Abstract
Objectives
We evaluated the antimicrobial susceptibility and ribotypes of Clostridium difficile isolates from participants in a Phase 2 study of ridinilazole, a novel targeted-spectrum agent for treatment of C. difficile infection.
Methods
Participants received ridinilazole (200 mg twice daily) or vancomycin (125 mg four times daily) for 10 days (ClinicalTrials.gov: NCT02092935). The MICs of ridinilazole and comparators for C. difficile isolates from stool samples were determined by agar dilution. Toxin gene profiling was performed by multiplex PCR and ribotype identification by capillary electrophoresis.
Results
Eighty-nine isolates were recovered from 88/100 participants (one participant had two strains at baseline). The median colony count (cfu/g stool) was 1.9 × 104 (range: 2.5 × 102–7.0 × 106). Twelve participants (three received ridinilazole and nine received vancomycin) experienced recurrence, confirmed by immunoassays for free toxin in stool samples. The ribotype of eight out of nine isolates obtained at recurrence matched those of the initial isolates. All isolates, including those obtained at recurrence, were susceptible to ridinilazole within the expected range [median (range) MIC: 0.12 (0.06–0.5) mg/L]. The median (range) vancomycin MIC was 1 (0.5–4.0) mg/L. At baseline, 13.6% and 13.3% of samples in the ridinilazole and vancomycin groups were positive for VRE, increasing to 23.7% and 29.7% by day 40, respectively. Common ribotypes included 014-20 (14 isolates), 027 (13), 106 (7), 002 (7), 078-126 (4), 001 (4), 087 (3) and 198 (3). Toxin gene profiling of nearly all baseline isolates (98.9%) revealed a binary toxin gene (cdtA/cdtB) prevalence of 35%.
Conclusions
Ridinilazole potently inhibited recovered C. difficile isolates. Recurrence was not associated with altered susceptibility.
Introduction
Clostridium difficile infection (CDI) is a leading cause of morbidity and mortality and has seen a significant increase in global incidence, driven partly by the emergence of fluoroquinolone-resistant NAP1 (ribotype 027) strains.1,2 Although the current mainstay antibiotics, vancomycin and metronidazole, are generally effective at treating initial infection, both agents are associated with unacceptably high rates of recurrent disease,3 with each recurrent episode being associated with increased morbidity and mortality as well as a heightened economic burden.4,5 Approximately 30% of patients experience a repeat infection following an initial episode, and the risk of recurrence doubles following a third episode.6
Ridinilazole (formerly SMT19969) is a novel targeted-spectrum oral antimicrobial under development for the treatment of CDI and for reducing the recurrence of CDI.7 Through fluorescent labelling, confocal microscopy and scanning electron microscopy studies, ridinilazole has been shown to impact cell division and septum formation.7,8 Preclinical efficacy studies have demonstrated its narrow spectrum of activity and potent bactericidal effect against a variety of clinical isolates of C. difficile, including less frequently isolated strains and isolates with varying antimicrobial resistance phenotypes.9–13 Ridinilazole has been shown in a hamster model to be effective both at treating primary infections and preventing recurrent infections.14 A Phase 1 study in healthy volunteers demonstrated single or multiple doses of ridinilazole to be safe and well tolerated, with negligible plasma concentrations and minimal impact on normal gut microbiota.15 A recent Phase 2 proof-of-concept clinical study compared the efficacy and safety of ridinilazole with that of vancomycin and demonstrated it to be non-inferior (15% margin) with regard to the primary efficacy measure, sustained clinical response (SCR), defined as clinical cure (≤3 unformed bowel movements within a 24 h period) at the test of cure (TOC) visit and lack of recurrence within 30 days of the end of treatment.16 Statistical superiority at the pre-specified 10% level was also demonstrated with ridinilazole. Improved SCR rates persisted across patient subgroups based on age, baseline severity, history of recurrence and use of concomitant antibiotics at baseline. Moreover, ridinilazole was well tolerated and had an adverse event profile similar to that of vancomycin. We now report the microbiological findings of this Phase 2 trial, focusing on the antimicrobial susceptibility and ribotyping of isolates from participants with primary and recurrent infections.
Patients and methods
Study design
This Phase 2, double-blind, randomized, active-controlled, parallel group design study (ClinicalTrials.gov: NCT02092935) was conducted at 33 centres in the USA and Canada, with study sites primarily consisting of hospitals and outpatient clinics.16
Ethics
Institutional review boards at each centre provided ethics approval. Ethical principles as set forth in the Declaration of Helsinki and all principles of good clinical practice were complied with. Written informed consent was obtained from all participants.
Study procedures
One hundred participants randomized in a 1:1 ratio received either ridinilazole (200 mg orally twice daily) or vancomycin (125 mg orally four times daily) for 10 days. A complete description of all study procedures has been published previously.16
Sample collection
Faecal samples collected at baseline, on days 5, 10, 25 and 40, and at recurrence were used for the culture and isolation of C. difficile vegetative cells and spores. Quantitative counts of spores and vegetative cells were conducted on all vegetative isolates and germinated spores as detailed below. Isolates underwent susceptibility testing against ridinilazole, vancomycin, fidaxomicin, metronidazole and other comparators. Isolates at baseline and at recurrence were ribotyped by capillary electrophoresis and were subjected to toxin gene profiling by multiplex PCR to detect the presence or absence of tcdA, tcdB and cdtA/B as well as tcdC deletions (see methods below).
C. difficile isolation
Stool samples were diluted and plated on pre-reduced selective medium cycloserine–cefoxitin–fructose agar.17,18 Plates were incubated for 48 h in an anaerobic chamber (5% CO2/10% H2/85% N2) at 35 ± 2°C and observed for characteristic growth and colonial morphology. Colonies that were ∼4 mm in diameter, yellow and had a ground-glass appearance were enumerated, with colony counts being reported as cfu/g stool. To determine the spore count, the stool sample was ethanol shocked prior to dilution and plated on cycloserine–cefoxitin–fructose agar with lysozyme.19 After enumeration, representative colonies were subcultured onto anaerobic blood agar (CDC) for further identification and preparation of frozen stock culture. A proline disc test (Remel Products, Lenexa, KS, USA) and Gram stain were performed.20 Isolates that were proline-positive, Gram-positive bacilli were further identified using rapid methodology with the API20A system (bioMérieux Inc., Durham, NC, USA). Identified isolates were kept frozen in skim milk at −80°C for susceptibility testing and/or reference.
Susceptibility testing
Susceptibilities of the isolates were assessed against a panel of antimicrobial agents that included ridinilazole, fidaxomicin, vancomycin, metronidazole, moxifloxacin, clindamycin, tigecycline, rifaximin, rifampicin, linezolid, imipenem and chloramphenicol. Susceptibility was determined by agar dilution methodology as described in CLSI M11-A8.21,22 Inocula were prepared using direct colony suspension to achieve a turbidity equivalent to that of a 0.5 McFarland standard (∼107 cfu/mL for C. difficile). The antibiotic-containing plates were prepared on the day of the test. A Steer replicator was used to inoculate the agar plates, resulting in a deposit of ∼104 cfu on the surface of the agar. The plates were incubated at 35 ± 2°C in an anaerobic chamber (5% CO2/10% H2/85% N2) for 48 h.21 The following reference organisms were included with each susceptibility testing run: C. difficile ATCC 700057, C. difficile ATCC 43255, Bacteroides thetaiotaomicron ATCC 29741 and Staphylococcus aureus ATCC 29213. Tests were repeated when the MICs of the control organisms were outside of the CLSI acceptable range.
PCR- and capillary electrophoresis-based ribotyping
Templates for amplicon generation were obtained by growing C. difficile on anaerobic blood agar medium in an atmosphere of 5% CO2/10% H2/85% N2 at 37°C for 48 h and then transferring 3–5 colonies to 200 μL of 10% Chelex 100 in 10 mM Tris–HCl/1 mM EDTA (pH 8) buffer. Samples were boiled for 15 min and centrifuged briefly (5 min at 3400 rpm). Template DNA was then diluted 1:10 in 10 mM Tris–HCl/1 mM EDTA (pH 8) buffer and stored at −20°C until ready for use. Ribotyping was then performed as previously described.23 Amplicons were stored at −20°C until ready to ship for fragment analysis. Plates were sent to the University of Michigan sequencing core facility for capillary electrophoresis (http://seqcore.brcf.med.umich.edu). Ribotypes were assigned as previously described,23 using an online analysis tool at the Walk Laboratory at Montana State University (http://thewalklab.com/tools).24 This database can identify 116 distinct C. difficile ribotypes and is cognate with the ribotyping database in the UK (Leeds Reference Laboratory) for 35 of the most clinically relevant ribotypes based on prevalence in the USA.
Toxin gene profiling
Using template DNA as isolated above for ribotyping, toxin gene profiles for C. difficile were determined using PCR methodology as described by Persson et al.25,26 Isolates were tested in duplicate and non-concordant tests were repeated. The following controls were included with each test: (i) a non-toxigenic strain of C. difficile VPI 11186 (ATCC 700057), with a PCR profile of tcdA−, tcdB−, cdtA−/B−, tcdC−, in which the entire PaLoc is absent and no functional cdt locus is present;27 (ii) C. difficile VPI 10463, with a PCR profile of tcdA+, tcdB+, cdtA−/cdtB−, with no detected deletion in the tcdC region;28,29 and (iii) C. difficile R20291, with a PCR profile of tcdA+, tcdB+, cdtA+/B+, with an 18 bp deletion in the tcdC hypervariable region.30 Loss of PaLoc was confirmed in non-toxigenic strains by PCR performed using Lok1/Lok3 primers as previously described.31
Identification and characterization of VRE
Samples were plated on selective-medium bile esculin azide agar with vancomycin (6 mg/L) for the isolation of VRE.32,33 Cultures were incubated at 35°C for up to 72 h in an atmosphere of 7% CO2 and then examined for a dark brown to black colour, indicating a positive esculin reaction. All esculin-positive, Gram-positive cocci were subcultured onto tryptic soy agar (Remel, Lenexa, KS, USA) for identification and preparation of frozen stock culture. A catalase test was then performed on all α- or non-haemolytic isolates. A test for confirmation of pyroglutamyl (PYR) aminopeptide activity and a test for confirmation of esculin hydrolysis (Visi-spot discs, Thermo Scientific Inc., Waltham, MA, USA) were performed on catalase-negative colonies. The PYR-positive isolates were presumptively identified as enterococci. Isolates were stored at −80°C for reference. Susceptibility of the enterococcal isolates to vancomycin was determined by agar dilution methodology as described in CLSI document M07-A10.34
Results
Vegetative isolates
Table 1 shows the rate of isolation and colony counts by treatment group of C. difficile isolated from stool at study entry (day −1), days 5, 10, 25 and 40, and at recurrence. Of the 100 randomized participants, 89 provided a baseline sample and C. difficile was successfully isolated from 43 participants at baseline in the ridinilazole group (2 strains from 1 participant) and 45 participants in the vancomycin group, for a total of 89 strains. Median counts and ranges at baseline were virtually identical in each arm. During the course of study drug dosing, there was a rapid decline in the proportion of participants from whom C. difficile could be isolated. By day 5, 12.2% of samples from the ridinilazole-treated participants had C. difficile recovered, compared with 16.7% of samples from vancomycin-treated participants. Day 5 median colony counts from the ridinilazole-treated participants from whom C. difficile was recovered were 3.85 log10 cfu/mL, compared with 4.60 log10 cfu/mL in the vancomycin arm. At day 40, 7.9% of the ridinilazole-treated participants had C. difficile isolated, compared with 10.8% of the vancomycin-treated participants.
Table 1.
Isolation of C. difficile vegetative cultures and spores over time comparing ridinilazole and vancomycin treatment groups
| timepoint | Vegetative cultures |
Spores |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| number of samples | number positive | % C. difficile isolated | log10 cfu/g stool |
number of samples | number positive | % C. difficile isolated | log10 cfu/g stool |
|||
| range | median | range | median | |||||||
| Ridinilazole treatment group | ||||||||||
| day −1 | 44 | 43 | 97.7 | ≤2.4–6.45 | 4.28 | 44 | 19 | 43.2 | 2.40–6.86 | 4.66 |
| day 5 | 41 | 5 | 12.2 | 2.70–5.36 | 3.85 | 41 | 7 | 17.1 | 2.88–5.51 | 4.11 |
| day 10 | 41 | 3 | 7.3 | 2.52–5.99 | 3.77 | 41 | 5 | 12.2 | 2.90–6.38 | 4.32 |
| day 25 | 41 | 3 | 7.3 | 3.23–5.18 | 3.70 | 41 | 12 | 29.3 | 2.66–5.72 | 3.55 |
| day 40 | 38 | 3 | 7.9 | 3.96–4.48 | 4.38 | 38 | 7 | 18.4 | 2.49–5.26 | 3.92 |
| recurrence | 3 | 3 | 100.0 | 3.56–4.20 | 3.72 | 3 | 0 | 0 | all ≤2.4 | NA |
| Vancomycin treatment group | ||||||||||
| day −1 | 45 | 45 | 100.0 | ≤2.4–6.85 | 4.26 | 44 | 16 | 36.4 | 2.43–6.98 | 4.38 |
| day 5 | 42 | 7 | 16.7 | 3.32–5.28 | 4.60 | 42 | 5 | 11.9 | 3.45–6.04 | 4.91 |
| day 10 | 42 | 4 | 9.5 | 2.56–4.18 | 3.64 | 41 | 4 | 9.8 | 3.23–6.23 | 3.61 |
| day 25 | 38 | 5 | 13.2 | ≤2.4–4.59 | 3.08 | 38 | 15 | 39.5 | ≤2.4–6.36 | 3.96 |
| day 40 | 37 | 4 | 10.8 | 3.08–5.85 | 4.57 | 36 | 11 | 30.6 | ≤2.4–5.32 | 3.96 |
| recurrence | 6a | 5 | 83.3 | 3.63–5.85 | 4.20 | 5b | 4 | 80 | 2.72–5.67 | 4.98 |
NA, not applicable.
There were nine recurrences, but only six recurrence samples were available for testing of vegetative cultures.
There were nine recurrences, but only five recurrence samples were available for spore testing.
Spore isolation
Data on isolation of C. difficile spores are also shown in Table 1. At baseline, 43.2% of samples from ridinilazole-treated participants had spores isolated, compared with 36.4% of samples from vancomycin-treated participants. The median counts were similar, although participants in the ridinilazole arm had marginally higher spore counts at baseline (4.66 versus 4.38 log10 cfu/mL for ridinilazole and vancomycin, respectively). The proportion of samples with detectable spore counts declined dramatically over the time of treatment to 12.2% and 9.8% on day 10 for the ridinilazole and vancomycin arms, respectively. By day 25 the proportion of spores increased in both arms (29.3% and 39.5% for ridinilazole and vancomycin, respectively). At day 40 the proportion of participants in the ridinilazole arm who had spores detected was 18.4%, compared with 30.6% in the vancomycin arm (P = 0.10).
Antimicrobial susceptibility
Table 2 presents the MICs of both study drugs and selected comparators for the C. difficile isolates recovered at baseline and at recurrence by treatment group for four selected agents. The distribution of MICs for ridinilazole was narrow, with an MIC90 of 0.25 mg/L and a range of 0.06–0.5 mg/L, consistent with the previously reported susceptibility of C. difficile to ridinilazole.10,12,13 The MIC90 for vancomycin was 2 mg/L, with a range of 0.5–4 mg/L. In Table S1 (available as Supplementary data at JAC Online) we list the susceptibility of all isolates against all agents tested.
Table 2.
Susceptibility of C. difficile isolates at baseline and at recurrence by treatment arm
| n | MIC (mg/L) | Ridinilazole | Vancomycin | Metronidazole | Fidaxomicin | |
|---|---|---|---|---|---|---|
| Ridinilazole group vegetative isolates | ||||||
| day −1 | 44 | MIC50 | 0.12 | 1 | 0.5 | 0.12 |
| MIC90 | 0.25 | 2 | 2 | 0.5 | ||
| range | 0.06–0.5 | 1–4 | 0.12–4 | 0.06–1 | ||
| recurrence | 3 | range | 0.12–0.25 | 2–4 | 0.25–2 | 0.12–0.5 |
| Vancomycin group vegetative isolates | ||||||
| day −1 | 45 | MIC50 | 0.12 | 1 | 0.25 | 0.25 |
| MIC90 | 0.5 | 2 | 1 | 0.5 | ||
| range | 0.06–0.5 | 0.5–2 | 0.12–2 | 0.06–1 | ||
| recurrence | 5 | MIC50 | 0.12 | 1 | 0.25 | 0.25 |
| range | 0.12–0.5 | 1–2 | 0.12–0.5 | 0.12–0.5 | ||
Ribotyping of baseline isolates
The distribution of ribotypes of isolates obtained at baseline is shown in Table 3, which presents the data by treatment arm. Ribotype 014-020 was most commonly seen, with eight isolates in the ridinilazole arm and six in the vancomycin arm belonging to this ribotype. Ribotype 027 was seen in 13 of the enrolled participants. There was a slightly higher proportion of 027 in the ridinilazole arm [8/44 (18%)] compared with the vancomycin arm [5/45 (11%)]. There was differential distribution of certain ribotypes between the vancomycin and ridinilazole arms: ribotype 001 (four in the vancomycin arm versus zero in the ridinilazole arm), 078-126 (four versus zero) and 106 (five versus two). Four patients (two in each arm) had ribotypes novel to our database (see the Patients and methods section) and there were 19 different ribotypes that were isolated only once, for a total of 37 different ribotypes.
Table 3.
Comparison of ribotype profiles of baseline isolates in the ridinilazole and vancomycin arms
| Ribotype | Ridinilazole (N = 44), n (%) | Vancomycin (N = 45), n (%) | Total (N = 89), n (%) |
|---|---|---|---|
| 014-020 | 8 (18.2) | 6 (13.3) | 14 (15.7) |
| 027 | 8 (18.2) | 5 (11.1) | 13 (14.6) |
| 106 | 2 (4.5) | 5 (11.1) | 7 (7.9) |
| 002 | 4 (9.1) | 3 (6.7) | 7 (7.9) |
| 001 | 0 (0.0) | 4 (8.9) | 4 (4.5) |
| 078-126 | 0 (0.0) | 4 (8.9) | 4 (4.5) |
| Novel | 2 (4.5) | 2 (4.4) | 4 (4.5) |
| 087 | 3 (6.8) | 0 (0.0) | 3 (3.4) |
| 198 | 2 (4.5) | 1 (2.2) | 3 (3.4) |
| Singletons (observed once) | 15 (34.1) | 15 (33.3) | 30 (33.7) |
Toxin gene profiles
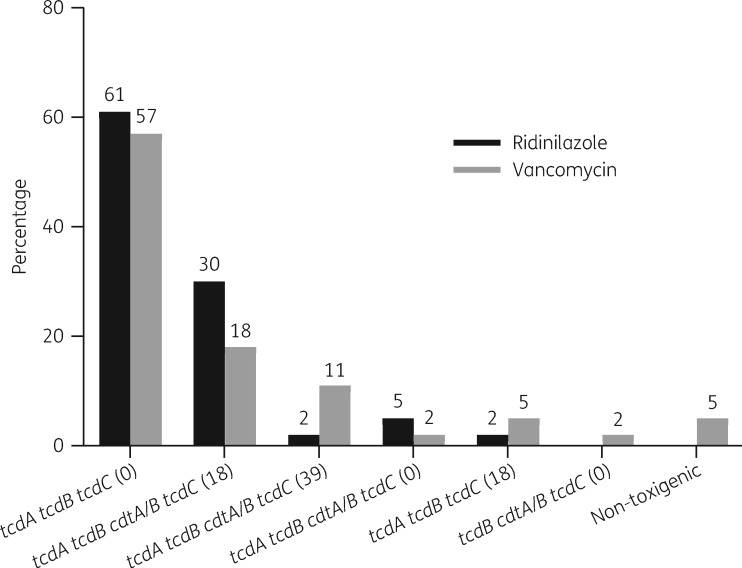
The toxin gene profiles by proportion at the time of diagnosis are shown in Figure 1. The most common toxin profiles observed were tcdA tcdB without (0 bp) any deletion in tcdC and tcdA tcdB cdtA/B with an 18 bp deletion in tcdC. The proportions were similar between the ridinilazole and vancomycin arms. There were two non-toxigenic strains, both of which were seen in the vancomycin arm. This may be due to infection with multiple strains, which has been shown to occur in 16% of CDI cases.35 Notable toxin profiles were a strain with no tcdA signal and six strains with ∼39 bp deletions in tcdC, four of which belonged to ribotype 078-126. There were no strains with only binary toxin, tcdB deletions or tcdC with ∼54 bp deletions.
Figure 1.
Percentage of toxin gene profiles at study entry by treatment group. Values in parentheses refer to the length of nucleotide base pairs missing/deleted in tcdC.
Identification and characterization of VRE
The rates of isolation of VRE at baseline were comparable in each treatment arm, with 13.6% and 13.3% of samples from ridinilazole- and vancomycin-treated participants being positive for VRE. Over time, the proportion of patients in whom VRE could be recovered increased in both arms, with 23.7% and 29.7% recovered at day 40 in the ridinilazole and vancomycin arms, respectively.
Recurrence isolates
A total of 12 participants (3 received ridinilazole and 9 received vancomycin) had a recurrence of CDI as diagnosed by a positive free-toxin enzyme immunoassay, with 9 of these participants providing a stool sample (3 received ridinilazole and 6 received vancomycin). Vegetative C. difficile was successfully isolated from samples from 8 of the 9 participants with recurrence from whom samples were available (3 received ridinilazole and 5 received vancomycin) (Table 1). Colony counts at the time of recurrence were similar to the baseline counts. All vegetative isolates recovered at recurrence maintained susceptibility to ridinilazole comparable with that of the baseline isolates. Of the 8 participants with strains isolated at baseline and at recurrence, ribotypes and toxin gene profiles were the same except for one ridinilazole-treated participant, in whom a recurrence isolate belonged to a novel ribotype (the baseline ribotype was 014-020).
No spores could be isolated from the ridinilazole-treated participants at the time of recurrence, whereas four of five participants in the vancomycin arm had spores detected (P = 0.07; Fisher’s exact test). One participant had insufficient sample to detect spores.
Discussion
In this small Phase 2 study, in which the two treatment groups were evenly matched at baseline from a microbiological perspective, C. difficile isolates obtained at baseline and at multiple timepoints during follow-up remained susceptible to both ridinilazole and vancomycin. There was no evidence for development of resistance to ridinilazole during the course of therapy and there was an absence of cross-resistance to ridinilazole and the other antimicrobial agents tested.
Ribotypes were evenly distributed between the two treatment groups. The overall percentage of patients with 027 (14.6%) was lower than that seen in most centres in a recent multicentre study in North America,36 and may reflect a declining incidence, as was reported in England and the USA.37,38 The exact reasons for this decline in incidence of 027 remain unclear. There was a wide array of ribotypes seen among participants enrolled in the trial, including a number of novel ribotypes. Where known, the majority of participants with recurrence were seen to have the same ribotype at recurrence as they did at baseline, suggesting that most recurrences were relapses of the initial infection rather than reinfection.
There were no significant differences between ridinilazole- and vancomycin-treated participants with respect to reductions in vegetative and spore counts during the course of therapy. At day 40 and at recurrence we noted only a small number of patients in the ridinilazole arm from whom spores could be isolated. This was unexpected, but with the small number of subjects studied, firm conclusions regarding this finding cannot be drawn. In addition, there were no significant differences in the presence or absence of VRE during therapy, although there was a trend towards an increasing presence of VRE in the post-dosing period in vancomycin-treated participants compared with ridinilazole-treated participants.
In summary, this study demonstrates the potent in vitro activity of the novel antimicrobial agent ridinilazole against C. difficile, with no emergence of resistance during treatment. Further clinical development of ridinilazole for the treatment of CDI is warranted.
Supplementary Material
Acknowledgements
These findings were presented in part at ASM Microbe, Boston, MA, USA, 2016 (Abstract 442).
We are grateful to the participants and investigators of this Phase 2 study (NCT02092935).
Funding
This work was supported by Summit Therapeutics Plc, Abingdon, UK and by a Translation Award from the Wellcome Trust (grant number 099444). The medical writing assistance (see below) was funded by Summit Therapeutics Plc, Abingdon, UK.
Transparency declarations
D. R. S. has received grants from Summit Therapeutics Plc, Actelion Pharmaceuticals and Merck, and is a consultant for Merck, Takeda and Sequiris. C. M. T. is a consultant for Summit Therapeutics Plc. R. J. V. is an employee of, and holds share options with, Summit Therapeutics Plc. All other authors: none to declare.
Medical writing assistance was provided by Prasad Kulkarni and Alexandra Rayser of the Healthcare Alliance Group, Voorhees, NJ, USA.
Author contributions
D. R. S. was involved in conceptualization of the study, funding acquisition, data analysis, project administration and writing of the manuscript. L. A. M. was involved in conceptualization of the study, data analysis and writing of the manuscript. C. M. T. was involved in conceptualization of the study, funding acquisition, data analysis, project administration and writing of the manuscript. J. C. and J. W. were involved in the investigation, methodology and writing of the manuscript. S. T. W. was involved in conceptualization of the study, data analysis and writing of the manuscript. R. J. V. was involved in conceptualization of the study, funding acquisition, data analysis and writing of the manuscript. All authors reviewed the manuscript and approved its submission for publication.
Supplementary data
Table S1 is available as Supplementary data at JAC Online.
References
- 1. Lessa FC, Mu Y, Bamberg WM. et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372: 825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He M, Miyajima F, Roberts P. et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 2013; 45: 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peterfreund GL, Vandivier LE, Sinha R. et al. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One 2012; 7: e46966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shields K, Araujo-Castillo RV, Theethira TG. et al. Recurrent Clostridium difficile infection: from colonization to cure. Anaerobe 2015; 34: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghantoji SS, Sail K, Lairson DR. et al. Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect 2010; 74: 309–18. [DOI] [PubMed] [Google Scholar]
- 6. Kelly CP. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin Microbiol Infect 2012; 18 Suppl 6: 21–7. [DOI] [PubMed] [Google Scholar]
- 7. Vickers RJ, Tillotson G, Goldstein EJ. et al. Ridinilazole: a novel therapy for Clostridium difficile infection. Int J Antimicrob Agents 2016; 48: 137–43. [DOI] [PubMed] [Google Scholar]
- 8. Bassères E, Endres BT, Khaleduzzaman M. et al. Impact on toxin production and cell morphology in Clostridium difficile by ridinilazole (SMT19969), a novel treatment for C. difficile infection. J Antimicrob Chemother 2016; 71: 1245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldstein EJ, Citron DM, Tyrrell KL. et al. Comparative in vitro activities of SMT19969, a new antimicrobial agent, against Clostridium difficile and 350 Gram-positive and Gram-negative aerobic and anaerobic intestinal flora isolates. Antimicrob Agents Chemother 2013; 57: 4872–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldstein EJ, Citron DM, Tyrrell KL.. Comparative in vitro activities of SMT19969, a new antimicrobial agent, against 162 strains from 35 less frequently recovered intestinal Clostridium species: implications for Clostridium difficile recurrence. Antimicrob Agents Chemother 2014; 58: 1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corbett D, Wise A, Birchall S. et al. In vitro susceptibility of Clostridium difficile to SMT19969 and comparators, as well as the killing kinetics and post-antibiotic effects of SMT19969 and comparators against C. difficile. J Antimicrob Chemother 2015; 70: 1751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freeman J, Vernon J, Vickers R. et al. Susceptibility of Clostridium difficile isolates of varying antimicrobial resistance phenotypes to SMT19969 and 11 comparators. Antimicrob Agents Chemother 2016; 60: 689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baines SD, Crowther GS, Freeman J. et al. SMT19969 as a treatment for Clostridium difficile infection: an assessment of antimicrobial activity using conventional susceptibility testing and an in vitro gut model. J Antimicrob Chemother 2015; 70: 182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sattar A, Thommes P, Payne L. et al. SMT19969 for Clostridium difficile infection (CDI): in vivo efficacy compared with fidaxomicin and vancomycin in the hamster model of CDI. J Antimicrob Chemother 2015; 70: 1757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vickers R, Robinson N, Best E. et al. A randomised phase 1 study to investigate safety, pharmacokinetics and impact on gut microbiota following single and multiple oral doses in healthy male subjects of SMT19969, a novel agent for Clostridium difficile infections. BMC Infect Dis 2015; 15: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vickers RJ, Tillotson GS, Nathan R. et al. Efficacy and safety of ridinilazole compared with vancomycin for the treatment of Clostridium difficile infection: a phase 2, randomised, double-blind, active-controlled, non-inferiority study. Lancet Infect Dis 2017; 17: 735–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. George WL, Sutter VL, Citron D. et al. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol 1979; 9: 214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jousimies-Somer H, Summanen P, Citron DM. et al. Wadsworth-KTL Anaerobic Bacteriology Manual. Belmont, CA, USA: Star Pub, 2002. [Google Scholar]
- 19. Wilcox MH, Fawley WN, Parnell P.. Value of lysozyme agar incorporation and alkaline thioglycollate exposure for the environmental recovery of Clostridium difficile. J Hosp Infect 2000; 44: 65–9. [DOI] [PubMed] [Google Scholar]
- 20. Fedorko DP, Williams EC.. Use of cycloserine-cefoxitin-fructose agar and l-proline–aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J Clin Microbiol 1997; 35: 1258–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clinical and Laboratory Standards Institute. Methods for Antimicrobial Testing of Anaerobic Bacteria—Eighth Edition: Approved Standard M11-A8. CLSI, Wayne, PA, USA, 2012. [Google Scholar]
- 22. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. CLSI, Wayne, PA, USA, 2015. [Google Scholar]
- 23. Walk ST, Micic D, Jain R. et al. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis 2012; 55: 1661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinson JN, Broadaway S, Lohman E. et al. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. J Clin Microbiol 2015; 53: 1192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Persson S, Torpdahl M, Olsen KE.. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 2008; 14: 1057–64. [DOI] [PubMed] [Google Scholar]
- 26. Persson S, Jensen JN, Olsen KE.. Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J Clin Microbiol 2011; 49: 4299–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moncrief JS, Zheng L, Neville LM. et al. Genetic characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates by PCR. J Clin Microbiol 2000; 38: 3072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carter GP, Lyras D, Allen DL. et al. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J Bacteriol 2007; 189: 7290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sullivan NM, Pellett S, Wilkins TD.. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun 1982; 35: 1032–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stabler RA, He M, Dawson L. et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 2009; 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Braun V, Hundsberger T, Leukel P. et al. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 1996; 181: 29–38. [DOI] [PubMed] [Google Scholar]
- 32. Sahm DF, Free L, Smith C. et al. Rapid characterization schemes for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol 1997; 35: 2026–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Isenberg HD. Clinical Microbiology Procedures Handbook. Washington, DC, USA: American Society of Microbiology, 1992. [Google Scholar]
- 34. Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Tenth Edition: Approved Standard M07-A10. CLSI, Wayne, PA, USA, 2015. [Google Scholar]
- 35. Behroozian AA, Chludzinski JP, Lo ES. et al. Detection of mixed populations of Clostridium difficile from symptomatic patients using capillary-based polymerase chain reaction ribotyping. Infect Control Hosp Epidemiol 2013; 34: 961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waslawski S, Lo ES, Ewing SA. et al. Clostridium difficile ribotype diversity at six health care institutions in the United States. J Clin Microbiol 2013; 51: 1938–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burnham CA, Carroll KC.. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 2013; 26: 604–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Snydman DR, McDermott LA, Jenkins SG. et al. Epidemiologic trends in Clostridium difficile isolate ribotypes in United States from 2010 to 2014. Open Forum Infect Dis 2017; 4 Suppl 1: S391. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.