Abstract
There is now ample evidence that endosymbionts can contribute to host adaptation to environmental challenges. However, how endosymbiont presence affects the adaptive trajectory and outcome of the host is yet largely unexplored. In Drosophila, Wolbachia confers protection to RNA virus infection, an effect that differs between Wolbachia strains and can be targeted by selection. Adaptation to RNA virus infections is mediated by both Wolbachia and the host, raising the question of whether adaptive genetic changes in the host vary with the presence/absence of the endosymbiont. Here, we address this question using a polymorphic D. melanogaster population previously adapted to DCV infection for 35 generations in the presence of Wolbachia, from which we removed the endosymbiont and followed survival over the subsequent 20 generations of infection. After an initial severe drop, survival frequencies upon DCV selection increased significantly, as seen before in the presence of Wolbachia. Whole-genome sequencing, revealed that the major genes involved in the first selection experiment, pastrel and Ubc-E2H, continued to be selected in Wolbachia-free D. melanogaster, with the frequencies of protective alleles being closer to fixation in the absence of Wolbachia. Our results suggest that heterogeneity in Wolbachia infection status may be sufficient to maintain polymorphisms even in the absence of costs.
Keywords: defensive symbiosis, host–pathogen interactions, Wolbachia, DCV virus, evolve and resequence
Introduction
Endosymbionts can impact strongly the fitness of their hosts, thus constituting a putatively important factor for the evolution of species (Moran et al. 2008; Engelstädter and Hurst 2009; Brucker and Bordenstein 2012; Martins et al. 2013). For example, the presence of heritable endosymbionts can increase host fitness through protection against pathogens (Oliver et al. 2003; Kaltenpoth et al. 2005; Scarborough et al. 2005; Teixeira et al. 2008; Gerardo and Parker 2014) or nutritional changes (Sabree et al. 2009; Gibson and Hunter 2010; Hosokawa et al. 2010). These effects, alone or in combination, may play a crucial role in the invasion and spreading of facultative endosymbionts in host populations (Engelstädter and Hurst 2009; Brucker and Bordenstein 2012; Faria and Sucena 2015). In fact, several studies demonstrated that the increase in host fitness favors endosymbiont spreading (Oliver et al. 2008; Jaenike et al. 2010; Himler et al. 2011).
In arthropods, the endosymbiont Wolbachia is widespread (Jeyaprakash and Hoy 2000; Weinert et al. 2015), but its infection frequency varies among Drosophila species and populations (Riegler et al. 2005; Corby-Harris et al. 2007; Richardson et al. 2012; Kriesner et al. 2013; Turelli et al. 2018). In addition, within populations of Drosophila, endosymbiont variants of the same strain can fluctuate in frequency throughout time or in the course of experimental evolution (Riegler et al. 2005; Miller and Riegler 2006; Kriesner et al. 2013; Versace et al. 2014; Faria et al. 2016). Drosophila melanogaster is infected with one Wolbachia strain, wMel, for which several variants have been described and clustered in two major monophyletic groups: wMel-like and wMelCS-like confer different levels of protection against DCV and FHV infection (Riegler et al. 2005; Nunes et al. 2008; Chrostek et al. 2013; Martinez et al. 2014; Faria et al. 2016). The wMelCS-like variants provide a more effective protection, which correlates with its higher load inside the host (Chrostek et al. 2013; Martinez et al. 2014; Faria et al. 2016). This variation in protective effect of Wolbachia against viral infection may constitute the mechanism behind the different incidence of Wolbachia infection observed within and across populations (Hedges et al. 2008; Teixeira et al. 2008; Moreira et al. 2009; Hughes et al. 2011). Importantly, a recent study revealed that evolution in the host genome in the presence of DCV operated at a slower pace in presence of Wolbachia, driving different plateaus of survival and pastrel protective-allele frequencies (Martinez et al. 2016).
In a previous study, we have performed experimental evolution of four replicates of a Wolbachia-infected outbred D. melanogaster population in presence of DCV (VirSys), with their respective control, that is, four replicates exposed to a systemic mock infection (ContSys) (Martins et al. 2014). After 20 generations of selection, we observed an increase in survival 10 days after infection from 33% to almost 80% in the populations selected against DCV (Martins et al. 2014). We showed that Drosophila adaptation to DCV relied on two major loci (containing three functionally validated genes: pastrel, CG8492, and Ubc-E2H) with different cross-resistance properties (Martins et al. 2014). Subsequently, we demonstrated that the most protective Wolbachia substrain, a wMelCS-like strain, was fixed by selection in populations evolving in presence of DCV and contributed to host adaptation (Faria et al. 2016). However, it is still unclear whether the genomic changes observed in the host are contingent upon the presence of Wolbachia or whether such changes would evolve also in the absence of the symbiont.
We asked whether DCV adapted populations with Wolbachia could further increase resistance after removal of the endosymbiont, and if so, whether the genetic basis of this adaptation was similar to that identified in the presence of the symbiont (Martins et al. 2014). To this end, we removed Wolbachia from newly derived populations, VirSys-tet and ContSys-tet, and performed in parallel with Wolbachia-infected populations, a further 20 generations of selection with DCV or with a mock infection.
We found that these previously adapted outbred populations of Drosophila without Wolbachia increased their immunocompetence against DCV to similar levels as in the presence of the symbiont. Resequencing the Wolbachia-free populations after 20 generations indicated that the increase in survival was based on the frequency increase of the same SNPs identified in the presence of Wolbachia and associated to the validated genes, pastrel and Ubc-E2H.
Materials and Methods
Fly Populations
We used an outbred population of D. melanogaster founded, expanded, and maintained as described elsewhere (Martins et al. 2013, 2014; Faria and Sucena 2017).
Wolbachia-Free Populations
Wolbachia-free populations VirSys-tet and ContSys-tet, generated from of the VirSys and ContSys, respectively, were derived at generation 35, by raising the progeny for two generations on food with tetracycline (0.05 mg/mL). Sequencing of initial and final populations (G0 and G20) showed absence of Wolbachia (supplementary table S1, Supplementary Material online). Two generations after tetracycline treatment, flora standardization was performed by direct contact of nontreated males with the food of treated populations: 100 males of each population replicate stayed over 24 h in contact with the food (inside the population cages) that posteriorly received the first generation of correspondent treated-populations, at generation 39–40. Each replicate population of VirSys-tet was systemically infected with DCV and ContSys-tet selection regimes kept as control.
Experimental Evolution
Starting from the base population, we derived 12 lines evolving under three different regimes (four replicates per treatment). In the VirSys treatment, adult flies were pricked in the thoracic region with DCV (2 × 107 tissue culture ID50) at each generation. A second treatment consisted of a control for pricking, in which the needle was dipped in sterile medium (ContSys). Finally, a second group of control lines consisted of flies kept in standard food without being pricked (control). The dose of DCV that was used caused an average mortality of 66% in the initial population 10 days after infection.
These treatments were administrated to 310 males and 310 females (4–6 days after eclosion). Selection lines were kept in large population cages and surviving individuals mated randomly; reproduction took place at days 5–7 after infection by providing fresh oviposition substrate. The number of individuals in the control populations was always reduced to the initial number of infected individuals (i.e., 600).
Egg density was limited to 400 per cup, a density determined experimentally to enable optimal larval development. Each generation cycle lasted 3 weeks. Before the beginning of the experiment, absence of vertical transmission of the parasite to the progeny was verified.
To monitor survival across generations, we infected at each generation additional sample males and female flies from each replicate of each treatment and monitored their survival in vials for at least 10 days.
Survival Analysis
To compare survival across generations in the different selection regimes—mock (ContSys) and DCV infected (VirSys), with or without (-tet) Wolbachia—the proportion of individuals surviving at days 6 and 10 after infection in each vial was first estimated using the Kaplan–Meier method. Subsequently, a generalized linear mixed model (GLMM) was fitted to the survival data at each time-point, assuming a binomial distribution and an underlying logit link function. The proportion of survivors, weighted by the number of individuals in each vial as dependent variable, was fitted in a model with sex, generation, selection regime and Wolbachia infection status as fixed factors. Line nested within selection regime, Wolbachia status and sex at each generation was considered a random factor. This base model was then used to then test for differences in survival between lines, both overall and across generations. We determined: 1) differences in the absolute survival for each selection regime/Wolbachia status combination, 2) changes in survival across generations, 3) changes in the survival differential between VirSys and ContSys selection regimes with or without Wolbachia, using the difference in the initial generation as a reference, and 4) if there was a linear trend for change (increase or decrease) across generations in the mean survival of the different regimes, by considering Generation an ordered factor. All analyses were done either averaging the results for both sexes (i.e., considering sex as a factor but averaging the estimates for both sexes) and independently for each sex.
All statistical analyses were done in R (version 3.4.2). Generalized linear mixed models were fitted with the glmer function, in the “lme4” package in R. The effects of the fixed factors and of the hierarchical interaction terms were compared using Type II Wald χ2 tests (ANOVA function in the “car” package). Contrasts of least-square means estimates, survival differentials and trends across generations were done on the most parsimonious model, that is, in models including only significant (P < 0.05) factors and interactions, using the emmeans and contrast function in the “emmeans” package. The reported P values for tests involving multiple comparisons were adjusted using a sequential Bonferroni correction.
Whole-Genome Sequencing
Genomic DNA preparation and sequencing were done as in Orozco-terWengel et al. (2012). Briefly, a pool of 200 individuals of each selection line was homogenized with an Ultraturrax T10 (IKA-Werke), and DNA was extracted from the homogenate using a high-salt extraction protocol. Genomic DNA was sheared using a Covaris S2 device (Covaris, Inc.) and paired-end libraries were prepared using the TruSeq v2 DNA Sample Prep Kit (Illumina). Libraries were size-selected for a mean insert size of 300 bp on agarose gels and amplified with ten PCR cycles, and 2 × 100-bp paired-end reads were sequenced on a HiSeq 2000 (Illumina). About 16 groups of populations were sequenced: four replicates of ContSys-tet and VirSys-tet, both at generation 0 and 20.
Read Quality Control and Mapping
Reads were mapped following the previously described pipeline for Pool-Seq analysis. Briefly, 125-bp paired-end reads were filtered for a minimum average base quality score of 18 and trimmed using PoPoolation (Kofler et al. 2011). Reads with a minimum length ≥50 bp were then mapped against a reference containing the FlyBase D. melanogaster genome r6.01 (http://flybase.org). We used the following parameters: seeding of the reads disabled (−l 110), 1% missing alignments assuming an error rate of 2% (−n 0.01), maximum number of two gap openings (−o 2) and a maximum gap extension of 12 bases (−e 12, −d 12). Paired-end data were merged to single files in SAM format with the “sampe” option of bwa. Files were converted to BAM format with SAMtools v1.3 (Li et al. 2009) and filtered for a minimum mapping quality of 20 and properly paired reads. BAM files were transformed to pileup files using SAMtools. We identified repetitive elements in the reference genome using RepeatMasker (http://www.repeatmasker.org) and removed them from the pileup files using PoPoolation. Pileup files were converted to the “synchronized” allele frequency format using PoPoolation2 mpileup2sync.jar application. Synchronized files from Martins et al.(2014), aligned to release r5.04 of the D. melanogaster genome were lifted over to r6.01 using CrossMap v0.2.7 (Zhao et al. 2014) and the chain file rep.mod_dm3ToDm6.over.chain available at https://zenodo.org/record/155396.
Coverage Analysis
Average Drosophila genome coverage was calculated across each major chromosomal arm, using sambamba (v0.6.6), filtering for properly paired reads, base (≥20) and mapping quality (≥20).
The average sequence coverage for the genome of the analyzed populations ranged from 32- to 82-fold. In subsequent analyses, allele counts were normalized to 30 (to allow comparisons with previously published results), by scaling the raw allele frequencies. The minimum allowed coverage per position was 15, and coverage <30 was left unscaled.
Efficiency of the tetracycline treatment to remove Wolbachia was assessed by calculating coverage across the Wolbachia genome. For this trimmed fastq reads were independently remapped to the wMel reference genome (AE017196.1) using bwa-mem (v0.7.17), and coverage was calculated across the whole genome, using identical parameters (supplementary table S1, Supplementary Material online).
SNP Calling
Only SNPs that met the following quality criteria were considered: 1) occurrence in at least two replicate populations, 2) the minor allele was covered by at least ten reads across all populations analyzed, and 3) the maximum coverage did not exceed 500.
Identification of Candidate SNPs
We used the Cochran–Mantel–Haenszel (CMH) test, as implemented in PoPoolation2 (Kofler et al. 2011) to identify SNPs with changes in allele frequencies between the different regimes that were consistent among replicates as described elsewhere (Orozco-terWengel et al. 2012). The CMH test is used to test 2×2×k contingency tables (where k is the number of independent replicates) for independence of marginal sums across k replicates. Under the null hypothesis, odds ratios for each replicate are not different from one (i.e., if the allele frequencies between two regimes, are the same). The statistic asymptotically follows a χ2 distribution with one degree of freedom. CMH tests were performed on a SNP-wise basis for the comparisons across groups of populations. The 99.95 percentile of the P value of this statistics, both at chromosome-wide and genome-wide levels, was used as an empirical false discovery rate for calling a significant SNP.
Regions of significantly differentiated SNPs were defined as groups of ten or more differentiated SNPs within 50 kb of each other. This interval provided a good balance between size, number of regions and number of SNPs contained in the region, and was the minimum distance at which recombination was observed after 50 generations of experimental evolution (Teotónio et al. 2009).
Effective population sizes (Ne) were calculated from the allele frequencies of all putatively neutral SNPs (i.e., nonsignificantly differentiated in any comparison) between coordinates 5–10 Mb in the five major chromosomal arms at two time points. Estimates were calculated using the function estimateNe of the package “poolSeq” (Taus et al. 2017), using the Np plan II estimator. This estimator corrects for the two-step sampling process inherent to poolSeq (individuals from a population and reads from a pool), and assumes that sampled individuals do not contribute for the next generation. The pattern of Ne’s obtained by the other estimators available in the package was globally consistent with the ones shown.
Selection coefficients for the candidate SNPs were calculated from the allele frequency trajectories using the function estimateSH of the package “poolSeq” (Taus et al. 2017), assuming codominance and an effective population size of 1000 individuals.
Results
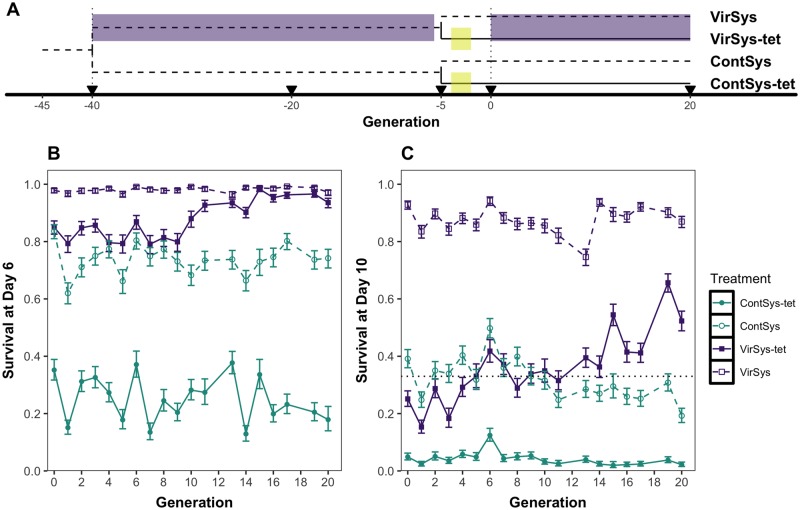
Previously, for 34 generations, four population replicates were challenged at each generation with a DCV systemic infection (VirSys) and four other replicates were used as control, pricked with a buffer solution (ContSys) (Martins et al. 2014). At generation 35, Wolbachia-free replicates for each of the four adapted and four control populations were derived, by administration of a tetracycline treatment for two generations (VirSys-tet and ContSys-tet). These populations were maintained in shared control conditions for three generations, to allow mitochondrial recovery and to standardize the microbiota between infected and noninfected populations. At generation 40, we began a new round of experimental evolution using the same DCV infection protocol as before (Martins et al. 2014), hereafter considered as generation 0 for these Wolbachia-free populations (fig. 1A). In brief, DCV infection was imposed at every generation using the ancestral virus strain and survival was measured daily until 10-day postinfection.
Fig. 1.—
Evolution of increased resistance to DCV in Wolbachia-free population. (A) Diagram representing the different selection regimes used in this study. VirSys populations were challenged with DCV at the generations indicated by the purple shaded area. ContSys population was mock infected. From each ContSys or VirSys population, a Wolbachia-free line (-tet) was derived by tetracycline treatment for two generation (indicated by the yellow boxes). Populations were sequenced at the generations indicated by arrowheads. (B and C) Experimental evolution trajectories over 20 generations of Wolbachia-positive populations (VirSys and ContSys) and Wolbachia-free populations (VirSys-tet and ContSys-tet) for survival to DCV infection at days 6 (B, left) and 10 (C, right) postinfection. Generation 0 represents the first generation of selection in the VirSys-tet populations after Wolbachia removal (40 generations after selection with DCV started) (Martins et al. 2014). Lines with circles represent populations exposed to the virus. Lines with squares represent control lines. Solid lines with filled symbols represent Wolbachia-free and dashed lines with open symbols Wolbachia-positive populations. Vertical bars correspond to the SEM survival among the four replicates.
The survival after DCV infection of Wolbachia positive populations remained virtually the same from G-10 to G0 (∼90% in evolved and 33% in control populations, supplementary fig. S1, Supplementary Material online) (Martins et al. 2014). After Wolbachia removal, survival of the treated populations dropped significantly relative to their parental, Wolbachia positive, counterparts (|z|> 5.200, P < 0.001, fig. 1B and C).
Throughout the ensuing 20 generations of exposure to DCV the VirSys-tet populations experienced a progressive increase in survival at 6- and 10-day postinfection (dpi), both in absolute terms (test for trend, |z|> 11.065, P < 0.0001) and relative to individuals from the ContSys-tet lines (difference in linear trends, |z|> 8.439, P < 0.0001).
Using generation 0 as reference, the increase in survival differential started to be significant after 14 and 15 generations of selection, for survival at 6 and 10 dpi, respectively, and remained significant in all subsequent generations (|z|> 3.167, P < 0.05). Some sex-specific differences were observed in the selection dynamics (supplementary text and fig. S2, Supplementary Material online), but overall there were no differences in the response trend between males and females (|z|< 1.690, P > 0.1822).
At generation 20, the survival of VirSys-tet populations at 6 dpi was similar to that of VirSys populations (z = 2.026, P = 0.1709). At 10 dpi, the populations without Wolbachia still had a significantly lower survival (z = 8.236, P < 0.0001) than their Wolbachia positive counterparts (fig. 1B). When the whole trajectory over 20 generations of selection is considered, resistance is significantly increased in VirSys-tet (z > 3.737, P < 0.0002).
Throughout this further selection with DCV, VirSys and ContSys maintained approximately the same survival after infection. VirSys maintained a very high resistance (test for trend, |z|= 0.277, P = 0.781) whereas ContSys fluctuated around the baseline before the start of the first adaptation experiment (on an average 66% mortality at 10 dpi), but with a slight, but significant, trend for decrease at 10 dpi (test for trend, z= −5.586, P < 0.0001).
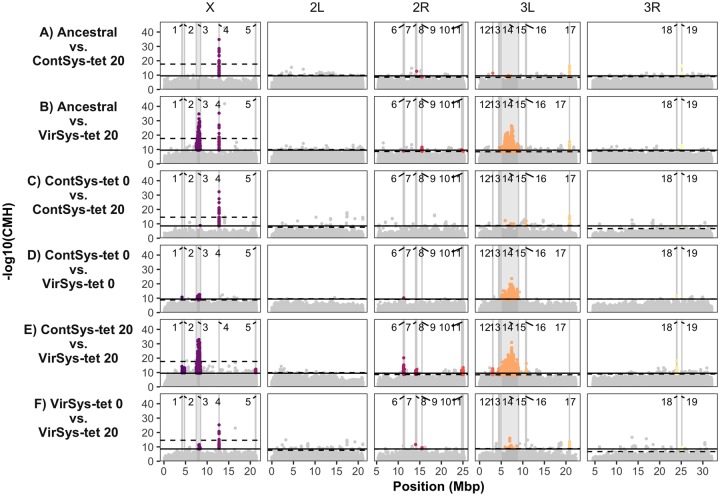
To uncover the genetic basis of the increased DCV resistance of Drosophila Wolbachia-free populations, we performed genome-wide sequencing of DNA pools (Pool-Seq) (Schlötterer et al. 2014) of all replicates of the two Wolbachia free treatments, VirSys-tet and ContSys-tet, at generations 0 and 20. We compared the allele frequencies of these populations against each other and against the founding, Wolbachia positive, populations at generation 40 (Ancestral population, fig. 2). Additionally, to determine if 1) the tetracycline treatment itself and/or 2) continued DCV selection in the VirSys populations before Wolbachia removal, led to significant allele frequency changes, we compared generations -20 and -5 before tetracycline treatment (i.e., generations 20 and 35 in Martins et al. 2014) (supplementary fig. S3, Supplementary Material online) with generation 0 after the tetracycline treatment. For all comparisons, we used the P values of the comparisons between Ancestral and ContSys(-tet) populations at a given generation as null distributions to define genome- and chromosome-wide cutoffs for significance (99.99% quantile across all comparisons), as in our previous work (Martins et al. 2014).
Fig. 2.—
Differentiation between Wolbachia-free selection regimes. −log10 values of the CMH statistic for every polymorphic SNP, across the five major chromosomal arms through pairwise comparison of allele frequencies between the indicated selection regimes: (A and B) Ancestral versus Wolbachia-free Control (ContSys-tet 20) and DCV exposed (VirSys-tet 20) populations after 20 generations of selection; (C) Wolbachia-free Control at generation 0 (ContSys-tet 0) and 20 (ContSys-tet 20); (D and E) Wolbachia-free Control (ContSys-tet) and DCV exposed (VirSys-tet) at generations 0 and 20; (F) Wolbachia-free DCV-exposed at generation 0 (VirSys-tet 0) and 20 (VirSys-tet 20). The solid and dotted lines represent the 99.99% quantile of the P values in the controls comparison at genome-wide and chromosome-wide levels, respectively. Positions within chromosomal regions corresponding to groups of differentiated SNPs (see Materials and Methods) are numbered based on their genomic coordinates, shaded and shown in different colors.
Between generations -20 and -5, ContSys populations showed very little differentiation, with relatively uniform CMH landscapes (supplementary fig. S3A, Supplementary Material online). The tetracycline treatment did not cause consistent allele frequency changes (supplementary fig. S3B and D, Supplementary Material online). However, it probably imposed a significant bottleneck, given the steep increase in the cutoff thresholds in the -5 versus 0 and -20 versus 0 comparisons (supplementary fig. S4, Supplementary Material online), and a decrease in estimated effective population sizes, especially in the ContSys populations (supplementary fig. S5, Supplementary Material online, comparison between generation -5 and generation 0). The comparison between the VirSys populations at generations -20 and -5 (supplementary fig. S3C, Supplementary Material online) yielded fewer nominally significant positions than the control comparisons (250 vs. 312), indicating that the lack of phenotypic changes, in the presence of Wolbachia, between generations 20 and 35 of DCV selection (Martins et al. 2014), were mirrored at the genomic level. As expected by drift, differentiation between ContSys populations increased with the duration of the experiment (supplementary fig. S4, Supplementary Material online).
Using the Drosophila melanogaster genome release R6.01 we identified two regions, one in the X chromosome (region 4, positions X: 12759143–12764257) and another in the left arm of the third chromosome (region 17, positions 3L: 20816331–20818821) showed parallel changes between generation 0 and 20 both in the ContSys-tet and VirSys-tet treatment (fig. 2C and F). While this suggests an adaptation of the host to the absence of Wolbachia, we cannot rule out that this selection signature does not reflect other changes associated with tetracycline treatment.
Two regions comprised most of the significantly differentiated SNPs between the ContSys-tet versus VirSys-tet populations, both at generation 0 and 20 (fig. 2B, D, and E). Consistent with our previous results, these were in a region spanning ∼1 Mb on the X chromosome (region 3, X: 7469749–8489733) and across a 4-Mb region on chromosome 3 L (region 14, 3L: 4975419–9021907). While the CMH values for the 3L SNPs remained relatively constant between generation 0 and 20, the differentiation peak on the X increased both in magnitude and broadness (contrast fig. 2D with fig. 2E). At generation 20, the most significantly differentiated SNPs in the X region corresponded to position X: 8101516, mapping to the gene CG2258, in close vicinity to the fourth most differentiated SNP, on the previously reported Ubc-E2H gene (X: 8090522), which increased in frequency from 0.67 to 0.92 (supplementary data set S2 and fig. S6, Supplementary Material online). Other highly differentiated SNPs were in the X: 7862409–7862410 (supplementary data set S2 and fig. S6, Supplementary Material online), mapping to a region of three loci, CG1409, Ir7b, and dec-1. As before, the most significantly differentiated SNPs in 3 L were located nearby pastrel, where the protective allele (commonly characterized by the SNPs 3L: 7357795) increased in frequency, from initially 0.73 to 0.88 (supplementary data set S2 and fig. S6, Supplementary Material online).
Similarly to the survival frequency after DCV infection, the frequencies of the protective alleles of pastrel and Ubc-E2H reached a plateau after the first 20 generations and remained stable in the subsequent 15 generations of selection (from Generation -20 to Generation -5) (supplementary figs. S6 and S7, Supplementary Material online) (s = 0.246 and 0.208 in the first 20 generations of selection for pst and Ubc-E2H vs. s = 0.022 and s = 0.078 between Generation -20 and 0). Upon Wolbachia removal, the frequencies for both protective alleles resumed their increase in frequency to reach the highest levels recorded over the 60 generations recorded (supplementary fig. S6, Supplementary Material online), with the biggest changes happening in the X-linked positions (s = 0.090 and 0.195 for pst and Ubc-E2H, respectively).
Discussion
We showed that once Wolbachia is removed survival upon DCV infection decreases strongly but reaches, after 20 generations, similar survival rates to the ones found in the presence of this protective endosymbiont. Using Pool-Seq, we mapped the selection targets to the previously identified genes pastrel and Ubc-E2H but the selected alleles in the Drosophila host do not reach fixation.
As previously observed (Martins et al. 2014), eliminating Wolbachia with tetracycline strongly reduces survival upon DCV infection in both adapted and control populations. The average survival at day 10 dropped by 71.8% in VirSys-tet and by 76.25% in ContSys-tet populations. Importantly, the VirSys-tet population maintained its higher degree of survival relative to ContSys-tet (fig. 1, generation 0), confirming that adaptation prior to tetracycline treatment was largely based on the host genome. Further, this suggests that the roles of Wolbachia and the host genome in the response to DCV infection are predominantly independent.
The comparison between ContSys-tet 0 versus 20 (fig. 2C) and VirSys-tet 0 versus 20 (fig. 2F), as well as between the Ancestral populations and the Wolbachia-free populations at generation 20 (fig. 2A and B), showed two consistent and specific differentiation peaks (X: 12759143–12764257 and 3L: 20816631–20818821, regions 4 and 17). This signature probably reflects the consequences of cleaning with tetracycline and could thus be caused by, 1) adapting to the absence of Wolbachia itself; 2) microbiota changes throughout treatment; 3) mitochondrial destabilization after antibiotic exposure or; 4) a mapping artefact from the last sequencing round. We have taken precautions to minimize some of these confounding effects. Regarding (2) and (3), we allowed the populations to recover from treatment for three generations and transferred gut microbiota from Wolbachia-positive individuals (see Materials and Methods) to normalize all treatments before starting the new round of DCV selection. Mapping artefacts (4) could not be completely excluded, although no evidence points to this situation when comparing generations 0 and 20. The same allele consistently increased in frequency in all tetracycline treated populations, with the major allele in these positions at generation 0 matching the former monomorphic allele present in the ancestral population (supplementary fig. S6, Supplementary Material online). Indeed, these positions were only detected as being polymorphic after the indel realignment step in our pipeline and with an increase in coverage of the differentiated region of the X chromosome, relative to nearby regions. Altogether this suggests adaptation to the absence of Wolbachia (i), and points to testable Drosophila candidate genes involved in the interaction with Wolbachia, and to the mechanisms through which the presence or absence of Wolbachia influences host populations.
Comparing whole genome sequences from the initial (0) and final (20) generations shows that the biggest difference between ContSys-tet and VirSys-tet still resides in high peaks of differentiated SNPs in loci that have previously been identified and validated as conferring resistance to DCV (Martins et al. 2014). First, we observed an increase in the frequency of SNPs mapping to pastrel. Although the previously reported protective allele of pastrel had a high frequency at the end of the first round of adaptation (0.733 vs. 0.158 in ContSys), the frequency change continued after removing Wolbachia. Pastrel is one major player in DCV response, as shown through GWAS (Magwire et al. 2012), multiparent advanced intercross (Cogni et al. 2016) and Evolve and Resequence approaches (Martins et al. 2014). Thus, after a decrease in viral immunocompetence by the removal of Wolbachia, the frequency rise of the pst protective allele constitutes a privileged mechanism for adaptation against viral infection. The same effect could be observed for SNPs in Ubc-E2H, another gene identified in the first round of selection and functionally validated for its role in protection against DCV infection (Martins et al. 2014). For instance, the SNPs X: 8090292/T and X: 8090522/T, increased in frequency between generations 0 and 20, from 0.66 to 0.86 (s = 0.114) and 0.67 to 0.92, respectively. Thus, adaptation upon removal of the protective endosymbiont Wolbachia is very likely to depend mostly on the same genes as before, pastrel and Ubc-E2H (Martins et al. 2014).
However, possibly as a consequence of the stronger selection exerted, one pair of SNPs (X: 7862409–7862410), located in the intergenic region between CG1409 and Ir7b overcame our detection level. These genes have no immune role described, but the putative protective allele(s) associated with this SNP increases from a frequency of 0.56 to 0.90 (s = 0.172). In the first round of selection in the presence of the endosymbiont this SNP increases modestly, but steadily, from 0.276 to 0.542 (Martins et al. 2014). In absence of LD information, we cannot rule out that linkage with Ubc-E2H caused the frequency increase of this region after the removal of Wolbachia. Nevertheless, we think that it is much more likely that in the current experimental evolution small effect alleles also contributed to adaptation (Martins et al. 2014; Faria et al. 2016). Functional tests of candidate genes will distinguish between these scenarios, potentially uncovering novel players and establishing how the effects are partitioned between different loci involved in the Drosophila–Wolbachia–DCV interaction.
It has been shown that Wolbachia can impact the selective process operating on the host (Faria et al. 2016; Martinez et al. 2016). In particular, Martinez, Cogni and colleagues showed that two populations evolving in parallel with or without Wolbachia will reach distinct survival plateaus (Martinez et al. 2016). This difference is strongly correlated with the frequency of the protective pastrel allele (Magwire et al. 2012) which increased in Wolbachia-containing populations to only half the frequency attained by its endosymbiont-free counterpart (Martinez et al. 2016). In addition, our approach was able to identify the role of Ubc-E2H as an antiviral protection locus. This suggests that the protective variant of the gene may not be present in the Drosophila panels and population used in the works of Magwire et al. (2012), Cogni et al. (2016) and Martinez et al. (2016). An alternative explanation lies in the strength and duration of our experimental evolution regime as necessary conditions to uncover the antiviral protective function of Ubc-E2H.
Contrasting these two approaches provides insight into the nature of the constraints imposed on the host by the presence of Wolbachia when it precedes or follows exposure to viral infection. Eventual differences in the results of these two approaches would have revealed the extent to which antiviral allelic variants are not recruited throughout the adaptation process due to the present or past presence of Wolbachia. In contrast, in both approaches, the increase of frequency of the major antiviral alleles drove the adaptive process. This seems to be independent of the Wolbachia presence and of the adaptive history of the population, as shown herein. Moreover, the question becomes why is there parallel evolution of this response? Firstly, our results suggest that the mechanisms of resistance induced by Wolbachia and by the host genes are independent. Secondly, this consistency in the genetic basis of the evolved response using different conditions, methods and populations/lines of Drosophila, is consistent with or in other cases described (ffrench-Constant et al. 1998; Sucena et al. 2003; Cresko et al. 2004; Jost et al. 2008; McGlothlin et al. 2014) and in contrast to others (Teotónio and Rose 2000; Wilkens and Strecker 2003; Hoekstra et al. 2006; Kawecki and Mery 2006; Simões et al. 2008). However, it is important to consider that the potential differences between viral doses and infection recurrence in the laboratory or in nature, is likely to change the way individuals respond against infection and, consequently, the pace and architecture of the adaptive process.
Consistent with what we showed before (Martins et al. 2014), Wolbachia bearing populations do not fix the alleles responsible for increased survival. The observed heterogeneity in Wolbachia infection status (Riegler et al. 2005; Corby-Harris et al. 2007; Richardson et al. 2012) and consequent variation in susceptibility within a population, even when the cost of resistance is not detectable (Faria et al. 2015), may help explain the maintenance of polymorphisms for survival to infection (Antonovics and Thrall 1994).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Viola Nolte and Tânia F. Paulo for help in experimental and analysis procedures, Sara Magalhães for critical comments and discussions that improved the manuscript. This work was supported by Instituto Gulbenkian de Ciência/Fundação Calouste Gulbenkian; FCT—Fundação para a Ciência e a Tecnologia (Portugal) (SFRH/BPD/62964/2009) to N.E.M. and (SFRH/BD/82299/2011 to V.G.F.); and Austrian Science Funds (FWF P27630) to C.S. CONGENTO, project LISBOA-01-0145-FEDER-022170, cofinanced by Lisboa Regional Operational Programme (Lisboa 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF), and Foundation for Science and Technology (Portugal).
Literature Cited
- Antonovics J, Thrall PH.. 1994. The cost of resistance and the maintenance of genetic polymorphism in host-pathogen systems. Proc R Soc B Biol Sci. 2571349:105–110. [Google Scholar]
- Brucker RM, Bordenstein SR.. 2012. Speciation by symbiosis. Trends Ecol Evol. 278:443–451. [DOI] [PubMed] [Google Scholar]
- Chrostek E, et al. , . 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 912:e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogni R, Cao C, Day JP, Bridson C, Jiggins FM.. 2016. The genetic architecture of resistance to virus infection in Drosophila. Mol Ecol. 2520:5228–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, et al. , . 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol. 7311:3470–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, et al. , . 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci USA. 10116:6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstädter J, Hurst GDD.. 2009. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst. 401:127–149. [Google Scholar]
- Faria VG, et al. , . 2015. Evolution of Drosophila resistance against different pathogens and infection routes entails no detectable maintenance costs. Evolution 6911:2799–2809. [DOI] [PubMed] [Google Scholar]
- Faria VG, et al. , . 2016. Drosophila adaptation to viral infection through defensive symbiont evolution. PLoS Genet. 129:e1006297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria VG, Sucena É.. 2015. Novel endosymbioses as a catalyst of fast speciation. In: Gontier N, editor. Reticulate evolution – symbiogenesis, lateral gene transfer, hybridization and infectious heredity, Dordrecht: Springer. p. 107–120. [Google Scholar]
- Faria VG, Sucena É.. 2017. From nature to the lab: establishing Drosophila resources for evolutionary genetics. Front Ecol Evol. 5:61. [Google Scholar]
- ffrench-Constant RH, Pittendrigh B, Vaughan A, Anthony N.. 1998. Why are there so few resistance-associated mutations in insecticide target genes? Philos Trans R Soc B Biol Sci. 3531376:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo NM, Parker BJ.. 2014. Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Curr Opin Insect Sci. 4:8–14. [DOI] [PubMed] [Google Scholar]
- Gibson CM, Hunter MS.. 2010. Extraordinarily widespread and fantastically complex: comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol Lett. 132:223–234. [DOI] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O’Neill SL, Johnson KN.. 2008. Wolbachia and virus protection in insects. Science 3225902:702.. [DOI] [PubMed] [Google Scholar]
- Himler AG, et al. , . 2011. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 3326026:254–256. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP.. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 3135783:101–104. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T.. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA. 1072:769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL.. 2011. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 75:e1002043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ.. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 3295988:212–215. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA.. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 94:393–405. [DOI] [PubMed] [Google Scholar]
- Jost MC, et al. , . 2008. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol Biol Evol. 256:1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M, Göttler W, Herzner G, Strohm E.. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 155:475–479. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Mery F.. 2006. Genetically idiosyncratic responses of Drosophila melanogaster populations to selection for improved learning ability. J Evol Biol. 194:1265–1274. [DOI] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C.. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 2724:3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P, Hoffmann A. a, Lee SF, Turelli M, Weeks AR.. 2013. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 99:e1003607.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. , . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, et al. , . 2012. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 811:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, et al. , . 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 109:e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, et al. , . 2016. Addicted? Reduced host resistance in populations with defensive symbionts. Proc R Soc B Biol Sci. 2831833:20160778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins NE, et al. , . 2014. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc Natl Acad Sci USA. 11116:5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins NE, Faria VG, Teixeira L, Magalhães S, Sucena É.. 2013. Host adaptation is contingent upon the infection route taken by pathogens. PLoS Pathog. 99:e1003601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW, et al. , . 2014. Parallel evolution of tetrodotoxin resistance in three voltage-gated sodium channel genes in the garter snake Thamnophis sirtalis. Mol Biol Evol. 3111:2836–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Riegler M.. 2006. Evolutionary dynamics of wAu-Like Wolbachia variants in neotropical Drosophila spp. Appl Environ Microbiol. 721:826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42:165–190. [DOI] [PubMed] [Google Scholar]
- Moreira LA, et al. , . 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 1397:1268–1278. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Neumeier H, Schlötterer C. 2008. Contrasting patterns of natural variation in global Drosophila melanogaster populations. Mol. Ecol. 17:4470–4479. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Campos J, Moran NA, Hunter MS.. 2008. Population dynamics of defensive symbionts in aphids. Proc R Soc B Biol Sci. 2751632:293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS.. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 1004:1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-terWengel P, et al. , . 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles: genomic signatures of adaptation to new environment. Mol Ecol. 2120:4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MF, et al. , . 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 812:e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler M, Sidhu M, Miller WJ, O’Neill SL.. 2005. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol. 1515:1428–1433. [DOI] [PubMed] [Google Scholar]
- Sabree ZL, Kambhampati S, Moran NA.. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci USA. 10646:19521–19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HC.. 2005. Aphid protected from pathogen by endosymbiont. Science 3105755:1781.. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Tobler R, Kofler R, Nolte V.. 2014. Sequencing pools of individuals—mining genome-wide polymorphism data without big funding. Nat Rev Genet. 1511:749–763. [DOI] [PubMed] [Google Scholar]
- Simões P, et al. , . 2008. How repeatable is adaptive evolution? The role of geographical origin and founder effects in laboratory adaptation. Evolution 628:1817–1829. [DOI] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL.. 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 4246951:935–938. [DOI] [PubMed] [Google Scholar]
- Taus T, Futschik A, Schlötterer C.. 2017. Quantifying Selection with Pool-Seq Time Series Data. Mol Biol Evol. 3411:3023–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L, Ferreira Á, Ashburner M.. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 612:e1000002. 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD.. 2009. Experimental evolution reveals natural selection on standing genetic variation. Nat Genet. 412:251–257. [DOI] [PubMed] [Google Scholar]
- Teotónio H, Rose MR.. 2000. Variation in the reversibility of evolution. Nature 4086811:463–466. [DOI] [PubMed] [Google Scholar]
- Turelli M, et al. , . 2018. Rapid Global Spread of wRi-like Wolbachia across Multiple Drosophila. Curr Biol. 286:963–971.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versace E, Nolte V, Pandey RV, Tobler R, Schlötterer C.. 2014. Experimental evolution reveals habitat specific fitness dynamics among Wolbachia clades in Drosophila melanogaster. Mol Ecol. 234:802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ.. 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc R Soc B Biol Sci. 2821807:20150249–20150249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens H, Strecker U.. 2003. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc. 804:545–554. [Google Scholar]
- Zhao H, et al. , . 2014. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 307:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.