Abstract
Homologous glycosyltransferases GTA and GTB perform the final step in human ABO(H) blood group A and B antigen synthesis by transferring the sugar moiety from donor UDP-GalNAc/UDP-Gal to the terminal H antigen disaccharide acceptor. Like other GT-A fold family 6 glycosyltransferases, GTA and GTB undergo major conformational changes in two mobile regions, the C-terminal tail and internal loop, to achieve the closed, catalytic state. These changes are known to establish a salt bridge network among conserved active site residues Arg188, Asp211 and Asp302, which move to accommodate a series of discrete donor conformations while promoting loop ordering and formation of the closed enzyme state. However, the individual significance of these residues in linking these processes remains unclear. Here, we report the kinetics and high-resolution structures of GTA/GTB mutants of residues 188 and 302. The structural data support a conserved salt bridge network critical to mobile polypeptide loop organization and stabilization of the catalytically competent donor conformation. Consistent with the X-ray crystal structures, the kinetic data suggest that disruption of this salt bridge network has a destabilizing effect on the transition state, emphasizing the importance of Arg188 and Asp302 in the glycosyltransfer reaction mechanism. The salt bridge network observed in GTA/GTB structures during substrate binding appears to be conserved not only among other Carbohydrate Active EnZyme family 6 glycosyltransferases but also within both retaining and inverting GT-A fold glycosyltransferases. Our findings augment recently published crystal structures, which have identified a correlation between donor substrate conformational changes and mobile loop ordering.
Keywords: glycosyltransferases, human ABO(H) blood group enzymes, X-ray crystallography
Introduction
The observed induction of anaphylaxis and death by mismatched blood transfusions (Milland and Sandrin 2006) led Landsteiner to discover the ABO(H) blood groups at the dawn of the 20th century (Landsteiner 1901). The human ABO(H) blood group antigens were eventually determined to be cell surface oligosaccharides conjugated to membrane glycoproteins and glycolipids present on many endothelial and epithelial tissues, including erythrocytes, and are also found as free oligosaccharides in the saliva, mucous, tears and other bodily fluids of “secretor-type” individuals (Henry et al. 1990). The ABO blood group is known widely for its involvement in or connection to numerous human diseases, including cholera, cardiovascular disorders (e.g. coronary heart disease, venous thromboembolism and von Willebrand disease) and cancers (e.g. gastric and pancreatic cancers) (Yamamoto et al. 2012; Liumbruno and Franchini 2013; Rummel and Ellsworth 2016).
Two homologous Carbohydrate Active EnZyme (CAZy) (Coutinho and Henrissat 1999; Lombard et al. 2014) family 6 glycosyltransferases (GTs) are involved in the final step of A and B antigen synthesis, where they catalyze the glycosyltransfer of GalNAc or Gal, respectively, to the H antigen acceptor terminal disaccharide (α-l-Fucp-(1→2)-β-d-Galp-O-R, where R is a glycolipid or glycoprotein; HA). The α-(1→3)-N-acetylgalactosaminyltransferase GTA (EC 2.4.1.40) transfers GalNAc from UDP-GalNAc to HA to produce the blood type A determinant (α-l-Fuc-(1→2)-[α-d-GalNAc-(1→3)]-β-d-Gal-O-R), while the α-(1→3)-galactosyltransferase GTB (EC 2.4.1.37) transfers Gal from UDP-Gal to HA to produce the blood type B determinant (α-l-Fuc-(1→2)-[a-d-Gal-(1→3)]-β-d-Gal-O-R). Interestingly, this means that the A and B antigen determinants, whose mismatch in a blood transfusion can be fatal, differ only in the C2 substituent of the terminal sugar, where A has an acetamido group and B has a hydroxyl (Hearn et al. 1968; Kobata et al. 1968, Yamamoto and Hakomori 1990; Yamamoto et al. 1990; Patenaude et al. 2002). Blood type O individuals typically have a mutant form of the gene encoding GTA and/or GTB and cannot express functional enzyme, and so biosynthesis halts at the H antigen.
GTA and GTB differ in only four “critical” amino acid residues out of 354 (GTA/GTB: Arg/Gly176, Gly/Ser235, Leu/Met266, Gly/Ala268) (Yamamoto and Hakomori 1990; Yamamoto et al. 1990) and are therefore the most homologous reported GTs that use distinct naturally occurring donor substrates. Of the four amino acid differences between GTA and GTB, only Leu/Met266 and Gly/Ala268 participate in donor sugar recognition (Yamamoto and Hakomori 1990; Seto et al. 1997,1999; Kamath et al. 1999; Patenaude et al. 2002; Monegal and Planas 2006), while Gly/Ser235 has been shown to affect acceptor binding (Patenaude et al. 2002; Letts et al. 2006), and Arg/Gly176 can influence internal loop ordering (Lee et al. 2005; Alfaro et al. 2008). Notably, GTA and GTB are two of the most-studied GTs with respect to structural characterization, mutagenesis and enzyme dynamics investigations. Much of this work utilized GTA/GTB chimeras described using four-letter codes where each letter corresponds to one of the four critical residues such that AAAA represents GTA, BBBB represents GTB and AABB represents a chimera with the first two critical residues of GTA and the last two critical residues of GTB.
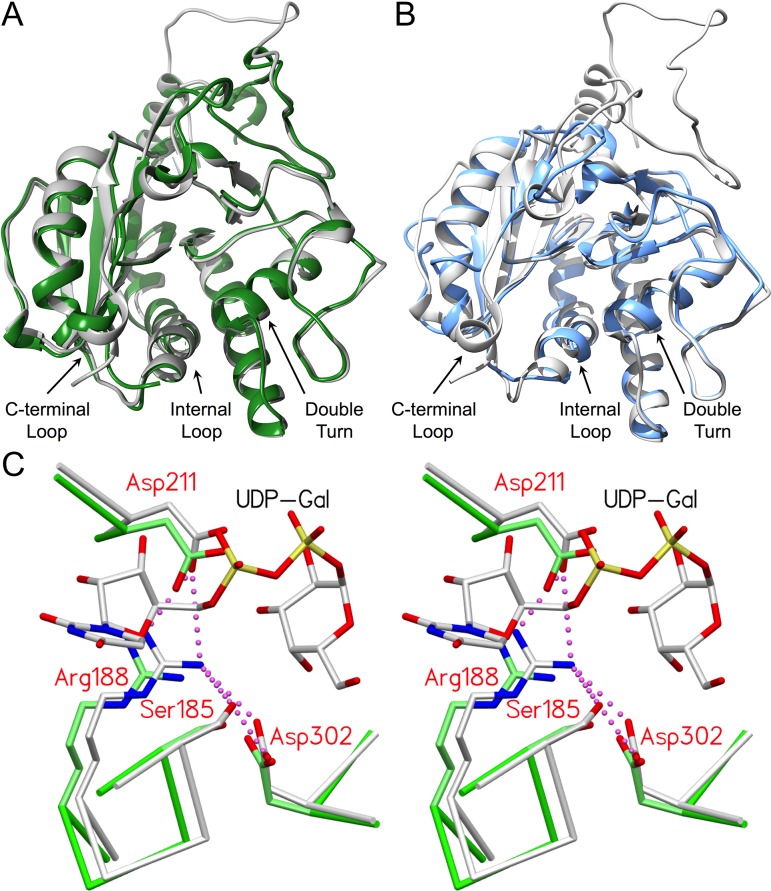
GTs are broadly classified based on reaction stereochemistry and fold type. GTA and GTB perform glycosyltransfer with conservation of the donor sugar α-linkage in the product and are classified as “retaining” enzymes. In contrast to inverting enzymes, the mechanism for this class of GTs remains uncertain. GTA and GTB have a GT-A fold type, which is characterized by two adjacent Rossmann fold-like domains and a conserved DXD motif within the active site. Additionally, they possess regions of mobile polypeptide that recognize and sequester substrate during catalysis and exhibit considerable structural homology with other family 6 enzymes, as shown in Figure 1 (Gastinel et al. 2001; Persson et al. 2001a; Patenaude et al. 2002; Qasba et al. 2005; Yazer and Palcic 2005; Letts et al. 2007; Alfaro et al. 2008; Schuman et al. 2010). Loop organization around substrate during catalysis is found in both retaining and inverting GTs, with the latter exemplified by β-1,4-galactosyltransferase I (β4Gal-T1; EC 2.4.1.38) (Qasba et al. 2002).
Fig. 1.
GTA and GTB are structurally homologous with family 6 GTs α3GalT and BoGT6a and share a number of conserved, equivalent residues critical to forming and stabilizing the salt bridge network. (A) Overlap of α3GalT (green, PDB code 1G93) with the catalytically competent closed state of the chimeric enzyme AABB (gray; PDB code 2RJ7; 0.77 Å RMSD). (B) Overlap of BoGT6a (blue; PDB code 4AYJ; 1.08 Å RMSD) with AABB (gray; PDB code 2RJ7). (C) Stereoview of the superposition of closed chimeric enzyme AABB (gray; PDB code 2RJ7) and α3GT (green; PDB code 1G93) showing the equivalent positions of active site residues Ser185/199, Arg188/202, Asp211/225 and Asp302/316 (GTB side chains labeled) with UDP-Gal bound in the catalytically competent “tucked under” conformation #1. Salt bridge interactions are depicted as magenta dashed spheres. Distances for all interactions are in Tables IV and V.
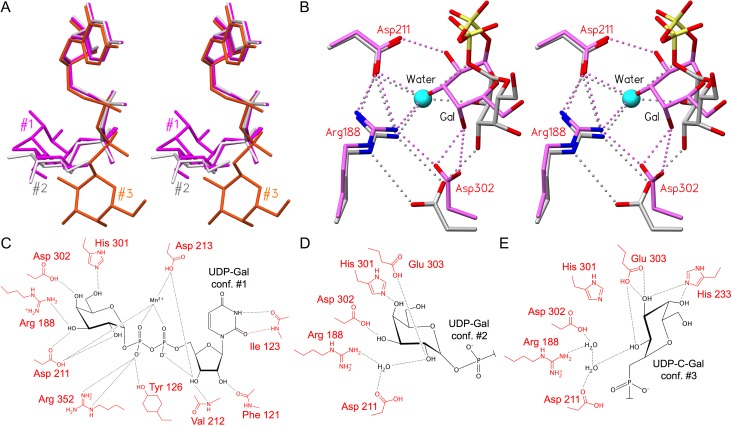
The mobile internal loops (aa 176–195) and C-terminal tails (aa 345–354) of the wild-type and mutant enzymes are observed to transition from a disordered open state to a highly ordered, catalytically competent closed state when both donor and acceptor substrates are bound (Patenaude et al. 2002; Alfaro et al. 2008; Gagnon et al. 2015). Recently, it was shown that GTA and GTB can recognize and bind lower energy donor conformations, as identified in solution (Angulo et al. 2006; Blume et al. 2006) and shift them through a series of discrete conformations of comparable free energy (Figure 2A), each stabilized by a set of hydrogen bonds, to the “tucked under” conformation associated with catalysis (Gagnon et al. 2015) (Figure 2B–E).
Fig. 2.
GTA and GTB donors adopt the same series of intermediate and catalytic conformations, each with a distinct set of hydrogen bonds. (A) Overlap of three donor conformations with tucked under conformation #1 in magenta (PDB code 2RJ7), intermediate conformation #2 in light gray (PDB code 5C1L) and “extended” conformation #3 in orange (PDB code 5C3D) (Alfaro et al. 2008; Gagnon et al. 2015). (B) Stereoview of the superposition of AABB (PDB code 2RJ7; conformation #1; magenta) and GTB (PDB code 5C1L; conformation #2; carbon atoms colored light gray) crystal structures depicting salt bridge and hydrogen bond interactions among Arg188, Asp211, Asp302, a water molecule (cyan sphere) and donor galactose. Interactions include a single/bidentate salt bridge between Arg188 and Asp302. In donor conformation #2, a water molecule occupies the same position as donor Gal-O3 in conformation #1. Except where otherwise indicated, atoms are colored by element with oxygen red, nitrogen blue and phosphorous yellow. (C–E) Schematic depictions of hydrogen bond interactions (black lines) between (C) UDP-Gal in tucked under conformation #1 and chimeric enzyme AABB (PDB code 2RJ7), (D) the substrate Gal moiety in the intermediate conformation #2 (PDB code 5C1L; GTB) and (E) in the extended conformation #3 (PDB code 5C3D; ABBB). Key active site resides are labeled in red and the donor sugar in black.
Several active site residues conserved in family 6 enzymes have been observed to make the same pattern of hydrogen bond contacts to the donor sugar in both the intermediate and tucked under conformations (Figure 2B), among these Arg188, Asp211 and Asp302 in GTA/GTB (Gagnon et al. 2015). Family 6 GT β-galactosyl α-1,3-galactosyltransferase (α3GT; EC 2.4.1.87) from Bos taurus (Zhang et al. 2003) possesses active site residues Ser199, Arg202, Asp225 and Asp316, which correspond to GTA/GTB residues Ser185, Arg188, Asp211 and Asp302, respectively (Figure 1C), and form the corresponding hydrogen bond contacts to the glycosyl moiety of the UDP-Gal donor in both GTs. A fourth GT, Bacteroides ovatus BoGT6a (EC 2.4.1.40), a metal-independent enzyme with an NXN motif instead of the DXD motif typically observed for family 6 GTs (Tumbale and Brew 2009) possesses corresponding residues Thr70, Arg73, Asn95 and Asp191 and displays similar interactions (Tumbale and Brew 2009; Thiyagarajan et al. 2012; Pham et al. 2014).
Given the central function this set of conserved residues plays in family 6 GTs, a detailed investigation into their contribution to structure and function is overdue. Here, we explore the kinetic and structural contributions of Arg188 and Asp302 using mutant enzymes GTB/R188K, GTA/D302A, GTA/D302C, GTA/D302L, GTB/D302A, GTB/D302C, GTB/D302E and GTB/D302L.
Results
Kinetics
Kinetic constants were determined for each purified mutant at a high concentration of the alternate substrate and are presented in Table I alongside published kinetic data for bovine α3GT (Zhang et al. 2003) All mutants had decreased activity compared to the corresponding wild-type enzymes. GTA/GTB R188K, D302A and D302L mutants exhibited nearly complete loss of activity. The GTB/D302A and GTB/R188K mutants had elevated Km values for UDP-Gal donor. The GTB/D302E mutant had the highest activity of the 302 mutants with a kcat that was 47% that of wild-type GTB. The Km for UDP-Gal donor was elevated with no change in the Km for acceptor. GTA/D302C and GTB/D302C kcat values were 9.1 and 9.0% of wild type, respectively (Table I). The Km values for donor and acceptor substrates were elevated for both D302C mutants.
Table I.
Kinetic constants for GTA/GTB wild-type and Asp302 & Arg188 mutants alongside kinetic constants for chimeric enzyme AABB and Asp316 mutants of bovine α3GT
| Enzyme/donor | GTA/UDP-GalNAc | GTB/UDP-Gal | AABB/UDP-GalNAc | AABB/UDP-Gal | α3GT/UDP-Gal | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KAcc (μM)a | KDon (μM)a | kcat (s−1) | KAcc (μM)a | KDon(μM)a | kcat (s−1) | KAcc (μM)b | KDon (μM)b | kcat (s−1) | KAcc (μM)b | KDon (μM)b | kcat (s−1) | KAcc (mM) | KDon (μM) | kcat (s−1) | |
| Wild type | 9.9 | 8.7 | 17.5 | 88 | 27 | 5.1 | 13 | 37 | 0.60 | 1.6 | 1.8 | 2.2 | 19.9c | 430c | 6.4c |
| 12.5d | 1.3d | ||||||||||||||
| D302A/D316A | LAf | LA | LA | 117 ± 6g | 170 ± 40 | 0.002 ± 0.0001 | – | – | – | – | – | – | – | 70d | 0.0013d |
| D302E/D316E | – | – | – | 100 ± 20 | 112 ± 18 | 2.4 ± 0.5 | – | – | – | – | – | – | 6.4e | 160e | 2.48e |
| D316N | – | – | – | – | – | – | – | – | – | – | – | – | Inactivee | ||
| D302C | 45 ± 5 | 22 ± 2 | 1.6 ± 0.4 | 332 ± 24 | 53 ± 4 | 0.46 ± 0.02 | – | – | – | – | – | – | – | – | – |
| D302L | LA | LA | LA | LA | LA | LA | – | – | – | – | – | – | – | – | – |
| R188K | – | – | – | 101 ± 20 | 140 ± 30 | 0.0006 ± 0.0006 | – | – | – | – | – | – | – | – | – |
| R188S | – | – | – | – | – | LAh | – | – | – | – | – | – | – | – | – |
aThe KAcc and KDon Michaelis–Menten constants are the Km for acceptor and donor, respectively.
bAs reported by Alfaro et al. (2008).
cAs reported by Zhang et al. (2003).
dAs reported by Monegal and Planas (2006).
eAs reported by Tumbale et al. (2008).
fLA = low activity enzymes with activities measured as less than 1/10,000 of wild-type enzyme.
gKm errors are from curve fitting while for kcat they are the average of kcats from donor and acceptor kinetics.
hAs reported inby Yazer and Palcic (2005).
*Boxes contain dashes when the specified mutation does not apply to a given enzyme. For example, Asp316 is a residue belonging to α3GT (not GTA/GTB or AABB), and so the kinetics for the D316N mutant are displayed appropriately for α3GT but not GTA, GTB or AABB.
Structure determination
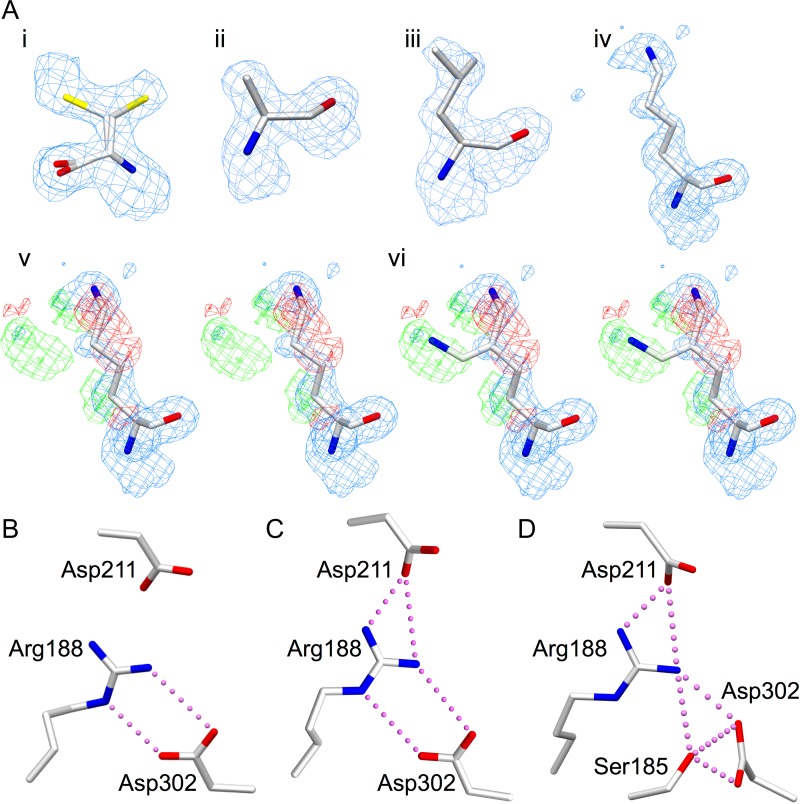
One GTA mutant and four GTB mutant enzymes formed diffraction-quality crystals: GTA/D302C, GTB/D302A, GTB/D302C, GTB/D302L and GTB/R188K. Data collection and refinement results for these mutant enzymes are summarized in Table II. Diffraction data were collected to a maximum resolution of 2.18–1.45 Å, with a final Rwork of 18.4–20.1% and Rfree of 20.2–24.6%. With the exception of disorder between atoms Cε and Nζ in the R188K structure, there is unambiguous electron density about the mutation sites (Figure 3A). While the internal loop and C-terminal tail were largely disordered, none of the mutant structures display significant perturbation of main chain or side chain polypeptide electron density from the wild-type structure. Appropriate electron density is observed for the mutant side chain in each Asp302 and Arg188 mutant structure (Figure 3A). Cys302 is disordered over two side chain conformations in both GTA/D302C and GTB/D302C structures (Figure 3A). The degree of internal loop disorder varied across all structures (Table III), while the C-terminal tail was uniformly disordered. The loop disorder observed in the mutant structures is similar to unliganded wild-type GTB (PDB code 1LZ7), and superposition of the ordered residues of 1LZ7 with any one of the structures reported here gives RMSD displacement values under 0.30 Å.
Table II.
Data collection and refinement statistics for the mutant enzymes. All crystals belong to space group C2221 with approximate unit cell dimensions of a = 52.6, b = 150.2, and c = 78.9 Å
| GTA/D302C | GTB/D302A | GTB/D302C | GTB/D302L | GTB/R188K | |
|---|---|---|---|---|---|
| Resolution (Å) | 20–1.54 | 20–1.45 | 20–2.18 | 20–1.69 | 20–1.45 |
| Rsym (%)a,b | 3.8 (32.0) | 2.9 (30.3) | 8.8 (29.4) | 4.6 (32.0) | 3.1 (28.3) |
| Completeness (%)b | 98.7 (100.0) | 99.5 (100.0) | 95.3 (98.8) | 98.3 (100.0) | 96.6 (96.2) |
| Unique reflections | 46,171 | 55,649 | 17,135 | 34,748 | 53,903 |
| Multiplicity | 4.44 | 4.28 | 4.68 | 4.71 | 3.84 |
| <I/σ(I)> | 17.2 | 21.1 | 9.1 | 15.1 | 21 |
| Refinement | |||||
| Reflections used in refinement | 43,837 | 52,822 | 16,261 | 32,996 | 51,167 |
| Rfree reflections | 2329 | 2825 | 874 | 1749 | 2733 |
| Wilson B-factor (Å2) | 20.4 | 21.7 | 32.1 | 22.1 | 20.4 |
| Rwork (%)c | 19.5 | 18.6 | 20.1 | 18.4 | 18.9 |
| Rfree (%)c,d | 21.9 | 20.2 | 24.6 | 22.2 | 20.5 |
| No. non-hydrogen protein atoms | 2247 | 2154 | 2139 | 2229 | 2269 |
| No. water molecules | 270 | 266 | 122 | 229 | 218 |
| r.m.s. angle (°)e | 1.41 | 1.43 | 1.33 | 1.44 | 1.47 |
| r.m.s. bond (Å)e | 0.0092 | 0.0093 | 0.0078 | 0.0093 | 0.0098 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 | 0 |
| Protein Data Bank code | 6BJI | 6BJJ | 6BJK | 6BJL | 6BJM |
aRsym = ∑hkl∑i|Ihkl, i – [Ihkl]|/∑hkl ∑iIhkl, i, where [Ihkl] is the average of Friedel-related observations (i) of a unique reflection (hkl).
bValues in parentheses represent highest resolution shell.
cRwork, Σ ||Fo| − |Fc||/Σ |Fo|.
d5% of reflections were omitted for Rfree calculations.
er.m.s., root-mean-square.
Fig. 3.
(A) Electron density for mutated key active site residues (i) D302C, with the sulfhydryl group modeled in two positions each with 50% occupancy, (ii) D302A, (iii) D302L, (iv) R188K modeled in one conformation, (v) the stereoview of R188K modeled in one conformation with the difference map included and (vi) the stereoview of R188K modeled in two conformations with the same maps as (v) demonstrating the flexibility of the Lys188 side chain. Electron density diagrams are 2Fo−Fc maps contoured at 1.0 σ (blue) and Fo−Fc difference maps contoured at −3.0 (red) and 3.0 σ (green). (B–D) Salt bridge interactions among Ser185, Arg188, Asp211 and Asp302 in (B) unliganded GTB in the open state (PDB code 2RIT), (C) GTB in the liganded intermediate state (conformation #2, PDB code 5C1L) and (D) chimeric enzyme AABB in the liganded closed state (conformation #1, PDB code 2RJ7). Atoms are colored by element with oxygen red, nitrogen blue and sulfur yellow.
Table III.
Internal loop disorder observed in wild-type GTB and Arg188 and Asp302 mutant structures. All residues in these internal loops lie in conformations corresponding to the “open” state described by Alfaro et al. (2008)
| Enzyme | Internal loop | |||
|---|---|---|---|---|
| 175 | 180 | 185 | 190 | |
| GTBa | VGayk | rWQDV | SMRRM | EMISD |
| GTA/D302C | VRayk | rwqDV | SMRRM | EMISD |
| GTB/D302A | VGayk | rwqdv | smrrm | emisd |
| GTB/D302C | Vgayk | rwqdv | smrrm | emiSD |
| GTB/D302L | Vgayk | rwqdV | SMRRM | EMISD |
| GTB/R188K | VGayk | rwqdV | SMRKM | EMISD |
aPDB code: 2RIT (Alfaro et al. 2008).
One-letter amino acid codes with upper case black letters correspond to unambiguous electron density for main chain and side chain atoms, blue corresponds to unambiguous electron density for main chain atoms only; red letters correspond to weak or ambiguous electron density for main chain and side chain atoms. Residues with one-letter amino acid codes in lower case have not been included in the refined models.
Discussion
An intricate hydrogen bond/salt bridge network drives formation of the catalytically competent state
Liganded and unliganded GTA/GTB structures have shown that binding of the donor sugar via Arg188, Asp211 and Asp302 is crucial to formation of the catalytically active closed conformation (Alfaro et al. 2008; Schuman et al. 2010), as these residues form a hydrogen bond network (which includes a number of salt bridges) with each other and the donor substrate (Table IV; Figure 2B).
Table IV.
Hydrogen bond interactions (with distances between 2.5–3.5 Å) between catalytically competent UDP-Gal donor substrate and key active site residues of GTA/GTB and CAZy family 6 homologs α3GalT and BoGT6a (Figures 1 and 2)
| GTA/GTB residuea | UDP-Gal | α3GalT residueb | UDP-Gal | BoGT6a residuec | UDP-Gal |
|---|---|---|---|---|---|
| R188 [NH1] | O3′ | R202 [NH1] | O3′ | R73 [NH2] | O3′ |
| R188 [NH2] | None | R202 [NH2] | None | R73 [NH1] | None |
| D211 [OD1] | O3′ | D225 [OD1] | O3′ | N95 [ND2] | O3′ |
| D211 [OD2] | O2′ | D225 [OD2] | O2′ | N95 [ND2] | O2′ |
| H301 [ND1] | O6′ | H315 [ND1] | O6′ | H190 [NE2] | O6′ |
| D302 [OD1] | O4′ | D316 [OD2] | O4′ | D191 [OD1] | O4′ |
| D302 [OD2] | None | D316 [OD1] | None | D191 [OD2] | None |
| E303 | Acceptor | E317 | O4′ | E192 | Acceptor |
aLiganded AABB, PDB code: 2RJ7 (Alfaro et al. 2008).
bLiganded α3GalT, PDB code: 1O7Q (Zhang et al. 2003).
cAcceptor-bound BoGT6a, PDB code: 4AYJ (Thiyagarajan et al. 2012), superposed with liganded AABB, PDB code: 2RJ7, where UDP-Gal substrate is oriented as described byThiyagarajan et al. (2012).
In wild-type GTA and GTB, Asp302 forms a bidentate salt bridge to Arg188 in unliganded open structures (Figure 3B) as well as in structures containing the donor in an intermediate conformation identified previously as conformation #2 (Gagnon et al. 2015) (Figures 2A and B and 3C), where it also forms a hydrogen bond to donor Gal-O3. Upon rotation of the donor into the tucked under conformation (conformation #1; Figure 2A) and concomitant enzyme closure, the side chain of Asp302 loses the Nε hydrogen bond with Arg188 and switches from binding Gal-O3 to Gal-O4 (Table IV; Figures 2B and 3D) (Alfaro et al. 2008; Schuman et al. 2010; Gagnon et al. 2015).
In conformation #2, Arg188 makes a single contact to the donor via a water bridge to Gal-O2. Donor hydroxyl groups move by as much as 4.0 Å during the shift to conformation #1, and both Arg188 and Asp211 side chains make direct hydrogen bonds to Gal-O3 (Figure 2B) (Alfaro et al. 2008; Gagnon et al. 2015). During the shift to conformation #1, the donor Gal-O6 is displaced by 2.7 Å. This prevents a steric clash from occurring between Trp181 and Gal-O6 of donor conformation #2 and promotes a stabilizing interaction between Trp181 and Arg352 of the C-terminal tail. When UDP-Gal shifts to conformation #1, the internal loop becomes more ordered, and Arg188 is anchored via a direct interaction with the donor glycosyl moiety as well as salt bridge interactions with Asp211 and Asp302.
The salt bridges formed among Arg188, Asp211 and Asp302 appear not only to mediate the stepwise shift of donor from the predominant solution conformation to the tucked under catalytically competent conformation but also to drive the organization of the enzyme to the closed state. Glycosyltransfer is thought to occur after the internal loop and C-terminal tail organizes about the substrates to achieve the closed state (Alfaro et al. 2008), a common theme in GTs (Rini et al. 2009), but it is clear that the formation of the closed state itself is promoted by the new and reordered active site interactions that form as the donor binds.
In addition to participating in the ordering of the internal loop upon donor binding, Asp302 resides in the active site within an identified “double-turn” motif (aa 298–303), a conformationally unstable structural feature consisting of two interpenetrating hairpin turns. The double-turn motif is found in family 6 enzymes, including α3GT, BoGT6a and GTA/GTB, as well as the family 8 α-1,4-galactosyltransferase LgtC and has a known catalytic role in GTA and GTB (Tumbale et al. 2008; Blackler et al. 2017). The residues in the hydrogen bond/salt bridge network serve in part to link the double turn with the internal loop. A corresponding set of interactions, as outlined in Table V, occurs in unliganded and liganded versions of structurally homologous family 6 GTs, α3GT and BoGT6a.
Table V.
Comparison of salt bridge interactions in unliganded and liganded GTB with UDP-Gal in alternate conformation #2 and liganded AABB with UDP-Gal in the catalytically competent conformation (#1) (black). Interactions are shown alongside those observed for α3GalT (blue) and BoGT6a (red) with relevant residues grouped with their GTA/GTB equivalents
| Atoms from conserved salt bridge residues | [NH1] from R188, R202, R73 | [NH2] from R188, R202, R73 | [NE] from R188, R202, R73 | [O] from S185, S199, T70 |
|---|---|---|---|---|
| Partnering atom(s) from GTB unligandeda | D302 [OD1], 3.4 Å | D302 [OD1], 3.1 Å | ||
| D302 [OD2], 3.5 Å | ||||
| Partnering atom(s) from GTB + UDP-Gal #2b | D211 [OD1], 3.6 Å | D211 [OD1], 2.8 Å | D302 [OD2], 3.7 Å | |
| D302 [OD1], 3.5 Å | ||||
| D302 [OD2], 3.8 Å | ||||
| Partnering atom(s) from GTB + UDP-Gal #1c | D211 [OD1], 3.4 Å | D211 [OD1], 2.8 Å | S185 [O], 3.0 Å | D302 [OD1], 4.0 Å |
| D302 [OD1], 2.9 Å | D302 [OD2], 3.4 Å | |||
| Partnering atom(s) from α3GalT unligandedd | D225 [OD1], 2.6 Å | S199 [O], 4.0 Å | S199 [O], 3.1 Å | R202 [NH2], 4.0 Å |
| D225 [OD1], 3.1 Å | R202 [NE], 3.1 Å | |||
| D316 [OD2], 3.6 Å | D316 [OD2], 3.7 Å | |||
| Partnering atom(s) from α3GalT + UDP + GalNAce | D225 [OD1], 2.8 Å | S199 [O], 4.0 Å | S199 [O], 3.0 Å | R202 [NH2], 4.0 Å |
| D225 [OD1], 3.2 Å | R202 [NE], 3.0 Å | |||
| D316 [OD2], 2.9 Å | D316 [OD2], 3.7 Å | |||
| Partnering atom(s) from BoGT6a unligandedf | D191 [OD1], 3.0 Å | N95 [OD1], 3.9 Å | ||
| D191 [OD1], 3.9 Å | ||||
| Partnering atom(s) from BoGT6a + FALg | N95 [OD1], 4.0 Å | |||
| D191 [OD1], 3.2 Å | N95 [OD1], 2.6 Å | T70 [O], 3.8 Å | R73 [NE], 3.8 Å | |
| D191 [OD2], 3.5 Å | D191 [OD2], 3.9 Å | D191 [OD2], 3.4 Å | D191 [OD1], 3.2 Å |
aUnliganded GTB, PDB code: 2RIT (Alfaro et al. 2008).
bLiganded GTB, PDB code: 5C1L (Gagnon et al. 2015).
cLiganded AABB, PDB code: 2RJ7 (Alfaro et al. 2008).
dUnliganded α3GalT, PDB code: 5NR9 (Albesa-Jove et al. 2017).
eLiganded α3GalT, PDB code: 1O7Q (Zhang et al. 2003).
fUnliganded BoGT6a, PDB code: 4AYL (Thiyagarajan et al. 2012).
gAcceptor-bound BoGT6a, PDB code: 4AYJ (Thiyagarajan et al. 2012), superposed with liganded AABB, PDB code: 2RJ7, where UDP-Gal is oriented as described by Thiyagarajan et al. (2012).
The crystal structure of unliganded α3GT (PDB code: 5NR9) (Albesa-Jove et al. 2017) contains two molecules in the asymmetric unit, with only one having comparable salt bridge interactions. This is likely due to an observed glycerol molecule derived from the crystallization conditions providing stabilizing interactions normally provided by the donor sugar moiety. The second molecule in the asymmetric unit does not have glycerol in the donor-binding site and does not possess the corresponding salt bridge network.
Although α3GT and BoGT6a share only ~43 and ~33% sequence identity with GTA/GTB, respectively, structural overlap with the chimeric enzyme AABB shows a striking similarlity. AABB/α3GT residues Ser185/Ser199, Arg188/Arg73, Asp211/Asn95 and Asp302/Asp191 are in near-identical orientations (Figure 1C). Corresponding BoGT6a residues Thr70, Arg73, Asn95, Asp191 achieve the closed enzyme state in the same fashion.
Salt bridge network conserved in retaining and inverting GT-A fold GTs
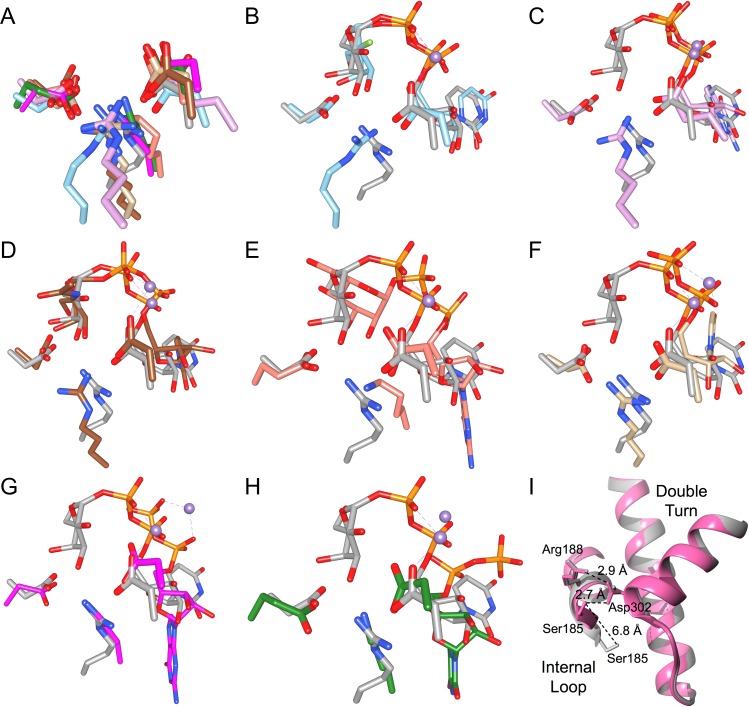
The GTA/GTB Arg188, Asp211 and Asp302 salt bridge network observed for family 6 GTs α3GT and BoGT6a may represent a conserved feature among other GT-A fold-type GTs. Overlap of chimeric AABB (PDB code: 2RJ7) with published GT-A fold structures shows that nearly all families contain at least one representative structure with equivalent interacting salt bridge residues involved in donor binding (Table VI, Figure 4A–H). The salt bridge network is conserved in the published structures of GT-A fold family 7, 13, 15, 27, 55, 64, 78 and 81 GTs, while family 2, 8 and 43 GTs do not show complete conservation. For example, the active sites of some family 2 GTs cannot be assessed due to ambiguous electron density or lack of intact donor-bound complexes: PF0058 from Pyrococcus furiosus, BF2801 from Bacteroides fragilis and Os07g0208500 from Oryza sativa. As well, the unliganded structures of family 2 GT SpsA do not show salt bridge conservation, though the cognate substrate for this enzyme is unknown and thus there exist no structural determinations of the natural donor complexes. Further, the quality of the electron density around the active sites for family 8 GTs Apre_0416 from Anaerococcus prevotii, Gtf2 from Streptococcus gordonii and the glycogenin α-glucosyltransferase from Caenorhabditis elegans does not allow an unambiguous determination of salt bridges, though conservation is seen for the remaining members of family GT-8. The final exception to salt bridge conservation is family 43 GT β-1,3-glucuronosyltransferase 3, which features Leu280 in the position corresponding to GTA/GTB residue Asp302.
Table VI.
Representative structures deposited in the Protein Data Bank of retaining and inverting GT-A fold GTs with salt bridge residues corresponding to GTA/GTB (family GT-6) residues Arg188, Asp211 and Asp302
| Reaction stereochemistry | Family | Example enzyme | Donor analog | Acceptor analog | PDB | Arg188 equivalent | Asp211 equivalent | Asp302 equivalent |
|---|---|---|---|---|---|---|---|---|
| Retaining | GT-8 | α-1,4-galactosyltransferase, LgtCa | UDP 2-deoxy-2-fluoro-galactose | 4′deoxylactose | 1GA8 | Arg86 | Asp103 | Asp188 |
| GT-15 | α1,2-mannosyltransferase, Kre2p/Mnt1pb | GDP | O1-methyl-mannose | 1S4P | Arg245* | Glu247 | Asp361 | |
| GT-27 | polypeptide GalNAc-transferase 2, GalNAc-T2c | UDP-N-acetylgalactosamine | EA2 peptide | 4D0T | Arg208 | Asp224 | Glu334 | |
| GT-55 | mannosyl-3-phosphoglycerate synthased | GDP-α-D-mannose | - | 2ZU8 | Lys145 | Asp168 | Glu271 | |
| GT-64 | α-1,4-N-acetylhexosaminyltransferase, EXTL2e | UDP | [glucuronic acid]β1–3[galactose]β1-O-naphthalenemethanol | 1ON8 | Arg135 | Asp151 | Asp245 | |
| GT-78 | mannosylglycerate synthase, MGSf | GDP | Malonate ion | 2Y4L | Lys76 | Asp100 | Asp192 | |
| GT-81 | glucosyl-3-phosphoglycerate synthaseg | UDP | 3-phosphoglyceric acid | 4DEC | Lys114 | Asp134 | Glu232 | |
| Inverting | GT-2 | Teichoic acid β-glycosyltransferaseh | UDP-N-acetylgalactosamine | - | 5TZJ | Arg75 | Asp91 | Glu177 |
| GT-7 | β1–4-galactosyltransferase-I, β4Gal-T1i | UDP | [N-acetylglucosamine] β1–6[galactose]β1-4[glucose] | 4EEA | Arg224 | Asp248 | Glu313 | |
| GT-13 | O-linked mannose β1,2-N-acetylglucosaminyltransferase 1, POMGnT1j | UDP | O-mannosylated peptide | 5GGI | Tyr372 | Glu393 | Trp475 | |
| GT-43 | Glucuronyltransferase, GlcAT-Pk | UDP | N-acetyllactosamine | 1V84 | Arg170 | Asp195 | Gln283 |
dKawamura et al. to be published.
*Unlike GTA/GTB Arg188 and the corresponding residues of the other GT-A fold GTs, Arg160 in this family 15 enzyme does not reside within an ordered alpha helix, though it appears to play the same or similar role in salt bridge formation.
Fig. 4.
The salt bridge interactions formed in the closed enzyme state represent a structurally conserved feature found in representative structures from each retaining GT-A fold family. (A) Overlap of salt bridge residues Arg188, Asp211 and Asp302 from chimeric AABB in gray (PDB code 2RJ7) with corresponding residues (listed in Table VI) in representative structures from each retaining GT-A fold family using the following color scheme: GT-8 in sky blue (PDB code 1GA8), GT-15 in plum (PDB code 1SP4), GT-27 in brown (PDB code 4D0T), GT-55 in salmon (PDB code 2ZU8), GT-64 in tan (PDB code 1ON8), GT-78 in deep pink (PDB code 2Y4L) and GT-81 in forest green (PDB code 4DEC). In (B-H) AABB again is shown in gray (PDB code 2RJ7) and is separately overlapped with each GT-A fold family representative, which collectively are colored with cyan for carbon, phosphorous and manganese. Overlaps are between AABB and a representative structure (listed in Table VI) of (B) GT-8, (C) GT-15, (D) GT-27, (E) GT-55, (F) GT-64, (G) GT-78 and (H) GT-81. Atoms are colored by element with oxygen red, nitrogen blue, fluorine light green, carbon as indicated above, phosphorous orange except where otherwise indicated and manganese medium-purple except where otherwise indicated. Color naming is consistent with the program USCF Chimera. (I) Overlap of AABB (pink, PDB code 2RJ7) with the semi-open GTB (gray, PDB code 5C1L) showing the >6 Å shift of internal loop residue Ser185 that occurs upon substrate binding and salt bridge reorganization.
The residue Ser185 is not strictly conserved across all GT-A fold GTs, and its presence is approximately correlated to internal loop length. For example, the relatively shorter internal loop of retaining GT-A fold family 27 polypeptide GalNAc-transferase 2 (GalNAc-T2) may require fewer contacts to achieve the ordered, closed catalytic state, meaning Leu204 may suffice in the position occupied by a hydrogen-bonding Ser185 in GTA/GTB. This trend also extends to inverting GT-A fold GTs, where residues equivalent to GTA/GTB Arg188, Asp211 and Asp302 are present in representative structures of each family (Table VI).
Conserved residue Ser185 stabilizes the closed state
In addition to Arg188, another residue of the internal loop, Ser185 shifts considerably as the donor makes the final transition from conformation #2 into the tucked under conformation #1 (Gagnon et al. 2015). Ser185 moves over 6 Å to form a hydrogen bond with Asp302 of the double-turn motif (Figure 4I) to stabilize the closed state. Notably, Nakahara et al. (2006) determined the kinetic constants for GTB S185N and S185C mutants of this residue and observed a decrease in kcat values (0.52 s−1 and 0.092 s−1, respectively) relative to their reported value for wild-type GTB (5.2 s−1) (Nakahara et al. 2006). These data underscore the importance of Ser185 in catalytic turnover in these enzymes.
Loop ordering and kinetics of Asp302 and Arg188 mutants
Of the five mutant enzyme structures, only GTA/D302C and GTB/R188K show any electron density for the side chain of residue 188 (Table III). This is not unexpected as the side chains of the corresponding residues in GTB/D302A and GTB/D302L cannot form hydrogen bonds, take part in the salt bridge network or stabilize the closed conformation, which is consistent with the near-zero activities of these mutants (Table I). Although D302C in both GTA/D302C and GTB/D302C is disordered over two conformations (Figure 5A) and sulfur is a poor hydrogen bond acceptor, the side chain may still be able to interact with the donor to participate in the hydrogen bond network interactions that stabilize the closed conformation to yield ~10% of wild-type activity (Table I).
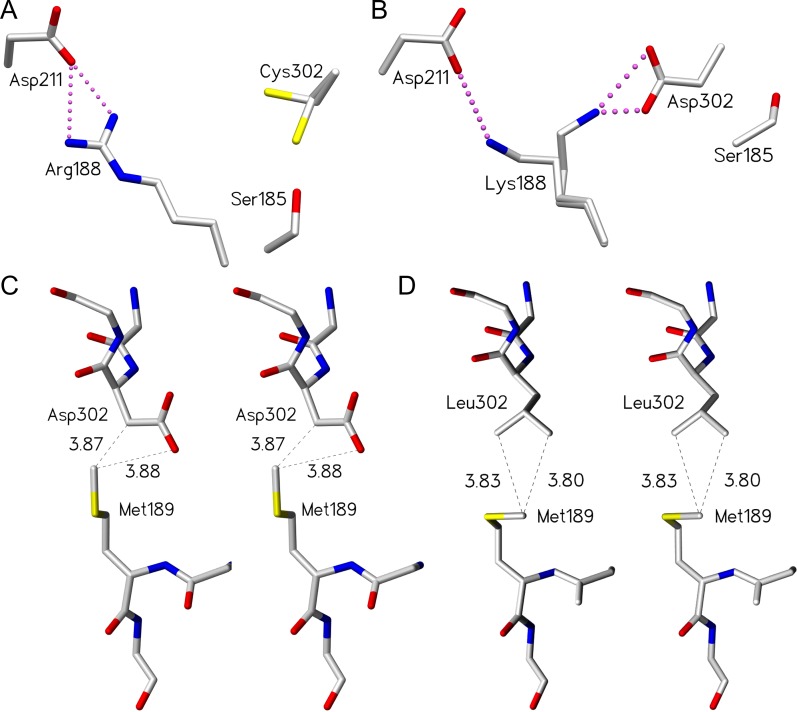
Fig. 5.
Certain mutants of residue 302 and 188 are able to compensate for the loss of critical hydrogen bonds within the salt bridge network by establishing alternative interactions. D302C and R188K mutants maintain the salt bridge network by adopting multiple conformations, while the D302L mutant maintains partial loop order via van der Waal’s contacts. (A) Cys302 side chain observed in two conformations in unliganded GTA/D302C. (B) Lys188 side chain observed in two conformations in unliganded GTB/R188K. Stereoview of (C) wild-type GTB residues Asp302 and Met189 (PDB code 2RIT) and (D) GTB/D302L residues Leu302 and Met189 with van der Waal’s interactions shown as dashed lines (distances shown in Ångstrom) between the leucine side chain carbon atoms and the methionine side chain methyl group. The dashed lines between the Met189 methyl group and the Asp302 carbonyl group represent a measured distance, not a van der Waal’s interaction. Atoms are colored by element with carbon gray, oxygen red, nitrogen blue and sulfur yellow. Salt bridge interactions are depicted as pink dashed spheres.
Critical residue Arg/Gly176 has also been associated with internal loop stability, with Arg176 imparting greater loop order due to more restricted main chain dihedral angles compared to Gly176 (Patenaude et al. 2002; Lee et al. 2005; Letts et al. 2006; Alfaro et al. 2008; Johal et al. 2012). This trend is again observed in the structures of the D302C mutants, the only Asp302 mutants that yielded diffraction quality crystals for both enzymes. The GTA/D302C structure displays unambiguous electron density for 11 additional internal loop residues over GTB/D302C (Table III).
An exception to the trend is the GTB/D302L mutant. Leu302, which cannot form a salt bridge interaction with Asp211 yet achieves ordering of the internal loop in the open position (i.e. not in the catalytic position) via the additional van der Waals contacts formed between Leu302 and Met189, as observed in the crystal structure of this mutant (Figure 5B and C).
Among the crystal structures determined for GTB mutants, GTB/R188K has the greatest internal loop order in the open conformation and partially maintains the salt bridge network (Table III), albeit with a significantly different pattern of hydrogen bonds in the active site from the wild type. The multivalency and higher pKa of the guanidinium group (Sokolowski et al. 1998) of the arginine side chain allow for more and stronger electrostatic interactions relative to lysine. This is consistent with the kinetic data for GTB R188K, which has higher donor and acceptor Km values relative to wild-type enzyme and displays a reduction in kcat of nearly four orders of magnitude (Table I). The structure of GTB/R188K suggests that Lys188 forms a disordered salt bridge with Asp211 and Asp302 (Figures 3A and 5B), a weaker set of interactions than seen for the wild type, indicating that both the length and multivalent nature of the Arg188 side chain are required to achieve the catalytic state.
The wild-type structures show that the closed catalytic conformation is achieved with formation of salt bridges from Asp211 and Asp302 to Arg188 (Figure 2B). The extent of internal loop disorganization observed in mutants of Arg188 or Asp302 underscores their importance. More conservative mutations (GTA/D302C, GTB/R188K) reduce loop ordering (Table III), though the salt bridge is approximately maintained (Figure 5A and B), while other mutations disrupt this network. Such mutants either maintain order through unconventional contacts (GTB/D302L; Figure 5D) or display a disordered internal loop (GTB/D302A, GTB/D302C) (Table III).
Acidic amino acid residue 302 is required for efficient catalysis
Kinetic and structural data demonstrate that Asp302 strongly influences enzyme efficiency (Table I). Mutation of Asp302 to nonpolar alanine and leucine nearly abolishes enzyme activity, while the conservative mutation to glutamate preserves almost 50% of wild-type activity and a mutation to cysteine preserves ~10% (Table I). In the case of D302C mutants, a comparable percentage of cysteine side chains are expected to be in the ionized state at pH 7.4, the pH used in these kinetic studies, which supports the notion that a negative charge at residue 302 is catalytically important. These data are consistent with the homologous bovine α3GT, which retains 40% of wild-type activity when the corresponding residue Asp316 (Figure 1C) is mutated to a glutamate (Tumbale et al. 2008) but loses nearly all detectable activity when mutated to alanine (Monegal and Planas 2006) or asparagine (Tumbale et al. 2008) (Table I). As with GTA/GTB, the loss of activity was attributed to α3GT’s requirement for a charged residue at this position (Tumbale et al. 2008).
Conclusions
Conserved active site residues Arg188 and Asp302 are critical for catalysis in GTA and GTB, and disruption of their hydrogen bond network through mutation can dramatically decrease enzymatic activity. The contributions of these residues stem not only from direct contact recognition of donor but also from their ability to stabilize through salt bridges and hydrogen bonds the internal loop in the closed conformation. Taken together, these data suggest that Arg188 and Asp302, in addition to Ser185 and Asp211, are critical to the generation of the catalytic state in GTA and GTB. Asp302 and Arg188 mutant enzymes are unable to achieve this state due to a weakened or abolished hydrogen bond network, where Ser185 cannot anchor the internal loop to the double-turn motif, impeding loop organization and by extension, catalysis.
Materials and methods
Generation of mutants, protein expression and purification
Site-directed mutagenesis for the Asp302 mutants was carried out as described previously (Marcus et al. 2003) using recombinant PCR with wild-type GTA or GTB (aa 63–354) in the plasmid vector pCWΔlac as template. The primers used for mutagenesis with codon substitutions in bold were as follows: D302A 5′ forward primer TGG CAC GCT GAA TCC CAC CTG AAC AAA TAC CTG; 5′ reverse primer GTG GGA TTC AGC GTG CCA AAC AGC TTC GAT ACC; D302C 5′ forward primer GTT TGG CAC TGC GAA TCC CAC CTG AAC AAA TAC; 5′ reverse primer GTG GGA TTC GCA GTG CCA AAC AGC TTC GAT ACC; D302L 5′ forward primer TGG CAC CTG GAA TCC CAC CTG AAC AAA TAC CTG; 5′ reverse primer GTG GGA TTC CAG GTG CCA AAC AGC TTC GAT ACC; D302E 5′ reverse primer GTG GGA TTC CTC GTG CCA AAC AGC TTC GAT ACC; 5′ forward primer TGG CAC GAG GAA TCC CAC CTG AAC AAA TAC CTG.
Site-directed mutagenesis for the Arg188 mutant was carried out using a QuikChange II XL site-directed mutagenesis kit (Strategene) and GTB (aa 63–354) in the plasmid vector pCWΔlac as previously described (Letts et al. 2007). The primers used for mutagenesis were as follows: R188K 5′ forward primer G CAG GAC GTT TCC ATG CGT AAA ATG GAA ATG ATC AGC GAC; 5′ reverse primer GTC GCT GAT CAT TTC CAT TTT ACG CAT GGA AAC GTC CTG CC. All mutant enzymes were purified according to a two-step protocol using ion-exchange and UDP-hexanolamine affinity chromatography (Marcus et al. 2003; Seto et al. 1999).
Kinetics
A radiochemical assay with hydrophobic acceptor Fuc-α-1→2-Gal-β-(CH2)7CH3 and radiolabeled sugar donors was used to characterize the mutants (Palcic et al. 1988; Alfaro et al. 2008). This assay is based on the removal of radiolabeled reaction products from unreacted donor on reverse-phase C18 cartridges. Assays were carried out at 37°C in a total volume of 12–15 μL with incubation times of 30–40 min using at least five different concentrations of acceptor or donor substrate with 1 mM of the nonvaried substrate. Enzyme concentrations ranged from 2.9 nM (GTA D302C) to 6.8 μM (GTB D302A) in 50 mM MOPS pH 7.0 with 20 mM MnCl2 and 1 mg/mL bovine serum albumin. Initial rate conditions were linear with no more than 10–20% of the substrate consumed. The kinetic parameters kcat and Km were obtained by nonlinear regression analysis of the Michaelis–Menten equation with GraphPad PRISM 3.0. Km errors are from curve fitting while for kcat they are the average of kcats from donor and acceptor kinetics.
Crystallography
Spontaneous crystals were recovered from concentrated stock solutions of GTB/D302A (68 mg/mL), GTB/D302L (40–50 mg/mL), GTB/D302C (55 mg/mL) and GTA/D302C (95–100 mg/mL) stored in 50 mM MOPS pH 7.0, 0.1 M NaCl, 1 mM DTT, 5 mM MnCl2 and kept at 4°C. GTB/R188K crystals were obtained by hanging drop vapor diffusion at 4°C, where 3 μL of concentrated stock (66–68 mg/mL) was mixed with 1 μL of reservoir solution containing 0.3 M sodium acetate and 0.3 M NaCl.
Prior to freezing, crystals were washed in Mother liquor initial (MLo) containing 6.8% PEG 4000, 40 mM sodium acetate pH 4.6, 30 mM ADA pH 7.5, 20 mM MES pH 6.5, 40 mM ammonium sulfate, 9 mM MnCl2, 30% glycerol.
Data were collected on a Rigaku RAXIS IV++ area detector and processed with d*trek (Pflugrath 1999). For data collection the detector distance was 72 mm and exposure times varied between 4.0 and 5.0 min for 0.5° oscillations. X-rays were produced by a MM-002 generator (Rigaku/MSC) coupled to Osmic “Blue” confocal X-ray mirrors at 30 W (Osmic). Crystals were frozen and maintained under cryogenic conditions at a temperature of −160°C using a CryoStream 700 crystal cooler (Oxford). Although the structures appeared to be isomorphous with the known structures of the wild-type enzymes, for completeness all data sets were solved using molecular replacement techniques (the MolRep module of the CCP4 suite) with wild-type GTA or GTB (Protein Data Bank (PDB) accession codes 1LZ0 and 1LZ7, respectively) (Patenaude et al. 2002) as starting models (Murshudov et al. 1997; Vagin and Teplyakov 1997; Vagin and Isupov 2001; Mccoy et al. 2007; Vagin and Teplyakov 2010) and subsequently refined using REFMAC5 (Murshudov et al. 1997) of the CCP4 program suite (Bailey 1994). Figures were generated using SetoRibbon (Evans, unpublished) as well as Chimera (Pettersen et al. 2004). Marvin was used for drawing two-dimensional hydrogen bond diagrams (Marvin version 5.9.0, 2012, ChemAxon (http://www.chemaxon.com)).
Supplementary Material
Acknowledgements
The authors are grateful to Ryan Huskins (University of Alberta) who carried out the isolation and characterization of GTB E302A.
Abbreviations
- CAZy
Carbohydrate Active EnZyme
- GTs
glycosyltransferases.
Supplementary data
Funding
This work was supported in whole or part by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.M.P. and the Canadian Institutes of Health Research to S.V.E. S.M.L.G. is the recipient of Natural Sciences and Engineering Research Council of Canada Graduate Scholarship D. M.S.G.L. is the recipient of a Natural Sciences and Engineering Research Council of Canada Graduate Scholarship M. J.A.L. is the recipient of a Marie Skłodowská-Curie Fellowship.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Albesa-Jove D, Sainz-Polo MA, Marina A, Guerin ME. 2017. Structural snapshots of alpha-1,3-galactosyltransferase with native substrates: Insight into the catalytic mechanism of retaining glycosyltransferases . Angew Chem Int Ed Engl. 56:14853–14857. [DOI] [PubMed] [Google Scholar]
- Alfaro JA, Zheng RB, Persson M, Letts JA, Polakowski R, Bai Y, Borisova SN, Seto NOL, Lowary TL, Palcic MM et al. 2008. ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes. J Biol Chem. 283:10097–10108. [DOI] [PubMed] [Google Scholar]
- Angulo J, Langpap B, Blume A, Biet T, Meyer B, Krishna NR, Peters H, Palcic MM, Peters T. 2006. Blood group B galactosyltransferase: Insights into substrate binding from NMR experiments. J Am Chem Soc. 128:13529–13538. [DOI] [PubMed] [Google Scholar]
- Bailey S. 1994. The CCP4 suite—Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 50:760–763. [DOI] [PubMed] [Google Scholar]
- Blackler RJ, Gagnon SM, Polakowski R, Rose NL, Zheng RB, Letts JA, Johal AR, Schuman B, Borisova SN, Palcic MM et al. 2017. Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site. Glycobiology. 27:370–380. [DOI] [PubMed] [Google Scholar]
- Blume A, Angulo J, Biet T, Peters H, Benie AJ, Palcic M, Peters T. 2006. Fragment-based screening of the donor substrate specificity of human blood group B galactosyltransferase using saturation transfer difference NMR. J Biol Chem. 281:32728–32740. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B. 1999. Carbohydrate-active enzymes: An integrated database approach In: Recent advances in carbohydrate bioengineering. Cambridge: The Royal Society of Chemistry. p. 3–12. [Google Scholar]
- Gagnon SM, Meloncelli PJ, Zheng RB, Haji-Ghassemi O, Johal AR, Borisova SN, Lowary TL, Evans SV. 2015. High resolution structures of the human ABO(H) blood group enzymes in complex with donor analogs reveal that the enzymes utilize multiple donor conformations to bind substrates in a stepwise manner. J Biol Chem. 290:27040–27052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastinel LN, Bignon C, Misra AK, Hindsgaul O, Shaper JH, Joziasse DH. 2001. Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J. 20:638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn VM, Smith ZG, Watkins WM. 1968. An a-N-acetyl-D-galactosaminyltransferase associated with the human blood-group A character. Biochem J. 109:315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SM, Benny AG, Woodfield DG. 1990. Investigation of Lewis phenotypes in polynesians: Evidence of a weak secretor phenotype. Vox Sang. 58:61–66. [DOI] [PubMed] [Google Scholar]
- Johal AR, Schuman B, Alfaro JA, Borisova S, Seto NOL, Evans SV. 2012. Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases. Acta Crystallogr D Biol Crystallogr. 68:268–276. [DOI] [PubMed] [Google Scholar]
- Kakuda S, Shiba T, Ishiguro M, Tagawa H, Oka S, Kajihara Y, Kawasaki T, Wakatsuki S, Kato R. 2004. Structural basis for acceptor substrate recognition of a human glucuronyltransferase, GlcAT-P, an enzyme critical in the biosynthesis of the carbohydrate epitope HNK-1. J Biol Chem. 279:22693–22703. [DOI] [PubMed] [Google Scholar]
- Kamath VP, Seto NOL, Compston CA, Hindsgaul O, Palcic MM. 1999. Synthesis of the acceptor analog alpha Fuc(1 - 2)alpha Gal-O(CH2)(7) CH3: A probe for the kinetic mechanism of recombinant human blood group B glycosyltransferase. Glycoconj J. 16:599–606. [DOI] [PubMed] [Google Scholar]
- Kobata A, Grollman EF, Ginsburg V. 1968. An enzymatic basis for blood type B in humans. Biochem Biophys Res Commun. 32:272–277. [DOI] [PubMed] [Google Scholar]
- Kuwabara N, Manya H, Yamada T, Tateno H, Kanagawa M, Kobayashi K, Akasaka-Manya K, Hirose Y, Mizuno M, Ikeguchi M et al. 2016. Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan. Proc Natl Acad Sci USA. 113:9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsteiner K. 1901. Über Agglutinationserscheinungen normalen menschlichen Blutes. Wien Klin Wochenschr. 46:1132–1134. [PubMed] [Google Scholar]
- Lee HJ, Barry CH, Borisova SN, Seto NOL, Zheng RXB, Blancher A, Evans SV, Palcic MM. 2005. Structural basis for the inactivity of human blood group O-2 glycosyltransferase. J Biol Chem. 280:525–529. [DOI] [PubMed] [Google Scholar]
- Letts JA, Persson M, Schuman B, Borisova SN, Palcic MM, Evans SV. 2007. The effect of heavy atoms on the conformation of the active-site polypeptide loop in human ABO(H) blood-group glycosyltransferase B. Acta Crystallogr D Biol Crystallogr. 63:860–865. [DOI] [PubMed] [Google Scholar]
- Letts JA, Rose NL, Fang YR, Barry CH, Borisova SN, Seto NOL, Palcic MM, Evans SV. 2006. Differential recognition of the type I and II H antigen acceptors by the human ABO(H) blood group A and B glycosyltransferases. J Biol Chem. 281:3625–3632. [DOI] [PubMed] [Google Scholar]
- Lira-Navarrete E, Iglesias-Fernandez J, Zandberg WF, Companon I, Kong Y, Corzana F, Pinto BM, Clausen H, Peregrina JM, Vocadlo DJ et al. 2014. Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide N-acetylgalactosaminyltransferase 2. Angew Chem Int Ed Engl. 53:8206–8210. [DOI] [PubMed] [Google Scholar]
- Liumbruno GM, Franchini M. 2013. Beyond immunohaematology: The role of the ABO blood group in human diseases. Blood Transfus. 11:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobsanov YD, Romero PA, Sleno B, Yu B, Yip P, Herscovics A, Howell PL. 2004. Structure of Kre2p/Mnt1p: A yeast alpha1,2-mannosyltransferase involved in mannoprotein biosynthesis. J Biol Chem. 279:17921–17931. [DOI] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SL, Polakowski R, Seto NO, Leinala E, Borisova S, Blancher A, Roubinet F, Evans SV, Palcic MM. 2003. A single point mutation reverses the donor specificity of human blood group B-synthesizing galactosyltransferase. J Biol Chem. 278:12403–12405. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr. 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milland J, Sandrin MS. 2006. ABO blood group and related antigens, natural antibodies and transplantation. Tissue Antigens. 68:459–466. [DOI] [PubMed] [Google Scholar]
- Monegal A, Planas A. 2006. Chemical rescue of alpha 3-galactosyltransferase. Implications in the mechanism of retaining glycosyltransferases. J Am Chem Soc. 128:16030–16031. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 53:240–255. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Hindsgaul O, Palcic MM, Nishimura SI. 2006. Computational design and experimental evaluation of glycosyltransferase mutants: Engineering of a blood type B galactosyltransferase with enhanced glucosyltransferase activity. Protein Eng Des Sel. 19:571–578. [DOI] [PubMed] [Google Scholar]
- Nielsen MM, Suits MD, Yang M, Barry CS, Martinez-Fleites C, Tailford LE, Flint JE, Dumon C, Davis BG, Gilbert HJ et al. 2011. Substrate and metal ion promiscuity in mannosylglycerate synthase. J Biol Chem. 286:15155–15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palcic MM, Heerze LD, Pierce M, Hindsgaul O. 1988 The use of hydrophobic synthetic glycosides as acceptors in glycosyltransferase assays. Glycoconjugate J. 5:49–63.
- Patenaude SI, Seto NOL, Borisova SN, Szpacenko A, Marcus SL, Palcic MM, Evans SV. 2002. The structural basis for specificity in human ABO(H) blood group biosynthesis. Nat Struct Biol. 9:685–690. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Dong J, Taniguchi F, Kitagawa H, Krahn JM, Pedersen LG, Sugahara K, Negishi M. 2003. Crystal structure of an alpha 1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J Biol Chem. 278:14420–14428. [DOI] [PubMed] [Google Scholar]
- Persson K, Buynak JD, Strynadka NCJ. 2001. a. Crystallographic, modeling, and SAR studies of 2 ‘-substituted-6-alkylidenepenam sulfones as inhibitors of the TEM-1 beta-lactamase. Abstr Pap Am Chem Soc. 221:U50–U50. [Google Scholar]
- Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NCJ. 2001. b. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol. 8:166–175. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera—A visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 55:1718–1725. [DOI] [PubMed] [Google Scholar]
- Pham TTK, Stinson B, Thiyagarajan N, Lizotte-Waniewski M, Brew K, Acharya KR. 2014. Structures of complexes of a metal-independent glycosyltransferase GT6 from Bacteroides ovatus with UDP-N-acetylgalactosamine (UDP-GalNAc) and its hydrolysis products. J Biol Chem. 289:8041–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba PK, Ramakrishnan B, Balaji PV. 2002. Oligosaccharide binding site of beta 1,4-galactosyltransferases (Gal-Ts) defined by docking various glycan substrates in the binding site. Abstr Pap Am Chem Soc. 223:U496–U496. [Google Scholar]
- Qasba PK, Ramakrishnan B, Boeggeman E. 2005. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem Sci. 30:53–62. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Boeggeman E, Qasba PK. 2012. Binding of N-acetylglucosamine (GlcNAc) beta1-6-branched oligosaccharide acceptors to beta4-galactosyltransferase I reveals a new ligand binding mode. J Biol Chem. 287:28666–28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini J, Esko J, Varki A. 2009. Chapter 5, Glycosyltransferases and Glycan-processing Enzymes In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. NY: Cold Spring Harbor. https://www.ncbi.nlm.nih.gov/books/NBK1921/. [PubMed] [Google Scholar]
- Rummel SK, Ellsworth RE. 2016. The role of the histoblood ABO group in cancer. Future Sci OA. 2:FSO107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman B, Persson M, Landry RC, Polakowski R, Weadge JT, Seto NOL, Borisova SN, Palcic MM, Evans SV. 2010. Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: Structural insights into cooperative substrate binding. J Mol Biol. 402:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto NO, Compston CA, Evans SV, Bundle DR, Narang SA, Palcic MM. 1999. Donor substrate specificity of recombinant human blood group A, B and hybrid A/B glycosyltransferases expressed in Escherichia coli. Eur J Biochem. 259:770–775. [DOI] [PubMed] [Google Scholar]
- Seto NOL, Palcic MM, Compston CA, Li H, Bundle DR, Narang SA. 1997. Sequential interchange of four amino acids from blood group B to blood group A glycosyltransferase boosts catalytic activity and progressively modifies substrate recognition in human recombinant enzymes. J Biol Chem. 272:14133–14138. [DOI] [PubMed] [Google Scholar]
- Sobhanifar S, Worrall LJ, King DT, Wasney GA, Baumann L, Gale RT, Nosella M, Brown ED, Withers SG, Strynadka NC. 2016. Structure and mechanism of Staphylococcus aureus TarS, the wall teichoic acid beta-glycosyltransferase involved in methicillin resistance. PLoS Pathog. 12:e1006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski T, Haselhorst T, Scheffler K, Weisemann R, Kosma P, Brade H, Brade L, Peters T. 1998. Conformational analysis of a Chlamydia-specific disaccharide alpha-Kdo-(2 - 8)-alpha-Kdo-(2 - 0)-allyl in aqueous solution and bound to a monoclonal antibody: Observation of intermolecular transfer NOEs. J Biomol NMR. 12:123–133. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan N, Pham TT, Stinson B, Sundriyal A, Tumbale P, Lizotte-Waniewski M, Brew K, Acharya KR. 2012. Structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen. Sci Rep. 2:940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbale P, Brew K. 2009. Characterization of a metal-independent CAZy family 6 glycosyltransferase from Bacteroides ovatus. J Biol Chem. 284:25126–25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbale P, Jamaluddin H, Thiyagarajan N, Brew K, Acharya KR. 2008. Structural basis of UDP-galactose binding by alpha-1,3-galactosyltransferase (alpha 3GT): Role of negative charge on aspartic acid 316 in structure and activity. Biochemistry. 47:8711–8718. [DOI] [PubMed] [Google Scholar]
- Urresti S, Albesa-Jove D, Schaeffer F, Pham HT, Kaur D, Gest P, van der Woerd MJ, Carreras-Gonzalez A, Lopez-Fernandez S, Alzari PM et al. 2012. Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria. J Biol Chem. 287:24649–24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Isupov MN. 2001. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 57:1451–1456. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. 1997. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 30:1022–1025. [Google Scholar]
- Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 66:22–25. [DOI] [PubMed] [Google Scholar]
- Yamamoto F, Cid E, Yamamoto M, Blancher A. 2012. ABO research in the modern era of genomics. Transfus Med Rev. 26:103–118. [DOI] [PubMed] [Google Scholar]
- Yamamoto F, Clausen H, White T, Marken J, Hakomori S. 1990. Molecular genetic basis of the histo-blood group ABO system. Nature. 345:229–233. [DOI] [PubMed] [Google Scholar]
- Yamamoto F, Hakomori S. 1990. Sugar-nucleotide donor specificity of histo-blood group A and B transferases is based on amino acid substitutions. J Biol Chem. 265:19257–19262. [PubMed] [Google Scholar]
- Yazer MH, Palcic MM. 2005. The importance of disordered loops in ABO glycosyltransferases. Transfus Med Rev. 19:210–216. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Swaminathan GJ, Deshpande A, Boix E, Natesh R, Xie Z, Acharya KR, Brew K. 2003. Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity. Biochemistry. 42:13512–13521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.