Abstract
Influenza A virus subtypes are determined based on envelope proteins encoded by the hemagglutinin (HA) gene and the neuraminidase (NA) gene, which are involved in attachment to the host, pathogenicity, and progeny production. Here, we evaluated such differences through co-evolution analysis between the HA and NA genes based on subtype and host. Event-based cophylogeny analysis revealed that humans had higher cospeciation values than avian. In particular, the yearly ML phylogenetic trees for the H1N1 and H3N2 subtypes in humans displayed similar topologies between the two genes in humans. Substitution analysis was verifying the strong positive correlation between the two genes in the H1N1 and H3N2 subtypes in humans compared with those in avian and swine. These results provided a proof of principle for the further development of vaccines according to hosts and subtypes against Influenza A virus.
Keywords: Co-evolution, influenza A virus, hemagglutinin, neuraminidase
Introduction
Influenza viruses are negative-sense RNA viruses that belong to the Orthomyxoviridae family. Influenza viruses A, B, C, and D, as well as Isavirus, Quaranjavirus, and Thogotovirus, are members of this family.1 Of these, influenza A virus has long been the cause of pandemic infections in humans, from H1N1 Spanish influenza in 1918 and H2N2 Asian influenza in 1958 to H3N2 Hong Kong influenza in 1968 and H1N1 pandemic in 2009.2 Furthermore, different subtypes of influenza A virus infect various hosts, including avian, humans, swine, dogs, horses, and bats.3 The viral envelope proteins of influenza A virus, encoded by the hemagglutinin (HA) and neuraminidase (NA) genes, play key roles in attachment of the viruses to host receptors involved in pathogenicity and release of progeny virus in specific hosts. These genes are also determining the subtypes of the viruses. A total of 198 influenza A virus subtypes will be produced with 18 HA and 11 NA genes, of which only 7% have been reported to infect humans.4 The HA gene, the major surface glycoprotein of influenza A virus, infects the host by binding to the host receptor protein, whereas the NA gene, a receptor destroying enzyme, is involved in release of the virus from the host cell.5 These two envelope proteins are the targets of antiviral drugs and cause the hosts antibodies to induce immune responses.
Different subtypes of influenza A virus infect different hosts. For example, subtypes with the HA1, HA2, HA3, NA1, and NA2 envelope proteins generally infect humans rather than other species.6 Although these proteins have identical functions in human and animal influenza viruses, they have some genetic differences. For example, avian influenza HA protein binds the alpha 2-3 sialic acid receptor, human influenza HA binds the 2-6 sialic acid receptor, and swine influenza HA can bind both receptors.7 The H5N1 subtype has a Glu in avian and a Lys in human influenza A virus at position 627 of the amino acid sequence of the PB2 protein, which is a subunit of the RNA polymerase complex.6 In addition, this subtype shows co-evolution patterns at the N-glycosylation site in both glycoproteins.8 In H5N1 virus extracted from human specimens, the HA gene was found to exhibit less than 1% diversity when compared to the gene in the avian virus during the same period, and the viral envelope genes were similar across hosts.9
The HA and NA genes of influenza A virus have shown intergenic interactions and postreassortment substitutions of charged amino acids in the HA proteins of different subtypes.10 In this study, we therefore selected 10 influenza A virus subtypes of avian, human, and swine influenza viruses to perform a co-evolution analysis using the HA and NA genes. Co-evolutionary traits of the two envelope proteins were identified, and sequence substitution analyses were performed to find correlations between subtypes and hosts.
Materials and Methods
Preparation of HA and NA sequence data for influenza A virus
Nucleotide sequence data were obtained from the NCBI Influenza Virus Resource (https://www.ncbi.nlm.nih.gov/genome/viruses/variation/flu/). We examined the HA and NA gene types of 198 subtypes of influenza A virus for co-evolution analysis (n ⩾ 4). Next, we selected full-length HA and NA gene sequences from each subtype of human, avian, and swine influenza virus isolated in the same year. Identical sequences were collapsed and the most recent representative sequence selected. By this process, the following 10 subtypes were selected: H1N1, H1N2, H2N2, H3N2, H5N1, H7N2, H7N7, H7N9, H7N10, and H9N2. The accession numbers for each nucleotide sequence and the nucleotide sequence data for the HA and NA genes were saved as “>accession_year.” The collected nucleotide data for each subtype are described in Supplemental Tables 1-10.
Co-evolution analysis between HA and NA genes
First, for multiple sequence alignment (MSA), alignment output files for the HA and NA genes were generated in the nexus format for each of the 10 subtypes according to host using the Clustal Ω program.11 Based on the MSA results, yearly co-evolution analysis was performed on the genes. A co-evolution analysis generally examines the evolutionary relationship between two different species in a host–parasite relationship. However, this study matched the HA and NA genes of influenza A virus by year to examine their associations over time using Jane 4, an event-based program.12 Costs for individual events were set to: cospeciation = 0, duplication = 1, switches = 2, losses = 1, failures to diverge = 1. HA switches/NA and cospeciation/NA were calculated using the collected data to measure the associations between the HA and NA genes for each subtype and host. Furthermore, to visualize the phylogenetic relationship between the genes, TreeMap 3.0 was used to pair data extracted during a given year for each subtype and host.13 Tanglegrams were created by matching each of the 10 subtypes with the hosts.
Substitution correlations between HA and NA genes
Two types of sequence analyses were performed to measure the similarities in phylogenetic topology and correlations according to substitutions in the nucleotide sequences. First, correlation analysis was conducted using the yearly pairwise alignment score values between the HA and NA genes for each subtype and host. MEGA6 was used to perform ClustalW alignment with a default gap opening penalty of 15, a gap extension penalty of 6.66, a DNA weight matrix of IUB, and a transition weight of 0.5. Pairwise distance was set to pairs of taxa and maximum composite likelihood was followed. Because the HA and NA gene sequences for the same year were extracted, pairwise alignment scores with the same conditions can be obtained. Based on the pairwise distances, the correlations between the HA and NA genes for each subtype and host were analyzed. SPSS version 24.0 (IBM Corp., Armonk, NY, USA) was used for Pearson’s correlation analysis. A P-value of less than 0.05 was considered to indicate statistical significance. BEAST v1.8.3 which is based on the relaxed-clock Bayesian Markov chain Monte Carlo method, was used to measure the evolution rate and divergent times of nucleotide substitutions in the HA and NA genes of influenza A virus.14 This method enables the measurement of lineage-specific variable amino acid substitution rates based on the posterior probability of sampling phylogenies. The substitution rates were calculated using the years of HA and NA gene extractions. In particular, to reduce error in relation to the number of input data, only data between the same HA and NA gene extracted from the same year were used. The substitution mode was based on the Blosum62 model, and the gamma model was used for the site heterogeneity model.15 Next, constant-size coalescent model was used for the strict clock model and tree prior.16 Chain length was set to 10,000,000 and the echo state was set to screen. The log parameter was set to every 1,000 generations. The BEAST v1.8.3 results were visualized using a tracer v1.6 through Bayesian phylogenetic inference.17
Results
Cophylogeny mapping of HA and NA genes by subtypes and hosts
The co-evolution analysis showed that humans had higher cospeciation values for the H1N1, H1N2, H2N2, H3N2, H5N1, and H9N2 subtypes than did avians, and that the values for humans were the same or similar to those for swine. In the H1N1 subtype, the cospeciation values for HA and NA genes/number of NA genes were 0.63, 0.19, and 0.27 in humans, avians, and swine respectively. The value in the H1N2 subtype was also the highest in humans (0.75) and lowest in avians (0.43) and swine (0.45). In the H3N2 subtype, the values were 0.50, 0.26, and 0.56 in humans, avians, and swine, respectively. In contrast, in the H5N1 and H9N2 subtypes, humans and swine had higher cospeciation values than the genetic switch value, whereas the opposite was true in avians. Because sequence data from other hosts could not be obtained for the H7N2, H7N7, H7N9, or H7N10 subtypes, the cospeciation values were only compared in avians. Cospeciation was highest in the H7N10 subtype (0.59) and lowest in the H7N2 subtype (0.42). The event values are detailed in Table 1.
Table 1.
Results from event based cophylogeny according to subtypes and hosts (default cost settings of 0, 1, 2, 1, 1 in Jane).
| Sub type | Host | Cospeciations | Duplications | Duplications and gene switch | Losses | Total cost | # of HA | HA switch/NA | Cospeciations/NA |
|---|---|---|---|---|---|---|---|---|---|
| H1N1 | Avian | 7 | 0 | 28 | 0 | 56 | 36 | 0.78 | 0.19 |
| Human | 34 | 0 | 19 | 15 | 53 | 54 | 0.35 | 0.63 | |
| Swine | 14 | 0 | 37 | 4 | 78 | 52 | 0.71 | 0.27 | |
| H1N2 | Avian | 6 | 0 | 7 | 0 | 14 | 14 | 0.50 | 0.43 |
| Human | 3 | 0 | 0 | 0 | 0 | 4 | 0 | 0.75 | |
| Swine | 10 | 0 | 11 | 3 | 25 | 22 | 0.50 | 0.45 | |
| H2N2 | Avian | 7 | 0 | 9 | 0 | 18 | 17 | 0.53 | 0.41 |
| Human | 6 | 0 | 5 | 2 | 12 | 12 | 0.42 | 0.50 | |
| H3N2 | Avian | 8 | 0 | 22 | 1 | 45 | 31 | 0.71 | 0.26 |
| Human | 24 | 0 | 24 | 9 | 57 | 49 | 0.49 | 0.50 | |
| Swine | 5 | 0 | 25 | 1 | 51 | 31 | 0.81 | 0.16 | |
| H5N1 | Avian | 9 | 0 | 14 | 1 | 29 | 24 | 0.58 | 0.38 |
| Human | 8 | 0 | 5 | 2 | 12 | 14 | 0.36 | 0.57 | |
| Swine | 5 | 0 | 3 | 0 | 6 | 9 | 0.33 | 0.56 | |
| H7N2 | Avian | 8 | 0 | 10 | 1 | 21 | 19 | 0.53 | 0.42 |
| H7N7 | Avian | 13 | 0 | 12 | 5 | 29 | 26 | 0.46 | 0.50 |
| H7N9 | Avian | 6 | 0 | 6 | 1 | 13 | 13 | 0.46 | 0.46 |
| H7N10 | Avian | 20 | 0 | 13 | 7 | 33 | 34 | 0.38 | 0.59 |
| H9N2 | Avian | 13 | 0 | 18 | 4 | 40 | 32 | 0.56 | 0.41 |
| Human | 3 | 0 | 2 | 2 | 6 | 6 | 0.33 | 0.50 | |
| Swine | 4 | 0 | 3 | 2 | 8 | 8 | 0.38 | 0.50 |
Bold font was used for cospeciations of modest effect size (>0.5).
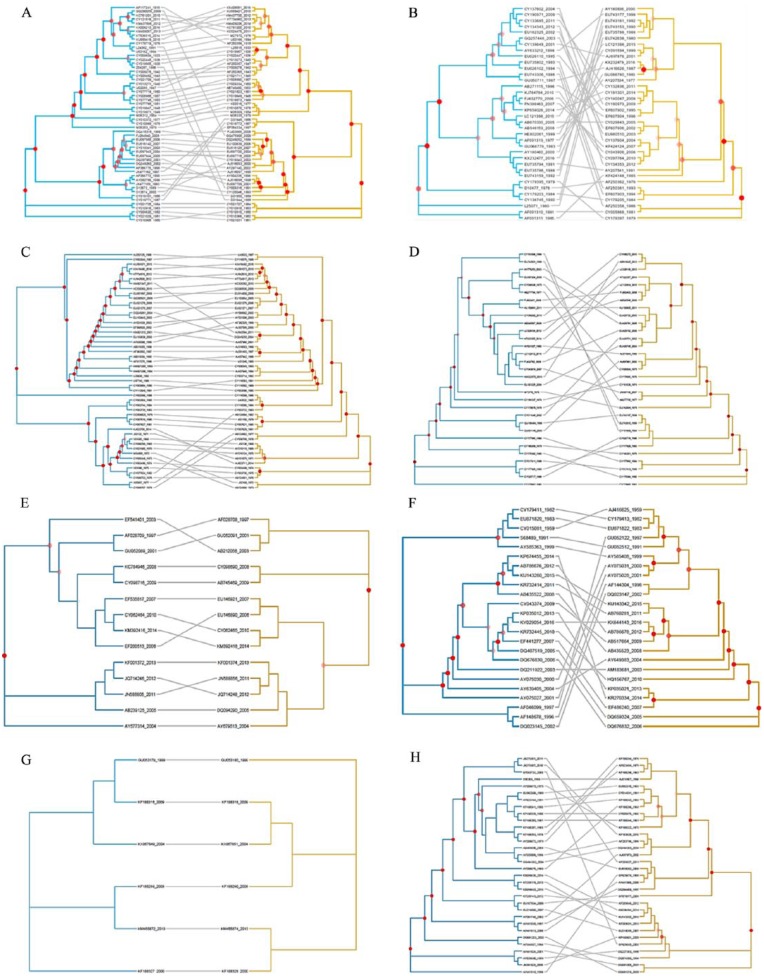
To visualize the phylogenetic relationship between the HA and NA genes, tanglegrams were created using TreeMap 3.0. As shown in Figure 1, there was a difference in phylogenetic congruence by subtype between humans and avian. In each tanglegram, nodes with bootstrap values of at least 50 were labeled in red. The results verified that the yearly ML phylogenetic tree for humans showed similar topology between the HA and NA genes in the H1N1 and H3N2 subtypes. In contrast, tanglegrams for avian viruses showed different topologies between the HA and NA genes in the yearly ML phylogeny. In particular, as shown in Figure 1C, the phylogenetic trees for each of the HA and NA genes in the H3N2 subtype in humans were similarly matched yearly from 1981 to 2016, with the exception of a few years. However, the avian showed no parallel matched pairs between each taxon.
Figure 1.
Tanglegram describing the source phylogenies used in reconciliation analysis of HA gene (left) and NA gene (right) among the subtypes and hosts. (A) H1N1 subtype from human, (B) H1N1 subtype from avian, (C) H3N2 subtype from human, (D) H3N2 subtype from avian, (E) H5N1 subtype from human, (F) H5N1 subtype from avian, (G) H9N2 subtype from human, and (H) H9N2 subtype from avian.
Comparison of substitutions of influenza A virus by subtype and host
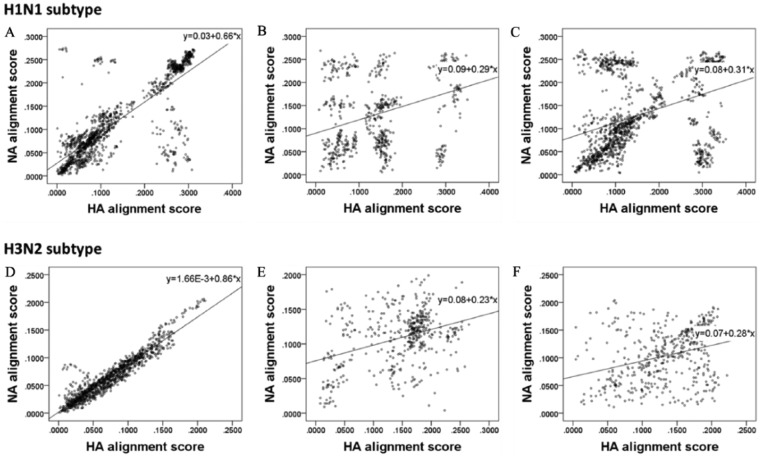
The results of Pearson’s correlation coefficient analysis were mostly similar to those of the reconciliation analysis, with differences in some subtypes. In the H1N1 subtype, there was a high positive correlation between the yearly pairwise alignment scores for the HA and NA genes in humans (r = 0.80, P < .001). The positive correlations were weaker in avians (r = 0.36, P < .001) and swine (r = 0.38, P < .001). In particular, humans showed a very strong positive correlation in the H3N2 subtype (r = 0.94, P < .001), whose tanglegram indicated highly similar topology between the phylogenetic trees of the HA and NA genes. The results of the correlation analysis for each subtype and host are outlined in Figure 2. Scatter plots and linear models were created for the H1N1 and H3N2 subtypes for each host with a correlation effect size of r > 0.8 (P < .001, n > 1000; Figure 2). In both subtypes, the pairwise alignment scores of the HA and NA genes were highly correlated in humans, but not significantly correlated in avians or swine.
Figure 2.
Correlation coefficient scatter plots of sequence alignment score between HA and NA gene. (A) H1N1 subtype from human, (B) H1N1 subtype from avian, (C) H1N1 subtype from swine, (D) H3N2 subtype from human, (E) H3N2 subtype from avian, and (F) H3N2 subtype from swine.
Next, substitution rates for each subtype and host were computed using the nucleotide sequences of the HA and NA genes for the 10 influenza A virus subtypes. For the H1N1, H1N2, H4N3, and H5N1 subtypes, the substitution rate for the NA gene was higher than that for the HA gene in avians, whereas the opposite was true in humans. There were differences in substitution rates between the HA and NA genes in all subtypes. In the H1N1 subtype, the substitution rate for the HA gene was 2.03 × 10-3 substitutions/site/year (95% highest posterior density [HPD], 1.37 × 10−3 to 2.83 × 10−3), whereas that for the NA gene was 1.79 × 10−3 substitutions/site/year (95% HPD, 1.33 × 10−3 to 2.18 × 10−3). In avian H1N1, the substitution rate for the HA gene was 1.63 × 10−3 substitutions/site/year (95% HPD, 1.30 × 10−3 to 1.96 × 10−3), whereas that for the NA gene was 2.02 × 10−3 substitutions/site/year (95% HPD, 1.77 × 10−3 to 2.28 × 10−3). In human H1N1, the substitution rate for the HA gene was 1.80 × 10−3 substitutions/site/year (95% HPD, 1.33 × 10−3 to 2.28 × 10−3), whereas that for the NA gene was 1.46 × 10−3 substitutions/site/year (95% HPD, 1.31 × 10−3 to 1.62 × 10−3). For the H3N2 subtype, the substitution rates were similar for the HA and NA genes that for the HA gene was 2.78 × 10−3 substitutions/site/year (95% HPD, 4.24 × 10-4 to 4.33 × 10−3), and that for the NA gene was 2.62 × 10−3 substitutions/site/year (95% HPD, 1.93 × 10−3 to 3.19 × 10−3). In avian H3N2, the substitution rate for the HA gene was 8.38 × 10−4 substitutions/site/year (95% HPD, 3.29 × 10-4 to 1.28 × 10−3), whereas that for the NA gene was 2.14 × 10−3 substitutions/site/year (95% HPD, 1.85 × 10−3 to 2.43 × 10−3). In humans, the substitution rate for the HA gene was 3.49 × 10−3 substitutions/site/year (95% HPD, 3.25 × 10−3 to 3.73 × 10−3), whereas that for the NA gene was 2.99 × 10−3 substitutions/site/year (95% HPD, 2.77 × 10−3 to 3.23 × 10−3). Other results are described in Table 2.
Table 2.
Mean nucleotide substitution rate of HA and NA gene according to subtypes and hosts.
| Subtype | Host | HA Substitution rate (sub/site/year) |
NA Substitution rate (sub/site/year) |
||||
|---|---|---|---|---|---|---|---|
| Mean | 95% HPD Lower | 95% HPD Higher | Mean | 95% HPD Lower | 95% HPD Higher | ||
| H1N1 | Avian | 1.63E-03 | 1.30E-03 | 1.96E-03 | 2.02E-03 | 1.77E-03 | 2.28E-03 |
| Human | 1.80E-03 | 1.33E-03 | 2.28E-03 | 1.46E-03 | 1.31E-03 | 1.62E-03 | |
| Swine | 2.66E-03 | 2.45E-03 | 2.86E-03 | 1.91E-03 | 1.75E-03 | 2.07E-03 | |
| Total | 2.03E-03 | 1.37E-03 | 2.83E-03 | 1.79E-03 | 1.33E-03 | 2.18E-03 | |
| H1N2 | Avian | 1.07E-05 | 6.05E-38 | 1.59E-05 | 3.74E-05 | 8.29E-33 | 1.23E-04 |
| Human | 1.76E-05 | 2.01E-66 | 1.22E-19 | 7.54E-23 | 9.26E-107 | 3.19E-39 | |
| Swine | 1.70E-03 | 9.36E-04 | 2.39E-03 | 2.56E-03 | 1.60E-03 | 3.67E-03 | |
| Total | 5.74E-04 | 2.01E-66 | 2.11E-03 | 8.66E-04 | 9.26E-10 | 3.09E-03 | |
| H2N2 | Avian | 4.07E-06 | 1.07E-62 | 2.84E-06 | 1.69E-04 | 6.33E-56 | 1.03E-03 |
| Human | 4.76E-07 | 6.64E-94 | 6.70E-12 | 2.87E-03 | 1.98E-03 | 3.85E-03 | |
| Total | 2.27E-06 | 6.64E-94 | 3.54E-10 | 1.52E-03 | 6.32E-56 | 3.48E-03 | |
| H3N2 | Avian | 8.38E-04 | 3.29E-04 | 1.28E-03 | 2.14E-03 | 1.85E-03 | 2.43E-03 |
| Human | 3.49E-03 | 3.25E-03 | 3.73E-03 | 2.99E-03 | 2.77E-03 | 3.23E-03 | |
| Swine | 4.01E-03 | 3.55E-03 | 4.46E-03 | 2.74E-03 | 2.37E-03 | 3.15E-03 | |
| Total | 2.78E-03 | 4.24E-04 | 4.33E-03 | 2.62E-03 | 1.93E-03 | 3.19E-03 | |
| H5N1 | Avian | 2.33E-06 | 3.12E-33 | 1.09E-05 | 3.01E-03 | 1.93E-03 | 3.89E-03 |
| Human | 1.55E-02 | 1.07E-02 | 2.13E-02 | 3.12E-03 | 1.94E-03 | 4.33E-03 | |
| Swine | 8.94E-14 | 3.72E-71 | 1.29E-17 | 1.63E-10 | 2.19E-69 | 1.26E-13 | |
| Total | 5.17E-03 | 3.71E-71 | 0.01 | 2.04E-03 | 2.19E-69 | 3.84E-03 | |
| H7N2 | Avian | 4.57E-03 | 3.53E-03 | 5.62E-03 | 2.50E-03 | 8.71E-04 | 3.96E-03 |
| H7N7 | Avian | 1.73E-12 | 2.16E-71 | 5.19E-27 | 5.97E-08 | 7.08E-62 | 2.64E-14 |
| H7N9 | Avian | 2.54E-03 | 1.30E-03 | 3.79E-03 | 1.93E-06 | 2.05E-70 | 2.54E-07 |
| H7N10 | Avian | 1.49E-03 | 1.23E-03 | 1.75E-03 | 2.68E-03 | 2.40E-03 | 2.94E-03 |
| H9N2 | Avian | 3.85E-03 | 3.52E-03 | 4.16E-03 | 2.58E-03 | 2.22E-03 | 2.93E-03 |
| Human | 1.48E-03 | 3.06E-44 | 5.50E-03 | 1.50E-06 | 4.59E-88 | 9.33E-14 | |
| Swine | 6.05E-04 | 1.15E-55 | 4.19E-03 | 6.69E-05 | 1.44E-131 | 6.19E-06 | |
| Total | 1.97E-03 | 1.14E-88 | 4.64E-03 | 8.81E-04 | 1.44E-131 | 2.77E-03 | |
Discussion and Conclusions
In this study, we compared the co-evolution patterns and correlations between HA and NA genes of influenza A virus according to subtypes and hosts. The results revealed that humans indicated higher cospeciation values than avian in the subtypes of H1N1, H1N2, H2N2, H3N2, H5N1, and H9N2. Reconciliation analysis showed that humans have higher cospeciation values than switch values for HA and NA. On the other hand, avian show higher switch values than cospeciation values in the subtypes. Especially, H1N1 and H1N2 subtype cospeciation distance was higher than that of the other subtypes. H1N1, H1N2, and H3N2 are known subtypes of the swine influenza A virus and are currently circulating among humans. HA plays an important role in determining the host range of influenza viruses, and an optimal balance between the activities of HA and NA is required for efficient viral replication and transmission.18 Thus, the HA and NA functional balance due to compensatory mutations may exert selective pressure on hosts.
Comparisons between substitution rates of the HA and NA genes in each subtype also showed that evolution rates differed among avian, humans, and swine. In particular, among the subtypes that showed high cospeciation values in our previous analysis, H1N1, H1N2, H3N2, and H5N1 had higher substitution rates for the NA gene than for the HA gene in avian. On the other hand, the substitution rate for the HA gene was higher than that for the NA gene in humans. There were significant positive correlations between the two genes in the H1N1 and H3N2 subtypes in humans compared with those in avian and swine, confirming that the HA and NA genes co-evolved in some subtypes and hosts. Following these results, the HA and NA genes, which encode envelope proteins that play key roles in viral attachment to hosts.
Reassortment exposes influenza HA to significant changes in selective pressure through genetic interactions with NA.19 Glycosylation of the receptor binding site is limited to HA and is often a result of antibody escape as antibodies are targeted against the entire globular head domain of HA. The argument has been made that glycosylation at the receptor binding site that reduces substrate affinity necessitates a change in NA to maintain viral fitness.20 Moreover, the strong receptor binding affinity of HA benefits viral replication in cells given that the HA and NA balance plays an important role in the viral life cycle.21 Other internal gene influenced the evolution to HA gene. Avian and human influenza viruses typically have a different sialic acid binding preference and only few amino acid changes in the HA protein can cause a switch from avian to human receptor specificity.22 In the hemadsorption assay, the presence of oligosaccharide side chains in the vicinity of the receptor binding site was shown to induce negative effects that interrupt the efficient HA and SA interaction.21 Also, as a result of distance correlation of protein in Influenza A genomes by the MirrorTree method shows that HA and NA have high correlation distance than other gene.23
In this study, we demonstrated the influenza A virus from the human has higher cospeciation value than avian and swine in H1N1, H1N2 and H3N2 subtypes. These subtypes are the known swine influenza A virus and currently circulating among human. The influenza glycoprotein HA plays an important role in determining the host range of influenza viruses. An optimal balance between the activities of HA and NA is required for efficient viral replication and transmission.18 So, HA and NA functional balance compensatory mutation may have selective pressure to hosts. We performed an event-based cophylogeny analysis with the HA and NA genes of influenza A virus to identify the differences among hosts. Correlations analyzed using sequence alignment scores to verify co-evolution based on sequences. However, as only published genetic sequences were examined, the small set of sequence data resulted in error rates in some subtypes. Therefore, we plan to analyze the co-evolution of the HA and NA genes in all subtypes and hosts by collecting novel data in a future study. Based on these findings pertaining to the phylogenetic evolution in relation to the co-evolution of the HA and NA genes in avian, humans, and swine and sequence variations caused by differences in substitution rates. The results of our phylogenetic co-evolution and sequence variation analyses provide a proof of principle for influenza virus vaccine design and antibody-mediated therapies based on cospeciated regions of the HA and NA genes.
Supplemental Material
Supplemental material, Supplementary_Tables for Comparative Co-Evolution Analysis Between the HA and NA Genes of Influenza A Virus by Jinhwa Jang and Se-Eun Bae in Virology: Research and Treatment
Footnotes
Funding:This work was supported by the “Solving grand-challenge problems in science and engineering to expand utilization of supercomputing (K-18-L12-C08)” funded by Korea Institute of Science and Technology Information.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JJ and SEB conceptualised the study design, literature search. JJ conducted the review processes as well as developed the initial draft manuscript. JJ and SEB critically revised the manuscript before publication. All authors reviewed and approved the final manuscript.
Disclosures and Ethics: The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Supplemental Material: Supplemental tables for this article are available online.
References
- 1. Couch RB. Orthomyxoviruses. In: Baron S, ed. Medical Microbiology. 4th ed. Galveston, TX: University of Texas Medical Branch; 1996; 723–730. [PubMed] [Google Scholar]
- 2. Kilbournf ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cauldwell AV, Long JS, Moncorge O, Barclay WS. Viral determinants of influenza A virus host range. J Gen Virol. 2014;95:1193–1210. [DOI] [PubMed] [Google Scholar]
- 4. Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaverin NV, Gambaryan AS, Bovin NV, et al. Postreassortment changes in influenza A virus hemagglutinin restoring HA–NA functional match. Virology. 1998;244:315–321. [DOI] [PubMed] [Google Scholar]
- 6. Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suzuki Y, Ito T, Suzuki T, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen W, Zhong Y, Qin Y, Sun S, Li Z. The evolutionary pattern of glycosylation sites in influenza virus (H5N1) hemagglutinin and neuraminidase. PLoS ONE. 2012;7:e49224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donis RO. Evolution of H5N1 avian influenza viruses in Asia. Emerg Infect Dis. 2005;11:1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaverin NV, Matrosovich MN, Gambaryan AS, et al. Intergenic HA-NA interactions in influenza A virus: postreassortment substitutions of charged amino acid in the hemagglutinin of different subtypes. Virus Res. 2000;66:123–129. [DOI] [PubMed] [Google Scholar]
- 11. Sievers F, Wilm A, Dineen DG, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Bio. 2011;7:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conow C, Fielder D, Ovadia Y, Libeskind-Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol. 2010;5:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charleston MA, Perkins SL. Lizards, malaria, and jungles in the Caribbean. In: Page RDM, ed. Tangled Trees: Phylogeny, Cospeciation, and Coevolution. Chicago, IL: The University of Chicago Press; 2003:65–92. [Google Scholar]
- 14. Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci. 1992;89:10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kingman JFC. Origins of the Coalescent: 1974-1982. Stoch Proc Appl. 1982;13:235–248. [Google Scholar]
- 17. Rambaut A, Suchard MA, Xie D, et al. Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer Up-dated 2014. Accessed 3 January 2017.
- 18. Cauldwell AV, Long JS, Moncorgé O, et al. Viral determinants of influenza A virus host range. J Gen Virol. 2014;95:1193–1210. [DOI] [PubMed] [Google Scholar]
- 19. Ward MJ, Lycett SJ, Avila D, et al. Evolutionary interactions between haemagglutinin and neuraminidase in avian influenza. BMC Evol Biol. 2013;13:222–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baigent SJ, McCauley JW. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001;79:177–185. [DOI] [PubMed] [Google Scholar]
- 21. Kim JL, Park MS. N-linked glycosylation in the hemagglutinin of influenza A viruses. Yonsei Med J. 2012;53:886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Graaf M, Fouchier RAM. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin C, Yau SS. A coevolution analysis for identifying protein-protein interactions by Fourier transform. PLoS ONE. 2017;12:e0174862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Tables for Comparative Co-Evolution Analysis Between the HA and NA Genes of Influenza A Virus by Jinhwa Jang and Se-Eun Bae in Virology: Research and Treatment