164-11-530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M. L., Mohapatra B., Shirley C. R. and Zhao M. (2003). Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111, 483-488. 10.1007/s00412-002-0227-z [DOI] [PubMed] [Google Scholar]
- Nantel F., Monaco L., Foulkes N. S., Masquilier D., LeMeur M., Henriksén K., Dierich A., Parvinen M. and Sassone-Corsi P. (1996). Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380, 159-162. 10.1038/380159a0 [DOI] [PubMed] [Google Scholar]
- Oliva R. (2006). Protamines and male infertility. Hum. Reprod. Update 12, 417-435. 10.1093/humupd/dml009 [DOI] [PubMed] [Google Scholar]
- Osaki E., Nishina Y., Inazawa J., Copeland N. G., Gilbert D. J., Jenkins N. A., Ohsugi M., Tezuka T., Yoshida M. and Semba K. (1999). Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 27, 2503-2510. 10.1093/nar/27.12.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M. P. and Sette C. (2010). Role of RNA-binding proteins in mammalian spermatogenesis. Int. J. Androl. 33, 2-12. 10.1111/j.1365-2605.2009.00959.x [DOI] [PubMed] [Google Scholar]
- Perry J., Palmer S., Gabriel A. and Ashworth A. (2001). A short pseudoautosomal region in laboratory mice. Genome Res. 11, 1826-1832. 10.1101/gr.203001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raverot G., Weiss J., Park S. Y., Hurley L. and Jameson J. L. (2005). Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol. 283, 215-225. 10.1016/j.ydbio.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Reuter M., Berninger P., Chuma S., Shah H., Hosokawa M., Funaya C., Antony C., Sachidanandam R. and Pillai R. S. (2011). Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264-267. 10.1038/nature10672 [DOI] [PubMed] [Google Scholar]
- Hernandez N. (1993). TBP, a universal eukaryotic transcription factor? Genes Dev. 7, 1291-1308. [DOI] [PubMed] [Google Scholar]
- Sarkar A. and Hochedlinger K. (2013). The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15-30. 10.1016/j.stem.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk J. A. C., Dietrich A. J. J., Vink A. C. G., Offenberg H. H., van Aalderen M. and Heyting C. (1998). Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma 107, 540-548. 10.1007/s004120050340 [DOI] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X. et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399-402. 10.1038/nmeth.2857 [DOI] [PubMed] [Google Scholar]
- Soper S. F. C., van der Heijden G. W., Hardiman T. C., Goodheart M., Martin S. L., de Boer P. and Bortvin A. (2008). Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15, 285-297. 10.1016/j.devcel.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Vourekas A., Zheng Q., Alexiou P., Maragkakis M., Kirino Y., Gregory B. D. and Mourelatos Z. (2012). Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 19, 773-781. 10.1038/nsmb.2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. (1999). From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 27, 1409-1420. 10.1093/nar/27.6.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Hu X., Li Z., Wang M., Li S., Wang X., Lin X., Liao S., Zhang Z., Feng X. et al. (2016). Transcription factor RFX2 is a key regulator of mouse spermiogenesis. Sci. Rep. 6, 20435 10.1038/srep20435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Fuente R. D. L., Leu N. A., Baumann C., McLaughlin K. J. and Wang P. J. (2006). Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173, 497-507. 10.1083/jcb.200603063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Liu J.-G., Zhao J., Brundell E., Daneholt B. and Höög C. (2000). The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5, 73-83. 10.1016/S1097-2765(00)80404-9 [DOI] [PubMed] [Google Scholar]
- Zhang P., Kang J.-Y., Gou L.-T., Wang J., Xue Y., Skogerboe G., Dai P., Huang D.-W., Chen R., Fu X.-D. et al. (2015). MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 25, 193-207. 10.1038/cr.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K. and Wang P. J. (2012). Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 8, e1003038 10.1371/journal.pgen.1003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Xiol J., Reuter M., Eckardt S., Leu N. A., McLaughlin K. J., Stark A., Sachidanandam R., Pillai R. S. and Wang P. J. (2010). Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. USA 107, 11841-11846. 10.1073/pnas.1003953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Grubisic I., Zheng K., He Y., Wang P. J., Kaplan T. and Tjian R. (2013). Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc. Natl. Acad. Sci. USA 110, 16886-16891. 10.1073/pnas.1317034110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wang M., Yuan Y., Wang X., Fu R., Wan H., Xie M., Liu M., Guo X., Zheng Y. et al. (2016). Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell 18, 330-340. 10.1016/j.stem.2016.01.017 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Sun Z., Means A. R., Sassone-Corsi P. and Bernstein K. E. (1996). cAMP-response element modulator tau is a positive regulator of testis angiotensin converting enzyme transcription. Proc. Natl. Acad. Sci. USA 93, 12262-12266. 10.1073/pnas.93.22.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. and Kleckner N. (1999). Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603-754. 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.