Abstract
Homeostasis of nutrient metabolism is critical for maintenance of the normal physiologic status of the cell and the integral health of humans and mammals. In vivo, there is a highly efficient and precise process involved in nutrient recycling and organelle cleaning. This process is named autophagy, and it can be induced in response to the dynamic change of nutrients. When cells face nutritional stress, such as stress caused by nutrient deficiency or nutrient excess, the autophagy pathway will be activated. Generally, when nutrients are withdrawn, cells will sense the signs of starvation and respond. AMP-activated protein kinase and the mammalian target of rapamycin, two of the major metabolic kinases, are responsible for monitoring cellular energy and the concentration of amino acids, respectively. Nutrient excess also induces autophagy, mainly via the reactive oxygen species and endoplasmic reticulum stress pathway. When nutritional stress activates the autophagy pathway, the nutrients or damaged organelles will be recycled for cell survival. However, if autophagy is overwhelmingly induced, autophagic cell death will possibly occur. The balance of the autophagy induction is the crucial factor for cell survival or death. Herein, we summarize the current knowledge on the induction of autophagy, the autophagy response under nutritional stresses, and autophagic cell death and related diseases, which will highlight the process of nutritional stress-induced autophagy and its important physiologic and/or pathologic roles in cell metabolism and diseases, and shed light on the research into the mechanism and clinical applications of autophagy induced by nutritional stresses.
Keywords: nutritional stress, autophagy, amino acids, lipophagy, autosis
Introduction
Cells have developed a way to adapt to changes in the environment, especially under nutrient starvation and stress conditions (1). Autophagy is a degradation pathway which can recycle nutrients and organelles to enable cells to adapt to undesirable surroundings. It is also a highly conserved cellular pathway. When cells sense nutritional stress, autophagy will be induced very quickly. At the beginning of autophagy initiation, an autophagic membrane will be isolated to form a phagophore. The phagophore membrane will then extend and curve to engulf the targeted cargo to form a closed double-membrane organelle, called an autophagosome. The autophagosome will eventually fuse with a lysosome upon maturation (2). During autophagy induction, many autophagy-related genes (ATGs) participate in the origination, extension, and fusion of autophagosomes. In general, nutritional stress is primarily induced by a nutrient imbalance. Autophagy can be induced by nutritional stresses, including nutritional starvation and excess nutrient stress. Starvation of nutrients such as amino acids or energy starvation will trigger autophagy activation through the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) (3) or the AMP-activated protein kinase (AMPK) signaling pathway (4), respectively. However, nutrient excess, especially high concentrations of glucose, will also induce autophagy. This process is mainly activated through the reactive oxygen species (ROS) pathway (5). When an animal is born, the transplacental nutrient supply is disrupted. During this period, the neonate needs to adapt to starvation by recycling nutrients (6). In many instances, autophagy is the key regulator that maintains the normal physiology of cells and animals. However, autophagy can also cause cell death as a result of its overwhelming induction. The term “autosis” defines autophagic cell death that is characterized by a large accumulation of autophagosomes during cell death (Figure 1) (7). This review aims to summarize the basic autophagy induction process, autophagy associated with nutrient starvation and excess, the autophagic cell death and its related diseases.
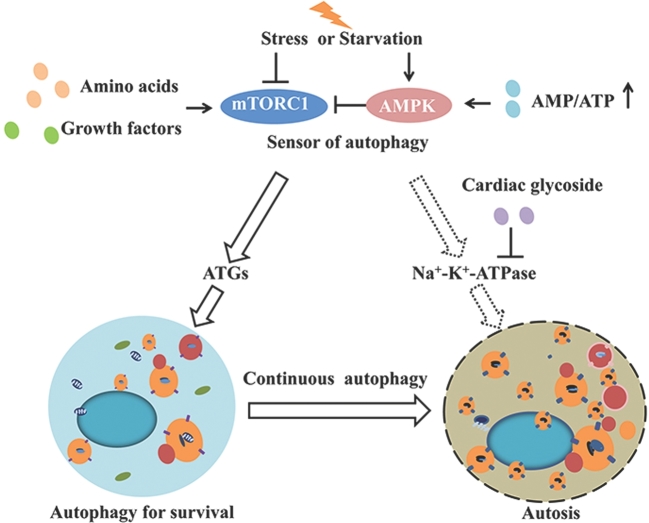
FIGURE 1.
Trade-off between autophagy and autosis induced by starvation and stress. When cells face starvation or stress, mTORC1 and AMPK will sense the signals to induce the autophagy pathway from amino acid or growth factor withdrawal and energy deficiency, respectively. Once autophagy is induced, ATGs will be recruited to assemble autophagosomes, which function to recycle nutrients and organelles to maintain the normal function of the cell. However, when the tradeoff of the degradation process is out of balance, overwhelming autophagic activity will kill the cells with overactivated Na+-K+-ATPase. AMPK, AMP-activated kinase; ATG, autophagy-related gene; mTORC1, mammalian target of rapamycin complex 1.
Mechanisms of Autophagy Induction
Autophagy initiation
At the beginning of autophagy initiation, a double-membrane phagophore forms and many ATGs are recruited to help the membrane expand. However, the source of the early double membrane is still unclear. A few studies indicate that the autophagosome may contain some of the lipids of Golgi and plasma membranes (8, 9). With the continuous recruitment of ATGs to the phagophore platform, the membrane expands to form a cup-shaped precursor of the autophagosome, called an omegasome (10).
As the autophagosome continues to expand, ATGs are increasingly recruited into the autophagic machinery. The autophagic machinery consists of three major systems. The first is the ATG1, ATG2, ATG9, and ATG18 system. The second is the vacuolar protein sorting 34 (VPS34) complex. The last is the system containing ATG8, ATG12, ATG7, ATG10, ATG3, ATG4, and ATG16 (11). ATG9 is a membrane protein in the autophagic machinery (12) which carries the membrane to expand the phagophore (13). ATG 1 complex, ATG 2, and ATG 18 are required for the retrieval of ATG9 from the pre-autophagosomal structure (PAS), which is important for recycling ATG9 and the autophagosome membrane extension. The VPS34 complex, which is located on the PAS, can recruit phosphatidylinositol-(3)-phosphate-binding proteins, which in turn recruit lipids (14, 15). When autophagy is induced, the ATG8-phosphatidylethanolamine complex will be cut by ATG4 (16). ATG12 binds to ATG5 and forms a complex with ATG16 (17). ATG12 coordinates with ATG8-phosphatidylethanolamine to expand the membrane (18, 19). Upon the PAS formation, the ATG11 recruits ATG proteins to the PAS when the autophagosomes close and fuse with a lysosome (20).
Autophagosome maturation
When the autophagosome double membrane enlarges and engulfs its cargo to form a closed structure, the autophagosome finishes its transformation to maturity. The process of autophagosome maturation is defined as the fusion of the autophagosome with a lysosome. During its maturation, a large amount of proteins are recruited to promote the contact and fusion of the autophagosome and the lysosome membrane. Many proteins play crucial roles in the membrane-membrane fusion, anchoring the membrane of the autophagosome to the lysosome. For example, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, including vesicle-associated membrane proteins (VAMP7, VAMP3, and VAMP8), Qa-SNARE syntaxin 17 (STX17), soluble NSF attachment protein 29 (SNAP-29), and vesicle transport interacting t-SNAREs homologue 1B, are all ligation proteins that mediate the fusion of the autophagosome and lysosome membrane (21–24). STX17 is a membrane protein which is localized at the outer membrane of the closed autophagosome and interacts with SNAP-29 and VAMP8. Depletion of STX17 disrupts the membrane fusion process of the autophagosome and lysosome, which leads to the accumulation of autophagosome in cytosol when autophagy is induced (24). In addition, during the autophagosome maturation, ATG14 binds to STX17-SNAP29 complex on the autophagosome and can promote the fusion process. ATG14 also promotes the interaction with VAMP8, which is located in the lysosome membrane (25). The CoCrMo metal particles (26) and endosomal complex required for the transport III complex are also necessary for the fusion and degradation of mature autophagosomes (27). The transport III complex is very important in the process of multivesicular body sorting. The homotypic fusion and protein sorting complex is also involved in the membrane-membrane fusion (28). Lysosome-associated membrane protein 2, which anchors to the lysosome membrane, seems to be involved in the fusion process. Mice deficient in lysosome-associated membrane protein 2 are also deficient in autophagosome maturation in neutrophils (29). Rab7 GTPase belongs to the small GTPase Rab protein family and also functions in the maturation process of both autophagosomes and endosomes. Inhibition of Rab7 will block autophagic vacuole fusion, and finally lead to the accumulation of autophagic vacuoles (30).
The autophagic machinery also participates in the maturation process of autophagosomes, even though its primary function is in the autophagy initiation process. The complexation of VPS34 with Beclin1/Atg6 and UV Radiation Resistance Associated Gene (UVRAG) contributes to the fusion of autophagosomes with late endosomes or lysosomes via the elevation of Rab7 activity (31). Another VPS34 complex contains Beclin1. Rubicon also contributes to the maturation of autophagosome. Inhibition of the complex activity owing to Rubicon deficiency results in a dramatic increase in autophagosome accumulation in the cytoplasm because of degradation failure (32). Tectonin beta-propeller repeat-containing protein 1 (TECPR1), which interacts with ATG5, may promote the maturation of autophagosomes (33).
Cellular Autophagy-Related Pathways
Under normal physiologic conditions, cellular autophagy activity is retained at a very low level. However, nutritional imbalance or stress, such as endoplasmic reticulum (ER) stress or oxidative stress, will robustly induce autophagy through various pathways.
mTORC1 and autophagy
The mTOR, as the nutrient master sensor, links amino acid signals to autophagy. The mTOR mediates cell growth by accelerating the synthesis pathway. Autophagy is a precise process that is controlled by mTOR activity in response to the concentration of amino acids and growth factors. The mTOR has two different complexes in mammals: mTORC1 and mTORC2. However, only mTORC1 can be activated by both amino acids and growth factors (34). In general, mTORC1 consists of mTOR, regulatory-associated protein of mTOR (raptor), mammalian lethal with SEC13 protein 8 (mLST8), proline-rich Akt substrate of 40 kDa (PRAS40) and DEP-domain-containing mTOR-interacting protein (Deptor) subunits (35) (Figure 2A). PRAS40 and Deptor repress mTORC1 activity in the dephosphorylated state. When activated, mTOR can phosphorylate both PRAS40 and Deptor, causing their disassociation from the remaining part of mTORC1 to increase the kinase activity (36). mTORC1 activation requires amino acid signals, which differ from those for mTORC2. The tuberous sclerosis (TSC)1/2 complex delivers the nutrient signal to mTORC1 by controlling the concentration of Ras homolog enriched in brain (Rheb) (Figure 2B). If Rheb binds to GTP, it will bind directly to mTORC1 and activate it (37). The Rag GTPases can activate mTORC1 by directly binding to and activating mTORC1 (Figure 2C) (38–40). RagA or RagB can bind to RagC or RagD to form the Rag complex (41), which is located on the lysosome membrane (42).
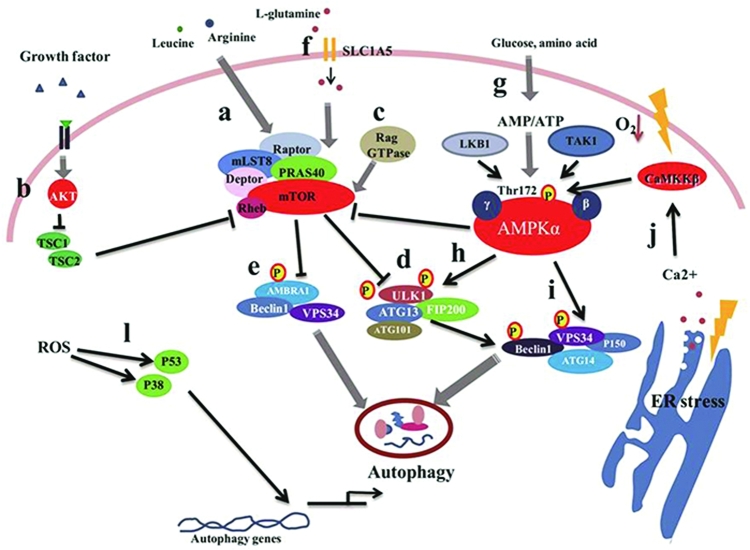
FIGURE 2.
Autophagy induced by nutrient and nutrition stresses via the mTOR and AMPK signal pathways. In response to nutrient deficiency or stress conditions, mTOR is activated following the suppression of the autophagy pathway, whereas energy withdrawal stimulates AMPK activation. This then inhibits mTOR activity by phosphorylation, which promotes the induction of autophagy. ER stresses, hypoxia, and ROS can induce autophagy by enhancing autophagy gene transcription or stimulating the AMPK pathway. (a) The highly conserved Ser/Thr protein kinase TOR is a key sensor and integrator of the amino acid pool signaling. mTORC1 comprises mTOR, Raptor, mLST8, PRAS40, and Deptor. (b) The growth factors regulate mTORC1 through the AKT-TSC1/TSC2 pathway. When growth factors exist, AKT is activated to inhibit TSC1/TSC2 to activate mTORC1, which inhibits the autophagy pathway. (c) The Rag GTPases can activate mTORC1 in response to amino acid signaling by directly binding and activating mTORC1. (d) ATG13 and ULK1, which form complexes with FIP200, can be directly phosphorylated by mTOR to inhibit ULK1 complex activity. (e) AMBRA1, with its ULK1-dependent pattern, induces autophagosome nucleation by promoting Beclin 1 interaction with VPS34. (f) The flux of L-glutamine has been found to be controlled by 2 transporters: SLC1A5 and SLC7A5/SLC3A2. (g) Environmental AMP, ADP, or ATP can directly bind AMPK via the adenine nucleotide-binding sites of the γ subunit. (h) Under glucose deficiency, activation of AMPK promotes autophagy induction by increased ULK1 autophosphorylation. (i) Upon glucose deficiency, the activity of different VPS34 complexes, which include VPS34, p150, ATG14, and Beclin 1 or UVRAG, is regulated. (j) The ER is the site of cellular Ca2+ storage, and continuous removal of Ca2+ from the ER lumen to the cytosol can induce ER stress. An increase in cytosolic Ca2+ has been shown to lead to initiation of autophagy. The ROS-mediated autophagy induction is P53 and P38 dependent. When ROS accumulate, P53 and P38 pathways activate to upregulate autophagic gene expression, which initiates autophagy. AMBRA1, autophagy and beclin regulator 1; AMPK, AMP-activated kinase; ATG, autophagy-related gene; CaMMKβ, calcium/calmodulin-dependent protein kinase kinase beta; Deptor, DEP-domain-containing mTOR-interacting protein; ER, endoplasmic reticulum; FIP200, FAK-family interacting protein of 200 kDa; LKB1, liver kinase B-1; mLST8, mammalian lethal with Sec13 protein 8; mTOR, mammalian target of rapamycin; PRAS40, proline-rich AKT substrate 40 kDa; Raptor, regulatory-associated protein of mTOR; Rheb, Ras homolog enriched in brain; ROS, reactive oxygen species; SLC1A5, solute carrier family 1 member 5; TAK1, transforming growth factor beta-activated kinase 1; TSC, tuberous sclerosis complex; ULK1, Unc-51 like autophagy activating kinase 1; UVRAG, UV radiation resistance associated gene; VPS34, vacuolar protein sorting 34.
A high concentration of amino acids inhibits autophagy. Amino acid deficiency also induces autophagy (43). Amino acid withdrawal activates the autophagy pathway by inhibiting mTORC1 and integrating signals from various metabolic stimuli (44). The relation between mTORC1 and autophagy has been studied for decades. In yeast, ATG1/Unc-51 like autophagy activating kinase (ULK1), which is a protein essential for autophagy, has been reported to be located downstream of TOR (45). However, when autophagy initiates, the ATG1 will form complexes with ATG13 and ATG17. The activity of this complex can be mediated by TOR to take part in the initiation of phagophores (46). In mammalian cells, ATG13and ULK1, which form complexes with FIP200, can be inhibited by mTORC1 (Figure 2D) (47). In addition, the site of phosphorylation of ULK1 by mTORC1 has been identified (757 serine) (48). Moreover, mTORC1 regulates autophagy initiation by affecting both complexes of ULK1 and VPS34 [Beclin1-VPS34-autophagy and beclin regulator 1 (AMBRA1)]. Once autophagy has been initiated, the ULK1 and VPS34 complexes are recruited to the isolated membranes (49). The ULK1 will then activate the VPS34 complex to phosphorylate the phosphatidylinositol to form phosphatidylinositol-(3)-phosphate. AMBRA1, in a ULK1-dependent pattern, induces autophagosome nucleation by promoting the interaction between Beclin1 and VPS34 (Figure 2E) (50, 51). In addition, mTORC1 phosphorylates AMBRA1 at serine 52, which decreases the phosphorylation of AMBRA1 by mTORC1, thereby inhibiting the modification of ULK1 (52).
AMPK and autophagy
Cells have evolved a system to adapt to dynamic nutrient status and quiescence by sensing the amino acid pool or energy source; this process is regulated by mTOR and AMPK. Extensive studies have reported that AMPK functioning as a major energy regulator is efficient at sensing and regulating cell physiology. AMPK is composed of three subunits: subunits α, β, and γ. Subunit α is a catalytic subunit, whereas β and γ are the regulatory units responding to AMPK activity. AMP, ADP, or ATP can directly bind the γ subunit of AMPK (Figure 2G). When the AMP/ATP or ADP/ATP ratio is elevated, AMPK will be activated by phosphorylation (53). When glucose deficiency occurs, AMPK promotes autophagy induction by increasing ULK1 autophosphorylation (54) (Figure 2H). Overexpression of wild-type AMPK-α induces a shift in ULK1 mobility, even under glucose-rich conditions. Moreover, ULK1 can be phosphorylated at serine 317 and serine 777 by AMPK, which is dependent on ULK1 phosphorylation under glucose deficiency. The interaction between ULK1-AMPK and ULK1-ATG13-FIP200 does not seems to be affected by glucose deficiency. Interactions between ULK1 and AMPK can be mediated by mTORC1. The overexpression of Rheb, an mTORC1 activator, decreases ULK1-AMPK binding. The interaction between endogenous ULK1 and AMPK will consistently be increased when mTORC1 is inhibited (48).
Recently, it was found that the autophagy induced by glucose deficiency is associated with both ULK1 and VPS34. When there is a deficiency of glucose, the activity of different VPS34 complexes, which include VPS34, P150, ATG14, and Beclin 1 or UVRAG, are regulated (Figure 2I) (55). Glucose deficiency leads to energy exhaustion and AMPK activation. Therefore, the activation of AMPK may be related to VPS34 complexes. AMPK can regulate the activity of VPS34 complexes both in vivo and in vitro. Moreover, AMPK can directly phosphorylate Beclin1 and VPS34. By using mass spectrometry, two phosphorylation sites of Beclin1 and VPS34 have been identified. As reported, Beclin1 with the S91/94A mutant failed to be activated by AMPK. However, the VPS34 complex composed of Beclin1 is also phosphorylated by AMPK. The Beclin1 S91/94 phosphorylation site is very important for autophagy initiation. The S91/94A mutant of Beclin1 compromises the autophagy flux when it replaces wild-type Beclin1 (55).
After autophagy has been induced, the autophagy initiation requires the activity of SIRT1 (56), which is activated under the condition of nutrient deficiency and then forms molecular complexes with ATG5, ATG7, and ATG8 (57). Glyceraldehyde-3-phosphate dehydrogenase directly regulates AMPK to affect the SIRT1 activity. Under conditions of energy deficiency, cytoplasmic glyceraldehyde-3-phosphate dehydrogenase can be phosphorylated on serine 122 by the activated AMPK to promote the initiation of autophagy (58).
ER stress and autophagy
ER stress induces numerous pathways that play important roles in autophagy. Generally, ER stress will occur when unfolded or misfolded proteins accumulate and the folding capacity of the ER chaperones exceeds the ER lumen (59). When cells are exposed to an overload of nutrients such as glucose, ER stress will be induced. The ER is the storage site of cellular Ca2+. The continuous removal of cellular Ca2+ from the ER lumen to the cytosol can also induce ER stress. As reported (60), a robust increase in cytosolic Ca2+ can directly induce autophagy (Figure 2J). The ER stress derived from Ca2+ signaling is mediated by calmodulin-dependent kinase-β, which is activated by increased cytosolic Ca2+. Ca2+ can also activate AMPK, resulting in activation of the autophagy pathway via the suppression of mTORC1 and the direct phosphorylation of ULK1, VPS34, and Beclin1 (48). The correct spatiotemporal availability of Ca2+ is critical in determining the role of Ca2+ in autophagy and cell fate. The balance between Ca2+ and autophagy controls cellular homeostasis and survival during several physiologic and pathologic conditions (61, 62). The unfolded proteins in the lumen of the ER can induce ER stress (63), which strongly induces autophagy. The unfolded protein stress pathway is regulated by three kinds of ER membrane-associated proteins: protein kinase R-like kinase (PERK), activating transcription factor (ATF)-6, and inositol-requiring enzyme 1. Specifically, PERK and ATF6 are identified as autophagy activators, whereas inositol-requiring enzyme 1 is a negative regulator of autophagy. PERK controls the mRNA level of microtubule-associated protein 1A/1B-light chain 3 (LC3) and ATG5 in the hypoxic responses caused by ATF4 and CHOP (64). The PERK/IkBα/NF-κB pathway can also contribute to autophagy (65). PERK directly phosphorylates the eukaryotic translation initiation factor 2α (eIF2α) on residue serine 51; this process is affected by many stresses (66). The eIF2α can also be phosphorylated by ATF4 (67). This transcription superfamily is responsible for the expression of many stress-response genes (66–68). Recently, the connection between ATF4 and autophagy induction was reported. When cells suffer ER stress, LC3B will be cleaved to form LC3-II, which promotes the elongation of the autophagic membrane (69). After being induced by ER stress, mTORC1 activity is repressed by TSC1/2 (70).
ROS and autophagy
ROS are reative oxygen species (71), which are mainly generated from the oxidative respiratory chain. ROS include oxygen anions, free radicals, and peroxides (72). ROS accumulation can oxidize cell components, including proteins and lipids, which can cause damage to the DNA and organelles, and can even lead to cell death (73). ROS accumulation will trigger oxidative stress involving many biological processes, such as apoptosis, necrosis and autophagy (74). Thus, autophagy will be induced by the accumulation of ROS and lipid peroxidation. One study revealed that ROS accumulation is an essential element of autophagy induced by nutrient deficiency or rapamycin treatment (75). ROS derived from mitochondria can oxidize lipids, then destroy the mitochondrial membrane structure, which will ultimately lead to cell death (76). Studies have also reported that the induction of autophagy in TNFα-treated cells via the NF-κB pathway requires the accumulation of ROS. Furthermore, both TNFα and ROS can promote Beclin 1 expression (77). ATGs up-regulation is associated with P38 and P53 activation induced by ROS (Figure 2L) (78). Conversely, p38 mitogen-activated protein kinases are also involved in the generation of ROS (79). Studies have shown that the ROS concentration can be dramatically elevated by inhibited p53 targeted gene expression, and will then induce autophagy (78). ROS can also initiate autophagy by activating the GSK-3β pathway (80).
Autophagy in Lipid and Carbohydrate Metabolism
Autophagy is initiated by starvation or other stress signals. The autophagosome membrane engulfs proteins or other organelles, which are then degraded in a lysosome, to complete the nutrient recycling. In addition to the metabolism of circulating proteins, autophagy is involved in the metabolism of lipids and glycogen as substrates.
Lipophagy
Lipids are very important substrates for cellular biomembrane construction and are also used as energy fuel. The homeostasis of lipids is regulated by complicated pathways, including lipid synthesis and lipolysis. However, excessive release of FAs is toxic to cells. Cells can transform excess FAs into triacylglycerol (TAG), which is stored in adipose tissue in the form of lipid droplets (LDs). TAGs are primarily stored in adipocytes, which can be degraded into glycerol and FAs to supply energy in response to energy depletion. The enzymatic hydrolysis process that TAGs undergo is called lipolysis. Hormone-sensitive lipase and monoacylglycerol lipase are two important lipases involved in the triglyceride hydrolysis process (81). Mobilization of triglycerides in LDs is a precisely regulated process. Lipolysis is regulated by many nutrients and hormones such as fatty acids and epinephrine. When the fatty acids and energy supply is deficient, cytoplasmic lipase will be recruited to the surface of LDs to start the lipolysis (82). At the beginning of lipolysis, adipocyte triglyceride lipase is recruited to LDs and its activity is regulated by perilipins and comparative gene identification-58. Then lipase will hydrolyze TAGs to diacylglycerols and FAs (83). Next, hormone-sensitive lipase activates the degradation of diacylglycerols to monoacylglycerols (MAGs) and FAs. Finally, MAGs are converted to glycerol and FAs by MAG lipase and then undergo β-oxidation to generate energy and heat (83).
In addition to the lipolysis pathway, autophagy also occurs in LD degradation in lysosomes by lysosomal acid lipase. The process of autophagic LD degradation is called lipophagy. Notably, the cytoplasmic lipases are catalyzed at pH 7, for neutral lipolysis. However, lysosomal acid lipase works at pH 4.5, for acid lipolysis (81). Lipophagy has been regarded as another important TAG utilization pathway for rapid LD degradation. Lipophagy occurs when LDs are surrounded and engulfed by double-membrane phagophores and are fused with a lysosome. The lysosome lumen contains hydrolases and lipases that help catalyze the degradation of LDs (84). The early phase of lipophagy has been observed in mouse hepatocytes during starvation. By treating with 3-methyladenine, an autophagy inhibitor or silence, the autophagy gene ATG5 will restrain lipophagy by blocking the TAG breakdown pathway. When autophagy is repressed, the concentration of TAGs and the size of LDs in the hepatocytes increase (85). When lipophagy induces, LC3 will surround the LDs to recruit ATGs. As expansion of autophagosome, LDs will be engulfed then degraded in autolysosome (84, 85).
Glycophagy
Autophagy is a very effective and precisely controlled system for nutrient recycling and metabolic homeostasis. Glycogen can be recognized and engulfed by autophagosomes and then transferred to lysosomes to be degraded. This process is called “glycophagy.” When glucose depletion occurs in liver or muscle cells, both gluconeogenesis and the autophagy pathway will be activated to maintain normal energy recycling. In general, when newborns are delivered from the uterus, nutrient starvation will occur. At this period, stored glycogen will be broken down quickly to release glucose into the blood stream for the body to use. In this specific period, autophagy plays a vital role (86). Glycogen exists in the cytosol and is stored in vacuolar form in cells (87). When in an autophagosome, glycogen can be converted by the glycogen-hydrolyzing acid glucan 1,4-α-glucosidase to nonphosphorylated glucose (87). Many myopathies are associated with glycophagy defects. However, the precise relation between myopathies and glycophagy, as well as their basic mechanism, remains unclear. Scientists specialized in myopathy, use a Drosophilamelanogaster model to study glycogen autophagy in skeletal muscles. When exposed to chloroquine, autophagic myopathy occurs because of the repression of autophagy. Under starvation, glycogen is engulfed by autophagosomes, and this process can be suppressed via the TOR pathway (88). In the myopathy model, glycogen has been identified as a major substrate of autophagy in the larval muscle. The glycophagy can be totally blocked in D. melanogaster muscle by blocking the autophagy pathway, which proves the requirement for the autophagy machinery in glycophagy (88).
Nutritional Stress in Autophagy
Amino acid starvation
Cells can adapt to excess or deficiency of nutrients through many nutritional sensors and regulators. Amino acids, as the resources for protein synthesis, affect cell growth and survival. When amino acids are deficient in the cell surroundings, cellular protein synthesis and mitosis will cease immediately. More importantly, with a shortage in amino acids, the autophagic signaling pathway will be activated to release amino acids by degrading proteins in order to maintain the availability of amino acids pool for vital protein synthesis. Certain amino acids can regulate the activity of mTORC1. Leucine, arginine and glutamine are the major amino acids, all of which can directly activate mTORC1 (Figure 3). These amino acids have intense effects on mTORCl. Their withdrawal will strongly inhibit mTORC1, which will trigger the induction of autophagy. Moreover, leucine belongs to the BCAAs, and also play important role as nitrogen donor for AAs, such as Glu, and Gln, which may function to regulate PI3K-AKT-mTOR pathway (89).
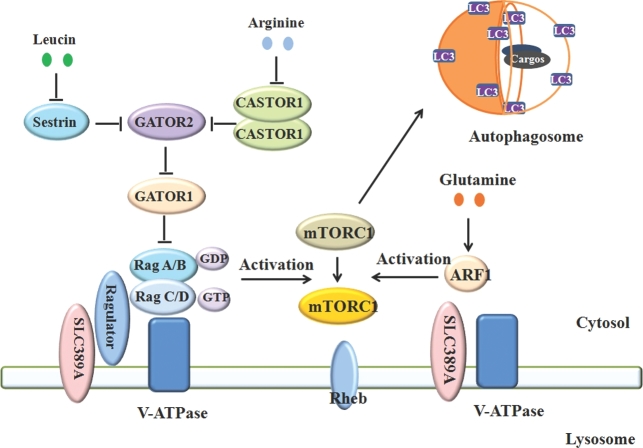
FIGURE 3.
Amino acid signals mediate the mTORC1 pathway and autophagy. Amino acids control the metabolic status of cells via the mTORC1 pathway. Withdrawal of amino acids will eliminate mTORC1 activity, which will trigger autophagy. Arginine, leucine, and glutamine are 3 amino acids that strongly regulate mTORC1 and autophagy through different pathways. Arginine can bind to CASTOR1 homodimer, which will cause CASTOR1 to be released from the GATOR2 complex to activate the GATOR2 complex. The activated GATOR2 will inhibit the GATOR1 complex, which will activate mTORC1. Leucine activates mTORC1 by activating the GATOR2 complex via inhibition of Sestrin associated with GATOR2. Glutamine has a different way to activate mTORC1, via ARF1 other than GATOR2/Rag GTPase and Ragulator. ARF1, adenosine diphosphate ribosylation factor-1 GTPase; CASTOR1, cellular arginine sensor for mTORC1; GATOR, GTPase activating proteins toward Rags; LC3, microtubule-associated protein 1A/1B-light chain 3; mTORC1, mammalian target of rapamycin complex 1; Rheb, Ras homolog enriched in brain; SLC, solute carrier; V-ATPase, vacuolar type H+-ATPase.
Arginine controls mTORC1 activity, which contributes to the control of cell growth and autophagy activity. Arginine has been reported to improve protein synthesis and cell growth of brown adipocyte via the activation of the mTORC1 (90). However, until last year, the mechanism by which arginine controls the mTORC1 pathway had yet to be identified (Figure 3). A cellular arginine sensor called cellular arginine sensor for mTORC1 (CASTOR1), has been found that can directly bind to arginine to activate the mTORC1 pathway. Under normal conditions, CASTOR1 can form a homodimer and a heterodimer with CASTOR2, both of which interact with the upstream activator of mTORC1 [GTPase activating proteins toward Rags 2 (GATOR2)] complex to inhibit the function of mTORC1. When arginine is enriched in the cell, the arginine can directly bind the two CASTOR1 complexes, which leads to the release of the CASTOR1 complexes from the GATOR2 complex, which then activates the mTORC1 (91). The crystal structure of an arginine-CASTOR1 binding model has been dissected. This crystal structure illustrates that the homodimer of CASTOR1 binds to arginine at the interface of its two ACT domains, and allosterically controls the adjacent GATOR2 binding site to trigger the dissociation from GATOR2 and activate the pathway of mTORC1 (92). In 2015, SLC38A9, a lysosomal transmembrane protein, was identified as amino acid transporter, especially arginine. The study revealed that SLC38A9 can transport arginine with a high Michaelis constant. A deficiency of this transporter inhibits the mTORC1 activity that is activated by arginine (93). These two arginine-activated mTORC1 models supply the arginine signaling network in autophagy control.
Leucine, a brand-chain amino acid, can activate mTORC1. Coincidently, the leucine binding protein Sestrin2 has also been identified as a GATOR2 complex inhibitor (Figure 3). Leucine can bind to Sestrin2 to dissociate the Sestrin2 from GATOR2, thereby activating GATOR2 and mTORC1. A mutation at the sestrin2 leucine binding site prevents sestrin2 from binding to leucine, which persistently inhibits mTORC1 activity (94). To better understand the dynamic structure of sestrin2-leucine, the authors dissected the crystal structure of leucine-Sestrin2. The structure shows that one Sestrin2 protein contains two structurally homologous domains (termed N-terminal and C-terminal, respectively). The leucine-binding pocket is localized in the C-terminal domain. The two domains are necessary for the binding of Sestrin2 to leucine and GATOR2 (95). Besides the leucine-GATOR2 model, another possible mechanism is Sestrin2 phosphorylation, which has been reported to be enhanced by leucine depletion. In contrast, leucine exposure immediately dephosphorylates Sestrin2, which activates mTORC1. Three Sestrin2 phosphorylation sites were identified by mass spectrometry (96). However, the mechanism of leucine mediated Sestrin2 phosphorylation and the upstream kinase still needs elucidating.
Glutamine is a nonessential amino acid that is able to activate mTORC1. Exposure to glutamine alone can activate mTORC1 after total amino acid withdrawal. mTORC1 activation is mediated by amino acids via the activated Rag GTPases, the Ragulator complex, and vacuolar H+-ATPase. Unlike leucine, the activation of mTORC1 by glutamine is in a Rag-independent manner. Moreover, glutamine can promote mTORC1 translocation to the lysosome membrane. This process depends on vacuolar H+-ATPase other than in the Ragulator complex. Furthermore, adenosine diphosphate ribosylation factor-1 GTPase is found to participate in glutamine-mediated mTORC1 translocation and activation (97) (Figure 3). Additionally, glutamine is required for extracellular leucine to activate mTORC1 (98). The balance of the transformation between glutamine and leucine synthesis promotes mTORC1 activation. Conversely, inhibition of glutaminolysis can activate mTOR. Furthermore, glutaminolysis has been demonstrated to regulate autophagy and cell size through mTORC1 (99).
Energy deficiency-induced autophagy
Glucose/glycogen
When cells suffer glucose starvation, AMPK will be activated immediately to prevent ATP consumption and increase glucose intake to maintain energy homeostasis. At the same time, the autophagic pathway is activated to recycle nutrients. When AMP/ATP or ADP/ATP is elevated, AMPK is activated and directly binds AMP or ADP molecules.
High concentrations of glycogen prevent the activation of AMPK even under the stimulation of muscle contraction or treatment of 5-aminoimidazole-4-carboxyamide-1-β-d-ribofuranoside (AICAR), an AMPK activator. Glycogen can inhibit AMPK activity by binding to the β subunit carbohydrate-binding module (CBM) directly. This finding implies that AMPK can directly bind to ATP/ADP/AMP and glycogen to regulate energy metabolism (89). Studies have shown that oligosaccharides with single branch points have more strongly inhibitory effects on AMPK than linear-linked oligosaccharides. The branched oligosaccharides can even inhibit the phosphorylation of AMPK by liver kinase B1 and calmodulin-dependent kinase-β. Interestingly, the CBM of the β2 subunit has a higher affinity for oligosaccharides than the β1 subunit (87). A few years ago, the crystal structure of glycogen-AMPK was dissected. One study found that AMPK is composed of an α subunit with an N-terminal kinase domain (KD) and a regulatory C-terminus, a glycogen-binding β subunit, and an AMP/ADP/ATP-binding γ subunit. Although ATP/ADP/AMP controls the kinase activity by modulating the interaction of adjacent autoinhibitory domain with the region of the KD, glycogen binding and CBM phosphorylation regulate the interaction between the CBM and the KD to inhibit AMPK activity (100).
Lipids and autophagy
Lipids are essential substances for cells. They not only supply energy as storage fat, but also compose the biomembrane structure. Lipid oxidation occurs with the participation of ROS, which contain carbon-carbon double bonds, especially PUFAs (101). Cellular lipids, such as glycolipids, phospholipids, and cholesterol, are vulnerable to oxidation by ROS or oxidative enzymes. To maintain the lipid oxidation status, cells can defend against these toxic substrates via antioxidant enzyme systems or antioxidant substances. However, if the extent of lipid oxidation is too overwhelming to correct, organelles will be damaged, and this can strongly induce autophagy even cell death (102).
4-Hydroxy-2-nonenal (HNE), the main product of omega-6 FA lipid peroxidation, has been revealed to activate the autophagy pathway. In rat aortic smooth muscle cells, the exposure of HNE activates autophagy. HNE can increase the modification of ER-associated proteins, activating ER stress through the PERK pathway. HNE can also promote autophagy induction by increasing LC3-II formation in a c-jun N-terminal kinase (JNK)-dependent manner. Inhibition of ER stress by JNK inhibitor can lead to cell death in HNE-treated cells (103).
As a saturated FA, palmitate (PA) can give rise to steatosis. Recent studies have revealed that exposure to PA can directly induce autophagy by increasing LC3 and P62 flux as well as the number of autophagosomes (104, 105). Inhibition of autophagy flux could convert hepatocytes treated by PA to apoptosis. By contrast, induction of autophagy by rapamycin can effectively inhibit apoptosis, which implies that autophagy may be induced to prevent apoptotic cell death. PA induces autophagy in hepatocytes without activating mTORC1 and ER stress but enhancing the protein kinase C pathway (106). Another study, treating H9c2 rat embryonic cardiac myoblasts with PA, revealed that autophagy was induced in these cells by detecting the autophagy flux using an electron microscope. However, ER stress is activated by the activation of PERK (104).
Excess nutrients and autophagy
Autophagy as a nutrient recycling system can be induced by amino acid and energy starvation stress via mTORC1 and AMPK, respectively. Notably, autophagy can also be induced in an excess nutrient condition.
Glucose starvation can induce autophagy by activating AMPK. However, a high concentration of glucose induces autophagy via another pathway. One study showed that high glucose (30 mM) can promote autophagy in podocytes, where numerous autophagosomes are accumulated. High glucose can also elevate ROS production in a time-dependent manner (107). Another study reported that high glucose can induce autophagy as well as ROS production in the retinal pigment epithelium cells. Therefore, autophagy induced by high glucose possibly occurs via the ROS pathway. Interestingly, high glucose treatment can activate mTORC1. However, inhibition of mTORC1 activity by rapamycin fails to affect autophagy induced by high glucose, indicating that autophagy mediated by high glucose is independent of mTORC1. In addition, autophagy induction associated with high glucose may be mediated via phosphorylated JNK (108). Yao et al. also discovered that high glucose can robustly increase the amount of phosphorylation of eIF2α or PERK, which serves as an ER stress marker. However, the silence of eIF2α caused by small interfering RNA can eliminate autophagic flux, indicating that high glucose-induced autophagy is mainly regulated through ER stress signaling.
Overwhelming Nutritional Stress and Autosis
When cells face various stresses, such as nutrient withdrawal or oxidative stress, autophagy will be induced to protect these cells against damage. Autophagy is a conservative cellular mechanism and is regarded as a protective measure. Studies have found that autophagy can also induce cell death, which has been termed “autosis.” If cellular stresses are beyond control, overwhelming induction of autophagy will lead to autosis. Historically, programming cell death has been classified into three kinds: apoptosis, autosis, and necrosis (109). In 2013, a study authenticated the presence of autophagic cell death and named it autosis. Cells with autosis contain large numbers of autolysosomes in their cytoplasm. This distinguishes autosis from both apoptosis and necrosis, and can also be induced by starvation and autophagy-inducing peptides (110). Generally, autosis has been characterized as having three important features. The first feature of autosis has been identified as cellular matrix adherence and the presence of RE debris. The second is that induction may be caused by Tat-Beclin1 peptide, nutrient withdrawal, or hypoxia-ischemia. The last feature is that the abolition of Na+-K+-ATPase activity can inhibit the occurrence of autosis (111). Studies have revealed that the depletion of ATG5 prevents cell death induced by interferon-γ. Consistently, the overexpression of ATG5 triggers cell death (112). In addition, in cancer cell lines, the depletion of ATGs can significantly block cell death induced by ROS (113). Moreover, in apoptotic cell death induction, inhibiting the activity of caspase can induce autophagic cell death without the occurrence of apoptosis (114).
Autosis has been found to exist in a Drosophila model faced the ischemia (115). Apoptosis occurs when cells is exposed to apoptosis inducing drugs. However, treatment with inhibitor of apoptosis after apoptosis induction will trigger the autosis (116). Studies have found that autosis can be blocked by the depletion of ATGs or the autophagy blocker 3-methyladenine. The cell death feature induced by Tat-beclin1 peptide differs from apoptosis and necrosis (110). More importantly, a type of cell death caused by nutrient deficiency has a similar phenotype to Tat-Beclin1 treatment. In clinical patients, cell death caused by permanent brain ischemia appears to show an autotic feature (117). In patients with severe anorexia nersova and liver damage, their hepatocyte morphology displays typical autosis. One study has found apathology similar to autosis in the liver of a patient (118). A high-output drug screen experiment has screened out an autosis target gene–the gene coding for Na+-K+-ATPase–which consumes a large amount of cellular ATP and contributes to autosis (Figure 1). In addition, autosis can be blocked by cardiac glycosides, which block Na+-K+-ATPase. In clinical treatment, cardiac glycosides can cure the brain damage caused by ischemia, as shown in a neonatal rat model (119).
Autosis-Related Diseases
Autosis occurs when autophagy induction is too strong to be controlled. Numerous autophagosomes accumulating in the cytoplasm are characteristic of autosis, which results in cell death. However, the symptom linking autosis to human disease has rarely been reported.
In 2008, a study found that four patients were suffering from acute liver insufficiency associated with severe anorexia nersova. However, no significant necrosis or apoptotic cells were found in their hepatocytes. In contrast, the accumulation of numerous autophagosomes was found in the hepatocytes, which seems to be an autosis-related symptom. In those patients, features of phase 1a and 1b autosis were apparent. The characteristics of phase 1a autosis include convoluted nucleus, moderately condensed chromatin, electron-dense mitochondria, dilated ER, and the presence numerous autophagosomes, autolysosomes, and empty vacuoles. The definition of Phase 1b is that the perinuclear space of hepatocytes is swollen in discrete regions surrounding the inner nuclear membrane. These swollen areas contain membrane-binding regions with a similar density and granularity to the cytosol. However, the features of phase 2 autosis were not observed. In addition, a significant increase of serum transaminases was observed in these patients (118).
In neonatal ischemic brain damage, many types of cell death occur. However, neurons in the CA3 region of the hippocampus display an early autophagic feature in the absence of apoptosis or necrosis. Prevention of autophagy by a pharmacologic inhibitor, or the knocking down of essential genes involved in autophagy, can significantly protect against ischemia-induced neuronal death. This result indicates that autophagy mainly contributes to this kind of cell death. However, autosis has not yet been found in the CA3 region of the hippocampus in humans (111).
Conclusions
The homeostasis of nutritional metabolism is vital for the maintenance of cell survival and normal physiologic functions. mTORC1 and AMPK have been well documented as sensors of primary cellular amino acids and energy status, respectively. When the amino acid pool is absent, autophagy will be induced by mTORC1 inactivation. AMPK serving as an energy status sensor can directly discriminate the concentration of AMP/ATP and ADP/ATP, resulting in its activation to initiate autophagy. By contrast, nutrient excess will also induce autophagy, mainly via ROS or ER stress. In most cases, autophagy protects cells against severe environmental stresses through recycling limited nutrients or degrading damaged organelles. This process is an efficient way to maintain the homeostasis of cell physiology. However, when nutritional stress is beyond control, the activation of overwhelming autophagy will also induce cell death via autosis. Retaining the proper nutrition level and autophagy status is critical for cell survival under conditions of nutritional stress.
Acknowledgments
The authors’ responsibilities were as follows—XM: was the main designer of the review; LH: was the main writer of the manuscript; J Zhang, NM, J Zhao, SWK, and XM: edited the manuscript; and all authors: read and approved the final manuscript.
Notes
This work was supported by the National Key R&D Program of China (2017YFD0500501), the National Natural Science Foundation of China (31722054, 31472101, and 31528018), the College of Animal Science and Technology “Young Talents Cultivation Program” in China Agricultural University (2017DKA001), the 111 Project (B16044), the National Department Public Benefit Research Foundation (201403047), and the Developmental Fund for Animal Science of Shenzhen Jinxinnong Feed Co., Ltd.
Author disclosures: LH, JZ, JZ, SWK and XM, no conflicts of interest.
Abbreviations used:
- AMBRA1
autophagy and beclin regulator 1
- AMPK
AMP-activated protein kinase
- ATF
activating transcription factor
- ATG
autophagy-related gene
- CASTOR1
cellular arginine sensor for mammalian target of rapamycin complex 1
- CBM
carbohydrate-binding module
- Deptor
DEP-domain-containing mTOR-interacting protein
- eIF2α
eukaryotic translation initiation factor 2α
- ER
endoplasmic reticulum
- GATOR
GTPase activating proteins toward Rags
- HNE
4-hydroxy-2-nonenal
- JNK
c-jun N-terminal kinase
- KD
kinase domain
- LC3
microtubule-associated protein 1A/1B-light chain 3
- LD
lipid droplet
- MAG
monoacylglycerol
- mTOR
mammalian target of rapamycin
- mTORC1(2)
mammalian target of rapamycin complex 1(2)
- PA
palmitate
- PAS
pre-autophagosomal structure
- PERK
protein kinase R-like kinase
- PRAS40
proline-rich Akt substrate of 40 kDa
- Rheb
Ras homolog enriched in brain
- ROS
reactive oxygen species
- SNAP-29
soluble NSF attachment protein 29
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- STX17
syntaxin 17
- TAG
triacylglycerol
- ULK1
Unc-51 like autophagy activating kinase 1
- VAMP
vesicle-associated membrane protein
- VPS34
vacuolar protein sorting 34
References
- 1. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. Mammalian autophagy: how does it work? Annu Rev Biochem 2016;85:685–713. [DOI] [PubMed] [Google Scholar]
- 3. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013;14:133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 2011;25:1895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J, Guo R, Yan H, Tian L, You Q, Li S, Wu K. Naringin inhibits ROS-activated MAPK pathway in high glucose-induced injuries in H9c2 cardiac cells. Basic Clin Pharmacol Toxicol 2014;114: 293–304. [DOI] [PubMed] [Google Scholar]
- 6. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032–6. [DOI] [PubMed] [Google Scholar]
- 7. Liu Y, Shoji-Kawata S, Sumpter RM Jr, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, et al. . Autosis is a Na+, K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A 2013;110:20364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010;12: 747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamamoto A, Masaki R, Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J Histochem Cytochem 1990;38:573–80. [DOI] [PubMed] [Google Scholar]
- 10. Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010, 12(9): 831–5. [DOI] [PubMed] [Google Scholar]
- 11. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007;9:1102–9. [DOI] [PubMed] [Google Scholar]
- 12. Noda T, Kim J, Huang W, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000;148:465–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005;1:101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reggiori F1, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004;6:79–90. [DOI] [PubMed] [Google Scholar]
- 15. Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomycescerevisiae. Gene 1997;192:245–50. [DOI] [PubMed] [Google Scholar]
- 16. Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000;151:263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 2012;31:4304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001;152:657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 2001;153:381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takáts S, Nagy P, Varga Á, Pircs K, Kárpáti M, Varga K, Kovács AL, Hegedűs K, Juhász G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 2013;201:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Atlashkin V, Kreykenbohm V, Eskelinen EL, Wenzel D, Fayyazi A, von Mollard GF. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol 2003;23:5198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 2009;1793: 1901–16. [DOI] [PubMed] [Google Scholar]
- 24. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012;151:1256–69. [DOI] [PubMed] [Google Scholar]
- 25. Ma X, Zhang S, He L, Rong Y, Brier LW, Sun Q, Liu R, Fan W, Chen S, Yue Z, et al. . MTORC1-mediated NRBF2 phosphorylation functions as a switch for the class III PtdIns3K and autophagy. Autophagy 2017;13:592–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Razi M, Chan E, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol 2009;185:305–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JA, Beigneux A, Ahmad ST, Young SG, Gao F. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 2007;17:1561–7. [DOI] [PubMed] [Google Scholar]
- 28. Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol 2009;21:543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell 2002;13:3355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhan L, Chen S, Li K, Liang D, Zhu X, Liu L, Lu Z, Sun W, Xu E. Autophagosome maturation mediated by Rab7 contributes to neuroprotection of hypoxic preconditioning against global cerebral ischemia in rats. Cell Death Dis 2017;8:e2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. . Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008;10: 776–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. . Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009;11:385–96. [DOI] [PubMed] [Google Scholar]
- 33. Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 2012;45:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng X, Liang Y, He Q, Yao R, Bao W, Bao L, Wang Z. Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids. Int J Mol Cell Med 2014;15:20753–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002;110:163–75. [DOI] [PubMed] [Google Scholar]
- 36. Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. . The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem 2007;282:20329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009;122:3589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 2005;19:15–26. [DOI] [PubMed] [Google Scholar]
- 39. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008;10:935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev 2011;25:1668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 1977;270:174–6. [DOI] [PubMed] [Google Scholar]
- 44. He L, Eslamfamc S, Ma X, Li D. Autophagy and the nutritional signaling pathway. Front Agr Sci Eng 2016;3:222–30. [Google Scholar]
- 45. Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000;150:1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004;306:990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009;20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008;181:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009;11:1433–7. [DOI] [PubMed] [Google Scholar]
- 51. Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. . Ambra1 regulates autophagy and development of the nervous system. Nature 2007;447:1121–5. [DOI] [PubMed] [Google Scholar]
- 52. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. . mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013;15:406–16. [DOI] [PubMed] [Google Scholar]
- 53. Hardie DG, Schaffer BE, Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol 2016;26:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liang J, Shao S, Xu Z, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. . The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 2007;9:218–24. [DOI] [PubMed] [Google Scholar]
- 55. Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013;152:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 2010;107:1470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008;105:3374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chang C, Su H, Zhang D, Wang Y, Shen Q, Liu B, Huang R, Zhou T, Peng C, Wong C, et al. . AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol Cell 2015;60:930–40. [DOI] [PubMed] [Google Scholar]
- 59. Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–29. [DOI] [PubMed] [Google Scholar]
- 60. Gao W, Ding W, Stolz DB, Yin X. Induction of macroautophagy by exogenously introduced calcium. Autophagy 2008;4:754–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Filippi-Chiela EC, Viegas MS, Thomé MP, Buffon A, Wink MR, Lenz G. Modulation of autophagy by calcium signalosome in human disease. Mol Pharmacol 2016;90:371–84. [DOI] [PubMed] [Google Scholar]
- 62. Decuypere JP, Kindt D, Luyten T, Welkenhuyzen K, Missiaen L, De Smedt H, Bultynck G, Parys JB. mTOR-controlled autophagy requires intracellular Ca(2+) signaling. PLoS One 2013;8:e61020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 2010;40:238–52. [DOI] [PubMed] [Google Scholar]
- 64. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. . The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010;120:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deng J, Lu P, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, Ron D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol 2004;24:10161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wek RC, Jiang H, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 2006;34:7–11. [DOI] [PubMed] [Google Scholar]
- 67. Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol 2008;40:14–21. [DOI] [PubMed] [Google Scholar]
- 68. Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099–108. [DOI] [PubMed] [Google Scholar]
- 69. B'chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013;41:7683–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 2010;6:239–47. [DOI] [PubMed] [Google Scholar]
- 71. Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 2007;17:422–7. [DOI] [PubMed] [Google Scholar]
- 72. Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res 2003;531:37–80. [DOI] [PubMed] [Google Scholar]
- 73. Farré JC, Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol 2004;14:515–23. [DOI] [PubMed] [Google Scholar]
- 74. He L, He T, Farrar S, Ji L, Liu T, Ma X. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell Physiol Biochem 2017;44:532–553. [DOI] [PubMed] [Google Scholar]
- 75. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007;26:1749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kirkland RA, Adibhatla RM, Hatcher JF, Franklin JL. Loss of cardiolipin and mitochondria during programmed neuronal death: evidence of a role for lipid peroxidation and autophagy. Neuroscience 2002;115:587–602. [DOI] [PubMed] [Google Scholar]
- 77. Kissová I, Deffieu M, Samokhvalov V, Velours G, Bessoule JJ, Manon S, Camougrand N. Lipid oxidation and autophagy in yeast. Free Radic Biol Med 2006;41:1655–61. [DOI] [PubMed] [Google Scholar]
- 78. McClung JM, Judge AR, Powers SK, Yan Z. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol 2010;298:C542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med 2008;44:1529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang S, Shih YL, Kuo TC, Ko WC, Shih CM. Cadmium toxicity toward autophagy through ROS-activated GSK-3beta in mesangial cells. Toxicol Sci 2009;108:124–31. [DOI] [PubMed] [Google Scholar]
- 81. Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol 2017. [DOI] [PubMed] [Google Scholar]
- 82. Wang H, Bell M, Sreenevasan U, Hu H, Liu J, Dalen K, Gong D. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem 2011;286:15707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang C. Lipid droplets, lipophagy, and beyond. Biochim Biophys Acta 2016;1861:793–805. [DOI] [PubMed] [Google Scholar]
- 84. Iancu TC, Manov I, Shaoul R, Haimi M, Lerner A. What's in a name?—“lipolysosome”: ultrastructural features of a lipid-containing organelle. Ultrastruct Pathol 2013;37:293–303. [DOI] [PubMed] [Google Scholar]
- 85. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009;458:1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kalamidas SA, Kotoulas OB, Kotoulas AO, Maintas DB. The breakdown of glycogen in the lysosomes of newborn rat hepatocytes: the effects of glucose, cyclic 3′, 5′-AMP and caffeine. Histol Histopathol 1994;9:691–8. [PubMed] [Google Scholar]
- 87. Ha J, Guan K, Kim J. AMPK and autophagy in glucose/glycogen metabolism. Mol Aspects Med 2015;46:46–62. [DOI] [PubMed] [Google Scholar]
- 88. Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 2013;11:e1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nie C, He T, Zhang W, Zhang G, Ma X. Branched chain amino acids: Beyond nutrition metabolism. Int J Mol Sci 2018;19:E954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ma X, Han M, Li D, Hu S, Gilbreath KR, Bazer FW, Wu G. L-Arginine promotes protein synthesis and cell growth in brown adipocyte precursor cells via the mTOR signal pathway. Amino Acids 2017;49:957–964. [DOI] [PubMed] [Google Scholar]
- 91. Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Sabatini DM. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 2016;165:153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 2016;536: 229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Wang T. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015;347:188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016;351:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Sabatini DM. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016;351:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kimball SR, Gordon BS, Moyer JE, Dennis MD, Jefferson LS. Leucine modulates mTORC1 signaling by acting specifically to alter the phosphorylation status of sestrin2. FASEB J 2016;30:1244–3. [Google Scholar]
- 97. Jewell JL, Kim YC, Russell RC, Yu F, Park HW, Plouffe SW, Guan K. Differential regulation of mTORC1 by leucine and glutamine. Science 2015;347:194–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. . Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009;136:521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Durán RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 2012;47:349–58. [DOI] [PubMed] [Google Scholar]
- 100. Koay A, Woodcroft B, Petrie EJ, Yue H, Emanuelle S, Bieri M, Ralph S. AMPK β subunits display isoform specific affinities for carbohydrates. FEBS Lett 2010;584:3499–503. [DOI] [PubMed] [Google Scholar]
- 101. Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol 2015;4:193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yin H, Xu L, Porter NA. Free radical lipid peroxidation: mechanisms and analysis. Chem Rev 2011;111:5944–72. [DOI] [PubMed] [Google Scholar]
- 103. Haberzettl P, Hill BG. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol 2013;1:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Park M, Sabetski A, Kwan CY, Turdi S, Sweeney G. Palmitate induces ER stress and autophagy in H9c2 cells: implications for apoptosis and adiponectin resistance. J Cell Physiol 2015;230:630–9. [DOI] [PubMed] [Google Scholar]
- 105. Yin J, Gu L, Wang Y, Fan N, Ma Y, Peng Y. Rapamycin improves palmitate-induced ER stress/NF κ B pathways associated with stimulating autophagy in adipocytes. Mediators Inflamm 2015;2015:272313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cai N, Zhao X, Jing Y, Sun K, Jiao S, Chen X, Wei L. Autophagy protects against palmitate-induced apoptosis in hepatocytes. Cell Biosci 2014;4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ma T, Zhu J, Chen X, Zha D, Singhal PC, Ding G. High glucose induces autophagy in podocytes. Exp Cell Res 2013;319:779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yao J, Tao Z, Li C, Li X, Cao G, Jiang Q, Yan B. Regulation of autophagy by high glucose in human retinal pigment epithelium. Cell Physiol Biochem 2014;33:107–16. [DOI] [PubMed] [Google Scholar]
- 109. Yang L, Wu K, Chiu W, Wang S, Shih CM. The cadmium-induced death of mesangial cells results in nephrotoxicity. Autophagy 2009;5:571–2. [DOI] [PubMed] [Google Scholar]
- 110. Fitzwalter BE, Thorburn A. Recent insights into cell death and autophagy. FEBS J 2015;282:4279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ 2015;22:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005;122:927–39. [DOI] [PubMed] [Google Scholar]
- 113. Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ 2008;15:171–82. [DOI] [PubMed] [Google Scholar]
- 114. Lamy L, Ngo VN, Emre NC, Shaffer AL, Yang Y, Tian E, Nair V, Kruhlak MJ, Zingon A, Landgren O, et al. . Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013;23:435–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang X, Yan H, Yuan Y, Gao J, Shen Z, Cheng Y, Shen Y, Wang R, Wang X, Hu W, et al. . Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy 2013;9:1321–33. [DOI] [PubMed] [Google Scholar]
- 116. Munoz-Pinedo C, Martin SJ. Autosis: a new addition to the cell death tower of babel. Cell Death Dis 2014;5:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Descloux C, Ginet V, Clarke PG, Puyal J, Truttmann AC. Neuronal death after perinatal cerebral hypoxia-ischemia: focus on autophagy-mediated cell death. Int J Dev Neurosci 2015;45:75–85. [DOI] [PubMed] [Google Scholar]
- 118. Kheloufi M, Boulanger CM, Codogno P, Rautou PE. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology 2015;62:657–8. [DOI] [PubMed] [Google Scholar]
- 119. Wang J, Portbury S, Thomas MB, Barney S, Ricca DJ, Morris DL, Warner DS, Lo DC. Cardiac glycosides provide neuroprotection against ischemic stroke: discovery by a brain slice-based compound screening platform. Proc Natl Acad Sci U S A 2006;103: 10461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]