Summary
• The plant hormone auxin regulates many aspects of plant growth and development. Auxin signaling involves hormone perception by the TRANSPORT INHIBITOR RESPONSE/AUXIN F-BOX (TIR1/AFB)–Aux/IAA co-receptor system, and the subsequent degradation of the Aux/IAA transcriptional repressors by the ubiquitin proteasome pathway. This leads to the activation of downstream gene expression and diverse physiological responses. Here, we investigate how the structural elements in the Aux/IAAs determine their function in Auxin perception and transcriptional repression.
• We took advantage of the facile genetics of the moss Physcomitrella patens to determine the activity of wild-type and mutant PpIAA1a proteins in a Δaux/iaa null background. In this way, Aux/IAA function was characterized at the molecular and physiological levels without the interference of genetic redundancy.
• We identified and characterized degron variants in Aux/IAAs that affect their stability and Auxin response. We also demonstrated that neither the Aux/IAA EAR motif nor Aux/IAA oligomerization is essential for the repressive function of Aux/IAA.
• Our study demonstrates how key elements within the Aux/IAA proteins fine tune stability and repressor activity, as well as the long-term developmental outcome.
Keywords: Aux/IAA, auxin, Physcomitrella, plant development, plant hormone
Introduction
The plant hormone auxin regulates a wide range of processes during plant development and response to the environment (Woodward & Bartel, 2005; Lavy & Estelle, 2016; Strader & Zhao, 2016; Weijers & Wagner, 2016). Although the chemical structure of Auxin is quite simple, the hormone triggers a complex set of downstream responses with different temporal and spatial patterns (Bargmann et al., 2013). Many studies have been devoted to the understanding of the mechanisms that produce this complexity at the cellular and molecular level.
Based on our current knowledge, Auxin acts through a transcriptional de-repression mechanism. The major repressors in the pathway are the Aux/indole-3-acetic acid (Aux/IAA) proteins, which are also components of the Aux/IAA–TRANSPORT INHIBITOR RESPONSE/AUXIN F-BOX (TIR1/AFB) co-receptor system for Auxin perception. The TIR1/AFB proteins are F-box proteins and subunits of SKP1 CULLIN F-BOXTIR1/afb (SCFTIR1/AFB) ubiquitin protein ligases (E3s). In the presence of Auxin, SCFTIR1/afb promotes the degradation of the Aux/IAAs, resulting in the activation of Auxin-responsive gene transcription (Salehin et al., 2015; Lavy & Estelle, 2016). A number of genetic studies have identified stabilized forms of Aux/IAAs that escape this degradation, revealing a core degron motif within Aux/IAA Domain II (Liscum & Reed, 2002). Structural studies later confirmed that this domain is the interaction surface with TIR1 in the presence of Auxin (Tan et al., 2007). There are 29 Aux/IAA proteins in Arabidopsis with varying stabilities. At least some of this variation can be attributed to the degron sequence (Dreher et al., 2006; Havens et al., 2012; Guseman et al., 2015; Pierre-Jerome et al., 2016). In addition, because the Aux/IAA protein contributes to Auxin binding, changes in the degron sequence may impact the affinity of the co-receptor pair for Aux. Indeed, we have demonstrated that different co-receptor pairs exhibit distinct Kd values for Auxin (Calderon Villalobos et al., 2012). Thus, variation in the degron sequence may affect the Auxin concentration at which degradation occurs, as well as the rate of degradation.
The Aux/IAAs repress the activity of a family of transcription factors called AUXIN RESPONSE FACTORs (ARFs), which share homology with Aux/IAAs in their C-terminus and are thought to control Auxin-responsive gene transcription. Repression may be achieved by recruiting a co-repressor called TOPLESS (TPL) through the Aux/IAA EAR motif (Weijers & Wagner, 2016). However, it is not clear whether Aux/IAA repression depends solely on TPL function.
Extensive yeast two-hybrid studies have shown that the Aux/IAAs and ARFs form homo-and heterodimers through their homologous C-terminal domains (Vernoux et al., 2011). A recent structural study has proposed that the C-terminal region constitutes a PB1 (Phox and Bem 1) domain (Korasick et al., 2014). The interaction between PB1 domains involves a conserved lysine residue and an acidic DxD/ExD (OPCA) motif (Fig. 1), which form two complementary interaction centers. As each PB1 domain has both acidic and basic interaction centers, the ARFs and Aux/IAA may form complex oligomers. The importance of oligomerization has been addressed in two studies. In one case, the phenotype conferred by overexpression of stabilized AtIAA16 was not maintained when either of the interaction centers was mutated to neutral alanine, indicating that, in this specific background, Aux/IAA oligomerization is required for repression by stabilized AtIAA16 (Korasick et al., 2014). By contrast, a similar study with AtIAA14 revealed that a mutant that was unable to form oligomers was still able to function as a repressor (Pierre-Jerome et al., 2016). In both instances, the gain-of-function nature of the stabilized Aux/IAA protein can be a complicating factor.
Fig. 1.
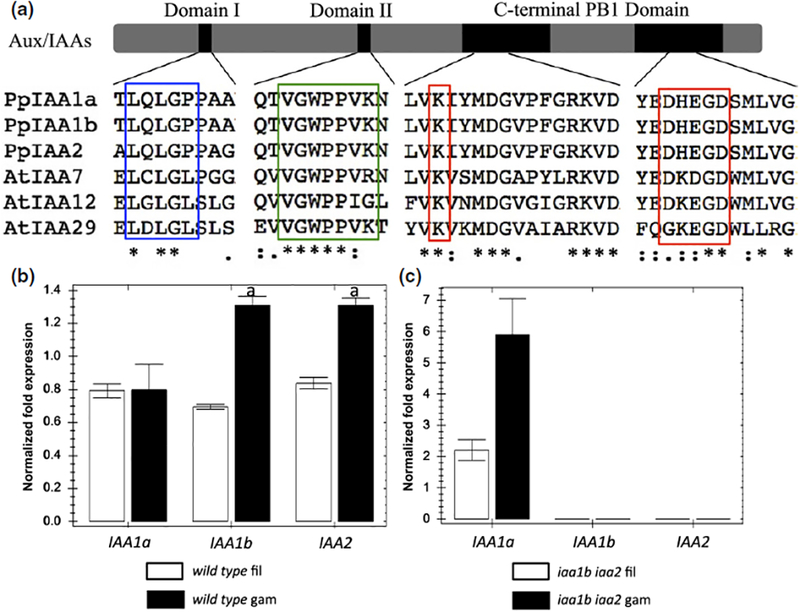
The Arabidopsis and moss auxin/indole-3-acetic acid (Aux/IAA) proteins share conserved elements in all three functional domains. Blue box, the EAR motif in Domain I; green box, the core degron motif in Domain II; red box, K and OPCA motifs in the PBI domain, which are highly conserved not only among Aux/IAAs, but also among the AUXIN RESPONSE FACTOR (ARF) transcription factors. (b) Expression of the three Aux/IAA genes in moss filaments and leafy gametophores determined by quantitative PCR. (c) Aux/IAA expression in iaa1b iaa2 double knockout mutant. Black, gametophores (gam); white, remaining filaments (fil). Normalized expression (ΔΔC(t) method using PpEF1a as a reference gene) is plotted as relative values. Error bars represent ± SEM. ‘a’ indicates that the difference between two tissues is significant at P < 0.05 (Student’s t-test), n = 4.
Genetic studies of the Aux/IAA genes in Arabidopsis have been hampered by extensive genetic redundancy amongst the family members (Overvoorde et al., 2005). Although stabilized forms of Aux/IAAs produce clear phenotypes, the gain-of-function nature of these mutations can complicate their analysis. Further, loss of Aux/IAA gene function typically does not confer a phenotype, limiting the genetic analysis of these genes (Overvoorde et al., 2005).
By contrast, the moss Physcomitrella patens has only three Aux/IAA genes (Lavy et al., 2016). Recently, we have reported a Δaux/iaa null mutant that displays a strong Auxin-constitutive phenotype. In this study, we used the facile genetics of Physcomitrella to investigate the biological significance of conserved elements in the Aux/IAA proteins, including the degron motif, the EAR motif and the conserved motifs in the C-terminus. The results provide unique insights into the function of the Aux/IAAs, both as co-receptors and repressors of Auxin signaling.
Materials and Methods
Moss strains and growth conditions
Wild-type ‘Gransden-2004’ and mutant P. patens strains were grown at 25°C under continuous light at an intensity of 40–70 μmol m−2 s−1. When grown for phenotypic and gene expression analysis, BCD medium was used. When grown for propagation purposes, BCDAT (BCD supplemented with 5 mM ammonium tartrate) was used.
Molecular cloning of gene expression constructs
To generate the construct for the expression of luciferase-tagged IAA1a, the following DNA fragments were PCR amplified with primers containing the corresponding restriction enzyme (RE) cutting sequences and cloned into the RE sites of the pBHRF2 backbone (Lavy et al., 2016) by enzyme digestion and ligation: the luciferase-coding sequence was cloned into the PstI and XbaI sites; the cDNA of IAA1a was cloned into the PstI site; c. 1 kilo-base (kb) of the genomic sequence upstream (5′) and downstream (3′) of the IAA1a gene coding region was cloned into the HindIII and SpeI sites. This creates the IAA1a 5′ genomic:cIAA1a: luciferase:HygR (from backbone, with LoxP sites):IAA1a 3′ genomic cassette for the expression of luciferase-tagged IAA1a in replacement of the native IAA1a gene. To generate the constructs for the expression of the mutant versions of IAA1a-luciferase, site-directed mutagenesis was performed with the construct above containing wild-type IAA1a. The primer sequences used for cloning and mutagenesis are listed in Supporting Information Table S1.
Moss transformation and screening of transgenic lines
Protoplast isolation and polyethylene glycol (PEG) -mediated transformation of the above constructs into the iaa1b iaa2 double mutant (Lavy et al., 2012) were performed as described by Nishiyama et al. (2000). After 5 d of regeneration, transformants were moved to BCDAT medium supplemented with 20 mg1−1 hygromycin for selection. The transformants that survived selection were screened by PCR for the presence of both left and right transgene-endogenous sequence junctions to verify the insertion of the transgene to the native locus. Reverse transcription-polymerase chain reaction (RT-PCR) with a pair of primers flanking an IAA1a intron was performed to confirm the replacement of genomic IAA1a with the cIAA1a-Luc transgene. The primer sequences are listed in Table S1.
To remove the possible tandem repeats generated by homologous recombination, the pActin:CRE plasmid was transformed into the confirmed lines above. This removes the possible repeats as well as the hygromycin resistant marker. Therefore, the transformants that cannot survive on hygromycin-containing medium were used for further analysis.
Phenotypic characterization
For phenotypic observations, small pieces of fresh protonemal tissue were inoculated on BCD mock or 1-naphthaleneacetic acid (NAA)-supplemented medium and grown for 1 month into well-developed colonies. Photographs were taken for each colony and typical representations are shown.
RNA isolation and quantitative RT-PCR
For the detection of tissue-specific Aux/IAA gene transcripts, protonemal tissue was grown on BCD plates with cellophane overlays for 3 wk. Leafy gametophores were cut from the remaining filamentous tissue and the two types of tissue were collected individually for RNA isolation. For the detection of Auxin-responsive marker gene transcripts, protonemal tissue was grown on BCDAT plates with cellophane overlays for 1 wk. The colonies were then transferred into liquid BCD medium containing either 10 μM IAA or the equivalent amount of solvent (ethanol). After incubation under moss growth conditions, the colonies were collected individually for RNA isolation.
Total RNA was isolated using an RNeasy Plant Mini Kit (Qia-gen); 500 ng of RNA were reverse transcribed using the Superscript III First Strand cDNA Synthesis System (Life Technologies, Waltham, MA, USA). A 20-μl RT reaction was diluted to a final volume of 200 μl; 4 μl of the diluted cDNA were used for detection by the CFX Connect™ Real-Time PCR Detection System (Bio-Rad). The Auxin-responsive marker genes used have been described in Lavy et al. (2016). Normalized expression (ΔΔC(t) method) was calculated using the Bio-Rad CFX manager software employing PpEF1a as a reference gene, and plotted as relative values ± SEM. Four biological replicates and four technical replicates were included in each analysis. t-test was used for statistical inference. The primer sequences used are listed in Table S1.
Luciferase-based degradation assay
Fresh protonemal tissue blended in sterile water with a homogenizer was spread and grown on BCDAT plates with cellophane overlays for 4 d. For each replicate of the transgenic line of interest, two tissue samples of a fixed amount (25 mm × 25 mm area on the plate) were taken and each was blended in 50 μl of 10 mM D-luciferin in a well of a 96-well plate, and their chemiluminescence was measured by an ImageQuant LAS 4000 Mini biomolecular imager (GE Healthcare Life Sciences, Marlborough, MA, USA) under super-resolution and quantified by ImageJ. This serves as the time point 0 reference for mock/Auxin treatment.
After either IAA stock solution or an equivalent volume of the solvent was added to the wells, the chemiluminescence was quantified in the same way at different time points. The ratio of the signal strength of treated vs mock was calculated and normalized by the ratio of time point 0 for adjustment of the difference in starting amount between the mock/treated samples. The normalized ratio represents the fold signal that remains after the corresponding time of Auxin treatment. The degradation curve was generated by plotting this normalized ratio against the treatment time. Three replicates were used for each data point, and the error bar represents the standard error (SE).
Results
Tissue-specific expression pattern of the moss Aux/IAA genes reveals gene redundancy and compensation at the molecular level
Auxin is known to play an important role in the vegetative growth of P. patens. At the protonemal stage, Auxin promotes the differentiation of chloroplast-rich filaments, called chloronemata, into elongated filaments with fewer chloroplasts, called caulonemata, whereas, at the stage of leafy gametophore formation, Auxin promotes stem elongation and rhizoid development (Ashton et al., 1979; Eklund et al., 2010; Prigge & Bezanilla, 2010). All three Physcomitrella Aux/IAA proteins possess the three conserved domains characteristic of the Aux/IAA family (Fig. 1a). IAA1a and IAA1b are 93% identical, whereas both of these proteins are 69% identical to IAA2 (Prigge et al., 2010). In a recent study, we have shown that the iaa1b iaa2 double mutant is indistinguishable from the wild-type in appearance and Auxin response, indicating that IAA1a is sufficient for normal development (Lavy et al., 2016). To explore this issue further, we determined the expression level of the three genes in protonemal filaments and gametophores in a wild-type and iaa1b iaa2 line.
Our results revealed that, in wild-type moss, IAA1a was expressed at a similar level in filaments and gametophores, whereas both IAA1b and IAA2 were expressed at a higher level in gametophores relative to filaments (Fig. 1b). Interestingly, in the absence of IAA1b and IAA2, the level of IAA1a transcript was higher in gametophores relative to filaments (Fig. 1c). Indeed, the level of IAA1a transcript in iaa1b iaa2 was roughly the same as the sum of IAA1a, IAA1b and IAA2 transcripts in the wild-type line, in both gametophores and filamentous tissue. This suggests that, in the absence of IAA1b and IAA2, IAA1a expression is up-regulated, compensating for the loss of the other two genes. As IAA1a is Auxin regulated (Fig. S1), this probably reflects the activity of a negative feedback loop involving the ARFs and the Aux/IAAs (Prigge et al., 2010; Salehin et al., 2015).
Because the iaa1b iaa2 line has a wild-type phenotype, it provides an excellent background for further studies of Aux/IAA function. In the following experiments, we used homologous recombination to generate a series of iaa1b iaa2 lines expressing wild-type and mutant versions of IAA1a (Fig. 2). These lines were used to study the function of Aux/IAA domains without the interference of other native Aux/IAAs.
Fig. 2.
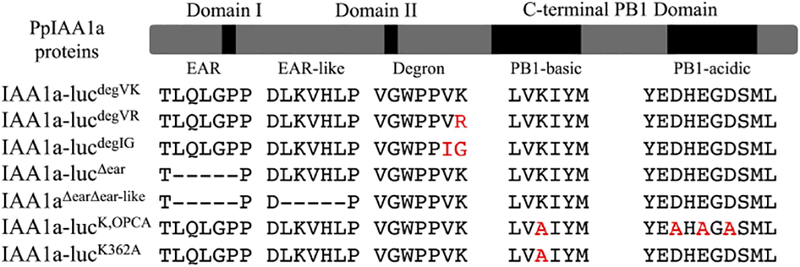
The mutant Physcomitrella PpIAA1a proteins described in this study. Red text indicates site of an amino acid substitution. (−) represents deleted residue.
Studies of the Aux/IAA degron motif
Because the Aux/IAA degron is the interaction surface for the TIR1/AFB protein and Auxin, it is a key element in Auxin signaling. In Arabidopsis, the canonical core degron sequence is VGWPPVR, which is shared by multiple family members, including IAA7, IAA14 and IAA28. There are also several variants of this sequence, including VGWPPIG (IAA12) and VGWPPVK (IAA29) (Fig. 1). All three moss Aux/IAA proteins have the VGWPPVK degron, like AtIAA29. In an earlier study, we showed that the TIR1-IAA7 co-receptor had a higher affinity for Auxin than TIR1-IAA12 in vitro, and that this difference was determined by the degron sequence (Calderon Villalobos et al., 2012). However, because neither the loss of the IAA7 or IAA12 genes results in a phenotype, it was not possible to test the significance of this difference in Arabidopsis (Overvoorde et al., 2005).
To address this question, we generated luciferase-tagged PpIAA1a constructs with degron motifs corresponding to AtIAA7 (IAA1a-lucdegVR), AtIAA12 (IAA1a-lucdegIG) and the PpIAAs (IAA1a-lucdegVK) (Fig. 2). Each construct was introduced into the iaa1b iaa2 background by homologous recombination, replacing the endogenous IAA1a gene.
The three lines showed a similar appearance when grown on medium without Auxin. However, when grown on medium containing the Auxin NAA, both IAA1a-lucdegVR and IAA1a-lucdegIG displayed resistance relative to IAA1a-lucdegVK. In the wild-type (IAA1a-lucdegVK), Auxin treatment resulted in reduced colony size, disruption of leafy gametophore formation and increased brown-pigmented rhizoids, as expected (Ashton et al., 1979). Each of these effects was reduced in the other two lines. IAA1a-lucdegVR showed increased leafy gametophore formation and colony size when treated with NAA, relative to IAA1a-lucdegVK, whereas IAA1a-lucdegIG was even less sensitive to Auxin treatment, with near-normal colony size and gametophore formation, even after treatment with 0.5 μM NAA. By contrast, we have demonstrated previously that a line lacking all three Aux/IAAs (Δaux/iaa) displays this phenotype in the absence of Auxin treatment (Fig. 3a) (Lavy et al., 2016).
Fig. 3.
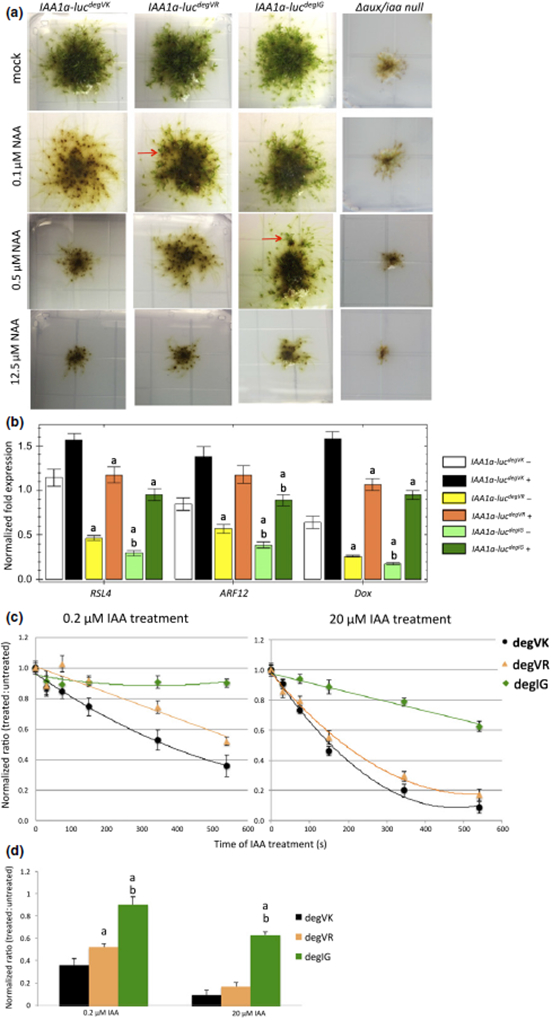
Characterization of natural variation in the Physcomitrella auxin/indole-3-acetic acid (Aux/IAA) degron motif. (a) The developmental phenotypes of PpIAA1a-luc/Δaux/iaa lines expressing wild-type and mutant IAA1a proteins grown on BCD medium with different auxin (1-naphthaleneacetic acid, NAA) levels, with the Δaux/iaa null mutant as a control. Leafy gametophore formation, which indicates resistance to external auxin, is blocked in IAA1adegVK on 0.1 μM NAA, but still occurs in IAA1adegVR (red arrow); IAA1adegIG is resistant to higher NAA concentrations relative to the other two lines. (b) Quantitative PCR analysis of auxin-responsive marker genes in the wild-type and mutant lines treated with mock (−) or 10 μM IAA (+) for 1 h. White and black, IAA1adegVK; yellow, IAA1adegVR; green, IAA1adegIG; darker colors represent 10 μM IAA-treated samples. Normalized expression (ΔΔC(t) method using PpEF1a as a reference gene) is plotted as relative values. Error bar represents ± SEM. ‘a’ indicates that difference from IAA1adegVK is significant at P < 0.05 (Student’s t-test), n = 4; ‘b’ indicates that difference between IAA1adegVK and IAA1degIG is significant at P < 0.05 (Student’s t-test), n = 4. (c) In vivo degradation curve of different IAA1a versions on 0.2 μM and 20 μM IAA treatments measured by luciferase chemiluminescence. The normalized luminescence ratio (treated : untreated) at each time point is plotted. Error bar represents ± SE. (d) Comparison of normalized ratio from (c) at 550 s of IAA treatment. ‘a’ indicates that difference between mutants and wild-type is significant at P< 0.5 (Student’s t-test), n = 3. ‘b’ indicates that difference from IAA1adegVR is significant at P < 0.05 (Student’s t-test), n = 3.
To determine whether Auxin resistance was accompanied by changes in Auxin-regulated gene expression, we determined the transcript level of three Auxin-regulated genes by quantitative PCR. Our results showed that, in both mock- and Auxin-treated conditions, expression of the marker genes in IAA1a-lucdegVR and IAA1a-lucdegIG lines was lower than in the wild-type. In addition, the expression of these genes was lower in IAA1a-lucdegIG than in IAA1a-lucdegVR (Fig. 3b), consistent with their phenotypes.
We then checked whether these defects were related to differences in IAA1a degradation. IAA1a-luc protein levels were determined by measurement of chemiluminescence at different time points after mock or Auxin treatment. The normalized ratio, which is the chemiluminescence from the Auxin-treated sample relative to the mock sample, was plotted against the treatment time (Fig. 3c). We found that, with 0.2 μM IAA, IAA1a-lucdegIG remained stable, whereas IAA1a-lucdegVR degraded more slowly than IAA1a-lucdegVK. At 20 μM IAA, the IAA1a-lucdegIG level began to decrease, but much more slowly than for the other two proteins. Thus, the difference in Auxin binding observed in vitro appears to result in a corresponding shift in the concentration of Auxin required to promote degradation.
AtIAA29 has the same degron as the moss Aux/IAAs (degVK). Based on previously published yeast two-hybrid data, IAA29 has a relatively low Auxin affinity. In moss, this degron contributes to a higher rate of Auxin-induced degradation than degVR or degIG, suggesting that the behavior of AtIAA29 is a result of features outside of the degron.
The EAR motif is not essential for Aux/IAA function
Aux/IAA repression is thought to require recruitment of the co-repressor TPL through an interaction with the EAR motif. The EAR domain is conserved in the moss and Arabidopsis Aux/IAA proteins (Fig. 1a), and previous yeast two-hybrid assays have shown that the moss Aux/IAAs interact with two moss TPL proteins through the EAR domain (Causier et al., 2012). We utilized our PpIAA1a-luc system to study the role of the EAR motif in vivo. We generated an EAR motif-deleted PpIAA1aΔear transgene and introduced this into Δiaa1b iaa2 plants by homologous recombination. In addition, we verified that IAAaΔear does not interact with moss TPLs in a yeast two-hybrid assay (Fig. S2a).
Phenotypic characterization showed that the IAA1aΔear line displayed increased caulonemata formation and reduced numbers of leafy gametophores relative to the wild-type, consistent with a reduction in IAA1aΔear repression. However, the appearance of the mutant was clearly different from the Δaux/iaa line, suggesting that IAA1aΔear retained some function. Further, when treated with different concentrations of Auxin, IAA1aΔear displayed an increasingly severe Auxin phenotype that correlated with the Auxin level (Fig. 4a). We then checked the expression level of the Auxin-responsive marker genes in the mutant and wild-type lines (Fig. 4b). In the absence of external Auxin, the IAA1aΔear mutant had a higher level of Auxin-responsive gene transcription than the wild-type. Moreover, external Auxin treatment resulted in increased gene expression relative to the mock condition, which correlated with the phenotypes above and suggested that IAA1aΔear is still able to repress transcription.
Fig. 4.
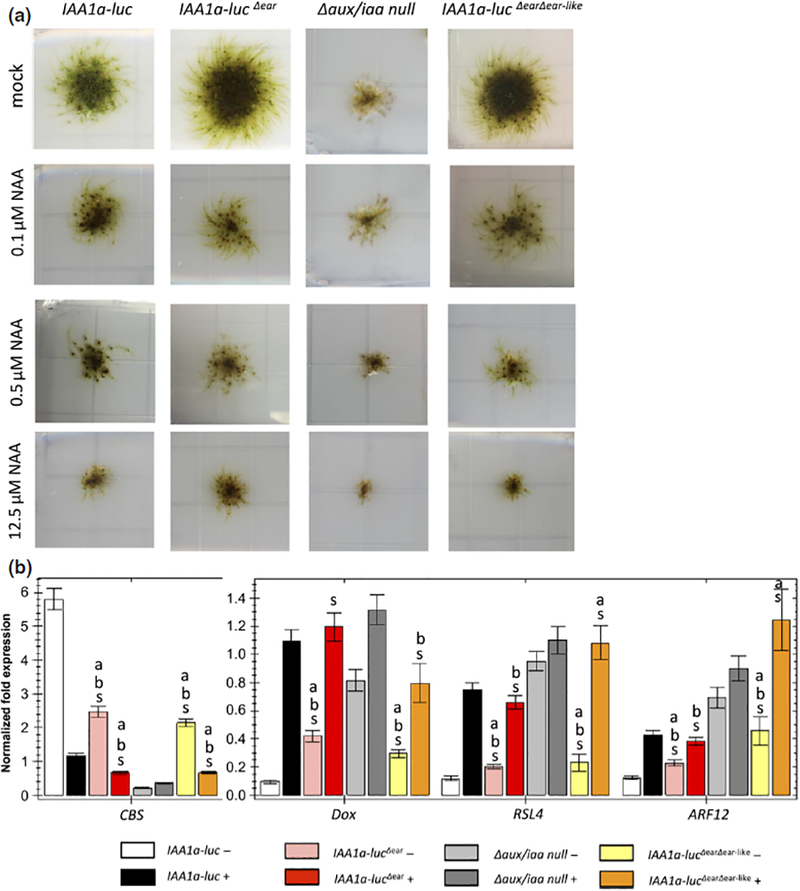
The EAR motif in the Physcomitrella auxin/indole-3-acetic acid (Aux/IAA) contributes to repression, but is not essential. (a) The developmental phenotypes of the PpIAA1a-lucΔear/Δaux/iaa mutant relative to the wild-type and Δaux/iaa null mutant grown on medium supplemented with different levels of auxin. IAA1a-lucΔear displays reduced leafy gametophore formation on unsupplemented medium relative to the wild-type, but is much more robust than the null mutant, and retains responsiveness to auxin treatments. (b) Quantitative PCR of auxin-responsive marker genes in the above lines treated with mock and 10 μΜ IAA for 1 h. White and black, IAA1a-luc; pink, IAA1a-lucΔear; gray, Δaux/iaa null mutant; darker colors represent 10 μM IAA-treated samples. Normalized expression (ΔΔC(t) method using PpEF1a as a reference gene) is plotted as relative values. Error bar represents ± SE. ‘a’ indicates that difference from IAA1a-luc is significant at P < 0.05 (Student’s t-test), n = 4. ‘b’ indicates that difference from Δaux/iaa is significant at P < 0.05 (Student’s t-test), n = 4. ‘s’ indicates that difference between conditions is significant at P < 0.05 (Student’s t-test), n = 4.
When aligning the moss and Arabidopsis Aux/IAA sequences, we discovered another leucine-rich region between Domains I and II in PpIAA1a (LKVHL) which is similar to the EAR motif. We generated another transgenic line expressing a version of IAA1a lacking both the EAR motif and this second EAR-like sequence (IAA1aΔearΔear-like). This line was similar to the IAA1aΔear line in appearance (Fig. 4a), indicating that the EAR-like sequence did not contribute to repression.
These results indicate that one or more EAR-independent repression mechanisms exist in the moss Auxin signaling pathway. As complete knockout of IAA1a in the Δaux/iaa null background results in fully constitutive Auxin responses, these mechanisms must be mediated by some elements within the IAA1a protein.
Both monomeric and oligomeric Aux/IAAs repress transcription in moss
In addition to the EAR motif, it has been proposed that Aux/IAA oligomerization through the C-terminal PB1 domain is required for repression. To explore this possibility, we generated an iaa1b iaa2 line expressing a mutant IAA1a protein in which key amino acids in the basic (K362) and acidic (D435 and D439) centers of the PB1 domain were replaced with alanine (IAA1aK,OPCA). Yeast two-hybrid assay showed that these mutations disrupted IAA1a self-interaction as well as interaction with ARFs, as expected (Fig. S2b). Phenotypic characterization revealed that the IAA1aK,OPCA line had an Auxin constitutive phenotype similar to that of the Δaux/iaa null mutant, with stunted, undifferentiated and leafless filaments. It was also completely insensitive to exogenous Auxin with respect to both morphology and gene expression (Fig. 5a,b). Therefore, the three conserved amino acids in the IAA1a PB1 domain are required for its repressive function in vivo. This result confirms that the repressive function of the Aux/IAA protein is totally dependent on interactions involving the PB1 domain.
Fig. 5.
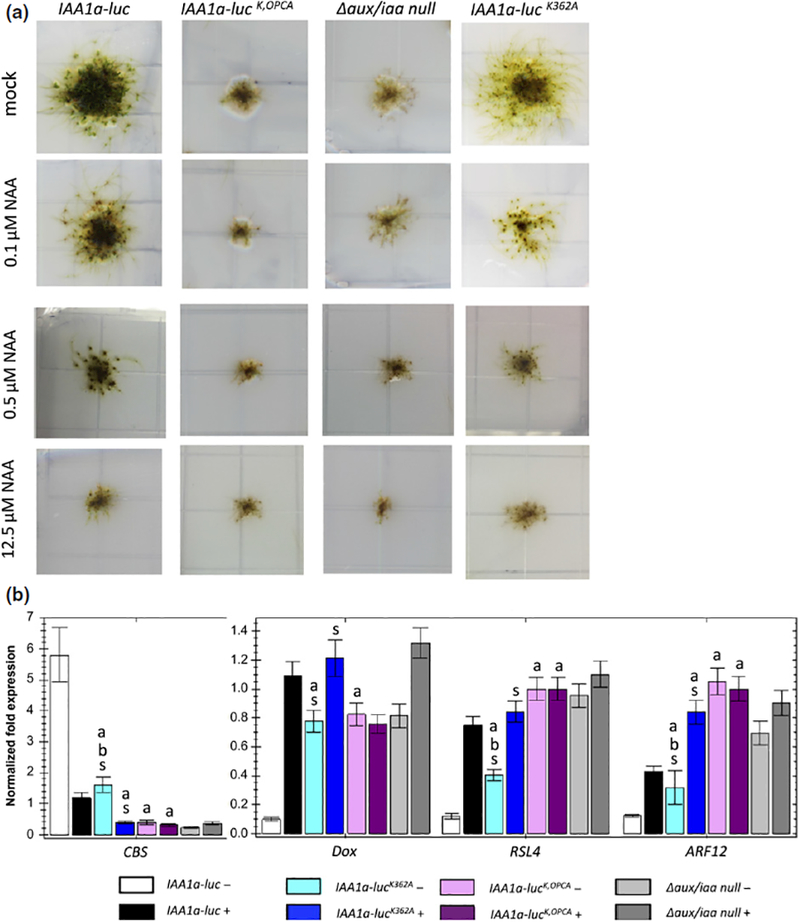
The PB1 domain plays an essential role in auxin/indole-3-acetic acid (Aux/IAA) function in Physcomitrella, but oligomerization is not required for repression. (a) The developmental phenotypes of K and opca single and double mutants relative to the wild type and Δaux/iaa null mutant grown on medium with different auxin levels. (b) Quantitative PCR of auxin-responsive marker genes in the above lines treated with mock and 10 μM IAA for 1 h. White and black, IAA1a-luc; blue, IAA1a-lucK362A; purple, IAA1a-lucK,OPCA; gray, Δaux/iaa null mutant; darker colors represent 10 μM IAA-treated samples. Error bar represents ± SE. ‘a’ indicates that difference from IAA1a-luc is significant at P < 0.05 (Student’s t-test), n = 4. ‘b’ indicates that difference from Δaux/iaa is significant at P < 0.05 (Student’s t-test), n = 4. ‘s’ indicates that difference between conditions is significant at P < 0.05 (Student’s t-test), n = 4.
We then characterized a line carrying a mutation in only one of the two interaction centers, IAA1aK362A. Again, yeast two-hybrid assays confirmed that this single mutation disrupts the IAA1a self-interaction, but not the ability to interact with an ARF protein (Fig. S2c). When grown on medium without Auxin, the IAA1aK362A mutant exhibited increased Auxin-related phenotypes relative to wild-type plants, including increased branching of caulonemata filaments, reduced leafy gametophores and darker brown-pigmented rhizoids. The expression level of the Auxin-responsive genes also indicated hypersensitivity to Auxin, correlating with the phenotypes (Fig. 5a,b).
These results confirm that oligomerization of the Aux/IAAs contributes to repressive function, as reported previously (Korasick et al., 2014; Pierre-Jerome et al., 2016). However, the phenotype of PpIAA1aK362A is much less severe than that of the Δaux/iaa mutant. When grown on Auxin medium, the PpIAA1aK362A mutant displays a further increase in rhizoid formation, the complete elimination of leafy gametophores and induction of marker gene expression, indicating that an IAA1a monomer represses transcription. These results are consistent with an earlier study in Arabidopsis (Pierre-Jerome et al., 2016), and demonstrate that, although Aux/IAA oligomerization is required for full activity as an Auxin signaling repressor, an Aux/IAA protein that is unable to oligomerize is also able to act as a repressor of the pathway.
Discussion
The clear Auxin-related phenotypes and reduced Aux/IAA gene redundancy make Physcomitrella an excellent system for the study of Auxin signaling. The fact that the Aux/IAA proteins in moss and Arabidopsis share conserved structural motifs makes it possible to combine the insights gained from previous Arabidopsis studies with the advantage of a simple genetic background in moss.
Our analysis indicates that the three members of the moss Aux/IAA family function redundantly. Although the IAA1a/b proteins are only 70% identical to IAA2, the loss of IAA1b and IAA2 has little effect on phenotype. Further, we found that the expression of IAA1a increased when the other two genes were deleted. As all three genes are regulated by Auxin and the ARF proteins, this compensation probably reflects negative feedback regulation of the Aux/IAA genes. Compensatory behavior may also partially explain the apparent redundancy of the Aux/IAA genes in Arabidopsis (Overvoorde et al., 2005).
The Aux/IAA proteins have two major roles in Auxin signaling; they function in Auxin perception as part of the co-receptor complex, and they act to repress transcription through an interaction with the ARF proteins. A number of studies have established that variation in the Aux/IAA degron sequence affects the stability of the protein (Dreher et al., 2006; Pierre-Jerome et al., 2013), and that this variation has an important role in the plant (Guseman et al., 2015). In addition, in a previous study, we have shown that different Arabidopsis AFB–Aux/IAA co-receptor pairs have distinct affinities for Auxin in vitro that are largely determined by the Aux/IAA protein (Calderon Villalobos et al., 2012). In particular, we found that the Kd of TIR1-IAA7 for Auxin was 10-fold lower than that of TIR1-IAA12, and that much of this difference was determined by variation in the degron motif. We proposed that the difference in Kd might act to expand the effective concentration range of the hormone. However, it was difficult to test this idea in Arabidopsis because of redundancy in the Aux/IAA family. The results described here reveal that differences in Auxin affinity between the IAA7 and IAA12 co-receptors are likely to have biological significance. At low Auxin levels (0.2 μM), IAA1a-lucdegVR is degraded, implying that the AFB-IAA1a-lucdegVR co-receptor pair, analogous to AFB–IAA7, binds Auxin at this concentration. By contrast, IAA1a-lucdegIG is stable, suggesting that AFB–IAA1a-lucdegIG, analogous to AFB–IAA12, has a lower affinity for Aux. As Auxin levels increase (20 μM), the AFB–IAA1a-lucdegIG–Auxin complex forms and degradation commences. Thus, in cells expressing both of these proteins, the active hormone concentration range is increased, relative to just one isoform. Importantly, these lines also differ in their Auxin transcriptional response and in long-term developmental outcomes. Although further studies in Arabidopsis are required to establish the importance of differences in Auxin affinity, our results suggest that co-receptor pairs with distinct biochemical properties can contribute to the complexity of Auxin signaling.
Based on previous studies, it is thought the Aux/IAA repression requires oligomerization and recruitment of the co-repressor TPL through the EAR domain (Szemenyei et al., 2008; Korasick et al., 2014). Because the moss system allows us to directly test the function of different Aux/IAA domains in the native context, we are able to obtain a more sophisticated understanding of Aux/IAA activity. Our results show that both the EAR motif and oligomerization contribute to repression, but neither are essential. Indeed, it is possible that Aux/IAA binding to ARF is sufficient for repression. Consistent with this model, a recent study has shown that activity of the Arabidopsis ARF protein MONOPTEROS requires binding of SWI/SNF chromatin remodeling ATPases (Wu et al., 2015). Further, the accumulation of the stabilized Aux/IAAs AXR3 and BDL reduces this interaction, suggesting that Aux/IAAs may repress transcription simply by binding to the ARF and inhibiting SWI/SNF binding. TPL may then sustain repression through chromatin modification, perhaps by recruiting histone deacetylases (Szemenyei et al., 2008).
Our results are also reminiscent of a recent study of the SMAX1-LIKE (SMXL) family of proteins (Liang et al., 2016). The SMXLs are repressors of strigolactone signaling that are degraded in response to the hormone strigolactone (Lumba et al., 2017). Because the SMXLs have an EAR domain, it has been proposed that they are transcriptional repressors. However, like the moss Aux/IAAs, the EAR domain contributes to SMXL function, but is not essential, suggesting that SMXLs either repress transcription through a different mechanism and/or repress the strigolactone response independently of their effects on transcription (Liang et al., 2016).
The complexity of the Aux/IAA gene family has hampered genetic studies of this important group of genes. By generating Physcomitrella lines that have a single mutant copy of the IAA1a gene, we have addressed the function of various subdomains within the protein. Our results provide a novel and more nuanced view of Aux/IAA function.
Supplementary Material
Acknowledgements
The authors would like to thank Drs Meirav Lavy and Michael Prigge for yeast two-hybrid plasmids and helpful advice. This work was supported by grants from the National Institutes of Health (NIH) (GM43644), the Gordon and Betty Moore Foundation and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Author contributions
This work was conceived by S.T. and M.E. S.T. carried out all the experiments. S.T. and M.E. wrote the manuscript.
Supporting Information
Additional Supporting Information may be found online in the Supporting Information tab for this article:
References
- Ashton NW, Grimsley NH, Cove DJ. 1979. Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427—435. [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M et al. 2013. A map of cell type-specific auxin responses. Molecular Systems Biology 9: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing ofauxin. Nature Chemical Biology 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiology 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Thelander M, Landberg K, Staldal V, Nilsson A, Johansson M, Valsecchi I, Pederson ER, Kowalczyk M, Ljung K et al. 2010. Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development 137: 1275–1284. [DOI] [PubMed] [Google Scholar]
- Guseman JM, Hellmuth A, Lanctot A, Feldman TP, Moss BL, Klavins E, Calderón Villalobos LI, Nemhauser JL. 2015. Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. 2012. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiology 160: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proceedings of the National Academy of Sciences, USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tao S, Shain S, Kuo A, Kirchsteiger K, Estelle M. 2016. Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. eLife 5: 13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tigyi K, Estelle M. 2012. The cyclophilin DIAGEOTROPICA has a conserved role in auxin signaling. Development 139: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O. 2016. SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28: 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49: 387–400. [PubMed] [Google Scholar]
- Lumba S, Holbrook-Smith D, McCourt P. 2017. The perception of strigolactones in vascular plants. Nature Chemical Biology 13: 599–606. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M. 2000. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Research 7: 9–17. [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H et al. 2005. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Lanctot A, Hageman A, Nemhauser JL. 2016. Functional analysis of molecular interactions in synthetic auxin response circuits. Proceedings of the National Academy of Sciences, USA 113: 11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Nemhauser JL. 2013. Tuning the auxin transcriptional response. Journal of Experimental Botany 64: 2557–2563. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Bezanilla M. 2010. Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137: 3535–3543. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M. 2010. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Current Biology 20: 1907–1912. [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Zhao Y. 2016. Auxin perception and downstream events. Current Opinion in Plant Biology 33: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annual Review of Plant Biology 67: 539. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Annals of Botany 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. 2015. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: 09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


