Abstract
Gene expression analysis was carried out in Jurkat cells in order to identify candidate genes showing significant gene expression alterations allowing robust discrimination of the Auger emitter 123I, incorporated into the DNA as 123I-iododeoxyuridine (123IUdR), from α- and γ-radiation. The γ-H2AX foci assay was used to determine equi-effect doses or activity, and gene expression analysis was carried out at similar levels of foci induction. Comparative gene expression analysis was performed employing whole human genome DNA microarrays. Candidate genes had to show significant expression changes and no altered gene regulation or opposite regulation after exposure to the radiation quality to be compared. The gene expression of all candidate genes was validated by quantitative real-time PCR. The functional categorization of significantly deregulated genes revealed that chromatin organization and apoptosis were generally affected. After exposure to 123IUdR, α-particles and γ-rays, at equi-effect doses/activity, 155, 316 and 982 genes were exclusively regulated, respectively. Applying the stringent requirements for candidate genes, four (PPP1R14C, TNFAIP8L1, DNAJC1 and PRTFDC1), one (KLF10) and one (TNFAIP8L1) gene(s) were identified, respectively allowing reliable discrimination between γ- and 123IUdR exposure, γ- and α-radiation, and α- and 123IUdR exposure, respectively. The Auger emitter 123I induced specific gene expression patterns in Jurkat cells when compared with γ- and α-irradiation, suggesting a unique cellular response after 123IUdR exposure. Gene expression analysis might be an effective tool for identifying biomarkers for discriminating different radiation qualities and, furthermore, might help to explain the varying biological effectiveness at the mechanistic level.
Keywords: gene expression, biomarker for high-LET radiation, α-radiation, Auger electron emitter, 123I-iododeoxyuridine exposure, microarray analysis, quantitative real-time PCR
INTRODUCTION
The rapid determination of individual radiation doses post-exposure is of importance when individuals are accidentally exposed to ionizing radiation, in order to ensure adequate treatment after high-dose exposure and long-term health follow-up in cases of low-dose exposure [1, 2].
The gold standard for radiation biodosimetry up to now has been the lymphocyte dicentric assay. However, this method is very time-consuming and not suitable for screening large populations, e.g. after nuclear terrorist attacks or radiation accidents [3]. In particular, the established biodosimetric methods do not allow discrimination between different radiation qualities. Therefore, it is necessary to develop high-throughput methods for biodosimetric purposes, permitting the analysis of many samples within a short period of time. Biodosimetry methods based on gene expression are a promising approach to overcoming the known limitations of established methods and will give additional insights into the complexity of cellular radiation responses. DNA microarray studies analyzing global gene expression in human peripheral blood lymphocytes after low-LET irradiation already allow the identification of radiation-responsive gene signatures suitable for a highly accurate prediction of the radiation dose [3–5]. Additionally, gene expression analysis might also allow the discrimination of different radiation qualities. To date, only a few studies have profiled genome-wide gene expression alterations in response to exposure to different radiation types, including Auger electron emission. There are only two studies that analyze global gene expression changes after exposure to Auger electrons and low-LET radiation. Sokolov et al. exposed human lung fibroblasts to 125IUdR, which was directly incorporated into the DNA, and compared the gene response to external γ-radiation delivered at a high dose rate with that delivered at a low dose rate [6, 7]. With regard to gene expression changes, they found no dose rate effect but a different gene response after 125IUdR exposure compared with that after γ-irradiation, suggesting that the different distribution of energy depositions within the cell caused different cellular responses. Ding et al. analyzed the gene expression in human bronchial epithelial cells after exposure to γ-rays, silicon ions and iron ions [8]. Their data indicated that cells elicit distinct gene expression patterns that are specific to the radiation type, e.g. they found one, two and three genes were exclusively regulated after exposure to iron ions, γ-rays and silicon ions, respectively. Additionally, they developed a gene signature comprising 73 genes capable of discriminating the radiation type. Chauhan et al. remarked that great efforts have been expended in developing strategies for radiation biodosimetry, with a specific focus on photon radiation, but these strategies may not provide adequate dose estimates for exposure to α-particles [9]. They employed genomic strategies in order to identify biomarkers in peripheral human blood mononuclear cells after α-irradiation and X-ray exposure. Subsequent comparison showed that both radiation types elicited similar gene responses with varying degrees of fold induction, but no α-particle–exclusive gene modulations were identified. However, a previous microarray study by Chauhan et al. identified five differently regulated genes in primary human keratinocytes after exposure to X-rays and α-particles [10]. However, the quantitative real-time PCR (qRT-PCR) validation only showed a significant change in gene expression after α-irradiation for one gene. The overarching goal of Chauhan et al. was the development of a forensic tool for identifying individuals, e.g. perpetrators who are planning a terrorist attack, who have handled special nuclear materials or have had dermal exposure to α-particle–emitting isotopes [10]. Another study by Chauhan et al. analyzed the gene expression in human lung fibroblasts after exposure to X-rays and α-particles [11]. Interestingly, six genes that showed a significant expression after α-irradiation were not significantly regulated after X-ray exposure. Furthermore, Danielsson et al., who irradiated primary human foreskin fibroblast cells with α-particles and γ-rays, observed different gene expression profiles for three genes as a function of radiation quality [12]. The above-mentioned gene expression studies indicated that the cellular response based on gene expression differs in relation to the radiation quality. However, since radiation dose alone modulates the gene expression, it is of major importance to carry out experiments at equi-effect doses.
In this study, human lymphoblastoid Jurkat cells were used because they have the same cellular origin as human lymphocytes that are predominantly used for gene expression–based biodosimetry studies [3–5, 13–15]. Contrarily to lymphocytes, proliferating Jurkat cells can incorporate 123I-iododeoxyuridine very efficiently into the DNA.
The biological effects of ionizing radiation are known to depend on dose and radiation quality. Low-LET γ-radiation induces in cells relatively uniformly distributed damage to all cellular structures, caused mostly by reactive oxygen and nitrogen species [16]. In contrast, high-LET α-particles deposit a large amount of energy over a short distance along the α-particle track. At low doses, only a small number of cell nuclei are hit, which means that the resulting biological effects depend on the cellular hit rate rather than on the energy absorbed by individual cells [17]. High-LET radiation induces complex DNA lesions, compared with low-LET radiation [18–22], which is probably the reason for the slower DNA repair observed after high-LET irradiation [23–25]. Auger electron emitters (AEEs) decay by electron capture and/or internal conversion, resulting in a cascade of mostly low-energy electrons, leading to high energy deposition in a very small volume around the decay site. Thus, damage on cellular level depends largely on the intracellular distribution of the nuclide. AEEs located exclusively in the cytoplasm cause, for example, low-LET-type cell survival, whereas DNA-associated AEEs cause high-LET-type cell survival [26, 27]. It is postulated that AEEs incorporated in the DNA cause complex DNA lesions [28], which is probably the reason for the rather high biological effectiveness of DNA-associated AEEs.
The aim of the present study was to identify biomarker genes in human T-lymphoma Jurkat cells at equi-effect doses/activities, allowing robust discrimination of the three different radiation qualities of Auger electrons (123I-iododeoxyuridine), low-LET γ-radiation (137Cs) and high-LET α-radiation (241Am). Such specific gene expression profiles might be of great value in gene-expression–based biodosimetry applications such as those we have recently developed for low-LET radiation [4, 5]. Additionally, gene expression analysis allows us to identify key signaling pathways, which can then help explain the varying biological effectiveness at the mechanistic level.
MATERIALS AND METHODS
Cell line and culture conditions
Human acute T-cell leukemia Jurkat cells were obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures, Germany). Cells were grown as a suspension cell culture in RPMI (Roswell Park Memorial Institute) 1640 medium (PAA Laboratories, Pasching, Austria) supplemented with 10% inactivated fetal bovine serum (Biochrom, Berlin, Germany), 100 units/ml penicillin and 100 μg/ml streptomycin (PAA Laboratories). Cells were routinely cultured in 75 cm2 flasks (TPP, Trasadingen, Switzerland) in a humidified atmosphere of 5% CO2 at 37°C.
γ-irradiation
The γ-irradiation was performed using 137Cs-irradiation equipment (Gammacell 40, Atomic Energy of Canada Limited, Mississauga, Canada) at a dose rate of ~0.71 Gy/min. Cells were irradiated with 0–10 Gy in T-75 flasks (TPP) in a heated chamber. Controls were sham-irradiated.
α-irradiation
α-particle exposure was carried out at Physikalisch-Technische Bundesanstalt (PTB). The α-irradiator has a relatively simple design and consists of a 241Am source (with a nominal activity of 195 kBq), which is mounted inside a stainless steel vacuum system. The irradiation port is sealed with a 2.5 μm–thick Mylar foil. The bottom of the specially designed cell dishes is formed by a 10 μm Mylar foil [29]. An air-filled safety gap of ~1 mm is left between the two Mylar foils. Due to the energy losses within the two Mylar foils, the α-particle energy at the center of the cells is reduced to ~3.35 MeV. This results in a LET of ~120 keV/μm averaged over the diameter of the cells. The fluence rate is (1 ± 0.15) × 107 h−1 cm−2, which corresponds to a dose rate of ~1.92 Gy/h. Prior to exposure, 80 000 Jurkat cells were transferred to the cell dish, mounted in a carrier system. The entire system was sealed with a flame-sterilized glass lid and placed above the α-source. Before the cells were irradiated with 0–1 Gy, the cells were allowed to settle on the Mylar foil as confirmed by phase-contrast microscopy (Axiovert 100 S, Zeiss, Göttingen, Germany). The irradiation set-up was heated by a 60 W bulb to ~37°C. Immediately after α-irradiation, the cells were transferred to a 6-well plate (TPP).
123IUdR synthesis
123IUdR synthesis was performed using a modified protocol from that of Baranowska-Kortylewicz et al. [30]. Na123I (T1/2 = 13.2 h) dissolved in 0.02 mol/l NaOH was provided by Zyklotron AG (Eggenstein-Leopoldshafen, Germany). At the time of calibration, the radionuclide purity was ~99.65% and the volume-specific activity was ~37 GBq/ml.
Synthesis of 5-(trimethylstannyl)-2’-deoxyuridine
5-iodo-2´-deoxyuridine (1.4 mmol, Fluka, Munich, Germany) was dissolved in 45 ml anhydrous dioxane (Fluka) in a three-necked flask at 60°C. The mixture was cooled to room temperature, and 6.1 mmol of hexamethylditin (Sigma-Aldrich, Munich, Germany) and 36 mmol bis(triphenylphosphine)palladium(II) dichloride (Fluka) were added. The mixture was boiled for 4 h at 112°C under reflux and then cooled to 40°C. The solvent was evaporated to dryness on a rotary evaporator. The solid residue was loaded onto a silica flash column and eluted with an 80:20 (v/v) mixture of CHCl3/CH3OH (Merck, Darmstadt, Germany) using flash chromatography (Axel Semrau, Sprockhövel, Germany) at a flow rate of 40 ml/min. The fractions containing the trimethylstannyl derivate were mixed and evaporated to a volume of 10 ml. For a second separation, 300 μl of the trimethylstannyl derivate was loaded again onto a silica column. The fraction containing only 5-(trimethylstannyl)-2'-deoxyuridine and no impurity was determined via thin-layer chromatography (TLC), and this fraction was subsequently evaporated to dryness.
Synthesis of 123IUdR
An aliquot of 100 MBq of Na123I solution was added to 100 μg of 5-(trimethylstannyl)-2'-deoxyuridine dissolved in 100 μl chloroform, and 5 μl of H2O2/CH3COOH (Merck) 1:3 (v/v) was added after brief mixing. The two-layer reaction mixture was sonicated (Cell Disruptor B15, Branson, USA) for 10 s, and 1 μl was spotted on a thin-layer chromatograph to determine the quality of the radio-iodination. Under a stream of nitrogen, the chloroform phase was evaporated to dryness, and 500 μl of phosphate-buffered saline (PBS, PAA Laboratories) was then added. The radioactivity of the solution was determined with an isotope calibrator (PTW Curiementor 2, Germany).
Cell cycle analysis, synchronization of cells, and incubation with 123IUdR
Cell cycle analysis based on DNA content was performed via 7-amino-actinomycin D (7AAD, BD Bioscience; Heidelberg, Germany) staining and flow cytometry. In detail, cells (2 × 105) were washed twice with PBS and fixed in ice-cold 70% ethanol for at least 30 min at −20°C. The cells were washed with PBS, resuspended in 200 μl PBS, and 5 μl of 7AAD staining solution was added. The cells were analyzed using the BD FACSDiva software and a BD FACSCanto II flow cytometer (BD Biosciences).
To maximize the incorporation of 123IUdR into the DNA, the Jurkat cells were synchronized in the G1/S boundary by aphidicolin (Sigma-Aldrich). Cells (5 × 105/ml) were incubated for 24 h with aphidicolin at a final concentration of 3 μg/ml, and the cells were then washed twice with PBS and cultured in growth medium at a final cell concentration of 3 × 105/ml. To reduce the potential cyto- and genotoxic effects of aphidicolin, the cells were allowed to progress through the S-phase and to complete the G2/M phase. At the second entry into the S-phase, Jurkat cells were incubated with 123IUdR for the following 20 h. Therefore, cells (3 × 105/ml) were grown in full cell culture medium and 0.5–50 kBq/ml 123IUdR was added. To maximize the uptake of 123IUdR into the DNA, 0.01–0.05 μM/ml fluorodeoxyuridine (FUdR, Fluka) and 0.01–0.05 μM/ml deoxycytidine (CdR, Fluka) was added to the cell culture medium. After 20 h of 123IUdR labeling, the cells were washed thoroughly three times with PBS to remove any unbound 123IUdR. For the gene expression analysis experiments, cells were then further cultivated in fresh medium for an additional 6 h prior to cell harvesting. In all experiments, non-irradiated controls were run in parallel with the experimental cell cultures, following identical protocols except that the radioactive 123IUdR was replaced with 0.05–0.1 μM/ml non-radioactive IUdR (Fluka).
Activity measurement and calculation of the accumulated decays of 123IUdR
After incubation with 123IUdR, the cells were washed twice in PBS. The cells were collected by centrifugation and the radioactivity of the cell pellet was measured in a gamma counter (1480 Wizard TM3, Perkin Elmer, Rodgau, Germany). To quantify the cellular uptake, cell numbers were determined with a cell counter (Casy Counter, Schärfe System, Reutlingen, Germany). The total accumulated decays per cell for a specific point in time were calculated using Excel (Microsoft, Redmond, USA), taking into account the half-life and time of measurement.
DNA incorporation of 123IUdR
After incubation with 123IUdR (50 kBq/ml), the cells were washed twice in PBS. Approximately 4 × 105 cells were used for DNA purification using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). DNA was extracted according to the manufacturer’s protocol, except that the DNA elution step was done three times to increase the overall DNA yield. Additionally, the optional step using RNase (100 mg/ml, Sigma-Aldrich) to get RNA-free genomic DNA was performed. Radioactivity in the DNA was measured in a gamma counter (1480 WizardTM 3, Perkin Elmer).
γ-H2AX-immunocytochemistry and fluorescence microscopy
Approximately 45 min after exposure, the cells were washed twice in PBS and then fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, UK) in PBS for 20 min at room temperature. Cells were washed twice in PBS, and then permeabilized with 1% Triton X-100 (Roth, Karlsruhe, Germany) in PBS for 20 min at room temperature. After washing with TBP buffer (0.2% Triton X-100, 1% bovine serum albumin (BSA), Sigma-Aldrich) in PBS, the cells were resuspended in 7.5% goat serum (PAA Laboratories) in PBS for 1 h at room temperature. The cells were washed with TBP buffer and, after centrifugation, the cell pellets were resuspended in TBP buffer containing anti-phospho-histone H2AX (Ser-139) mouse monoclonal antibody (mAb) (1:700, Millipore, Billerica, MA, USA) and incubated for 45 min at room temperature. The cells were washed twice with TBP buffer and, after centrifugation, the cell pellets were resuspended in TBP buffer containing Alexa Fluor 488 goat anti-mouse IgG (H+L) highly cross-adsorbed (1:700, Invitrogen, Karlsruhe, Germany) and incubated for 45 min at room temperature in the dark. Finally, the cells were washed twice with TBP buffer. For imaging by fluorescence microscopy, ~30 000 γ-H2AX–stained cells were cytospinned (Rotofix 32A, Hettich, Tuttlingen, Germany) and counter-stained with 4', 6-diamidine-2'-phenylindole dihydrochloride (DAPI) (1 μg/ml, Sigma). Images of γ-H2AX foci were taken with a fluorescence microscope (Zeiss Axioplan 2, Göttingen, Germany) with a Plan-Apochromat 63×/1.4 oil immersion objective (Zeiss).
γ-H2AX quantification by flow cytometry
The DNA of the γ-H2AX–stained cells was counter-stained with 7AAD staining solution. Sample data were acquired using BD FACSDiva software and a BD FACSCanto II flow cytometer (BD Biosciences) equipped with a 488 nm laser. Debris was excluded by gating events of forward scatter versus the side-scatter plot. Doublets were excluded using a plot of the linear 7AAD fluorescence area against the signal width. Apoptotic sub-G1 cells were excluded and only G1, S and G2/M phase cells were used for the analysis. FITC fluorescence was compensated versus linear 7AAD fluorescence. For each sample, 10 000 single non-debris events were recorded. Logarithmic FITC fluorescence (γ-H2AX) was plotted versus linear 7AAD fluorescence. Mean values of the FITC fluorescence were obtained.
Determination of equi-effect doses/activities for the gene expression analysis
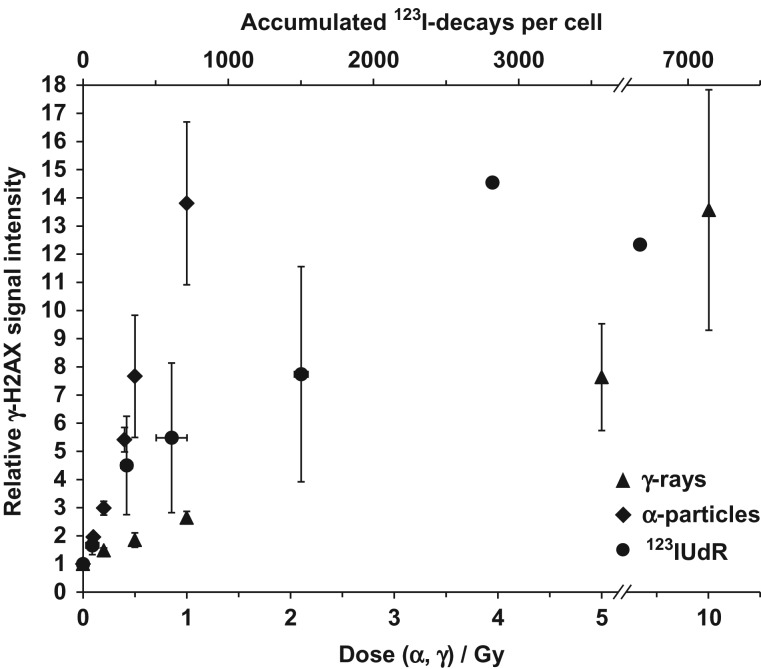
Equi-effect doses/activities, i.e. radiation doses and exposure conditions causing the same biological effect level, were determined for the three investigated radiation qualities with regard to γ-H2AX foci formation. As described above, the increase of the mean γ-H2AX signal of single irradiated cells was measured by flow cytometry. Suitable doses (γ-rays and α-particles) or accumulated decays (123IUdR) inducing almost identical increases in γ-H2AX signal intensity were determined (Fig. 3) and were subsequently used for all gene expression experiments.
Fig. 3.
Flow cytometric quantification of γ-H2AX signal intensity in Jurkat cells 45 min after withdrawal of 123IUdR and after exposure to γ-rays and α-particles. For each radiation quality, median values are expressed as the relative increase of the overall γ-H2AX signal intensity of the cell nuclei with respect to controls. Equi-effect doses and activity for all three radiation qualities were determined. The error bars represent the standard deviation of the mean (n = 3).
RNA isolation
The total RNA was isolated 6 and 24 h after exposure using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. RNA quantification was carried out using a NanoDrop-1000 spectrophotometer (Peqlab, Erlangen, Germany), and RNA quality was monitored with the Agilent 2100 Bioanalyzer (Agilent, Böblingen, Germany). All extracted RNA samples were found to be of good quality. RNA integrity numbers (RINs) ranged from 9 to 10. Three biological replicates were prepared for each dose and each point in time.
DNA microarray hybridization
DNA microarray experiments were performed according to the manufacturer’s manual and as previously described [4, 5]. Of the total RNA, 400 ng was transcribed into cDNA with an oligo-dT primer, followed by transcription into cRNA labeled with cyanine 3-CTP (Quick-Amp Labeling Kit, One-color, Agilent). cRNA purification was performed with the RNeasy Mini Kit (Qiagen). cRNA yields and the dye incorporation were measured with the NanoDrop-1000 spectrophotometer. Labeled cRNA samples were hybridized for 17 h to 44 k Whole Human Genome DNA microarrays (G4112F, Agilent) using a hybridization oven (Agilent). After hybridization and washing, DNA microarrays were scanned with the Microarray Scanner (G2505 B, Agilent) as recommended by Agilent.
Analysis of DNA microarray data
The images of the scanned microarrays were processed with the Agilent Feature Extraction software. The gene expression data were processed, normalized and analyzed using the Agilent GeneSpring GX software. By data filtering, non-uniform outliers were excluded, as well as signals that were not significantly above the background intensity in at least 25% of the samples. In order to indicate the significant regulated genes in Tables 1 and 2 as well as in Figs 4 and 5, the P values were adjusted using the method of Benjamini and Hochberg to calculate the false discovery rate (FDR). Genes with a fold change >1.5 and an FDR < 0.15 were considered as significantly regulated.
Table 1.
Significantly regulated genes after exposure to 123IUdR, α-particles and γ-rays with a fold change >1.5. (P values were adjusted using the method of Benjamini and Hochberg to calculate FDR values.)
| Significantly regulated genes (fold change > 1.5) | ||
|---|---|---|
| (FDR < 0.15)a | (P < 0.05)b | |
| 2600 accumulated 123I-decays per cell (26 h of exposure) | 344 | 554 |
| 10 Gy γ-irradiation (6 h) | 0 | 409 |
| 10 Gy γ-irradiation (24 h) | 1321 | 1827 |
| 1 Gy α-irradiation (6 h) | 1 | 200 |
| 1 Gy α-irradiation (24 h) | 596 | 653 |
aFDR-adjusted P-values.
bNon-corrected P-values.
Table 2.
Significantly up- or downregulated genes with the highest fold changes 6 h after withdrawal of 123IUdR (a) and 24 h after exposure to α-particles (b) and γ-rays (c) using the determined equi-effect doses/activities
| a) | 2600 accumulated 123I-decays per cell (26 h of exposure) | 1 Gy α-irradiation (24 h) | 10 Gy γ-irradiation (24 h) | |||||
|---|---|---|---|---|---|---|---|---|
| ProbeName | GeneSymbol | Description | P-value (FDR) | Fold change | P-value (FDR) | Fold change | P-value (FDR) | Fold change |
| A_32_P235274 | Synthetic construct Homo sapiens gateway clone IMAGE:100020693 3' read LUM mRNA. [CU675508] | 0.02 | 6.21 | |||||
| A_23_P211468 | oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence [AA837799] | 0.03 | 5.52 | 0.06 | 2.91 | 0.10 | 17.67 | |
| A_24_P326491 | MKX | Homo sapiens mohawk homeobox (MKX), transcript variant 1, mRNA [NM_173576] | 0.09 | 4.30 | 0.09 | 2.68 | 0.11 | 5.25 |
| A_32_P219135 | LOC401317 | PREDICTED: Homo sapiens hypothetical LOC401317 (LOC401317), partial miscRNA [XR_108761] | 0.06 | 4.28 | 0.14 | 2.83 | ||
| A_23_P257881 | 0.06 | 3.95 | ||||||
| A_23_P382775 | BBC3 | Homo sapiens BCL2 binding component 3 (BBC3), transcript variant 4, mRNA [NM_014417] | 0.00 | 3.53 | 0.07 | 1.94 | 0.11 | 3.40 |
| A_32_P228037 | PDE11A | Homo sapiens phosphodiesterase 11A (PDE11A), transcript variant 4, mRNA [NM_016953] | 0.07 | 3.42 | 0.06 | 3.26 | 0.09 | 4.47 |
| A_23_P99442 | FLT3 | Homo sapiens fms-related tyrosine kinase 3 (FLT3), mRNA [NM_004119] | 0.14 | 3.29 | 0.07 | 2.90 | 0.14 | 3.02 |
| A_23_P39799 | LOXL3 | Homo sapiens lysyl oxidase-like 3 (LOXL3), mRNA [NM_032603] | 0.09 | 3.13 | 0.14 | 2.87 | ||
| A_24_P119246 | Homo sapiens uncharacterized gastric protein ZG16P mRNA, partial cds. [AF264625] | 0.08 | 3.12 | |||||
| A_23_P93348 | LTB | Homo sapiens lymphotoxin beta (TNF superfamily, member 3) (LTB), transcript variant 1, mRNA [NM_002341] | 0.06 | −4.03 | ||||
| A_32_P224697 | AF150420 Human mRNA from cd34+ stem cells Homo sapiens cDNA clone CBNAZD12, mRNA sequence [AF150420] | 0.06 | −3.94 | |||||
| A_23_P82929 | NOV | Homo sapiens nephroblastoma overexpressed gene (NOV), mRNA [NM_002514] | 0.14 | −3.71 | ||||
| A_23_P401024 | NCRNA00304 | Homo sapiens non-protein coding RNA 304 (NCRNA00304), non-coding RNA [NR_024347] | 0.09 | −3.59 | 0.10 | −2.13 | 0.12 | −4.47 |
| A_24_P532478 | Homo sapiens full-length insert cDNA clone ZD79H01. [AF086429] | 0.02 | −3.11 | |||||
| A_24_P113131 | BZRAP1 | Homo sapiens benzodiazapine receptor (peripheral) associated protein 1 (BZRAP1), transcript variant 1, mRNA [NM_004758] | 0.03 | −3.06 | 0.08 | −1.66 | 0.09 | −2.97 |
| A_32_P34696 | BX101288 Soares_parathyroid_tumor_NbHPA Homo sapiens cDNA clone IMAGp998N074186, mRNA sequence [BX101288] | 0.01 | −3.04 | 0.15 | −1.74 | |||
| A_24_P886589 | 0.09 | −3.03 | ||||||
| A_23_P152559 | BZRAP1 | Homo sapiens peripheral benzodiazepine receptor interacting protein mRNA, complete cds. [AF039571] | 0.01 | −2.98 | 0.06 | −1.58 | 0.11 | −2.59 |
| A_23_P128375 | C12orf34 | Homo sapiens chromosome 12 open reading frame 34 (C12orf34), mRNA [NM_032829] | 0.03 | −2.95 | 0.07 | −1.67 | 0.09 | −3.19 |
| b) | 2600 accumulated 123I-decays per cell (26 h of exposure) | 1 Gy α-irradiation (24 h) | 10 Gy γ-irradiation (24 h) | |||||
|---|---|---|---|---|---|---|---|---|
| ProbeName | GeneSymbol | Description | P-value (FDR) | Fold change | P-value (FDR) | Fold change | P-value (FDR) | Fold change |
| A_32_P909570 | WDR72 | Homo sapiens WD repeat domain 72 (WDR72), mRNA [NM_182758] | 0.13 | 8.14 | ||||
| A_23_P30091 | UCP1 | Homo sapiens uncoupling protein 1 (mitochondrial, proton carrier) (UCP1), nuclear gene encoding mitochondrial protein, mRNA [NM_021833] | 0.07 | 5.49 | 0.13 | 6.72 | ||
| A_24_P847956 | LOC284561 | PREDICTED: Homo sapiens hypothetical LOC284561 (LOC284561), partial miscRNA [XR_112106] | 0.06 | 5.12 | ||||
| A_23_P41114 | CSTA | Homo sapiens cystatin A (stefin A) (CSTA), mRNA [NM_005213] | 0.12 | 1.82 | 0.08 | 4.73 | 0.11 | 4.62 |
| A_23_P423309 | PCDH12 | Homo sapiens protocadherin 12 (PCDH12), mRNA [NM_016580] | 0.06 | 4.34 | 0.13 | 5.52 | ||
| A_32_P158181 | TNFSF15 | Homo sapiens tumor necrosis factor (ligand) superfamily, member 15 (TNFSF15), transcript variant 1, mRNA [NM_005118] | 0.11 | 4.20 | 0.14 | 8.16 | ||
| A_23_P164057 | MFAP4 | Homo sapiens microfibrillar-associated protein 4 (MFAP4), transcript variant 2, mRNA [NM_002404] | 0.14 | 2.01 | 0.06 | 4.13 | 0.13 | 5.25 |
| A_32_P599 | 0.11 | 3.44 | ||||||
| A_32_P228037 | PDE11A | Homo sapiens phosphodiesterase 11A (PDE11A), transcript variant 4, mRNA [NM_016953] | 0.07 | 3.42 | 0.06 | 3.26 | 0.09 | 4.47 |
| A_23_P138194 | NCF2 | Homo sapiens neutrophil cytosolic factor 2 (NCF2), transcript variant 1, mRNA [NM_000433] | 0.09 | 3.12 | 0.12 | 3.93 | ||
| A_32_P188193 | 0.10 | −4.49 | ||||||
| A_32_P3342 | THC2676548 | ALU7_HUMAN (P39194) Alu subfamily SQ sequence contamination warning entry, partial (19%) [THC2676548] | 0.13 | −4.25 | ||||
| A_32_P90468 | 0.08 | −3.66 | ||||||
| A_32_P211048 | 0.09 | −3.29 | ||||||
| A_23_P3921 | FLJ11710 | Homo sapiens cDNA FLJ11710 fis, clone HEMBA1005149. [AK021772] | 0.12 | −3.29 | ||||
| A_32_P52519 | OTOA | Homo sapiens otoancorin (OTOA), transcript variant 2, mRNA [NM_170664] | 0.10 | −3.23 | ||||
| A_32_P194372 | Homo sapiens cDNA FLJ26036 fis, clone PRS00145. [AK129547] | 0.13 | −3.12 | |||||
| A_32_P113404 | 0.08 | −3.02 | ||||||
| A_32_P163594 | 0.08 | −2.83 | ||||||
| A_24_P915692 | PHLDA1 | Homo sapiens pleckstrin homology-like domain, family A, member 1 (PHLDA1), mRNA [NM_007350] | 0.10 | −2.78 | ||||
| c) | 2600 accumulated 123I-decays per cell (26 h of exposure) | 1 Gy α-irradiation (24 h) | 10 Gy γ-irradiation (24 h) | |||||
|---|---|---|---|---|---|---|---|---|
| ProbeName | GeneSymbol | Description | P-value (FDR) | Fold change | P-value (FDR) | Fold change | P-value (FDR) | Fold change |
| A_23_P211468 | oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence [AA837799] | 0.03 | 5.52 | 0.06 | 2.91 | 0.10 | 17.67 | |
| A_23_P17354 | GDAP1L1 | Homo sapiens ganglioside-induced differentiation-associated protein 1-like 1 (GDAP1L1), mRNA [NM_024034] | 0.12 | 2.95 | 0.12 | 10.97 | ||
| A_32_P226614 | PREDICTED: Homo sapiens hypothetical LOC100506544 (LOC100506544), partial miscRNA [XR_109720] | 0.08 | 2.88 | 0.12 | 10.50 | |||
| A_32_P87191 | FLJ21408 | Homo sapiens hypothetical LOC400512 (FLJ21408), non-coding RNA [NR_037184] | 0.11 | 8.62 | ||||
| A_32_P158181 | TNFSF15 | Homo sapiens tumor necrosis factor (ligand) superfamily, member 15 (TNFSF15), transcript variant 1, mRNA [NM_005118] | 0.11 | 4.20 | 0.14 | 8.16 | ||
| A_24_P350656 | NFASC | Homo sapiens neurofascin (NFASC), transcript variant 5, mRNA [NM_001005389] | 0.12 | 7.46 | ||||
| A_23_P104996 | BEST1 | Homo sapiens bestrophin 1 (BEST1), transcript variant 1, mRNA [NM_004183] | 0.12 | 7.44 | ||||
| A_24_P532232 | CREB5 | Homo sapiens cAMP responsive element–binding protein 5 (CREB5), transcript variant 1, mRNA [NM_182898] | 0.13 | 7.27 | ||||
| A_23_P30091 | UCP1 | Homo sapiens uncoupling protein 1 (mitochondrial, proton carrier) (UCP1), nuclear gene encoding mitochondrial protein, mRNA [NM_021833] | 0.07 | 5.49 | 0.13 | 6.72 | ||
| A_23_P58266 | S100P | Homo sapiens S100 calcium–binding protein P (S100P), mRNA [NM_005980] | 0.10 | 1.55 | 0.11 | 1.56 | 0.12 | 6.07 |
| A_23_P93027 | FGF18 | Homo sapiens fibroblast growth factor 18 (FGF18), mRNA [NM_003862] | 0.01 | −2.10 | 0.09 | −5.37 | ||
| A_23_P401024 | NCRNA00304 | Homo sapiens non-protein coding RNA 304 (NCRNA00304), non-coding RNA [NR_024347] | 0.09 | −3.59 | 0.10 | −2.13 | 0.12 | −4.47 |
| A_32_P150263 | XKR4_MOUSE (Q5GH67) XK-related protein 4, partial (3%) [THC2654127] | 0.02 | −1.93 | 0.07 | −1.81 | 0.09 | −4.39 | |
| A_23_P37685 | TMEM204 | Homo sapiens transmembrane protein 204, mRNA (cDNA clone MGC:111564 IMAGE:4692469), complete cds. [BC105785] | 0.06 | −2.57 | 0.14 | −4.03 | ||
| A_24_P524452 | Synthetic construct Homo sapiens gateway clone IMAGE:100023058 3’ read HIST2H2BE mRNA. [CU686711] | 0.06 | −2.52 | 0.11 | −3.70 | |||
| A_32_P107850 | Homo sapiens mRNA; cDNA DKFZp451M139 (from clone DKFZp451M139). [AL833303] | 0.13 | −3.66 | |||||
| A_32_P152348 | HIST1H2BD | AGENCOURT_8209273 NIH_MGC_112 Homo sapiens cDNA clone IMAGE:6265521 5’, mRNA sequence [BQ683489] | 0.05 | −2.48 | 0.11 | −3.64 | ||
| A_24_P479793 | 0.02 | −2.47 | 0.07 | −2.12 | 0.10 | −3.49 | ||
| A_24_P260639 | HIST1H1D | Homo sapiens histone cluster 1, H1d (HIST1H1D), mRNA [NM_005320] | 0.09 | −2.70 | 0.13 | −3.45 | ||
| A_23_P128375 | C12orf34 | Homo sapiens chromosome 12 open reading frame 34 (C12orf34), mRNA [NM_032829] | 0.03 | −2.95 | 0.07 | −1.67 | 0.09 | −3.19 |
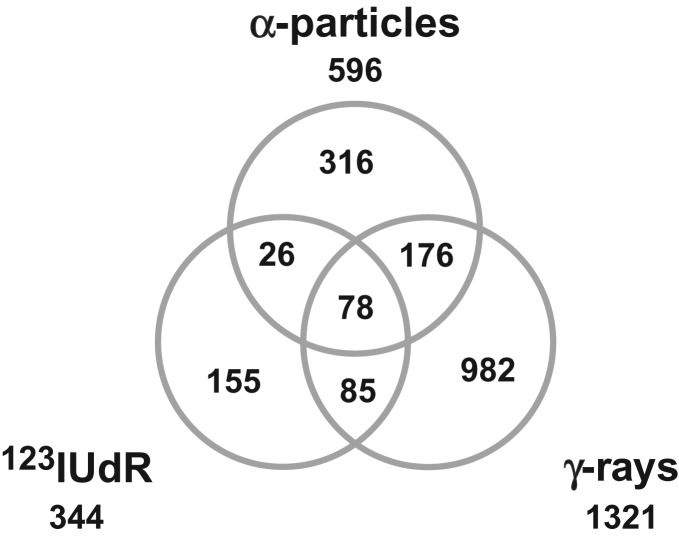
Fig. 4.
Venn diagram after exposure to 123IUdR, α-particles and γ-rays: 155, 316 and 982 genes were exclusively regulated 6 h after withdrawal of 123IUdR and 24 h after α- and γ-irradiation, respectively. (Ratio of gene expression >1.5-fold, FDR < 0.15; n = 3).
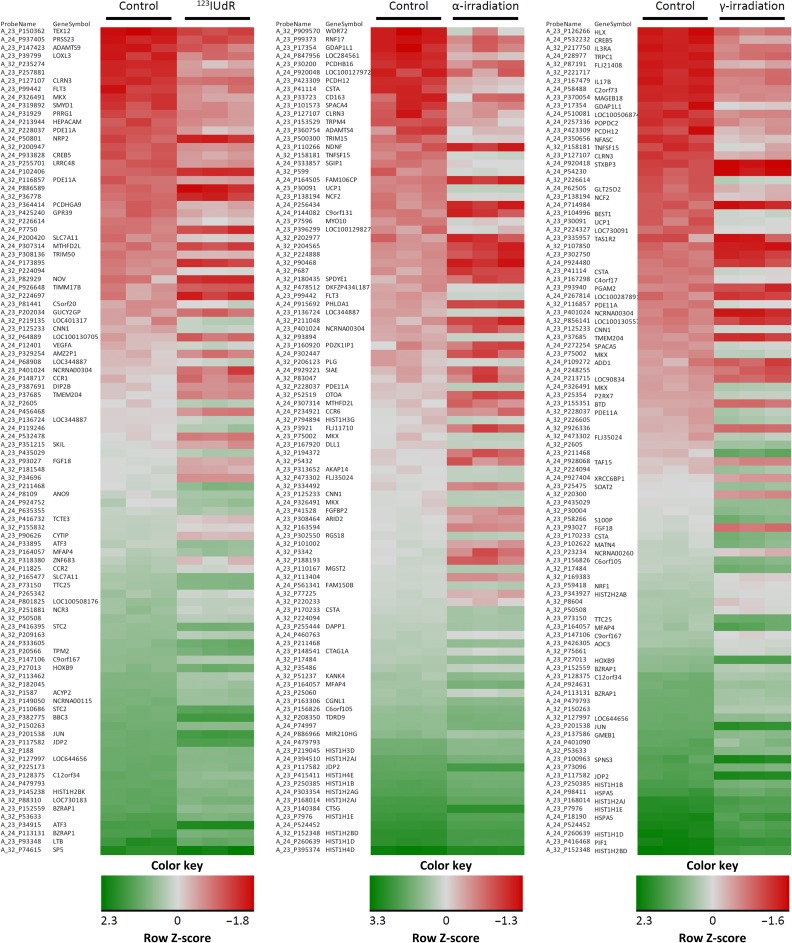
Fig. 5.
Heatmaps illustrating Z score–transformed expression levels of overall 100 significantly up- or downregulated genes with the highest fold changes 6 h after withdrawal of 123IUdR and 24 h after exposure to α-particles and γ-rays using the determined equi-effect doses/activities.
Functional analysis of significantly regulated genes
The significantly expressed genes 6 h after exposure to 123IUdR (20 h) and 24 h after α- or γ-irradiation were functionally categorized using the Database for Annotation Visualization and Integrated Discovery 6.8 (DAVID) [31, 32]. To assign the significantly altered genes to affected biological processes and pathways, we used the gene ontology analysis feature and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [33], respectively.
Filtering for candidate genes, allowing discrimination of radiation quality
Genes with non-FDR adjusted P-values were considered for the filtering process to increase the number of potential candidate genes. The criteria for candidate genes were: a significant expression change (>1.5 fold; P < 0.05) and no altered (1-fold ± 0.1) regulation, or even an opposing (>1.1-fold) regulation, in response to the other radiation qualities. Furthermore, the gene expression fold changes of the unirradiated control samples should be similar (1-fold ± 0.25) and not significantly different. Regarding 123IUdR exposure, the filtered genes were reviewed in the scientific literature, and all the genes with a function in cell cycle regulation were excluded, because the cells were synchronized in the cell cycle before 123IUdR exposure. After filtering, the gene expression of the candidate genes was validated by qRT-PCR.
Quantitative real-time PCR
The isolated RNA was DNase-treated using a Turbo DNA-free Kit (Applied Biosystems, Darmstadt, Germany). Gene-specific, intron-spanning primers were designed for qRT-PCR measurements (NCBI/Primer-BLAST). For one qRT-PCR reaction, the components of a Power SYBR® Green RNA-to-CTTM 1-Step Kit (Applied Biosystems, Darmstadt, Germany) and 30 ng of total RNA were mixed according to the manufacturer’s instructions. The qRT-PCR reactions were performed using the Applied Biosystems 7500 Real Time PCR System (Applied Biosystems). The results were evaluated with the Sequence Detection Software 1.3.1 (Applied Biosystems) and Excel (Microsoft). The detected signals were normalized by the internal control dye ROX, and the Ct of the samples was normalized in relation to the Ct of the endogenous control (GAPDH). Relative fold changes were calculated by the ΔΔCt method. For the presentation of the qRT-PCR data, the values were normalized to only one of the non-irradiated controls of the different radiation qualities, which allowed a comparison of the gene expression between the different controls. The same procedure was applied for the microarray.
RESULTS
DNA incorporation of 123IUdR
As a result of the short ranges of low-energy Auger electrons, the biological effectiveness depends on the intracellular localization of the nuclide [26, 27]. Thus, DNA-incorporated AEEs possess the highest biological effectiveness per decay. Therefore, it is important to determine the amount of DNA-incorporated 123IUdR. Approximately 4 × 105 cells were incubated with 50 kBq/ml 123IUdR for 20 h, and subsequently the DNA was extracted. A total activity of 31.7 ± 8.5 mBq was determined per cell, of which 29.7 ± 8.1 mBq was found in the DNA fraction.
Therefore, ~94% of the 123IUdR activity measured in Jurkat cells was located in the DNA (Fig. 1).
Fig. 1.
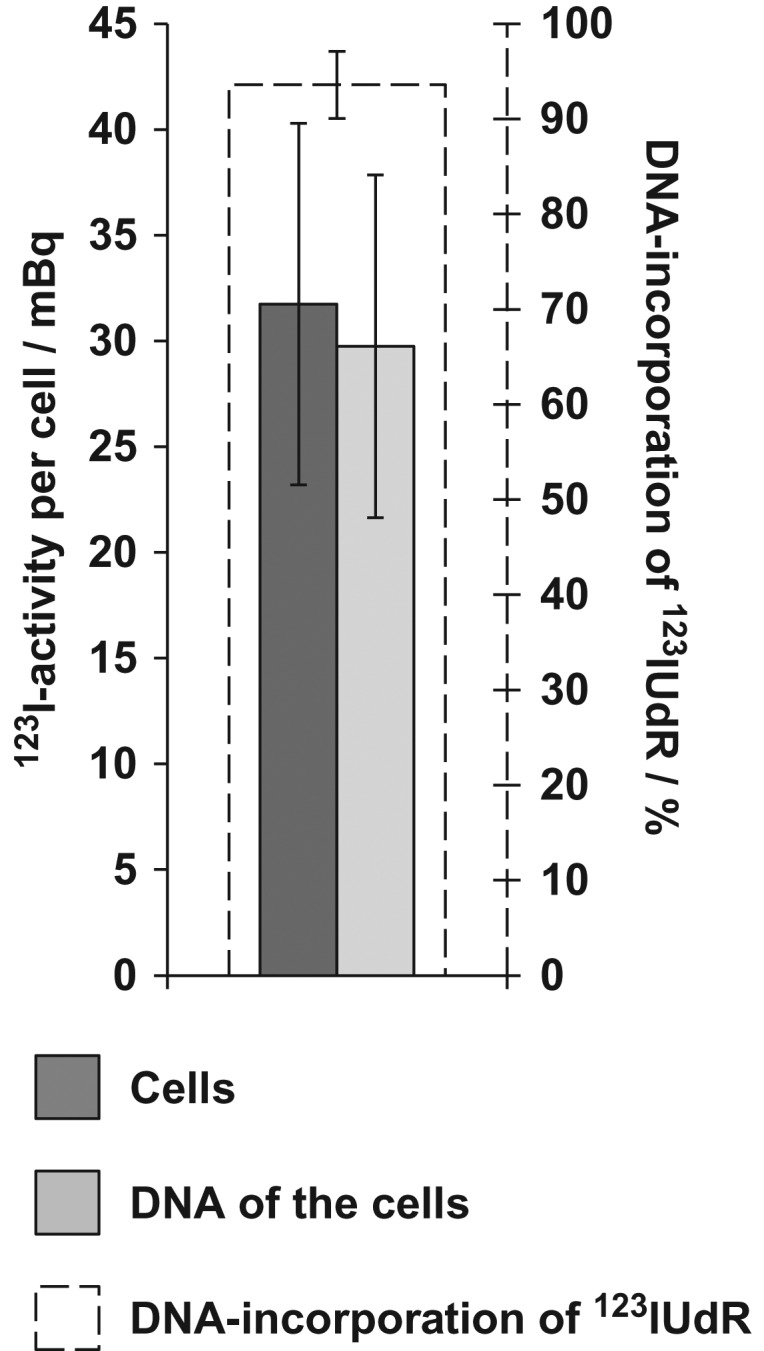
DNA uptake of 123IUdR in Jurkat cells after exposure to 50 kBq/ml 123IUdR for 20 h. Approximately 94% of the 123IUdR activity measured in Jurkat cells was found to be associated with the DNA. The error bars represent the standard deviation of the mean (n = 3).
Extent of DNA damage determined by γ-H2AX foci formation
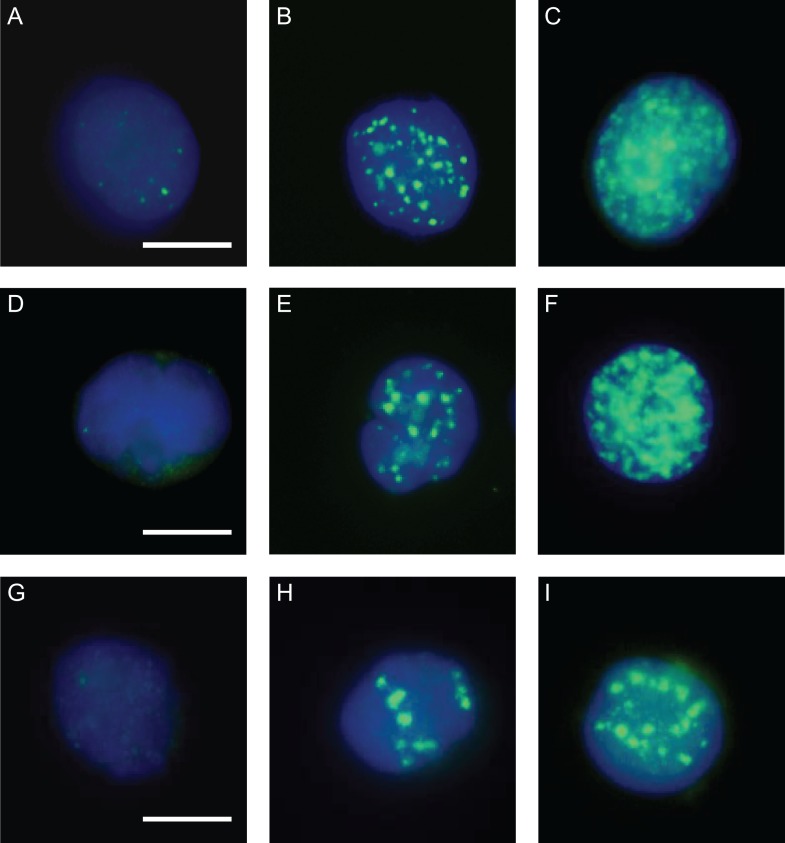
The formation of γ-H2AX foci after exposure to different radiation qualities was analyzed to determine equi-effect doses/activities, which then allowed comparative gene expression analysis at a very similar DNA damage/effect level for all three investigated radiation qualities. Phosphorylation of histone H2AX (γ-H2AX) occurs at sites flanking DNA double-strand breaks after exposure to ionizing radiation [34], so it can be used as a biomarker for DNA damage. Representative images of γ- and α-irradiated cells, as well as cells exposed to 123IUdR, are shown in Fig. 2.
Fig. 2.
Representative images of γ-H2AX immuno-stained Jurkat cells 45 min after withdrawal of 123IUdR and after exposure to γ-rays and α-particles. (B) 400 accumulated 123I-decays per cell; (C) 2600 accumulated 123I-decays per cell; (E) 1 Gy γ-rays; (F) 10 Gy γ-rays; (H) 0.1 Gy α-particles; (I) 1 Gy α-particles; (A, D and G) corresponding non-irradiated controls. DAPI counterstained; bar = 5 μm.
Compared with γ-rays and 123IUdR exposure, α-irradiation–induced γ-H2AX foci were much wider and larger and showed an overall brighter signal intensity on average. Additionally, clusters of α-irradiation–induced foci were observed, which could not be properly microscopically analyzed to count discrete foci. To overcome these limitations, flow cytometry was chosen to quantify the increase in the mean γ-H2AX signal of single irradiated cells compared with the respective control cells (see Materials and Methods). A similar DNA damage effect level (~14-fold increase in γ-H2AX signal intensity compared with control cells) was found 45 min after exposure to 10 Gy γ-rays and 1 Gy α-particles, and 45 min after 20 h exposure to 123IUdR, which corresponded to ~2600 accumulated 123I decays per cell (Fig. 3).
Gene expression changes following exposure to different radiation qualities
Gene expression was analyzed after adding 123IUdR for 20 h to cell cultures and an additional 6 h after withdrawal of 123IUdR as well as 6 h and 24 h after α- and γ-irradiation using the determined equi-effect doses/activities. Genes with a fold change >1.5 and an FDR < 0.15 were considered to be significantly deregulated. Except for one single gene, no significant alteration in gene expression was found 6 h after α- or γ-irradiation (Table 1).
In contrast, 344, 596 and 1321 genes were significantly deregulated 6 h after exposure to 123IUdR (20 h) and 24 h after α- and γ-irradiation, respectively. Whereas 155, 316 and 982 genes were exclusively regulated 6 h after 123IUdR exposure (20 h) and 24 h after α- and γ-irradiation, respectively, only 78 deregulated genes were shared between all radiation qualities (Fig. 4). A selection of significantly up- or downregulated genes with the highest fold changes 6 h after withdrawal of 123IUdR and 24 h after exposure to α-particles and γ-rays are presented in Table 2 and as heatmaps in Fig. 5.
Identification of candidate genes allowing the discrimination of different radiation qualities
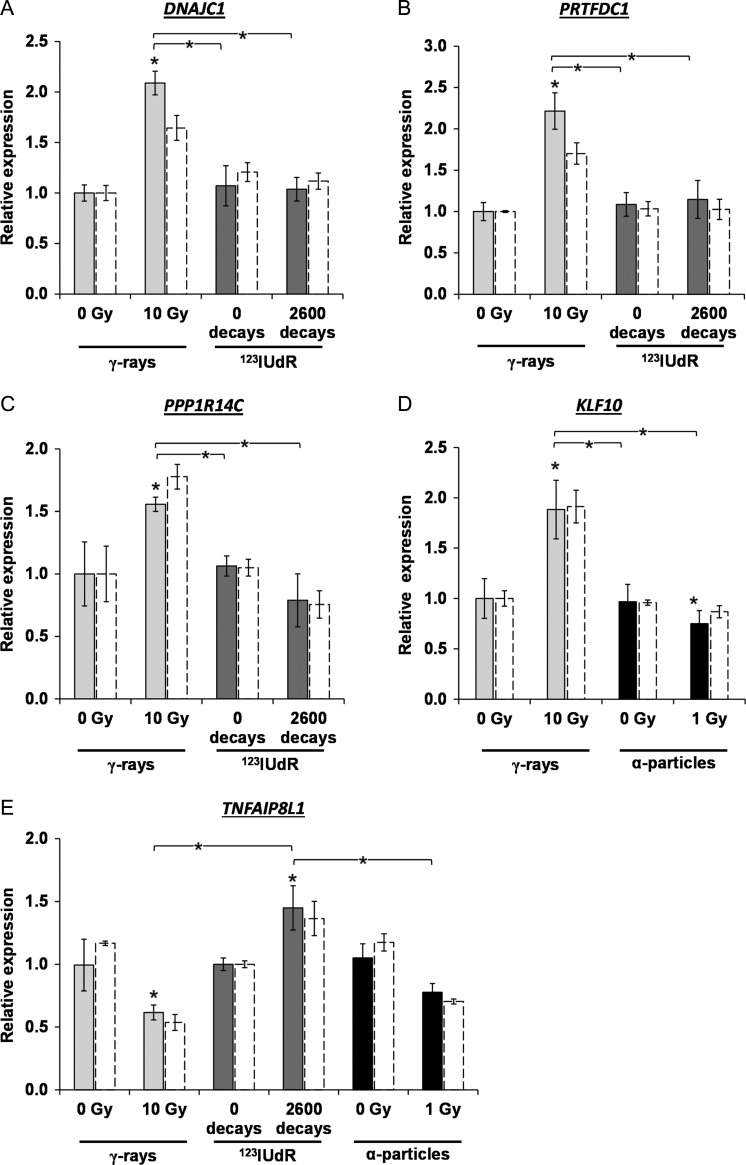
The DNA microarray data were filtered to identify suitable robust candidate genes allowing the discrimination of different radiation qualities. A candidate gene had to show a significant expression fold change of >1.5 fold after exposure to a specific radiation quality, and display no altered (1-fold ± 0.1) or even opposing (>1.1-fold) regulation in response to the other radiation qualities. Concomitantly, we used P values, which were not adjusted using the FDR (Benjamini and Hochberg), for the analysis of the significance in order to consider more genes, as shown in Table 1. However, this loss of statistical stringency was justified because the gene expression level of the candidate genes were verified and validated by qRT PCR (Fig. 6). Accordingly, candidate genes allowing discrimination between the three radiation qualities (γ-radiation, α-radiation and AEE exposure) were identified: four genes for γ-radiation versus 123IUdR exposure (PPP1R14C, TNFAIP8L1, DNAJC1 and PRTFDC1), one gene for α-radiation versus 123IUdR exposure (TNFAIP8L1) and one gene for γ-radiation versus α-radiation (KLF10).
Fig. 6.
Identification of robust candidate genes allowing discrimination of different radiation qualities. (A, B) DNAJC1 and PRTFDC1 6 h after withdrawal of 123IUdR versus 24 h after γ-irradiation; (C) PPP1R14C 6 h after withdrawal of 123IUdR versus 6 h after γ-irradiation; (D) KLF10 24 h after γ-irradiation versus 24 h after α-irradiation; (E) TNFAIP8L1 6 h after withdrawal of 123IUdR versus 6 h after γ- and α-irradiation. Solid bars show the data of the quantitative real-time PCR in comparison with the microarray data (white bars with dashed lines). The error bars represent the standard deviation of the mean (n = 3, *P < 0.05).
The qRT-PCR data revealed that genes PRTFDC1, DNAJC1 and PPP1R14C showed a significantly increased gene expression with a fold change of >1.5 in comparison with the non-irradiated control 24 h and 6 h after γ-irradiation, respectively (Fig. 6a–c). In contrast, no altered regulation of PRTFDC and DNAJC1 and also a 1.35-fold downregulation of PPP1R14C were determined after 123IUdR exposure. The TNFAIP8L1 gene allowed discrimination between 123IUdR exposure versus γ-radiation, as well as 123IUdR exposure versus α-radiation (Fig. 6e). The qRT-PCR data showed that this gene was significantly 1.45-fold upregulated after 123IUdR exposure, whereas it was 1.6-fold (significant) and 1.35-fold (not significant) downregulated 6 h after γ- and α-irradiation, respectively. KLF10 allowed discrimination between γ- and α-radiation (Fig. 6d). According to the qRT-PCR data, this gene was 1.9-fold induced and 1.3-fold repressed (both significantly) 24 h after γ-and α-irradiation, respectively.
For all candidate genes, the displayed microarray and qRT-PCR data of the different radiation qualities were normalized to only one of the non-irradiated controls. However, the gene expression differences between the different controls that could be displayed in this way were small and not significantly different. The data of the DNA microarray analysis was validated with the qRT-PCR (Fig. 6).
Biological processes and pathways affected by different radiation qualities
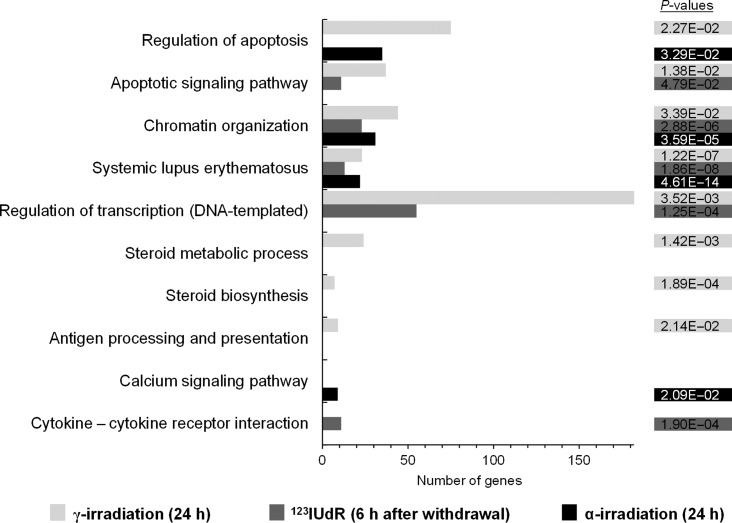
We found that 344, 596 and 1321 genes were significantly deregulated 6 h after exposure to 123IUdR (20 h) and 24 h after α- and γ-irradiation, using the determined equi-effect doses/activities. We functionally categorized these genes, using DAVID [31, 32], to examine biological processes and pathways affected by the three different radiation qualities (Fig. 7). The chromatin organization and the systemic lupus erythematosus pathway were regulated by all three radiation qualities. The regulation of apoptosis was affected by α- or γ-irradiation, and the apoptotic signaling pathway was induced after exposure to γ-radiation or 123IUdR. An effect on the regulation of transcription (DNA-templated) was seen after exposure to γ-radiation and 123IUdR. The steroid biosynthesis and steroid metabolic process, as well as the antigen processing and presentation, were exclusively regulated after γ-irradiation. Additionally, the cytokine – cytokine receptor interaction or the calcium signaling pathway were exclusively affected after exposure to 123IUdR or α-particles, respectively.
Fig. 7.
Genes significantly expressed 6 h after exposure to 123IUdR (20 h), 24 h after α- or γ-irradiation using the determined equi-effect doses/activities were functionally categorized. Affected biological processes and pathways as well as the respective P-values are shown.
DISCUSSION
In order to investigate whether exposure to different radiation qualities is reflected in a significantly different gene expression, a whole human gene expression analysis was carried out in Jurkat cells after exposure to the Auger electron emitter 123I, γ- and α-radiation at equi-effect doses or activities. The DNA microarray analysis revealed that a significant alteration in gene expression was detectable at 24 h, but not at 6 h, after γ- and α-irradiation, when using the FDR values for the statistics. However, the FDR adjustment was responsible for having no regulated genes at 6 h after γ- and α-irradiation, as is shown in Table 1. When considering the genes with P values (P < 0.05) not adjusted by FDR, Table 1 shows that fewer genes were deregulated 6 h after γ- or α-irradiation when compared with 24 h. It has previously been shown in human lymphoblastoid cells and lymphocytes after γ-irradiation [5, 14] as well as in human pulmonary epithelial cells after α-irradiation [35], that more genes were significantly regulated 24 h after irradiation when compared with earlier time points (3–6 h). However, significantly regulated genes were found 6 h after 123IUdR withdrawal from the cell cultures. Because the cells were synchronized and released into the S-phase before they were incubated with 123IUdR for 20 h, a large proportion of cells were assumed to incorporate 123IUdR in the first few hours. Thus, the time between irradiation and RNA isolation was much more similar to that for the cells that were harvested 24 h after α- and γ-irradiation. However, the fraction of cells that incorporated 123IUdR at the end of the 20 h 123IUdR incubation period, and which were more comparable with the cells harvested 6 h after α- and γ-irradiation, most probably had insufficient time to express pronounced alterations in gene expression. This might explain why the overall number of regulated genes 24 h after γ-irradiation was approximately 4-fold higher compared with that after 123IUdR exposure. Another reason for the smaller number of genes showing significantly altered gene expression after 123IUdR exposure might be a dose rate effect. Cells exposed to 123IUdR received the dose in a time window of ~20 h whereas the γ-irradiated cells were exposed only for 30 min. In contrast, experiments conducted by Sokolov et al. did not show any dose rate effect [7]. They exposed human lung fibroblasts to 125IUdR and 1 Gy of external γ-radiation delivered at a high (HDR) or low dose rate (LDR), so that approximately 21–23 γ-H2AX foci were counted 48 h after exposure to 125IUdR and after 1 Gy HDR γ-irradiation. Using this experimental design for the DNA microarrays, Sokolov et al. found no differences in the number of differentially expressed genes following HDR and LDR γ-radiation, but ten times fewer differentially regulated genes in the 125IUdR-treated cells [7]. Sokolov et al. suggested that the effect of ionizing radiation on genome-wide gene expression depends rather on the distribution of energy depositions within the cell than on dose rate [7]. Similar to our findings, Sokolov et al. reported that 80–90% of the 125I-associated radioactivity was DNA-bound, which means that the cell nucleus was irradiated almost exclusively [7].
The DNA microarray analysis revealed that the number of significantly regulated genes 24 h after 1 Gy α-irradiation was 2.2-fold lower than that 24 h after the 10-fold dose of 10 Gy γ-rays, even though the observed extent of DNA damage was similar. Similar to our findings, Kurpinski et al. showed in human mesenchymal stem cells that the alterations in gene expression 24 h after 0.1 Gy exposure to Fe ions were 4.4-fold lower compared with cells irradiated 24 h after 1 Gy X-rays [36]. In contrast, Danielsson et al. observed in human foreskin fibroblast cells a 1.4-fold stronger induction of gene expression 5 h after 0.25 Gy α-irradiation, compared with a 12-fold higher radiation dose of γ-rays [12].
The analysis of the significantly regulated genes regarding biological processes and/or pathways showed that apoptosis is regulated after exposure to 123IUdR, γ- and α-radiation. A significant deregulation of apoptotic relevant genes was also shown by Danielsson et al., who irradiated primary human foreskin fibroblast cells with α-particles and γ-rays [12]. Interestingly, Sokolov et al. showed that 125IUdR did not cause deregulation of apoptosis-associated genes, in contrast to γ-radiation delivered at high or low dose rates [6]. In our study, the chromatin organization and the systemic lupus erythematosus pathway were significantly regulated by all three radiation qualities. Here, a detailed analysis showed that mostly histone genes were involved, and that these were predominantly downregulated. This downregulation can be explained by a radiation-induced cell cycle arrest followed by an inhibition of DNA synthesis and histone gene downregulation, because no newly synthesized DNA had to be assembled by histones [37, 38]. A downregulation of histone genes was also shown in human fibroblasts after 125IUdR exposure [39], as well as in human lymphoblastoid cell lines after exposure to γ-rays and iron ions [40]. In addition to a regulation by all three radiation qualities, an exclusive regulation was found regarding the cytokine – cytokine receptor interaction after exposure to 123IUdR, which was already reported by Knops et al. in human lymphocytes after γ-irradiation [5]. The calcium signaling pathway was uniquely regulated after α-irradiation, which is similar to the data of Mezentsev et al. showing a regulation of three pathway-related genes in human keratinocytes after proton-irradiation [41]. Steroid biosynthesis and the steroid metabolic process were exclusively affected after γ-irradiation. Data showing a link between steroid-relevant genes and radiation are rare, although Lin et al. reported regulation of a steroid receptor gene in breast cancer cells after ionizing irradiation [42]. A further exclusive regulated pathway induced by γ-irradiation but not by 123IUdR exposure in our study was antigen processing and presentation. This is in very good accordance with the data of Sokolov et al. showing that this pathway was only regulated after γ-irradiation but not after exposure to 125IUdR [6]. Interestingly, the candidate genes allowing the discrimination between the different radiation qualities were not involved in any over-represented biological processes or pathways.
Microarray studies in irradiated Jurkat cells are very sparse. Only Park et al. and Mori et al. did partial genome microarray analyses, using arrays representing only 2400 or 4300 human cDNAs, respectively very much in contrast to our whole-genome microarray analyses including 41 000 genes and transcripts [43, 44].
When comparing the list of 38 genes regulated in Jurkat cells 24 h after 8 Gy γ-irradiation from Park et al. with the 1321 significantly regulated genes 24 h after 10 Gy γ-irradiation in our study, only IL3RA (interleukin 3 receptor, alpha) was identified to be upregulated in both analyses [44]. However, one has to keep in mind that the study of Park et al. was restricted to only 1/20 of the initial transcripts of our study. The list of 90 genes significantly regulated 24 h after 2 Gy X-ray exposure published by Mori et al. share some similarities with our data [43]. An upregulation of ANAPC1 (anaphase-promoting complex subunit 1), ATF3 (activating transcription factor 3) and DDB2 (damage-specific DNA-binding protein 2), as well as a downregulation of RAVER1 (ribonucleoprotein, PTB-binding 1) and ARHGDIA (Rho GDP dissociation inhibitor (GDI) alpha), was found in both studies. Again, Mori et al. determined the gene expression in only 4300 human cDNAs and irradiated the cells with 2 Gy X-rays only, both of which contribute to the overall small number of deregulated genes and the small overlap with our gene list.
The Venn diagram of the DNA microarray data showed that 155, 316 and 982 genes were exclusively differentially regulated 6 h after 123IUdR withdrawal and 24 h after α- and γ-irradiation, respectively (Fig. 4). However, only four (PPP1R14C, TNFAIP8L1, DNAJC1 and PRTFDC1), one (KLF10) and one (TNFAIP8L1) gene(s) identified, that enable a reliable discrimination of γ- versus 123IUdR exposure, γ- versus α-radiation and α- versus 123IUdR exposure, respectively (Fig. 6). Obviously, the number of potential candidate genes was considerably reduced due to the stringent requirements of the filtering process already mentioned in Results.
The gene TNFAIP8L1 enabled discrimination of 123IUdR exposure versus γ-radiation as well as 123IUdR exposure versus α-radiation. TNFAIP8L1, also known as TIPE1, plays a role in cell secretion, carcinogenesis, and cell death regulation and is associated with high levels of mRNA in most human carcinoma cell lines [45]. It has been reported that the expression of TNFAIP8L1 can already be induced 1 h after oxidative stress via ROS [46]. Due to the ongoing decay of 123IUdR until cell harvest, it can be assumed that ROS were continuously generated in Jurkat cells and might induce the upregulation of TNFAIP8L1. In contrast, after γ- and α-irradiation, ROS were induced at the time of exposure, probably leading to an immediate upregulation of TNFAIP8L1, but this was followed by a downregulation in the 6 h until cell harvest, mostly because of (i) absence of the short-lived ROS and (ii) the prevailing enhanced levels of the TNFAIP8L1 gene product, which could cause downregulation by a negative feedback loop.
The genes PPP1R14C, DNAJC1 and PRTFDC1 allowed differentiation between 123IUdR exposure and γ-radiation. According to present knowledge about the function of their gene products, it cannot be deduced why these genes showed differential gene expression regulation after γ-irradiation and 123IUdR exposure. PPP1R14C alias KEPI is a PKC-dependent protein phosphatase 1 inhibitor. Previous studies have noted that PPP1R14C is downregulated in melanoma and breast cancer cell lines compared with in normal cells [47, 48]. An association of PPP1R14C with ionizing radiation was only shown by Datta et al. in mice [49]. These authors observed a changed regulation 2 months after 2 Gy γ-irradiation in mammary gland cells. The gene product of DNAJC1, alias MTJ1 or HTJ1, is a transmembrane and heat shock protein, which plays a role in protein folding and trafficking, apoptosis, immune regulation [50, 51] and probably chromatin remodeling [52]. PRTFDC1 was identified as a potential tumor suppressor gene in cancer cells [53, 54], but the function of PRTFDC1 is still unknown.
The expression data showed that the gene KLF10 allowed discrimination between γ- and α-radiation. KLF10, also known as TIEG or TIEG1, plays a role in the TGFbeta/Smad pathway and has been implicated in apoptosis induction, inflammation inhibition, and cell differentiation and proliferation [55]. Previous studies showed that the TGFbeta/Smad pathway could be activated in human cells by the radiation-induced generation of ROS [56, 57]. Additionally, it was shown that the expression of KLF10 was induced during ROS-related apoptosis induction in hepatoma cells [58]. Why KLF10 was induced 24 h after γ-irradiation and repressed after α-irradiation cannot be conclusively deduced from the present stage of knowledge about the function of its gene product. Moreover, Cao et al. stated that the KLF10 gene product does not seem to have any function in Jurkat cells [59]. Interestingly, Cao et al. observed in Jurkat cells that after TGF-β treatment the expression level of KLF10 increased rapidly, reached a maximum after 90 min and decreased under the control level after 24 h [59]. Thus, it might be speculated that the α-particles used in our study might have induced KLF10 expression in a very similar way; however, the gene expression was still upregulated 24 h after γ-irradiation. A radiation-induced upregulation of KLF10, alias TIEG, was also shown by Sokolov et al. in human lung fibroblasts after exposure to 125IUdR and γ-rays delivered at a high dose rate [7]. However, the data from the present study showed only a long-term upregulation of KLF10 after γ-irradiation, but not after exposure to 123IUdR.
In this study, for the first time, AEEs were compared with high-LET as well as low-LET radiation in terms of differential gene expression for identifying robust gene signatures, allowing discrimination between radiation qualities. Therefore, a direct comparison with other studies is rather difficult.
Regarding a comparison between Auger electrons and other radiation qualities, the studies by Sokolov et al. are the only global microarray studies at present [6, 7]. They analyzed the gene expression of normal human lung fibroblasts after exposure to 125IUdR and external γ-radiation delivered at HDR or LDR. Among the genes that were exclusively expressed after γ-irradiation, Sokolov et al. mentioned, among other things, the proliferation-regulating genes RB1 and GADD45A, the cyclin-dependent kinase regulator gene CDK5R1, the cyclin gene CCNB1 and the mitosis-related kinase gene CDK [7]. None of these genes showed a significant regulation in our study.
Ding et al. irradiated normal human bronchial epithelial cells with γ-rays, silicon and iron ions and employed DNA microarrays using equi-effect doses, similar to the design of our study [8]. Ding et al. developed a 73-gene signature set capable of predicting the radiation quality to which cells had been exposed [8]. However, Chauhan et al. employed DNA microarrays and qRT-PCR technology to identifying biomarkers in peripheral human blood mononuclear cells after α-irradiation and X-ray exposure [9]. They showed that both radiation types elicited similar gene expression responses with a varying degree of fold change. Finally, however, they were unable to identify any biomarkers that would allow a robust discrimination between these two radiation qualities. Since we also only detected one suitable gene (KLF10) for the robust discrimination of α-irradiation and γ-rays, it seems that α-irradiation and low-LET exposure might be hard to distinguish on the gene expression level.
In a further study, Chauhan et al. examined α-particle radiation effects on gene expression changes in order to identify potential signaling pathways that may be involved in radon gas exposure and lung carcinogenesis using human primary lung fibroblasts [11]. Interestingly, in this study Chauhan et al. applied qRT-PCR and found that the genes GDF15, LOC387763, PDK4, ANGPTL4, NR4A2 and FGF2 showed a significant upregulation after α-irradiation and no significant regulation after X-ray exposure [11]. The expression of these genes was not significantly changed in our analysis.
For the first time, robust gene signatures were identified, allowing discrimination between exposure to Auger electrons, γ-radiation and α-radiation. Applying our very stringent requirements for marker genes, four, one and one gene(s) allowed discrimination between γ- and 123IUdR exposure, α- and 123IUdR exposure, and γ- and α-radiation, respectively, at equi-effect doses/activity.
The experiments should be expanded to include other cell types, especially human peripheral blood lymphocytes; however, the implementation of such marker genes into gene expression–based biodosimetry might prove to be challenging. Nevertheless, the knowledge obtained is very valuable as it shows that the cellular response as observed on the gene expression level is not only significantly different as a function of radiation dose, but also with respect to radiation quality. Not only does discrimination between high-LET α-radiation and low-LET γ-radiation seem to be feasible, but also distinction between different types of high-LET radiation, since the AEE 123I used in this study has high-LET properties, especially with respect to the density of energy deposition inside a nano-scale target volume during decay [60–62].
ACKNOWLEDGEMENTS
The authors thank A. Heiske for excellent technical support in preparing the cell dishes and operating the α-irradiator at PTB, as well as D. Oskamp for support regarding 123IUdR synthesis.
CONFLICT OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
FUNDING
This work was supported by the German Federal Ministry of Education and Research [BMBF; grant numbers 02NUK005A and 02NUK043A].
REFERENCES
- 1. Jaworska A, Ainsbury EA, Fattibene P et al. Operational guidance for radiation emergency response organisations in Europe for using biodosimetric tools developed in EU MULTIBIODOSE project. Radiat Prot Dosimetry 2015;164:165–9. [DOI] [PubMed] [Google Scholar]
- 2. Romm H, Ainsbury E, Bajinskis A et al. Web-based scoring of the dicentric assay, a collaborative biodosimetric scoring strategy for population triage in large scale radiation accidents. Radiat Environ Biophys 2014;53:241–54. [DOI] [PubMed] [Google Scholar]
- 3. Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys 2008;71:1236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boldt S, Knops K, Kriehuber R et al. A frequency-based gene selection method to identify robust biomarkers for radiation dose prediction. Int J Radiat Biol 2012;88:267–76. [DOI] [PubMed] [Google Scholar]
- 5. Knops K, Boldt S, Wolkenhauer O et al. Gene expression in low- and high-dose-irradiated human peripheral blood lymphocytes: possible applications for biodosimetry. Radiat Res 2012;178:304–12. [DOI] [PubMed] [Google Scholar]
- 6. Sokolov M, Panyutin IG, Neumann R. Genome-wide gene expression changes in normal human fibroblasts in response to low-let gamma-radiation and high-let-like 125IUdR exposures. Radiat Prot Dosimetry 2006;122:195–201. [DOI] [PubMed] [Google Scholar]
- 7. Sokolov MV, Smirnova NA, Camerini-Otero RD et al. Microarray analysis of differentially expressed genes after exposure of normal human fibroblasts to ionizing radiation from an external source and from DNA-incorporated iodine-125 radionuclide. Gene 2006;382:47–56. [DOI] [PubMed] [Google Scholar]
- 8. Ding LH, Park S, Peyton M et al. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to gamma-rays and different elemental particles of high Z and energy. BMC Genomics 2013;14:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chauhan V, Howland M, Wilkins R. Identification of gene-based responses in human blood cells exposed to alpha particle radiation. BMC Med Genomics 2014;7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chauhan V, Howland M, Greene HB et al. Transcriptional and secretomic profiling of epidermal cells exposed to alpha particle radiation. Open Biochem J 2012;6:103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chauhan V, Howland M. Gene expression responses in human lung fibroblasts exposed to alpha particle radiation. Toxicol In Vitro 2014;28:1222–9. [DOI] [PubMed] [Google Scholar]
- 12. Danielsson A, Claesson K, Parris TZ et al. Differential gene expression in human fibroblasts after alpha-particle emitter 211At compared with 60Co irradiation. Int J Radiat Biol 2013;89:250–8. [DOI] [PubMed] [Google Scholar]
- 13. Rothkamm K, Beinke C, Romm H et al. Comparison of established and emerging biodosimetry assays. Radiat Res 2013;180:111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amundson SA, Bittner M, Meltzer P et al. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res 2001;156:657–61. [DOI] [PubMed] [Google Scholar]
- 15. Amundson SA, Do KT, Vinikoor LC et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res 2008;68:415–24. [DOI] [PubMed] [Google Scholar]
- 16. Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene 2003;22:5734–54. [DOI] [PubMed] [Google Scholar]
- 17. Goodhead DT. The initial physical damage produced by ionizing-radiations. Int J Radiat Biol 1989;56:623–34. [DOI] [PubMed] [Google Scholar]
- 18. Burkart W, Jung T, Frasch G. Damage pattern as a function of radiation quality and other factors. C R Acad Sci III 1999;322:89–101. [DOI] [PubMed] [Google Scholar]
- 19. Jenner TJ, deLara CM, O’Neill P et al. Induction and rejoining of DNA double-strand breaks in V79-4 mammalian-cells following gamma-irradiation and alpha-irradiation. Int J Radiat Biol 1993;64:265–73. [DOI] [PubMed] [Google Scholar]
- 20. Ottolenghi A, Merzagora M, Paretzke HG. DNA complex lesions induced by protons and alpha-particles: track structure characteristics determining linear energy transfer and particle type dependence. Radiat Environ Biophys 1997;36:97–103. [DOI] [PubMed] [Google Scholar]
- 21. Prise KM, Folkard M, Newman HC et al. Effect of radiation quality on lesion complexity in cellular DNA. Int J Radiat Biol 1994;66:537–42. [DOI] [PubMed] [Google Scholar]
- 22. Karlsson KH, Stenerlow B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat Res 2004;161:517–27. [DOI] [PubMed] [Google Scholar]
- 23. Takahashi A, Yamakawa N, Kirita T et al. DNA damage recognition proteins localize along heavy ion induced tracks in the cell nucleus. J Radiat Res 2008;49:645–52. [DOI] [PubMed] [Google Scholar]
- 24. Stenerlow B, Hoglund E, Carlsson J et al. Rejoining of DNA fragments produced by radiations of different linear energy transfer. Int J Radiat Biol 2000;76:549–57. [DOI] [PubMed] [Google Scholar]
- 25. Ritter S, Durante M. Heavy-ion induced chromosomal aberrations: a review. Mutat Res 2010;701:38–46. [DOI] [PubMed] [Google Scholar]
- 26. Howell RW, Rao DV, Hou DY et al. The question of relative biological effectiveness and quality factor for auger emitters incorporated into proliferating mammalian cells. Radiat Res 1991;128:282–92. [PubMed] [Google Scholar]
- 27. Rao DV, Narra VR, Howell RW et al. In-vivo radiotoxicity of DNA-incorporated 125I compared with that of densely ionising alpha-particles. Lancet 1989;2:650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pomplun E, Terrissol M, Hille R. Ratio of complex double strand break damage induced by 125IUdR and 123IUdR correlates with experimental in vitro cell killing effectiveness. Radiat Prot Dosimetry 2002;99:81–2. [DOI] [PubMed] [Google Scholar]
- 29. Greif KD, Brede HJ, Frankenberg D et al. The PTB single ion microbeam for irradiation of living cells. Nucl Instrum Meth Phys Res B 2004;217:505–12. [Google Scholar]
- 30. Baranowska-Kortylewicz J, Helseth L, Lai J et al. Radiolabeling kit/generator for 5-radiohalogenated uridines. J Labelled Compd Radiopharm 1994;34:513–21. [Google Scholar]
- 31. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 32. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rogakou EP, Pilch DR, Orr AH et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858–68. [DOI] [PubMed] [Google Scholar]
- 35. Chauhan V, Howland M, Mendenhall A et al. Effects of alpha particle radiation on gene expression in human pulmonary epithelial cells. Int J Hyg Environ Health 2012;215:522–35. [DOI] [PubMed] [Google Scholar]
- 36. Kurpinski K, Jang DJ, Bhattacharya S et al. Differential effects of x-rays and high-energy 56Fe ions on human mesenchymal stem cells. Int J Radiat Oncol Biol Phys 2009;73:869–77. [DOI] [PubMed] [Google Scholar]
- 37. Su C, Gao G, Schneider S et al. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J 2004;23:1133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao J. Coordination of DNA synthesis and histone gene expression during normal cell cycle progression and after DNA damage. Cell Cycle 2004;3:695–7. [PubMed] [Google Scholar]
- 39. Sokolov MV, Neumann RD, Panyutin IG. Effects of DNA-targeted ionizing radiation produced by 5-[125I]iodo-2'-deoxyuridine on global gene expression in primary human cells. BMC Genomics 2007;8:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meador JA, Ghandhi SA, Amundson SA. p53-independent downregulation of histone gene expression in human cell lines by high- and low-LET radiation. Radiat Res 2011;175:689–99. [DOI] [PubMed] [Google Scholar]
- 41. Mezentsev A, Amundson SA. Global gene expression responses to low- or high-dose radiation in a human three-dimensional tissue model. Radiat Res 2011;175:677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin DL, Chang C. p53 is a mediator for radiation-repressed human TR2 orphan receptor expression in MCF-7 cells, a new pathway from tumor suppressor to member of the steroid receptor superfamily. J Biol Chem 1996;271:14649–52. [DOI] [PubMed] [Google Scholar]
- 43. Mori M, Benotmane MA, Vanhove D et al. Effect of ionizing radiation on gene expression in CD4+ T lymphocytes and in Jurkat cells: unraveling novel pathways in radiation response. Cell Mol Life Sci 2004;61:1955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park W-Y, Hwang C-I, Im C-N et al. Identification of radiation-specific responses from gene expression profile. Oncogene 2002;21:8521. [DOI] [PubMed] [Google Scholar]
- 45. Cui J, Zhang G, Hao C et al. The expression of TIPE1 in murine tissues and human cell lines. Mol Immunol 2011;48:1548–55. [DOI] [PubMed] [Google Scholar]
- 46. Ha JY, Kim JS, Kang YH et al. Tnfaip8 l1/Oxi-β binds to FBXW5, increasing autophagy through activation of TSC2 in a Parkinson’s disease model. J Neurochem 2014;129:527–38. [DOI] [PubMed] [Google Scholar]
- 47. Daskalow K, Boisguerin P, Jandrig B et al. Generation of an antibody against the protein phosphatase 1 inhibitor KEPI and characterization of the epitope. Anticancer Res 2010;30:1573–8. [PubMed] [Google Scholar]
- 48. Liu QR, Zhang PW, Zhen Q et al. KEPI, a PKC-dependent protein phosphatase 1 inhibitor regulated by morphine. J Biol Chem 2002;277:13312–20. [DOI] [PubMed] [Google Scholar]
- 49. Datta K, Hyduke DR, Suman S et al. Exposure to ionizing radiation induced persistent gene expression changes in mouse mammary gland. Radiat Oncol 2012;7:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Misra UK, Gonzalez-Gronow M, Gawdi G et al. The role of MTJ-1 in cell surface translocation of GRP78, a receptor for alpha 2-macroglobulin-dependent signaling. J Immunol 2005;174:2092–7. [DOI] [PubMed] [Google Scholar]
- 51. Papalas JA, Vollmer RT, Gonzalez-Gronow M et al. Patterns of GRP78 and MTJ1 expression in primary cutaneous malignant melanoma. Mod Pathol 2010;23:134–43. [DOI] [PubMed] [Google Scholar]
- 52. Kroczynska B, King-Simmons L, Alloza L et al. BIP co-chaperone MTJ1/ERDJ1 interacts with inter-alpha-trypsin inhibitor heavy chain 4. Biochem Biophys Res Commun 2005;338:1467–77. [DOI] [PubMed] [Google Scholar]
- 53. Cai LY, Abe M, Izumi SI et al. Identification of PRTFDC1 silencing and aberrant promoter methylation of GPR150, ITGA8 and HOXD11 in ovarian cancers. Life Sci 2007;80:1458–65. [DOI] [PubMed] [Google Scholar]
- 54. Suzuki E, Imoto I, Pimkhaokham A et al. PRTFDC1, a possible tumor-suppressor gene, is frequently silenced in oral squamous-cell carcinomas by aberrant promoter hypermethylation. Oncogene 2007;26:7921–32. [DOI] [PubMed] [Google Scholar]
- 55. Subramaniam M, Hawse JR, Rajamannan NM et al. Functional role of KLF10 in multiple disease processes. BioFactors 2010;36:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang M, Saha J, Hada M et al. Novel Smad proteins localize to IR-induced double-strand breaks: interplay between TGFβ and ATM pathways. Nucleic Acids Res 2013;41:933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kirshner J, Jobling MF, Pajares MJ et al. Inhibition of transforming growth factor-β1 signaling attenuates ataxia telangiectasia mutated activity in response to genotoxic stress. Cancer Res 2006;66:10861–9. [DOI] [PubMed] [Google Scholar]
- 58. Ribeiro A, Bronk SF, Roberts PJ et al. The transforming growth factor β1-inducible transcription factor, TIEG1, mediates apoptosis through oxidative stress. Hepatology 1999;30:1490–97. [DOI] [PubMed] [Google Scholar]
- 59. Cao Z, Wara AK, Icli B et al. Kruppel-like factor KLF10 targets transforming growth factor-β1 to regulate CD4+CD25− T cells and T regulatory cells. J Biol Chem 2009;284:24914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boyd M, Ross SC, Dorrens J et al. Radiation-induced biologic bystander effect elicited in vitro by targeted radiopharmaceuticals labeled with alpha-, beta-, and auger electron-emitting radionuclides. J Nucl Med 2006;47:1007–15. [PubMed] [Google Scholar]
- 61. Boyd M, Sorensen A, McCluskey AG et al. Radiation quality-dependent bystander effects elicited by targeted radionuclides. J Pharm Pharmacol 2008;60:951–8. [DOI] [PubMed] [Google Scholar]
- 62. Mairs RJ, Fullerton NE, Zalutsky MR et al. Targeted radiotherapy: microgray doses and the bystander effect. Dose Response 2007;5:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]