Abstract
Infectious pancreatic necrosis virus (IPNV) is a naked double-stranded RNA virus with a bi-segmented genome that is classified within the family Birnaviridae, genus Aquabirnavirus. IPNV was first detected in Italian trout farms in the late 1970s and ultimately became endemic. To characterize the evolution of IPNV circulating in Italy, particularly whether there is a link between evolutionary rate and virulence, we obtained and analyzed the VP1 (polymerase) and the pVP2 (major capsid protein precursor) sequences from 75 IPNV strains sampled between 1978 and 2017. These data revealed that the Italian IPNV exhibit relatively little genetic variation over the sampling period, falling into four genetic clusters within a single genogroup (group 2 for VP1 and genogroup V for pVP2) and contained one example of inter-segment reassortment. The mean evolutionary rates for VP1 and pVP2 were estimated to be 1.70 and 1.45 × 10−4 nucleotide substitutions per site, per year, respectively, and hence significantly lower than those seen in other Birnaviruses. Similarly, the relatively low ratios of non-synonymous (dN) to synonymous (dS) nucleotide substitutions per site in both genes indicated that IPNV was subject to strong selective constraints, again in contrast to other RNA viruses infecting salmonids that co-circulate in the same area during the same time period. Notably, all the Italian IPNV harbored a proline at position 217 (P217) and a threonine at position 221 (T221) in pVP2, both of which are associated with a low virulence phenotype. We therefore suggest the lower virulence of IPNV may have resulted in reduced rates of virus replication and hence lower rates of evolutionary change. The data generated here will be of importance in understanding the factors that shape the evolution of Aquabirnaviruses in nature.
Keywords: IPNV, Italy, trout, evolution, phylogeny, virulence
1. Introduction
Infectious pancreatic necrosis virus (IPNV) is the type species of the genus Aquabirnavirus, family Birnaviridae, and appears as a nude, single-shelled icosahedral viral particle with a diameter of approximately 65 nm (Delmas, Mundt, and Vakharia 2012). The IPNV genome comprises two linear double-stranded RNA (dsRNA) molecules termed segment A and B (Dobos 1976). Segment A possesses two open reading frames (ORFs), the larger of which encodes the polyprotein NH2-pVP2-VP4-VP3-COOH that is cleaved during translation to produce the VP2 precursor (pVP2), the VP4 protease and the minor capsid component VP3. pVP2 is further processed at its C terminal to generate the mature capsid protein VP2 (Dobos and Rowe 1977; Huang et al. 1986; Duncan et al. 1987; Magyar and Dobos 1994) that induces neutralizing antibody production and is related to virulence (Heppell et al. 1995; Santi, Vakharia, and Evensen 2004; Shivappa et al. 2004). Segment A also contains a minor ORF that overlaps the N terminal of the larger ORF and encodes a non-structural protein with anti-apoptotic properties (VP5) (Havarstein et al. 1990; Hong, Gong, and Wu 2002). The RNA-dependent RNA polymerase (RdRp), also known as VP1, is encoded by segment B and can be found either free or covalently associated to the genomic RNA (Duncan et al. 1991; Dobos 1995).
Aquabirnaviruses are traditionally classified into two serogroups (A and B) comprising 10 different serotypes, namely A1–A9 and B1, also known as West Buxton (WB), Sp, Ab, Hecht (He), Tellina (Te), Canada 1 (Can1), Canada 2 (Can2), Canada 3 (Can3), Jasper and TV-1. The serotype subdivision partially reflects geographical origins. The A1 serotype is largely found in the USA, A2-A5 and B1 are typically found in Europe and Asia, and A6-A9 include isolates from Canada (Hill and Way 1995). Blake et al. (2001) classified IPNV based on a phylogenetic analysis of VP2 sequences, and identified six different genogroups (I–VI). More recently, a seventh genogroup (VII) comprising Japanese aquabirnaviruses was established based on an analysis of the VP2/VP4 junction (Nishizawa, Kinoshita, and Yoshimizu 2005). An additional study of complete VP1 sequences has identified three different IPNV genotypes (Groups 1, 2 and MABV) (Barrera-Mejía et al. 2010).
IPNV triggers serious disease in salmonids reared under intensive conditions, causing sudden increases in mortality in fry and fingerlings (Wolf 1988; Jarp et al. 1995; Johans et al. 2002). Importantly, IPNV pathogenicity is variable, and mortality can range from 10 per cent to 90 per cent depending on the viral strain, host, stress factors, infectious dose, environmental conditions and, ultimately, fish age (Dorson and Touchy 1981; Dobos and Roberts 1983; Okamoto et al. 1984; McAllister and Owens 1995; Taksdal et al. 1998; Bruslind and Reno 2000; Moen et al. 2009; Houston et al. 2010; Gadan et al. 2013). Indeed, adult fish surviving the infection remain chronically infected for life, exhibiting neither clinical signs nor mortality (Bootland, Dobos, and Stevenson 1991) and intermittently shed virus through biological fluids (Hill 1982).
In Italy, IPN was reported for the first time in farmed rainbow trout (Oncorhynchus mykiss) (Bovo and Giorgetti 1979) in the late 1970s and eventually became endemic with the advent of intensive farming. The prevalence of the infection at the farm level is approximately 40 per cent, and the estimated mortality in hatched fry ranges between 10 per cent and 30 per cent. To date, little is known about the diversity and evolution of IPNV circulating in Italian salmonid farms, particularly the rate at which these viruses evolve, nor about the factors that shape genetic diversity. To fill these gaps, we describe, for the first time, the phylogenetic history of IPNV since its introduction into Italy and provide new information on the evolutionary dynamics of the genus Aquabirnaviridae.
2. Methods
2.1 Viruses and cell culture isolation
Fish samples were collected in the period 1978–2017 through active and passive surveillance activities already in place for infectious haematopoietic necrosis (IHN) and viral haemorrhagic septicaemia (VHS) (Decision 2006/88/EC) carried out by the Istituto Zooprofilattico Sperimentale delle Venezie (Italy). Hence, the IPNV samples characterized here derive from both monitoring and clinical outbreaks reported by farmers. Animals originated from 46 small-medium size facilities (farms with broodstock, hatcheries and on-growing farms) yielding <100 tons/year and located in the northeastern part of Italy (Friuli Venezia Giulia, Trentino Alto Adige and Veneto regions). These regions account for approximately 65 per cent of the national salmonid production. Viral isolates were obtained after a maximum of two passages in blue gill fry (BF-2) and/or epithelioma papulosum cyprini (EPC) cell lines (Wolf and Quimby 1962; Fijan et al. 1983) following standard procedures. IPNV strains (n = 75 in total) were isolated from rainbow trout (Oncorhynchus mykiss) (n = 61), char (Salvelinus spp.) (n = 8), brown trout (Salmo trutta) (n = 5) and Arctic char (Salvelinus alpinus) (n = 1) specimens. Detailed information on the region and species of origin, sampling date and genetic data accessibility are available in Supplementary Table S1.
2.2 RNA isolation and RT-PCR
Total RNA was purified from 140 μl of cell culture supernatant using the QIAamp Viral RNA Mini kit (Qiagen) and subsequently quantified spectrophotometrically with Nanodrop One ™ (ThermoFisher Scientific). The nucleotide regions corresponding to the viral polymerase (VP1) and the precursor of the outer capsid protein (pVP2) were amplified using the SuperScript III One-Step RT-PCR system with Platinum Taq High Fidelity (ThermoFisher Scientific) and specific primer sets (Table 1). Specifically, the 25 μl reverse transcription polymerase chain reaction (RT-PCR) mix contained 1X reaction mix, 5 mM MgSO4, 0.4 μM of each primer, 0.5 μl enzyme mix, 5 μl template RNA, 10 U RNasin Plus Ribonuclease Inhibitor (Promega) and distilled water to volume. The following thermal profile was applied: 30 min retrotranscription at 55°C, 2 min pre-denaturation at 94°C, 40 cycles of 15 sec denaturation at 94°C, 30 sec annealing at 56°C, 3 min elongation at 68°C and a final elongation step at 68°C for 5 min.
Table 1.
Primers used for the amplification of the complete VP1 and pVP2 nucleotide sequences of Italian IPNV.
| Primer | Sequence (5′ → 3′) | Target (bp) | References |
|---|---|---|---|
| FA5′NC | GGAAAGAGAGTTTCAACG | pVP2 (1758) | Romero-Brey et al. (2009) |
| P12 | TGCACCACAGGAAAGATGACTC | Cutrín et al. (2004) | |
| B-B5′NC | GGAAACAGTGGGTCAACG | VP1 (2777) | Santi, Vakharia, and Evensen (2004) |
| B-Bgl3′NC | GGGGTCCCTGGCGGAAC |
2.3 Genome sequencing
PCR products were purified with Agencourt AMPure XP (Beckman Coulter) and quantified with Qubit dsDNA HS assay kit (ThermoFisher Scientific). Libraries were prepared using the Nextera XT DNA Sample Preparation kit (Illumina), and Agencourt AMPure XP (Beckman Coulter) was used for fragments selection. Fragment quality and size were checked with High Sensitivity DNA Analysis kit (Agilent). Libraries were pooled in equimolar ratios and finally sequenced with the Miseq v3 Reagent Kit (600 cycles) using the Illumina MiSeq platform.
The quality of the reads was assessed using FastQC v0.11.2 (Andrews 2014). Raw data were filtered and processed by removing: (i) reads with more than 10 per cent of undetermined bases, (ii) reads with more than 100 bases with Q score <7, (iii) potential duplicate reads, (iv) Illumina adapter sequences using Scythe v0.991 (https://github.com/vsbuffalo/scythe) and (v) low-quality ends with Sickle v1.33 (https://github.com/najoshi/sickle). Reads shorter than 80 bases or unpaired after previous filters were also discarded. High-quality reads were aligned against the reference sequences under the GenBank accession numbers M58757 (VP1) and EF493156 (VP2) using BWA v0.7.12 (Li and Durbin 2010) and standard parameters. Alignments were converted into .bam format with SAMtools v0.1.19 (Li et al. 2009) and sorted by position. Variants were called using LoFreq v2.1.2 (Wilm et al. 2012) by editing potential alignments errors, re-aligning reads in close proximity of insertions and deletions (indels) and re-calibrating base quality with Picard-tools v2.1.0 (http://picard.sourceforge.net) and GATK v3.5 (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013). LoFreq was then run on edited alignments with the option ‘–call-indels’ to produce a .vcf file containing both single nucleotide polymorphisms (SNPs) and indels. Indels with a frequency <50 per cent and SNPs with a frequency <25 per cent were discarded. To obtain consensus sequences using a ‘reference-based’ assembly, the following criteria were adopted: (i) given a position ‘j’, if coverage is <10× an undetermined nucleotide ‘N’ is assigned, (ii) given a position ‘j’, if coverage is ≥10× and no SNPs are observed the nucleotide of the reference sequence is assigned, (iii) given a position ‘j’, if coverage is ≥10× and at least one SNPs is detected the nucleotide corresponding to the observed variants is assigned, according to the IUPAC code (http://www.bioinformatics.org/sms/iupac.html). Finally, the terminal sequences and low coverage regions of the consensus sequences produced were manually checked with Tablet v1.14.10.21 (Milne et al. 2010).
2.4 Phylogenetic analysis
The VP1 and pVP2 consensus sequences obtained here were aligned and compared to reference sequences available in GenBank representative of the known IPNV genogroups using MEGA7 (Kumar, Stecher and Tamura 2016). Phylogenetic trees of these sequences were inferred using the maximum likelihood (ML) method available in the PhyML program version 3.1 (Guindon et al. 2010). These analyses utilized the general time-reversible (GTR) model of nucleotide substitution with a gamma distribution of among-site rate variation (with four rate categories, Γ4) and an SPR branch-swapping search procedure (Darriba et al. 2012). The best-fit model of nucleotide substitution was determined using MEGA7 (Kumar, Stecher, and Tamura 2016). Five hundred bootstrap replicates were performed to assess the robustness of each node using the same substitution model as above. Phylogenetic trees were visualized using the FigTree v1.4 package (http://tree.bio.ed.ac.uk/software/figtree/). Detection of possible recombination events was performed with the GARD method available in the Datamonkey/HyPhy package (Delport et al. 2010), employing the GTR nucleotide substitution model and neighbor-joining phylogenetic trees as input. Pairwise nucleotide (nt) and amino acid (aa) distances (p-distance method) among the Italian IPNV were estimated for both VP1 and pVP2 using MEGA7 (Kumar, Stecher, and Tamura 2016).
2.5 Analysis of rates of nucleotide substitution
To assess whether IPNV evolves according to a molecular clock, a necessary prerequisite for accurately estimating evolutionary rate, we performed a regression of root-to-tip genetic distances against sampling date using TempEst v1.5.1 (Rambaut et al. 2016). This analysis utilized both the VP1 and pVP2 Italian sequences, with the ML phylogenetic trees described above as input.
Given the presence of sufficient clock-like structure (see Section 3) (Figs 3 and 4), mean rates of nucleotide substitution per site, per year (subs/site/year) and times to the most recent common ancestor were estimated for the Italian IPNV using the Markov Chain Monte Carlo method implemented in BEAST version 1.8.4 (Drummond et al. 2012). This analysis employed the GTR model of nucleotide substitution, as determined previously by MEGA7 (Kumar, Stecher, and Tamura 2016), estimated base frequencies, a gamma model for site heterogeneity and no partition into codon positions. To select the best-fit molecular clock model (uncorrelated relaxed clock or strict clock) (Drummond et al. 2006), the estimators of the marginal likelihood—path sampling (PS) and stepping-stone (SS) sampling (Baele, Lemey, and Vansteelandt 2013)—were calculated. A constant population size coalescent model was used as a tree prior. For each parameter estimate, statistical uncertainty was expressed in values of the 95 per cent highest posterior density (HPD) credible interval. To achieve convergence assessed with Tracer v1.6 (Rambaut 2014), chains were run for 50 million iterations. Finally, TreeAnnotator v1.8.1 (Drummond and Rambaut 2007) was employed to summarize Maximum Clade Credibility (MCC) phylogenetic trees from the posterior distribution of trees, after the removal of an adequate burn-in (10% of the samples). Nodes with high posterior support values (≥0.9) were annotated with age intervals (height 95% HPD). MCC trees were visualized with FigTree v1.4.
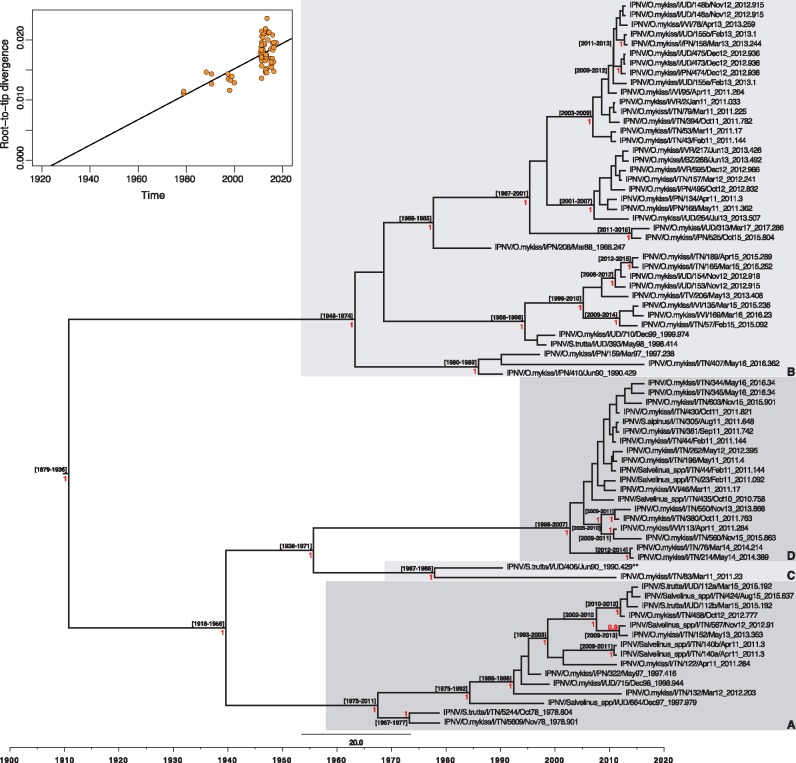
Figure 3.
Maximum clade credibility (MCC) tree for the VP1 sequence (2535 bp) of Italian IPNV strains. The tree was scaled to real time under a strict molecular clock. Mean tMRCAs (95% HPD range) are shown at the nodes. The red numbers at branch points represent posterior probability values (only values ≥ 0.9 are reported). Monophyletic clusters arbitrarily named A, B, C and D are highlighted with grey boxes. The putative reassortant strain, IPNV/S.trutta/I/UD/406/Jun90, is labeled with two asterisks (**). The upper-left panel represents the root-to-tip regression of the genetic distance against sampling date.
2.6 Analysis of selection pressures
Selection pressures in the VP1 and pVP2 gene sequences of the Italian IPNV were estimated using the ratio of non-synonymous (dN) to synonymous (dS) nucleotide substitutions per site (depicted in the ratio dN/dS). Posterior probabilities (PP) values ≥0.9 and P ≤ 0.05 were considered significant. These ratios were estimated using the Single Likelihood Ancestor Counting (SLAC), the Fixed Effects Likelihood (FEL), the Internal Fixed Effects Likelihood (IFEL) and the Fast Unconstrained Bayesian Approximation (FUBAR) methods (Kosakovsky Pond, and Frost 2005; Murrell et al. 2013) available in the Datamonkey/HyPhy package (Delport et al. 2010). These analyses employed the best-fit nucleotide substitution model identified for each data set and neighbor-joining phylogenetic trees as input.
3. Results
3.1 Sequencing and data accessibility
We determined the consensus sequences of the VP1 and the pVP2 genes of 75 IPNV samples from Italy. Average coverage ranged between 197-6882× and was therefore sufficient for reliable variant calling and for the production of high-quality consensus sequences. MiSeq raw data were submitted to the NCBI Sequence Read Archive (SRA) under the accession numbers SRR6302464–SRR6302538. The VP1 (2,535 bp) and pVP2 (1,536 bp) consensus sequences have also been deposited in GenBank under the accession numbers MG543492–MG543566 (VP1) and MG543567–MG543641 (pVP2).
3.2 Molecular characterization of Italian IPNV
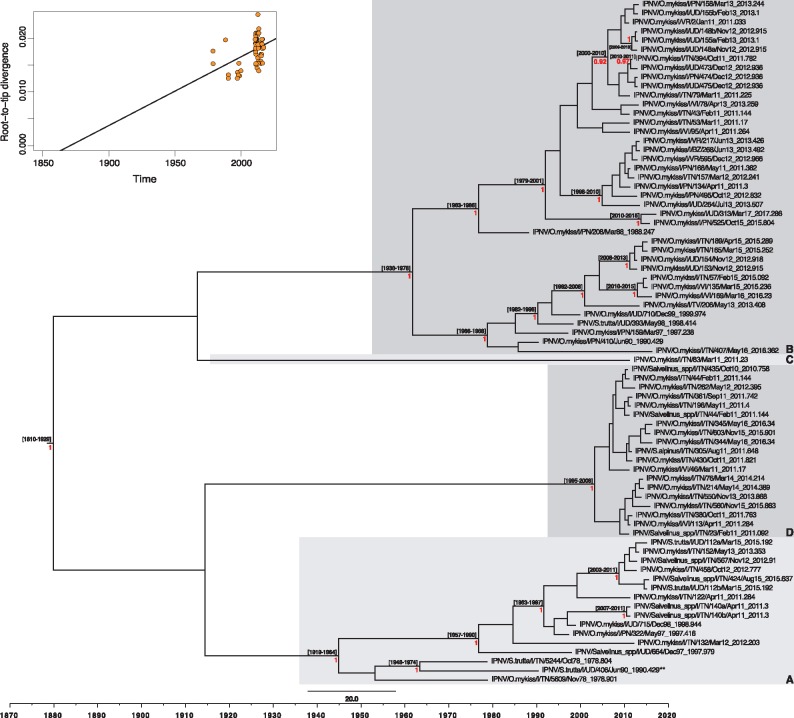
According to the classification proposed by Barrera-Mejía et al. (2010), the ML phylogenetic tree based on the VP1 sequences showed that all the Italian IPNV belonged to genetic group 2 that contains viruses from Europe and Canada (Fig. 1). Similarly, the analysis performed on the pVP2 sequences placed all the Italian IPNV within genogroup V based on the genotype subdivision developed by Blake et al. (2001), which corresponds to the Sp serotype (Fig. 2) that is common in Europe. In both the VP1 and pVP2 phylogenies, the Italian IPNV were distributed into four well-supported genetic clusters, arbitrarily named A–D. All the isolates were consistently placed within groups in both trees, with the exception of virus IPNV/S.trutta/I/UD/406/Jun90 that fell in clusters C and A in the VP1 and pVP2 phylogenies, respectively, reflecting the occurrence of inter-segment reassortment. Group A comprised virus isolates collected during the period 1978–2015, group B was formed by IPNV retrieved between 1988 and 2017, group C comprised two viruses from 1990 and 2011 (VP1) and one from 2011 (pVP2), and group D comprised recent strains collected during the period 2010–16 (Figs 1 and 2). No evidence of intra-segment recombination was found in either data set.
Figure 1.
ML phylogenetic tree of the complete VP1 sequence of IPNV (2535 bp). The Italian IPNV strains are shown in bold. Reference strains sequences are shown in grey. The genogroup subdivision (1, 2, MABV) according to Barrera-Mejía et al. (2010) is reported. Monophyletic clusters containing Italian strains and arbitrarily named A, B, C and D are highlighted with grey boxes. The putative reassortant strain, IPNV/S.trutta/I/UD/406/Jun90, is labeled with two asterisks (**). The numbers at nodes represent bootstrap values (only values ≥ 70% are reported). Branch lengths are scaled according to the number of nucleotide substitutions per site. The tree is mid-point rooted for clarity only.
Figure 2.
ML phylogenetic tree of the pVP2 sequence of IPNV (1536 bp). The Italian IPNV strains are shown in bold. Reference strains sequences are shown in grey. The genogroup subdivision (I-VII) according to Blake et al. (2001) and Nishizawa et al. (2005) is reported. Monophyletic clusters containing Italian strains and arbitrarily named A, B, C and D are highlighted with grey boxes. The putative reassortant strain, IPNV/S.trutta/I/UD/406/Jun90, is labeled with two asterisks (**). The numbers at nodes represent bootstrap values (only values ≥ 70% are reported). Branch lengths are scaled according to the number of nucleotide substitutions per site. The tree is mid-point rooted for clarity only.
The deduced amino acid sequences of the capsid protein precursor of the Italian IPNV were scrutinized for the presence of amino acid signatures previously found to be associated with putative virulence determinants identified in experimental challenge studies (Bruslind and Reno 2000; Santi, Vakharia, and Evensen 2004; Shivappa et al. 2004; Song et al. 2005) (Table 2). Intriguingly, all the Italian IPNV harbored a proline at position 217 (P217) and a threonine at position 221 (T221) that have been reported previously to be associated with a moderate/low virulent IPNV phenotype (Santi, Vakharia and Evensen 2004; Shivappa et al. 2004; Song et al. 2005).
Table 2.
VP2 amino acid signatures putatively involved in IPNV virulence.
| Amino acid signatures |
||||
|---|---|---|---|---|
| Site | Highly virulent | Moderately virulent | Avirulent | Italian IPNV (No.) |
| 199 | Ta | Ia | Ia | T (1/75), I (74/75) |
| 217 | Ta–d | Ab, Pa,d | Pa,c | P (75/75) |
| 221 | Ac,d, Ta | Aa, d | Tc, d Aa | T (75/75) |
| 247 | Ta | Aa | Aa | A (75/75) |
| 286 | Kb | Ab | – | G (28/75), R (47/75) |
| 288 | Va | Aa | Va | V (75/75) |
| 500 | Ya | Ha | Ha | H (3/75), Y (72/75) |
The specific amino acid residue and the associated phenotype are shown for each site. The number of Italian IPNV strains harboring specific amino acid signatures are reported in the last column.
3.3 Selection pressures in IPNV
The mean dN/dS ratios estimated for the polymerase and the capsid protein precursor sequences of Italian IPNV were 0.069 and 0.096, respectively. The lower rate of non-synonymous compared to synonymous substitution per site indicates that these sequences are subject to relatively strong purifying selection over the sampling period. Indeed, the only evidence for site-specific positive selection in the Italian VP1 and pVP2 sequences in any of the methods used (SLAC, FEL, IFEL and FUBAR) was at residue 248 of the pVP2 (IFEL, P = 0.034; FUBAR, PP = 0.98).
3.4 Rates and dates of IPNV evolution in Italy
The Italian IPNV strains analyzed here were characterized by low levels of genetic diversity: 0–3.23 per cent for VP1 and 0–2.93 per cent for pVP2. Similarly, at amino acid level, the distances estimated were 0–1.9 per cent for the viral polymerase and 0–2.34 per cent for the capsid protein precursor. The distances calculated between the oldest (IPNV/S.trutta/I/TN/5244/Oct78) and the youngest (IPNV/O.mykiss/I/UD/313/Mar17) samples of the data set were 2.13 per cent (nt) and 0.36 per cent (aa) for VP1 and 2.8 per cent (nt) and 1.37 per cent (aa) for pVP2.
A root-to-tip regression of the genetic distance against sampling date showed the existence of some temporal structure (i.e. clock-like evolution) in both data sets (Figs 3 and 4), with correlation coefficients of 0.63 and 0.41 for VP1 and pVP2, respectively. This allowed a more detailed estimation of the rates of evolution with a Bayesian Markov chain Monte Carlo approach (Drummond et al. 2012). Accordingly, the estimators of the marginal likelihood (PS and SS) identified the strict and the uncorrelated lognormal relaxed molecular clocks as the best-fit molecular clock models for the polymerase gene and the capsid protein precursor sequence, respectively. Under these models, the mean evolutionary rates of the Italian IPNV were 1.7 × 10−4 subs/site/year (95% HPD, 1.24–2.2 × 10−4 subs/site/year) for VP1 and 1.45 × 10−4 subs/site/year (95% HPD, 0.91–2.03 × 10−4 subs/site/year) for pVP2. These rates are consistently lower than those observed in other birnaviruses (see Section 4). Finally, we used these rates to estimate the times to the common ancestry of the Italian IPNV, which dated to 1878–1936 (95% HPD values; mean 1909) and 1814–1930 (95% HPD values; mean 1875) for the polymerase and the capsid protein genes, respectively, as shown in the MCC trees (Figs 3 and 4).
Figure 4.
Maximum clade credibility (MCC) tree for the pVP2 sequence (1536 bp) of Italian IPNV strains. The tree was scaled to real time under an uncorrelated lognormal relaxed molecular clock. Mean tMRCAs (95% HPD range) are shown at the nodes. The red numbers at branch points represent posterior probability values (only values ≥ 0.9 are reported). Monophyletic clusters arbitrarily named A, B, C and D are highlighted with grey boxes. The putative reassortant strain, IPNV/S.trutta/I/UD/406/Jun90, is labeled with two asterisks (**). The upper-left panel represents the root-to-tip regression of the genetic distance against sampling date.
4. Discussion
We analyzed, for the first time, the evolutionary history and dynamics of IPNV isolated from farmed Italian salmonids between 1978 and 2017. There is currently some discordance in the classification of the infectious pancreatic necrosis virus due to diverse genogroups numbering adopted by different authors. Following the recommendation of Bain, Gregory, and Raynard (2008), we subdivided the data into six genogroups (I–VI) for the pVP2 sequences as proposed by Blake et al. (2001), with the addition of the seventh genogroup (VII) identified by Nishizawa et al. (2005). To enhance our study, we also employed the VP1 classification adopted by Barrera-Mejía et al. (2010). Based on these criteria, we found that all Italian IPNV belonged to a single genogroup in each gene: genogroup V in pVP2, corresponding to the Sp serotype, and genogroup 2 in VP1, and are further divided into four genetic clusters. With the exception of group D, which only contained recent IPNV strains (2010–16), groups A, B and C were formed by viral isolates recovered from wider time periods (1978–2015, 1988–2017 and 1990–2011, respectively) (Figs 1 and 2).
The phylogenetic analysis of both segments enabled us to identify a single reassortment event. Segmented RNA viruses are known to undergo genetic/antigenic shift by exchanging genomic molecules (McDonald et al. 2016), and this has been hypothesized previously also for IPNV by Romero-Brey et al. (2009) and was recently demonstrated by Lago et al. (2017). In the present study, we identified a single reassortant strain (IPNV/S.trutta/I/UD/406/Jun90) that clustered with group C in VP1 and with group A in pVP2. Given the long sampling period (1978–2017) of our study, such reassortment events may be relatively rare. Clearly, additional investigations are needed to determine the mechanisms that prevent or facilitate the occurrence of reassortment in IPNV, including differential replication and selection of viral genes, preferential linkage between segments A and B, selective packaging, and adaptation to different host species (Lago et al. 2017).
Little is known about evolutionary processes in IPNV in particular, or in the genus Aquabirnavirus in general. Using a data set of 75 samples isolated between 1978 and 2017 from Italian farmed salmonids, we provide the first assessment of evolutionary dynamics in IPNV, the type species of Aquabirnaviridae. Most notably, the viruses sampled here exhibited relatively low levels of genetic diversity (0–3.23% for VP1 and 0–2.93% for pVP2) over a period of almost 40 years, reflecting relatively low rates of nucleotide substitution. Indeed, we observed mean rates of only 1.70 × 10−4 subs/site/year for the polymerase gene and 1.45 × 10−4 subs/site/year for the major capsid protein precursor gene. These rates are lower, and less variable, than those observed previously in infectious bursal disease virus (IBDV), another member of the Birnaviridae family infecting poultry: 2.98 × 10−4 and 7.09 × 10−4 subs/site/year (95% HPD, 2.56–13.7 × 10−4 subs/site/year) for VP1 and 3.29 × 10−4 and 8.63 × 10−4 subs/site/year (95% HPD, 4.14–14.1 × 10−4 subs/site/year) for VP2 (Alfonso-Morales et al. 2013, 2015; Silva et al. 2013). More notably, it was also reported that the evolutionary rate of very virulent IBDV (vvIBDV) was higher than that of non-vvIBDV: 4.80 × 10−3 subs/site/year versus 1.88 × 10−4 subs/site/year (Alfonso-Morales et al. 2015). These authors suggested that the low rate in non-vvIBDV reflects its circulation and co-evolution within a restricted host population and, consequently, a different host–virus relationship. Similarly, we speculate that the lower evolutionary rate observed in Italian IPNV might also reflect the seemingly low virulence nature of the viruses sampled here, apparent in both the low mortality recorded in the field and by the presence of amino acid signature associated with low virulent phenotypes (Table 2). The relatively strong purifying selection observed in IPNV—with mean dN/dS ratios of only 0.069 for VP1 and 0.096 for pVP2—similarly supports this idea. Notably, an analogous correlation between low mortality and specific amino acid signatures has been noted elsewhere (Eriksson-Kallio et al. 2016; Mutoloki et al. 2016; Büyükekiz et al. 2017; Holopainen, Eriksson-Kallio, and Gadd 2017), although the multi-factorial nature of IPN and of virulence in general means that such relationship is not always clear (Ruane et al. 2009; Smail et al. 2006; Ruane et al. 2015; Julin, Mennen, and Sommer 2013).
In a previous study, we determined the molecular evolution of two salmonid rhabdoviruses in farmed Italian trout over the past 30 years: the IHN virus (IHNV) and the VHS virus (VHSV) (Abbadi et al. 2016). Their mean substitution rates were 11 × 10−4 subs/site/year and 7.3 × 10−4 subs/site/year, with dN/dS ratios of 0.38 and 0.21, respectively, and hence significantly different from that of IPNV observed here. This is notable as in the context of Italian trout farming IPNV, IHNV and VHSV are subject to common environmental conditions as they affect the same fish population. Indeed, these viruses can frequently co-circulate in the same farm or even in the same individuals. However, our study shows that these three viruses exhibit very different evolutionary dynamics and selection pressures. We hypothesize that the higher substitution rates of IHNV and VHSV might reflect elevated rates of virus replication and, possibly, a higher virulence during disease outbreaks, which in turn might increase positive selection pressure on the virus. Indeed, the mortality due to IHNV and VHSV is very high in the earliest developmental stages of fish (80–100%) and can also heavily affect adult animals. Conversely, IPNV causes moderate mortality (10–30%) in fry only, while survivors act as healthy lifelong carriers (Bootland et al. 1991). Interestingly, the threonine to alanine reversion at position 221 (T221A) of the VP2 protein enhances the replication capacity of IPNV in vitro and results in a higher virulence in vivo (Gadan et al. 2013). It is noteworthy that all the Italian IPNV herein characterized contained the T221 signature (Table 2), which is consistent with the lower replication efficiency of these viruses postulated by Gadan et al. (2013). We therefore propose that lower rates of replication also reduce evolutionary rates, and that lower virulence diminishes the extent of host selection on the virus so that selection pressures on the virus are also lower. However, additional work is needed to determine if the low rate of nucleotide substitution in IPNV reflects viral generation time and assess the relative extent of inter- versus intra-host evolution. It is also possible that the different molecular evolutionary dynamics in IPNV, IHNV and VHSV might be shaped by the diverse management approaches adopted for these pathogens. Indeed, as both IHNV and VHSV are the etiological agents of two serious OIE and EU notifiable diseases (World Organization of Animal Health/OIE, 2016; Decision 2006/88/CE) they are actively eradicated, or at least confined to avoid their spread, while no sanitization is generally applied for IPNV. The effects of the absence of control measures against IPNV are worsened by the under-estimation of its prevalence as adult fish act as healthy carriers, which in turn increases the endemicity of IPNV infection.
There is a general consensus that the onset and the development of infectious pancreatic necrosis is the result of a complex interplay between fish species, host immunological status, environmental conditions, disease management and stress factors (Bain, Gregory, and Raynard 2008; Houston et al. 2010; Skjesol et al. 2011; Dadar et al. 2013; Gadan et al. 2013; Tapia et al. 2015; Mutoloki et al. 2016; Büyükekiz et al. 2017). Additionally, the observation of specific amino acid signatures of VP2 related to virulence during in vivo experiments indicates that disease outcome might also depend on intrinsic virological features (Bruslind and Reno 2000; Santi, Vakharia, and Evensen 2004; Shivappa et al. 2004; Song et al. 2005). Obviously, the identification of such signatures is complicated by the fact that IPNV pathogenicity is multifactorial, such that inconsistent data exist in the literature. However, if the VP2 amino acid changes truly are virulence markers, then it is clear that our samples harbored residues associated with moderate/low virulence (Table 2). Indeed, all 75 viruses exhibited the P217 and T221 signatures that might explain the low mortality percentages recorded in the field and are consistent with previous observations (Eriksson-Kallio et al. 2016; Holopainen, Eriksson-Kallio, and Gadd 2017; Büyükekiz et al. 2017). However, it must be emphasized that the retrospective association between genomic fingerprints and in vivo virulence phenotype is extremely difficult, mainly because of the lack of systematic record keeping. This underscores that the molecular characterization of IPNV together with the methodical collection of field information are of utmost importance for the study of the mechanisms orchestrating IPNV virulence. Additionally, the limited genetic diversity and low rates of evolutionary change in our samples mean that molecular epidemiological inferences may be poorly informative in the identification of outbreak origins or tracking the spread of the infection among farms. The collection of field information and the application of classic epidemiological analyses will therefore continue to play a key role in understanding the risk factors determining IPNV diffusion and for the implementation of proper control measures.
Supplementary Material
Acknowledgements
This study was funded by the Italian Ministry of Health, RC IZSVe 17/13. ECH was funded by an ARC Australian Laureate Fellowship (FL170100022). GZ activity was supported by the RC IZSVe 05/14. Michele Gastaldelli is kindly acknowledged for graphical assistance. The authors wish to thank Andrea Fabris for providing important field information and Alice Fusaro for fruitful discussion. Special thanks also go to the official veterinary inspectors and to the fish farmers of the regions Friuli Venezia Giulia, Trentino Alto Adige and Veneto for fish sampling.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Abbadi M. et al. (2016) ‘Molecular Evolution and Phylogeography of Co-Circulating IHNV and VHSV in Italy’, Frontiers in Microbiology, 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Morales A. et al. (2013) ‘Spatiotemporal Phylogenetic Analysis and Molecular Characterisation of Infectious Bursal Disease Viruses Based on the VP2 Hyper-Variable Region’, PLoS One, 8: e65999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Morales A. et al. (2015) ‘Evaluation of a Phylogenetic Marker Based on Genomic Segment B of Infectious Bursal Disease Virus: Facilitating a Feasible Incorporation of This Segment to the Molecular Epidemiology Studies for This Viral Agent', PLoS One, 10: e0125853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2014) ‘FastQC, A quality control tool for high throughput sequence data’ <http://www.bioinformatics.babraham.ac.uk/projects/fastqc>.
- Baele G., Lemey P., Vansteelandt S. (2013) ‘Make the Most of Your Samples: Bayes Factor Estimators for High-Dimensional Models of Sequence Evolution’, BMC Bioinformatics, 14: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain N., Gregory A., Raynard R. S. (2008) ‘Genetic Analysis of Infectious Pancreatic Necrosis Virus from Scotland’, Journal of Fish Diseases, 31: 37–47. [DOI] [PubMed] [Google Scholar]
- Barrera-Mejía M. et al. (2010) ‘Molecular Characterization of the VP1 Gene of a Mexican Isolate of Infectious Pancreatic Necrosis Virus’, Canadian Journal of Veterinary Research, 74: 218–22. [PMC free article] [PubMed] [Google Scholar]
- Blake S. et al. (2001) ‘Phylogenetic Relationships of Aquatic Birnaviruses Based on Deduced Amino Acid Sequences of Genome Segment a cDNA’, Diseases of Aquatic Organisms, 45: 89–102. [DOI] [PubMed] [Google Scholar]
- Bootland L. M., Dobos P., Stevenson R. M. W. (1991) ‘The IPNV Carrier State and Demonstration of Vertical Transmission in Experimentally Infected Brook Trout’, Diseases of Aquatic Organisms, 10: 13–21. [Google Scholar]
- Bovo G., Giorgetti G.. 1979, ‘Isolamento Su Colture Cellulari Di Tessuto Ed Identificazione Sierologica Del Virus Della Necrosi Pancreatica Infettiva (NPI) Delle Trote Di Allevamento’, Atti Società Italiana delle Scienze Veterinarie, 33: 295. [Google Scholar]
- Bruslind L. D., Reno P. W. (2000) ‘Virulence Comparison of Three Buhl-Subtype Isolates of Infectious Pancreatic Necrosis Virus in Brook Trout Fry’, Journal of Aquatic Animal Health, 12: 301–15. [Google Scholar]
- Büyükekiz A. G. et al. (2017) ‘Infectious Pancreatic Necrosis Virus (IPNV) Serotype Sp Is Prevalent in Turkish Rainbow Trout Farms’, Journal of Fish Diseases, 41: 95–104. [DOI] [PubMed] [Google Scholar]
- Cutrín J. M. et al. (2004) ‘Restriction Fragment Length Polymorphisms and Sequence Analysis: An Approach for Genotyping Infectious Pancreatic Necrosis Virus Reference Strains and Other Aquabirnaviruses Isolated from Northwestern Spain’, Applied and Environmental Microbiology, 70: 1059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar M. et al. (2013) ‘Sequence Analysis of Infectious Pancreatic Necrosis Virus Isolated from Iranian Reared Rainbow Trout (Oncorhynchus mykiss) in 2012’, Virus Genes, 47: 574–8. [DOI] [PubMed] [Google Scholar]
- Darriba D. et al. (2012) ‘jModelTest 2: More Models, New Heuristics and Parallel Computing’, Nature Methods, 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas D., Mundt E., Vakharia V. N. (2012) ‘Family Birnaviridae’, in King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. (eds.) Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press, 499–507. [Google Scholar]
- Delport W. et al. (2010) ‘Datamonkey 2010: A Suite of Phylogenetic Analysis Tools for Evolutionary Biology’, Bioinformatics, 26: 2455–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A. et al. (2011) ‘A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data’, Nature Genetics, 43: 491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos P. (1976) ‘Size and Structure of the Genome of Infectious Pancreatic Necrosis Virus’, Nucleic Acids Research, 3: 1903–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos P. (1995) ‘Protein-Primed RNA Synthesis in Vitro by the Virion-Associated RNA Polymerase of Infectious Pancreatic Necrosis Virus’, Virology, 208: 19–25. [DOI] [PubMed] [Google Scholar]
- Dobos P., Rowe D. (1977) ‘Peptide Map Comparison of Infectious Pancreatic Necrosis Virus-Specific Polypeptides’, Journal of Virology, 24: 805–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos P., Roberts T. E. (1983) ‘The Molecular Biology of Infectious Pancreatic Necrosis Virus: A Review’, Canadian Journal of Microbiology, 29: 377–84. [DOI] [PubMed] [Google Scholar]
- Dorson M., Touchy C. (1981) ‘The Influence of Fish Age and Water Temperature on Mortalities of Rainbow Trout, Salmo Gairdneri Richardson, Caused by a European Strain of Infectious Pancreatic Necrosis Virus’, Journal of Fish Diseases, 4: 213–21. [Google Scholar]
- Drummond A. J. et al. (2006) ‘Relaxed Phylogenetics and Dating with Confidence’, PLoS Biology, 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian Evolutionary Analysis by Sampling Trees’, BMC Evolutionary Biology, 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A.. et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R. et al. (1987) ‘Synthesis of the Infectious Pancreatic Necrosis Virus Polyprotein, Detection of a Virus-Encoded Protease, and Fine Structure Mapping of Genome Segment a Coding Regions’, Journal of Virology, 61: 3655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R. et al. (1991) ‘Sequence Analysis of Infectious Pancreatic Necrosis Virus Genome Segment B and Its Encoded VP1 Protein: A Putative RNA-Dependent RNA Polymerase Lacking the Gly-Asp-Asp Motif’, Virology, 181: 541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson-Kallio A. M. et al. (2016) ‘Infectious Pancreatic Necrosis Virus (IPNV) Strain with Genetic Properties Associated with Low Pathogenicity at Finnish Fish Farms’, Diseases of Aquatic Organisms, 118: 21–30. [DOI] [PubMed] [Google Scholar]
- Fijan N. et al. (1983) ‘Some Properties of the Epithelioma Papulosum Cyprini (EPC) Cell Line from Carp Cyprinus carpio’, Annales De L’Institut Pasteur/Virologie, 134: 207–20. [Google Scholar]
- Gadan K. et al. 2013, ‘Stress-Induced Reversion to Virulence of Infectious Pancreatic Necrosis Virus in Naïve Fry of Atlantic Salmon (Salmo salar L.)’, PLoS One, 8: e54656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0.’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Havarstein L. S. et al. (1990) ‘Sequence of the Large Double-Stranded RNA Segment of the N1 Strain of Infectious Pancreatic Necrosis Virus: A Comparison with Other Birnaviridae’, Journal of General Virology, 71: 299–308. [DOI] [PubMed] [Google Scholar]
- Heppell J. et al. (1995) ‘Strain Variability and Localization of Important Epitopes on the Major Structural Protein (VP2) of Infectious Pancreatic Necrosis Virus1’, Virology, 214: 40–9. [DOI] [PubMed] [Google Scholar]
- Hill B. J. 1982, ‘Infectious Pancreatic Necrosis Virus and Its Virulence’, in Roberts R.J. (ed.) Microbial Diseases of Fish, pp. 91–114. Oxford: Academic Press. [Google Scholar]
- Hill B. J., Way K. (1995) ‘Serological Classification of Infectious Pancreatic Necrosis (IPN) Virus and Other Aquatic Birnaviruses’, Annual Review of Fish Diseases, 5: 55–77. [Google Scholar]
- Holopainen R. et al. (2017) ‘Molecular Characterisation of Infectious Pancreatic Necrosis Viruses Isolated from Farmed Fish in Finland’, Archives of Virology, 162: 3459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.-R., Gong H.-Y., Wu J.-L. (2002) ‘IPNV VP5, a Novel anti-Apoptosis Gene of the Bcl-2 Family, Regulates Mcl-1 and Viral Protein Expression’, Virology, 295: 217–29. [DOI] [PubMed] [Google Scholar]
- Houston R. D. et al. (2010) ‘The Susceptibility of Atlantic Salmon Fry to Freshwater Infectious Pancreatic Necrosis Is Largely Explained by a Major QTL’, Heredity, 105: 318–27. [DOI] [PubMed] [Google Scholar]
- Huang M. T. F. et al. (1986) ‘A Physical Map of the Viral Genome for Infectious Pancreatic Necrosis Virus Sp: Analysis of Cell-Free Translation Products Derived from Viral cDNA Clonest’, Journal of Virology, 60: 1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarp J. et al. (1995) ‘Risk Factors for Furunculosis, Infectious Pancreatic Necrosis and Mortality in Post-Smolt of Atlantic Salmon, Salmo solar L.’, Journal of Fish Diseases, 18: 67–78. [Google Scholar]
- Johans R. et al. (2002) ‘Pathological Changes in Juvenile Atlantic Halibut Hippoglossus Hippoglossus Persistently Infected with Nodavirus’, Diseases of Aquatic Organisms, 50: 161–9. [DOI] [PubMed] [Google Scholar]
- Julin K., Mennen S., Sommer A. I. (2013) ‘Study of Virulence in Field Isolates of Infectious Pancreatic Necrosis Virus Obtained from the Northern Part of Norway’, Journal of Fish Diseases, 36: 89–102. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Frost S. D. W. (2005) ‘A Simple Hierarchical Approach to Modeling Distributions of Substitution Rates’, Molecular Biology and Evolution, 22: 223–34. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago M. et al. (2017) ‘In Vitro Reassortment between Infectious Pancreatic Necrosis Virus (IPNV) Strains: The Mechanisms Involved and Its Effect on Virulence’, Virology, 501: 1–11. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010) ‘Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform’, Bioinformatics, 26: 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R.. et al. , 1000 Genome Project Data Processing Subgroup. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar G., Dobos P. (1994) ‘Evidence for the Detection of the Infectious Pancreatic Necrosis Virus Polyprotein and the 17-kDa Polypeptide in Infected Cells and of the NS Protease in Purified Virus’, Virology, 204: 580–9. [DOI] [PubMed] [Google Scholar]
- McAllister P. E., Owens W. J. (1995) ‘Assessment of the Virulence of Fish and Mouuscan Isolates of Infectious Pancreatic Necrosis Virus for Salmonid Fish by Challenge of Brook Trout, Salvelinus fontinalis (Mitchill)’, Journal of Fish Diseases, 18: 97–103. [Google Scholar]
- McDonald S. M. et al. (2016) ‘Reassortment in Segmented RNA Viruses: Mechanisms and Outcomes’, Nature Reviews Microbiology, 14: 448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. (2010) ‘The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data’, Genome Research, 20: 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I. et al. (2010) ‘Tablet–Next Generation Sequence Assembly Visualization’, Bioinformatics, 26: 401–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T. et al. (2009) ‘Confirmation and Fine-Mapping of a Major QTL for Resistance to Infectious Pancreatic Necrosis in Atlantic Salmon (Salmo salar): Population-Level Associations between Markers and Trait’, BMC Genomics, 10: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B. et al. (2013) ‘FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection’, Molecular Biology and Evolution, 30: 1196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoloki S. et al. (2016) ‘Infectious Pancreatic Necrosis Virus Causing Clinical and Subclinical Infections in Atlantic Salmon Have Different Genetic Fingerprints’, Frontiers in Microbiology, 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T., Kinoshita S., Yoshimizu M. (2005) ‘An Approach for Genogrouping of Japanese Isolates of Aquabirnaviruses in a New Genogroup, VII, Based on the VP2/NS Junction Region’, Journal of General Virology, 86: 1973–8. [DOI] [PubMed] [Google Scholar]
- Okamoto N. et al. (1984) ‘The Relation between the Change of Quantities of Infectious Pancreatic Necrosis Virus in Infected Rainbow Trout Fry and the Disease Process’, Fish Pathology, 19: 1–4. [Google Scholar]
- Rambaut A. et al. (2014). Tracer v1.6. <http://tree.bio.ed.ac.uk/software/tracer/>.
- Rambaut A. et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen)’, Virus Evolution, 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I. et al. (2009) ‘Genetic Analysis of Aquabirnaviruses Isolated from Wild Fish Reveals Occurrence of Natural Reassortment of Infectious Pancreatic Necrosis Virus’, Journal of Fish Diseases, 32: 585–95. [DOI] [PubMed] [Google Scholar]
- Ruane N. M. et al. (2009) ‘Molecular Differentiation of Infectious Pancreatic Necrosis Virus Isolates from Farmed and Wild Salmonids in Ireland’, Journal of Fish Diseases, 32: 979–87. [DOI] [PubMed] [Google Scholar]
- Ruane N. M. et al. (2015) ‘Phylogenetic Analysis of Infectious Pancreatic Necrosis Virus in Ireland Reveals the Spread of a Virulent Genogroup 5 Subtype Previously Associated with Imports’, Archives of Virology, 160: 817–24. [DOI] [PubMed] [Google Scholar]
- Santi N., Vakharia V. N., Evensen Ø. (2004) ‘Identification of Putative Motifs Involved in the Virulence of Infectious Pancreatic Necrosis Virus’, Virology, 322: 31–40. [DOI] [PubMed] [Google Scholar]
- Shivappa R. B. et al. (2004) ‘Molecular Characterization of Sp Serotype Strains of Infectious Pancreatic Necrosis Virus Exhibiting Differences in Virulence’, Diseases of Aquatic Organisms, 61: 23–32. [DOI] [PubMed] [Google Scholar]
- Silva F. M. F. et al. (2013) ‘Tracking the Molecular Epidemiology of Brazilian Infectious Bursal Disease Virus (IBDV) Isolates’, Infection, Genetics and Evolution, 13: 18–26. [DOI] [PubMed] [Google Scholar]
- Skjesol A. et al. (2011) ‘IPNV with High and Low Virulence: Host Immune Responses and Viral Mutations during Infection’, Virology Journal, 8: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smail D. A. et al. (2006) ‘Infectious pancreatic necrosis virus in Atlantic salmon, Salmo salar L., post-smolts in the Shetland Isles, Scotland: viruses identification, histopathology, immunohistochemistry and genetic comparison with Scottish mainland isolates’, Journal of Fish Diseases, 29: 31–41. [DOI] [PubMed] [Google Scholar]
- Song H. et al. (2005) ‘Molecular Determinants of Infectious Pancreatic Necrosis Virus Virulence and Cell Culture Adaptation’, Journal of Virology, 79: 10289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taksdal T. et al. (1998) ‘Induction of Infectious Pancreatic Necrosis (IPN) in Covertly Infected Atlantic Salmon, Salmo salar L., Post-Smolts by Stress Exposure, by Injection of IPN Virus (IPNV) and by Cohabitation’, Journal of Fish Diseases, 21: 193–204. [DOI] [PubMed] [Google Scholar]
- Tapia D. et al. (2015) ‘Detection and Phylogenetic Analysis of Infectious Pancreatic Necrosis Virus in Chile’, Diseases of Aquatic Organisms, 116: 173–84. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. et al. (2013) ‘From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline’, Current Protocols in Bioinformatics, 43: 11.10.1–.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A. et al. (2012) ‘LoFreq: A Sequence-Quality Aware, Ultra-Sensitive Variant Caller for Uncovering Cell-Population Heterogeneity from High-Throughput Sequencing Datasets’, Nucleic Acids Research, 40: 11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. (1988). Fish Viruses and Fish Viral Diseases. Ithaca: Cornell University Press. [Google Scholar]
- Wolf K., Quimby M. C. (1962) ‘Established Eurythermic Line of Fish Cells in Vitro’, Science (New York, N.Y.), 135: 1065–6. [DOI] [PubMed] [Google Scholar]
- World Organisation for Animal Health [OIE] (2016) Manual of Diagnostic Tests for Aquatic Animals. <http://www.oie.int/international-standard-setting/aquatic-manual/access-online/>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We determined the consensus sequences of the VP1 and the pVP2 genes of 75 IPNV samples from Italy. Average coverage ranged between 197-6882× and was therefore sufficient for reliable variant calling and for the production of high-quality consensus sequences. MiSeq raw data were submitted to the NCBI Sequence Read Archive (SRA) under the accession numbers SRR6302464–SRR6302538. The VP1 (2,535 bp) and pVP2 (1,536 bp) consensus sequences have also been deposited in GenBank under the accession numbers MG543492–MG543566 (VP1) and MG543567–MG543641 (pVP2).