Abstract
Streptococcus gordonii and Streptococcus sanguinis are typically found among the normal oral microbiota but can also cause infective endocarditis. These organisms express cell surface serine-rich repeat adhesins containing “Siglec-like” binding regions (SLBRs) that mediate attachment to α2-3-linked sialic acids on human glycoproteins. Two known receptors for the Siglec-like adhesins are the salivary mucin MG2/MUC7 and platelet GPIbα, and the interaction of streptococci with these targets may contribute to oral colonization and endocarditis, respectively. The SLBRs display a surprising diversity of preferences for defined glycans, ranging from highly selective to broader specificity. In this report, we characterize the glycoproteins in human plasma recognized by four SLBRs that prefer different α2-3 sialoglycan structures. We found that the SLBRs recognize a surprisingly small subset of plasma proteins that are extensively O-glycosylated. The preferred plasma protein ligands for a sialyl-T antigen-selective SLBR are proteoglycan 4 (lubricin) and inter-alpha-trypsin inhibitor heavy chain H4. Conversely, the preferred ligand for a 3′sialyllactosamine-selective SLBR is glycocalicin (the extracellular portion of platelet GPIbα). All four SLBRs recognize C1 inhibitor but detect distinctly different glycoforms of this key regulator of the complement and kallikrein protease cascades. The four plasma ligands have potential roles in thrombosis and inflammation, and each has been cited as a biomarker for one or more vascular or other diseases. The combined results suggest that the interaction of Siglec-like adhesins with different subsets of plasma glycoproteins could have a significant impact on the propensity of streptococci to establish endocardial infections.
Keywords: C1-INH, GspB, ITIH4, platelet GPIb, PRG4
Introduction
Infective endocarditis (IE) is a life-threatening cardiovascular disease in which microbes colonize and persist in platelet–fibrin thrombi on cardiac valve surfaces. Endocardial infection is initiated when bacteria in the blood stream attach to a platelet–fibrin matrix on the surface of damaged cardiac valves (Sullam and Sande 1992; Fitzgerald et al. 2006). The subsequent binding of circulating platelets and plasma components to bacteria on infected valves, in conjunction with bacterial growth, contributes to the progression of disease, resulting in macroscopic endocardial lesions (vegetations) (Durack 1975; Rouzet et al. 2008). Although the viridans group streptococci, which include Streptococcus gordonii, Streptococcus sanguinis and Streptococcus mitis, are a leading cause of IE, only a small number of streptococcal adhesins that contribute to IE have been identified. Among the best characterized are GspB and Hsa, two semi-conserved, serine-rich repeat (SRR) glycoproteins of S. gordonii.
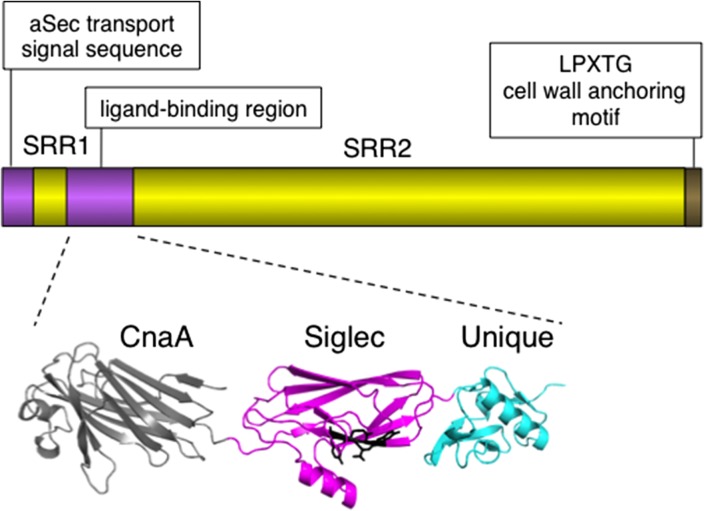
The SRR glycoproteins comprise a unique family of adhesins expressed by streptococci and staphylococci and bind a wide range of ligands, having a significant impact on colonization and virulence in a variety of infections (Wu et al. 1998; Siboo et al. 2005; Samen et al. 2007; Rose et al. 2008; Xiong et al. 2008; Shivshankar et al. 2009; van Sorge et al. 2009; Seo et al. 2012, 2013). Although the overall domain organization of the SRR glycoproteins is conserved (Figure 1), the ligand-binding regions are highly divergent and modular (Ramboarina et al. 2010; Pyburn et al. 2011; Schulte et al. 2014; Yang et al. 2014). The “Siglec-like” binding regions (SLBRs) of the SRR glycoproteins expressed by oral streptococci such as S. gordonii and S. sanguinis bind sialylated carbohydrate epitopes on the human salivary mucin MG2/MUC7, which likely contributes to colonization of tooth surfaces by these bacterial species (Plummer and Douglas 2006; Takamatsu et al. 2006; Bensing, Khedri, et al. 2016). Upon the entry of streptococci into the blood stream through lesions in the oral epithelium, the Siglec-like SRR adhesins can also mediate attachment to the same or similar sialylated glycans on red blood cells, extracellular matrix proteins or platelet GPIbα (Takahashi et al. 1997, 2004; Plummer et al. 2005; Deng et al. 2014; Bensing, Khedri, et al. 2016; Haworth et al. 2017). The latter interaction has been shown to enhance virulence within the endovascular space, thus contributing to the pathogenesis of IE (Takahashi et al. 2006; Xiong et al. 2008; Pyburn et al. 2011).
Fig. 1.
Features of the SRR glycoprotein GspB. The upper portion of the figure indicates the conserved domain organization for the family of adhesins. The ligand-binding regions are diverse and modular. The structural domains of the ligand-binding region of the S. gordonii GspB are shown. The Siglec and Unique domains are required for sialoglycan binding, and the conserved YTRY motif residues are indicated in black. The function of the CnaA domain is unknown. Many SLBRs lack a CnaA domain, and some have different modules adjoining the Siglec and Unique domains.
We have characterized extensively the structures and functions of nearly a dozen SLBRs from a variety of oral and endocarditis strains of streptococci. Binding to sialoglycans is dependent on a conserved YTRY motif in the SLBRs, and high-resolution co-crystal structures indicate that the threonine and arginine residues in this motif make important contacts with sialic acid (Bensing, Loukachevitch, et al. 2016; Bensing, Khedri, et al. 2016). However, the overall amino acid sequences of the SLBRs can vary by over 50% (Bensing, Khedri, et al. 2016). When tested on microarrays containing defined glycans, and in quantitative enzyme-linked assays, we found that the ligand repertoires of the adhesins range from being highly selective for a single type of sialylated trisaccharide (e.g. sialyl-T antigen or 3′sialyllactosamine), to a broad set of related sialoglycans that may include sialyl Lewis antigens (fucosylated glycans) and sulfated glycans.
The SLBRs also show differences in binding to platelet GPIbα, with those that are selective for sTa displaying relatively low affinity for GPIbα and platelets as compared with SLBRs that recognize 3′SLn (Bensing, Khedri, et al. 2016). This is perhaps not surprising since, although sTa is a major glycan on the salivary mucin MG2/MUC7, it is a relatively minor glycan on GPIbα (Korrel et al. 1984; Prakobphol et al. 1998; Karlsson and Thomsson 2009). Paradoxically, the SLBRs derived from IE strains of streptococci tend to be sTa-selective, whereas SLBRs of oral streptococcal isolates more typically recognize 3′SLn or related glycans (Bensing, Khedri, et al. 2016). This suggests there might be additional sialylated glycoprotein targets beyond GPIbα that either enhance the virulence of sTa-binding strains or diminish the virulence of 3′SLn-binding strains.
One confounding issue in determining the role of sialoglycan binding in the pathogenesis of IE is that the spatial and temporal distribution of specific sialoglycan structures in human tissues is largely unknown. This is especially true for O-linked glycans including sTa, which are notoriously difficult to characterize. Plasma is potentially a rich source of additional targets for the Siglec-like adhesins, as it contains a number of sialylated proteins that could contribute to pathogenesis, as well as minor amounts of glycoproteins shed from other tissues. In this report, we identify plasma protein ligands for four SLBRs that have different α2-3 sialoglycan repertoires. We show that the SLBRs recognize different subsets of O-glycosylated plasma proteins, or in some cases different glycoforms of the same plasma protein. The results support binding of the SLBRs to O-glycans rather than N-glycans and indicate that the glycosylation of several plasma proteins is much more extensive and heterogeneous than previously reported. The findings have important implications not only for cardiovascular infections by oral streptococci but also for the characterization of plasma protein biomarkers.
Results
The sTa- and 3′SLn-selective SLBRs recognize different subsets of plasma glycoproteins
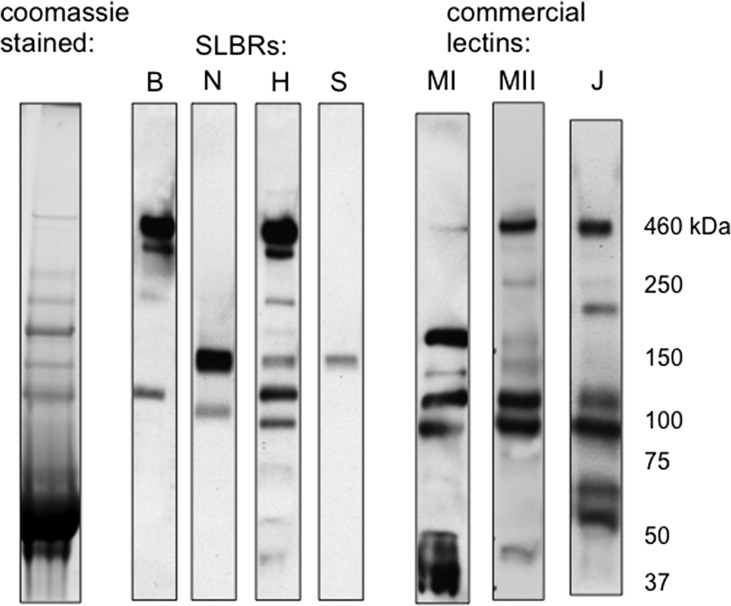
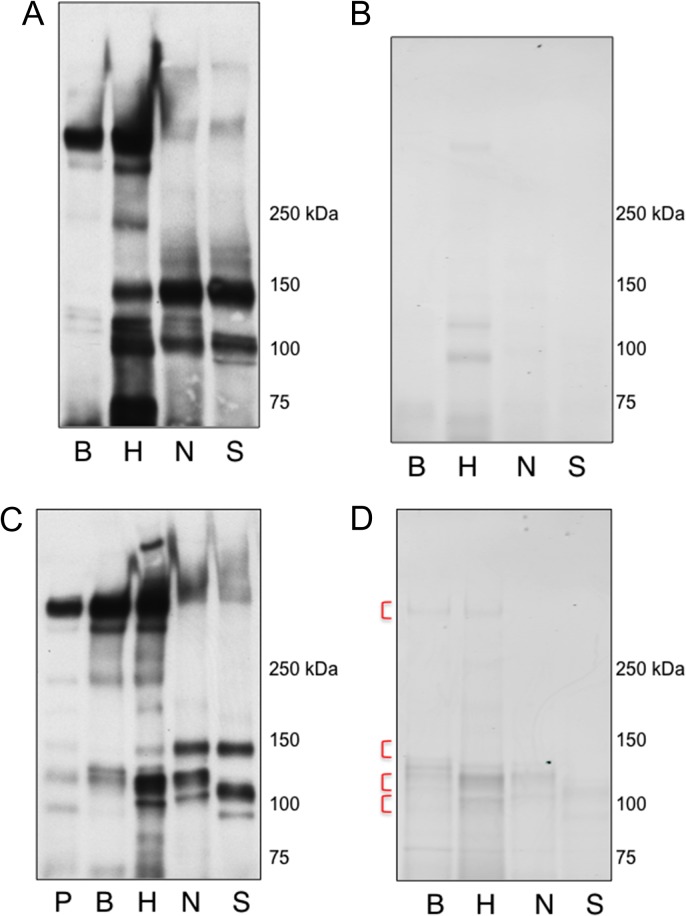
We previously reported the sialoglycan ligand preferences for seven SLBRs, as well as the comparative binding to salivary MG2/MUC7 and platelet GPIbα (Deng et al. 2014; Bensing, Khedri, et al. 2016). Based on those results, we chose four SLBRs with distinctly different binding properties (summarized in Figure 2) and examined whether they show differential recognition of plasma glycoproteins. To identify potential ligands, the four SLBRs were first used as probes in far-western (lectin) blotting of human plasma from a single donor (Figure 3). The sTa-selective SLBR-B (derived from the SRR adhesin GspB) interacted with two sialylated glycoproteins, with apparent molecular masses of approximately 460 and 120 kDa. In comparison, the 3′SLn-preferring SLBR-N also recognized two plasma proteins, but with different apparent masses (150 and 100 kDa) compared with those recognized by SLBR-B. The SLBR-H (derived from the SRR adhesin Hsa), which can bind 3′SLn in addition to sTa, recognized proteins of the same size as those recognized by SLBR-B and SLBR-N but also showed relatively minor detection of several other proteins ranging from 40 to 220 kDa. The SLBR-S binds neither sTa nor 3′SLn and showed little or no affinity for more than 40 other defined sialoglycans but is thought to prefer a di-sialylated glycan based on comparative analysis of high-resolution crystal structures (Deng et al. 2014; Loukachevitch et al. 2016). This SLBR showed relatively weak reactivity with a single plasma protein of 150 kDa. These patterns of plasma glycoprotein recognition were consistent when using plasma from different donors, or when using a commercial preparation of pooled human plasma, although the signal intensity of the 120 kDa protein detected with SLBR-B and SLBR-H tended to vary (see below). Thus, the differences in binding to defined tri- and tetrasaccharides are reflected in the recognition of distinctly different subsets of plasma glycoproteins.
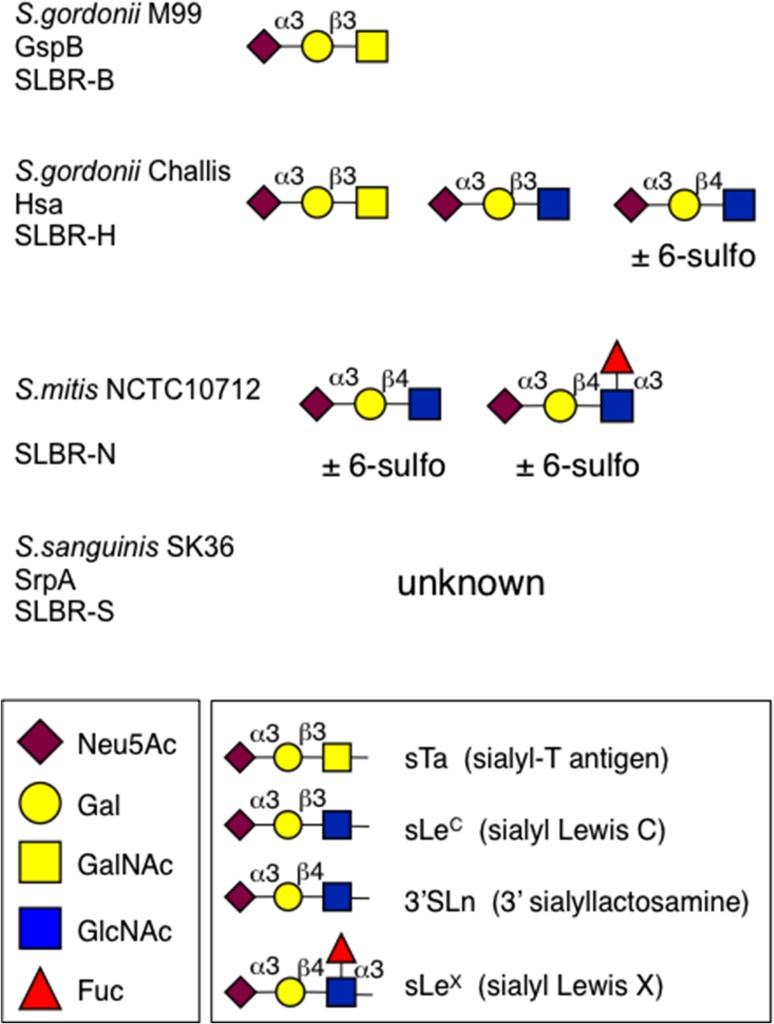
Fig. 2.
Previously defined high-affinity ligands for four SLBRs. Streptococcal strains and the SRR glycoproteins from which the SLBRs were derived are indicated. The ligand preferences were determined using a custom microarray and by quantitative enzyme-linked assays (Deng et al. 2014; Bensing, Khedri, et al. 2016). The 6-sulfo modification corresponds to HSO3 at carbon 6 of the GlcNAc residue. SLBR-S did not show high affinity for any of more than 50 sialoglycans tested but is suspected to prefer a di-sialylated compound (Deng et al. 2014; Loukachevitch et al. 2016). Monosaccharide symbols follow the Symbol Nomenclature for Glycans system (Varki et al. 2015).
Fig. 3.
Differential recognition of plasma glycoproteins by four SLBRs. SLBR binding is compared with that of commercial lectins MAL-I (MI), MAL-II (MII) and Jacalin (J). Lanes were loaded with 0.4 μL plasma (22 μg of total protein). Proteins were separated on a 3–8% polyacrylamide gradient, transferred to nitrocellulose and then probed with the indicated SLBR or lectin.
The preferential binding of SLBR-B to the 460 and 120 kDa proteins, combined with the lack of binding by SLBR-N, suggested that these plasma glycoproteins are decorated predominantly with sTa. Conversely, the selective binding of SLBR-N (and not SLBR-B) to the 150 kDa glycoprotein indicated that it may have more 3′SLn or sLeX epitopes (see Figure 2). These findings were supported by the pattern of reactivity seen when lectin blotting with Mal-I, Mal-II and Jacalin. The results indicate that the SLBRs show different degrees of selectivity and different specificities compared with each other and with more common sialoglycan-recognizing lectins.
Affinity capture of sialylated plasma glycoproteins
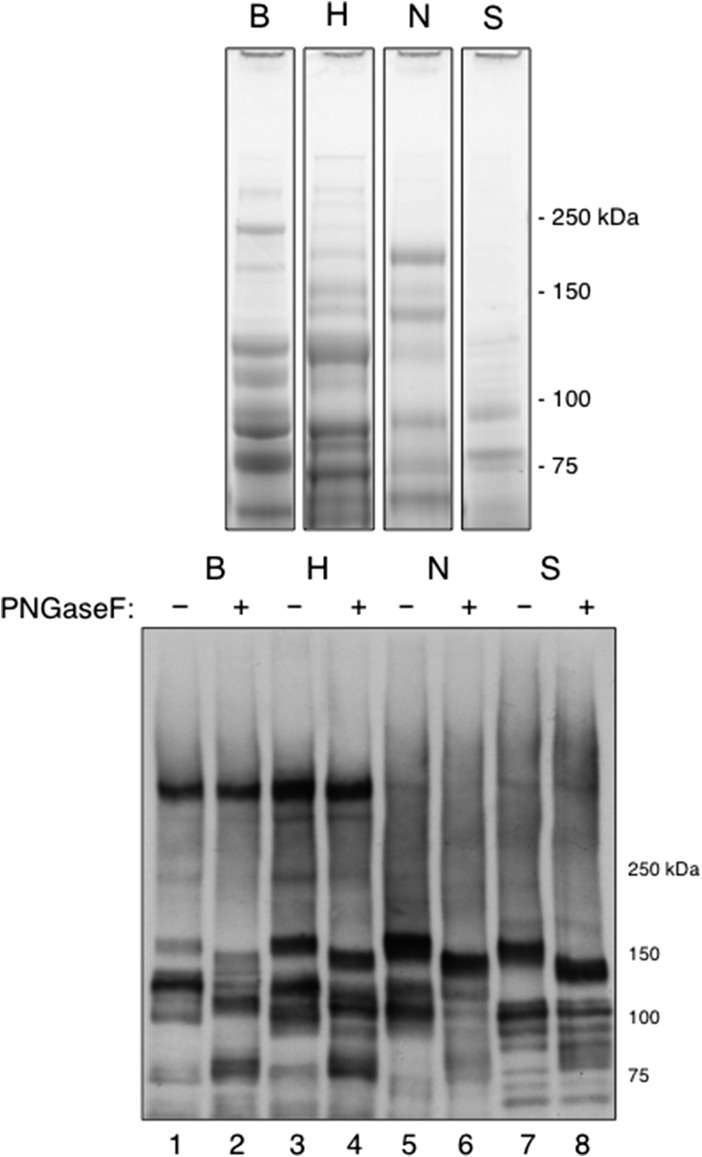
We next sought to characterize in more detail the plasma proteins recognized by each of the four streptococcal SLBRs, as well as the glycans associated with those proteins. Initially, low-stringency wash conditions were employed to enable capture of both high- and low-affinity targets. The four SLBRs captured different subsets of plasma proteins, as determined by Coomassie staining and by far-western blotting (Figure 4). Each of the four SLBRs captured glycoproteins of the size detected by far-western blotting of human plasma, as seen in Figure 3. For example, SLBR-B captured 460 and 120 kDa glycoproteins (lower panel, lane 1). Surprisingly, the SLBR-B also captured some of the 150 and 100 kDa glycoproteins, although the amount was apparently lower compared with those captured by SLBR-H (lane 3). Likewise, SLBR-N captured a 120 kDa sialoglycoprotein, in addition to the expected 150 and 100 kDa proteins (lane 5), and SLBR-S captured a 100 kDa sialoglycoprotein, in addition to the expected 150 kDa glycoprotein (lane 7). Neither SLBR-N nor SLBR-S captured the 460 kDa glycoprotein. Thus, four different but overlapping subsets of proteins from a single-donor plasma sample were recognized and captured by the four SLBRs.
Fig. 4.
Affinity capture of plasma glycoproteins by immobilized SLBRs. Proteins were captured from single-donor plasma under relatively low-stringency conditions. Upper: Captured proteins were separated on a 4–12% polyacrylamide gradient and stained with Coomassie. Lower: Captured proteins ± PNGaseF treatment were separated on a 3–8% polyacrylamide gradient gel, transferred to nitrocellulose and then probed with the broad-specificity SLBR-H.
We next examined whether the SLBR-captured plasma proteins were decorated with N-glycans and whether N-glycans had a role in SLBR binding. A portion of the captured glycoproteins were treated with PNGase F (to release N-glycans) prior to electrophoresis and then probed with SLBR-H. Treatment resulted in a shift in apparent mass of the 150, 120 and 100 kDa proteins, as compared with untreated samples, indicating that N-glycans were released from those proteins. However, PNGase F treatment did not eliminate SLBR-H reactivity (Figure 4, lanes 2, 4, 6 and 8). De-N-glycosylation also had no effect on the binding of the other three SLBRs, as measured by far-western blotting (data not shown). These results support binding of the SLBRs to O-linked vs. N-linked glycans.
Identification of plasma glycoprotein ligands for the SLBRs
In order to identify the sialylated plasma proteins directly recognized by each of the four SLBRs, we repeated the protein capture experiments several times, using moderately higher stringency conditions to enrich for high affinity ligands. We also compared different sources of plasma, to assess the reproducibility of our findings. When using a commercial preparation of pooled human plasma, the capture of sialylated glycoproteins was nearly identical to results with single-donor plasma (Figure 4) as determined by far-western blotting (Figure 5A). However, only the 120 and 100 kDa proteins captured by SLBR-H were readily detected on the Coomassie-stained gel (Figure 5B). We also examined the capture of proteins from plasma of a second donor. Again, far-western blotting demonstrated the capture of the expected glycoproteins (Figure 5C), although a slightly different appearance of the stained proteins was evident (Figure 5D vs. 5B). Thus, a relatively small number of plasma protein ligands were reproducibly captured by the four SLBRs, regardless of the plasma source and purification conditions.
Fig. 5.
Affinity capture of glycoproteins from pooled human plasma or single-donor plasma, using moderately high stringency. Proteins captured from pooled human plasma were separated by SDS PAGE, transferred to nitrocellulose and probed with a combination of SLBR-H and SLBR-N (A), or were stained with Coomassie (B). Proteins captured from single-donor plasma were separated by SDS PAGE, transferred to nitrocellulose and then probed with SLBR-H (C). The unfractionated plasma (P) is also shown. Alternatively, the captured proteins were separated by SDS PAGE and then stained with Coomassie (D). The regions of the gel that were excised and used for O-glycan profiling and glycopeptide analysis are indicated with red brackets.
To identify the ligands that were captured by each SLBR and that were detected by far-western blotting, corresponding bands were excised from Coomassie-stained gels, digested with trypsin and analyzed by mass spectrometry (Table I). The most abundant protein detected in the 460 kDa region of proteins captured by SLBR-B and SLBR-H was proteoglyan 4 (PRG4), a paralog of vitronectin that contains a mucin-like core and is also known as lubricin, superficial zone protein or megakaryocyte-stimulating factor (Estrella et al. 2010). Western blot analysis of the SLBR-captured proteins vs. whole plasma confirmed the binding of PRG4 by SLBR-B and -H but not by SLBR-N or -S (Figure 6). Significant amounts of ApoB100 were also found in this region. However, this latter protein is not reportedly O-glycosylated and was not detected by western blotting (data not shown), confirming minimal amounts compared with PRG4.
Table I.
Identification by mass spectrometry of SLBR-captured plasma glycoproteins
| Glycoprotein | SLBR | Accession number | Sequence coverage (%) | Theoretical mass (kDa) | Apparent mass range (kDa) |
|---|---|---|---|---|---|
| PRG4 | B | Q92954 | 22 | 151 | 460 |
| H | 26 | ||||
| GPIbαa | N | P07359 | 15 | 72 | 140–150 |
| ITIH4 | B | Q14624 | 67 | 103 | 120 |
| H | 69 | ||||
| C1-INH | B | P05155 | 33 | 55 | 90–120 |
| H | 58 | ||||
| N | 21 | ||||
| S | 36 |
aThe identified peptides span residues 25–503 of 562 and correspond to the extracellular domain.
Fig. 6.
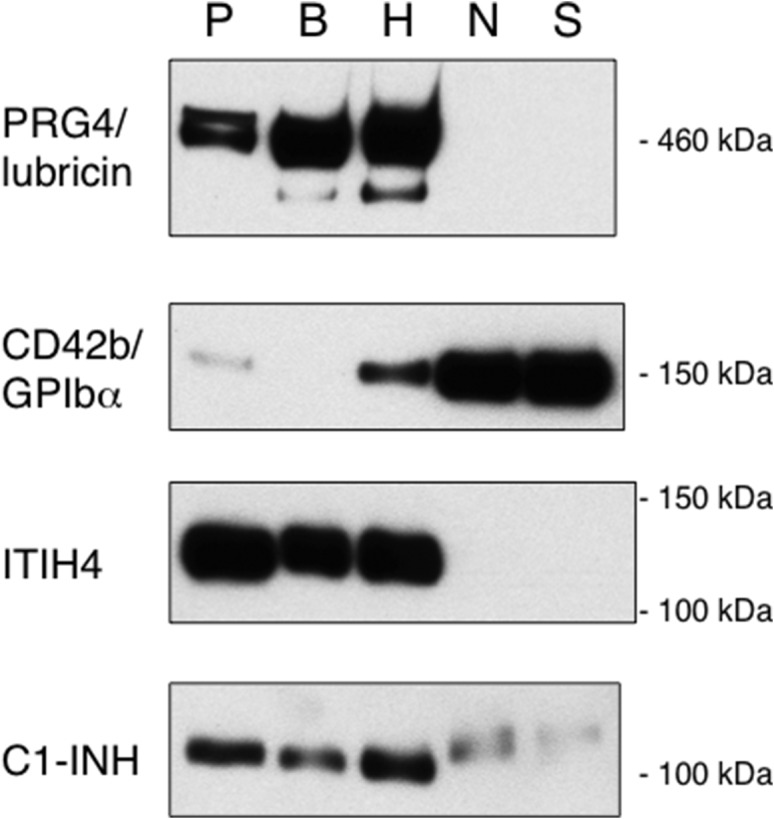
Western blot analysis of plasma glycoproteins captured by four SLBRs. The SLBR-captured plasma proteins, or unfractionated plasma from which the glycoproteins were captured (P), were separated by electrophoresis on 3-8% polyacrylamide, transferred to nitrocellulose and then probed with the indicated antibody. For each antibody, no signals were detected outside of the cropped regions.
The 150 kDa region of proteins captured by SLBR-N had an extremely low yield of tryptic peptides. The identified proteins included ceruloplasmin, aminopeptidase N (also known as CD13) and platelet GPIbα. Although the latter two are membrane proteins, fragments corresponding to the extracellular domains are known to circulate in plasma. Accordingly, only peptides derived from the extracellular domains of aminopeptidase N and GPIbα were identified by mass spectrometry (Table I and data not shown). Of the three proteins identified by mass spectrometry, only GPIbα was also detected by western blotting (Figure 6). In addition, the western blot analysis indicated that GPIbα was more readily captured by SLBR-N and SLBR-S, as compared with SLBR-H and SLBR-B.
Inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) was the most abundant protein identified in the 120 kDa region of proteins captured by both SLBR-B and SLBR-H. Capture of ITIH4 by SLBR-B and -H, but not -N or -S, was verified by western blotting (Figure 6). Of note, the affinity-captured ITIH4 displayed a slightly higher apparent mass compared with the ITIH4 in unfractionated plasma. This suggests that minor, more extensively glycosylated forms of ITIH4 were captured, as compared with the main glycoforms of ITIH4 in plasma.
The 100 kDa region of proteins captured by all four SLBRs contained the C1 esterase inhibitor (C1-INH), also known as Serpin G1, as well as relatively minor amounts of several other plasma proteins. Western blot analysis confirmed the capture of C1-INH by each of the SLBRs, but differences in electrophoretic mobility were evident. This suggests that the SLBRs may recognize distinctly different glycoforms of C1-INH.
The combined results indicate that PRG4 and ITIH4 are the principal ligands for SLBR-B and SLBR-H, whereas glycocalicin (the shed, extracellular portion of platelet GPIbα) was most readily captured by SLBR-N and SLBR-S. All four SLBRs captured C1-INH, but displaying different apparent molecular masses, suggestive of different glycoforms of this protein. Thus, the different glycan preferences of the four SLBRs are manifested as binding to different subsets of plasma glycoproteins and glycoforms.
O-glycan profiling of SLBR affinity-captured plasma proteins
We also analyzed the O-glycans associated with the SLBR-captured glycoproteins, using mass spectrometry of glycans released by β-elimination (Table II and Supplementary data, Tables S1–S11). For SLBR-B, we characterized the glycans associated with the 460, 120 and 100 kDa protein regions shown in Figure 5D. The most abundant O-glycans released from the 460 kDa region were sialylated core 1 glycans (41% sTa and 32% di-sialylated T antigen). In the 120 and 100 kDa regions, sTa was again the most abundant glycan detected (39 and 50%, respectively, of the released glycan population). Relatively minor amounts of other sialylated or fucosylated O-glycans, including core 2 and other structures, were detected in each of the three regions of proteins captured by SLBR-B (Supplementary data, Tables S1–S3). These results confirm that the plasma proteins recognized by SLBR-B display sTa as a major O-glycan and are consistent with SLBR-B binding directly to sTa on these proteins.
Table II.
Identification by mass spectrometry of major O-glycans on captured glycoproteins
| SLBR/target glycoprotein | Mass | Composition (Neu5Ac-Hex-HexNAc-Fuc) | Absolute abundance | Relative abundance (%) | Putative structure |
|---|---|---|---|---|---|
| B/460 | 674.24 | 1-1-1-0 | 1,596,976 | 41 | core 1 sTa |
| B/120 | 674.24 | 1-1-1-0 | 104,747 | 39 | core 1 sTa |
| B/100 | 674.24 | 1-1-1-0 | 292,375 | 50 | core 1 sTa |
| H/460 | 1039.37 | 1-2-2-0 | 978,243 | 50 | core 2 pentasaccharide |
| H/150 | 674.24 | 1-1-1-0 | 414,423 | 42 | core 1 sTa |
| H/120 | 674.24 | 1-1-1-0 | 354,376 | 35 | core 1 sTa |
| H/100 | 674.24 | 1-1-1-0 | 1,811,453 | 62 | core 1 sTa |
| N/150 | 1330.47 | 2-2-2-0 | 1,584,866 | 58 | core 2 hexasaccharide |
| N/100 | 1330.47 | 2-2-2-0 | 317,081 | 32 | core 2 hexasaccharide |
| S/150 | 1330.47 | 2-2-2-0 | 1,220,413 | 34 | core 2 hexasaccharide |
| S/100 | 1330.47 | 2-2-2-0 | 528,549 | 36 | core 2 hexasaccharide |
For SLBR-H, the O-glycans associated with proteins in all four of the mass regions indicated in Figure 5D were characterized (Table II and Supplementary data, Tables S4–S7). The most abundant glycan released from the 460 kDa region was a mono-sialylated core 2 pentasaccharide (50%), with sTa being the second most abundant (24%). The same two sialoglycans were the most abundant glycans released from the 150 kDa region, although sTa was more abundant than the core 2 pentasaccharide (42% vs. 9%). sTa was also the major O-glycan released from the 120 and 100 kDa regions (35 and 62% of the total, respectively). The remainder of the O-glycans detected in the 120 and 100 kDa regions were again a mix of core 2 and more complex sialylated or fucosylated glycans that were notably different from those on the 120 and 100 kDa proteins captured by SLBR-B. These results are consistent with sTa as a preferred ligand for SLBR-H but do not exclude direct binding to other structures, such as the mono-sialylated core 2 pentasaccharide. The results also suggest the capture of different glycoproteins or protein glycoforms by SLBR-H vs. SLBR-B.
For SLBR-N and -S, we characterized the O-glycans associated with proteins in the 150 and 100 kDa regions. The 150 kDa glycoproteins, which did not stain with Coomassie but were readily detected by far-western blotting, had an abundance of O-glycans, which consisted mainly of core 2, and only a minor amount of any core 1 glycans (Table III, Supplementary data, Tables S8 and S10). The most abundant O-glycan on the 100 kDa glycoproteins captured by SLBR-N and -S was the di-sialylated core 2 hexasaccharide (32 and 36% of the O-glycan population, respectively). However, the SLBR-N-captured population included 30% core 1 glycans (21% sTa and 9% di-sialylated T antigen), whereas the SLBR-S-captured proteins had just 3% core 1 glycans. These results indicate that SLBR-N and -S have a strong preference for proteins bearing sialylated core 2 glycans and are consistent with SLBR-N binding to 3′SLn, and with the possibility that SLBR-S may recognize a di-sialylated core 2 glycan.
Table III.
Glycopeptide analysis of SLBR-captured plasma proteins
| Captured glycoprotein (SLBR/kDa) | Protein name | Accession number | Peptidea | O-glycan (Neu5Ac-Hex-HexNAc-Fuc) |
|---|---|---|---|---|
| B/460 | PRG4 | Q92954 | 320-DLAPTSKV | 1-1-1-0 |
| 327-VLAKPTPK | 0-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 1-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 2-1-1-0, 2-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 0-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 1-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 2-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 2-2-2-0 | |||
| H/460 | PRG4 | Q92954 | 252-ITTAKPNPINPR | 1-1-1-0 |
| 327-VLAKPTPK | 1-1-1-0 | |||
| b -EPAPTTPK- | 1-1-1-0 | |||
| 623-KLTPTTPEK | 1-1-1-0 | |||
| 623-KLTPTTPEK | 1-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 1-1-1-0, 1-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 2-1-1-0 | |||
| 872-SPDESTPELSAEPTPK | 2-2-2-0 | |||
| B/120 | ITIH4 | Q14624 | 634-IPKPEASFSPR | 0-1-1-0 |
| 634-IPKPEASFSPR | 1-1-1-0 | |||
| 690-LAILPASAPPATSNPDPAVSR | 2-2-2-0, 2-3-3-2 | |||
| H/150 | ITIH4 | Q14624 | 634-IPKPEASFSPR | 1-1-1-0 |
| H/120 | ITIH4 | Q14624 | 634-IPKPEASFSPR | 2-2-2-0 |
| 634-IPKPEASFSPR | 1-1-1-0 | |||
| 690-LAILPASAPPATSNPDPAVSR | 2-2-2-0, 2-3-3-0 | |||
| 690-LAILPASAPPATSNPDPAVSR | 2-3-3-2 | |||
| 716-IEETTMTTQTPAPIQAPSAILPLPGQSVER | 1-3-2-1 | |||
| B/100 | C1-INH | P05155 | 45-VATTVISK | 1-1-1-0 |
| 45-VATTVISK | 1-1-1-0 | |||
| H/100 | C1-INH | P05155 | 45-VATTVISK | 1-1-1-0 |
| 45-VATTVISK | 0-0-1-0 | |||
| 45-VATTVISK | 0-1-1-0 | |||
| 45-VATTVISK | 2-2-2-0 | |||
| N/100 | C1-INH | P05155 | 45-VATTVISK | 2-2-2-0 |
We also performed O-glycan profiling of proteins from a different plasma sample, purified under slightly lower stringency conditions (depicted in Figure 4). In general, those results corroborated the findings described above, with more core 1 vs. core 2 glycans on proteins captured by SLBR-B, nearly equal amounts of core 1 and core 2 glycans on proteins captured by SLBR-H, and an abundance of core 2 but little or no core 1 structures in the SLBR-N- and SLBR-S-captured samples (Supplementary data, Tables S12–17). The combined results of O-glycan profiling of the SLBR-captured plasma proteins support the previously defined preferences for each SLBR and indicate that SLBR-N and SLBR-S recognize proteins decorated with core 2 glycans, whereas SLBR-B preferentially binds proteins modified with the mono-sialylated core 1 glycan sTa. The SLBR-H may prefer sTa but can apparently recognize proteins with either type of O-glycan.
Identification of glycopeptides derived from the affinity-captured plasma proteins
In order to confirm the covalent attachment of one or more of the liberated O-glycans (Table II and Supplementary data, Tables S1–S17) to specific plasma proteins, we next sought to characterize intact tryptic glycopeptides derived from the SLBR-captured plasma proteins. Duplicate samples of the 11 gel regions indicated in Table II, as well as selected regions from the gel shown in Figure 4, were analyzed. No tryptic glycopeptides were detectably released from the 150 kDa region of proteins captured by SLBR-N, or -S, or from the 100 kDa region of proteins captured by SLBR-S, consistent with the possibility that extensive glycosylation inhibited trypsinolysis, or that the proteins themselves were protease inhibitors (e.g. C1-INH). Results for the remaining regions are summarized in Table III. In brief, a number of PRG4 glycopeptides were identified in the 460 kDa region of proteins captured by SLBR-B and -H, with slight differences between the two. The glycopeptides derived from PRG4 captured by SLBR-B included sTa modification of S325, T877 and T885, whereas those captured by SLBR-H had sTa modification of T253, T332, T625, T628, S881 and T885. There were further differences in the occupancy and glycans linked to T332, T877, S881 and T885, indicative of both macro- and micro-heterogeneity in the plasma PRG4 population. The results indicate that different sub-populations (i.e. glycoforms) of PRG4 were captured by SLBR-B and -H and support the possibility that SLBR-B may recognize sTa in a different context, as compared with SLBR-H.
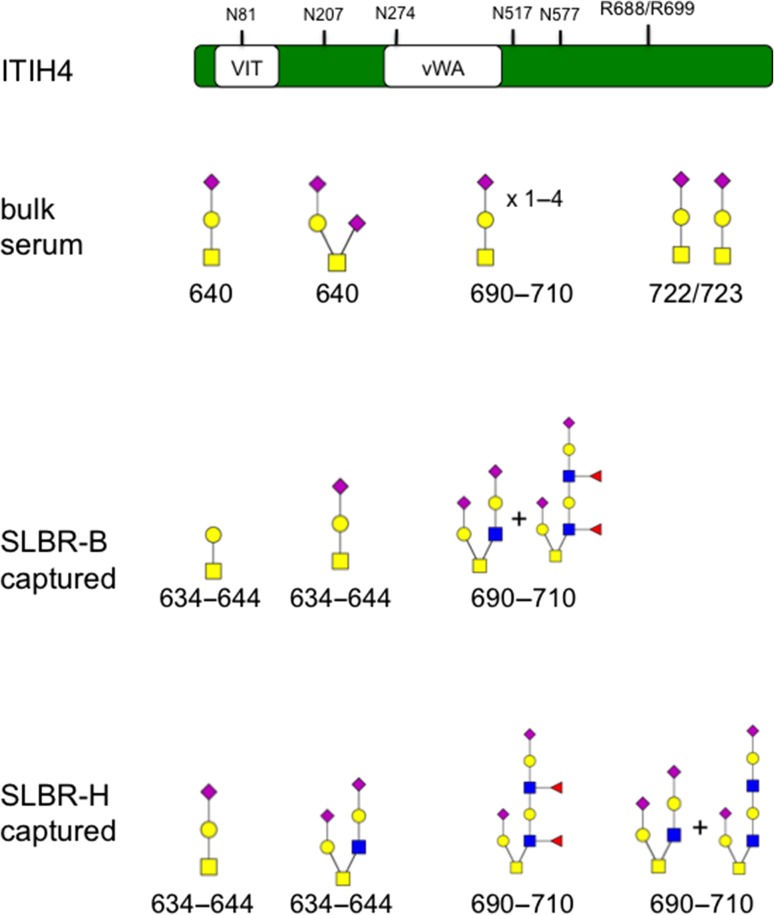
The 120 kDa region captured by SLBR-B had either T antigen or sTa on ITIH4 residues 634–644 and had a core 2 hexasaccharide along with a fucosylated decasaccharide on residues 690–710. The 120 kDa region captured by SLBR-H had either sTa or the di-sialylated core 2 hexasaccharide on ITIH4 residues 634–644. In addition, either a core 2 hexasaccharide along with an octasaccharide or a fucosylated decasaccharide was detected on ITIH4 residues 690–710. In previous glycopeptide analyses, only sTa or di-sialylated T antigen modification of ITIH4 residue S640, the region spanning residues 690–710 and the residues T722/T723 have been reported (Figure 7). These results suggest that SLBR-B and SLBR-H captured differently glycosylated populations of ITIH4, including minor glycoforms of the total ITIH4 present in human plasma. This is consistent with the results shown in Figure 6, where the SLBR-captured ITIH4 displayed slightly higher apparent mass compared with bulk plasma.
Fig. 7.
Glycopeptides of ITIH4 from serum vs. SLBR-enriched fractions from plasma. ITIH4 is a 963-amino acid protein containing vault protein (VIT) and von Willebrand factor type A (vWA) domains. Sites of N-glycosylation (N) and kallikrein cleavage (R688/R699) are indicated. The bulk serum analysis was done elsewhere (Chandler et al. 2014). Monosaccharide symbols follow the Symbol Nomenclature for Glycans system (Varki et al. 2015).
For the 100 kDa regions, a C1-INH peptide spanning residues 45–52 and modified with sTa was identified in the SLBR-B captured material, whereas the same C1-INH peptide was modified with a core 2 hexasaccharide in the SLBR-N-captured band. In the SLBR-H-captured samples, this C1-INH peptide was found to be modified with either the core 2 hexasaccharide, nonsialylated T antigen or a single HexNAc moiety. In some cases, S51 was determined to bear the glycan modification. Although the sTa modification of C1-INH residues T47 and T48 has been reported previously (discussed below), modification of this region with a core 2 glycan, and modification of S51, has not. Thus, the results of glycopeptide analysis confirm the findings of protein ID by mass spectrometry and western blotting of the captured proteins, which indicated PRG4 and ITIH4 as the primary ligands in the 460 and 120 kDa ranges of proteins captured by SLBR-B and -H. Moreover, the results show that different and previously unreported glycoforms of ITIH4 and C1-INH, as well as PRG4, appeared to be captured by the SLBRs.
Discussion
The search for additional ligands for the Siglec-like streptococcal adhesins produced several unexpected findings. First, the differences in affinity of SLBRs for defined α2-3 sialoglycans result in their interaction with distinctly different subsets of sialylated plasma glycoproteins. This includes the recognition of entirely different proteins and, in some cases, different glycoforms of the same protein. Of more than 20 known O-glycosylated plasma proteins, some of which are highly abundant, only one or two were found to be reproducibly strong ligands for the SLBRs. The one exception is that the broad-specificity SLBR-H binds at least four plasma ligands. A second important finding is that the SLBR-captured proteins display a rich assortment of O-glycans, as suggested by MS profiling of released glycans, and confirmed in part by glycopeptide analysis. This indicates that the target plasma proteins are much more extensively and heterogeneously O-glycosylated than has been previously reported. The latter finding may be significant in that each of the plasma proteins recognized by the SLBRs (i.e. PRG4, GPIbα, ITIH4 and C1-INH) has been cited as a biomarker for one or more human diseases, including rheumatoid arthritis, chronic obstructive pulmonary disease, obesity, type 2 diabetes, various cancers, stroke, depression, hepatic fibrosis, thrombocytopenia, pre-eclampsia and hereditary angioedema (Kawle et al. 2015; Lee, Feng, et al. 2015; Lee, Chuang, et al. 2015; Geyer et al. 2016). Although the level of each SLBR ligand has been correlated with disease, it is presently unclear how changes in the extent of glycosylation may impact not only the stability in plasma but also the function of the protein in health or disease. A brief description of each ligand, and the known roles in inflammation or thrombosis, will be provided before presenting the possible contribution to IE.
The sTa-selective SLBR-B and the broader specificity SLBR-H most readily and consistently captured PRG4 from various plasma preparations. The product of the prg4 gene has been extensively characterized as a mucin-like component of synovial fluid, commonly referred to as lubricin. The O-glycans are important for its function as a friction-reducing lubricant on articular cartilage surfaces and prevent cell adhesion in synovial joints (Jay et al. 2001). Recent reports have indicated that recombinant human lubricin can modulate inflammatory responses via interaction with CD44 and Toll-like receptors (Al-Sharif et al. 2015; Iqbal et al. 2016). These combined properties may explain why it is undergoing evaluation as potential therapeutic for treatment of chronic and acute cartilage damage (Larson et al. 2017; Waller et al. 2017). PRG4 has been detected in tissues beyond synovial fluid, including blood. However, the O-glycan composition of PRG4 has previously only been determined for the form isolated from synovial fluid. In an analysis of acidic proteins extracted from joint fluid of rheumatoid arthritis and osteoarthritis patients, PRG4 had 168 O-glycosylation sites in the central serine/threonine/proline-rich region, and the sites were mainly modified with nonsialylated or mono-sialylated core 1 glycans (Ali et al. 2014). Minor amounts of di-sialylated core 1- or core 2-modified glycopeptides have also been detected, as well as sulfated and fucosylated core 2 glycans, including sLeX epitopes (Jin et al. 2012). Although the O-glycan composition of plasma PRG4 had not been characterized prior to this study, an increased sialic acid content, as compared with the form in synovial fluid was noted previously (Solka et al. 2016). The results presented here indicate that plasma PRG4 is indeed laden with sialylated glycans, O-linked to several residues that had not been identified as glycosylation sites in synovial lubricin (i.e. S325, T625, T628, T877, S881 and T885). This suggests that the source of plasma PRG4 is different from synovial lubricin. Although the function in plasma is unknown, PRG4 has been implicated in lipid metabolism, since levels are strongly correlated with LDL, triglyceride and cholesterol levels (Geyer et al. 2016).
ITIH4 is another plasma component consistently captured by SLBR-B and -H. ITIH4 is one of five inter-alpha-trypsin inhibitor heavy chains, a family of acute-phase proteins involved in stabilization of the extracellular matrix. However, ITIH4 is unlike other ITIH proteins in that it lacks a consensus sequence for covalent linkage to bikunin (the “light chain” peptide protease inhibitor) and thus lacks trypsin-inhibitory activity. Instead, the C-terminal region includes a bioactive peptide that is generated by kallikrein cleavage (Figure 7). ITIH4 has five reported N-glycosylation sites, and at least four confirmed O-glycosites (S640, S696, T701 and T709) modified predominantly with simple core 1 glycans but with minor amounts of more complex O-glycans (Chandler et al. 2014; Hoffmann et al. 2016). The fraction of plasma proteins captured by SLBR-B and -H that was enriched for ITIH4 (i.e. the 120 kDa range) was laden with sTa but also had a variety of core 2 and more complex O-glycans. The linkage of at least some of the larger O-glycans to ITIH4 was confirmed by glycopeptide analysis of the SLBR-captured proteins (Table III and Figure 7). The ITIH4 captured by SLBR-H had at least one glycopeptide different from the ITIH4 captured by SLBR-B, and the total O-glycan composition indicated a slightly lower percentage of sTa (35% vs. 39%). It is likely that the extent of glycosylation affects the susceptibility to kallikrein cleavage, and thus the potential activity of ITIH4.
In contrast to SLBR-B, the 3′SLn-preferring SLBR-N most readily captured GPIbα. This heavily sialylated platelet membrane protein is the well-characterized receptor for von Willebrand factor, which is exposed on the subendothelium at regions of vascular damage. GPIbα is known to be shed from activated platelets and is therefore considered to be a marker of platelet activation. The GPIbα-enriched fraction of plasma proteins captured by SLBR-N and SLBR-S contained an abundance of the di-sialylated core 2 hexasaccharide. This structure was reported as the major O-glycan on GPIbα extracted from platelets, although minor amounts of mono- or di-sialyted core 1 were also noted (Korrel et al. 1984). The 150 kDa region of proteins captured by SLBR-H had an abundance of sTa, which suggests that a population of plasma GPIbα might be different from platelet GPIbα. Because we were unable to liberate any GPIbα glycopeptides and one ITIH4 glycopeptide was detected in the 150 kDa region (Table III), at least some of the sTa in the SLBR-H sample could derive from ITIH4. Moreover, the minimal capture by SLBR-H and especially SLBR-B is consistent with little or no sTa on plasma GPIbα. Therefore, it is not yet possible to determine whether the plasma GPIbα has a broader variety of glycoforms as compared with GPIbα in platelet membranes.
C1-INH was captured by all four streptococcal SLBRs. C1-INH is a member of the Serine protease inhibitor (serpin) family known as Serpin G1 and has long been used for the diagnosis and treatment of hereditary angioedema. C1-INH has been extensively characterized for its role in inhibition of complement, coagulation and fibrinolytic pathways. In addition to the complement pathway proteins C1r and C1s, it can inhibit the activity of kallikrein and coagulation factor XIIa. C1-INH is known to be extensively glycosylated, with estimates up to ~50% carbohydrate content by mass, including six N-linked and at least nine (but possibly as many as 26) O-linked glycans. The reported O-glycosylated residues include S31, T47, T48, S63, S64, T67, T71, T72 and T76, but additional evidence indicated that up to 17 additional serine and threonine residues may bear O-glycan modifications, and that these residues are most often modified with sTa (Stavenhagen et al. 2017). Of note, Ghannam et al. found that the removal of simple O-glycans from C1-INH affected the interaction with the contact-phase protease kallikrein but not with the complement protease C1s (Ghannam et al. 2016). Thus, the O-glycans impart pathway-specific functions.
Our results are consistent with the recent findings that C1-INH can display extreme glycoform heterogeneity. Direct linkage of sTa or the core 2 hexasaccharide to C1-INH residue S51 was supported by glycopeptide analysis (Table III). To our knowledge, this represents a novel C1-INH O-glycosite and novel glycopeptides. In addition, the profile of O-glycans released from the 100 kDa region (C1-INH fraction) of proteins captured by the four SLBRs indicated more core 2 vs. core 1 on the SLBR-S- vs. -N-captured C1-INH, and especially compared with the C1-INH captured by SLBR-H and -B (roughly 12:1 vs. 2:1 or 1:3, respectively). Based in part on western blot analysis (Figure 6), we suspect the C1-INH fraction captured by SLBR-N and SLBR-S likely spans the 100–120 kDa regions. The identification of additional C1-INH glycopeptides will help to resolve this issue.
It is presently unclear whether the SLBR-captured glycoforms of C1-INH represent minor populations of the total plasma C1-INH, or glycoforms that are abundant but not readily detected using standard analytical methods (i.e. not amenable to typical purification, proteolysis or ionization strategies). The ability to detect different glycoforms of C1-INH renders the SLBRs potentially valuable tools for defining specific sialoglycan modifications on this important biomarker and therapeutic, as well as for other glycoprotein biomarkers. The SLBRs could also be useful for the large-scale affinity purification of different glycoforms of C1-INH, in order to assess the functional consequences of different O-glycan modifications on C1-INH activity.
Regarding the role of the Siglec-like streptococcal adhesins in the pathogenesis of IE, we anticipate that the interaction of blood-borne streptococci with one or more of the newly identified ligands may tip the balance from clearance of the microbes to persistence in a cardiac valve niche. Each of the plasma ligands has a known or putative role in coagulation or thrombolysis, and the interaction of streptococci with any of the four targets could potentially impact these processes. One possibility is that the interaction of sTa-binding streptococcal strains with PRG4 or ITHI4 may augment the colonization of platelet–fibrin thrombi on cardiac valve surfaces. Alternatively, binding of strains to plasma GPIbα could prevent attachment to platelets that have deposited on damaged cardiac valves, thereby mitigating infection. It is also possible that sequestration of different forms of C1-INH could impact the formation and dissolution of infected thrombi, and thus the survival of streptococci within mature macroscopic lesions.
Materials and methods
Reagents
Dulbecco’s phosphate-buffered saline (DPBS), horseradish peroxidase-conjugated streptavidin and horseradish peroxidase-conjugated antibodies were purchased from Sigma (St. Louis, MO). Biotinylated lectins were from Vector Laboratories (Burlingame, CA). Glutathione Sepharose was from GE Healthcare (Uppsala, Sweden). Rabbit polyclonal anti-GST, polyacrylamide gradient gels, electrophoretic running and transfer buffers, LDS sample buffer and SimplyBlue SafeStain were from Life Technologies (Carlsbad, CA).
GST-SLBR expression and purification
The recombinant SLBRs used in these studies have been described previously as GST-GspBBR∆cnaA (SLBR-B), GST-HsaBR (SLBR-H), GST-NCTC10712BR (SLBR-N) and GST-SrpABR (SLBR-S) (Bensing, Loukachevitch, et al. 2016; Bensing, Khedri, et al. 2016). The GST-SLBR fusion proteins were expressed in Escherichia coli strain BL21 and purified using glutathione Sepharose as described previously (Bensing, Khedri, et al. 2016). For use in affinity capture experiments, the purified GST-SLBRs were not eluted from the glutathione Sepharose, but instead the resin-bound SLBRs were stored at −20°C in DPBS.
Far-western and lectin blotting of human plasma
Pooled citrated human plasma was purchased from Innovative Research (Novi, MI), and single-donor plasma was prepared from citrated whole blood obtained from individual donors. Plasma was diluted 1:10 into TE buffer (10 mM Tris, 1 mM EDTA, pH 8), combined with LDS sample buffer and loaded (5 μL) to wells of 3–8% polyacrylamide gradient gels. Following electrophoresis, proteins were transferred to BioTraceNT nitrocellulose (Pall Corporation, Pensacola, FL) and probed with GST-SLBRs as described (Bensing, Khedri, et al. 2016). When probed with biotinylated lectins, membranes were treated with Synthetic Block (Invitrogen) in DPBS prior to addition of biotinylated Jacalin, MAL-I or MAL-II at a final concentration of 0.2 μg/mL. After incubation for 1 h at RT, membranes were washed three times with DPBS and then incubated with horseradish peroxidase-conjugated streptavidin at 0.05 μg/mL in 1× Blocking Reagent (Roche Diagnostics, Indianapolis, IN) in DPBS. Membranes were again rinsed three times with DPBS and then developed with SuperSignal West Pico (Thermo Scientific, Rockford, IL).
Affinity capture of plasma glycoproteins
One milliliter of plasma was diluted 1:10 in TE buffer, filtered through a 0.45-μM membrane and combined with approximately 500 μg GST-SLBR immobilized on 100 μL glutathione Sepharose. After tumbling 2 h at 4°C, the resin was washed three times with 3 mL TE (for Figure 4) or TE plus 150 mM NaCl (for Figures 5 and 6). The affinity-captured proteins were held on ice prior to further processing.
Removal of N-glycans from affinity-captured plasma glycoproteins
The SLBR-captured proteins were treated with PNGaseF (New England BIolabs, Ipswich, MA) as recommended by the manufacturer. In brief, 25 μL of resin carrying 125 μg GST-SLBR and captured plasma proteins was suspended in 25 μL of denaturing buffer (5% SDS, 0.4 M DTT), held at 100°C for 10 min, cooled to RT, combined with 25 μL NP-40 (2% in 0.1 M sodium phosphate, pH 7.5) and incubated with or without 2500 units PNGaseF for 1 h at 37°C.
Identification of captured plasma glycoproteins
The resin-bound GST-SLBRs and affinity-captured plasma proteins were co-eluted into LDS sample buffer supplemented with DTT (50 mM final concentration), separated by electrophoresis in 4–12% polyacrylamide gradient gels and then stained with SimplyBlue SafeStain. Selected protein bands were excised from the gel and submitted for protein identification by nanoflow LC–MS/MS of tryptic digests (MSBioworks, Ann Arbor, MI). Western blot analysis was then used to confirm the capture of the most abundant proteins identified by MS. Five microliters of the captured proteins in LDS sample buffer (approximately 14 μg total combined proteins) were separated by electrophoresis in 3–8% polyacrylamide gradient gels and then transferred to BioTraceNT. Membranes were blocked for 1 h at RT in 1 × Blocking Reagent in DPBS and then incubated with the indicated mouse or rabbit antibodies for 1 h. Membranes were washed three times with DPBS and then incubated for 1 h at RT with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies diluted 1:20,000 in DPBS. Membranes were again rinsed three times with DPBS and then developed with SuperSignal West Pico (Thermo Scientific).
O-glycan profiling and O-glycopeptide analysis
Excised gel slices were minced, treated by four cycles of rinsing with 100 mM ammonium bicarbonate and dehydration in 100% acetonitrile and then dried to completion in a vacuum evaporator. For the analysis of O-glycans linked to the 100 and 120 kDa glycoproteins captured under relatively low stringency (Figure 4), the gel pieces were immersed in a mixture of 100 mM NaOH and 2 M NaBH4 and incubated at 45°C for 18 h to release the O-glycans. The supernatant was collected and placed on ice, and the remaining gel pieces were washed with water and sonicated for 30 min to extract the remaining O-glycans. The initial and secondary extracts were combined and acidified to pH 4–6 by dropwise addition of 10% acetic acid. For the glycoproteins captured under higher stringency (Figure 5D), N-glycans were first released from the samples by treatment with PNGaseF for 18 h at 37°C, and the secondary extraction of O-glycans was facilitated with a solution of 60% acetonitrile, 5% formic acid in water. The O-glycan samples were then enriched using porous graphitized carbon cartridges (Agilent, Santa Clara, CA) and dried prior to analysis by mass spectrometry.
For the O-glycopeptide analysis, 1 μg of trypsin was added to the dehydrated gel pieces and samples were incubated at 37°C for 18 h. After the removal of the supernatant, the gel pieces were sonicated in a solution of 5% formic acid, 60% acetonitrile for 30 min for complete extraction of O-glycopeptides. Any N-glycans were then released using 2 μL of PNGase F, and the glycopeptide samples were dried prior to mass spectrometry.
Both the O-glycans and the O-glycopeptides were analyzed using an Agilent 1200 Series HPLC system coupled to an Agilent 6520 Quadrupole Time-of-Flight mass spectrometer. The O-glycans were separated with porous graphitized carbon stationary phase (250 Å, 5 μm) and identified by their tandem MS spectra, using MassHunter Qualitative Analysis B.07.00 and referring to a mammalian O-glycan library. Short polymers of hexose were omitted from the O-glycan profile tables. The O-glycopeptides were separated with Zorbax C18 stationary phase (300 Å, 5 μm), and the data collected for the O-glycopeptides were searched against the human proteome database and an in-house established O-glycan library with Byonic™ software (Protein Metrics, San Carlos, CA).
Supplementary Material
Abbreviations
- SLBRs
“Siglec-like” binding regions
- IE
infective endocarditis
- SRR
serine-rich repeat
- PRG4
proteoglyan 4
- ITIH4
inter-alpha-trypsin inhibitor heavy chain H4
- C1-INH
C1 esterase inhibitor
- DPBS
Dulbecco’s phosphate-buffered saline
Supplementary data
Funding
This work was supported by the American Heart Association [17SDG33660424] and the National Institutes of Health [R01AI106987; U01CA199792, U01OD024857 and R21DE025826].
Conflict of interest statement
None declared.
References
- Al-Sharif A, Jamal M, Zhang LX, Larson K, Schmidt TA, Jay GD, Elsaid KA. 2015. Lubricin/proteoglycan 4 binding to cd44 receptor: A mechanism of the suppression of proinflammatory cytokine-induced synoviocyte proliferation by lubricin. Arthritis Rheumatol. 67:1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali L, Flowers SA, Jin C, Bennet EP, Ekwall AK, Karlsson NG. 2014. The O-glycomap of lubricin, a novel mucin responsible for joint lubrication, identified by site-specific glycopeptide analysis. Mol Cell Proteomics. 13:3396–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Khedri Z, Deng L, Yu H, Prakobphol A, Fisher SJ, Chen X, Iverson TM, Varki A, Sullam PM. 2016. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology. 26:1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Loukachevitch LV, McCulloch KM, Yu H, Vann KR, Wawrzak Z, Anderson S, Chen X, Sullam PM, Iverson TM. 2016. Structural basis for sialoglycan binding by the Streptococcus sanguinis SrpA adhesin. J Biol Chem. 291:7230–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KB, Brnakova Z, Sanda M, Wang S, Stalnaker SH, Bridger R, Zhao P, Wells L, Edwards NJ, Goldman R. 2014. Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J Proteome Res. 13:3314–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. 2014. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 10:e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack DT. 1975. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol. 115:81–89. [DOI] [PubMed] [Google Scholar]
- Estrella RP, Whitelock JM, Packer NH, Karlsson NG. 2010. The glycosylation of human synovial lubricin: Implications for its role in inflammation. Biochem J. 429:359–367. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JR, Foster TJ, Cox D. 2006. The interaction of bacterial pathogens with platelets. Nat Rev Microbiol. 4:445–457. [DOI] [PubMed] [Google Scholar]
- Geyer PE, Wewer Albrechtsen NJ, Tyanova S, Grassl N, Iepsen EW, Lundgren J, Madsbad S, Holst JJ, Torekov SS, Mann M. 2016. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol Syst Biol. 12:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannam A, Sellier P, Fain O, Martin L, Ponard D, Drouet C. 2016. C1 Inhibitor as a glycoprotein: The influence of polysaccharides on its function and autoantibody target. Mol Immunol. 71:161–165. [DOI] [PubMed] [Google Scholar]
- Haworth JA, Jenkinson HF, Petersen HJ, Back CR, Brittan JL, Kerrigan SW, Nobbs AH. 2017. Concerted functions of Streptococcus gordonii surface proteins PadA and Hsa mediate activation of human platelets and interactions with extracellular matrix. Cell Microbiol. 19:e12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Marx K, Reichl U, Wuhrer M, Rapp E. 2016. Site-specific O-glycosylation analysis of human blood plasma proteins. Mol Cell Proteomics. 15:624–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal SM, Leonard C, Regmi SC, De Rantere D, Tailor P, Ren G, Ishida H, Hsu C, Abubacker S, Pang DS et al. 2016. Lubricin/proteoglycan 4 binds to and regulates the activity of Toll-like receptors in vitro. Sci Rep. 6:18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay GD, Harris DA, Cha CJ. 2001. Boundary lubrication by lubricin is mediated by O-linked beta(1-3)Gal-GalNAc oligosaccharides. Glycoconj J. 18:807–815. [DOI] [PubMed] [Google Scholar]
- Jin C, Ekwall AK, Bylund J, Bjorkman L, Estrella RP, Whitelock JM, Eisler T, Bokarewa M, Karlsson NG. 2012. Human synovial lubricin expresses sialyl Lewis x determinant and has L-selectin ligand activity. J Biol Chem. 287:35922–35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson NG, Thomsson KA. 2009. Salivary MUC7 is a major carrier of blood group I type O-linked oligosaccharides serving as the scaffold for sialyl Lewis x. Glycobiology. 19:288–300. [DOI] [PubMed] [Google Scholar]
- Kawle AP, Nayak AR, Lande NH, Kabra DP, Chandak NH, Badar SR, Raje DV, Taori GM, Daginawala HF, Kashyap RS. 2015. Comparative evaluation of risk factors, outcome and biomarker levels in young and old acute ischemic stroke patients. Ann Neurosci. 22:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrel SA, Clemetson KJ, Van Halbeek H, Kamerling JP, Sixma JJ, Vliegenthart JF. 1984. Structural studies on the O-linked carbohydrate chains of human platelet glycocalicin. Eur J Biochem. 140:571–576. [DOI] [PubMed] [Google Scholar]
- Larson KM, Zhang L, Elsaid KA, Schmidt TA, Fleming BC, Badger GJ, Jay GD. 2017. Reduction of friction by recombinant human proteoglycan 4 in IL-1alpha stimulated bovine cartilage explants. J Orthop Res. 35:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Chuang HC, Chen TT, Liu WT, Su CL, Feng PH, Chiang LL, Bien MY, Ho SC. 2015. Proteoglycan 4 is a diagnostic biomarker for COPD. Int J Chron Obstruct Pulmon Dis. 10:1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Feng PH, Ho SC, Chuang KJ, Chen TT, Su CL, Liu WT, Chuang HC. 2015. Inter-alpha-trypsin inhibitor heavy chain 4: A novel biomarker for environmental exposure to particulate air pollution in patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 10:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukachevitch LV, Bensing BA, Yu H, Zeng J, Chen X, Sullam PM, Iverson TM. 2016. Structures of the Streptococcus sanguinis SrpA binding region with human sialoglycans suggest features of the physiological ligand. Biochemistry. 55:5927–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer C, Douglas CW. 2006. Relationship between the ability of oral streptococci to interact with platelet glycoprotein Ibalpha and with the salivary low-molecular-weight mucin, MG2. FEMS Immunol Med Microbiol. 48:390–399. [DOI] [PubMed] [Google Scholar]
- Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Ian Douglas CW. 2005. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br J Haematol. 129:101–109. [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Thomsson KA, Hansson GC, Rosen SD, Singer MS, Phillips NJ, Medzihradszky KF, Burlingame AL, Leffler H, Fisher SJ. 1998. Human low-molecular-weight salivary mucin expresses the sialyl lewisx determinant and has L-selectin ligand activity. Biochemistry. 37:4916–4927. [DOI] [PubMed] [Google Scholar]
- Pyburn TM, Bensing BA, Xiong YQ, Melancon BJ, Tomasiak TM, Ward NJ, Yankovskaya V, Oliver KM, Cecchini G, Sulikowski GA et al. 2011. A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 7:e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboarina S, Garnett JA, Zhou M, Li Y, Peng Z, Taylor JD, Lee WC, Bodey A, Murray JW, Alguel Y et al. 2010. Structural insights into serine-rich fimbriae from Gram-positive bacteria. J Biol Chem. 285:32446–32457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, Orihuela CJ. 2008. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 198:375–383. [DOI] [PubMed] [Google Scholar]
- Rouzet F, Dominguez Hernandez M, Hervatin F, Sarda-Mantel L, Lefort A, Duval X, Louedec L, Fantin B, Le Guludec D, Michel JB. 2008. Technetium 99m-labeled annexin V scintigraphy of platelet activation in vegetations of experimental endocarditis. Circulation. 117:781–789. [DOI] [PubMed] [Google Scholar]
- Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. 2007. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun. 75:5405–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T, Lofling J, Mikaelsson C, Kikhney A, Hentrich K, Diamante A, Ebel C, Normark S, Svergun D, Henriques-Normark B et al. 2014. The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold. Open Biol. 4:130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM. 2012. Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 8:e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Xiong YQ, Sullam PM. 2013. Role of the serine-rich surface glycoprotein Srr1 of Streptococcus agalactiae in the pathogenesis of infective endocarditis. PLoS One. 8:e64204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. 2009. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 73:663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboo IR, Chambers HF, Sullam PM. 2005. Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect Immun. 73:2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solka KA, Miller IJ, Schmid TM. 2016. Sialidase unmasks mucin domain epitopes of lubricin. J Histochem Cytochem. 64:647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen K, Kayili HM, Holst S, Koeleman C, Engel R, Wouters D, Zeerleder S, Salih B, Wuhrer M. 2017. N- and O-glycosylation analysis of human C1-inhibitor reveals extensive mucin-type O-glycosylation. Mol Cell Proteomics. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullam PM, Sande MA. 1992. Role of platelets in endocarditis: Clues from von Willebrand disease. J Lab Clin Med. 120:507–509. [PubMed] [Google Scholar]
- Takahashi Y, Sandberg AL, Ruhl S, Muller J, Cisar JO. 1997. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to alpha2-3-linked sialic acid-containing receptors. Infect Immun. 65:5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Takashima E, Shimazu K, Yagishita H, Aoba T, Konishi K. 2006. Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1. Infect Immun. 74:740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Yajima A, Cisar JO, Konishi K. 2004. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect Immun. 72:3876–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. 2006. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 74:1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. 2009. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis. 199:1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller KA, Chin KE, Jay GD, Zhang LX, Teeple E, McAllister S, Badger GJ, Schmidt TA, Fleming BC. 2017. Intra-articular recombinant human proteoglycan 4 mitigates cartilage damage after destabilization of the medial meniscus in the Yucatan minipig. Am J Sports Med. 45:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 28:487–500. [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. 2008. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog. 45:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Jiang YL, Zhang J, Wang L, Bai XH, Zhang SJ, Ren YM, Li N, Zhang YH, Zhang Z et al. 2014. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 10:e1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.