Abstract
Non-coding RNAs are increasingly recognized not only as regulators of various biological functions but also as targets for a new generation of RNA therapeutics and biomarkers. We hereby review recent insights relating to non-coding RNAs including microRNAs (e.g. miR-126, miR-146a), long non-coding RNAs (e.g. MIR503HG, GATA6-AS, SMILR), and circular RNAs (e.g. cZNF292) and their role in vascular diseases. This includes identification and therapeutic use of hypoxia-regulated non-coding RNAs and endogenous non-coding RNAs that regulate intrinsic smooth muscle cell signalling, age-related non-coding RNAs, and non-coding RNAs involved in the regulation of mitochondrial biology and metabolic control. Finally, we discuss non-coding RNA species with biomarker potential.
This article is part of the Mini Review Series from the Varenna 2017 meeting of the Working Group of Myocardial Function of the European Society of Cardiology.
Keywords: Non-coding RNA, Vascular disease, Biomarker
1. Introduction
Manifestations of vascular diseases are the leading causes of morbidity and mortality.1 Endothelial dysfunction is a key initiator of vascular disease. Proliferation, migration, and the phenotype switches of smooth muscle cells are further hallmarks of vascular disease. Inflammatory cells aggravate vascular disease by release of secreted growth factors and cytokines, as well as cell/cell interactions that perpetuate the response to injury. Relatively recently, non-coding RNA has been discovered as new regulators of vascular function and angiogenesis. Non-coding RNAs include microRNAs (miRs, miRNAs, and short non-coding RNAs of about 20 nt length), long non-coding RNAs (lncRNAs) (length of >200 nt), and circular RNAs, a specific subtype of lncRNAs that form circular structures2 through back-splicing events. Herein, we focus on recent new insights how non-coding RNAs constitute regulatory therapeutic targets and biomarkers in vascular disease, with a special focus on cardiac disease-associated factors (e.g. hypoxia, ageing, smooth muscle cell biology, and metabolism).
2. Hypoxia-regulated non-coding RNAs
Hypoxia is a key trigger for angiogenic events and has a substantial impact on the non-coding transcriptome. Oxygen depletion alters endothelial expression of a wide range of lncRNAs, as indicated by next-generation RNA sequencing and microarray approaches in endothelial cells subjected to hypoxia.3 Validation experiments confirmed strong hypoxia-dependent activation of two intergenic lncRNAs (LINC00323 and MIR503HG). Silencing of these lncRNA transcripts led to angiogenic defects, including repression of growth factor signalling and/or the key endothelial transcription factor GATA2. Endothelial loss of these hypoxia-driven lncRNAs impaired cell-cycle control and inhibited capillary formation. The potential clinical importance of identified endothelial lncRNAs to vascular structural integrity was demonstrated in an ex vivo model of human-induced pluripotent stem cell-based engineered heart tissue showing that pharmacological inhibition of these lncNRAs impaired vascular structure appearance. Interestingly, research in the non-coding RNA field discovered the well-known endothelial (and protein-coding RNA) transcription factor GATA2 as a common target for many non-coding RNAs.4 GATA2 orchestrates the expression of many endothelial-specific genes, illustrating its crucial importance for endothelial cell function.5 In addition to being regulated through the actions of endothelial lncRNAs LINC00323 and MIR503HG, GATA2 was recently identified to be also a master switch for several key microRNAs. Using profiling approaches, the GATA2-dependent miR transcriptome was identified.6 Indeed, global miRNAnome-screening identified several GATA2-regulated miRNAs, including miR-126 and miR-221. Specifically, proangiogenic miR-126 was regulated by GATA2 transcriptionally and targeted anti-angiogenic SPRED1 and FOXO3a contributing to GATA2-mediated formation of normal vascular structures, whereas GATA2 deficiency led to vascular abnormalities. In contrast to GATA2 deficiency, supplementation with miR-126 normalized vascular function and expression profiles of cytokines contributing to proangiogenic paracrine effects. GATA2 silencing resulted in endothelial DNA hypomethylation leading to induced expression of anti-angiogenic miR-221 by GATA2-dependent demethylation of a putative CpG island in the miR-221 promoter. Mechanistically, a reverted GATA2 phenotype by endogenous suppression of miR-221 was mediated through direct proangiogenic miR-221 target genes ICAM1 and ETS1. Of therapeutic importance was the finding that in a mouse model of carotid injury with endothelial-specific repressed GATA2, systemic supplementation of miR-126-coupled nanoparticles enhanced miR-126 availability in the carotid artery. MiR-126 improved re-endothelialization of injured carotid arteries in vivo, thus proving a therapeutic strategy for treatment of GATA2-deficient vascular diseases.
An additional screen for hypoxia-regulated lncRNAs revealed that the long non-coding antisense transcript of GATA6 (GATA6-AS) is induced by hypoxia in endothelial cells as well.7 Silencing of GATA6-AS in endothelial cells in vitro diminished TGF-β2-induced endothelial–mesenchymal transition. Transplantation of GATA6-AS modulated human umbilical vein endothelial cells (HUVECs) via application of an antisense oligonucleotide (GapmeR) promoted the formation of human blood vessels in immune-deficient mice. Mechanistically, GATA6-AS interacted with the known deaminase LOXL2, which can remove activating H3K4me3 chromatin marks, and controlled a set of angiogenesis-related genes that are inversely regulated by LOXL2 and GATA6-AS silencing. Specifically, GATA6-AS silencing reduces H3K4me3 methylation of two of these genes, periostin and cyclooxygenase-2, suggesting that GATA6-AS acts as negative regulator of nuclear LOXL2 function. Interestingly—at least in endothelial cells in vitro—the levels of secreted LoxL2, which are known to regulate collagen cross-linking and are implicated in cardiac fibrosis,8 were not affected.
Non-coding RNAs are also molecular targets in therapeutic revascularization. It was recently demonstrated that endothelial cells can be derived via both directed differentiation and haematopoietic origin.9 Expression of the lncRNA SENCR, a lncRNA already known to be expressed in vascular smooth muscle cells (SMC),10 was up-regulated upon differentiation to endothelial cells, and manipulation of SENCR during differentiation affected endothelial cell appearance. Interestingly, SENCR modulation modified the angiogenic phenotype of endothelial cells, suggesting that lncRNAs have important regulatory functions for vascular cell types. Such studies are consistent with others in the field, assessing different lncRNAs in endothelial cells.11–13 Circulating levels of SENCR are also an independent predictor of diastolic function and remodelling in patients with Type 2 diabetes.14
Table 1.
Non-coding RNA associated biomarker studies
Apart from linear lncRNAs, circular forms of RNA species exist and are differentially regulated in the context of cardiovascular diseases.2,15 CircRNAs lack polyadenylation, are resistant to RNase R digestion, and localized to the cytoplasm. Boeckel et al.16 explored the expression and function of circular RNAs in endothelial cells. Using a modified computational analysis pipeline,17 RNA sequencing data of ribo-minus RNA from HUVECs cultured under normoxic or hypoxic conditions were analysed. cZNF292, cAFF1, and cDENND4C were shown to be up-regulated by hypoxia. Silencing of cZNF292 inhibited cZNF292 expression and reduced tube formation and spheroid sprouting of endothelial cells in vitro. Since circRNAs were previously suggested to act as microRNA sponges,17 the authors also explored whether this mechanism of action accounts for the biological function of endothelial circRNAs, by merging the RNA sequencing data with Argonaute HITS-CLIP data. Herein, the majority of circRNAs were shown to not possess a microRNA binding site, and only a small number have more than one binding site, suggesting that the majority of circRNAs are not acting as miRNA sponges, although this requires further exploration.
3. Cardiovascular ageing-associated non-coding RNAs
The role of ageing associated non-coding RNAs has been recently reviewed.18 Herein, we focus on novel lncRNAs that were not covered in the aforementioned review. The hypoxia-sensitive nuclear-localized lncRNA Meg3 was induced highly in endothelial cells of aged mice in vivo compared to controls and its levels correlate with ageing in human heart tissue.19In vitro, Meg3 was increased in replicative senescent HUVECs. Silencing of Meg3 using locked nucleic acid gapmeRs induced angiogenic sprouting and proliferation and repressed senescence as evidenced by the reduction of SA-β-galactosidase activity of endothelial cells in vitro. Conversely, lentiviral overexpression of Meg3 inhibited sprouting angiogenesis and cell-cycle progression, although splicing isoforms of Meg3 show differential effects. In vivo, silencing of Meg3 in aged mice using gapmeRs in combination with hind limb ischaemia significantly repressed Meg3 levels in the hind limb and increased recovery of perfusion compared to control mice. These results demonstrate that silencing Meg3 may be a potential strategy to reduce endothelial senescence and increase regenerative angiogenesis. Of note, Meg3 is also expressed in other non-endothelial cells such as cardiac fibroblasts and its silencing was effective in reducing cardiac fibrosis showing importance of this lncRNA broadly as a potential target in the treatment of cardiovascular diseases.20
Figure 1.
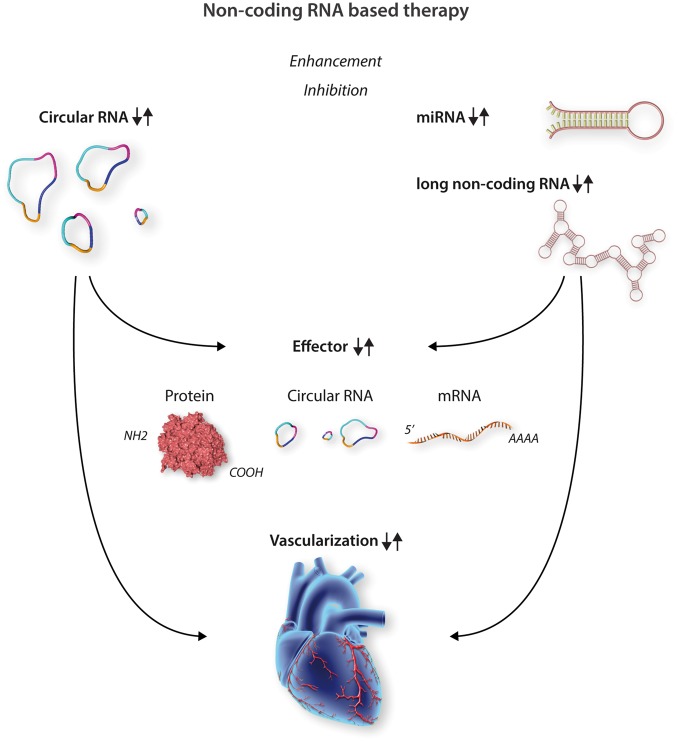
Non-coding RNAs as angiogenic therapeutic entry points. Enhancement or inhibition strategy can be followed for modulation of vascular non-coding RNAs. Circular RNA, miRNA, or long non-coding RNA are target structures for therapeutic intervention. Modulation of RNA subtypes triggers expression changes (up or down) of interacting effectors (e.g. proteins such as chromatin modifiers or ribosomal factors or different RNA species). Collectively cardiac vascularization is positively or negatively influenced dependent on the chosen non-coding RNA therapy.
A novel approach for aged patients with cardiovascular diseases using non-coding RNAs as targets was recently presented. Ageing populations show higher incidences of myocardial infarction (MI) and heart failure (HF). With regard to miRNAs, miR-22 was shown to be strongly increased during ageing in murine and human hearts and was identified as an abundant and strong inhibitor of cardioprotective autophagy.21 Inhibition of miR-22 in ageing cardiomyocytes activated autophagy and inhibited cellular hypertrophy. Pharmacological inhibition of miR-22 post-MI in older mice activated cardiac autophagy, prevented post-infarction remodelling, and improved cardiac function compared with control subjects. Interestingly, similar effects were less pronounced in younger mice with significantly lower cardiac miR-22 expression levels. In addition, circulating levels of miR-22 in 154 patients with systolic HF were highly associated with early mortality. Thus, miR-22 seems to be an important regulator of cardiac autophagy and a potential therapeutic target, especially in the older myocardium. Clearly, targeting therapeutics to the aged or diseased myocardium in human is challenging and requires sophisticated delivery strategies to be developed.
4. Non-coding RNAs in smooth muscle cell biology
The expression and function of lncRNA in smooth muscle cells remains relatively poorly defined. Aside from SENCR,10 a recent study identified a single transcript (three exons) lncRNA called SMILR (Smooth Muscle cell Induced LncRNA) that was activated following exposure of basal vSMC to pro-proliferative signals. Following exposure to a combination of platelet-derived growth factor and interleukin-1α, SMILR was induced.22 Interestingly, SMILR was localized both in the nuclear and cytoplasmic compartments, suggestive of differential modes of action within the cell. Further, an accurate lncRNA quantification assay for secretion from cells and human plasma samples was developed. Indeed, plasma levels of SMILR were elevated in patients with higher C-reactive protein levels compared with patients with lower levels, albeit in a small population sample set. Using a siRNA approach, an anti-proliferative effect following efficient down-regulation of SMILR was identified, with effects on the neighbouring gene HAS2. Further, levels of SMILR were higher in patients with advanced atherosclerosis compared with stable patient samples, suggesting relevance of human disease. These studies clearly show the importance of lncRNA expression on function of vSMC. Since vSMC are centrally important in vascular health and disease, this suggests a much greater understanding of both required and essential lncRNA characteristics. Several questions remain regarding the function of SMILR. These include the mode of action with respect to vSMC proliferation. Further, how this is consistent across vascular beds and vasculoproliferative diseases, as well as refining the therapeutic potential of SMILR inhibition to block proliferation. Notably, in the context of vein graft failure, an anti-proliferative strategy would provide likely efficacy when considering the predominant role of vSMC in vein graft neointima formation. Other studies have also demonstrated the importance of lncRNA in vSMC function,23,24 consistent with the notion that they hold important regulatory potential in vascular health and disease.
Figure 2.
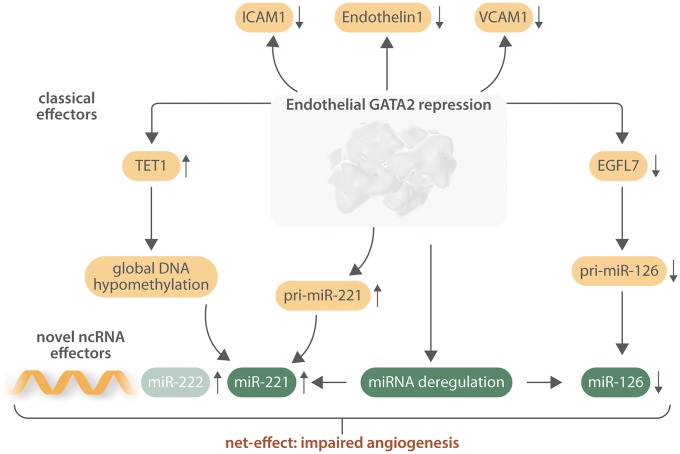
GATA2 as a central player for angiogenic non-coding RNAs. Besides the regulation of GATA2-dependent coding genes (e.g. ICAM1, VCAM1), endothelial transcription factor GATA2 directly controls transcription of miR-126/miR-221 locus. Interestingly, loss of GATA2 causes DNA hypomethylation thereby activating miR-221 expression. Next to that, GATA2 repression lowers miR-126 expression levels causing an up-regulation of anti-angiogenic factors. Overall, downstream modulation leads to anti-angiogenic outcome and imbalanced endothelial cell biology. Modified from Ref.6
5. Non-coding RNAs in the control of mitochondrial function and energy metabolism
Recently, miRNAs emerged as central regulators of mitochondrial function and energy metabolism in diabetes, hypertension, ischaemia, atherosclerosis, and cardiotoxicity. Mitochondrial miRNAs—also mitomiRs—are enriched in those diseases. In diabetes mellitus, mitomiRs are enriched in spatially distinct compartments,25 whereas in hypertensive-hearts their expression differs in the early and later stage of HF.26 In general, miR-146a, miR-181c, and miR-378 act as important therapeutic targets affecting mitochondrial function in cardiovascular diseases. MiR-146a was first reported to affect cardiac metabolism during peripartum cardiomyopathy.27 Uptake by cardiomyocytes of endothelial cell-released miR-146a decreased the metabolic activity of cardiomyocytes during pregnancy, with down-regulation of Erbb4, Notch1, and Irak1. Inhibition of miR-146a is thereby protective. Its suppression is not only beneficial in peripartum cardiomyopathy, but also in pressure overload-induced cardiomyopathy,28 and in atherosclerosis.29 Inhibition of miR-146a in pressure overload—either with aortic banding or with angiotensin-II infusion-blunted the cardiac hypertrophic response and protected against systolic dysfunction.29 MiR-146a decreased dihydrolipoyl succinyl transferase (DLST) levels, a rate-controlling enzyme in the tricarboxyl acid cycle in the failing heart, thereby impairing cardiac oxidative metabolism. Both fatty acid and glucose oxidation decreased upon pressure overload in wild-type mice, but were preserved upon miR-146a inhibition. Increase in DLST upon loss of miR-146a helped to preserve these oxidative fluxes, protecting against maladaptive hypertrophy, and dysfunction. As in peripartum cardiomyopathy27 and in atherosclerosis,29 miR-146a seems to be mainly derived from endothelial cells. In line, also in atherosclerosis, deficiency of miR-146a in those endothelial cells tempered the chronic inflammatory response to the atherogenic high-fat diet, thereby protecting against atheroma formation.29 MiR-181c is another detrimental mitomiR involved in mitochondrial function. Its inhibition increases Bcl2, a key-player in mitochondrial apoptosis and morphology, and thereby protects against cardiomyocyte apoptosis in vitro.30 In doxorubicin-induced toxicity in vivo, miR-181c inhibition decreases the reactive oxygen species production and reduces basal mitochondrial respiration.31 In ischaemic hearts, miR-181c targets mitochondrial COX1, and its deficiency thereby resulted in decreased infarct size, emphasizing the overall cardio-protective effect of miR-181c inhibition. Further, presence of miR-378 attenuated ischaemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes32 and blunted cardiac hypertrophy and dysfunction upon cardiac overload by targeting Ras signalling.33 In the diabetic heart, antagomiR blockade of this mitomiR-378 increased ATP6 protein production and thereby also improved cardiac function.25 In a human infarct study, miR-378 modulated the proangiogenic capacity of CD34+ progenitor cells after MI, with clear stimulatory effects on endothelial cells as confirmed in vitro and in vivo.34
In conclusion, diverse mitomiRs modulate mitochondrial function in cardiovascular diseases caused by ischaemia, the metabolic syndrome—diabetes, hypertension, and hyperlipidaemia—and cardiotoxic agents. Although inhibition of the mitomiRs-146a is beneficial in hypertensive and peripartum cardiomyopathy, and in atherosclerotic disease, and inhibition of miR-181c in ischaemic and toxic cardiomyopathy, the presence of miR-378 is needed to protect against cardiac dysfunction caused by ischaemic injury and maladaptive hypertrophy.
6. Circulating microRNAs as novel cardiovascular biomarkers
Previous studies have highlighted the presence of endogenous circulating miRNAs that are not cell-associated. Zampetaki et al.35,36 have performed the first systematic analysis of circulating miRNAs in a large community-based study and revealed a diagnostic potential of miRNA changes associated with Type 2 diabetes and cardiovascular disease. In subsequent studies, it has become apparent that platelets have abundant amounts of miRNAs,37 and that circulating miRNAs reflect platelet activation.38,39 As platelets are anucleate and do not perform transcription, it was initially thought that circulating miRNAs are unlikely to be platelet-derived. However, surprisingly many abundant plasma and serum miRNAs, including miRNAs like miR-126 that were previously thought to be endothelial specific, can originate from platelets.37–39 YRNAs is another species of circulating non-coding RNAs that is platelet-derived.39 There still remains a gap in our understanding of how changes in platelet biology relate to circulating miRNAs. In contrast, miR-122 is a liver-specific miRNA that is readily detectable in the circulation.40 Notably, circulating levels of miR-122 are strongly associated with the risk of developing metabolic syndrome.41 The presence of circulating tissue-derived miRNAs provides the possibility of a cross-organ communication by miRNAs.
7. Conclusion and outlook
The discovery of miRNAs and other non-coding RNAs such as lncRNAs and circRNAs that are involved in transcriptional and other functional regulation of the vasculature have transformed our understanding of biological processes and disease development, especially in cardiovascular diseases. This might lead to new therapeutics and diagnostics. The non-coding RNAome offers promising opportunities for treating and assessing cardiovascular disease, but many obstacles still need to be overcome. A major point to address in therapeutic use of non-coding RNAs is to develop tailored drug delivery with e.g. heart specificity. Next to that, lncRNA and circRNAs are relatively new areas of research, thus it is paramount to better understand their biological function.
Conflict of interest: T.T. filed and licensed patents about non-coding RNAs. T.T. holds shares of Cardior Pharmaceuticals GmbH. A.H.B. filed a patent on the lncRNA SMILR. M.M. filed and licensed patents on miRNAs as cardiovascular biomarkers.
Funding
This study was supported by the German Ministry for Education and Research (IFB-Tx to T.T., 01EO1302) and the Leducq Fondation (MIRVAD); British Heart Foundation Chair of Translational Cardiovascular Sciences and a European Research Council Advanced grant (VASCMIR) (to A.H.B.). M.M. is a British Heart Foundation (BHF) Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/16/14/32397), support by the National Institute of Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London in partnership with King’s College Hospital, and an excellent initiative [Competence Centers for Excellent Technologies (COMET)] of the FFG (Austrian Research Promotion Agency): Research Center of Excellence (K-Project Nr. 843536) funded by BMVIT (Federal Ministry for Transport, Innovation and Technology), BMWFW (federal Ministry of Science, Research and Economy), the Wirtschaftsagentur Wien, and Standortagentur Tirol.
References
- 1. GBD 2013 Mortality and Causes of Death Collaborators . Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beermann J, Piccoli MT, Viereck J, Thum T.. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- 3. Fiedler J, Breckwoldt K, Remmele CW, Hartmann D, Dittrich M, Pfanne A, Just A, Xiao K, Kunz M, Muller T, Hansen A, Geffers R, Dandekar T, Eschenhagen T, Thum T.. Development of long noncoding RNA-based strategies to modulate tissue vascularization. J Am Coll Cardiol 2015;66:2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, Galuppo P, Kneitz S, Pena JT, Sohn-Lee C, Loyer X, Soutschek J, Brand T, Tuschl T, Heineke J, Martin U, Schulte-Merker S, Ertl G, Engelhardt S, Bauersachs J, Thum T.. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011;124:720–730. [DOI] [PubMed] [Google Scholar]
- 5. Thum T, Haverich A, Borlak J.. Cellular dedifferentiation of endothelium is linked to activation and silencing of certain nuclear transcription factors: implications for endothelial dysfunction and vascular biology. FASEB J 2000;14:740–751. [DOI] [PubMed] [Google Scholar]
- 6. Hartmann D, Fiedler J, Sonnenschein K, Just A, Pfanne A, Zimmer K, Remke J, Foinquinos A, Butzlaff M, Schimmel K, Maegdefessel L, Hilfiker-Kleiner D, Lachmann N, Schober A, Froese N, Heineke J, Bauersachs J, Batkai S, Thum T.. MicroRNA-based therapy of GATA2-deficient vascular disease. Circulation 2016;134:1973–1990. [DOI] [PubMed] [Google Scholar]
- 7. Neumann P, Jae N, Knau A, Glaser SF, Fouani Y, Rossbach O, Kruger M, John D, Bindereif A, Grote P, Boon RA, Dimmeler S.. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun 2018;9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang J, Savvatis K, Kang JS, Fan P, Zhong H, Schwartz K, Barry V, Mikels-Vigdal A, Karpinski S, Kornyeyev D, Adamkewicz J, Feng X, Zhou Q, Shang C, Kumar P, Phan D, Kasner M, López B, Diez J, Wright KC, Kovacs RL, Chen P-S, Quertermous T, Smith V, Yao L, Tschöpe C, Chang C-P.. Targeting LOXL2 for cardiac interstitial fibrosis and heart failure treatment. Nat Commun 2016;7:13710.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, Brittan M, Hunter A, McBride M, McClure J, Miano JM, Emanueli C, Mills NL, Mountford JC, Baker AH.. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther 2016;24:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM.. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 2014;34:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S.. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389–1397. [DOI] [PubMed] [Google Scholar]
- 12. Uchida S, Dimmeler S.. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737–750. [DOI] [PubMed] [Google Scholar]
- 13. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q.. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143–1156. [DOI] [PubMed] [Google Scholar]
- 14. de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T.. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep 2016;6:37354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta SK, Garg A, Bar C, Chatterjee S, Foinquinos A, Milting H, Streckfuss-Bomeke K, Fiedler J, Thum T.. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res 2018;122:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S.. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res 2015;117:884–890. [DOI] [PubMed] [Google Scholar]
- 17. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 18. Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW 2nd, Thum T, Heymans S; Cardiolinc network. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol 2015;12:415–425. [DOI] [PubMed] [Google Scholar]
- 19. Boon RA, Hofmann P, Michalik KM, Lozano-Vidal N, Berghäuser D, Fischer A, Knau A, Jaé N, Schürmann C, Dimmeler S.. Long noncoding RNA Meg3 controls endothelial cell aging and function: implications for regenerative angiogenesis. J Am Coll Cardiol 2016;68:2589–2591. [DOI] [PubMed] [Google Scholar]
- 20. Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T.. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res 2017;121:575–583. [DOI] [PubMed] [Google Scholar]
- 21. Gupta SK, Foinquinos A, Thum S, Remke J, Zimmer K, Bauters C, de Groote P, Boon RA, de Windt LJ, Preissl S, Hein L, Batkai S, Pinet F, Thum T.. Preclinical development of a microRNA-based therapy for elderly patients with myocardial infarction. J Am Coll Cardiol 2016;68:1557–1571. [DOI] [PubMed] [Google Scholar]
- 22. Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, Joshi N, Dweck MR, Miano JM, McBride MW, Newby DE, McDonald RA, Baker AH.. Smooth Muscle Enriched Long Noncoding RNA (SMILR) regulates cell proliferation. Circulation 2016;133:2050–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C.. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014;130:1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, Schones DE, Natarajan R.. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 2013;113:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE, Martinez I, Hollander JM.. Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart. Circ Cardiovasc Genet 2015;8:785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Song C, Zhou X, Han X, Li J, Wang Z, Shang H, Liu Y, Cao H.. Mitochondria associated microRNA expression profiling of heart failure. Biomed Res Int 2017;2017:4042509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Halkein J, Tabruyn SP, Ricke-Hoch M, Haghikia A, Nguyen NQ, Scherr M, Castermans K, Malvaux L, Lambert V, Thiry M, Sliwa K, Noel A, Martial JA, Hilfiker-Kleiner D, Struman I.. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest 2013;123:2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heggermont WA, Papageorgiou AP, Quaegebeur A, Deckx S, Carai P, Verhesen W, Eelen G, Schoors S, van Leeuwen R, Alekseev S, Elzenaar I, Vinckier S, Pokreisz P, Walravens AS, Gijsbers R, Van Den Haute C, Nickel A, Schroen B, van Bilsen M, Janssens S, Maack C, Pinto Y, Carmeliet P, Heymans S.. Inhibition of microRNA-146a and overexpression of its target dihydrolipoyl succinyltransferase protect against pressure overload-induced cardiac hypertrophy and dysfunction. Circulation 2017;136:747–761. [DOI] [PubMed] [Google Scholar]
- 29. Cheng HS, Besla R, Li A, Chen Z, Shikatani EA, Nazari-Jahantigh M, Hammoutene A, Nguyen MA, Geoffrion M, Cai L, Khyzha N, Li T, MacParland SA, Husain M, Cybulsky MI, Boulanger CM, Temel RE, Schober A, Rayner KJ, Robbins CS, Fish JE.. Paradoxical suppression of atherosclerosis in the absence of microRNA-146a. Circ Res 2017;121:354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H, Li J, Chi H, Zhang F, Zhu X, Cai J, Yang X.. MicroRNA-181c targets Bcl-2 and regulates mitochondrial morphology in myocardial cells. J Cell Mol Med 2015;19:2084–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Das S, Kohr M, Dunkerly-Eyring B, Lee DI, Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA, Steenbergen C.. Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J Am Heart Assoc 2017;6:e004694.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, Ren AJ, Yuan WJ, Lin L.. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 2012;17:410–423. [DOI] [PubMed] [Google Scholar]
- 33. Nagalingam RS, Sundaresan NR, Gupta MP, Geenen DL, Solaro RJ, Gupta M.. A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J Biol Chem 2013;288:11216–11232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Templin C, Volkmann J, Emmert MY, Mocharla P, Müller M, Kraenkel N, Ghadri J-R, Meyer M, Styp-Rekowska B, Briand S, Klingenberg R, Jaguszewski M, Matter CM, Djonov V, Mach F, Windecker S, Hoerstrup SP, Thum T, Lüscher TF, Landmesser U.. Increased proangiogenic activity of mobilized CD34+ progenitor cells of patients with acute ST-segment-elevation myocardial infarction: role of differential microRNA-378 expression. Arterioscler Thromb Vasc Biol 2017;37:341–349. [DOI] [PubMed] [Google Scholar]
- 35. Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M.. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817. [DOI] [PubMed] [Google Scholar]
- 36. Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S, Mayr M.. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290–299. [DOI] [PubMed] [Google Scholar]
- 37. Sunderland N, Skroblin P, Barwari T, Huntley RP, Lu R, Joshi A, Lovering RC, Mayr M.. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ Res 2017;120:418–435. [DOI] [PubMed] [Google Scholar]
- 38. Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I, Langley SR, Sivaprasad S, Markus HS, Mitchell JA, Warner TD, Kiechl S, Mayr M.. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res 2013;112:595–600. [DOI] [PubMed] [Google Scholar]
- 39. Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, Sunderland NP, Willeit K, Morton AC, Armstrong PC, Chan MV, Lu R, Yin X, Gracio F, Dudek K, Langley SR, Zampetaki A, de Rinaldis E, Ye S, Warner TD, Saxena A, Kiechl S, Storey RF, Mayr M.. Association of microRNAs and YRNAs with platelet function. Circ Res 2016;118:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Willeit P, Skroblin P, Kiechl S, Fernandez-Hernando C, Mayr M.. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Eur Heart J 2016;37:3260–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramirez CM, Goedeke L, Rotllan N, Bonora E, Hughes AD, Santer P, Fernandez-Hernando C, Tilg H, Willeit J, Kiechl S, Mayr M.. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017;66:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]