Acclimation of wheat to waterlogging is mediated by regulation of hormonal metabolism and transport in adventitious roots and during their emergence.
Keywords: Adventitious roots, aerenchyma, gene expression, plant hormones, root elongation/growth, stem node, waterlogging
Abstract
To gain insights into the molecular mechanisms underlying hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat, the present study investigated transcriptional regulation of genes related to hormone metabolism and transport in the root and stem node tissues. Waterlogging-induced inhibition of axile root elongation and lateral root formation, and promotion of surface adventitious and axile root emergence and aerenchyma formation are associated with enhanced expression levels of ethylene biosynthesis genes, ACS7 and ACO2, in both tissues. Inhibition of axile root elongation is also related to increased root indole acetic acid (IAA) and jasmonate (JA) levels that are associated with up-regulation of specific IAA biosynthesis/transport (TDC, YUC1, and PIN9) and JA metabolism (LOX8, AOS1, AOC1, and JAR1) genes, and transcriptional alteration of gibberellin (GA) metabolism genes (GA3ox2 and GA2ox8). Adventitious root emergence from waterlogged stem nodes is associated with increased levels of IAA and GA but decreased levels of cytokinin and abscisic acid (ABA), which are regulated through the expression of specific IAA biosynthesis/transport (TDC, YUC1, and PIN9), cytokinin metabolism (IPT5-2, LOG1, CKX5, and ZOG2), ABA biosynthesis (NCED1 and NCED2), and GA metabolism (GA3ox2 and GA2ox8) genes. These results enhance our understanding of the molecular mechanisms underlying the adaptive response of wheat to waterlogging.
Introduction
Wheat (Triticum aestivum L.) is one of the most economically important cereal crops in the world; however, its production is challenged by a variety of abiotic stresses including waterlogging, which is reported to cause yield loss in ~15–20% of the wheat crop annually (Herzog et al., 2016). Waterlogging causes significant reduction in gas diffusion and thereby creates low oxygen conditions in the root zone (Armstrong, 1979), and this reduces the synthesis of ATP and availability of carbohydrate resources for root growth and its functioning, leading to a substantial decrease in crop growth and yield (Herzog et al., 2016).
Plants can acclimate to waterlogging-induced low oxygen conditions in the soil through morphological, anatomical, and metabolic responses. The major morphological/anatomical responses of roots of dryland plant species to oxygen-deficient conditions include the development of a shallow root system involving short and thick roots, and formation of aerenchymatous adventitious roots (Bailey-Serres et al., 2012; Yamauchi et al., 2018). Low oxygen conditions also cause inhibition of root growth/elongation and reduction in the emergence and elongation of lateral roots in wheat (Yamauchi et al., 2014). Several studies have demonstrated that these root traits are controlled by plant hormones. For example, ethylene, auxin, and abscisic acid (ABA) are involved in the inhibition of primary root elongation in plant species such as rice and wheat under normal/non-stress conditions (Konishi et al., 2005; McSteen, 2010; Yu et al., 2016), while gibberellin (GA) is implicated in promoting root elongation (Komatsu and Konishi, 2005). Previous reports, however, have shown the requirement of ABA for maintenance of primary root growth under stress conditions such as drought/low water potential (Saab et al., 1990; Sharp et al., 1994). Other plant hormones such as cytokinin and jasmonates (JAs) are also reported to inhibit primary root growth (Stenlid, 1982; Liu et al., 2015). Previous studies have also implicated ethylene in inducing aerenchyma formation in the root of dryland plant species such as wheat and maize under oxygen-deficient conditions through enhancing cortical cell death (Jackson et al., 1985; Jackson and Armstrong, 1999; He et al., 1994, 1996; Yamauchi et al., 2014).
Adventitious roots that emerge from stem nodes in response to waterlogging often develop aerenchyma connected to the shoot, improving gas diffusivity along the root (Sauter, 2013; Steffens and Rasmussen, 2016). Several studies have shown that auxin plays a key role in adventitious root formation (Delarue et al., 1998; Falasca and Altamura, 2003; Vidoz et al., 2010). Adventitious roots emerge through the nodal epidermis, and this process is enhanced by ethylene through its action of inducing nodal epidermal cell death (Mergemann and Sauter, 2000). Ethylene-induced death of epidermal cells has been shown to be mediated by reactive oxygen species (ROS) (Steffens et al., 2012; Gonzali et al., 2015; Mittler, 2017). Furthermore, ethylene-induced death of cortical cells, which leads to aerenchyma formation, is reported to be mediated by ROS (Yamauchi et al., 2017). Other plant hormones such as cytokinin, JA, ABA, and GA are also implicated in regulating adventitious root formation (Verstraeten et al., 2014), highlighting the significance of finely regulated hormonal crosstalk. For example, auxin regulates adventitious root formation by modulating JA homeostasis through auxin-inducible Gretchen Hagen 3 (GH3)-like proteins (Gutierrez et al., 2012), while the role of ethylene in regulating adventitious root formation can also be associated with its regulation of auxin transport (Vidoz et al., 2010). Cytokinin is reported to act as an auxin antagonist in the formation of adventitious roots, probably through inducing Aux/IAA (auxin/indole acetic acid), and thereby repressing PIN proteins and auxin flow (Della Rovere et al., 2013). It has also been shown that GA promotes ethylene-induced adventitious root formation while its antagonist ABA represses ethylene-induced and GA-promoted emergence of adventitious roots (Steffens et al., 2006).
Despite these reports, the molecular mechanisms underlying the regulation of hormonal metabolic pathways in adventitious roots and during their emergence in response to waterlogging are poorly understood in dry land crop species, especially in the polyploid wheat for which genomic/genetic resources to undertake such studies have been limited. To this end, the present study integrated physiological, histological, molecular, and metabolomics approaches to investigate changes in root anatomy/morphology, expression patterns of hormonal metabolic genes, and the levels of the respective plant hormones in wheat root and stem node tissues exposed to waterlogging during the stem elongation growth phase, which is reported to be very sensitive to waterlogging-induced yield losses (Marti et al., 2015).
Materials and methods
Plant growth and waterlogging treatment
Plants of common wheat cv. Harvest were grown in 3 liter plastic pots (one plant per pot) containing a mixture of locally sourced top soil and sand (2:1, v:v) supplied with fertilizers at 22 °C/20 °C (day/night) under a 16 h/8 h photoperiod as described previously (Nguyen et al., 2016). The top soil comprised 39% sand, 17% silt, and 44% clay, while the sand soil comprised 98% sand, 1% silt, and 1% clay. Thirty-day-old plants were subjected to waterlogging by submerging each pot in a 6 liter pot filled with water in order to maintain the water level at ~2 cm above the soil surface, and more water was added to the bigger pot as needed to maintain the water level. Control plants were subjected to regular watering (0.5 liter per pot every other day). At 1, 7, 14, and 28 days after waterlogging (DAWL), each pot containing the waterlogged or control plant was completely immersed in a large (63 liter) plastic box filled with tap water to loosen and remove the soil from the pot. Roots were separated gently from the soil while still in the water (for a maximum of 10 min), followed by a final wash of roots with a gentle stream of tap water to clean any remaining soil particles (for a maximum of 5 min). Control and waterlogged basal stem nodes, located within the 2 cm of water above the soil surface and from which new adventitious roots (hereafter referred to as ‘surface adventitious roots’) emerged, were excised from the main stem and tillers. Stem nodes were collected from plants waterlogged for 7, 14, and 28 d and their respective controls as they were not detected in most of the tillers/stems at 1 DAWL. To determine gene expression and hormone levels, three independent biological replicates of the whole root (roots of an individual plant/replicate), which consists of a mixture of the various root types described above (hereafter referred to as ‘root’), and stem node (stem nodes of individual plant/replicate) tissues were collected from both control and waterlogged plants. Tissues were immediately fixed (root tissues), or frozen in liquid nitrogen (roots and stem nodes) and then stored at –80 °C until further use. Root and shoot dry weights, number of tillers, number and length of axile (primary and nodal) roots, number of branch/lateral roots (see Chochois et al., 2012 for a description of root types), and number of surface adventitious roots were determined manually using freshly harvested roots (roots of three individual plants per replicate, three replicates).
Identification of wheat hormone metabolic genes
The hormone metabolism genes analyzed in this study were selected from our preliminary GenChip analysis based on the differential expression (≥2-fold) of their respective probe sets between control and waterlogged roots at 14 and/or 28 DAWL. To annotate the probe sets, sequences of the hormone metabolic genes of Arabidopsis were first identified from The Arabidopsis Information Resource database and used as queries to search for their homologs in the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/) using an E- value of <10−10. The resulting rice gene sequences were then blast searched against the wheat 61 k microarray platform in the Plant Expression Database (PLEXdb; http://www.plexdb.org/) to identify the corresponding probe sets and wheat unigenes using an E-value of 10–20. Sequences of the wheat unigenes were then blast searched against the wheat genome data in Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/) to identify the respective full-length wheat genes and proteins, which were used as queries to search for orthologs in Arabidopsis and other cereal species (Supplementary Table S1 at JXB online). Genes/homologs were named based on their orthologs in Arabidopsis and/or other cereal species (Supplementary Table S1), and the naming of the cytokinin metabolism and IAA transport genes has taken into account previously published reports (Song et al., 2012; Talboys et al., 2014). Genes related to ROS production, and ABA and GA (GA3ox2 and GA3ox3) metabolism have been reported previously (see Supplementary Table S2 for the references). Primer information of the genes analyzed in this study is presented in Supplementary Table S2.
RNA extraction and quantitative (q) RT-PCR analysis
Total RNA was extracted from the root and the stem node tissues using Trizol (Invitrogen, Carlsbad, CA, USA), and the cDNA samples for qPCR assays were prepared as described previously (Nguyen et al., 2016). Relative transcript levels were determined using the Livak and Schmittgen (2001) method after normalization with 18SrRNA as a reference gene. Transcript levels were compared across gene homologs, tissues, and growth conditions using the average transcript levels of one of the homologs in control roots at 1 DAWL as a reference sample (see figure legends for the details). For accurate and reproducible comparisons of the expression levels among the gene homologs, qRT-PCR efficiency of each gene was determined using the Ct slope method as described previously (Yao et al., 2012), and amplification efficiencies within the 90–105% acceptable range were obtained for all the genes studied (Supplementary Table S2).
Anatomical analysis of aerenchyma in the root
Nodal root segments (at 4 cm from the root apex) from plants waterlogged for 28 d and the respective controls were collected directly in a fixative containing 2.5% glutaraldehyde and 1.6% paraformaldehyde in 0.05 M phosphate-buffered saline (PBS). The samples were subjected to a vacuum for 30 min at 25 mm of Hg and then stored overnight at 4 °C. The fixed tissues were then serially dehydrated in 50, 70, 95, and 100% ethanol for 90 min in each solution at 4 °C. The dehydrated tissues were transferred to a mixture of absolute ethanol:historesin (2:1, v/v) containing the activator (5 g in 50 ml) and incubated for 24 h followed by a transfer to a mixture of absolute ethanol:historesin (1:2, v/v) and incubation for 24 h, and finally to 100% historesin for 48 h in which a freshly prepared solution was used after the first 24 h. After embedding in glycol methacrylate, sections (4 μm) were prepared using a JB-4 Sorvall microtome (Thermo Scientific, Waltham, MA, USA) and then stained for 2 min in 0.05% (w/v) toluidine blue O in benzoate buffer at pH 4.4. After destaining and drying, the samples were examined with an Olympus CX41 bright field microscope (Olympus Life Science, Tokyo, Japan) at ×4. The images of the sections were captured using a Lumenera Infinity HD camera (Lumenera Corporation, Ottawa, ON, Canada).
Hormone measurement
Lyophilized root and stem node tissues harvested from control and waterlogged plants were homogenized with acetonitrile containing 1% (v/v) acetic acid and internal standards as described previously (Son et al., 2016; Izydorczyk et al., 2018). Extraction of acidic and basic hormones from the homogenates and subsequent analyses of their levels with LC-ESI-MS/MS (Agilent 1260-6430) were performed as described in Yoshimoto et al. (2009).
Statistical analysis
Statistically significant differences between control and waterlogged samples were tested by Student’s t-test at a probability of P<0.05.
Results
Root and shoot growth, anatomy, and morphology
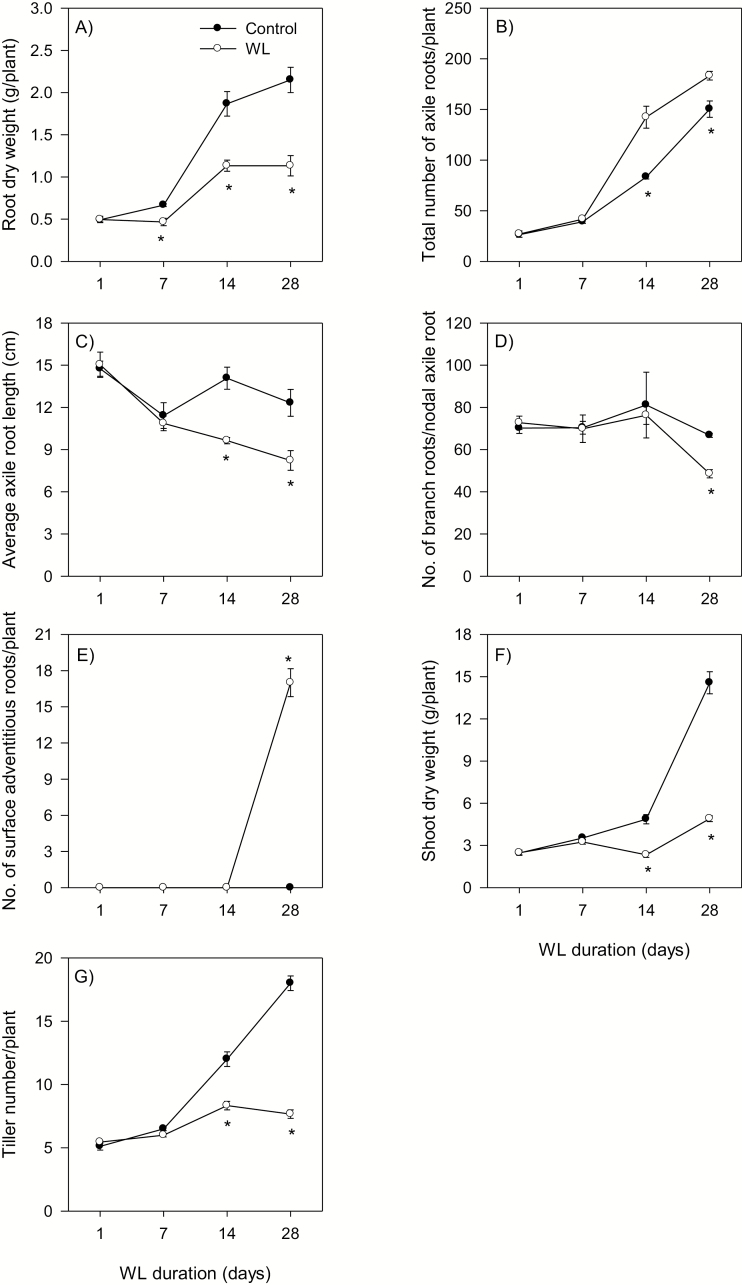
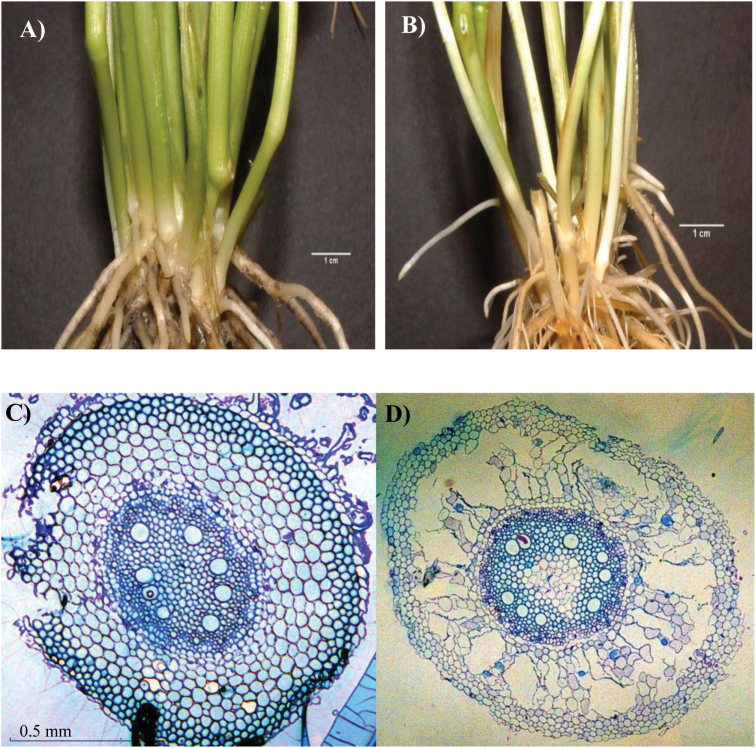
Waterlogging for 7 d caused a 30% reduction in root dry weight, and this effect was more pronounced as waterlogging continued for 14 d and 28 d (40–48%) (Fig. 1A). Waterlogging for 14 d and 28 d increased the total number of axile roots (primary and nodal) per plant but decreased their average length (Fig. 1B, C). Reduction in the number of branch roots per nodal axile root, as determined from the five longest nodal axile roots, was observed only after 28 d of waterlogging (Fig. 1D). Formation of surface adventitious roots (17 per plant) was also evident only after 28 d of waterlogging (Figs 1E, 2B), and this was accompanied by aerenchyma formation in the nodal axile roots (Fig. 2D). Waterlogging for 14 d and 28 d also decreased tiller number (1.5- to 2.3-fold) and shoot dry weight (52–66%) (Fig. 1F, G).
Fig. 1.
Root and shoot phenotype under control and waterlogged conditions. Root dry weight (A), total number of axile roots per plant (B), average length of axile root (C), number of branch roots per nodal axile root (D), number of surface adventitious roots per plant (E), shoot dry weight (F), and tiller number (G) of plants waterlogged for 1, 7, 14, and 28 d, and their respective controls. Data are means of three independent replicates (roots of three individual plants per replicate) ±SE. Asterisks denote a statistically significant difference between samples derived from control and waterlogged plants within each duration of waterlogging (P<0.05; Student t-test). WL, waterlogged.
Fig. 2.
Emergence of surface adventitious root and aerenchyma formation. Comparison of surface adventitious root emergence in control (A) and waterlogged (B) plants at 28 d after waterlogging. Lateral sections of control (C) and 28 d waterlogged (D) nodal root segments (4 cm from the root apex) at ×4 magnification. Aerenchyma formation is evident in the cortical cells of roots waterlogged for 28 d.
Transcriptional regulation of genes related to ethylene biosynthesis
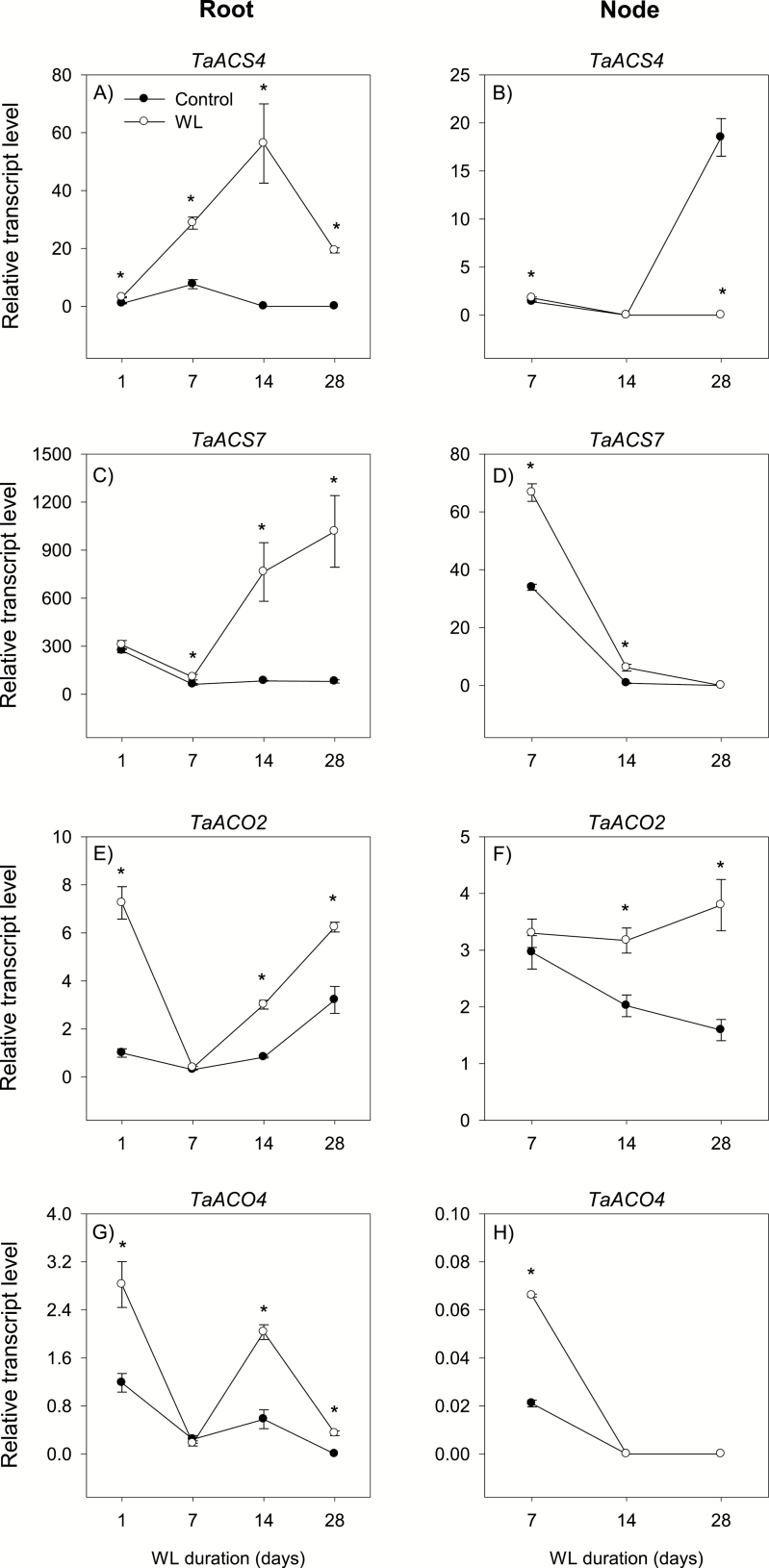
Ethylene is synthesized from S-adenosylmethionine, which is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by the action of ACC synthase (ACS) (Yang and Hoffman, 1984). This reaction represents the first committed and rate-limiting step of ethylene synthesis. Subsequently a reaction catalyzed by ACC oxidase (ACO) converts ACC to ethylene. Our study examined the expression patterns of ACS genes (ACS2, ACS4, and ACS7) and ACO genes (ACO2, ACO4, and ACO5) in both root and stem node tissues under control and waterlogged conditions. The expression levels of ACS4, ACS7, ACO2, and ACO4 were relatively higher than those of the other homologs in the root and/or stem node tissues under both growth conditions (Supplementary Table S3).
Root
The expression levels of ACS4 and ACS7 in the root decreased to low/below detectable levels during the course of the experiment under control conditions but increased in response to waterlogging for 1 d (ACS4; >3-fold) or 7 d (ACS7; >1.7-fold) (Fig. 3A, C). The expression levels of ACO2 and ACO4 were constantly very weak under control conditions, except for the transient increase of ACO2 expression at 28 DAWL (Fig. 3E, G). Waterlogging for 1, 14, and 28 d increased the expression levels of both genes (~2-fold or more). Although the expression of ACO5 was very weak in the root under control conditions, waterlogging for 14 d and 28 d enhanced its expression level (Supplementary Table S3). Under both growth conditions, the expression levels of ACS7 and ACO2 were higher than those of the other homologs.
Fig. 3.
Expression of ethylene biosynthesis genes. Relative transcript levels of ACS4 (A and B), ACS7 (C and D), ACO2 (E and F), and ACO4 (G and H) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, and G) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, H), and their respective controls. Transcript levels of the ACS and ACO genes in both tissues were determined using Ta18SrRNA as the reference gene and expressed relative to the transcript levels of ACS4 and ACO2 in control roots at 1 d after the start of waterlogging, respectively, which were arbitrarily set a value of 1. Data are means of three biological replicates ±SE. Asterisks denote a statistically significant difference in transcript levels between tissue samples derived from control and waterlogged plants within each duration of waterlogging (P<0.05; Student t-test). WL, waterlogging. ACS2 and ACO5 exhibited either very minimal or undetectable levels of expression irrespective of the tissue type or growth conditions, as shown in Supplementary Table S3.
Stem node
The expression of ACS4 and ACS7 in the stem node decreased below detectable levels by the end of the experiment under both control and waterlogged conditions, except for the marked increase of ACS4 expression in control stem nodes at 28 DAWL (Fig. 3B, D). Both genes, however, exhibited higher levels of expression in stem nodes waterlogged for 7 d and/or 14 d (ACS4, 1.3-fold; ACS7, ~2-fold or more) than the respective controls. Stem nodes waterlogged for 7 d and the corresponding controls exhibited a similar level of ACO2 expression, and this level was maintained with further waterlogging but decreased under control conditions (1.9-fold) (Fig. 3F). The expression level of ACO4 in the stem node was either very minimal or below the detectable level under both growth conditions (Fig. 3H). Overall, ACS7 and ACO2 exhibited the highest levels of expression in the stem node irrespective of growth conditions.
Transcriptional control of genes related to ROS production
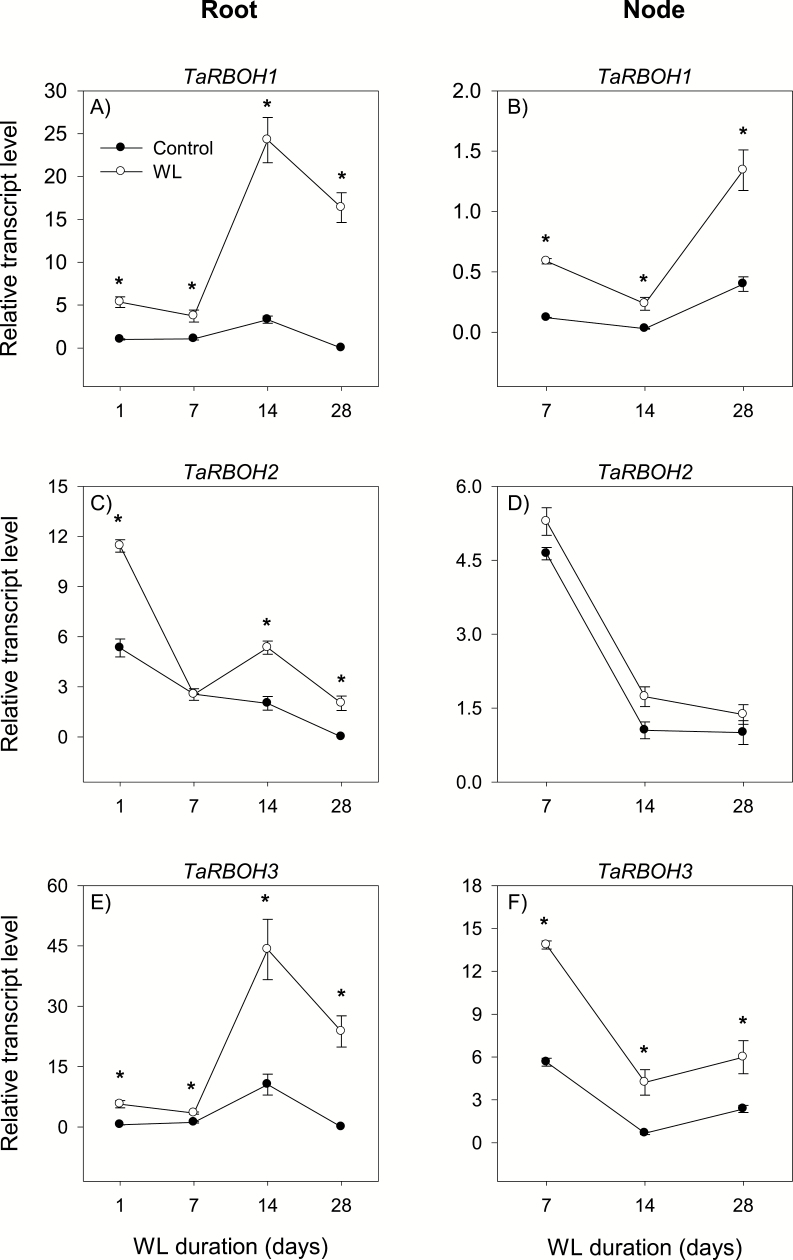
ROS are produced as by-products of basic cellular processes such as photosynthesis and respiration, and through the action of several enzymes. Respiratory burst oxidase homologs (RBOHs), also known as NADPH oxidases, have been reported to be key players in this regard (Suzuki et al., 2011). Our study examined the expression patterns of three RBOH genes (RBOH1, RBOH2, and RBOH3) in the root and stem node tissues under control and waterlogged conditions.
Root
The expression of all RBOH genes in the root decreased below detectable levels by the end of the experiment under control conditions despite the transient increases observed for RBOH1 and RBOH3 at 14 DAWL (Fig. 4A, C, E). Waterlogging, however, increased the expression levels of RBOH genes (from undetectable levels or >2-fold), with the major effects observed at 14 and 28 DAWL.
Fig. 4.
Expression of respiratory burst oxidase homolog genes. Relative transcript levels of RBOH1 (A and B), RBOH2 (C and D), and RBOH3 (E and F) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, and E) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, and F), and their respective controls. Transcript levels of each gene in both tissues were determined as described in Fig. 3, and are expressed relative to the transcript level of RBOH1 in control roots at 1 d after the start of waterlogging, which was arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3.
Stem node
The expression levels of all RBOH genes in the stem node either decreased to low levels or remained constantly low during the course of the experiment under control conditions (Fig. 4B, D, F). Waterlogging increased the expression levels of RBOH1 and RBOH3 (>2.5-fold) but did not affect the expression level of RBOH2.
Regulation of IAA metabolism and transport
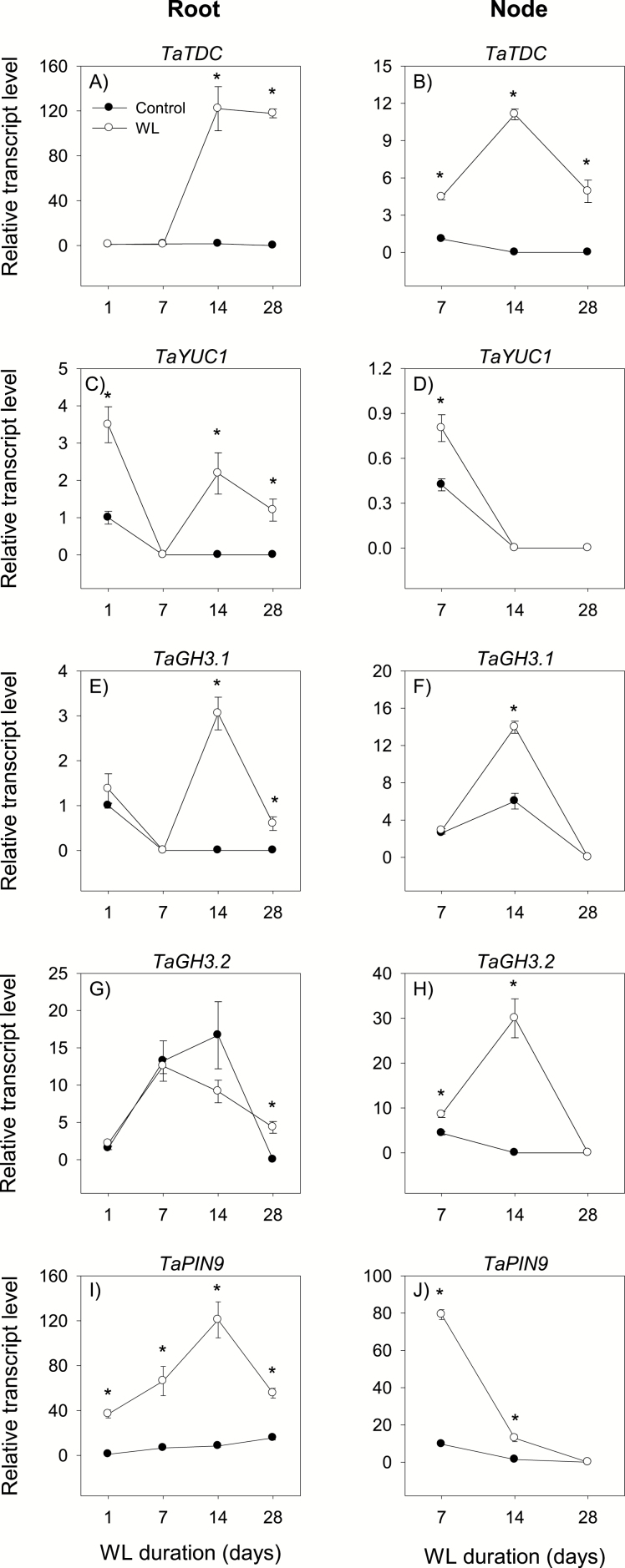
IAA is the major bioactive auxin in plants, and it is produced from tryptophan via indole-3-pyruvate by the actions of tryptophan aminotransferase (TAA) and YUCCAs (YUCs), respectively (Kasahara, 2016). Alternatively, tryptophan is proposed to be converted to IAA via tryptamine and indole-3-acetaldehyde by tryptophan decarboxylase (TDC) and aldehyde oxidase (AAO), respectively (Quittenden et al., 2009). IAA is inactivated mainly through its conjugation with amino acids by the action of GH3 proteins. The IAA level in plants is also regulated by its transport, which is mediated by the PIN FORMED (PIN) auxin efflux carriers that have a polar cellular distribution. This study examined the expression patterns of IAA biosynthesis (TDC, AAO, TAA1, YUC1, and YUC10), conjugation (GH3; GH3.1, and GH3.2), and transport (PIN; PIN5, and PIN9) genes in waterlogged root and stem node tissues and their respective controls. Our analysis revealed that the TDC, YUC1, GH3.1, GH3.2, and PIN9 are expressed in the root and/or stem node tissues under both control and waterlogged conditions, while no transcript of TAA1, AAO, YUC10, and PIN5 was detected irrespective of tissue type or growth conditions (Supplementary Table S3).
Root
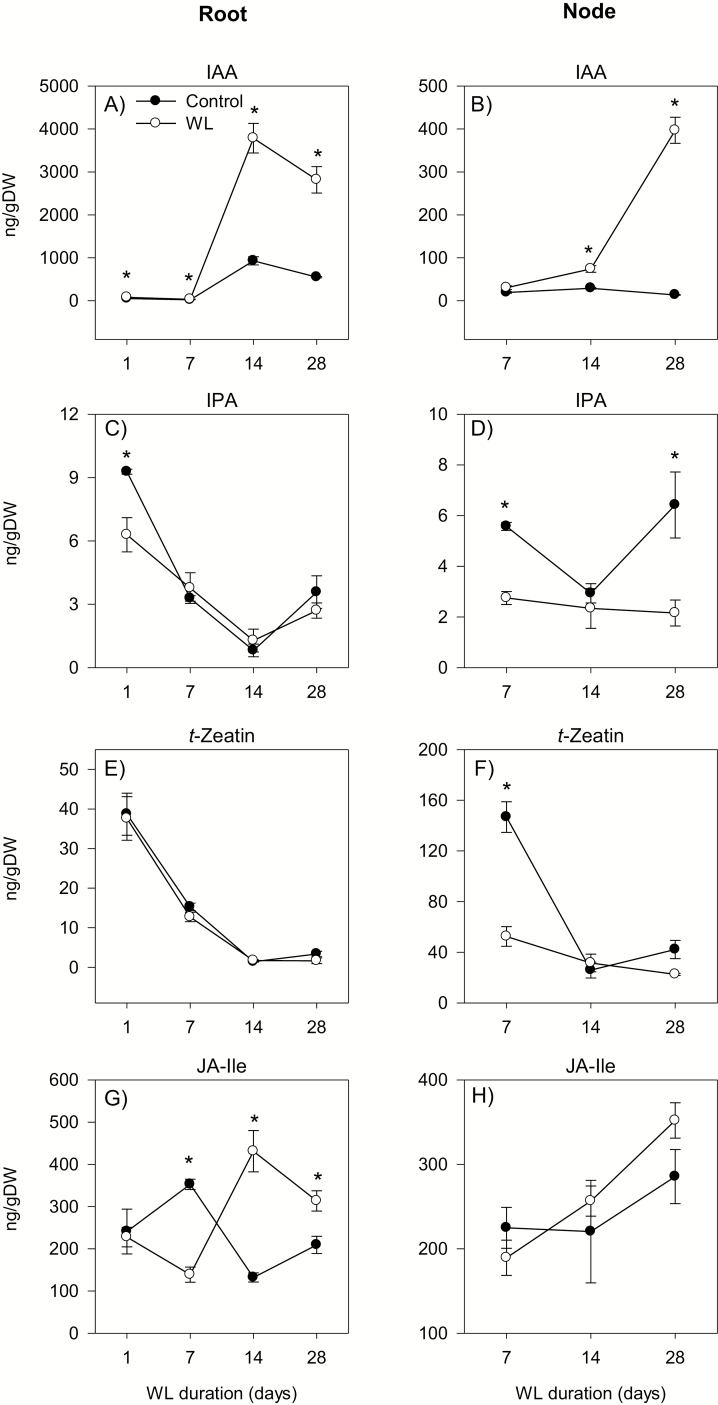
The expression of TDC and YUC1 in the root decreased below detectable levels during the course/by the end of the experiment under control conditions but increased by waterlogging for 14 d and 28 d (Fig. 5A, C). The expression level of YUC1 also increased by waterlogging for 1 d (3.5-fold). The expression level of PIN9 was constantly very weak under control conditions but increased (3.5- to 37-fold) by waterlogging irrespective of the duration (Fig. 5I). Waterlogging for 14 d and/or 28 d also increased the expression levels of GH3.1 and GH3.2 (from undetectable levels) (Fig. 5E, J). The root IAA level increased under both control and waterlogged conditions; however, roots waterlogged for 14 d and 28 d exhibited a higher (>4-fold) level of IAA than that observed in the respective controls (Fig. 6A).
Fig. 5.
Expression of auxin metabolism and transport genes. Relative transcript levels of TDC (A and B), YUC1 (C and D), GH3.1 (E and F), GH3.2 (G and H), and PIN9 (I and J) in root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, G, and I) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, H, and J), and their respective controls. Transcript levels of TDC, YUC1, GH3, and PIN9 genes in both tissues were determined as described in Fig. 3, and are expressed relative to the respective transcript levels of TDC, YUC1, GH3.1, and PIN9 in control roots at 1 d after the start of waterlogging, respectively, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3. No transcript of TAA1, AAO, YUC10, and PIN5 was detected irrespective of tissue type or growth conditions, as shown in Supplementary Table S3.
Fig. 6.
Auxin, cytokinin, and jasmonate levels. Indole acetic acid (IAA; A and B), N6-isopentenyladenine (IPA; C and D), trans-zeatin (t-zeatin, E and F), and jasmonoyl-l-isoleucine (JA-Ile; G and H) levels in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, and G) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, and H), and their respective controls. Data are means of three biological replicates ±SE. Asterisks denote a statistically significant difference in hormone levels between samples derived from control and waterlogged plants within each duration of waterlogging (P<0.05; Student t-test). WL, waterlogging.
Stem node
The expression of TDC in the stem node decreased below a detectable level during the course of the experiment under control conditions but increased (from undetectable levels or >4-fold) by waterlogging irrespective of the duration (Fig. 5B). Stem nodes waterlogged for 7 d and/or 14 d exhibited higher expression levels of YUC1 and PIN9 (~2-fold or more) than that observed in the corresponding controls (Fig. 5D, J). The expression of both genes decreased below detectable levels by 28 DAWL under both growth conditions. The expression of GH3.1 and GH3.2 also decreased below detectable levels under both growth conditions, except for the strong transient increases of their expression by waterlogging for 14 d (Fig. 5F, H). An increase in the expression level of GH3.2 was also observed by waterlogging for 1 d. The stem node IAA level remained constant under control conditions, but increased in response to waterlogging for 14 d and 28 d (2.5- to 29-fold) (Fig. 6B).
Regulation of cytokinin metabolism
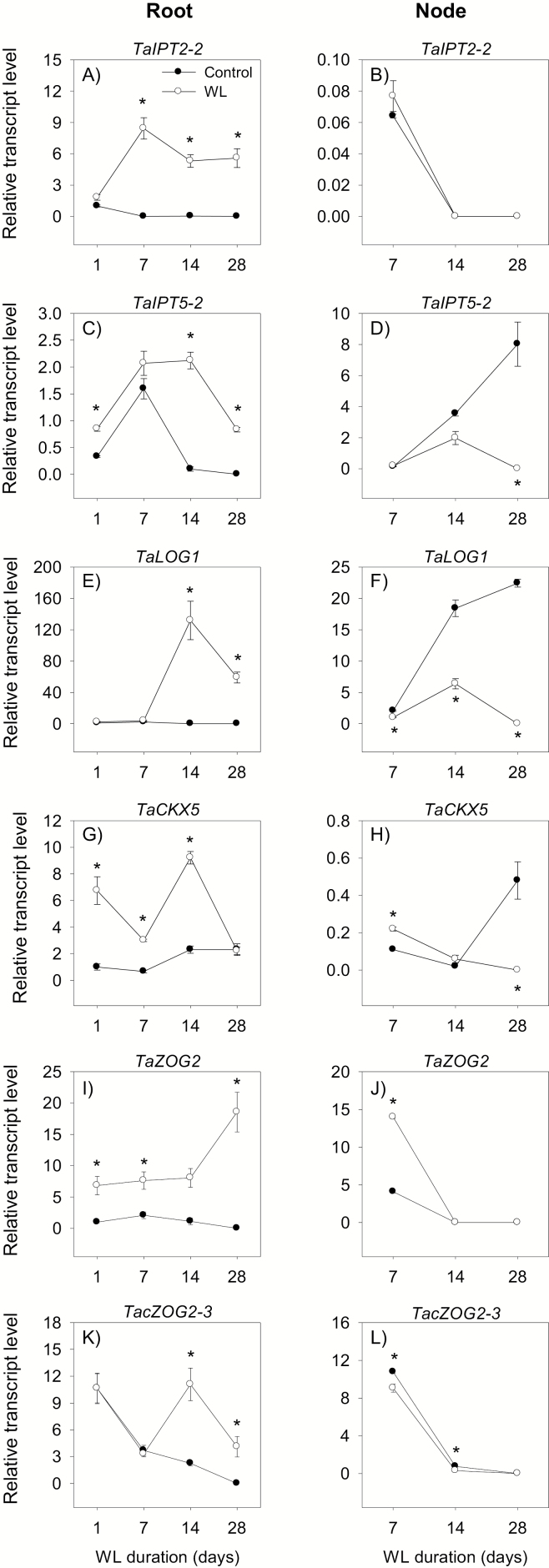
The first and rate-limiting step of cytokinin biosynthesis involves the formation of isopentenyladenine (iP) nucleotides from dimethylallyl diphosphate and adenine nucleotides, preferably ATP and ADP, by the action of adenosine phosphate-isopentenyltransferase (IPT) (Hirose et al., 2008). Cytochrome P450 monooxygenases (CYP735As) convert the iP nucleotides to trans-zeatin (t-zeatin) nucleotides; both the iP and t-zeatin nucleotides can be directly activated to free base bioactive cytokinins by cytokinin nucleoside 5′-monophosphate phosphoribohydrolase, also known as lonely guy (LOG) (Kurakawa et al., 2007). Inactivation of cytokinin is catalyzed by cytokinin oxidases/dehydrogenases (CKXs) and/or cytokinin-conjugating O-glucosyltransferases (ZOGs). In this study, we analyzed the expression patterns of cytokinin biosynthesis (IPT2-2 and IPT5-2), activation (LOG1, LOG3, and LOG5), catabolism (CKX4-2 and CKX5), and conjugation (ZOG2 and cZOG2-3) genes in waterlogged root and stem node tissues and their respective controls. Our data revealed that all the cytokinin metabolic genes studied are expressed in the root and stem node tissues under both control and waterlogged conditions, except that the expression levels of LOG3 in both tissues, and IPT2-2 and LOG5 in the stem node tissue were very minimal/undetectable irrespective of growth conditions (Supplementary Table S3).
Root
The expression levels of IPT2-2, IPT5-2, LOG1, and LOG5 in the root decreased below detectable levels during the course or by the end of the experiment under control conditions but strongly increased in response to waterlogging for seven and/or more days (Fig. 7A, C, E; Supplementary Table S3). Between the two IPT genes, IPT2-2 exhibited the highest level of waterlogging-induced expression. Under control conditions, the expression level of the cytokinin inactivation gene, CKX5, increased (>2-fold) while the expression levels of CKX4-2, ZOG2, and cZOG2-3 decreased below detectable levels (Fig. 7G, I, K; Supplementary Table S3). Waterlogging, however, led to temporally distinct increases in the expression levels of these genes; CKX5 expression level was induced by waterlogging up to 14 d while the expression levels of the other genes were induced by waterlogging irrespective of the duration (ZOG2) or mainly at 14 and 28 DAWL (CKX4-1 and ZOG2-3). Root N6-isopentenyladenine (IPA) and t-zeatin levels decreased under both control and waterlogged conditions, and were not affected by waterlogging (Fig. 6C, E).
Fig. 7.
Expression of cytokinin metabolism genes. Relative transcript levels of IPT2-2 (A and B), IPT5-2 (C and D), LOG1 (E and F), CKX5 (G and H), ZOG2 (I and J), and cZOG2-3 (K and L) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, G, I, and K) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, H, J, and L), and their respective controls. Transcript levels of IPT genes, LOG1, CKX5, and ZOG genes in both tissues were determined as described in Fig. 3, and are expressed relative to the transcript levels of IPT2-2, LOG1, CKX5, and ZOG2 in control roots at 1 d after the start of waterlogging, respectively, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3. Expression levels of LOG3 in both tissues, and IPT2-2 and LOG5 in the stem node were either minimal or undetectable irrespective of growth conditions, as shown in Supplementary Table S3.
Stem node
The expression level of IPT2-2 in the stem node was very minimal or decreased below a detectable level during the course of the experiment under both control and waterlogged conditions (Fig. 7B). The expression levels of IPT5-2 and LOG1, however, increased under control conditions but decreased (>1.8-fold or to undetectable levels) by waterlogging for 14 d and 28 d (IPT5-2) or irrespective of the duration (LOG1) (Fig. 7D, F). Between the two IPT genes, IPT5-2 showed the highest level of expression under both growth conditions. Expression levels of cytokinin-inactivating genes (CKX4-2, CKX5, ZOG2, and cZOG2-3) decreased to lower or below detectable levels under both growth conditions, except for the increase of CKX5 expression in control stem nodes at 28 DAWL (Fig. 7H, J, L; Supplementary Table S3). However, stem nodes waterlogged for 7 d exhibited higher expression levels of CKX4-2, CKX5, and ZOG2 (>2-fold) than that observed in their respective controls. The levels of IPA and t-zeatin in the stem node decreased/slightly decreased under both growth conditions, except for the increase of IPA level in control stem nodes at 28 DAWL (to a level similar to that observed at 7 DAWL) (Fig. 6D, F). Stem nodes waterlogged for 7 d and 28 d, however, contained a lower amount of IPA and t-zeatin (>2-fold) than that found in their respective controls.
Regulation of jasmonate metabolism
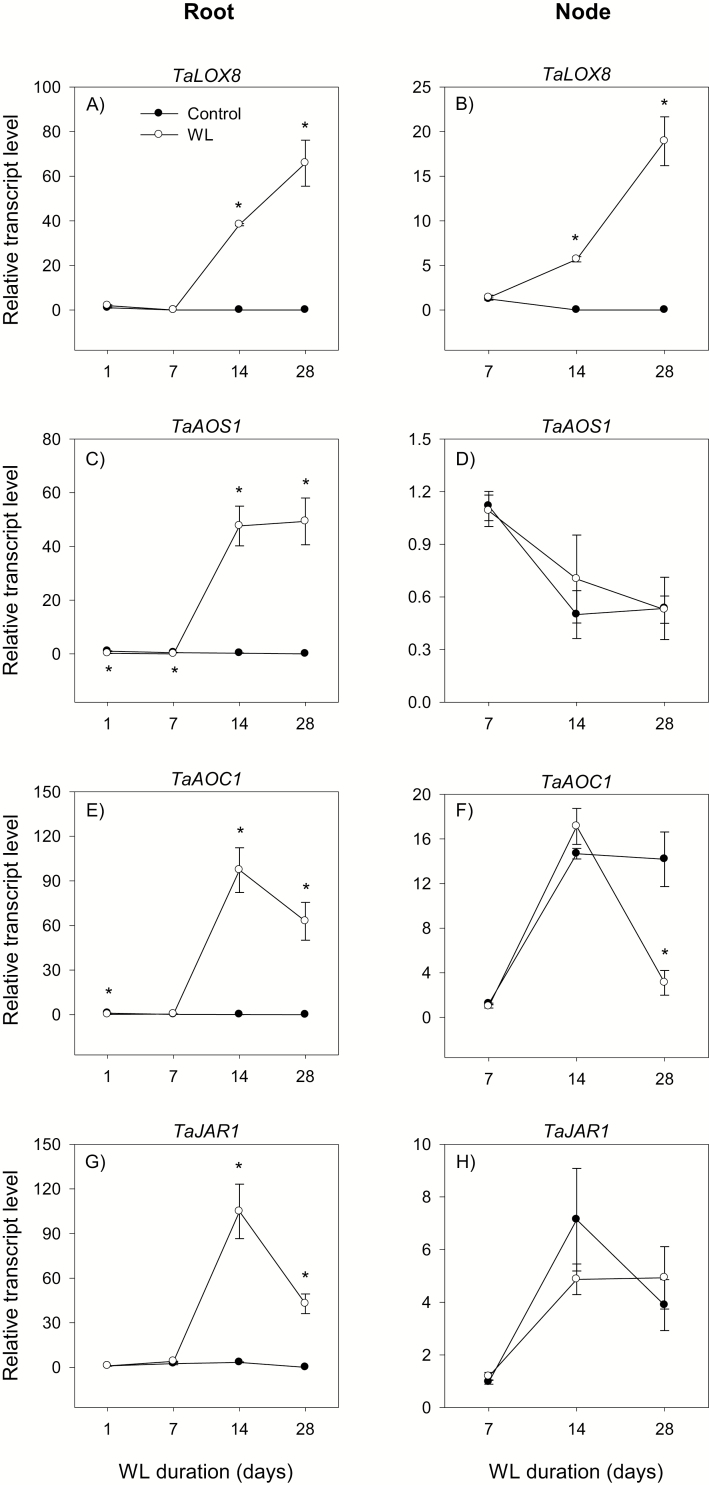
The synthesis of JA involves the conversion of polyunsaturated fatty acids to 12-oxo-phytodienoic acid (OPDA) by a series of reactions catalyzed by lipoxygenase (LOX), allen oxide synthase (AOS), and allen oxide cyclase (AOC) (Schaller and Stintzi, 2009). The OPDA is then converted to jasmonic acid by the action of OPDA reductase (OPR) and three cycles of β-oxidation. Jasmonate resistant 1 (JAR1), which is a jasmonate-amido synthetase, catalyzes the conversion of jasmonic acid to jasmonyl-l-isoleucine conjugate (JA-Ile), a type of JA that is crucial for JA signaling (Staswick, 2008). This study analyzed the expression patterns of genes involved in JA biosynthesis including LOX (LOX2 and LOX8), AOS (AOS1 and AOS2), and AOC (AOC1 and AOC2), and JA conjugation (JAR; JAR1) in waterlogged root and stem node tissues and their respective controls. Our analysis revealed that all the JA metabolic genes studied are expressed in both root and stem node tissues under control and/or waterlogged conditions except that the expression of LOX2 and AOS2 in the root and AOC2 in the stem node was either below detectable levels or relatively lower than the levels of their other homologs (LOX8, AOS1, and AOC1) irrespective of growth conditions (Supplementary Table S3).
Root
The expression levels of all the JA metabolic genes expressed in the root were either constantly very weak or decreased below detectable levels by the end of the experiment under control conditions but increased (>3-fold) by waterlogging for 14 d and 28 d (Fig. 8A, C, E, G; Supplementary Table S3). Despite its transient increase and decrease at 7 DAWL in control and waterlogged roots, respectively, the JA-Ile level decreased under control conditions but increased by waterlogging for 14 d and 28 d, leading to the detection of a higher amount of JA-Ile (1.5- to 3.3-fold) in waterlogged than control roots (Fig. 6G).
Fig. 8.
Expression of jasmonate metabolism genes. Relative transcript levels of LOX8 (A and B), AOS1 (C and D), AOC1 (E and F), and JAR1 (G and H) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, and G) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, and H), and their respective controls. Transcript levels of each gene in both tissues were determined as described in Fig. 3, and are expressed relative to their respective transcript levels in control roots at 1 d after the start of waterlogging, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3. LOX2 in both tissues, AOS2 in the root, and AOC2 in the stem node exhibited either relatively lower levels of expression than their respective homologs or undetectable levels of expression irrespective of growth conditions, as shown in Supplementary Table S3.
Stem node
The expression of LOX8 and AOS1 decreased to low or below detectable levels during the course of the experiment under control conditions, whereas the expression levels of AOC1 and JAR1 increased (Fig. 8B, D, F, H). However, the expression level of LOX8 increased markedly (from an undetectable level) by waterlogging for 14 d and 28 d, while that of AOC1 decreased in response to waterlogging for 28 d (>4-fold). Waterlogging did not affect the expression levels of AOS1 and JAR1. The stem node JA-Ile level increased under both control and waterlogged conditions, and it was not affected by waterlogging (Fig. 6H).
Regulation of abscisic acid metabolism
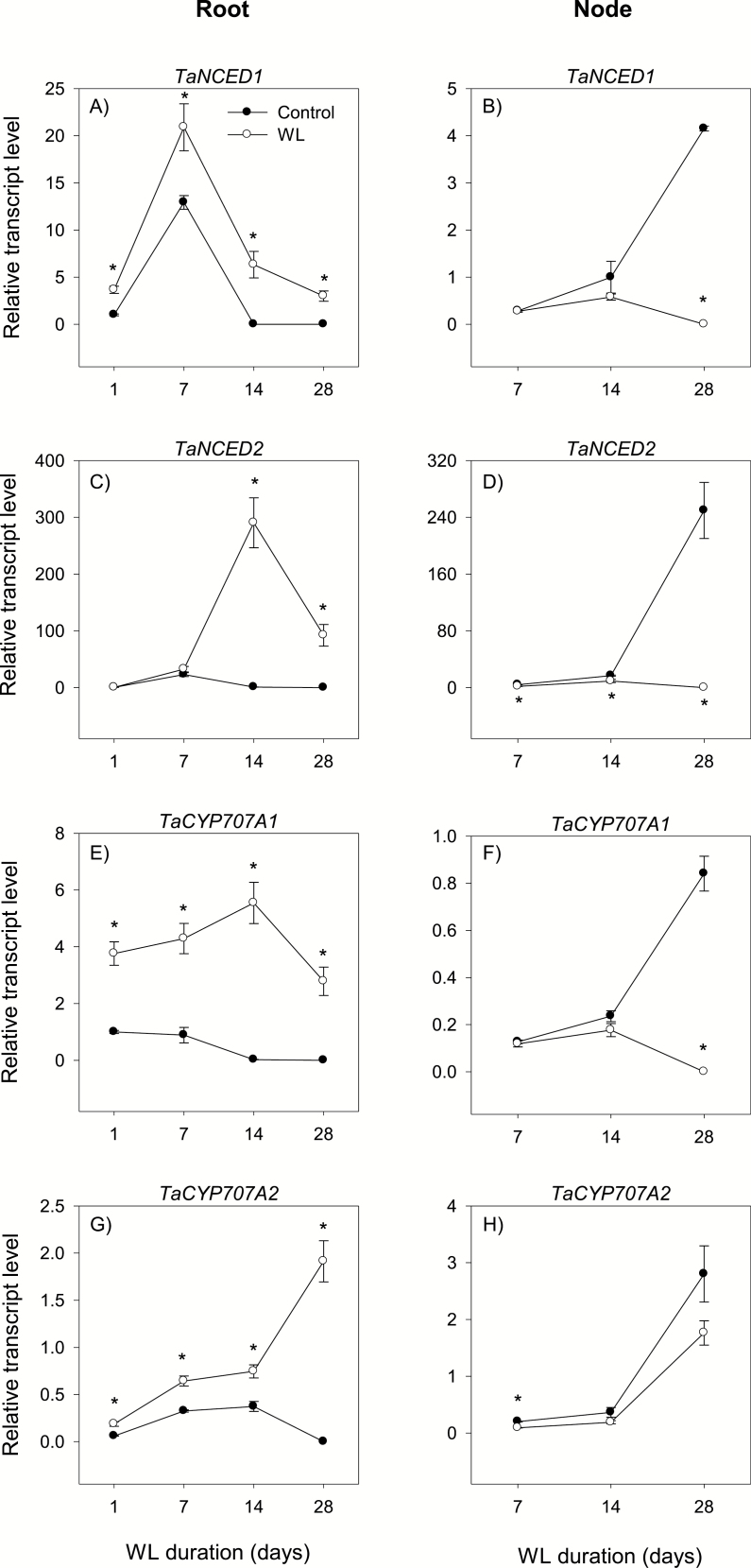
ABA is synthesized from zeaxanthin involving several reactions, and the reaction catalyzed by 9-cis-epoxycarotenoid dioxygenase (NCED) is considered to be rate limiting, while the ABA 8′-hydroxylation reaction, which is catalyzed by ABA 8'-hydroxylase (ABA8′OH), represents the major mechanism of ABA inactivation (Nambara and Marion-Poll, 2005). Thus, genes encoding NCED and ABA8′OH (encoded by CYP707A genes) play key roles in regulating ABA levels in plant tissues. This study examined the expression patterns of NCED (NCED1 and NCED2) and CYP707A (CYP707A1 and CYP707A2) genes in root and stem node tissues under control and waterlogged conditions.
Root
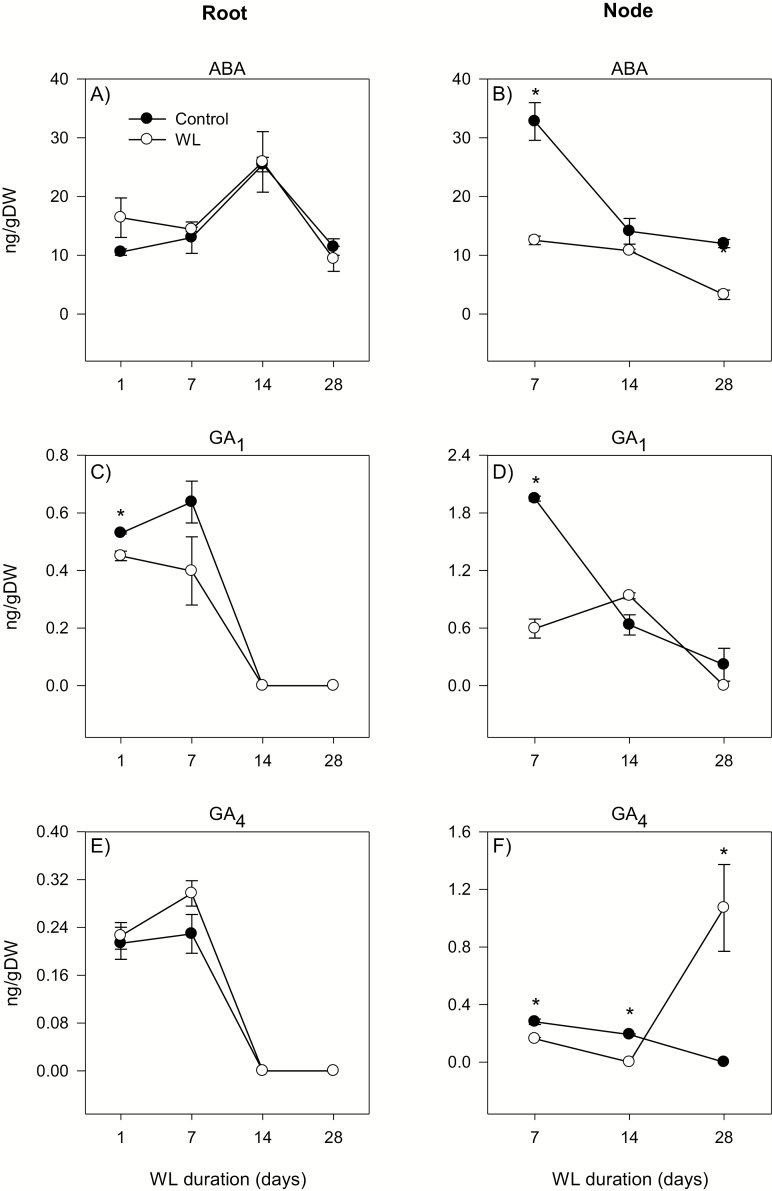
The expression of NCED and CYP707A genes in the root decreased below detectable levels by the end of the experiment under control conditions, except for the transient increases of NCED1 and NCED2 expression at 7 DAWL (Fig. 9A, C, E, G). Waterlogging increased the expression level of NCED1 (from an undetectable level or >1.6-fold) irrespective of the duration and that of NCED2 (from very low/undetectable level) only after 14 d and 28 d (Fig. 9A, C). Between the two NCED genes, NCED2 exhibited the highest level of waterlogging-induced expression. Waterlogging also increased the expression levels of CYP707A1 and CYP707A2 (from undetectable levels or >2-fold) irrespective of the duration (Fig. 9E, G). The root ABA level remained almost constant under both control and waterlogged conditions, except for the transient increase observed at 14 DAWL, and it was not affected by waterlogging (Fig. 10A).
Fig. 9.
Expression of abscisic acid metabolism genes. Relative transcript levels of NCED1 (A and B), NCED2 (C and D), CYP707A1 (E and F), and CYP707A2 (G and H) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, and G) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, and H), and their respective controls. Transcript levels of the NCED and CYP707A genes in both tissues were determined as described in Fig. 3, and are expressed relative to the transcript levels of NCED1 and CYP707A1 in control roots at 1 d after the start of waterlogging, respectively, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3.
Fig. 10.
Abscisic acid and gibberellin levels. ABA (A and B), GA1 (C and D), and GA4 (E and F) levels in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, and E) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, and F), and their respective controls. Other data descriptions are as indicated in Fig. 6.
Stem node
The expression levels of NCED and CYP707A genes in the stem node increased during the course of the experiment under control conditions (Fig. 9B, D, F, H). Waterlogging for 28 d, however, decreased the expression of all genes below detectable levels except that the CYP707A2 expression level showed only a slight reduction (1.6-fold). The stem node ABA level decreased under both control and waterlogged conditions; however, waterlogged stem nodes contained only 27–76% of the ABA found in the corresponding controls (Fig. 10B).
Regulation of gibberellin metabolism
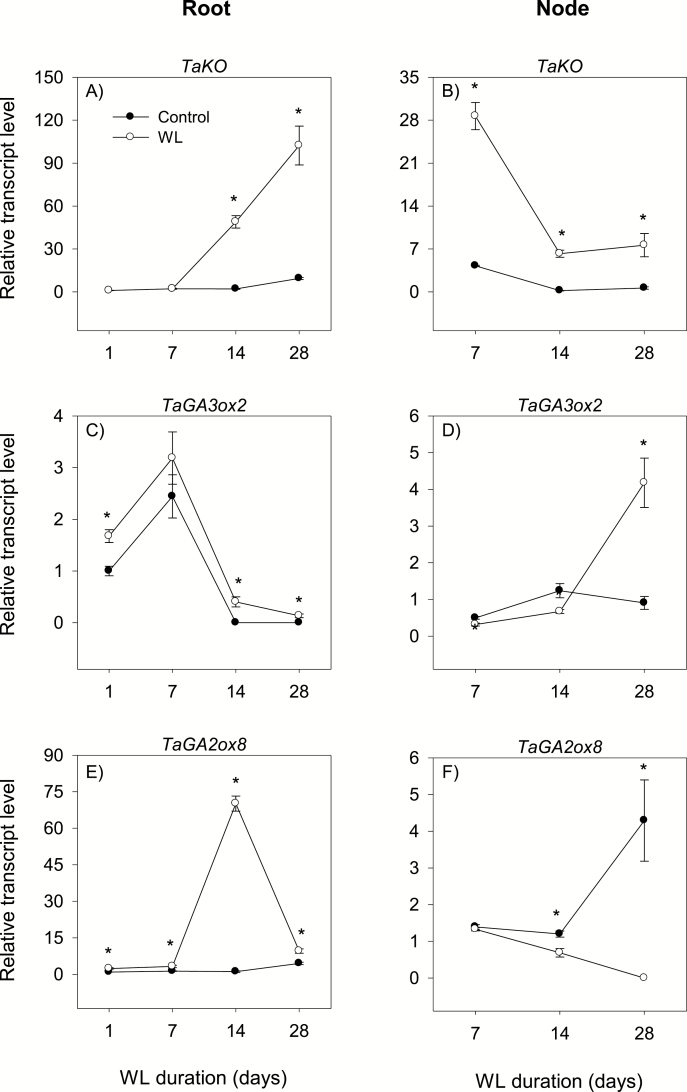
Gibberellins are synthesized from geranyl geranyl diphosphate, which is converted to ent-kaurene by the consecutive actions of ent-copalyl diphosphate synthase and ent-kaurene synthase (Yamaguchi, 2008). Subsequent oxidation of ent-kaurene by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) produces GA12. The synthesis of bioactive GAs, mainly GA4 and GA1, involves a series of oxidations of GA12 or its 13-hydroxylated form (GA53) by the actions of GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), and the bioactive GAs are inactivated mainly through GA 2-oxidation, which is catalyzed by GA 2-oxidase (GA2ox) (Hedden and Phillips, 2000). These enzymes play key roles in regulating bioactive GA levels in plant tissues (Yamaguchi, 2008). This study investigated the expression patterns of GA biosynthesis (KO, GA3ox2, and GA3ox3) and catabolism (GA2ox8) genes in waterlogged root and stem node tissues and their respective controls. Our analysis showed that all the GA metabolic genes studied except GA3ox3 are expressed in both tissues under control and/or waterlogged conditions (Supplementary Table S3).
Root
The expression levels of KO, GA3ox2, and GA2ox8 in the root either remained constantly very weak or decreased below detectable levels during the course of the experiment under control conditions, except for the transient increases of KO and GA2ox8 expression at 28 DAWL (Fig. 11A, C, E). Waterlogging for 14 d and 28 d increased the expression levels of KO (10- to 24-fold), GA3ox2 (from an undetectable level), and GA2ox8 (2.1- to 62-fold). The waterlogging-induced expression level of GA2ox8 was, however, much higher than that of GA3ox2 (Supplementary Table S4). Root GA1 and GA4 decreased below detectable levels by 14 DAWL under both control and waterlogged conditions, and were not affected by waterlogging (Fig. 10C, E).
Fig. 11.
Expression of gibberellin metabolism genes. Relative transcript levels of KO (A and B), GA3ox2 (C and D), and GA2ox8 (E and F) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, and E) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, and F), and their respective controls. Transcript levels of each gene in both tissues were determined as described in Fig. 3, and are expressed relative to their respective transcript levels in control roots at 1 d after the start of waterlogging, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3. No transcript of TaGA3ox3 was detected irrespective of tissue type or growth conditions, as shown in Supplementary Table S3.
Stem node
The expression levels of KO, GA3ox2, and GA2ox8 in the stem node either decreased or remained constantly weak during the course of the experiment under control conditions, except for the transient increase of GA2ox8 expression at 28 DAWL (Fig. 11B, D, F). Relative to that observed in the control stem node, waterlogging increased the expression level of KO irrespective of the duration (6.7- to 31-fold), while the expression level of GA3ox2 increased only at 28 DAWL (4.6-fold). In contrast, waterlogging for 14 d and 28 d decreased the expression level of GA2ox8 (1.8-fold or to an undetectable level). The levels of GA1 and GA4 in the stem node decreased under both control and waterlogged conditions, except for the transient increase of the GA4 level (from an undetectable level to 1 ng g DW–1) in response to waterlogging for 28 d (Fig. 10D, F).
Discussion
Waterlogging decreased the dry weight of the whole root, and this occurred earlier than its effect on the other aspects of root morphology (Fig. 1). It also inhibited elongation of axile roots and emergence of branch/lateral roots while promoting the formation of surface adventitious roots and aerenchyma (Figs 1, 2). Similar effects of waterlogging have also been reported in wheat seedlings and adult plants (Malik et al., 2002; Yamauchi et al., 2014; Herzog et al., 2016). Waterlogging increased the number of axile roots per plant; however, this result contrasts with a previous report that has shown waterlogging-induced reduction in the number of these roots (Malik et al., 2002). This difference could be due to the fact that the two studies used different genotypes or it could arise as a consequence of the growth stages at which the waterlogging treatment was initiated, when the plants were 21 d old (Malik et al., 2002) vs. 30 d old (this study), which potentially affects the establishment and final number of the tillers. Consistently, tiller number was reduced only by 1.5- to 2.3-fold in our study (Fig. 1G) but by up to 4-fold in Malik et al. (2002).
The effects of waterlogging on root growth, emergence and elongation of axile and lateral roots, and aerenchyma formation are associated with enhanced expression levels of ethylene biosynthetic genes, mainly ACS7 and ACO2 (Fig. 3), suggesting increased ethylene synthesis in waterlogged roots. Consistently, increases in the ACS expression level and ethylene synthesis have been reported to be associated with inhibition of root elongation, and the emergence and elongation of branch/lateral roots and promotion of aerenchyma formation in wheat seedlings under oxygen-deficient conditions (Yamauchi et al., 2014). Our results therefore imply the importance of transcriptional control of ethylene synthesis in the root, mainly through the expression of ACS7 and ACO2, in modulating the adaptive response of wheat to waterlogging. Since ethylene also promotes adventitious root emergence in wheat under oxygen-deficient conditions (Yamauchi et al., 2014), our gene expression data in the stem node suggest the significance of transcriptional regulation of ethylene synthesis, mainly via the expression of ACS7 and ACO2, in the formation of surface adventitious roots (Figs 1E, 3).
Previous studies in rice have shown the significance of RBOH-mediated ROS production in ethylene-induced death of epidermal cells during adventitious root emergence from stem nodes (Steffens et al., 2012; Gonzali et al., 2015; Mittler, 2017) and ethylene-induced death of cortical cells during aerenchyma formation under oxygen-deficient conditions (Yamauchi et al., 2017). Consistently, co-induction of the expression levels of ethylene biosynthesis and RBOH genes in response to waterlogging is associated with the formation of surface adventitious roots and aerenchyma (Figs 1E, 2–4). It is important to note here that ethylene signaling can also induce epidermal cell death and thereby adventitious root emergence through a ROS-independent signaling pathway (Steffens and Sauter, 2009). Furthermore, the mechanisms underlying the regulation of ethylene synthesis in wheat, one of the dryland crop species, can be different from that of rice (Yamauchi et al., 2015, 2016). Since the role of ethylene in mediating adaptations to flooding/waterlogging/oxygen-deficient conditions has been studied extensively in different crop species including wheat (Jackson et al., 1985; Jackson and Armstrong, 1999; Yamauchi et al., 2013, 2014), our study was focused mainly on the other hormones that are much less studied with respect to waterlogging.
Auxin has been implicated in the inhibition of primary root elongation and promotion of adventitious root formation in cereal crops such as rice (Zhuang et al., 2006; Liu et al., 2009; McSteen, 2010). Therefore, the association of a higher root IAA level (4.1- to 5.1-fold) and shorter axile root phenotype in waterlogged plants with enhanced expression levels of TDC, YUC1, and PIN9 (Figs 1, 5, 6) suggests the importance of IAA biosynthesis and transport in regulating the IAA level and root elongation under waterlogged conditions. An increase in IAA level has also been reported in the roots of Citrus spp. under long-term flooding (Arbona and Gómez-Cadenas, 2008). The prevalence of a higher IAA level (2.6- to 29-fold) and enhanced expression levels of TDC, YUC1, and PIN9 in waterlogged stem nodes is associated with the formation of surface adventitious roots from the stem nodes at 28 DAWL. This, along with the observation of more belowground axile roots in waterlogged than control plants, supports that increased synthesis/transport of auxin positively regulates adventitious root emergence (Boerjan et al., 1995; Liu et al., 2009; Vidoz et al., 2010). Consistently, chemical treatments or mutations that alter auxin transport and synthesis affect adventitious root formation (Liu et al., 2009; Vidoz et al., 2010; Della Rovere et al., 2013; Sauter and Steffens, 2014). Although the PIN genes of Arabidopsis act jointly in regulating auxin distribution and thereby determination of root stem cell specification/patterning and growth of primary roots (Blilou et al., 2005), our expression data suggest that PIN9 acts as a major player in this regard. Increases in the expression levels of IAA-conjugating genes, GH3.1 and GH3.2, in waterlogged roots and stem nodes (Fig. 5) might suggest their negative feedback regulation due to waterlogging-induced IAA accumulation in order to maintain IAA homeostasis. Auxin synthesis in the root and its transport towards the elongation zone where it acts to inhibit cell elongation are stimulated by ethylene (Růžička et al., 2007). Furthermore, auxin transport and adventitious root formation under waterlogged conditions are enhanced by increased ethylene synthesis (Vidoz et al., 2010). Therefore, co-inductions in the expression levels of specific auxin biosynthesis and/or transport and ethylene biosynthesis genes in waterlogged roots and stem nodes (Figs 3, 5) might suggest the importance of auxin–ethylene interplay in regulating root elongation and the formation of surface adventitious roots from the stem nodes.
Cytokinin has been reported to inhibit root elongation in wheat under normal conditions (Stenlid, 1982). Waterlogging increased the expression levels of genes involved in cytokinin synthesis, activation, and inactivation, leading to no significant change in root IPA and t-zeatin levels (Figs 6, 7). Thus, inhibition of root elongation under waterlogging conditions with no apparent change in root IPA and t-zeatin levels might suggest that the regulation of this root trait in waterlogged wheat is not dependent on the level of cytokinin. Cytokinins are mainly produced in the root and are transported to the shoot (Müller and Leyser, 2011). Consistently, increasing cytokinin synthesis specifically in the root has been shown to improve shoot growth under stress conditions (Ghanem et al., 2011). Given that waterlogging did not affect root IPA and t-zeatin levels, the reduced shoot growth in waterlogged plants (Fig. 1F) might be due to impaired root to shoot transport of cytokinin. In agreement with this, short-term flooding has been shown to decrease the level of cytokinin in the xylem sap of sunflower plants (Burrows and Carr, 1969). Cytokinin has also been implicated as a negative regulator of adventitious root formation (Ramírez-Carvajal et al., 2009). Thus, the association of waterlogging-induced reductions in stem node IPA and t-zeatin levels with decreased expression levels of IPT5-2 and LOG1, and enhanced expression levels of CKX5 and ZOG2 at the earlier stage of waterlogging (by 7 DAWL) (Figs 1E, 2, 6, 7) might suggest the importance of transcriptional regulation of cytokinin biosynthesis, activation, and inactivation in the formation of surface adventitious roots. Given that inhibition of adventitious root formation by cytokinin is linked to its role in repressing the expression levels of auxin transport genes (Della Rovere et al., 2013), the lower cytokinin level in waterlogged stem nodes (Fig. 6D, F) might contribute to the higher expression level of PIN9 and thereby promoting the formation of surface adventitious roots (Figs 1E, 5J).
Increasing the JA level through mutations or exogenous JA application has been shown to lead to inhibition of root growth in cereals such as rice (reviewed in Liu et al., 2015). Consistently, waterlogging-induced inhibition of root growth is accompanied by an increase in root JA-Ile level, which is associated with enhanced expression levels of JA biosynthetic genes (Figs 6G, 8). It has been shown previously that JA enhances adventitious root formation in rice under normal/aerobic conditions (Moons et al., 1997). However, the emergence of surface adventitious roots from stem nodes waterlogged for 28 d in spite of the absence of significant effects of waterlogging on the expression level of JAR1, a gene encoding an enzyme catalyzing the final step of JA-Ile synthesis, and JA-Ile level in the stem node (Figs 1E, 6H, 8H) suggests that this root morphological acclimation to waterlogging in wheat is not regulated by the JA level.
ABA is also implicated in the inhibition of primary root elongation in cereal crops such as rice under normal conditions (Konishi et al., 2005), although it has the opposite effect under stress conditions such as drought/low water potential (Saab et al., 1990; Sharp et al., 1994). Waterlogging enhanced the expression levels of both ABA biosynthetic (NCED) and catabolic (CYP707A) genes in the root relative to that observed under control conditions, leading to no change in ABA level. The prevalence of shorter roots in waterlogged wheat with no apparent effect on root ABA level (Figs 1, 10A) suggests that inhibition of wheat root elongation under waterlogging conditions is not dependent on the ABA level. Previous studies with other plant species such as alfalfa and Arabidopsis have also shown that the root ABA level is not affected by long-term flooding/oxygen-deficient conditions (Castonguay et al., 1993; de Bruxelles et al., 1996). The emergence of surface adventitious roots from waterlogged stem nodes along with reductions in the expression levels of ABA biosynthetic genes and stem node ABA content (Figs 1E, 9B, D, 10B) support that ABA acts as a negative regulator of adventitious root formation (Steffens and Sauter, 2005).
Although root bioactive GA decreased below detectable levels in the course of the experiment under both control and waterlogged conditions (Fig. 10C, E), the higher expression level of the GA catabolic gene, GA2ox8, relative to that observed for the regulatory GA biosynthetic gene, GA3ox2, in roots waterlogged for 14 d and 28 d (Supplementary Table S4) might suggest lower root GA levels that cause inhibition of root elongation as observed in rice (Komatsu and Konishi, 2005). Based on the detection of a reduced GA level in the xylem sap, Reid et al. (1969) also suggested decreased GA synthesis in waterlogged roots of tomato. Previous reports have implicated GA in promoting ethylene-induced adventitious root formation (Suge, 1985; Steffens and Sauter, 2005). Thus, the association of waterlogging-induced accumulation of bioactive GA (GA4) with the expression patterns of GA biosynthesis (KO and GA3ox2) and inactivation (GA2ox8) genes in stem nodes that produce surface adventitious roots (Figs 1E, 10F, 11) implies the importance of transcriptional control of GA metabolism in regulating this morphological acclimation of wheat to waterlogging.
In summary, the results of our study suggest that morphological and anatomical adaptive responses of wheat to waterlogging involve modulation of the levels of different plant hormones in root and stem node tissues through transcriptional regulation of specific genes in their respective metabolic pathways, and the findings of this study provide important insights into the molecular mechanisms underlying the acclimation of wheat to waterlogging.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Sequence similarity of genes analyzed in this study with their orthologs.
Table S2. Gene-specific primers used for expression analysis.
Table S3. Expression levels of hormone metabolic genes analyzed in this study.
Table S4. Comparison of expression levels across all gibberellin metabolism genes.
Acknowledgements
This work was supported by grants from the Networks of Centers of Excellence Program-BioFuelNet Canada, Manitoba Wheat and Barley Growers Association, Natural Sciences and Engineering Research Council of Canada to BTA, and the University of Manitoba Graduate Fellowship to T-NN. The authors would like to thank Andre Dufresne for his technical assistance with histological analysis.
Glossary
Abbreviations:
- ABA
abscisic acid
- DAWL
days after waterlogging
- GA
gibberellin
- IAA
indole acetic acid
- JA
jasmonate
- JA-Ile
jasmonoyl-l-isoleucine
- ROS
reactive oxygen species.
References
- Arbona V, Gómez-Cadenas A. 2008. Hormonal modulation of citrus responses to flooding. Journal of Plant Growth Regulation 27, 241. [Google Scholar]
- Armstrong W. 1979. Aeration in higher plants. In: Woolhouse HW, ed. Advances in botanical research. London: Academic Press, 225–332. [Google Scholar]
- Bailey-Serres J, Lee SC, Brinton E. 2012. Waterproofing crops: effective flooding survival strategies. Plant Physiology 160, 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, et al. . 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, et al. . 1995. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. The Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows WJ, Carr DJ. 1969. Effects of flooding the root system of sunflower plants on the cytokin in content in the xylem sap. Physiologia Plantarum 22, 1105–1112. [DOI] [PubMed] [Google Scholar]
- Castonguay Y, Nadeau P, Simard RR. 1993. Effects of flooding on carbohydrate and ABA levels in roots and shoots of alfalfa. Plant, Cell and Environment 16, 695–702. [Google Scholar]
- Chochois V, Vogel JP, Watt M. 2012. Application of Brachypodium to the genetic improvement of wheat roots. Journal of Experimental Botany 63, 3467–3474. [DOI] [PubMed] [Google Scholar]
- de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R. 1996. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiology 111, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen VH, Caboche M, Bellini C. 1998. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. The Plant Journal 14, 603–611. [DOI] [PubMed] [Google Scholar]
- Della Rovere F, Fattorini L, D’Angeli S, Veloccia A, Falasca G, Altamura MM. 2013. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of arabidopsis. Annals of Botany 112, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca G, Altamura MM. 2003. Histological analysis of adventitious rooting in Arabidopsis thaliana (L.) Heynh seedlings. Plant Biosystems 137, 265–273. [Google Scholar]
- Ghanem ME, Albacete A, Smigocki AC, et al. . 2011. Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) plants. Journal of Experimental Botany 62, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Cardarelli F, et al. . 2015. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nature Plants 1, 15151. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, et al. . 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. The Plant Cell 24, 2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CJ, Drew MC, Morgan PW. 1994. Induction of enzymes associated with lysigenous aerenchyma formation in roots of Zea mays during hypoxia or nitrogen starvation. Plant Physiology 105, 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. 1996. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiology 112, 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. 2000. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Herzog M, Striker GG, Colmer TD, Pedersen O. 2016. Mechanisms of waterlogging tolerance in wheat—a review of root and shoot physiology. Plant, Cell and Environment 39, 1068–1086. [DOI] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. 2008. Regulation of cytokinin biosynthesis, compartmentalization and translocation. Journal of Experimental Botany 59, 75–83. [DOI] [PubMed] [Google Scholar]
- Izydorczyk C, Nguyen T-N, Jo S, Son S, Tuan PA, Ayele BT. 2018. Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signalling mediates the effects of suboptimal and supraoptimal temperatures on seed germination in wheat (Triticum aestivum L.). Plant, Cell and Environment 41, 1022–1037. [DOI] [PubMed] [Google Scholar]
- Jackson MB, Armstrong W. 1999. Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biology 1, 274–287. [Google Scholar]
- Jackson MB, Fenning TM, Drew MC, Saker LR. 1985. Stimulation of ethylene production and gas-space (aerenchyma) formation in adventitious roots of Zea mays L. by small partial pressures of oxygen. Planta 165, 486–492. [DOI] [PubMed] [Google Scholar]
- Kasahara H. 2016. Current aspects of auxin biosynthesis in plants. Bioscience, Biotechnology, and Biochemistry 80, 34–42. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Konishi H. 2005. Proteome analysis of rice root proteins regulated by gibberellin. Genomics, Proteomics and Bioinformatics 3, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Kitano H, Komatsu S. 2005. Identification of rice root proteins regulated by gibberellin using proteome analysis. Plant, Cell and Environment 28, 328–339. [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. [DOI] [PubMed] [Google Scholar]
- Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P, Shou H. 2009. Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Research 19, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang S, Sun N, Liu H, Zhao Y, Liang Y, Zhang L, Han Y. 2015. Functional diversity of jasmonates in rice. Rice 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Malik AI, Colmer TD, Lambers H, Setter TL, Schortemeyer M. 2002. Short-term waterlogging has long-term effects on the growth and physiology of wheat. New Phytologist 153, 225–236. [Google Scholar]
- Marti J, Savin R, Slafer GA. 2015. Wheat yield as affected by length of exposure to waterlogging during stem elongation. Journal of Agronomy and Crop Science 201, 473–486. [Google Scholar]
- McSteen P. 2010. Auxin and monocot development. Cold Spring Harbor Perspectives in Biology 2, a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergemann H, Sauter M. 2000. Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiology 124, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. 2017. ROS are good. Trends in Plant Science 22, 11–19. [DOI] [PubMed] [Google Scholar]
- Moons A, Prinsen E, Bauw G, Van Montagu M. 1997. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. The Plant Cell 9, 2243–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Annals of Botany 107, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Nguyen T-N, Son S, Jordan MC, Levin DB, Ayele BT. 2016. Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biology 16, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quittenden LJ, Davies NW, Smith JA, Molesworth PP, Tivendale ND, Ross JJ. 2009. Auxin biosynthesis in pea: characterization of the tryptamine pathway. Plant Physiology 151, 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Dervinis C, Davis JM. 2009. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiology 150, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DM, Crozier A, Harvey BMR. 1969. The effects of flooding on the export of gibberellins from the root to the shoot. Planta 89, 376–379. [DOI] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. The Plant Cell 19, 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab IN, Sharp RE, Pritchard J, Voetberg GS. 1990. Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiology 93, 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Steffens B. 2014. Biogenesis of adventitious roots and their involvement in the adaptation to oxygen limitations. In: van Dongen JT, Licausi F, eds. Low-oxygen stress in plants. Heidelberg: Springer, 299–312. [Google Scholar]
- Sauter M. 2013. Root responses to flooding. Current Opinion in Plant Biology 16, 282–286. [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. 2009. Enzymes in jasmonate biosynthesis—structure, function, regulation. Phytochemistry 70, 1532–1538. [DOI] [PubMed] [Google Scholar]
- Sharp RE, Wu Y, Voetberg GS, Saab IN, LeNoble ME. 1994. Confirmation that abscisic acid accumulation is required for maize primary root elongation at low water potentials. Journal of Experimental Botany 45, 1743–1751. [Google Scholar]
- Son S, Chitnis VR, Liu A, Gao F, Nguyen T-N, Ayele BT. 2016. Abscisic acid metabolic genes of wheat (Triticum aestivum L.): identification and insights into their functionality in seed dormancy and dehydration tolerance. Planta 244, 429–447. [DOI] [PubMed] [Google Scholar]
- Song J, Jiang L, Jameson PE. 2012. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biology 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. 2008. JAZing up jasmonate signaling. Trends in Plant Science 13, 66–71. [DOI] [PubMed] [Google Scholar]
- Steffens B, Kovalev A, Gorb SN, Sauter M. 2012. Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. The Plant Cell 24, 3296–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Rasmussen A. 2016. The physiology of adventitious roots. Plant Physiology 170, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M. 2005. Epidermal cell death in rice is regulated by ethylene, gibberellin, and abscisic acid. Plant Physiology 139, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M. 2009. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. The Plant Cell 21, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Wang J, Sauter M. 2006. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223, 604–612. [DOI] [PubMed] [Google Scholar]
- Stenlid G. 1982. Cytokinins as inhibitors of root growth. Physiologia Plantarum 56, 500–506. [Google Scholar]
- Suge H. 1985. Ethylene and gibberellin: regulation of internodal elongation and nodal root development in floating rice. Plant and Cell Physiology 26, 607–614. [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Talboys PJ, Healey JR, Withers PJA, Jones DL. 2014. Phosphate depletion modulates auxin transport in Triticum aestivum leading to altered root branching. Journal of Experimental Botany 65, 5023–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten I, Schotte S, Geelen D. 2014. Hypocotyl adventitious root organogenesis differs from lateral root development. Frontiers in Plant Science 5, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoz ML, Loreti E, Mensuali A, Alpi A, Perata P. 2010. Hormonal interplay during adventitious root formation in flooded tomato plants. The Plant Journal 63, 551–562. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annual Review of Plant Biology 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Colmer TD, Pedersen O, Nakazono M. 2018. Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress. Plant Physiology 176, 1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Shimamura S, Nakazono M, Mochizuki T. 2013. Aerenchyma formation in crop species: a review. Field Crops Research 152, 8–16. [Google Scholar]
- Yamauchi T, Shiono K, Nagano M, et al. . 2015. Ethylene biosynthesis is promoted by very-long-chain fatty acids during lysigenous aerenchyma formation in rice roots. Plant Physiology 169, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Tanaka A, Mori H, Takamure I, Kato K, Nakazono M. 2016. Ethylene-dependent aerenchyma formation in adventitious roots is regulated differently in rice and maize. Plant, Cell and Environment 39, 2145–2157. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Watanabe K, Fukazawa A, Mori H, Abe F, Kawaguchi K, Oyanagi A, Nakazono M. 2014. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. Journal of Experimental Botany 65, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Yoshioka M, Fukazawa A, Mori H, Nishizawa NK, Tsutsumi N, Yoshioka H, Nakazono M. 2017. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. The Plant Cell 29, 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 35, 155–189. [Google Scholar]
- Yao Z, Liu L, Gao F, Rampitsch C, Reinecke DM, Ozga JA, Ayele BT. 2012. Developmental and seed aging mediated regulation of antioxidative genes and differential expression of proteins during pre- and post-germinative phases in pea. Journal of Plant Physiology 169, 1477–1488. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant Cell 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jin C, Sun C, Wang J, Ye Y, Zhou W, Lu L, Lin X. 2016. Inhibition of ethylene production by putrescine alleviates aluminium-induced root inhibition in wheat plants. Scientific Reports 6, 18888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Jiang J, Li J, Ma Q, Xu Y, Xue Y, Xu Z, Chong K. 2006. Over-expression of OsAGAP, an ARF-GAP, interferes with auxin influx, vesicle trafficking and root development. The Plant Journal 48, 581–591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.