Abstract
Background
Mitochondrial acyl-CoA dehydrogenase family member 9 (ACAD9) is essential for the assembly of mitochondrial respiratory chain complex I. Disease causing biallelic variants in ACAD9 have been reported in individuals presenting with lactic acidosis and cardiomyopathy.
Results
We describe the genetic, clinical and biochemical findings in a cohort of 70 patients, of whom 29 previously unpublished. We found 34 known and 18 previously unreported variants in ACAD9. No patients harbored biallelic loss of function mutations, indicating that this combination is unlikely to be compatible with life. Causal pathogenic variants were distributed throughout the entire gene, and there was no obvious genotype-phenotype correlation.
Most of the patients presented in the first year of life. For this subgroup the survival was poor (50% not surviving the first 2 years) comparing to patients with a later presentation (more than 90% surviving 10 years). The most common clinical findings were cardiomyopathy (85%), muscular weakness (75%) and exercise intolerance (72%). Interestingly, severe intellectual deficits were only reported in one patient and severe developmental delays in four patients. More than 70% of the patients were able to perform the same activities of daily living when compared to peers.
Conclusions
Our data show that riboflavin treatment improves complex I activity in the majority of patient-derived fibroblasts tested. This effect was also reported for most of the treated patients and is mirrored in the survival data. In the patient group with disease-onset below 1 year of age, we observed a statistically-significant better survival for patients treated with riboflavin.
Electronic supplementary material
The online version of this article (10.1186/s13023-018-0784-8) contains supplementary material, which is available to authorized users.
Keywords: Complex I, Cardiomyopathy, Heart transplantation, Mitochondrial disorder, Lactic acidosis, Treatment, Prognosis, Neonatal, Vitamin, Activities of daily living
Background
Complex I of the mammalian mitochondrial respiratory chain is a large multimeric complex composed of 44 subunits encoded by the mitochondrial and nuclear genome. Beside the structural subunits, at least 19 complex I specific assembly factors are required to obtain fully assembled complex I [1].
One assembly factor is ACAD9. Beside its role in the proper assembly of complex I, ACAD9 exhibits acyl-CoA dehydrogenase (ACAD) activity [2, 3]. ACADs belong to a family of flavoenzymes involved in the ß-oxidation of acyl-CoA and amino acid catabolism. ACAD9 is most homologous (47% amino acid identity, 65% amino acid similarity) to very long-chain acyl-CoA dehydrogenase (VLCAD). Both ACAD9 and VLCAD function as homodimers associated with the inner mitochondrial membrane and catalyze the initial step of the fatty acid oxidation (FAO) cycle [4].
Mutations in ACAD9 have been related to human disease [5–7]. The clinical presentation of ACAD9 deficiency is dominated by cardiomyopathy. Other features are lactic acidosis, myopathy and developmental delay. Age of onset, severity of symptoms and progression are variable. We have shown that residual ACAD9 enzyme activity, and not complex I activity, correlates with the severity of clinical symptoms in ACAD9 deficient patients [3].
In anecdotal reports of patients with a predominance of myopathic features, alleviation of symptoms under riboflavin treatment has been reported [5, 7, 8]. Riboflavin is the precursor of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), which are cofactors for complex I and numerous dehydrogenases involved in FAO. The mode of action is unclear, previous studies suggested that riboflavin increases the mitochondrial FAD concentration thereby supporting FAD binding and consecutively improving ACAD9 folding and stability, thus promoting complex I assembly [9].
Bezafibrate, a peroxisome proliferator-activated receptor (PPAR)-alpha activator that controls the expression of many FAO genes, has been reported as a potential treatment for FAO disorders, with beneficial response in six patients [10]. Recently, this was weakened by a double-blind randomized crossover study of bezafibrate in five individuals with acyl-CoA dehydrogenase very long chain (ACADVL) deficiencies in whom no improvement could be detected [11].
In this study, we provide a comprehensive overview of the clinical, biochemical and genetic spectrum of 70 ACAD9 deficient individuals, of whom 29 are unpublished. We further evaluate the effect of riboflavin in patients and the effect of riboflavin and bezafibrate supplementation in patient-derived fibroblast cell lines.
Methods
Individuals
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Written informed consent was obtained from all individuals or caregivers. The clinical data were collected via an online survey completed by the respective physician. The online survey included 93 questions regarding age at presentation, current age or age at death, signs and symptoms during the fetal and neonatal period, at the beginning and during the course of disease, circumstances of death etc.. A special emphasis lay on the cardiac and neurological phenotype, daily life activities and the use of cardiac medication as well as vitmains and co-factors (e.g. riboflavin).
Kaplan Meier curves were created using the R project for statistical computing (survival package, https://www.r-project.org/).
Molecular genetic investigations
Exome sequencing, panel sequencing and Sanger sequencing was performed as described previously [7, 12–22].
Cell culture
Human fibroblast cells were grown in Dulbecco’s modified Eagle medium-high glucose supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin (Invitrogen) and 200 μM uridine (Sigma-Aldrich) at 37 °C in an atmosphere containing 5% CO2.
Riboflavin and bezafibrate treatment
The fibroblast cell lines were treated with 400 μM bezafibrate, 530 nM riboflavin or vehicle (DMSO) for 72 h as previously described [23, 24]. On the second day the cells were seeded at 20,000 cells/well in 80 μl DMEM in 96 well cell culture microplate and incubated overnight at 37 °C and 5% CO2. On the third day of the experiment the medium was changed to 180 μl unbuffered DMEM and incubated for at least 30 min at 37 °C without CO2.
Oxygen consumption measurement
Oxygen consumption rate (OCR) was measured using an XF96 extracellular flux analyzer (Seahorse Biosciences, North Billeric, MA, USA) as previously described [25, 26] under basal conditions, in the presence of oligomycin (1 μM, ATP synthase inhibitor), FCCP (0.4 μM, mitochondrial oxidative phosphorlyation system (OXPHOS) uncoupler) antimycin A (2.5 μM, complex III inhibitor) and/or rotenone (0.5 μM, complex-I inhibitor). Antimycin and/or rotenone blocked all mitochondrial respiration and were subtracted from all values. Data was normalized to DNA content with CyQuant (Invitrogen).
Western blot and BN-PAGE analysis
Western Blot analyses of different proteins were performed according to standard protocols [5, 27] ACAD9, VLCAD, MCAD, subunits of the respiratory chain complex I (NDUFS1, NDUFA9) and complex II (SDHA) were investigated and ß-actin was used as a loading control (Abcam, Sigma-Aldrich, MitoSciences 1:1000).
Electrophoresis and in-gel quantification of fluorescent-labeled proteins as well as supercomplex assembly are described in Additional file 1 [28, 29].
Results
Individuals
Seventy individuals (41 females) from 50 families were recruited, of which 29 were previously unreported (Additional file 2: Table S1 [30–32] and Additional file 3: Table S2). Individuals were numbered I1-I70, their respective fibroblast cell lines (if available) accordingly F1-F70. In the majority of patients investigated (n = 55) a complex I deficiency was found in sceletal (n = 44) or heart muscle (n = 7) and/or fibroblast cell line (n = 26).
Molecular genetic investigations
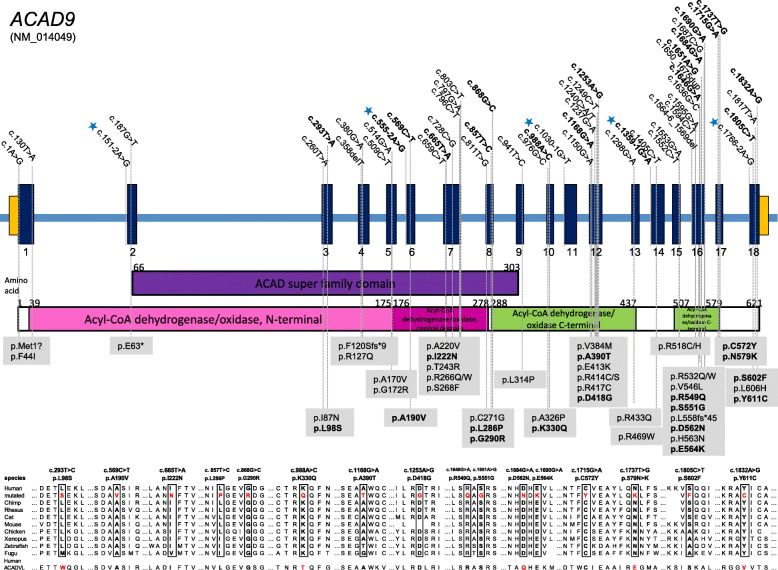
Figure 1 and Additional file 2: Table S1 present the 18 previously unreported variants (in bold) and 34 known variants in ACAD9 found in our cohort. Of these 42 were missense, one frame shift, one nonsense, seven splice site and one start codon mutation. No individual harbored two variants predicted to lead to a loss of protein function.
Fig. 1.
ACAD9 mutation status, gene structure and conservation of affected amino acid residues. Gene structure of ACAD9 with localization of mutations in 70 patients. Blue asterisks indicate splice site mutations. Newly identified mutations are shown in bold. Conservation of amino acid residues affected by missense variants
Based on the prevalence of deleterious ACAD9 alleles in the normal population (GnomAD, www.gnomad.broadinstitute.org, [33]) we estimated that approximately 59 children with ACAD9 deficiency will be born each year in Europe (for calculation see Additional file 3 Table S2).
Clinical spectrum
The data are summarized in Table 1 and Additional file 4: Table S3. Not all data were available for all patients, the denominator indicates the number of patients for which data were available. Currently 37 individuals are alive at a median age of 14 years (range 24 days – 44 years), the median age of patients deceased was 3 months (range 1 day – 44 years). Patients with a presentation in the first year of life (n = 50) show a significantly worse survival when compared to patients presenting later (n = 20, Fig. 2a). One individual (I18) was reported with fetal cardiomegaly, two were reported with fetal rhythm abnormalities, all passed away early, on day 1 (I18), 2 (I42) and 280 (I55), respectively.
Table 1.
Main clinical findings
| Number | n available | Percent | |
|---|---|---|---|
| Prenatal findings | |||
| Cardiomegaly | 1 | 60 | 2 |
| Rhythm abnormalities | 2 | 60 | 3 |
| Decreased child movements | 1 | 60 | 2 |
| Oligohydramnios | 4 | 60 | 7 |
| Intrauterine growth failure | 6 | 60 | 10 |
| Neonatal course | |||
| Lactic acidosis | 21 | 63 | 33 |
| Cardiomyopathy | 15 | 63 | 24 |
| Rhythm abnormalities | 4 | 54 | 7 |
| Respiratory failure necessitating artificial ventilation | 6 | 54 | 11 |
| Severe liver dysfunction/failure | 2 | 54 | 4 |
| Severe renal dysfuntion/failure | 2 | 54 | 4 |
| Most frequent clinical findings | |||
| Cardiomyopathy at presentation/during course | 44/56 | 66/66 | 67/85 |
| Muscular weakness at presentation/during course | 21/37 | 48/49 | 44/75 |
| Exercise intolerance at presentation/during course | 21/34 | 49/47 | 43/72 |
| Neurological findings | |||
| Severe intellectual disability (clinical impression) | 1 | 51 | 2 |
| Mild intellectual disability (clinical impression) | 14 | 48 | 29 |
| Severe developmental delay (clinical impression) | 4 | 52 | 8 |
| Mild developmental delay (clinical impression) | 23 | 51 | 45 |
| Optic atrophy, retinits pigmentosa | 0 | 70 | 0 |
| Neuroradiological findings | |||
| MRI: basal ganglia alterations | 4 | 24 | 17 |
| MRI: leukoencephalopathy | 5 | 21 | 24 |
| MRI: global brain atrophy | 2 | 20 | 10 |
| MRI: isolated cerebellar atrophy | 1 | 20 | 5 |
| MRS: lactate peak (any location) | 2 | 16 | 13 |
| Activities of daily living | |||
| Age adequate behaviour | 27 | 39 | 69 |
| Attending/finished regular school | 26 | 41 | 63 |
| Able to sit independently | 34 | 42 | 81 |
| Able to walk independently | 33 | 41 | 80 |
| Able to eat and drink independently | 33 | 41 | 80 |
| Able to perform personal hygiene independently | 30 | 41 | 73 |
| Able to communicate with words/sentences | 34/31 | 43/42 | 79/74 |
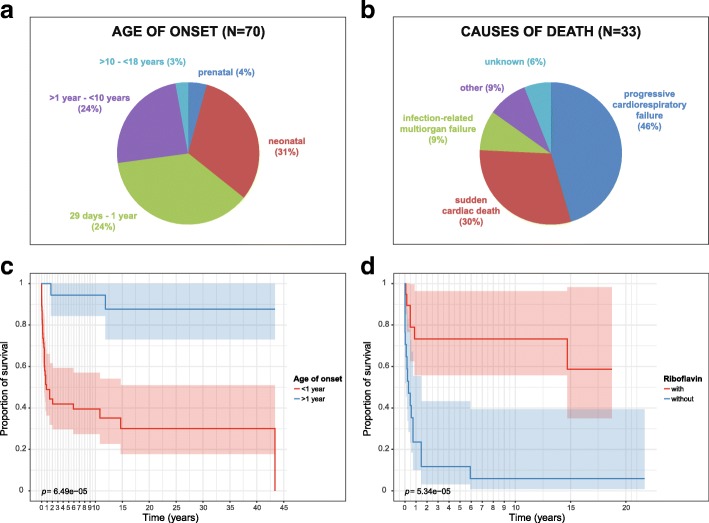
Fig. 2.
Age of onset, causes of death, survival and effect of riboflavin on survival of ACAD9 patients. a Age of onset of symptoms, (b) Causes of death, (c) Kaplan-Maier survival rates. In red, patients with a disease presentation in the first year of life. In blue, patients with a later presentation (p = 6.49e-05). b In red, patients with a disease presentation in the first year of life and treated with riboflavin. In blue, patients of the same age category but untreated with riboflavin (p = 5.34e-05, confidence 95%)
Cardiomyopathy and treatment
I20 presented with hypertrophic cardiomyopathy in the first year of life. Due to rapid deterioration, she received heart transplantation at 2 years. She died of cardiac failure 4 years later. I21 presented with hypertrophic cardiomyopathy at 18 months and subsequently developed neurological symptoms (ataxia and epilepsy), which were non-progressive and mild. She was successfully heart transplanted at the age of 9 years and is currently 15 years old. I22, currently 35 years old, presented with a progressive biventricular hypertrophic cardiomyopathy in childhood and was transplanted at the age of 18 years. After a follow up of six and 17 years, respectively, their cardiac function remained satisfactory. I30 showed tachycardia in the first days of life birth and signs of heart failure at 1 month. Despite undergoing cardioverter-defibrillator implantation and subsequent heart transplantation, he died at 3 months of age.
Regarding drug treatment, a positive effect on heart failure was reported for beta-blocking agents (14/44 = 32%), ACE inhibitors (6/40 = 15%), calcium-channel blockers (1/37 = 3%) and diuretics (3/39 = 8%). No patient received digitoxin or digoxin. A worsening effect was only reported for one patient on beta blockers.
Riboflavin and other oral vitamin treatment
Of the entire cohort of 67 patients, 20 patients were reported as not treated; data about treatment and/or effect were unavailable for 15 patients. Data on the general clinical effect of riboflavin as reported by the responsible physician were available for 31 patients. For 20 patients (20/31 = 65%) physicians reported a beneficial effect, for 11 (35%) no effect. No clinical deterioration or side effects were reported. Detailed data on onset of riboflavin treatment, dosage, duration and clinical effect were only available for a minority of patients and have not been investigated.
To analyze the effect of riboflavin treatment, we focused on the patients presenting during the first year of life as these was the biggest subgroup and the group with the shortest survival suggesting the most severe course. For 39 of these 50 patients, data on riboflavin treatment were available (n = 17 untreated, n = 22 treated). Figure 2b shows the Kaplan-Meier curve for both groups of patients and indicates a significantly better survival rate for patients with oral riboflavin treatment (deceased n = 7/22) in contrast to untreated patients (deceased n = 16/17).
Regarding other food supplements, several patients were reported as taking coenzyme Q10, biotin and L-carnitine with anecdotal positive effects.
Cell culture experiments
Effect of different ACAD9 mutations on ACAD9 protein level and respiratory chain complex I activity
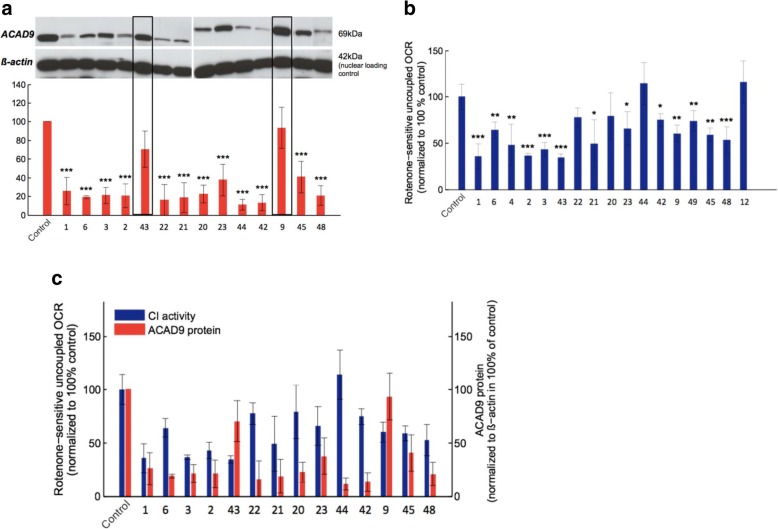
ACAD9 levels were significantly reduced in all but two of 14 examined patient fibroblast cell lines; both exceptional cell lines (F9, F43) carried a homozygous p.(Arg518His) variant and showed normal ACAD9 levels (Fig. 3a).
Fig. 3.
Measurements of ACAD9 protein level and complex I activity in patient derived fibroblasts. a Western blot and quantification of ACAD9 protein levels in patient derived fibroblasts and control. b Complex I activity in patient derived fibroblasts and control. c Comparison between remaining ACAD9 protein (red) and Complex I activity (blue). Data expressed as average of three independent western blots and average of > 10 technical replicates (oxygen consumption rate ± SD)
Complex I-dependent respiration was found to be significantly decreased in 13 of 17 evaluated patient cell lines. The cell lines F44 (homozygous p.[Leu98Ser]) and F12 (homozygous p.[Arg532Trp]) showed no complex I deficiency; F22 (p.[splice];[Arg433Gln]) and F20 (p.[Phe120Serfs*9];[Arg532Trp] showed only mildly reduced levels (Fig. 3b).
There was no correlation between complex I activity and residual ACAD9 protein levels. Interestingly, nearly normal complex I activity was recorded in the cell line of F42 (p.[Glu564Lys];[Tyr611Cys]) despite very low steady-state ACAD9 protein levels, indicating that the remaining ACAD9 chaperone activity might be high enough to correct assembly of complex I (Fig. 3c).
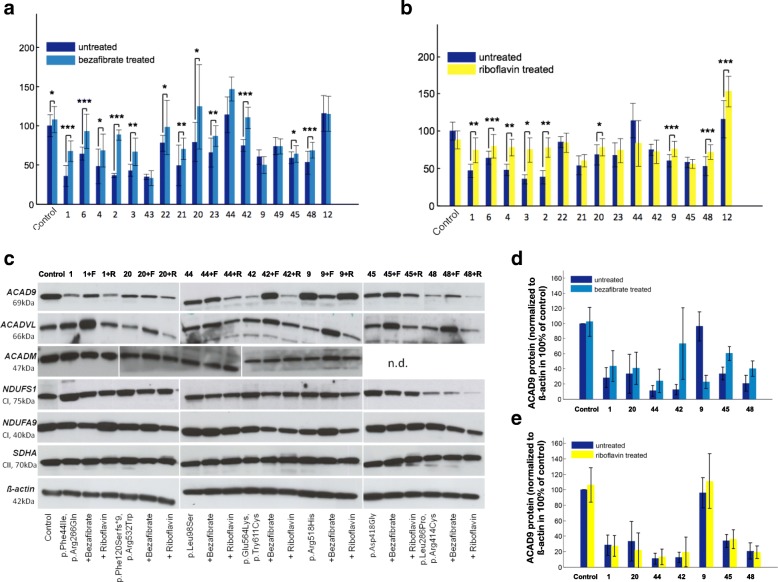
Respiratory chain complex I activity after bezafibrate and riboflavin supplementation
After 72 h of bezafibrate treatment, the complex I activity increased in the control and in all but three patients cell lines. The increase was significant in 12 out of 17 patient cell lines. Five patient cell lines reached almost normal levels (Fig. 4a). The already normal complex I activity of two cell lines (F44 and F12) remained unchanged.
Fig. 4.
Effect of bezafibrate and riboflavin supplementation on respiratory chain activities in fibroblast cell lines. Maximal oxygen consumption rate (OCR) was measured in pmol/(s*Mill) of ACAD9 patient and control fibroblasts with and without (a) bezafibrate (400 μM for 72 h) and (b) riboflavin (530 nM for 72 h) treatment. Data are expressed as the average of > 10 technical replicates and normalized to control. ± SD. ***P < 0.001, **P < 0.01, *P < 0.05. (c) Whole cell lysate of control and different ACAD9 deficient fibroblasts +/− bezafibrate/riboflavin visualized with antibodies against ACAD9, ACADVL, ACADM, SDHA, ß-actin (loading control), NDUFS1 and NDUFA9 (d, e) Quantification of ACAD9 and ACADVL protein levels
Riboflavin supplementation led to a significant improvement of complex I activity in nine out of 15 patient cell lines and had no effect in the remaining six cell lines. The increase ranged from 14 to 109% (Fig. 4b).
Supercomplex formation after bezafibrate treatment
All investigated cell lines had a clear reduction of assembled supercomplexes, however, there was no correlation between the amount of ACAD9 protein and the extent of supercomplex formation, (e.g. F23 with almost normal amount of ACAD9 presented a complete loss of supercomplexes whereas F44 with nearly absent ACAD9 protein presented a high amount of assembled supercomplexes). This indicates that small quantities of productive ACAD9 can fulfill assembly function. An increase in the assembly of supercomplexes was found in four out of five cell lines treated with bezafibrate (Additional file 5 Figure S1).
Findings in cell culture versus clinical effect in patients supplemented with riboflavin
The same mutation as in I12/F12 has previously been reported in I6/F6 (P2 in [7]). I6 was reported to benefit from oral riboflavin. Both cell lines, F6 and F12, consequently showed improving complex I activity under riboflavin treatment (Fig. 3b). Data for treatment of paired cells and patients were only available for eight patients. In seven pairs the effect was concordant in cells and patients. Six pairs (I/F2, 3, 4, 6, 9, 12) showed positive effects both in cell culture and clinical, one pair (I/F44) did not show any response. I48 did clinically not respond to treatment, whereas his cells did.
Discussion
Complex I deficiency is the most common biochemical signature of mitochondrial disorders. Given the number of ACAD9 deficient patients described here for a disorder genetically defined only in 2010, and based on the frequency of deleterious alleles described to date, ACAD9 is likely to be one of the more common causes of mitochondrial respiratory chain deficiency, with a conservative estimate of 59 new patients born every year in Europe, and 689 world-wide (Additional file 3 Table S2).
The mutations of the 70 patients from 50 families with ACAD9 deficiency were located across the coding sequence of the gene, with no founder mutations identified. However, no individual harbored two clear loss of function alleles, suggesting that a complete loss of ACAD9 function might be incompatible with life. This is also supported by the fact that the homozygous knock out mouse was found to be embryonic lethal (Schiff, Vockley, personal communication). No genotype-phenotype correlation for mutations could be identified based on specific regions of the gene or functional domains of the protein.
The vast majority of patients presented with hypertrophic cardiomyopathy, lactic acidosis, muscle weakness, and exercise intolerance. However, patients without cardiomyopathy also were identified our study. Although both ACAD9 and VLCAD deficiency can present with cardiomyopathy, the clinical phenotype is otherwise distinct, with hypoglycemia, rhabdomyolysis and liver failure, typically seen in VLCAD. These symptoms were infrequently seen in our ACAD9 deficient cohort.
Our data suggest, that there are two subgroups of ACAD9 deficient patients. Patients who presented in the first year of life often died early and, if surviving, did more poorly than those who presented later. In contrast to many other mitochondrial disorders, severe intellectual disability and developmental delay, as well as other neurological features, were seen in only a minority of (surviving) patients. Indeed, all patients with severe developmental delay (n = 4) or intellectual delay (n = 1) had early disease onset. Furthermore, most patients currently alive were able to perform routine activities of daily living.
This observation, is not only very important for providing anticipatory guidance, but might also influence a decision regarding heart transplantation. Four patients of our cohort underwent heart transplantation. Unfortunately, the two patients who presented within the first year died despite all efforts. In contrast, the two patients presenting after the age of 1 year developed normally and are currently aged 15 and 35 years, respectively. Additional longitudinal studies are warranted to better identify which patients with ACAD9 deficiency are appropriate heart transplant candidates.
Supplementation with riboflavin showed improvement in complex I activity in the vast majority of patient fibroblasts, and most patients similarly were reported to have clinical benefit with treatment. Most notably, patients presenting within the first year of life show a significant better survival when treated with riboflavin. One limitation of this observation could be that most of the deaths occurred at the end of the first year of life. This might indicate that our analysis is prone to survivor treatment selection bias. Detailed data about the starting point of riboflavin treatment, the dosage etc. in more patients are needed.
This observation supports anecdotal reports in the literature. In our cohort, families 1 and 33 are particularly instructive. In both families the first child (I1, I45) died within the first 2 years of life without riboflavin supplementation, whereas the younger affected siblings (I2, I45 and I46), in whom supplementation was begun immediately upon diagnosis, are currently still alive (aged 10, 1.5 and 11 years, retrospectively). Cases I5 and I6 were first reported with riboflavin responsive complex I deficiency before their molecular defect was known [7, 34]. Paired data on fibroblasts and patient riboflavin treatment were available for eight patients, six of which showed parallel beneficial effects and one no effect. Further cellular studies are necessary to define the mode of action of riboflavin in ACAD9 deficiency.
The PPAR promotor activator bezafibrate has been reported to be of benefit in other FAODs. In all cell lines examined in this study, bezafibrate improved the formation of respiratory chain super complexes, likely explaining the improved respiration of the patient cell lines as measured by whole cell oximetry. While only a limited number of cell lines were tested, these results suggest a potential role for bezafibrate or other PPAR activators in the treatment of ACAD9. However, similar effects for bezafibrate have been reported in cell models of other fatty acid oxidation defects, but were not proven in humans.
Our retrospective data provides additional description of the clinical and genetic spectrum of ACAD9 deficiency, and provides valuable insight for the development of future clinical trials of riboflavin, bezafibrate, or other therapies. While the current study was not designed to be a clinical trial, the anecdotal improvement of many ACAD9 deficient patients to riboflavin justifies a trial of riboflavin (20 mg/kg/day, maximum 200 mg/day) in every patient with this diagnosis. Given the high frequency of ACAD9 deficiency, we propose that it would be reasonable to consider riboflavin administration for phenotypically-consistent patients whilst their genetic investigations are underway [35]. This also underlines that in patients with suspicion of a mitochondrial disorder next generation sequencing techniques should be initiated promptly, in selected cases accompanied by studies in affected tissues. For these patients, early diagnosis and therapeutic intervention could be the difference between life and death.
Conclusions
ACAD9 typically presents with cardiomyopathy, exercise intolerance and muscular weakness and the clinical course might respond to riboflavin.
Additional files
Additional data. (DOCX 38 kb)
Table S1. Compound heterozygous and homozygous ACAD9 variants identified in 67 patients present in this study (DOCX 75 kb)
Table S2. Calculation of European incidence of ACAD9 deficiency (DOCX 36 kb)
Table S3. Clinical characteristics of the 67 patients present in this study (DOCX 72 kb)
Figure S1. Representative picture of Complex I assembly in fibroblasts of individual 1 (A, upper panels) Two-dimensional BN/SDS-PAGE separation and quantification of fluorescent-labelled mitochondrial complexes from 10 mg patient (left) and control fibroblasts (right) are shown. (A, lower panels) show silver stained 2 D gels. (B) Quantified Supercomplexes in 2D gels from control and patient fibroblast with and without bezafibrate treatment for 72 h. (C) Panoramaplots of 2D gels with assignment of signals used for quantification of complexes. Assignment of complexes: O, OGDC, oxoglutarate dehydrogenase complex; V, complex V or ATP synthase; III, complex III or cytochrome c reductase; IV, complex IV or cytochrome c oxidase; S, supercomplexes composed of respiratory chain complexes I, III, and IV. 2-D gels were scanned side by side for direct comparison and are shown as pseudocolors. (pdf). (PDF 1109 kb)
Acknowledgements
None.
Funding
This study was supported by the German Bundesministerium für Bildung und Forschung (BMBF) and Horizon2020 through the E-Rare project GENOMIT (01GM1603 and 01GM1207 for HP and FWF I 2741-B26 for JAM); the EU FP7 Mitochondrial European Educational Training Project (317433 to HP and JAM); the EU Horizon2020 Collaborative Research Project SOUND (633974 to HP); T.M., P.F., M.S., I.W. and H.P: were supported by a German Federal Ministry of Education and Research (BMBF, Bonn, Germany) grant to the German Network for Mitochondrial Disorders (mitoNET, 01GM1113A/B/C/D). J.A.M., S.B.W., W.S. were supported by the Vereinigung zur Förderung Pädiatrischer Forschung und Fortbildung Salzburg. I.W. was supported by the Cluster of Excellence “Macromolecular Complexes” at the Goethe University Frankfurt (EXC 115). JV was supported in part by a United States Public Health Grant NIH R01 DK78775. RWT and RM are supported by the Wellcome Centre for Mitochondrial Research (203105/Z/16/Z), the Medical Research Council (MRC) Centre for Translational Research in Neuromuscular Disease, Mitochondrial Disease Patient Cohort (UK) (G0800674), the Lily Foundation and the UK NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children. CLA was supported by a National Institute for Health Research (NIHR) doctoral fellowship (NIHR-HCS-D12–03-04). FF was supported by the national key research and developmental program of China (#2016YFC1306203).
Availability of data and materials
Please contact authors for data requests.
Abbreviations
- ACAD
Acyl-CoA dehydrogenase
- F
Patient derived fibroblast cell line
- FAO
Fatty acid oxidation
- I
Individual
- OCR
Oxygen consumption rate
- OXPHOS
Oxidative phosphorlyation system
- VLCAD
Very long-chain acyl-CoA dehydrogenase
Authors’ contributions
HP, SBW, BMR, EM conceived and designed the study, BMR. performed the experiments, all authors analysed and interpreted the clinical and diagnostic laboratory data, BMR, EM, HP, SBW. drafted the article, all authors critically revised it for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000.
Consent for publication
Written informed consent was obtained from all individuals or caregivers.
Competing interests
All authors declare no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13023-018-0784-8) contains supplementary material, which is available to authorized users.
Birgit M. Repp and Elisa Mastantuono are equal first authors.
Charlotte L. Alston, Manuel Schiff and Tobias B. Haack are equal co-authors.
Holger Prokisch and Saskia Wortmann jointly supervised this work.
Contributor Information
Birgit M. Repp, Email: birgit.repp@helmholtz-muenchen.de
Elisa Mastantuono, Email: elisa.mastantuono@helmholtz-muenchen.de.
Charlotte L. Alston, Email: charlotte.alston@ncl.ac.uk
Manuel Schiff, Email: manuel.schiff@aphp.fr.
Tobias B. Haack, Email: Tobias.Haack@med.uni-tuebingen.de
Agnes Rötig, Email: agnes.rotig@inserm.fr.
Anna Ardissone, Email: Anna.Ardissone@istituto-besta.it.
Anne Lombès, Email: anne.lombes@inserm.fr.
Claudia B. Catarino, Email: Claudia.Catarino@med.uni-muenchen.de
Daria Diodato, Email: daria.diodato@opbg.net.
Gudrun Schottmann, Email: gudrun.schottmann@charite.de.
Joanna Poulton, Email: joanna.poulton@obs-gyn.ox.ac.uk.
Alberto Burlina, Email: alberto.burlina@unipd.it.
An Jonckheere, Email: An.Jonckheere@uza.be.
Arnold Munnich, Email: arnold.munnich@inserm.fr.
Boris Rolinski, Email: boris@rolinski.net.
Daniele Ghezzi, Email: Daniele.Ghezzi@istituto-besta.it.
Dariusz Rokicki, Email: d.rokicki@ipczd.pl.
Diana Wellesley, Email: dgw@soton.ac.uk.
Diego Martinelli, Email: diego.martinelli@opbg.net.
Ding Wenhong, Email: xiaolei263@sina.com.
Eleonora Lamantea, Email: Eleonora.Lamantea@istituto-besta.it.
Elsebet Ostergaard, Email: elsebet.ostergaard@dadlnet.dk.
Ewa Pronicka, Email: e.pronicka@ipczd.pl.
Germaine Pierre, Email: germaine.pierre@uhbristol.nhs.uk.
Hubert J. M. Smeets, Email: Bert.Smeets@maastrichtuniversity.nl
Ilka Wittig, Email: wittig@med.uni-frankfurt.de.
Ingrid Scurr, Email: Ingrid.Scurr@uhbristol.nhs.uk.
Irenaeus F. M. de Coo, Email: mdecoo@planet.nl
Isabella Moroni, Email: isabella.moroni@istituto-besta.it.
Joél Smet, Email: Joel.Smet@ugent.be.
Johannes A. Mayr, Email: h.mayr@salk.at
Lifang Dai, Email: dailifang1111@126.com.
Linda de Meirleir, Email: Linda.DeMeirleir@uzbrussel.be.
Markus Schuelke, Email: markus.schuelke@charite.de.
Massimo Zeviani, Email: massimo.zeviani@mrc-mbu.cam.ac.uk.
Raphael J. Morscher, Email: morscher@protonmail.com
Robert McFarland, Email: robert.mcfarland@ncl.ac.uk.
Sara Seneca, Email: Sara.Seneca@uzbrussel.be.
Thomas Klopstock, Email: Thomas.Klopstock@med.uni-muenchen.de.
Thomas Meitinger, Email: meitinger@helmholtz-muenchen.de.
Thomas Wieland, Email: thomas.wieland@helmholtz-muenchen.de.
Tim M. Strom, Email: strom@helmholtz-muenchen.de
Ulrike Herberg, Email: ulrike.herberg@ukb.uni-bonn.de.
Uwe Ahting, Email: uwe.ahting@humangenetik.med.tum.de.
Wolfgang Sperl, Email: w.sperl@salk.at.
Marie-Cecile Nassogne, Email: marie-cecile.nassogne@uclouvain.be.
Han Ling, Email: hanl6610@sina.com.
Fang Fang, Email: 13910150389@163.com.
Peter Freisinger, Email: freisinger_p@klin-rt.de.
Rudy Van Coster, Email: rudy.vancoster@ugent.be.
Valentina Strecker, Email: VSTina@web.de.
Robert W. Taylor, Email: Robert.Taylor@newcastle.ac.uk
Johannes Häberle, Email: johannes.haeberle@kispi.uzh.ch.
Jerry Vockley, Email: vockleyg@upmc.edu.
Holger Prokisch, Email: prokisch@helmholtz-muenchen.de.
Saskia Wortmann, Phone: +49-(0) 89 4140 9889, Email: wortmann-hagemann@helmholtz-muenchen.de.
References
- 1.Sánchez-Caballero L, Guerrero-Castillo S, Nijtmans L. Unraveling the complexity of mitochondrial complex I assembly: a dynamic process. BBA - Bioenerg. 1857;2016:980–990. doi: 10.1016/j.bbabio.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 2.Ensenauer R, He M, Willard J-M, Goetzman ES, Corydon TJ, Vandahl BB, et al. Human acyl-CoA Dehydrogenase-9 plays a novel role in the mitochondrial β-oxidation of unsaturated fatty acids. J Biol Chem. 2005;280:32309–32316. doi: 10.1074/jbc.M504460200. [DOI] [PubMed] [Google Scholar]
- 3.Schiff M, Haberberger B, Xia C, Mohsen A-W, Goetzman ES, Wang Y, et al. Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum Mol Genet. 2015;24:3238–3247. doi: 10.1093/hmg/ddv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, et al. Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem Biophys Res Commun. 2002;297:1033–1042. doi: 10.1016/S0006-291X(02)02336-7. [DOI] [PubMed] [Google Scholar]
- 5.Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 6.Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Gerards M, van den Bosch BJC, Danhauser K, Serre V, van Weeghel M, Wanders RJA, et al. Riboflavin-responsive oxidative phosphorylation complex I deficiency caused by defective ACAD9: new function for an old gene. Brain. 2011;134:210–219. doi: 10.1093/brain/awq273. [DOI] [PubMed] [Google Scholar]
- 8.Aintablian HK, Narayanan V, Belnap N, Ramsey K, Grebe TA. An atypical presentation of ACAD9 deficiency: diagnosis by whole exome sequencing broadens the phenotypic spectrum and alters treatment approach. Mol Genet Metab rep. 2017;10:38–44. doi: 10.1016/j.ymgmr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas TG, Henriques BJ, Rodrigues JV, Bross P, Gregersen N, Gomes CM. Cofactors and metabolites as potential stabilizers of mitochondrial acyl-CoA dehydrogenases. Biochim Biophys Acta - Mol Basis Dis. 1812;2011:1658–1663. doi: 10.1016/j.bbadis.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Bonnefont JP, Bastin J, Laforêt P, Aubey F, Mogenet A, Romano S, et al. Long-term follow-up of Bezafibrate treatment in patients with the Myopathic form of carnitine Palmitoyltransferase 2 deficiency. Clin Pharmacol Ther. 2010;88:101–108. doi: 10.1038/clpt.2010.55. [DOI] [PubMed] [Google Scholar]
- 11.Orngreen MC, Madsen KL, Preisler N, Andersen G, Vissing J, Laforet P. Bezafibrate in skeletal muscle fatty acid oxidation disorders: a randomized clinical trial. Neurology. 2014;82:607–613. doi: 10.1212/WNL.0000000000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haack TB, Haberberger B, Frisch E-M, Wieland T, Iuso A, Gorza M, et al. Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J Med Genet. 2012;49:277–283. doi: 10.1136/jmedgenet-2012-100846. [DOI] [PubMed] [Google Scholar]
- 13.Garone C, Donati MA, Sacchini M, Garcia-Diaz B, Bruno C, Calvo S, et al. Mitochondrial encephalomyopathy due to a novel mutation in ACAD9. JAMA Neurol. 2013;70:1177–1179. doi: 10.1001/jamaneurol.2013.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouws J, te Brinke H, Nijtmans LG, Houten SM. ACAD9, a complex I assembly factor with a moonlighting function in fatty acid oxidation deficiencies. Hum Mol Genet. 2014;23:1311–1319. doi: 10.1093/hmg/ddt521. [DOI] [PubMed] [Google Scholar]
- 15.Lagoutte-Renosi J, Ségalas-Milazzo I, Crahes M, Renosi F, Menu-Bouaouiche L, Torre S, et al. Lethal neonatal progression of fetal cardiomegaly associated to ACAD9 deficiency. JIMD Rep. 2015;28:1. doi: 10.1007/8904_2015_499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wortmann SB, Koolen DA, Smeitink JA, van den Heuvel L, Rodenburg RJ. Whole exome sequencing of suspected mitochondrial patients in clinical practice. J Inherit Metab Dis. 2015;38:437–443. doi: 10.1007/s10545-015-9823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alston CL, Howard C, Oláhová M, Hardy SA, He L, Murray PG, et al. A recurrent mitochondrial p.Trp22Arg NDUFB3 variant causes a distinctive facial appearance, short stature and a mild biochemical and clinical phenotype. J Med Genet. 2016;53:634–641. doi: 10.1136/jmedgenet-2015-103576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collet M, Assouline Z, Bonnet D, Rio M, Iserin F, Sidi D, et al. High incidence and variable clinical outcome of cardiac hypertrophy due to ACAD9 mutations in childhood. Eur J Hum Genet. 2016;24:1112–1116. doi: 10.1038/ejhg.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohda M, Tokuzawa Y, Kishita Y, Nyuzuki H, Moriyama Y, Mizuno Y, et al. A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 2016;12:e1005679. doi: 10.1371/journal.pgen.1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legati A, Reyes A, Nasca A, Invernizzi F, Lamantea E, Tiranti V, et al. New genes and pathomechanisms in mitochondrial disorders unraveled by NGS technologies. Biochim Biophys Acta Bioenerg. 1857;2016:1326–1335. doi: 10.1016/j.bbabio.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Leslie N, Wang X, Peng Y, Valencia CA, Khuchua Z, Hata J, et al. Neonatal multiorgan failure due to ACAD9 mutation and complex I deficiency with mitochondrial hyperplasia in liver, cardiac myocytes, skeletal muscle, and renal tubules. Hum Pathol. 2016;49:27–32. doi: 10.1016/j.humpath.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 22.Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucińska-Więckowska A, et al. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric Centre. J Transl Med. 2016;14:174. doi: 10.1186/s12967-016-0930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastin J, Aubey F, Rötig A, Munnich A, Djouadi F. Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J Clin Endocrinol Metab. 2008;93:1433–1441. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 24.Cornelius N, Byron C, Hargreaves I, Fernandez Guerra P, Furdek AK, Land J, et al. Secondary coenzyme Q10 deficiency and oxidative stress in cultured fibroblasts from patients with riboflavin responsive multiple acyl-CoA dehydrogenation deficiency. Hum Mol Genet. 2013;22:3819–3827. doi: 10.1093/hmg/ddt232. [DOI] [PubMed] [Google Scholar]
- 25.Invernizzi F, D’Amato I, Jensen PB, Ravaglia S, Zeviani M, Tiranti V. Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion. 2012;12:328–335. doi: 10.1016/j.mito.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer LS, Prokisch H. Identification of disease-causing mutations by functional complementation of patient-derived fibroblast cell lines. New York: Humana Press; 2017. pp. 391–406. [DOI] [PubMed] [Google Scholar]
- 27.Haack TB, Gorza M, Danhauser K, Mayr JA, Haberberger B, Wieland T, et al. Phenotypic spectrum of eleven patients and five novel MTFMT mutations identified by exome sequencing and candidate gene screening. Mol Genet Metab. 2014;111:342–352. doi: 10.1016/j.ymgme.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Schägger H. Tricine–SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 29.Wittig I, Braun H-P, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 30.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouws J, Wibrand F, Van Den Brand M, Hanka V, Duno M, et al. A patient with complex I deficiency caused by a novel ACAD9 mutation not responding to riboflavin treatment. JIMD Rep. 2014;12:37–45. doi: 10.1007/8904_2013_242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fragaki K, Chaussenot A, Boutron A, Bannwarth S, Cochaud C, Richelme C, et al. Severe defect in mitochondrial complex I assembly with mitochondrial DNA deletions in ACAD9 -deficient mild myopathy. Muscle Nerve. 2017;55:919–922. doi: 10.1002/mus.25262. [DOI] [PubMed] [Google Scholar]
- 33.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholte HR, Busch HFM, Bakker HD, Bogaard JM, Luyt-Houwen IEM, Kuyt LP. Riboflavin-responsive complex I deficiency. Biochim Biophys Acta. 1995;1271:75–83. doi: 10.1016/0925-4439(95)00013-T. [DOI] [PubMed] [Google Scholar]
- 35.Distelmaier F, Haack TB, Wortmann SB, Mayr JA, Prokisch H. Treatable mitochondrial diseases: cofactor metabolism and beyond. Brain. 2017;140:e11. doi: 10.1093/brain/aww303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional data. (DOCX 38 kb)
Table S1. Compound heterozygous and homozygous ACAD9 variants identified in 67 patients present in this study (DOCX 75 kb)
Table S2. Calculation of European incidence of ACAD9 deficiency (DOCX 36 kb)
Table S3. Clinical characteristics of the 67 patients present in this study (DOCX 72 kb)
Figure S1. Representative picture of Complex I assembly in fibroblasts of individual 1 (A, upper panels) Two-dimensional BN/SDS-PAGE separation and quantification of fluorescent-labelled mitochondrial complexes from 10 mg patient (left) and control fibroblasts (right) are shown. (A, lower panels) show silver stained 2 D gels. (B) Quantified Supercomplexes in 2D gels from control and patient fibroblast with and without bezafibrate treatment for 72 h. (C) Panoramaplots of 2D gels with assignment of signals used for quantification of complexes. Assignment of complexes: O, OGDC, oxoglutarate dehydrogenase complex; V, complex V or ATP synthase; III, complex III or cytochrome c reductase; IV, complex IV or cytochrome c oxidase; S, supercomplexes composed of respiratory chain complexes I, III, and IV. 2-D gels were scanned side by side for direct comparison and are shown as pseudocolors. (pdf). (PDF 1109 kb)
Data Availability Statement
Please contact authors for data requests.