Repression of ABA degradation in transgenic barley alters the expression of genes involved in strigolactone biosynthesis, revealing cross-talk between ABA and strigolactone that affects the development of tillers.
Keywords: Abscisic acid, barley, cereals, hormone regulation, phytohormone cross-talk, shoot branching, strigolactone biosynthesis, tillering
Abstract
Strigolactones (SLs) represent a class of plant hormones that are involved in inhibiting shoot branching and in promoting abiotic stress responses. There is evidence that the biosynthetic pathways of SLs and abscisic acid (ABA) are functionally connected. However, little is known about the mechanisms underlying the interaction of SLs and ABA, and the relevance of this interaction for shoot architecture. Based on sequence homology, four genes (HvD27, HvMAX1, HvCCD7, and HvCCD8) involved in SL biosynthesis were identified in barley and functionally verified by complementation of Arabidopsis mutants or by virus-induced gene silencing. To investigate the influence of ABA on SLs, two transgenic lines accumulating ABA as a result of RNAi-mediated down-regulation of HvABA 8’-hydroxylase 1 and 3 were employed. LC-MS/MS analysis confirmed higher ABA levels in root and stem base tissues in these transgenic lines. Both lines showed enhanced tiller formation and lower concentrations of 5-deoxystrigol in root exudates, which was detected for the first time as a naturally occurring SL in barley. Lower expression levels of HvD27, HvMAX1, HvCCD7, and HvCCD8 indicated that ABA suppresses SL biosynthesis, leading to enhanced tiller formation in barley.
Introduction
Strigolactones (SLs) are a group of carotenoid-derived lactones that have been recently identified as regulators of plant growth and development. SLs were initially characterized as rhizosphere signals that stimulate seed germination in root parasitic plants (Xie et al., 2010; Ruyter-Spira et al., 2013). Later, they were also identified as signals for the establishment of symbiosis with arbuscular-mycorrhizal fungi, a process that improves phosphorus nutrition and water uptake in plants (Akiyama and Hayashi, 2006; Xie et al., 2010). Another major function of SLs is based on their translocation from roots to shoots, where they suppress lateral shoot branching by inhibiting the outgrowth of axillary buds (Gomez-Roldan et al., 2008; Umehara et al., 2008). Insights into the key components of SL biosynthesis and signaling have been gained from studies of mutants with different branching phenotypes in a variety of plant species, such as ramosus (rms) in pea, decreased apical meristem (dad) in Petunia, more axillary growth (max) in Arabidopsis, and dwarf (d)/high tillering dwarf (htd) in rice (Sorefan et al., 2003; Booker et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007; Simons et al., 2007; Drummond et al., 2009; Brewer et al., 2016). These studies confirmed a conserved role for SLs in the regulation of axillary bud outgrowth (Domagalska and Leyser, 2011; Ruyter-Spira et al., 2013). SLs are also involved in other aspects of development, including root architecture, leaf senescence, secondary growth, and drought tolerance (Woo et al., 2001; Snowden et al., 2005; Agusti et al., 2011; Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Rasmussen et al., 2012; Bu et al., 2014; Yamada et al., 2014; Ueda and Kusaba, 2015).
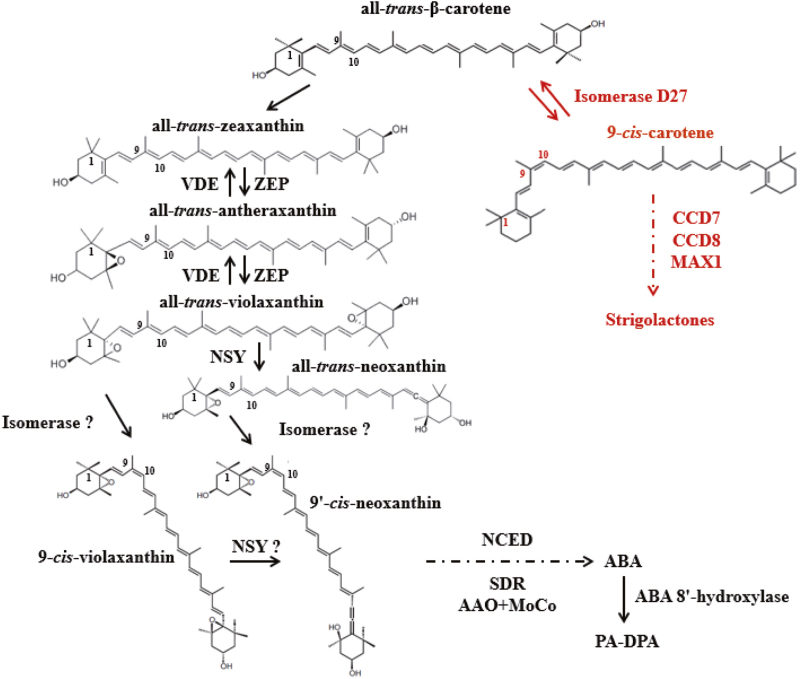
To date, more than 20SLs have been isolated and characterized from root exudates of various plant species. All canonical SLs contain a tricyclic lactone (ABC part) that connects via an enol ether linkage to a butenolide moiety (D-ring); they often carry different substituents on the A- and B-rings but have the same C–D moiety (Xie et al., 2013). 5-deoxystrigol and 4-deoxyorobanchol are thought to be the common precursors of the canonical strigol- and orobanchol-type SLs (Al-Babili and Bouwmeester, 2015). Strigolactones are derived from the carotenoid biosynthetic pathway, in which all-trans-β-carotene acts as the precursor for SL biosynthesis (Fig. 1). Several enzymes act sequentially in SL biosynthesis (Saeed et al., 2017): DWARF27 (D27) is a β-carotene isomerase, CAROTENOID CLEAVAGE DIOXYGENASE 7 (CCD7) and CCD8 sequentially cleave 9-cis-β-carotene to produce the carotenoid precursor carlactone (CL). In rice, CL is oxidized by two cytochrome P450 enzymes (carlactone oxidase and 4-deoxyorobanchol-4-hydroxylase), converting CL into 4-deoxyorobanchol and further into orobanchol (Zhang et al., 2015). MORE AXILLARY GROWTH 1 (MAX1) was identified as a factor upstream of these, providing the early catalytic activity, and both redundancy and functional diversification are associated with gene duplication in angiosperm lineages (Challis et al., 2013). MAX1 was identified as a class Ш cytochrome P450 enzyme in Arabidopsis (Waldie et al., 2014) that converts CL into carlactonoic acid (CLA), which is further converted in a species-specific way to either methyl carlactonoate (MeCLA) (Abe et al., 2014), 4-deoxyorobanchol, or orobanchol (Yoneyama et al., 2018). Finally, MeCLA is the substrate for LATERAL BRANCHING OXIDOREDUCTASE (LBO), resulting in the formation of an unidentified SL-like compound (Brewer et al., 2016). This core set of enzymes is mostly active in roots, and root-synthesized SLs are secreted by ABCG-type transporters such as PhPDR1 into the rhizosphere or transported to the shoot (Kretzschmar et al., 2012; Sasse et al., 2015). Perception and signaling of SLs in plants requires two major proteins, namely MAX2, an F-box protein of the Skp–cullin–F-box (SCF) E3 ubiquitin ligase complex (Stirnberg et al., 2007; Smith and Li, 2014), and DWARF14 (D14), an α/β-fold hydrolase protein (Arite et al., 2009; Hamiaux et al., 2012). The D14 protein has been shown to act as a SL receptor by hydrolysing SLs into a covalently linked intermediate, which initiates a conformational change of D14 that facilitates its interaction with MAX2 (de Saint Germain et al., 2016; Yao et al., 2016). In barley, the identification and functional analysis of D14 was achieved by analysis of tilling mutants (Marzec et al., 2016).
Fig. 1.
Biosynthetic pathways of abscisic acid and strigolactones. Solid arrows indicate one-step modification of an intermediate and dashed arrows represent multistep modifications of an intermediate. Enzyme names are given in bold. Abbreviations: ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; NSY, neoxanthin synthase; NCED, 9-cis-epoxycarotenoid dioxygenase; SDR, short-chain dehydrogenase/reductase; AAO, ABA aldehyde oxidase; MoCo, molybdenum cofactor synthase; D27, dwarf 27; CCD7/8, carotenoid cleavage dioxygenase 7/8; MAX1, more axillary growth 1.
Abscisic acid (ABA) is a key regulator of plant responses to abiotic stresses and is involved in numerous developmental processes (Vishwakarma et al., 2017). Both ABA and SLs are carotenoid-derived molecules (apocarotenoids) that share all-trans-β-carotene as a common biosynthetic precursor (Matusova et al., 2005). The precursor is derived from the isoprene derivative isopentenyl diphosphate (IPP) (Schwartz et al., 1997) and is converted by a series of epoxidation and hydroxylation reactions into two all-trans-epoxycarotenoids, namely all-trans-violaxanthin and all-trans-neoxanthin (Al-Babili and Bouwmeester, 2015). Violaxanthin and neoxanthin are then converted into 9-cis and 9’-cis isomers, respectively, by an unknown cis/trans isomerase. Alder et al. (2012) found that the rice SL biosynthetic enzyme D27 has isomerase activity, converting all-trans-β-carotene into 9-cis-β-carotene, and that this conversion is reversible. The structural similarity between β-carotene and epoxycarotenoids raises the possible involvement of D27 in ABA biosynthesis. Next, 9-cis-EPOXYCAROTENOID DIOXYGENASE (NCED) catalyses the oxidative cleavage of 9-cis-violaxanthin and/or 9’-cis-neoxanthin to produce xanthoxin, which is considered the rate-limiting step in ABA biosynthesis (Chernys and Zeevaart, 2000; Iuchi et al., 2001; Qin and Zeevaart, 2002). For ABA catabolism, the majority of ABA is converted to its inactive form via hydroxylation at the 8’-carbon atom, and this is carried out by the cytochrome P450-type protein ABA-8’-HYDROXYLASE. In barley, three members of the ABA8’-OH gene family have been described (Seiler et al., 2014). Spontaneous cyclization of hydroxylated ABA results in the formation of phaseic acid (PA), which is further reduced to dihydrophaseic acid (DPA) by a soluble reductase. The spontaneous cyclization to PA results in a significant reduction in the biological activity, and reduction to DPA results in a complete loss of activity (Zou et al., 1995; Zhou et al., 2004; Seiler et al., 2011). In line with its important role as a phytohormone, a fine-tuned balance between biosynthesis, inactivation, and degradation controls ABA homeostasis (Seiler et al., 2011).
Given its biosynthetic origin and the importance of ABA in abiotic stress responses, attention has been drawn to a possible interaction between it and SLs, and the consequences this would have for abiotic stress responses. In drought-stressed leaves, ABA levels usually rise due to increased NCED activity, which results in stomatal closure (Zhu, 2002). In Lotus japonicus, osmotic stress was found to decrease SL levels in root tissues and exudates, primarily by altering transcript levels of biosynthetic and transporter-encoding genes (Liu et al., 2015). Pre-treatment of plants with SLs inhibited the osmotic stress-induced ABA increase in roots by down-regulating the ABA biosynthetic gene LjNCED2. During osmotic stress in L. japonicus roots, only a decrease in SL levels allowed ABA to increase (Liu et al., 2015). A negative interaction between ABA and SLs was also observed in tomato (Ruiz-Lozano et al., 2016). In Arabidopsis mutants, a suppressive function of ABA was shown for axillary bud outgrowth that depended on the ratio of red to far-red light (Reddy et al., 2013; Yao and Finlayson, 2015). However, the molecular mechanism underlying the interaction of SLs and ABA remains unclear.
In this study, we investigated the interaction between ABA and SLs and its effect on shoot architecture in barley under non-stressed conditions. Employing HvABA 8’-hydroxylase RNAi plants (Seiler et al., 2014), we analysed phytohormone levels and tiller formation. As a prerequisite for this study, four barley genes involved in SL biosynthesis were identified and functionally characterized by genetic complementation and virus-induced gene silencing (VIGS). Our results indicate that enhanced levels of endogenous ABA cause a decrease in SL production via repression of SL biosynthesis genes during a late developmental phase in barley. Our results provide genetic evidence for an antagonistic interaction between SLs and ABA that takes place at the level of SL biosynthesis.
Materials and methods
Plant material and cultivation
Hordeum vulgare cv. Golden Promise (two-rowed spring barley cultivar, from the IPK Gatersleben gene bank) and the derived transgenic lines LOHi236 and LOHi272 (Seiler et al., 2014) were cultivated in a growth chamber. Seeds were sown directly in pots filled with two parts compost, two parts ‘Substrat 2’ (Klasmann-Deilmann), and one part quartz sand. Vernalization was performed for a period of 8 weeks from germination under a temperature regime of 11/7 °C day/ night with 10 h light. Growing conditions were divided into four phases: (1) 2 weeks at 14 /9 °C day/night with 12 h light; (2) 2 weeks at 16/9 °C day/night with 14 h light; (3) 2 weeks at 20/12 °C day/night with 16 h light; and (4) ongoing 20/14 °C day/night with 16 h light. The plants were fertilized regularly with Plantacote pluss (15 g pot-1; Aglukon) during the vegetative phase and with ‘Hakaphos Rot’ liquid fertilizer (once a week; 2–4%; Compo Expert) from the start of spike development. Total tiller number until maturity was counted for all plants every week (n≥20 plants). Hordeum vulgare cv. Black Hulless, a six-rowed spring barley cultivar, was used for the VIGS experiments (Scofield et al., 2005)) and grown in a climate chamber at 20–25 °C with a 16/8 h light/dark photoperiod until the two-leaf stage, then inoculated with Barley stripe mosaic virus (BSMV), kept for 2 weeks and transferred to a growth chamber (22/18 °C day/night with 16 h light).
Arabidopsis thaliana Columbia-0 (Col-0), the Atd27 mutant (GABI-Kat collection, GK-134E08.01) (Waters et al., 2012), and the Atmax1-1 mutant, kindly provided by Prof. Ottoline Leyser (Booker et al., 2005) were used for genetic complementation experiments. Arabidopsis seeds were surface-sterilized with 20% (v/v) bleach for 15 min, washed, and plated on Murashige and Skoog (MS) medium supplemented with 2% (w/v) sucrose for 10 d. Seedlings with the same size were transferred to soil under a 16/8 h light/dark, 22/18 °C long-day (LD) regime with a light intensity of 120–150 μmol m−2 s−1. Pots were placed in the growth room in a randomized design. Observations of primary shoot branching phenotype were repeated at least two times (n=15–20).
Phylogenetic analyses
Amino acid sequence alignments were performed using the ClustalW module in the MEGA 6.0 program (Tamura et al., 2013). Neighbor-joining trees and bootstrap analyses were also conducted using MEGA 6.0, and the following parameters were selected: model, Jones-Taylor-Thornton (JTT); bootstrap, 1000 replicates; and gap/missing data, pairwise deletion. The protein sequences utilized in this analysis as well as NCBI accession numbers and data sources are listed in Supplementary Table S3 at JXB online.
Generation of the BSMV-HvCCD7 and BSMV-HvCCD8 constructs and BSMV-mediated virus-induced gene silencing
HvCCD7 sequences (295 bp and 306 bp) were amplified using Pfu DNA polymerase (Thermo Scientific) with gene-specific primers harboring the BamHI and PacI restriction sites (see Supplementary Table S1), and then directionally cloned in antisense orientation into the BamHI- and PacI-digested BSMV-γ-MCS multiple cloning site (Bruun-Rasmussen et al., 2007) to generate BSMV-HvCCD7-1 and BSMV-HvCCD7-2, respectively (Supplementary Fig. S1a). Using the same strategy, the constructs BSMV-HvCCD8-1 (202 bp) and BSMV-HvCCD8-2 (338 bp) were also generated (Fig. S1b; Table S1). Virus-induced gene silencing (VIGS) was performed according to Bruun-Rasmussen et al. (2007). BSMV sub-genomic components α, β, and γ were linearized by MluI digestion (T7-α and T7-γ recombinant) or SpeI (T7-β) and used as templates for in vitro transcription using the AmpliCap-Max™ T7 High Yield Message Maker Kit (CELLSCRIPT™), following the manufacturer’s protocol. The three viral RNA components were mixed in a 1:1:1 ratio and 4.5 μl of combined transcripts were mixed with 10 μl of FES buffer (77 mM glycine, 60 mM K2HPO4, 22 mM Na4P2O7.10H2O, and 1% w/v bentonite) and applied with gentle strokes to Bentonite-dusted leaves ((Holzberg et al., 2002). Only plants showing virus symptoms 14 d post-inoculation (dpi) were included in further analysis. Presence of the virus was further confirmed by RT-PCR at 21 dpi. Experiments were repeated three times and n=12 plants per construct were evaluated for tiller number.
Vector constructions of pAtD27::HvD27 and 35S::HvMAX1
The AtD27 promoter was amplified from genomic DNA and ligated into the pCR™2.1-TOPO® vector (Invitrogen). Using SpeІ and XhoІ, the sequence was further constructed into the pd35S-Nos-AB-M vector (see Supplementary Fig. S14). The plasmid sequence pAtD27-Nos-AB-M was confirmed by digestion and sequencing. For sub-cloning of HvD27 under the control of the AtD27 promoter, the sequence was amplified from barley leaf cDNA and transferred into the TOPO-TA vector and confirmed by sequencing. The HvD27 sequence was amplified with gene-specific primers having the XhoІ and SalІ sites. To generate pAtD27::HvD27-Nos-AB-M, the PCR product was ligated via XhoІ and SalІ into pAtD27-Nos-AB-M. The pAtD27::HvD27-Nos-AB-M cassette was then transferred into the p6U10 binary vector for plant transformation (Supplementary Fig. S14).
For construction of the pd35S::HvMAX1-Nos-AB-M overexpression cassette, HvMAX1 was amplified from barley leaf cDNA and transferred into the pCR™2.1-TOPO® vector (Invitrogen). The plasmid sequence was confirmed by sequencing. The HvMAX1 TOPO clone was amplified with gene-specific primers having the XhoІ and HindIII sites. To generate pd35S::HvMAX1-Nos-AB-M, the PCR product was ligated via XhoІ and HindIII to pd35S-Nos-AB-M. The pd35S::HvMAX1-Nos-AB-M was then transferred into the p6U10 binary vector for plant transformation. The primers used are listed in Supplementary Table S1. The vectors were transformed into Agrobacterium tumefaciens strain EHA105 (Chetty et al., 2013) and used for floral dip transformation (Clough and Bent, 1998) of Col-0 and the respective mutants. Resistant plants were selected on half-strength MS medium supplemented with 50 μg ml–1 hygromycin. Plates were kept at 4 °C for 2 d and then transferred to growth chamber conditions. Selected plants (T1) were transferred to Arasystem trays (BetaTech bvba, Gent, Belgium) containing Soilrite MixTM. The segregation ratio was estimated from T2 plants. Ten lines (T2) showing a 3:1 segregation ratio were further selected on antibiotic-containing medium to obtain homozygous lines. Analysis was done on T3 homozygous transgenic lines, and their transgenic status was confirmed by RT-PCR analyses.
RNA extraction, RT-PCR, and qRT-PCR
Fresh plant tissues from independent or pooled biological replicates (e.g. manually dissected meristems) from the same treatment were ground to fine powder in liquid nitrogen and stored at –80 °C. Total RNA was isolated from 100 mg ground material using TRIZOL reagent (Invitrogen GmbH, Karlsruhe, Germany) and RNAeasy columns (Qiagen, Hilden, Germany), treated with RNAse-free DNase (Ambion), and finally dissolved in 50 μl RNase-free water. After performing an RNA quality test, concentrations were measured using a NanoDrop photometer (Peqlab). cDNA synthesis was carried out using SuperScriptTM III (Invitrogen GmbH) with 1 μg of total RNA, following the manufacturer’s instructions. For semi-quantitative RT-PCR, 1 μl of diluted cDNA (1:3) was used in a final PCR volume of 20 μl and run for 35 cycles. PCR products (12 μl) were separated on a 1% (w/v) agarose gel.
For real-time qRT-PCR, the reactions were performed in 384-well plates using an ABI PRISM 7900 HT Sequence Detection System and SYBR Green (Applied Biosystems). The PCR protocol consisted of an initial denaturation step (50 °C for 2 min, 95 °C for 10 min), followed by 45 cycles of 95 °C for 15 s and 60 °C for 60 s. Dissociation curves were recorded and data were analysed using SDS2.2.1 software (Applied Biosystems). CT values for each primer set were normalized to the CT value of the reference amplicon (serine/threonine protein phosphatase PP2A-4, catalytic subunit, EST clone HZ44D03).
PCR efficiency was calculated from the slope of the exponential phase of the amplification, following the suggestions made by Ramakers et al. (2003). The changes in relative expression levels of target genes in the qRT-PCR were calculated according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). Primer sequences can be found in Supplementary Table S1.
Extraction and quantification of SLs and ABA
ABA was extracted from fresh plant material using ethyl acetate (100%). Isotopically labelled D6-ABA was used as an internal standard and added to each sample during the extraction procedure. Extraction was carried out twice with 1 ml of ethyl acetate at 4 °C. The supernatant collected after centrifugation (13.000 g, 10 min, 4 °C) was evaporated to dryness at room temperature using a vacuum concentrator. The dried samples were dissolved in acetonitrile:methanol (1:1) and filtered using a 0.8-µm filter (Vivaclear). The filtrate (10 µl) was used for subsequent quantification by LC-MS/MS (Dionex Summit coupled to Varian 1200 L). Chromatographic separation was carried out on a C18 column (4 µm, 100 mm; GENESIS; Vydac/USA). Multiple reaction monitoring (MRM) and quantification were done using the mass traces 263/153 for ABA and 269/159 for D6-ABA. The validity of the extraction and measurement procedure was verified in recovery experiments (approx. 82–95%). Quantification was based on calibration with known ABA standards and individual recovery rates for the samples, as described previously (Kong et al., 2008).
For the collection of root exudates, 10 germinated seeds of each line were planted in a 3-l plastic pots filled with 1.5 l of silver sand. After 1 week, plants were thinned to five plants per pot. Each pot was supplemented with 500 ml of half-strength modified Hoagland’s nutrient solution containing 0.1 mM KH2PO4 (Supplementary Table S2) at 48-h intervals. The plants were allowed to grow in a growth chamber for 13 weeks, and in the 14th week they were watered with a P-deficient nutrient solution (Hoagland’s nutrient solution with 0 mM KH2PO4) to increase SL production: 3 l of P-deficient nutrient solution was added to the top of each pot and allowed to drain freely through the holes in the bottom to remove P from the sand. The plants were kept under P-deficiency for 1 week. In the 15th week, the same draining with 3 l of P-deficient nutrient solution was repeated to remove any accumulated SLs. Then, 48 h later, root exudates were collected in a 1-l plastic bottle by passing 1 l of nutrient solution without phosphate through each pot. The root exudates were passed through an SPE C18-fast column (500 mg), and SLs were eluted with 6 ml of 100% acetone. For root extracts, 1 g fresh weight of ground root tissue was extracted following the method described by Jamil et al. (2012). The resulting extracts were evaporated to dryness, taken up in hexane, loaded on pre-equilibrated Silica gel Grace Pure SPE (200 mg) columns, and eluted with 2 ml of hexane:ethyl acetate (1:9) for further purification. The solvent was evaporated, and the residue was dissolved in 200 μl of 25% (v/v) acetonitrile in water and filtered through Minisart SRP4 0.45-μm filters (Sartorius) before LC-MS/MS analysis.
SLs were analysed by comparing retention times and mass transitions with SL standards (2’-epi-5-deoxystrigol, 5-deoxystrigol, and D6-2’-epi-5-deoxystrigol) using a Waters Xevo TQ mass spectrometer equipped with an electrospray-ionization source and coupled to a Waters Acquity Ultraperformance LC system using the settings described by Jamil et al. (2012) with some modifications (Cardoso et al., 2014). D6-2’-epi-5-deoxystrigol served as an internal standard that was added before analysis. Detection and quantification of SLs were performed with three biological replicates.
Statistical analysis
Values derived from several biological replicates were used to calculate means (±SD), and statistical significance was assessed using Student’s t-test and one-way ANOVA across genotypes with Tukey’s multiple comparison test (P˂0.05). These calculations were performed using Genstat, 17th edition (VSN International Ltd, Hemel Hempstead, UK).
Results
Identification of barley genes homologous to those involved in strigolactone biosynthesis in Arabidopsis
Despite the importance of SLs, rather limited information is available for this class of plant hormones in barley. In order to identify genes involved in SL biosynthesis in barley, protein sequences from Arabidopsis and rice were used as templates to perform a protein homology search based on sequences derived from the barley genome (International Barley Genome Sequencing Consortium, 2012) (http://webblast.ipk-gatersleben.de/barley/index.php; see Supplementary Fig. S2, Table S3). For D27, two amino acid sequences were identified: BAJ90178.1 (Hordeum_10) with 79.2% similarity to rice OsD27 (Lin et al., 2009), and BAJ85620.1 (Hordeum_16) with 47.2% similarity (Supplementary Table S3). Phylogenetic relationships among protein sequences of orthologous genes were determined by arbitrarily rooted neighbor-joining algorithms (Supplementary Fig. S2a). The phylogenetic reconstitution included confirmed and putative D27 sequences from graminaceous and dicotyledonous species and showed that only BAJ90178.1 grouped closely with OsD27 in a sub-cluster of orthologous proteins. This sub-cluster also contained the D27 orthologues from Medicago truncatula (van Zeijl et al., 2015) and Arabidopsis, encoded by AT1G03055 (Waters et al., 2012). Thus, BAJ90178.1 (encoded by MLOC_67450, HORVU7Hr1G096930) was designated as HvD27.
In order to identify the orthologous sequences of CCD7 and CCD8 in barley, the amino acid sequences of AtCCD7 (NP_182026.5 encoded by AT2G44990) and AtCCD8 (NP_195007.2 encoded by AT4G32810) were used as references (Wang et al., 2011). The retrieved barley amino acid sequences encoded by MLOC_55474.1 (HORVU2Hr1G097730) and MLOC_66551.1 (HORVU3Hr1G071170) were most closely related to CCD7 and CCD8, respectively, showing 33.7% and 56.6% identity. Subsequent phylogenetic analysis divided the CCD7 and CCD8 proteins into two distinct clusters (Supplementary Fig. S2b). As MLOC_55474.1 grouped together with OsCCD7 and Arabidopsis MAX3/CCD7, it was designated as HvCCD7. Likewise, MLOC_66551.1 grouped together with OsCCD8 and Arabidopsis MAX4/CCD8. Based on their sequence identity, MLOC_55474.1 and MLOC_66551.1 were designated further as HvCCD7 and HvCCD8, respectively.
A BLAST search using AtMAX1 (NP_565617.2) encoded by AT2G26170 as the target (Abe et al., 2014) yielded two barley sequences. A reconstituted phylogeny including sequences from monocot and dicot plants inferred at least two separate clades, with the second clade harboring only a small number of orthologous sequences from graminaceous species, including BAJ97619.1 (Hordeum_2; Supplementary Fig. S2c). The barley sequence BAJ98237.1 (Hordeum_1, AK367034, encoded by HORVU4Hr1G079620) grouped closely to Arabidopsis MAX1 (57% sequence identity) and was therefore designated as HvMAX1. Based on their homology and phylogenetic relationships, the identified barley genes (see genome map in Supplementary Fig. S2d) were selected for further characterization of their putative functions in the SL biosynthesis pathway.
Functional characterization of HvCCD7 and HvCCD8 genes by virus-induced gene silencing in barley
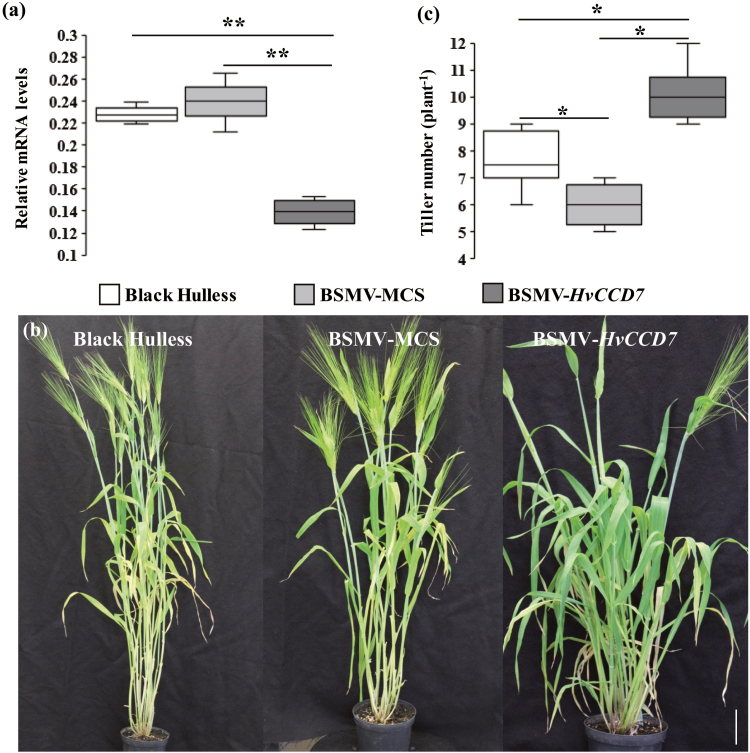
Gene-silencing using vectors derived from BSMV represents an efficient approach to study gene functions in barley (Pacak et al., 2010). For the functional characterization of HvCCD7 and HvCCD8 gene products, we implemented VIGS employing coding sequences that were re-confirmed by sequencing. The VIGS approach was chosen for two reasons: first, the promotor region of the identified genes was still unknown; and second, the expressions of HvCCD7 and HvCCD8 were weaker compared to HvD27 and HvMAX1 (Supplementary Fig. S7). The coding sequence of each target gene was subjected to an in silico analysis using the si-Fi software (siRNA Finder; http://labtools.ipk-gatersleben.de/) to select sequences of 200–340 nucleotides that were predicted to produce a large number of silencing-effective siRNAs. Two VIGS constructs were generated carrying antisense cDNA fragments from either HvCCD7 or HvCCD8 (Supplementary Fig. S1). The second leaf of a barley plant was infected with one of these constructs or with the empty BSMV vector (BSMV-MCS). At 9–11 dpi, leaf numbers two and three began to display typical mosaic-type necrosis and curling symptoms (Supplementary Fig. S3a). As confirmed for HvCCD7 by RT-PCR, harvested leaf material showing these symptoms contained viral DNA (Fig. S3b). The extent of the silencing response of HvCCD7 was monitored by qRT-PCR on RNA from leaves with the most pronounced symptoms. Relative to non-transformed wild-type plants, BSMV-MCS-infected plants showed a slightly (but not significantly) elevated mRNA level of HvCCD7. In contrast, BSMV-HvCCD7-infected plants showed a significant reduction in mRNA levels (Fig. 2a). After 15 weeks of growth, the BSMV-MCS-infected plants produced on average one tiller less than non-infected plants (Fig. 2b). BSMV-HvCCD7-infected plants were stunted in growth and developed on average three tillers more than the BSMV-infected control (Fig. 2c). Due to their late outgrowth, most tillers in BSMV-HvCCD7-silenced plants did not develop ears. This phenotype resembles the dit1 rice mutant defective in OsCCD7 (Kulkarni et al., 2014).
Fig. 2.
Virus-induced gene silencing (VIGS) of HvCCD7 in barley. (a) HvCCD7 transcript levels in leaf tissue as determined by qRT-PCR. Serine/threonine protein phosphatase PP2A-4 was used as the reference gene. The box-plots represent the results from three independent samples. (b) The tillering phenotypes of BSMV-MCS and BSMV-HvCCD7-silenced plants and the reference variety Black Hulless. The scale bar is 10 cm. (c) Total tiller number per plant. The box-plots represent of the results from 12 plants per genotype. Significant differences according to Student’s t-test are indicated: *P˂0.05, **P˂0.01.
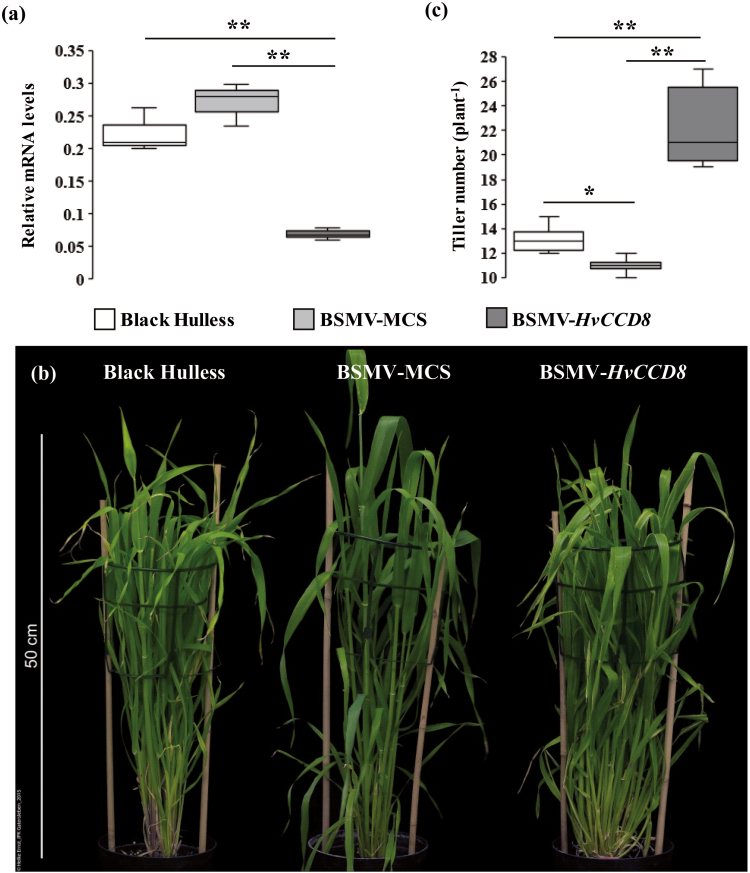
In parallel, barley plants infected with an antisense construct of HvCCD8 were found to severely suppress mRNA levels of HvCCD8 (Fig. 3a). These plants showed a bushy phenotype, which was accompanied by continuous tillering (Fig. 3b). After 11 weeks of growth under long-day conditions, these plants had produced twice as many tillers as BSMV-infected control plants (Fig. 3c). This phenotype is in accordance with the expectation of enhanced tiller formation caused by suppressed SL biosynthesis.
Fig. 3.
Virus-induced gene silencing (VIGS) of HvCCD8 in barley. (a) HvCCD8 transcript levels in leaf tissue as determined by qRT-PCR. Serine/threonine protein phosphatase PP2A-4 was used as the reference gene. The box-plots represent the results from three independent samples. (b) The tillering phenotype of BSMV-MCS and BSMV-HvCCD8-silenced plants and the reference variety Black Hulless. (c) Total tiller number per plant. The box-plots represent of the results from 12 plants per genotype. Significant differences according to Student’s t-test are indicated: *P˂0.05, **P˂0.01.
Functional characterization of HvD27 and HvMAX1 genes by genetic complementation analysis
In order to confirm a function of HvD27 and HvMAX1 in shoot branching, a heterologous complementation approach was employed. The Arabidopsis mutant d27 (Waters et al., 2012) and Col-0 wild-type plants were transformed with the cDNA of HvD27 under the control of the endogenous AtD27 promoter. For each transformation, three independent transgenic lines from the T3 generation that expressed the transgene were obtained (Supplementary Fig. S4a, b) and shoot branching was recorded at flowering. The Atd27 mutants produced eight branches compared to three or four observed for Col-0 plants (Supplementary Fig. S4a, c). This enhanced branching in the Atd27 mutant was reversed almost to the level of the untransformed controls in transgenic lines expressing the identified barley HvD27 gene. In pAtD27::HvD27-transformed lines, HvD27 expression resulted in an almost complete suppression of primary rosette-leaf branching in the Col-0 background (Supplementary Fig. S4a, c). ABA concentrations were analysed in leaves of 6-week-old Arabidopsis plants, and higher levels were found in the selected line pAtD27::HvD27 B16-5 than in Col-0 (Supplementary Fig. S5).
For functional verification of the putative HvMAX1 gene, the Atmax1 mutant (Booker et al., 2004) and Col-0 wild-type plants were transformed with the cDNA of HvMAX1 under the control of the CaMV35S promoter. As the HvMAX1 promotor sequence was yet not identified and AtMAX1 showed a relative strong and ubiquitous expression (Winter et al., 2007), the CaMV35S promotor was chosen for the complementation approach. Three independent homozygous lines were recovered from the T3 generation of each transformation event and verified for transgene expression (Supplementary Fig. S6a, b). Enhanced branching of the Atmax1 mutant was restored to Col-0 levels in the 35S::HvMAX1/Atmax1 transgenic lines, while in the Col-0 background HvMAX1 expression led to an almost complete suppression of primary rosette-leaf branching (Supplementary Fig. S6a, c). These results demonstrate that HvD27 and HvMAX1 are able to complement the shoot branching phenotype of the corresponding Arabidopsis mutants and are able to restore the function of AtD27 and AtMAX1, respectively.
Tissue-dependent expression of strigolactone biosynthesis genes
To determine tissue-dependent expression of the four identified SL biosynthesis genes, qRT-PCR was performed on RNA isolated from roots, leaves, stem bases, and axillary buds of barley plants grown in a growth chamber for 10 weeks. All four genes were substantially expressed in roots (Supplementary Fig. S7), and HvD27, HvCCD7, and HvCCD8 showed similar expression levels in the stem base. In contrast, HvD27 and HvMAX1 exhibited very high transcript levels in leaves, whereas HvCCD7 and HvCCD8 expression was found to be lower. In axillary buds, the mRNA levels of all four genes were low. These data are in agreement with a functional SL biosynthesis pathway in roots, where exceptionally high transcript levels of HvMAX1 may point to a non-limiting role of this gene.
Silencing HvABA 8’-hydroxylase genes induces a high-tillering phenotype
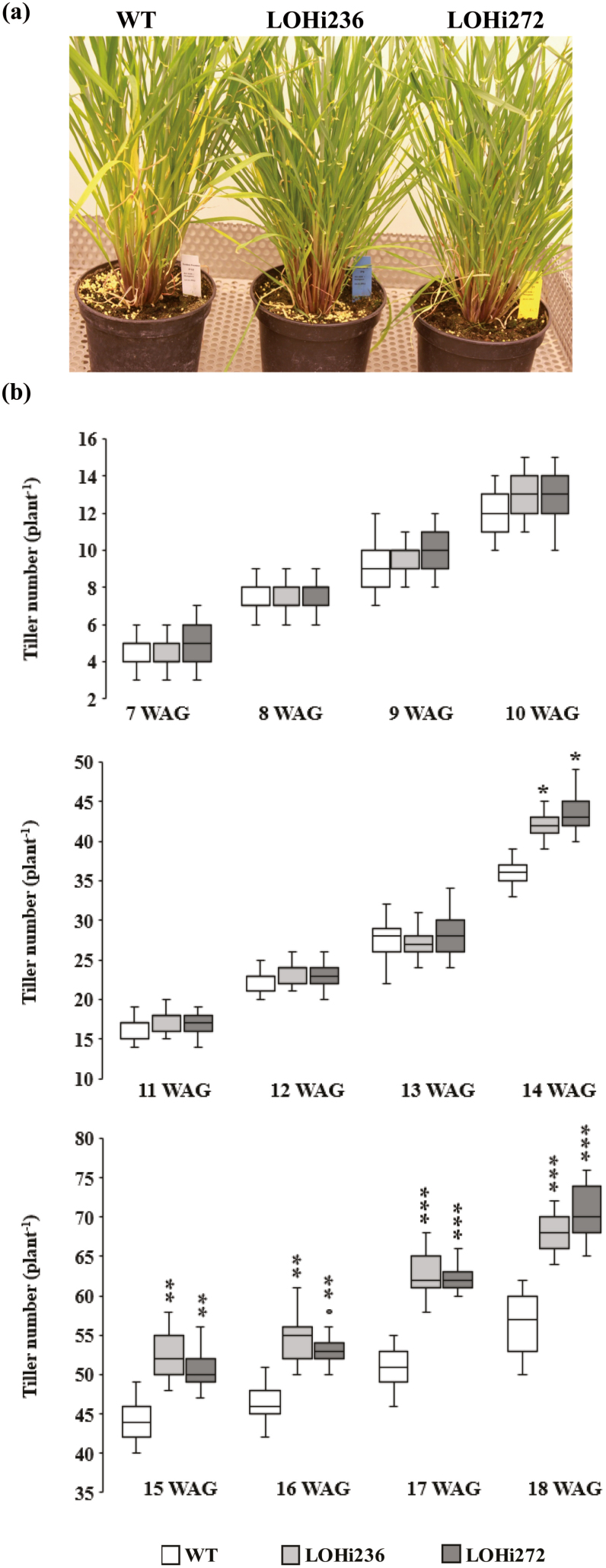
Given the importance of SLs in inhibiting shoot branching and the interplay of ABA and SLs under abiotic stress conditions (López-Ráez et al., 2010; Ha et al., 2014), we examined shoot architecture in previously characterized transgenic barley plants expressing an RNAi construct for HvABA 8’-hydroxylase LOHi236 and LOHi272 (Seiler et al., 2014). In these lines transgene expression is driven by the barley late embryogenesis abundant (Lea) B19.3 gene promoter, which is active during the vegetative growth phase in root and shoot tissues (Supplementary Fig. S8). In previous work (Seiler et al., 2014) it was shown that these plants have reduced expression of HvABA 8’-hydroxylase genes, which is responsible for ABA degradation, leading to altered levels of endogenous ABA. When cultured under controlled conditions in a growth chamber, shoot development of the transgenic lines was similar to the wild-type for the first 13 weeks of growth; however, from the 14th week onwards both transgenic lines produced significantly more tillers (Fig. 4). Due to continuous watering and nutrient supply, tillering continued until heading, when the LOHi236 and LOHi272 lines had produced approximately 10–15 tillers more than the wild-type.
Fig. 4.
Tiller number for RNAi lines grown in the growth chamber. (a) The tillering phenotype in the wild-type (WT, Golden Promise) and the HvABA 8’-hydroxylase RNAi lines (LOHi236, LOHi272) at 16 weeks after germination (WAG). (b) Total tiller number per plant in the LOHi236 and LOHi272 transgenic lines compared with the wild-type from 7 to 18 WAG. of the box-plots represent the results from 20 plants per genotype. Significant differences as determined by Student’s t-test are indicated: *P˂0.05, **P˂0.01, ***P˂0.001.
To investigate whether this increase in final tiller number might result from differential regulation of axillary bud outgrowth or increased numbers of axillary meristems, the axillary meristems were examined at an early developmental stage. Sections of axillary meristem were taken at around 3–4 weeks after germination (WAG) and inspected microscopically. No differences were found in the anatomy or number between the LOHi lines and the Golden promise plants (Supplementary Fig. S9). When grown under greenhouse conditions, the LOHi lines showed a similar difference with more tillers, although the average number of developed tillers was reduced in all genotypes (Supplementary Fig. S10). The differences between greenhouse and growth chamber might have been caused by the different light conditions (Supplementary Table S4). In both cases, the difference in tiller formation became apparent at a relatively late developmental stage (12–14 WAG). Thus, enhanced tiller formation in HvABA8’-hydroxylase-suppressed lines was robust and reminiscent of a SL-deficient phenotype. Furthermore, increased tiller formation resulted in a lower number of spikes per plant, lower thousand-grain weight, and hence lower yield of the LOHi plants (Supplementary Fig. S11).
ABA accumulation in the stem base and roots of LOHi lines
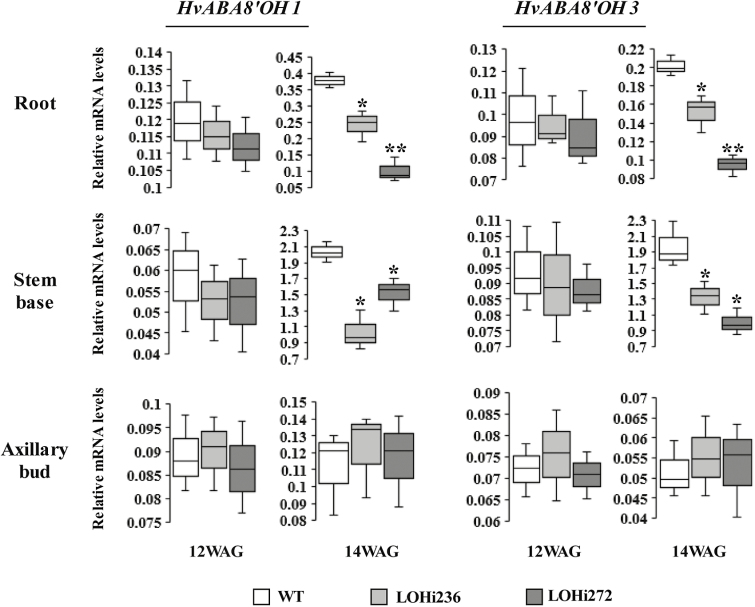
To investigate the relationship between ABA homeostasis and the late-tillering phenotype, mRNA levels of ABA-related genes and ABA concentrations were determined in LOHi236 and LOHi272 and compared to untransformed wild-type Golden Promise controls. As quantified by RT-qPCR, the mRNA levels of HvABA8’OH2 were relatively low in all tissues analysed, indicating that the encoded gene product might not significantly contribute to ABA degradation (Supplementary Fig. S12). The mRNA levels of the RNAi-targeted HvABA8’OH1 and HvABA8’OH3 genes were quantified in roots, stem bases, and axillary buds at 12 and 14 WAG (Fig. 5). At 12 WAG, when no significant difference in tiller formation was detectable, there was also no significant influence detectable of the RNAi construct on the relative mRNA levels of HvABA8’OH1 and HvABA8’OH3 in roots, stem bases, or axillary buds of LOHi plants. At 14 WAG, a clear increase of HvABA8’OH1 and 3 transcript levels was detectable, which was significantly lower in roots and stem bases of LOHi plants compared to controls.
Fig. 5.
Relative transcript levels of the genes HvABA8’OH1 and HvABA8’OH3 in the stem base, roots, and axillary buds in wild-type (WT) plants and transgenic lines (LOHi236, LOHi272) at 12 and 14 weeks after germination (WAG). The box-plots represent the results from three independent biological samples. Serine/threonine protein phosphatase PP2A-4 was used as the reference gene. Significant differences as determined by Student’s t-test are indicated: *P˂0.05, **P˂0.01.
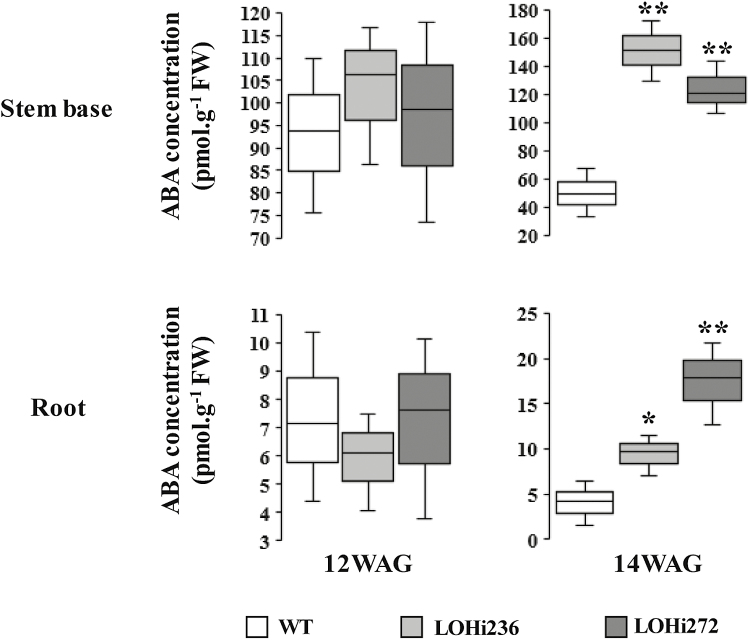
LC-MS/MS analysis was used to determine ABA concentrations in the stem base and roots, In wild-type plants, the highest ABA concentration was detectable at 12 WAG in the stem base and no differences with the LOHi lines were detectable at this time. At 14 WAG the ABA concentration in the stem bases decreased in the wild-type (from 94 to 50 pmol g–1 FW), whereas mean concentrations increased in the LOHi lines (Fig. 6). This decrease was absent in the LOHi lines. In agreement with the reduced mRNA levels of HvABA8’OH1 and 3, ABA levels were ~70 % higher in the LOHi lines compared to the wild-type at 14 WAG. In roots, the detectable ABA content was ~10 times lower compared to the stem base. In line with higher ABA levels in the LOHi stem base at 14 WAG, higher ABA levels were also observed in root tissues.
Fig. 6.
ABA concentrations in the stem base and roots of wild-type (WT) plants and LOHi antisense lines at 12 and 14 weeks after germination (WAG). The box-plots represent of the results from three independent biological replicates. Significant differences as determined by Student’s t-test are indicated: *P˂0.05, **P˂0.01.
5-deoxystrigol is lower in root exudates of LOHi lines
Although SLs have been detected in a variety of plant species, none have been identified in barley so far. To explore the identity of SLs in barley, LC-MS/MS analysis was performed. Barley plants were pre-cultured hydroponically, transferred to pots with sand, and subjected to phosphate starvation. Root exudates were collected, analysed, and the chromatograms were compared with retention times (RT) and mass transitions of available SL standards. As an example, a standard of 5-deoxystrigol was detected at RT 7.95 min in the MRM channel m/z 331.2>234.15, the internal standard D6-5-deoxystrigol was detected at RT 7.92 min in the MRM channel m/z 337.16>97.017, and 2’-epi-5-deoxystrigol was detected at RT 7.88 in the MRM channel m/z 331.2>234.0 (Supplementary Fig. S13). The chromatogram of a crude root exudate revealed an intense peak in the channel m/z 331.2>216.15 at RT 7.95 min, which matched the standard of 5-deoxystrigol.
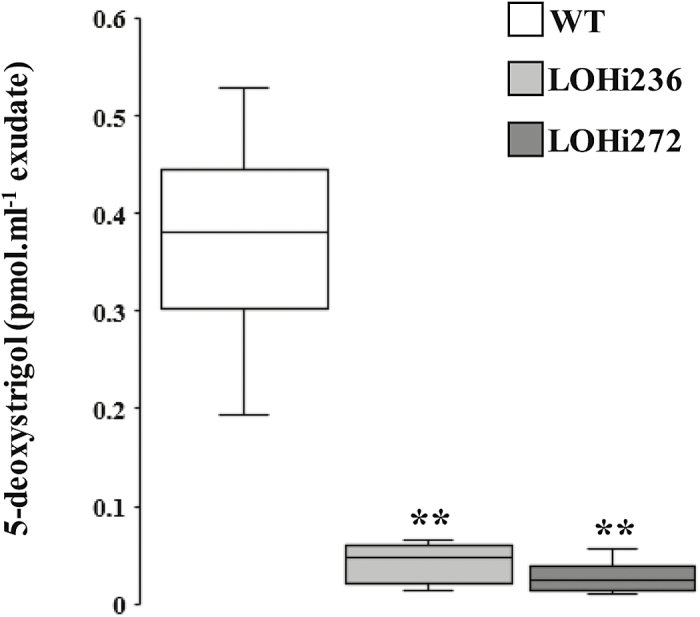
To investigate the abundance of detectable SLs in the LOHi lines, plants were pre-cultured hydroponically, transferred to pots with sand and subjected to phosphate starvation and root exudate analysis. 5-deoxystrigol was significantly (P<0.01) lower in root exudates of LOHi236 and LOHi272 compared with the wild-type at 14 WAG (Fig. 7). This lower abundance coincided with increased tiller formation in the LOHi lines.
Fig. 7.
Strigolactone concentrations in barley root exudates as determined by MRM-LC-MS/MS analysis. The data show the concentrations of 5-deoxystrigol in root exudates of LOHi (LOHi236 and LOHi272) and wild-type plants (WT) after 7 d of phosphate starvation, which commenced at 14 weeks after germination. The box-plots represent of the results from five independent biological replicates. Significant differences as determined by Student’s t-test are indicated: **P˂0.01.
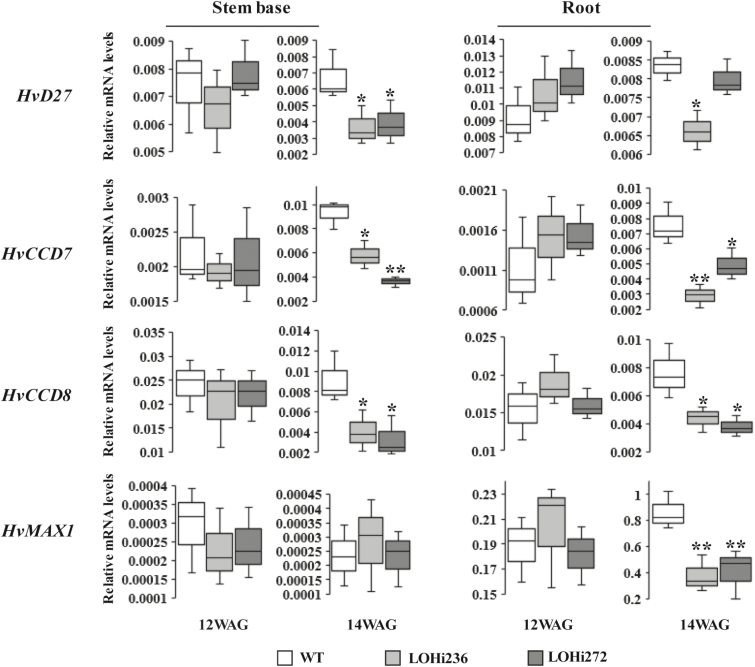
To corroborate a functional link between ABA and HvD27, HvMAX1, HvCCD7, and HvCCD8, gene expression was analysed using qRT-PCR. At 12 WAG, no differences between the LOHi lines and wild-type plants were observed for the mRNA abundance of all four genes in the stem base and root tissue (Fig. 8). At 14 WAG, the relative mRNA levels of HvD27, HvCCD7, and HvCCD8 were found to be significantly lower in the stem base and root tissue of the LOHi lines compared to the wild-type, although there was inconsistency in the LOHi lines in the root tissue. HvMAX1, which in contrast to the other genes was only clearly detectable in roots at later time points, showed a tissue-specific reduction of transcript levels in LOHi plants.
Fig. 8.
Transcript levels of genes from the SL biosynthetic pathway in stem bases and roots of wild-type (WT) plants and LOHi antisense lines at 12 and 14 weeks after germination (WAG). The box-plots represent of the results from three biologically independent samples. Serine/threonine protein phosphatase PP2A-4 was used as the reference gene. Significant differences as determined by Student’s t-test are indicated: *P˂0.05, **P˂0.01.
Discussion
ABA and SLs are plant hormones involved in the regulation of growth. Both share a common precursor (all-trans-β-carotene) but knowledge of the genes involved in SL biosynthesis in barley is limited. Here, we identified four genes involved in SL biosynthesis in barley and identified 5-deoxystrigol in root exudates. Based on analysis of transgenic plants with suppressed ABA degradation, we have shown that there is cross-talk between ABA and SL biosynthesis that controls tillering in barley. Thus, the present study provides for the first time genetic evidence for an antagonistic interaction between ABA and SLs in plants.
Identification of HvD27, HvCCD7, HvCCD8, and HvMAX1 and their involvement in SL biosynthesis
Despite the importance of SLs for shoot architecture in plants, little information is available for this class of plant hormones in cereal species other than rice. Based on amino acid similarities to homologous proteins in rice and Arabidopsis, HvD27, HvCCD7, HvCCD8, and HvMAX1 were identified in barley and confirmed by phylogenetic analysis to represent the closest homologs of their functionally characterized counterparts in these two species (Supplementary Fig. S2). Based on their tissue-specific mRNA abundance, HvD27, HvCCD7, and HvCCD8 were highly expressed in both the stem base and roots at the time points examined (Fig. 8). However, the newly identified gene HvMAX1 was only expressed in roots, suggesting a role in the root-specific conversion of carlactone to carlactonic acid and further to 5-deoxystrigol, which was subsequently detected in barley root exudates (Fig. 7). Such a pathway is in agreement with the reported ability of barley root exudates to stimulate germination of Striga (Steinkellner et al., 2007) and reminiscent of SL species found in Arabidopsis or Nicotiana benthamiana (Marzec and Muszynska, 2015). By contrast, orobanchol-type SLs have been identified as the predominant forms in rice (Xie et al., 2010).
To verify the proposed gene function in SL biosynthesis, virus-induced gene silencing was performed to silence HvCCD7 (Fig. 2) and HvCCD8 (Fig. 3) in planta. This post-transcriptional gene silencing resulted in increased tiller formation, which is in agreement with the proposed involvement of HvCCD7 and HvCCD8 in SL production. For functional analysis of HvD27 and HvMAX1, heterologous complementation was performed employing the corresponding Arabidopsis thaliana mutant lines (Booker et al., 2004; Waters et al., 2012). Functional complementation was achieved by ectopic expression of HvD27 and HvMAX1. Both barley genes were able to complement the highly branched phenotype of the respective Arabidopsis mutants back to wild-type levels (Supplementary Figs S4, S6). Due to the lack of sequence information this approach could not be applied to HvCCD7 and HvCCD8.
To further validate the function of the identified genes in tiller formation, transgenic LOHi plants with suppressed hydroxylation of ABA were examined. These plants showed a tissue-specific and development-specific suppression of HvD27, HvCCD7, HvCCD8, and HvMAX1 that temporally coincided with enhanced tiller formation (14 WAG, Fig. 4). This reduction in transcript levels of SL biosynthesis genes was accompanied by lower abundance of 5-deoxystrigol in root exudates of P-starved barley plants (Fig. 7). Taken together, these results support the physiological function of the identified genes in SL biosynthesis. According to previous results in Arabidopsis (Brewer et al., 2016), the genes identified here are likely to represent core components of the SL biosynthesis pathway in barley.
In addition to the SL biosynthesis genes discussed here, further genes are involved in SL signaling such as D14, MAX2, and D53 (Al-Babili and Bouwmeester, 2015). As pointed out by Brewer et al. (2016), knowledge of bioactive SLs is limited; therefore, identification of additional genes is required. Recently, HvD14 has been identified and characterized as a component involved in SL signaling in barley. A mutation in this gene causes a larger number of tillers than in wild-type plants (Marzec et al., 2016).
Involvement of 5-deoxystrigol in tiller formation in barley
5-deoxystrigol has been proposed as a common precursor of various SLs (Rani et al., 2008; Zwanenburg et al., 2009). It is widely distributed in the plant kingdom and has been detected in root exudates from both monocots (Awad et al., 2006) and dicots (Yoneyama et al., 2007). Using LC-MS/MS, we exclusively identified 5-deoxystrigol in root exudates of barley. Other known SLs could not be detected or were below the detection limits. Root exudates of barley have been reported to stimulate germination of Striga and Orobanche seeds (Steinkellner et al., 2007). However, we cannot rule out the possibility that other unknown SLs, including orobanchol-type, could be synthesized and released via root exudates in barley.
As SLs are of low abundance and difficult to detect even with sensitive LC-MS/MS devices, plants were subjected to P-starvation prior to collection of root exudate, since P-deficiency promotes SL biosynthesis and release (Al-Babili and Bouwmeester, 2015). In all studies reported so far, the amount of detectable SLs in root exudates (also sampled after P-starvation) has correlated with the inhibition of above-ground lateral organ outgrowth (Brewer et al., 2013; Borghi et al., 2016). We took the detectable amount of 5-deoxystrigol in root exudates as proxy for 5-deoxystrigol biosynthesis in the roots. Supported by the root-specific expression of HvMAX1, we propose that the formation of 5-deoxystrigol is confined to roots, as reflected by its occurrence in the root exudate. Whether 5-deoxystrigol or other bioactive SLs might be translocated to the stem base cannot be concluded based on the present results. To date, 5-deoxystrigol is the first member of this class of plant hormones identified in barley.
Interaction between abscisic acid and strigolactones occurs at the level of SL biosynthesis
In the present work, we choose a genetic approach to investigate ABA–SL interactions. We employed the transgenic barley lines LOHi236 and LOHi272, in which transcript levels of HvABA 8’-hydroxylase genes are post-transcriptionally suppressed by an RNAi construct (Seiler et al., 2014). In these lines, transcript levels of HvABA 8’-hydroxylase1 and 3 were strongly decreased in the stem base tissue at 14 WAG, confirming that silencing resulted in down-regulation of HvABA 8’-hydroxylase1 and 3 (Fig. 5). In the stem base, effective post-transcriptional gene silencing was most likely responsible for enhanced tiller formation, because suppression of these genes caused reduced degradation of ABA and resulted in three-fold higher ABA levels at 14 WAG (Fig. 6). In line with this, phenotypic differences were detected 14 WAG, but not earlier. This was probably due to (1) the Lea gene promoter that we used driving expression of the RNAi silencing inducer at this late developmental phase (Supplementary Fig. S8), and (2) the development-specific expression pattern of the target genes (Fig. 5). In several studies a negative association has been reported between ABA and axillary bud development. In Arabidopsis buds, increased expression of ABA-associated genes under light with a low R:FR ratio results in suppressed stem branching (González-Grandío et al., 2013). Therefore, we initially expected that elevated ABA levels would suppress tiller formation. However, elevated ABA accumulation in the roots and stem base of both LOHi lines clearly coincided with higher tiller numbers (Figs 4, 6, Supplementary Fig. S10). ABA is therefore unlikely to be directly involved in tiller formation. Instead, we conclude that lower SL biosynthesis, as a result of increased ABA, is responsible for the observed phenotype, since SLs have been characterized as suppressors of lateral shoot branching (Borghi et al., 2016). The possibility that SL biosynthesis is regulated by ABA has already been suggested by exogenous application of ABA to Arabidopsis plants, leading to a decrease in the transcript levels of the SL biosynthesis genes CCD7 and CCD8 (Ha et al., 2014). In the present transgenic approach, the major change in ABA levels was detected in the stem base, but also in roots, although to a lesser extent (Fig. 6). This coincided with a significant reduction of SL biosynthesis-related transcript levels (Fig. 8). Thus, we conclude that the elevated ABA levels in these tissues caused the transcriptional down-regulation of genes involved in SL biosynthesis. Interestingly, substantial expression of HvMAX1 was only detectable in wild-type roots. However, at 14 WAG transcript levels had increased in the wild-type but not in the LOHi lines. These genotype-, tissue-, and developmental-specific differences are in agreement with the detected differences in 5-deoxystrigol production (Fig. 7). Based on our findings here, we propose a model for the regulatory interaction between ABA and SLs in tiller formation (Fig. 9). In wild-type plants, homeostasis between ABA and SLs is important for reduced tiller formation after flowering. A transiently elevated endogenous level of ABA, as triggered by the silencing of the HvABA 8’-hydroxylase genes, represses SL biosynthesis mainly via down-regulating the transcript levels of HvD27, HvCCD7, HvCCD8, and HvMAX1, which results in less SL production. As a consequence, lower 5-deoxystrigol levels result in reduced tiller suppression, which causes the observed high-tillering phenotype of the LOHi lines at later developmental stages.
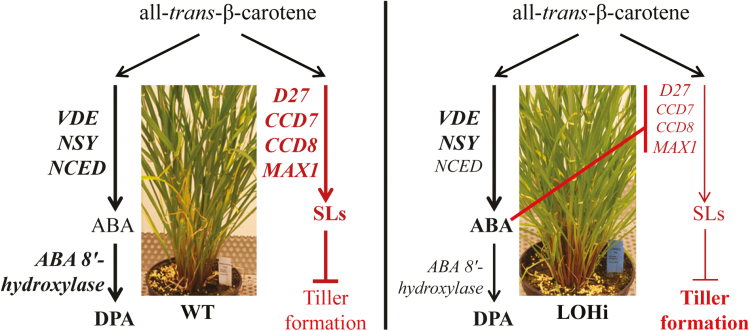
Fig. 9.
Schematic model of the regulatory interaction between ABA and SL in tiller formation. The wild-type (WT) plants maintain hormonal homeostasis between ABA and SL. In contrast, in the LOHi lines transiently high endogenous ABA levels triggered by a spatial-temporal silencing of the HvABA 8’-hydroxylase genes represses SL biosynthesis, mainly via down-regulation of HvCCD7/8, which results in low SL production. This subsequently releases the inhibition of SL, and finally causes a later, high-tillering phenotype. Abbreviations: VDE, violaxanthin de-epoxidase; NSY, neoxanthin synthase; NCED, 9-cis-epoxycarotenoid dioxygenase; D27, dwarf 27; CCD7/8, carotenoid cleavage dioxygenase 7/8; MAX1, more axillary growth 1; DPA, dihydrophaseic acid.
HvD27 as the target for ABA-dependent SL biosynthesis
Based on low SL levels of the corresponding mutant, OsD27 has been identified as an iron-containing enzyme involved in SL biosynthesis (Lin et al., 2009). The purified enzyme exerts isomerase activity that is required for the conversion of all-trans-β-carotene to 9-cis-β-carotene in a reversible manner (Alder et al., 2012). All-trans-β-carotene is the shared common precursor for ABA and SL production. The 9-cis/all-trans isomerase activity of D27 raised the question whether this enzyme might also be involved in ABA biosynthesis by pulling all-trans-β-carotene into ABA biosynthesis (Fig. 1). Recently, this activity was confirmed by a study on the substrate specificity of the rice SL biosynthesis enzyme DWARF27 in carotenoid-accumulating E. coli strains and in in vitro assays performed with the recombinant OsD27 enzyme (Bruno and Al-Babili, 2016) (Fig. 1). This finding is further supported by the observation that increasing HvD27 activity in Arabidopsis by ectopic overexpression resulted in an increase of ABA, modulating ABA homoeostasis (Supplementary Fig. S5). This increase was far below the ABA levels typically associated with drought stress (~20–40 ng ABA g–1 FW; Cho et al., 2008; Liu et al., 2014) and it was not accompanied by any visual phenotype, and may therefore be subject to the complex autoregulation of ABA homoeostasis. Considering that SLs are mostly synthesized in roots with minor synthesis in shoots (Al-Babili and Bouwmeester, 2015) and that in barley HvMAX1 is predominately expressed in roots (Fig. 8), we propose that the major site of the interaction between ABA and SL is the root tissue. To what extent ABA-mediated suppression of SL biosynthesis genes in the stem base contributed to enhanced tiller outgrowth requires further investigation.
The isomerase activity of HvD27 separates the SL pathway from ABA biosynthesis and is thus an ideal target for hormonal interaction. On the one hand, ABA may most effectively suppress the complete SL biosynthesis pathway by only regulating HvD27. On the other hand, suppression of HvD27 may favor ABA biosynthesis via higher availability of the precursor and thus become part of the auto-regulatory pathway in ABA homeostasis.
Taken together, the interaction between ABA and SLs identified here may play an important role in agricultural plant production. Tiller formation at later developmental stages is an undesired trait, because late tillers rarely contribute to grain yield. SL production especially at later developmental stages may help to avoid surplus tiller formation, in particular when ABA levels are low. The ABA–SL interaction may represent an important mechanism that suppresses the generation of unproductive tillers.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Genomic organization of BSMV RNA γ modified to express antisense gene-specific fragments.
Fig. S2. Phylogenetic trees of amino acid sequences of proteins in the SL biosynthesis pathway and barley genomic map with relative physical positions of the genes investigated in this study.
Fig. S3. Virus-induced gene silencing of HvCCD7 in barley.
Fig. S4. Genetic complementation of the Arabidopsis d27 mutant with the Hordeum vulgare D27 gene.
Fig. S5. ABA concentrations in 6-week-old Arabidopsis leaves of Col-0 and the transgenic line B16-5 expressing the pAtD27::HvD27 construct in the Col-0 background.
Fig. S6. Genetic complementation of the Arabidopsis max1 mutant with the Hordeum vulgare MAX1 gene.
Fig. S7. Expression of genes from the SL biosynthesis pathway across various tissues and organs in barley wild-type plants, together with selected reference genes.
Fig. S8. Expression levels of late embryogenesis-abundant (Lea) B19.3 in barley various tissues and across the life cycle of barley.
Fig. S9. Development of axillary meristems in wild-type and LOHi antisense lines.
Fig. S10. Variation in tiller numbers with time under greenhouse conditions.
Fig. S11. Spike number per plant and thousand-grain weight at maturity under greenhouse conditions.
Fig. S12. Expression levels of the genes HvABA8’OH 1, 2, and 3 in the roots, stem base, leaves, and axillary buds in wild-type plants at 10 WAG.
Fig. S13. MRM-LC-MS/MS analysis of strigolactones identity in barley root exudates.
Fig. S14. Plasmid maps of pd35S-Nos-AB-M and p6U10.
Table S1. List of primer sequences for PCR, RT-PCR, and quantitative RT-PCR.
Table S2. Details of the Hoagland’s nutrient solution.
Table S3. List of protein sequences used in the phylogenetic analysis.
Table S4. Details of growth conditions used for the tiller phenotyping of the LOHi lines.
Acknowledgements
We thank Jana Lorenz, Gabriele Einert, Mandy Pueffeld, Elke Liemann, and Sabine Skiebe for their excellent technical assistance. This research was supported by IZN (Interdisciplinary Centre for Crop Plant Research), Halle (Saale), Saxony-Anhalt, Germany and the IPK Graduate School. We thank Tadao Asami for the kind gift of 5-deoxystrigol and D6-5-deoxystrigol. We are thankful to Dr. Götz Hensel (RG Plant Reproductive Biology, IPK) for providing the plasmids pd35SNOS-AB-M and p6U10 required for successful plant transformation.
Author contributions
NS, MK, NvW, HB, CS, and HW designed the research; HW, TC, HB, KE, MRH, WC, and PS performed the experiments and analysed the data; and MK, NvW, and HW wrote the paper.
References
- Abe S, Sado A, Tanaka K, et al. 2014. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences, USA 111, 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Herold S, Schwarz M, et al. 2011. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences, USA 108, 20242–20247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Hayashi H. 2006. Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Annals of Botany 97, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant & Cell Physiology 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Awad AA, Sato D, Kusumoto D, Kamioka H, Takeuchi Y, Yoneyama K. 2006. Characterization of strigolactones, germination stimulants for the root parasitic plants Striga and Orobanche, produced by maize, millet and sorghum. Plant Growth Regulation 48, 221–227. [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. 2005. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Developmental Cell 8, 443–449. [DOI] [PubMed] [Google Scholar]
- Borghi L, Liu GW, Emonet A, Kretzschmar T, Martinoia E. 2016. The importance of strigolactone transport regulation for symbiotic signaling and shoot branching. Planta 243, 1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. 2013. Diverse roles of strigolactones in plant development. Molecular Plant 6, 18–28. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F, et al. 2016. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Al-Babili S. 2016. On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27. Planta 243, 1429–1440. [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Madsen CT, Jessing S, Albrechtsen M. 2007. Stability of Barley stripe mosaic virus-induced gene silencing in barley. Molecular Plant-Microbe Interactions 20, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Bu Q, Lv T, Shen H, et al. 2014. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiology 164, 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Zhang YX, Jamil M, et al. 2014. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proceedings of the National Academy of Sciences, USA 111, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty VJ, Ceballos N, Garcia D, Narvaez-Vasquez J, Lopez W, Orozco-Cardenas ML. 2013. Evaluation of four Agrobacterium tumefaciens strains for the genetic transformation of tomato (Solanum lycopersicum L.) cultivar Micro-Tom. Plant Cell Reports 32, 239–247. [DOI] [PubMed] [Google Scholar]
- Challis RJ, Hepworth J, Mouchel C, Waites R, Leyser O. 2013. A role for MORE AXILLARY GROWTH1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiology 161, 1885–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA. 2000. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiology 124, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SM, Kang BR, Han SH, Anderson AJ, Park JY, Lee YH, Cho BH, Yang KY, Ryu CM, Kim YC. 2008. 2R,3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Molecular Plant-Microbe Interactions 21, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA, et al. 2016. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology 12, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nature Reviews. Molecular cell biology 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Drummond RS, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. 2009. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P. 2013. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell 25, 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CV, Leyva-Gonzalez MA, Osakabe Y, et al. 2014. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences, USA 111, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP. 2002. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant Journal 30, 315–327. [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. 2001. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. The Plant Journal 27, 325–333. [DOI] [PubMed] [Google Scholar]
- Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ. 2012. Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta 235, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology 142, 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, et al. 2011. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Kong L, Abrams SR, Owen SJ, Graham H, von Aderkas P. 2008. Phytohormones and their metabolites during long shoot development in Douglas-fir following cone induction by gibberellin injection. Tree Physiology 28, 1357–1364. [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344. [DOI] [PubMed] [Google Scholar]
- Kulkarni KP, Vishwakarma C, Sahoo SP, et al. 2014. A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. Journal of Genetics 93, 389–401. [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, et al. 2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell 21, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He H, Vitali M, et al. 2015. Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: exploring the interaction between strigolactones and ABA under abiotic stress. Planta 241, 1435–1451. [DOI] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. 2014. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytologist 201, 1192–1204. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△CT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, et al. 2010. Does abscisic acid affect strigolactone biosynthesis?New Phytologist 187, 343–354. [DOI] [PubMed] [Google Scholar]
- Marzec M, Gruszka D, Tylec P, Szarejko I. 2016. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiologia Plantarum 158, 341–355. [DOI] [PubMed] [Google Scholar]
- Marzec M, Muszynska A. 2015. In silico analysis of the genes encoding proteins that are involved in the biosynthesis of the RMS/MAX/D pathway revealed new roles of strigolactones in plants. International Journal of Molecular Sciences 16, 6757–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. 2005. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiology 139, 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak A, Geisler K, Jørgensen B, Barciszewska-Pacak M, Nilsson L, Nielsen TH, Johansen E, Grønlund M, Jakobsen I, Albrechtsen M. 2010. Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA. 2002. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiology 128, 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Rani K, Zwanenburg B, Sugimoto Y, Yoneyama K, Bouwmeester HJ. 2008. Biosynthetic considerations could assist the structure elucidation of host plant produced rhizosphere signalling compounds (strigolactones) for arbuscular mycorrhizal fungi and parasitic plants. Plant Physiology and Biochemistry 46, 617–626. [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiology 158, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA. 2013. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiology 163, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lozano JM, Aroca R, Zamarreño ÁM, Molina S, Andreo-Jiménez B, Porcel R, García-Mina JM, Ruyter-Spira C, López-Ráez JA. 2016. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant, Cell & Environment 39, 441–452. [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. 2013. The biology of strigolactones. Trends in Plant Science 18, 72–83. [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, et al. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones?Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed W, Naseem S, Ali Z. 2017. Strigolactones biosynthesis and their role in abiotic stress resilience in plants: a critical review. Frontiers in Plant Science 8, 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J, Simon S, Gübeli C, Liu GW, Cheng X, Friml J, Bouwmeester H, Martinoia E, Borghi L. 2015. Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Current Biology 25, 647–655. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. 2005. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiology 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N. 2011. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. Journal of Experimental Botany 62, 2615–2632. [DOI] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Reddy PS, et al. 2014. Abscisic acid flux alterations result in differential abscisic acid signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiology 164, 1677–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. 2007. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiology 143, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Li J. 2014. Signalling and responses to strigolactones and karrikins. Current Opinion in Plant Biology 21, 23–29. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. 2005. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell 17, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes & Development 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner S, Lendzemo V, Langer I, Schweiger P, Khaosaad T, Toussaint JP, Vierheilig H. 2007. Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant–fungus interactions. Molecules 12, 1290–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. 2007. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal 50, 80–94. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Kusaba M. 2015. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiology 169, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- van Zeijl A, Liu W, Xiao TT, Kohlen W, Yang WC, Bisseling T, Geurts R. 2015. The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biology 15, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, et al. 2017. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Frontiers in Plant Science 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie T, McCulloch H, Leyser O. 2014. Strigolactones and the control of plant development: lessons from shoot branching. The Plant Journal 79, 607–622. [DOI] [PubMed] [Google Scholar]
- Wang RK, Lu JJ, Xing GN, Gai JY, Zhao TJ. 2011. Molecular evolution of two consecutive carotenoid cleavage dioxygenase genes in strigolactone biosynthesis in plants. Genetics and Molecular Research 10, 3664–3673. [DOI] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. 2012. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiology 159, 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PloS ONE 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kisugi T, Uchida K, Ito S, Akiyama K, Hayashi H, Yokota T, Nomura T, Yoneyama K. 2013. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Molecular Plant 6, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annual Review of Phytopathology 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M. 2014. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408. [DOI] [PubMed] [Google Scholar]
- Yao C, Finlayson SA. 2015. Abscisic acid is a general negative regulator of arabidopsis axillary bud growth. Plant Physiology 169, 611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RF, Ming ZH, Yan LM, et al. 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Mori N, Sato T, et al. 2018. Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytologist 218, 1522–1533. [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K. 2007. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227, 125–132. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ruyter-Spira C, Bouwmeester HJ. 2015. Engineering the plant rhizosphere. Current Opinion in Biotechnology 32, 136–142. [DOI] [PubMed] [Google Scholar]
- Zhou R, Cutler AJ, Ambrose SJ, et al. 2004. A new abscisic acid catabolic pathway. Plant Physiology 134, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Abrams GD, Barton DL, Taylor DC, Pomeroy MK, Abrams SR. 1995. Induction of lipid and oleosin biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L.cv Reston (biological responses in the presence of 8[prime]-hydroxyabscisic acid). Plant Physiology 108, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal 48, 687–698. [DOI] [PubMed] [Google Scholar]
- Zwanenburg B, Mwakaboko AS, Reizelman A, Anilkumar G, Sethumadhavan D. 2009. Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Management Science 65, 478–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.