Stronger Na+ extrusion and vacuolar sequestration are essential to confer better salt tolerance in bread wheat than in durum wheat. Removal of the root meristems increased salt sensitivity in wheat.
Keywords: Long-distance signalling, sodium extrusion, sodium sensing, tissue specificity, vacuolar sequestration
Abstract
The progress in plant breeding for salinity stress tolerance is handicapped by the lack of understanding of the specificity of salt stress signalling and adaptation at the cellular and tissue levels. In this study, we used electrophysiological, fluorescence imaging, and real-time quantitative PCR tools to elucidate the essentiality of the cytosolic Na+ extrusion in functionally different root zones (elongation, meristem, and mature) in a large number of bread and durum wheat accessions. We show that the difference in the root’s ability for vacuolar Na+ sequestration in the mature zone may explain differential salinity stress tolerance between salt-sensitive durum and salt-tolerant bread wheat species. Bread wheat genotypes also had on average 30% higher capacity for net Na+ efflux from the root elongation zone, providing the first direct evidence for the essentiality of the root salt exclusion trait at the cellular level. At the same time, cytosolic Na+ accumulation in the root meristem was significantly higher in bread wheat, leading to the suggestion that this tissue may harbour a putative salt sensor. This hypothesis was then tested by investigating patterns of Na+ distribution and the relative expression level of several key genes related to Na+ transport in leaves in plants with intact roots and in those in which the root meristems were removed. We show that tampering with this sensing mechanism has resulted in a salt-sensitive phenotype, largely due to compromising the plant’s ability to sequester Na+ in mesophyll cell vacuoles. The implications of these findings for plant breeding for salinity stress tolerance are discussed.
Introduction
Salinity stress is one of the major environmental constraints limiting agricultural crop production. However, physiological and genetic complexity of salinity stress tolerance has significantly handicapped the progress in crop breeding for salt-tolerant crops. One of the keys to overcoming this limitation is an understanding of the specificity of salt stress signalling and adaptation at the cellular and tissue levels.
Bread wheat (Triticum aestivum) is always considered as a more salt-tolerant species than durum wheat (Triticum turgidum) (Munns and James, 2003; Munns and Tester, 2008); this difference has been largely attributed to its superior ability to maintain lower Na+ accumulation in the leaf/shoot (Colmer et al., 2006; Cuin et al., 2010; James et al., 2011; Munns et al., 2012). However, it remains unclear whether this pattern is achieved by stronger Na+ exclusion traits from the shoot such as preventing Na+ loading into the xylem (Davenport et al., 2005; Läuchli et al., 2008; Zhu et al., 2016) or increased Na+ retrieval from the shoot (James et al., 2006; Davenport et al., 2007), or is due to reduced net root Na+ uptake per se. Also, some recent studies showed the lack of significant correlation between shoot Na+ exclusion ability in a broad range of plants (Arabidopsis, Rus et al., 2006; Jha et al., 2010; wheat, Genc et al., 2007; tomato, Almeida et al., 2014), indicating the importance of Na+ sequestration in the shoot (so-called tissue tolerance) as arguably the most essential component of salinity stress tolerance (Munns et al., 2016).
If the rate of unidirectional Na+ uptake in plants is about the same but they accumulate different quantities of Na+ in their tissues, then the difference in salinity stress tolerance may be attributed to the ability of a specific genotype to extrude Na+ actively after it has crossed the plasma membrane and entered the cytosol. This extrusion ability is mediated by the operation of the plasma membrane Na+/H+ exchangers encoded by SOS1 (Salt Overly Sensitive 1) in Arabidopsis (Shi et al., 2000, 2003). It is estimated that ~95% of all Na+ taken up by roots is pumped back into the rhizosphere (Munns and Tester, 2008) via this mechanism. To the best of our knowledge, the comparative functional analysis of the activity of SOS1-like Na+ efflux systems has never been conducted for a large number of plant accessions, and the question of whether the difference in salinity stress tolerance between durum and bread wheat is attributed to the difference in SOS1 activity has not been addressed. Meanwhile, insights into this issue might be beneficial for improving plant salt tolerance since in most cases the accumulated excessive Na+ in the cytosol causes the Na+ toxicity. Filling this knowledge gap was one of objectives of this study.
While the importance of cell type specificity in plant adaptive responses to salinity is widely accepted, most studies addressed this issue at the transcriptional (e.g. tissue-specific expression patterns of genes in plants under salt stress; Claes et al., 1990; Löw et al., 1996; Saijo et al., 2000; Jain et al., 2007; Dinneny et al., 2008; Iyer-Pascuzzi et al. ,2011) rather than the functional level. At the same time, there is a growing body of evidence suggesting that tissue specificity of stress sensing may be a key to understanding the complexity of the overall plant salt tolerance (Kiegle et al., 2000; Verdoy et al., 2004; Wu et al., 2015a; L. Shabala et al., 2016). In this context, knowledge of Na+ distribution between various cell compartments (i.e. the cytosol and vacuole) in different cell types may shed light on both mechanisms enabling plant adaptation to saline conditions and the nature of the elusive Na+ sensor. Indeed, the importance of vacuolar Na+ sequestration for overall salt tolerance is widely accepted (Apse et al., 1999; Zhang et al., 2001), and improved salt tolerance resulting from overexpressing the NHX1 K+(Na+)/H+ exchanger has been reported in many species (Apse et al., 1999; Xue et al., 2004; H. Chen et al., 2007; He et al., 2007; Zhang and Blumwald, 2001). However, most of the reported studies were focused on the essentiality of vacuolar Na+ sequestration in the shoot mesophyll cells, while the patterns of Na+ uptake, distribution, and sequestration in various types of root cells remain largely unexplored. At the same time, root cells confer the first line of defence and have to deal with the presence of high Na+ concentrations in the rhizosphere. Is vacuolar Na+ sequestration in roots essential for salinity stress tolerance? If yes, can this conclusion be extrapolated to all cell types? Answering these questions was another objective of this study.
While downstream targets and effectors mediating plant adaptive responses to salinity have been studied in sufficient detail, the nature of the putative salt sensor in plants remains a great mystery (Maathuis, 2014; Shabala et al., 2015). In our previous study (Wu et al., 2015a), we compared several bread wheat genotypes contrasting in salinity tolerance and found that the salt-tolerant group possesses a higher fluorescence Na+ signal from the cytosol in the root meristem zone compared with the salt-sensitive group. This (unexpected) finding led to the suggestion that the root meristem zone might participate in salt sensing, or execute a role as a salt sensor. Validating this hypothesis was the third objective of this study.
The main findings of our work are 2-fold. First, we provide direct evidence that higher Na+ extrusion ability in the root elongation zone and better vacuolar Na+ sequestration ability in the mature root zone may be essential for superior salinity tolerance in bread wheat as compared with durum wheat. Secondly, we provide new supporting evidence that root meristem cells might act as a tentative salt sensor or at least harbour the salt sensor components, and tampering with this sensing mechanism in roots affects a plant’s ability to sequester Na+ in the shoot mesophyll via some as yet unknown long-distance signal propagating from the root to the shoot.
Materials and methods
Plant material and growth conditions for MIFE and laser scanning confocal microscopy (LSCM) experiments
We used 27 bread wheat (Triticum aestivum) and 19 durum wheat (Triticum turgidum spp. durum) varieties. All seeds were obtained from the Australian Winter Cereal collection and multiplied in our laboratory. Twenty seeds for each variety were surface sterilized with 5% commercial bleach (containing 42 g l–1 NaClO) for 10 min, and then washed under running tap water for ~0.5 h. From 12 to 16 uniform seeds were grown in a l litre PVC container with distilled water. Good aeration was provided with aquarium stones connected to an air pump (Aqua One® 9500, Sydney, Australia). Seedlings were grown in darkness at 25 ± 2 °C for 3 d followed by the salinity treatment (100 mM NaCl) for 24 h. Salt-stressed seedlings were used to investigate the Na+ extrusion ability of wheat roots. The reasons why we used young wheat seedlings for microelectrode ion flux estimation (MIFE) experiments are 4-fold. First, root sensitivity to salt gradually declines with age (e.g. Chen et al., 2005). Thus, working with young seedlings has the advantage of achieving a better signal-to-noise resolution (hence, more accurate genotypic differentiation). Secondly, the ultimate aim of this work was to arm breeders with convenient high-throughput tools allowing a rapid screening of hundreds of lines. Working with younger seedlings is more convenient from this perspective. Thirdly, the MIFE measurements require roots to be exposed to the aqueous medium. From our past experience, prolonged hydroponic plant growth in nutrient-rich solution is confounded by a high probability of roots being contaminated. Growing plants in the soil and then washing roots to make them accessible for MIFE measurements is also a lengthy and highly complicated process that is further jeopardized by the possibility of the root tissue being damaged by such procedures. Finally, preliminary experiments in our laboratory have shown that the observed genotypic difference in ion flux patterns is largely invariant with plant age (data not shown).
Glasshouse experiments
Plants were grown in the glasshouse facilities at the University of Tasmania, essentially as described in Wu et al. (2015b). Briefly, 12–14 seeds of each variety were sown in a 4.5 litre PVC pot filled with standard potting mix (triplicates). After 6 d, pots were thinned to leave eight similar plants per pot, and salt treatment (300 mM NaCl) was applied for a further 5 weeks. A saucer was placed under each pot, and plants were irrigated twice a day by an automatic watering system with dripper outlets for both non-saline conditions and salinity treatment. The salt tolerance index (quantified on 0–10 scales and based on the extent of leaf chlorosis and the plant survival rate; Zhou et al., 2012) was scored before harvesting (see Supplementary Fig. S1A at JXB online). The higher numbers represent the lower salinity tolerance (e.g. 0, no visual stress symptoms; 10, dead plants). The above tolerance index measured at the vegetative stage showed significant (P<0.001) correlation with the relative grain yield of wheat cultivars reported in our earlier experiments (Supplementary Fig. S1B) and thus could be used as a convenient proxy for quantification of plant salinity stress tolerance.
Preparation of ion-selective microelectrodes for non-invasive ion flux measurement
Net Na+ and H+ fluxes were measured from excised roots using non-invasive MIFE (Shabala et al. 2006). Briefly, blank microelectrodes were pulled out from borosilicate glass capillaries (GC 150-10; Harvard Apparatus, Kent, UK), dried in an oven (at 225 ºC, overnight), and silanized with tributylchlorosilane (No 282707, Sigma-Aldrich, St. Louis, MO, USA). The silanized microelectrode blanks were filled from the back with an appropriate backfilling solution (for H+, 15 mM NaCl+40 mM KH2PO4, pH 6.0 adjusted using NaOH; and for Na+, 0.5 M NaCl) followed by front filling the tip with a corresponding liquid ion exchanger (LIX). A commercially available LIX was used for H+ (catalogue no. 95297, Sigma-Aldrich) while the LIX for Na+ was prepared in the laboratory as described in Jayakannan et al. (2011). The prepared microelectrodes were mounted in MIFE electrode holders and calibrated in an appropriate set of standards (see Shabala et al., 2006 for methodological details). Only microelectrodes with a slope >50 mV per decade and correlation >0.999 were used.
MIFE screening for root Na+ extrusion ability
All 46 bread and durum wheat genotypes were screened for their ability to extrude Na+ actively from the root, using the non-invasive MIFE technique. For this, we have used the protocol developed by Cuin et al. (2011) and validated in further studies (Zhu et al., 2016). In brief, roots of hydroponically grown seedlings treated with 100 mM NaCl for 24 h [see ‘Plant material and growth conditions for MIFE and laser scanning confocal microscopy (LSCM) experiments’] were gently rinsed in a beaker containing 10 mM CaCl2 for 1 min followed by gentle rinsing in ddH2O for another minute. About 3 cm long root segments were cut starting from the root tip. Excised root segments were then mounted in a fixed plastic holder in a Petri dish containing 10 ml of ddH2O. Na+ and H+ selective microelectrode tips were positioned 40 μm above the root epidermis in the middle of the elongation zone (~2 mm from the tip). Measurements started 30 min after immobilization. By this time, all transient responses related to Donnan exchange in the apoplast have subsided (see Cuin et al., 2011 for evidence) and the measured Na+ efflux from the root was mediated mainly by SOS1-like Na+/H+ exchangers in the root plasma membrane (see Supplementary Fig. S2 for supporting evidence). During measurements, electrodes were travelling forth and back between two positions, 40 μm and 110 μm from the root surface, in a 12 s square-wave cycle. The electrochemical gradient potential was recorded and then converted into net ion fluxes using the calibration values of the ion-selective microelectrodes and cylindrical diffusion geometry of the root (MIFEFLUX software; Newman, 2001; Shabala et al., 2006). At least six root segments were assessed for each treatment and variety. Each root was measured for at least 5 min, to ensure steady-state flux values.
Imaging Na+ distribution in vacuolar and cytosolic compartments in mesophyll cells and different root zones in wheat by LSCM
The green fluorescent Na+ dye CoroNa Green acetoxymethyl ester (cat. no. C36676, Invitrogen) was used to assess the amount of Na+ accumulated in the cytosol and vacuole in different root zones and also mesophyll cells using established protocols (Bonales-Alatorre et al., 2013 for mesophyll cells; Wu et al., 2015a for root cells). For root measurements, two 10 mm long segments were cut from seminal wheat roots—one in the mature zone (30–40 mm from the apex) and another one from the apex (the first 10 mm). Four-day-old wheat seedlings (grown hydroponically) under 100 mM NaCl for another 72 h were used for collecting root samples. For the leaf experiment, the youngest fully expanded leaves were excised, their abaxial epidermis peeled off with a pair of fine forceps to expose the mesophyll cells, and 5 × 8 mm segments were cut. Leaf samples were prepared from plants grown in the potting mix. Roots meristem tissue was removed from 4-day-old hydroponically grown seedlings. These seedlings were then transplanted to the potting mix, allowed to adapt for 2 d, and then treated with 200 mM NaCl for 2 weeks. Root and leaf samples were incubated in a solution containing 20 μM CoroNa Green for 2 h in the dark, rinsed in a buffer solution (5 mM MES, pH 6.3 adjusted with KOH), and examined using an upright Leica laser scanning confocal microscope SP5 (Leica Microsystems, Germany) equipped with a ×40 oil immersion objective (excitation wavelength, 488 nm; emission, 510–520 nm). LAS Lite software (Leica Microsystems, Germany) was used for image analyses (Supplementary Fig. S3).
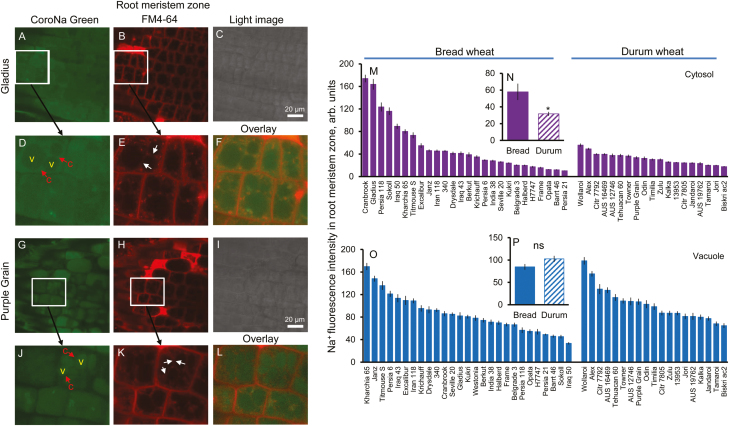
To validate the above protocol further and provide accurate quantification of the fluorescent signal in each intracellular compartment, roots were co-stained with CoroNa Green-AM and FM4-64, a dye that stains membrane structures including the plasma and tonoplast mebranes (Oh et al., 2010; Bassil et al., 2011). Roots were co-stained with 20 μM CoroNa Green-AM and 20 μM FM4-64 for 2 h in the dark as describe above. The samples were then rinsed with the buffer solution (5 mM MES, pH 6.3 adjusted with KOH) for 3 min and analysed using confocal imaging facilities (Confocal SP5). The 488 nm excitation line was used for FM4-64 fluorescence, and its signals were collected with a 615 nm long-pass filter. Figure 4 shows the results of the co-staining of roots with CoroNa Green dye and FM4-64 membrane dye.
Fig. 4.
Genetic variability of Na+ intensity in the cytosol and vacuole in the root meristem zone. (A and G) Representative images of root meristem zone cells stained with CoroNa Green dye in bread wheat Gladius (A) and durum wheat Purple Grain (G) cultivars. (B, H) and (C, I) The corresponding FM4-64 tonoplast staining and light images, respectively. (D–F) and (J–L) The representative area of Na+ staining in Gladius and Purple Grain, respectively. c, cytosol, v, vacuole. (M and O) Na+ content in the cytosolic (M) and vacuolar (O) compartments in the root meristem zone of 46 wheat varieties contrasting in their salinity tolerance quantified by the intensity of CoroNa Green fluorescence (arbitrary units). Mean ±SE (n=72–96 cells from at least six individual plants). (N and P) Averaged pooled values for Na+ intensity in the cytosol (N) and vacuole (P) in the root meristem zone for bread and durum wheat clusters. Mean ±SE [n=19 (durum wheat) and 27 (bread wheat)]. *Significant at P<0.05. Hydroponically grown 4-day-old wheat seedlings were treated with 100 mM NaCl for 72 h.
As CoroNa Green is not a ratiometric dye, the absolute Na+ concentration in cell compartments could not be quantified. Because of this, the mean fluorescence intensity values for cytosolic and vacuolar compartments were calculated (in arbitrary units) for each cell in each region. Light microscopy images (visualizing root cell morphology) and FM4-64 membrane dye were used to clarify the signal profile distribution in the cytosol and vacuole, and special attention was paid to ensure that all the imaging settings on the confocal microscope were identical, to allow such a comparison. Root meristem, elongation, and mature zones were distinguished by root morphology. Readings (72–96 cells for each genotype for the root experiment and 47–86 cells for the mesophyll experiment) were averaged and reported. This large sample size has minimized possible confounding effects of dye loading. It should also be mentioned that we have previously undertaken a comprehensive methodological study optimizing the loading time and dye concentration (Wu et al., 2015a) and showed that reported readings are not affected by any salt stress-induced fluctuation in either cytosolic pH or K+ (Wu et al., 2015a).
Before the above experiments were conducted, the possible impact of the loading time and dye concentration used was studied in dedicated methodological experiments. In one of them, we studied the effect of CoroNa Green dye concentration on observed patterns of Na+ distribution within the cell (using a dye concentration up to 80 μM). These results are shown in Supplementary Fig. S4. We have also studied the effects of incubation time (increasing the time of dye loading up to 5 h; Supplementary Fig. S5). Our results (Supplementary Figs S4, S5) showed that the patterns of Na+ signal distribution between the cytosol and vacuole in the root zones of two contrasting varieties were invariant in both dye concentration used and loading (incubation) time. Also, the pattern of Na+ intensity between the cytosol and vacuole in wheat root is not affected by a different depth of laser scanning (Wu et al., 2015a).
Studying physiological effects of removal of the root meristem
From 12 to 16 uniform seeds of salt-tolerant bread wheat (variety Kharchia 65) were grown hydroponically in l litre PVC containers. The container was filled with ddH2O (sterile double-distilled water) and seedlings were grown with aeration for 3–4 d in the dark. Seedling numbers were thinned to six in each container. Roots meristem (the first ~0.3 mm apical segment from the root tip) was surgically removed from seminal roots in each plant under a microscope before applying treatment (Supplementary Fig. S6). Plants were then transferred back into containers with 1/4 Hoagland solution and grown for a further 10 d in the presence and absence of 200 mM NaCl under ~150 μmol m–2 s–1 irradiance at 26/20 ± 1.0 °C day/night temperatures, 16 h/8 h day/night cycle. A number of physiological parameters were measured at the end of the experiment, including: leaf chlorophyll content (measured in arbitrary units by a chlorophyll meter SPAD-502, Konica Minolta, Tokyo, Japan), the maximal photochemical efficiency of PSII, Fv/Fm (quantified by a chlorophyll fluorometer OS-30p, Optisciences, Hudson, NH, USA), and shoot and root FW (from plants rinsed in ddH2O to remove the possible salt residues). Samples were then dried at 65 °C for 72 h in a Unitherm Dryer (Birmingham, UK) and the DW of shoots and roots was estimated. During salt stress treatment, lateral roots in salt-stressed plants were checked and eliminated every day.
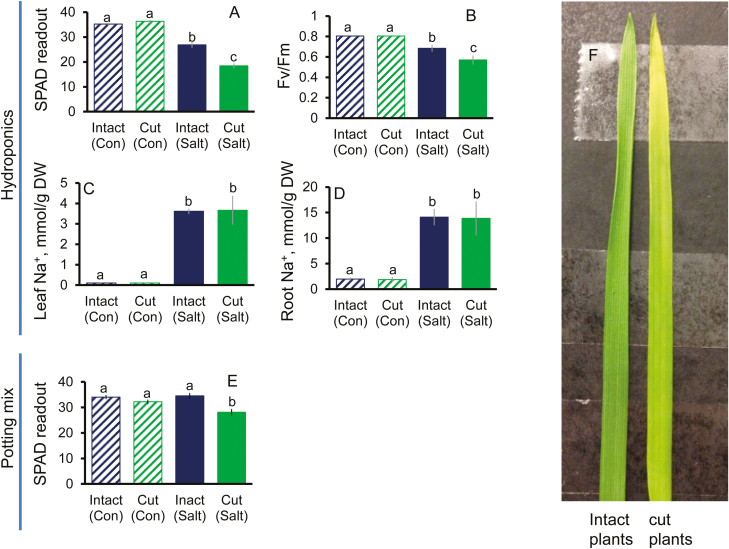
Another experiment with root meristem removal was conducted in plants grown in a potting mix under glasshouse conditions. Seedlings were grown in paper rolls in containers with ddH2O for 4 d. Root meristem segments were removed from each root of each plant, as described above. Then control plants (with intact roots) and cut plants (with the meristem zone removed from roots) were transplanted to a 1.7 litre pot filled with the standard potting mix [80% composted pine bark; 10% sand and 10% coir peat; plus complete N:P:K (8:4:10), 1 kg m–3; dolomite, 8 kg m–3; gypsum, 1 kg m–3; iron sulphate, 1 kg m–3; isobutylenediurea, 1 kg m–3; trace element mix, 0.75 kg m–3; wetting agent, 0.75 kg m–; zeolite, 0.75 kg m–3; pH 6.0]. Pots were hand-watered with tap water for 2 d to allow the transplanted plants to adapt to the potting mix. Then, plant numbers were thinned to six in each pot and salt stress (200 mM NaCl) was applied for a further 15 d. At the end of the experiment, SPAD values were measured.
Leaf and root Na+ content
Dry leaf and root samples were digested in a 120 ml Teflon digestion vessel in a microwave digester (Mars 6 Microwave Digestion System, CEM Corporation, Matthews, NC, USA) using 7 ml of 70% HNO3 per 0.1 g specimen. Digested samples were centrifuged at 5000 g for 10 min at room temperature (Avanti J-30I centrifuge, Beckman Coulter, Krefeld, Germany). After centrifugation, 0.2 ml of the digested solution was diluted with ddH2O to a final volume of 10 ml. Na+ content was then measured using a Flame Photometer (PFP7, Jenway, UK).
Quantitative real-time PCR analysis
Plants with intact and cut roots grown in 1/4 Hoagland solution in the absence and presence of 200 mM NaCl were used. The youngest fully expanded leaves (variety Kharchia 65) were harvested and snap-frozen in liquid nitrogen. The leaf RNA was extracted and synthesized to cDNA by using the RNeasy Mini Kit (QIAGEN) and the QuantiTect® Reverse Transciption Kit (QIAGEN) following the manufacturer’s instructions. Quantitative real-time PCR was performed using a RG6000 Rotor-Gene Q Real Time Thermal Cycler and SYBR green PCR reagent (QIAGEN) as described in Vandesompele et al. (2002). The 2−∆∆CT method (Livak and Schmittgen 2001) was used to analyse the relative expression levels of the studied genes. Primers with a single melting curve were designed by Primer Premier6 based on the conserved part of the gene sequence to determine the expression of TaSOS1, TaNHX1, TaVP, and TaHA1 (see Supplementary Table S1 for primer sequences). The control gene (TaActin) which shows a constant expression level in all samples was used for normalization of the test gene transcript. Experiments were repeated three times (each biological replicates had 3–4 technical replicates) with consistent results.
Data analysis
All data (given as mean ±SE, n=biological replicates) were analysed by using SPSS 20.0 for windows (SPSS Inc., Chicago, IL, USA). The comparisons of different parameters between bread and durum wheat were performed by independent samples t-test and one-way ANOVA based on Duncan’s multiple range test (two tailed). *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively; ns stands for P>0.05. Different lower case letters represent the significance at P<0.05. The significance of correlations between different parameters was determined by bivariate correlations based on Pearson correlation (two tailed).
Results
Bread wheat excludes more Na+ from the root elongation zone
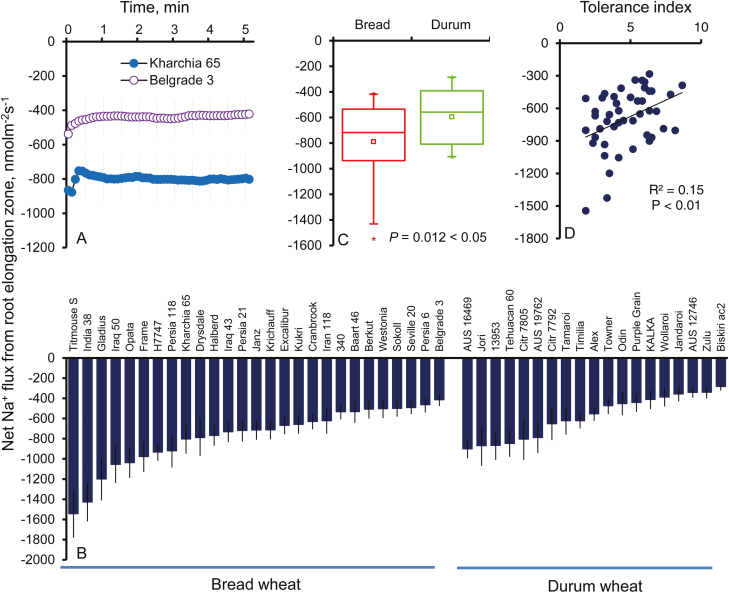
Confocal imaging of SOS1–green fluorescent fusion proteins in transgenic Arabidopsis plants indicated that SOS1 is localized in the plasma membrane, with the strongest signal detected in the root apex and, specifically, the elongation zone (Shi et al., 2002). Accordingly, we have undertaken the functional assessment of SOS1-like activity in epidermal cells in this zone in various durum and bread wheat cultivars using the MIFE technique. At 30 min after rinsing with CaCl2 solution, measured Na+ fluxes were steady (Fig. 1A) and thus reflected the functional activity of SOS1. Two lines of evidence support this claim: (i) such Na+ efflux showed high sensitivity to amiloride, a known inhibitor of Na+/H+ exchangers (Kleyman and Cragoe, 1988; Cuin et al., 2011); and (ii) unlike wild-type Arabidopsis plants, where Na+ efflux was detected, the process was absent from roots of the sos1 mutant lacking a functional SOS1 transporter (Supplementary Fig. S2). Over 5-fold variability was found among the 46 screened wheat varieties, with the highest Na+ efflux being –1584 ± 237 (in Titmouse S, bread wheat) and the lowest Na+ efflux being –284 ± 39 nmol m–2 s–1 (in Biskiri AC2, durum wheat) (Fig. 1B). On average, Na+ extrusion ability in bread wheat in the root elongation zone was 33% higher compared with durum wheat (mean flux values 773 ± 55 nmol m–2 s–1 and –580 ± 48 nmol m–2 s–1; significant at P<0.05) (Fig. 1C). A strong and positive correlation was observed between Na+ extrusion and plant overall salinity stress tolerance based on the damage index (r=0.39, P<0.01; Fig. 1D). Also, a significant (P<0.01) up-regulation of TaSOS1 in the root apex was found in bread wheat but not durum wheat under salt stress (Supplementary Fig. S7A).
Fig. 1.
Genetic variability of Na+ efflux in the root elongation zone after the removal of external salt. (A) Net steady-state Na+ efflux measured from the root elongation zone after the removal of external salt in two bread wheat genotypes (Kharchia 65, salt tolerant; and Belgrade 3, salt sensitive). The sign convention is ‘efflux negative’. Mean ±SE (n=6−13). (B) Net steady-state Na+ efflux measured from all 46 wheat varieties. Mean ±SE (n=6−13). (C) Pooled mean values of net Na+ efflux for bread and durum wheat clusters. Mean ±SE [n=19 (durum wheat) and 27 (bread wheat)]. *Significant at P<0.05. (D) Correlation between net Na+ efflux and tolerance index in the studied wheat varieties. Every single point represents one variety. Hydroponically grown 3-day-old wheat seedlings treated with 100 mM NaCl for 24 h were used.
Durum wheat compensates poor Na+ extrusion ability from the root elongation zone by more efficient vacuolar Na+ sequestration
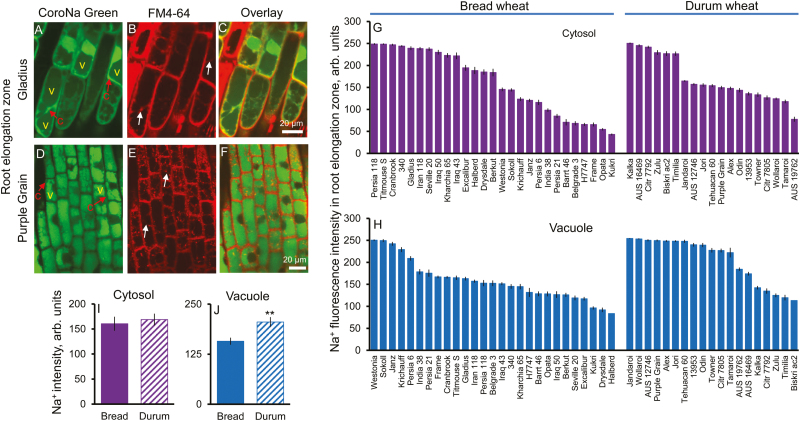
The profile of Na+ distribution between the cytosol and vacuole in the root elongation zone is shown in Fig. 2A–E. A large variability (5.8-fold) of cytosolic Na+ intensity was observed in all 46 wheat varieties tested, ranging from the highest value of 250 ± 2 (Kalka, durum wheat) to the lowest of 43 ± 2 (Kukri, bread wheat) (Fig. 2G). Similarly, a 3-fold variability was found for vacuolar Na+ intensity in the root elongation zone within the 46 screened wheat varieties (Fig. 2H). It ranged from the highest value of 254 ± 0.3 (Jandaroi, durum wheat) to the lowest of 83 ± 5 (Halberd, bread wheat) (Fig. 2H). No significant difference in cytosolic Na+ intensity in the root elongation zone was found between bread and durum wheat (mean fluorescence intensities 160 ± 14 and 169 ± 12, respectively; P>0.05; Fig. 2I). A significant negative correlation (P<0.05) was found between Na+ extrusion and cytosolic Na+ intensity in the root elongation zone (Supplementary Fig. S8A). At the same time, durum wheat showed an ~31% higher capacity for vacuolar Na+ sequestration (205 ± 12 and 157 ± 9, respectively; P<0.01; Fig. 2J) compared with bread wheat. A significant up-regulation (P<0.05) of TaNHX1 in the root apex was found in durum wheat, whereas no change was found in bread wheat under salt stress (Supplementary Fig. S7C). Also, a significant negative correlation (P<0.05) was found between cytosolic and vacuolar Na+ intensity in the root elongation zone (Supplementary Fig. S8B). Taken together with the finding that bread wheat has significantly higher Na+ extrusion in the root elongation zone than durum wheat (Fig. 1B, C), our results suggest that both species are capable of maintaining the same level of cytosolic Na+ in the elongation zone, although by different mechanisms: in bread wheat, Na+ removal is directed towards the apoplast, while in durum wheat this is achieved by the higher extent of Na+ sequestration in the vacuole.
Fig. 2.
Genetic variability of Na+ intensity in the cytosol and vacuole in the root elongation zone. (A, B) and (D, E) Representative images of root elongation zone cells stained with CoroNa Green dye and FM4-64 membrane dye in bread wheat Gladius (A, B) and durum wheat Purple Grain (D, E) cultivars. (C and F) The corresponding overlay images. c, cytosol, v, vacuole. (G and H) Na+ content in the cytosolic (G) and vacuolar (H) compartments in the root elongation zone of 46 wheat varieties contrasting in their salinity tolerance quantified by the intensity of CoroNa Green fluorescence (arbitrary units). Mean ±SE (n=72–96 cells from at least six individual plants). (I and J) Averaged pooled values for Na+ intensity in the cytosol (I) and vacuole (J) in the root elongation zone for bread and durum wheat clusters. Mean ±SE [n=19 (durum wheat) and 27 (bread wheat)]. **Significant at P<0.01. Hydroponically grown 4-day-old wheat seedlings were treated with 100 mM NaCl for 72 h.
Bread wheat possesses superior vacuolar Na+ sequestration ability in the root mature zone
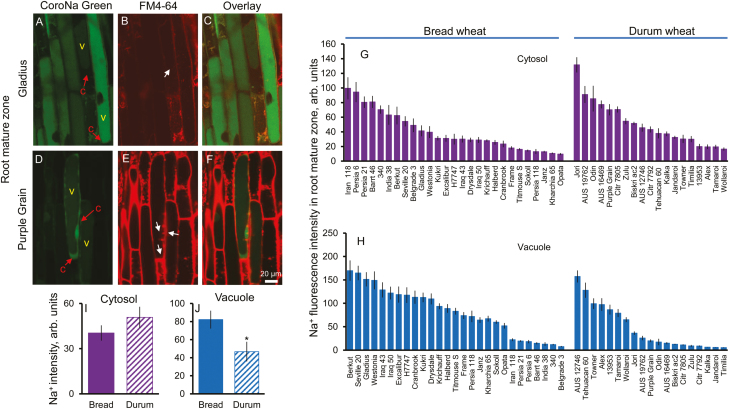
Given the rather limited volume of the root elongation zone, it is difficult to envisage that vacuolar Na+ sequestration in this zone will have a major impact on whole-plant Na+ relations and a control of its delivery to the shoot. Accordingly, we quantified profiles of the vacuolar and cytosolic Na+ distribution in the mature root zone (representing the major bulk of the root) (Fig. 3A–E). A large variability of Na+ intensity in the cytosol (13.6-fold, Fig. 3G) and the vacuole (31.4-fold, Fig. 3H) in the root mature zone was found in the screened varieties. Jori (durum wheat) showed the highest cytosolic Na+ intensity, 132 ± 10, whereas Opata (bread wheat) showed the lowest, 9.7 ± 1.4 (Fig. 3G). Furthermore, the highest vacuolar Na+ intensity was found in the variety Berkut (bread wheat), 170 ± 22, compared with the lowest, 5.4 ± 0.6, in the variety Timilia (durum wheat) (Fig. 3H). In the root mature zone, a significantly higher (75%) Na+ intensity in the vacuole was found in bread wheat than in durum wheat (82 ± 9 versus 47 ± 11, P<0.05; Fig. 3J), compared with no significant difference between bread and durum wheat in the cytosol (40.4 ± 5.1 versus 50.9 ± 7.0, P>0.05; Fig. 3I). A significant negative correlation (P<0.01) was found between cytosol and vacuolar Na+ intensity in the root mature zone (Supplementary Fig. S8C).
Fig. 3.
Genetic variability of Na+ intensity in the cytosol and vacuole in the root mature zone. (A, B) and (D, E) Representative images of root mature zone cells stained with CoroNa Green dye and FM4-64 membrane dye in bread wheat Gladius (A, B) and durum wheat Purple Grain (D, E) cultivars. (C and F) The corresponding overlay images. c, cytosol, v, vacuole. (G and H) Na+ content in the cytosolic (G) and vacuolar (H) compartments in the root mature zone of 46 wheat varieties contrasting in their salinity tolerance quantified by the intensity of CoroNa Green fluorescence (arbitrary units). Mean ±SE (n=72–96 cells from at least six individual plants). (I and J) Averaged pooled values for Na+ intensity in the cytosol (I) and vacuole (J) in the root mature zone for bread and durum wheat clusters. Mean ±SE [n=19 (durum wheat) and 27 (bread wheat)]. *Significant at P<0.05. Hydroponically grown 4-day-old wheat seedlings were treated with 100 mM NaCl for 72 h.
Cytosolic Na+ accumulation in the root meristem zone was significantly higher in bread wheat
To investigate further the intracellular Na+ distribution profiles in the root meristem, FM4-64 membrane dye was also used to co-stain wheat roots, allowing a clear visualization of Na+ distribution in various intracellular compartments. Representative images of Na+ distribution between the cytosol and vacuole (visualized by CornoNa Green dye and FM4-64 membrane dye) in the root meristem zone in bread (Gladius) and durum (Purple Grain) wheat are shown in Fig. 4A–L. A 16.7-fold variability in cytosolic Na+ intensity was observed within the 46 screened wheat varieties, ranging from the highest at 174 ± 7 (Cranbrook, bread wheat) to the lowest at 10.4 ± 0.4 (Persia 21, bread wheat) (Fig. 4M). Similarly, a large variability (5.4-fold) was found for vacuolar Na+ intensity in the root meristem zone, ranging from the highest value of 180 ± 7 (Wollaroi, durum wheat) to the lowest of 34 ± 2 (Iraq 50, bread wheat) (Fig. 4O). On average, a nearly 2-fold higher cytosolic Na+ intensity was found in bread wheat in this zone (57.9 ± 9.8 versus 31.7 ± 2.3, P<0.05; Fig. 4N). At the same time, no significant difference was found in vacuolar Na+ intensities between bread and durum wheat (85 ± 6 versus 103 ± 7, respectively, P>0.05; Fig. 4P). No significant (P>0.05) correlation was found between cytosol and vacuolar Na+ intensity in the root meristem zone Supplementary Fig. S8D).
Tampering with the root meristem results in a salt-sensitive phenotype in bread wheat
From the above data, we have hypothesized that the root meristem may operate as, or harbour, a salt stress sensor. According to this hypothesis, a stress-induced increase in the cytosolic Na+ in this zone may be essential to induce a cascade of signalling and adaptive responses, at both the tissue and the whole-plant level. To test this hypothesis, experiments were conducted using the salt-tolerant Kharchia 65 bread wheat variety in an attempt to modify salinity stress tolerance by tampering with salt sensing in root meristems. Under control conditions, the surgical removal of the meristematic tissue in seminal roots did not affect whole-plant physiological characteristics (Fig. 5A–D), as judged by leaf chlorophyll content (SPAD readings; Fig. 5A), maximal photochemical efficiency of PSII (Fig. 5B), and leaf (Fig. 5C) and root (Fig. 5D) Na+ content. At the same time, the same procedure has resulted in a salt-sensitive phenotype under stress conditions, with significantly higher SPAD (Fig. 5A, F) and Fv/Fm (Fig. 5B) values found in intact plants (with intact roots) compared with cut plants (with removal of the root meristem zone) in hydroponic experiments. Interestingly, no difference in the total Na+ content in either leaf or root was found between intact plants and those which had been ‘tampered with’ (Fig. 5C, D). This may be indicative that while the overall rate of Na+ uptake and transport to the shoot was not altered by the removal of the root meristem, the distribution of Na+ between the cytosol and vacuole in mesophyll cells might be different between the intact plants and cut plants. Needless to say, detrimental effects of salinity are not necessarily determined by the absolute amounts of salt accumulated in the shoot but rather its distribution between the intracellular compartments, as shown in numerous studies on halophytes.
Fig. 5.
Comparative analysis of selected agronomical and physiological characteristics of plants after the surgical removal of the root meristem tissue. Salt-tolerant bread wheat variety Kharchia 65 was used. Plants were grown either hydroponically (A–D; 200 mM NaCl, 10 d) or in a potting mix (E; 200 mM NaCl, 15 d) and treated with 200 mM NaCl for either 10 d or 15 d, respectively. Chlorophyll content (SPAD value) (A for hydroponic experiment, E for potting mix experiment), maximum photochemical efficiency of PSII (chlorophyll fluorescence Fv/Fm value, arbitrary units) (B), and leaf (C) and root (D) Na+ content were measured at the end of the experiment and compared between intact (with intact roots) and cut plants (in which root meristems were removed). Mean ±SE (n=24−36). (F) The comparison between leaves from intact and cut hydroponically grown plants after 10 d of salt stress. Different lower case letters denote significant difference between data at P<0.05. Con, non-saline condition.
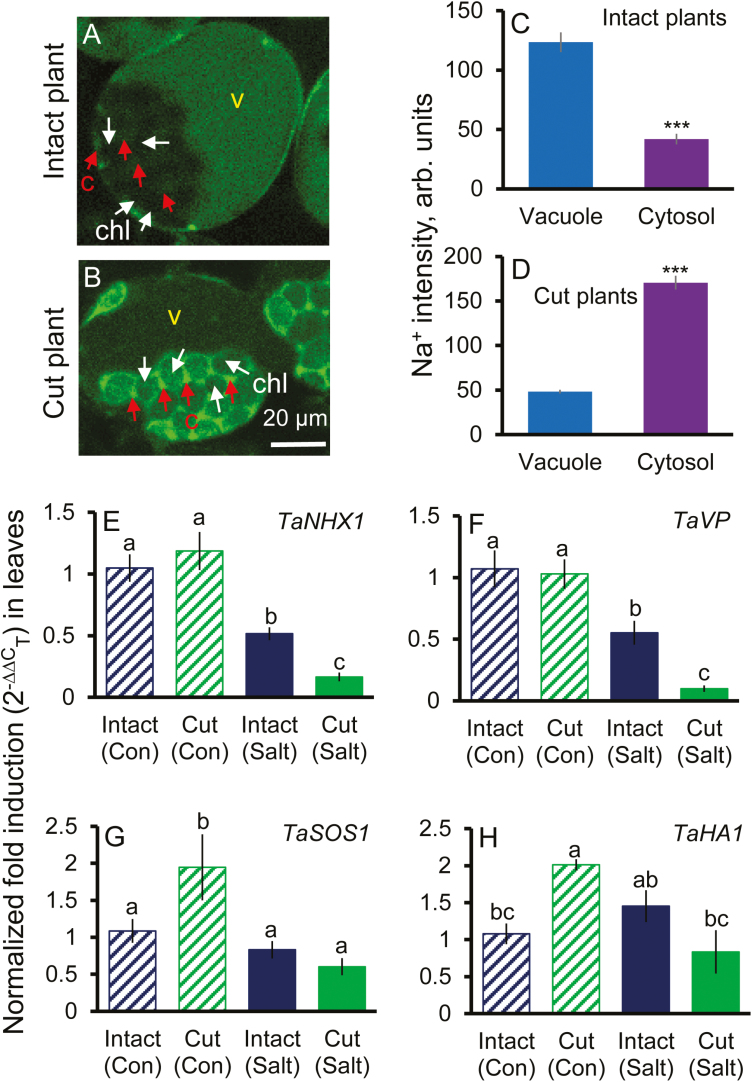
To confirm the above hypothesis, we repeated the same experiment in pot-grown (rather than hydroponically grown) plants exposed to more prolonged (15 d) salt stress. Similar to the results in the hydroponic experiment, significantly higher SPAD values were found in intact plants than in cut plants grown under salt stress conditions (Fig. 5E) but not in non-saline conditions. Staining mesophyll cells with CoroNa Green dye revealed that cells from intact plants possessed a 3- to 4- fold higher ability to remove Na+ from the cytosol and sequester it into the mesophyll cell vacuoles, compared with plants in which root meristems were removed (Fig. 6A–D). These might cause both higher Na+ toxicity and lower ability for osmotic adjustment executed by the vacuolar Na+, and thus result in higher sensitivity to salt stress in the plants which were tampered with. Consistent results demonstrating the salt-sensitive phenotype caused by the removal of the root meristems were also reported in experiments involving other bread and durum wheat genotypes and different durations and levels of saline stress (Supplementary Figs S9, S10). Taken together, these data strongly support the notion that tampering with root salt sensing by the meristem has major consequences for the long-distance stress signalling between the root and shoot and vacuolar Na+ sequestration in the shoot.
Fig. 6.
Na+ distribution and relative expression levels of several key genes related to Na+ transport in leaf mesophyll cells in intact plants and plants with the root meristems removed. Representative images showing CoroNa Green dye signal from leaf mesophyll cells in intact plants (with intact roots) (A) and cut plants (in which the root meristem tissues were removed) (B). (C and D) Intensity of CoroNa Green fluorescence (arbitrary units) in the cytosol and vacuole in the intact and cut plants, respectively. Mean ±SE (n=47−86 cells from at least three individual plants). c, cytosol; v, vacuole; chl, chloroplast. Plants were grown in potting mix under 200 mM NaCl for 15 d. (E– H) Comparison of transcriptional profiles of genes mediating Na+ removal from the cytosol [TaNHX1 (E), TaVP (F), TaSOS1 (G), and TaHA1 (H)] in the youngest fully expanded leaves of intact plants and plants in which root meristems were surgically removed. Plants were grown hydroponically in the presence of 200 mM NaCl for 10 d. Mean ±SE (n=3 biological replicates). The salt-tolerant bread wheat variety Kharchia 65 was used in experiments. ***Significant at P<0.001. Different lower case letters mean P<0.05.
Tampering with the root meristem affects transcription of genes conferring Na+ sequestration in the vacuole of mesophyll cells
We aimed to reveal if the above effect of root meristem stress sensing on Na+ sequestration in the vacuole of mesophyll cells was due to transcriptional or post-translational regulation. For this, we have compared transcriptional profiles of several key genes potentially mediating the above process. Theu included the TaNHX1 gene encoding the tonoplast Na+(K+)/H+ exchanger; genes for two tonoplast-based H+-pumps fuelling NHX1 activity (TaHA1 for H+-ATPase and TaVP for H+-PPase); and the TaSOS1 gene encoding the plasma membrane Na+/H+ antiporter. In both intact plants and those that were ‘tampered with’, salinity stress reduced transcript levels of TaNHX1 and TaVP (Fig. 6E, F), but in plants deprived of the ability to sense salt stress by the root meristem the effect was much more (2- to 3-fold stronger) dramatic. The transcript levels of TaHA1 and TaSOS1 were not affected by salinity in intact plants but were reduced 2- to 3-fold in plants with root meristems removed (Fig. 6G, H). Interestingly, transcript levels of TaSOS1 and TaHA1 were significantly higher in cut plants than in intact plants under non-saline condition (Fig. 6G, H).
Discussion
Na+ extrusion from the root elongation zone is essential for salt tolerance in wheat
Most authors have a tendency to use the term ‘sodium exclusion’ in a very broad sense, and often as a synonym for prevention of its accumulation in the shoot. As a result, the actual role of Na+ extrusion from the root, and its contribution to the differential salt tolerance between bread and durum wheat remains largely unknown. A comparison was done by monitoring 22Na+ uptake in radiotracer experiments (Davenport et al., 1997; Z. Chen et al., 2007); neither of these experiments has revealed any significant correlations between plant salinity tolerance and unidirectional Na+ uptake by roots, with all studied wheat and barley cultivars showing more or less the same rate of 22Na+ uptake.
Na+/K+-ATPase expels three Na+ ions from the cell in exchange for two K+ ions in animal cells (Yatime et al., 2011; Galva et al., 2012). In lower plants, such as Physcomitrella patens, extrusion of Na+ can be achieved by sodium ATPase (PpENA1) (Benito and Rodríguez-Navarro, 2003; Lunde et al., 2007). In higher plants, however, the SOS1 Na+/H+ antiporter remains the only known transporter to exclude Na+ from the cytosol into the apoplast. β-Glucuronidase (GUS) expression analysis showed that SOS1 was preferentially expressed in the root apex but not the mature zone in Arabidopsis (Shi et al., 2002). This was confirmed by our functional studies on wheat. While in the mature root zone Na+ efflux mostly ranged between –20 nmol m–2 s–1 and –70 nmol m–2 s–1 (Cuin et al., 2011), 10- to 20-fold higher Na+ extrusion ability was observed in this study (for the same conditions) in the root elongation zone, with net Na+ fluxes ranging from –284 ± 39 nmol m–2 s–1 to –1584 ± 237 nmol m–2 s–1 (Fig. 1). These Na+ fluxes measured by the MIFE Na+-selective microelectrodes were sensitive (>80% inhibition; Supplementary Fig. S2) to amiloride, a known blocker of the mammalian Na+/H+ exchanger (Kleyman and Cragoe, 1988) and were not observed in an Arabidopsis mutant lacking a functional SOS1 gene (Supplemenatary Fig. S2). As a known mammalian Na+/H+ exchanger blocker, amiloride is also widely used in different plant species (Blumwald et al., 1987; Fukuda et al., 1998; Qiu et al., 2003; Pardo et al., 2006; Cuin et al., 2011). Taken together, this implies that Na+ fluxes reported in this work were mediated mainly by SOS1-like plasma membrane Na+/H+ exchangers in epidermal cells in the wheat root elongation zone. Thus, the reported finding of significantly higher (by 33%; P<0.05; Fig. 1) Na+ extrusion ability of bread wheat measured by MIFE provides the first functional evidence for the role of the SOS1 extrusion mechanism as the factor conferring differential salt stress tolerance between bread and durum wheat species, as all the reported evidence to date has attributed this trait to shoot tissue (Cuin et al., 2010; Munns et al., 2012). Although the bulk of the root is comprised of mature root cells, the possibility of internal Na+ transport within the root cortex is highly likely. Thus, the scenario where Na+ taken up by the bulk of the root is trafficked to the apex and then extruded via SOS1 exchangers cannot be ruled out.
Root vacuolar Na+ sequestration in the mature zone explains superior bread wheat performance under saline conditions
While vacuolar Na+ sequestration is unequivocally accepted as a key trait conferring salinity tissue tolerance, the absolute majority of published papers referred to this mechanism operating in the shoot, both in halophytes (Flowers and Colmer, 2008; Shabala and Mackay, 2011) and in glycophytes (Apse et al., 1999; Munns and Tester, 2008). In this work, we provide the first large-scale evidence that such a trait is also applicable to plant roots and is essential to confer the difference in salinity stress tolerance between related species.
The physiological rationale behind this phenomenon may be the fact that the mature root zone represents the major bulk of the root and thus possesses enough capacity to reduce the Na+ load to the shoot. HKT-mediated retrieval of Na+ from the xylem has been widely reported to be linked to reduced Na+ accumulation in the shoot (Sunarpi et al., 2005; Huang et al., 2006; Byrt et al., 2007) and ultimately salt tolerance in wheat (Munns et al., 2012). However, there is not much physiological rationale to load Na+ actively in the shoot (see Shabala, 2013 for thermodynamic analysis and arguments) and then retrieve it back. This futile cycle can be avoided, and the same effect can be achieved at a lower cost by immediate deposition of salt load in the root cortex in the mature zone. More interestingly, vacuolar fluorescence Na+ signal intensity is much higher in the root elongation zone than in the mature zone in wheat (Figs 2, 3), suggesting that Na+ might be trafficked from the root mature zone to the elongation zone. This is in agreement with the finding that the root apex has a significantly higher sap Na+ content than the mature root tissue (Supplementary Fig. S11).
The above pattern and correlation between vacuolar Na+ concentration in root cells and overall plant salinity stress tolerance does not hold for the elongation zone (Fig. 2). Here, durum wheat compensates poor Na+ extrusion ability from the elongation zone by more efficient vacuolar Na+ sequestration. The ultimate result is the same: both species maintain the same Na+ concentrations in the cytosol in the root elongation zone, though by different mechanisms. It may also be tempting to suggest that this strategy may allow durum wheat to adjust osmotically and maintain cell elongation by mainly relying on (freely available) Na+. The energy cost of osmotic adjustment by means of inorganic ions is an order of magnitude lower than by de novo synthesis of compatible solutes (Shabala and Shabala, 2011; Munns and Gilliham, 2015). It also should be noted that a possible contribution of accumulation of compatible solutes in the cytoplasm to equilibrate increased vacuolar osmolality cannot be fully ruled out, especially in the elongation and mature root cells which have a central vacuole. As durum wheat possesses a much lower root K+ retention ability compared with bread wheat (Cuin et al., 2008), higher reliance of Na+ may be suggested as a plausible strategy to achieve osmotic adjustment without redirecting additional ATP resources for organic osmolyte biosynthesis.
Na+ sequestration in mesophyll cells was affected by the removal of the root meristem tissue
Plant adaptive responses to the environment require orchestrated regulation of a plethora of physiological mechanisms and thus imply efficient root to shoot communication (S. Shabala et al., 2016). The communication modes between the root and shoot are highly diverse and include a broad range of physical [electric and hydraulic signals, propagating Ca2+ and reactive oxygen species (ROS) waves], chemical (assimilates, hormones, and nutrients), and other molecular (peptides, proteins, and RNA) signals (Baxter et al., 2014; Gilroy et al., 2014; Van Bel et al., 2014; S. Shabala et al., 2016). Any of these signals may potentially be responsible for the observed salt-sensitive phenotype after tampering with the root meristem signalling.
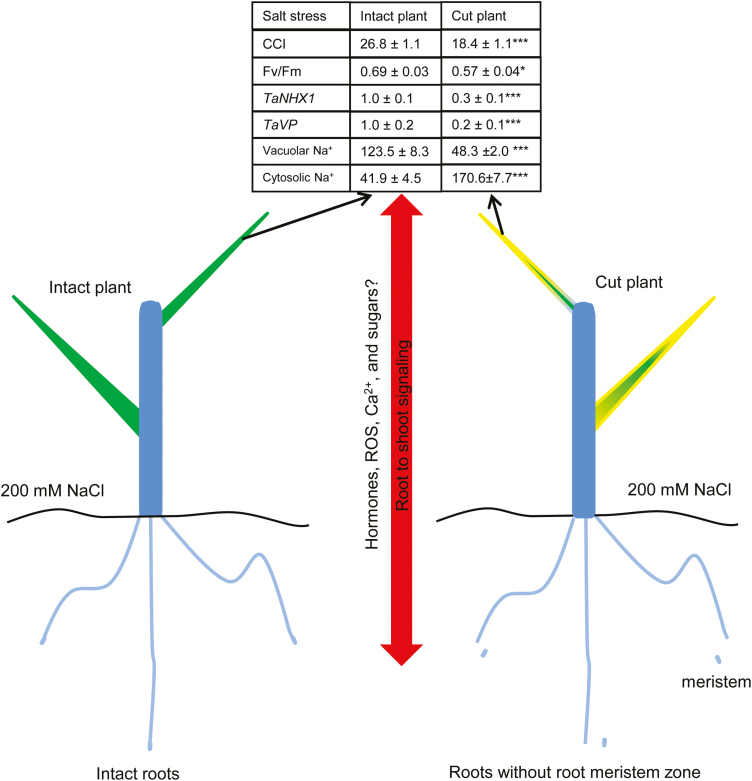
Our current knowledge of how salt stress is sensed by plant tissues is severely limited and, while several mechanisms have been considered as suitable to undertake the role of the putative salt sensor (for the most recent reviews, see Maathuis, 2014; Shabala et al., 2015), no explicit evidence has been provided in the literature. This work makes an important contribution to filling this knowledge gap. We first provide strong evidence that such a salt stress sensor is located in the root meristem. Indeed, removing the meristem tissue resulted in a salt-sensitive phenotype (Figs 5, 7; Supplementary Fig. S9) in otherwise highly salt-tolerant Kharchia 65 and Westonia wheat varieties. Secondly, we showed that meristem signalling to the shoot is essential in enabling vacuolar Na+ sequestration ability in leaf mesophyll cells, in a process that is at least partially conferred by differential expression of a set of Na+ transport-related genes at the tonoplast (Fig. 6). Thirdly, given the fact that both durum and bread wheat had experienced the same osmotic stress (and hence tension on the plasma membrane), our results rule out mechanosensing mechanisms (Monshausen and Haswell, 2013) as a key determinant of differential salinity stress tolerance between durum and bread wheat. Instead, taken together, our results point out the important role of elevated cytosolic Na+ in the signalling process, narrowing down the list of candidate mechanism to only a few. The first candidate may include the SOS1 Na+/H+ antiporter per se operating in the sensing mode (Zhu, 2003). This protein has a very long tail (700 amino acids) residing in the cytosol that potentially senses Na+. The second Na+ sensor candidate in systems may refer to the proteins which comprise the side chains of an aspartate and a histidine that form a DxR/KxxH motif (Thomson et al., 2015), and Maathuis (2014) has estimated that ~1200 candidate proteins containing this motif exist in Arabidopsis. Future studies therefore should focus on tissue-specific profiles of expression of some of these candidate genes. The third potential candidates are NCX Na+/Ca2+ exchangers (Shabala et al., 2015). A recent bioinformatics analysis (Wang et al., 2012) suggests the existence of a putative NCX gene in Arabidopsis, which shows increased transcript levels when plants are exposed to salinity.
Fig. 7.
The proposed model comparing major changes in gene expression profiles, Na+ ion distribution, and phenotyping responses in leaves of plants compromised in salt sensing by the meristem tissue. Data in the table are significant at *P<0.05 and ***P<0.001.
Although recent findings emphasized the essential role of AtNHX1 in K+ accumulation in the vacuole (Bassil et al., 2011; Barragán et al. ,2012), they do not completely disregard its involvement in vacuolar Na+ sequestration, and it is highly plausible that NHX1 has a dual role (Na+ and K+/H+ antiporter) (Maathuis, 2014; Liu et al., 2017). While the tonoplast NHX1 Na+(K+)/H+ exchanger is critical for vacuolar Na+ sequestration and thus plant salt tolerance (Apse et al., 1999), for its operation it should be fuelled by either vacuolar PPase or H+-ATPase (Beyenbach and Wieczorek, 2006; Silva and Gerós, 2009). Here we showed that preventing meristem-mediated root to shoot salt signalling resulted in a significant down-regulation of transcript levels in both tonoplast pumps (Fig. 6). This had a profound effect on vacuolar Na+ sequestration (Fig. 6) and overall plant performance under saline conditions. Salt stress-induced nuclear and DNA degradation and programmed cell death phenotypes were also found in root meristematic cells (Liu et al., 2000; Huh et al., 2002). Moreover, under salt stress, the NHX1 gene is up-regulated in Arabidopsis (Gaxiola et al., 1999). However, the story in wheat might be different (Fig. 6E). Mullan et al. (2007) showed that the transcript level of NHX1 in wheat was almost unchanged in the fourth leaf blade under 200 mM NaCl, whereas a clear decrease was observed in roots. Thus, the tissue-specific aspect should be accounted for. Other factors such as treatment time, leaf age, and salt stress concentration should also be considered.
Revealing the molecular nature of the root to shoot signal controlling vacuolar Na+ sequestration in the mesophyll (and ultimately salinity tissue tolerance) remains a challenging but highly important task for the future. Importantly, different signalling systems operate at very different time scales, and it was also suggested that some of these systems may operate in a priming mode(s), while others deliver more specific information about the precise nature of the signal (S. Shabala et al., 2016). So, what is the nature of the salt-induced root signals?
It is known that the cytokinin and auxin antagonistic interaction is important in controlling meristem activity (Del Ioio et al., 2007, 2008). Thus, the change at the hormonal level might be one explanation for the altered Na+ distribution in salinized plants after removal of the roots meristem zone. Other candidates such as brassinosteroid (Hacham et al., 2011), ROS (Gilroy et al., 2014), Ca2+ (Dodd et al., 2010), abscisic acid (Liang et al., 2007), and sugars (Rosa et al., 2009; Krasensky and Jonak, 2012) may participate in the long-distance salt stress signal transduction and thus should be considered in future work (Fig. 7). ROS were previously suggested to play a role in systematic signal transduction under abiotic stress (Miller et al., 2009; Mittler et al., 2011; Baxter et al., 2014). Ca2+ is known as a second messenger and is involved in a broad range of plant cell activities in regulating plant growth and development (Hepler, 2005). Recently, salt stress-induced Ca2+ waves were found to be associated with rapid, long-distance root to shoot signalling in plants (Choi et al., 2014). Furthermore, overexpression of ABF2 (ABRE-binding bZIP factor), an essential component of glucose signalling, improved salt stress tolerance in Arabidopsis (Kim et al., 2004). Taken together, the signalling events involved in this phenomenon may be a complex co-ordinated by different signalling molecules in a ‘fine tune’ and/or ‘coarse tune’ mode. Future studies should be conducted to investigate the identity of specific signalling molecules and their role in the phenomenon whereby leaf Na+ distribution in salinized plants was affected by the removal of its root meristem tissue.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Genetic variability in salinity stress tolerance (quantified as a tolerance index) amongst 46 wheat varieties and its correlation with relative grain yield under salt stress.
Fig. S2. Na+ efflux measured by the MIFE technique from plant roots using the ‘recovery protocol’ is mediated by SOS1 Na+/H+ exchangers.
Fig. S3. Quantification of intracellular Na+ distribution in the cytosol and vacuole.
Fig. S4. The effect of CoroNa Green dye concentration on the intensity of the fluorescent Na+ signal in wheat roots measured from different root zones.
Fig. S5. The effect of incubation time of CoroNa Green (20 µM) on the intensity of the fluorescent Na+ signal in wheat roots measured from different root zones.
Fig. S6. The procedure for removal of the root meristem tissue.
Fig. S7. The relative gene expression patterns in the root apex and mature root in bread (cv. Gladius) and durum wheat (cv. Purple Grain) after salt stress.
Fig. S8. Correlations between cytosolic and vacuolar Na+ intensity and Na+ efflux in wheat cultivars.
Fig. S9. Removal of the root meristem results in a salt-sensitive phenotype.
Fig. S10. Removal of the root meristem results in higher vulnerability of durum wheat (cv. Wallaroi) to salt stress.
Fig. S11. Sap Na+ content in the root apical and mature tissues.
Table S1. Primers used in this study.
Acknowledgements
We thank Dr Min Zhu, Mr Xin Huang, and Ms Feifei Wang for their technical help. This work was supported by the Grain Research and Development Corporation to SS and MZ and by the Australian Research Council to SS.
Abbreviations
- ABF2
ABRE-binding bZIP factor
- HKT
high affinity K+ transporter
- LIX
liquid ion exchanger
- LSCM
laser scanning confocal microscopy
- MIFE
microelectrode ion flux estimation
- NCX
Na+/Ca2+ exchanger
- ROS
reactive oxygen species
- SOS1
Salt Overly Sensitive 1
- TaHA1
bread wheat H+-ATPase
- TaVP
bread wheat H+-PPase
References
- Almeida P, Freon R, de Boer GJ, de Boer AH. 2014. Role of Na+, K+, Cl−, proline and sucrose concentrations in determining salinity tolerance and their correlation with the expression of multiple genes in tomato. AOB Plants 6, plu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. 1999. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 258, 1256−1258. [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM. 2012. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. The Plant Cell 24, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Ohto M, Esumi T, et al. 2011. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growthand development. The Plant Cell 23, 224−239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Benito B, Rodríguez-Navarro A. 2003. Molecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens. The Plant Journal 36, 382–389. [DOI] [PubMed] [Google Scholar]
- Beyenbach KW, Wieczorek H. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. Journal of Experimental Biology 209, 577–589. [DOI] [PubMed] [Google Scholar]
- Blumwald E, Cragoe EJ, Poole RJ. 1987. Inhibition of Na/H antiport activity in sugar beet tonoplast by analogs of amiloride. Plant Physiology 85, 30–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonales-Alatorre E, Shabala S, Chen ZH, Pottosin I. 2013. Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, Chenopodium quinoa. Plant Physiology 162, 940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R. 2007. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiology 143, 1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, An R, Tang JH, Cui XH, Hao FS, Chen J, Wang XC. 2007. Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Molecular Breeding 19, 215−225. [Google Scholar]
- Chen Z, Newman I, Zhou M, Mendham N, Zhang G, Shabala S. 2005. Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant, Cell and Environment 28, 1230–1246. [Google Scholar]
- Chen Z, Pottosin II, Cuin TA, et al. 2007. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiology 145, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S. 2014. Slat stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proceedings of the National Academy of Sciences, USA 111, 6497−6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A. 1990. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. The Plant Cell 2, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Flowers TJ, Munns R. 2006. Use of wild relatives to improve salt tolerance in wheat. Journal of Experimental Botany 57, 1059–1078. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Betts SA, Chalmandrier R, Shabala S. 2008. A root’s ability to retain K+ correlates with salt tolerance in wheat. Journal of Experimental Botany 59, 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S. 2011. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant, Cell and Environment 34, 947–961. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Parsons D, Shabala S. 2010. Wheat cultivars can be screened for NaCl salinity tolerance by measuring leaf chlorophyll content and shoot sap potassium. Functional Plant Biology 37, 656–664. [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R. 2005. Control of sodium transport in durum wheat. Plant Physiology 137, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M. 2007. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant, Cell and Environment 30, 497–507. [DOI] [PubMed] [Google Scholar]
- Davenport RJ, Reid RJ, Smith FA. 1997. Sodium–calcium interactions in two wheat species differing in salinity tolerance. Physiologia Plantarum 99, 323−327. [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17, 678–682. [DOI] [PubMed] [Google Scholar]
- Dell Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 17, 678−682. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, et al. 2008. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. 2010. The language of calcium signaling. Annual Review of Plant Biology 61, 593–620. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. 2008. Salinity tolerance in halophytes. New Phytologist 179, 945–963. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Yazaki Y, Ishikawa T, Koike S, Tanaka Y. 1998. Na+/H+ antiporter in tonoplast vesicles from rice roots. Plant and Cell Physiology 39, 196−201. [Google Scholar]
- Galva C, Artigas P, Gatto C. 2012. Nuclear Na+/K+-ATPase plays an active role in nucleoplasmic Ca2+ homeostasis. Journal of Cell Science 125, 6137–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink G. 1999. The Arabidopsis thaliana transporters, AtNHX1 and Avp1 can function in cation detoxification in yeast. Proceedings of the National Academy of Sciences, USA 96, 1480−1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genc Y, McDonald GK, Tester M. 2007. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant, Cell and Environment 30, 1486–1498. [DOI] [PubMed] [Google Scholar]
- Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, Mittler R. 2014. A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends in Plant Science 19, 623–630. [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. 2011. Brassinosteroid perception in the epidermis controls root meristem size. Development 138, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Pasapula SV, Luo S, et al. 2007. Ectopic expression of AtNHX1 in cotton (Gossypium hirsutum L.) increases proline content and enhances photosynthesis under salt stress conditions. Journal of Cotton Science 11, 266–274. [Google Scholar]
- Hepler PK. 2005. Calcium: a central regulator of plant growth and development. The Plant Cell 17, 2142–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R. 2006. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiology 142, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GH, Damsz B, Matsumoto TK, Reddy MP, Rus AM, Ibeas JI, Narasimhan ML, Bressan RA, Hasegawa PM. 2002. Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. The Plant Journal 29, 649–659. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. 2011. Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell 21, 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology 143, 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Blake C, Byrt CS, Munns R. 2011. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. Journal of Experimental Botany 62, 2939–2947. [DOI] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R. 2006. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiology 142, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakannan M, Babourina O, Rengel Z. 2011. Improved measurements of Na+ fluxes in plants using calixarene-based microelectrodes. Journal of Plant Physiology 168, 1045–1051. [DOI] [PubMed] [Google Scholar]
- Jha D, Shirley N, Tester M, Roy SJ. 2010. Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant, Cell and Environment 33, 793–804. [DOI] [PubMed] [Google Scholar]
- Kiegle E, Moore CA, Haseloff J, Tester MA, Knight MR. 2000. Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. The Plant Journal 23, 267–278. [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. 2004. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. The Plant Journal 40, 75–87. [DOI] [PubMed] [Google Scholar]
- Kleyman TR, Cragoe EJ Jr. 1988. Amiloride and its analogs as tools in the study of ion transport. Journal of Membrane Biology 105, 1–21. [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuchli A, James RA, Huang CX, McCully M, Munns R. 2008. Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant, Cell and Environment 31, 1565–1574. [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM. 2007. Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Developmental Biology 304, 297–307. [DOI] [PubMed] [Google Scholar]
- Liu T, van Staden J, Cress WA. 2000. Salinity induced nuclear and DNA degradation in meristematic cells of soybean (Glycine max (L.)) roots. Plant Growth and Regulation 30, 49–54. [Google Scholar]
- Liu X, Cai S, Wang G, et al. 2017. Halophytic NHXs confer salt tolerance by altering cytosolic and vacuolar K+ and Na+ in Arabidopsis root cell. Plant Growth Regulation 82, 333–351. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Löw R, Rockel B, Kirsch M, Ratajczak R, Hörtensteiner S, Martinoia E, Lüttge U, Rausch T. 1996. Early salt stress effects on the differential expression of vacuolar H+-ATPase genes in roots and leaves of Mesembryanthemum crystallinum. Plant Physiology 110, 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde C, Drew DP, Jacobs AK, Tester M. 2007. Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress. Plant Physiology 144, 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM. 2014. Sodium in plants: perception, signalling, and regulation of sodium fluxes. Journal of Experimental Botany 65, 849−858. [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. 2009. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science Signaling 2, ra45. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave?Trends in Plant Science 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Haswell ES. 2013. A force of nature: molecular mechanisms of mechanoperception in plants. Journal of Experimental Botany 64, 4663–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan DJ, Colmer TD, Francki MG. 2007. Arabidopsis–rice–wheat gene orthologues for Na+ transport and transcript analysis in wheat–L. elongatum aneuploids under salt stress. Molecular Genetics and Genomics 277, 199–212. [DOI] [PubMed] [Google Scholar]
- Munns R, Gilliham M. 2015. Salinity tolerance of crops—what is the cost?New Phytologist 208, 668–673. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA. 2003. Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253, 201–218. [Google Scholar]
- Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD. 2016. Tissue tolerance: an essetial but elusive trait for salt-tolerant crops. Functional Plant Biology 43, 1103−1113. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, et al. 2012. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30, 360–364. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Newman IA. 2001. Ion transport in roots: measurement of fluxes using ion-selective microelectrodes to characterize transporter function. Plant, Cell and Environment 24, 1–14. [DOI] [PubMed] [Google Scholar]
- Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ. 2010. Intracellular consequences of SOS1 deficiency during salt stress. Journal of Experimental Botany 61, 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ. 2006. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. Journal of Experimental Botany 57, 1181–1199. [DOI] [PubMed] [Google Scholar]
- Qiu QS, Barkla BJ, Vera-Estrella R, Zhu JK, Schumaker KS. 2003. Na+/H– exchange activity in the plasma membrane of Arabidopsis. Plant Physiology 132, 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal M, Prado FE. 2009. Soluble sugars—metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signaling and Behavior 4, 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. 2006. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genetics 2, e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. 2000. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. The Plant Journal 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Shabala L, Zhang J, Pottosin I, et al. 2016. Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity stress. Plant Physiology 172, 2445–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. 2013. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of Botany 112, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Demidchik V, Shabala L, Cuin TA, Smith SJ, Miller AJ, Davies JM, Newman IA. 2006. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+ -permeable channels. Plant Physiology 141, 1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Mackay A. 2011. Ion transport in halophytes. Advances in Botanical Research 57, 151−199. [Google Scholar]
- Shabala S, Shabala L. 2011. Ion transport and osmotic adjustment in plants and bacteria. Biomolecular Concepts 2, 407–419. [DOI] [PubMed] [Google Scholar]
- Shabala S, White R, Djorjevic M, Ruan YL, Mathesius U. 2016. Root to shoot signaling: integration of diverse molecules, pathways and functions. Functional Plant Biology 43, 87–104. [DOI] [PubMed] [Google Scholar]
- Shabala S, Wu H, Bose J. 2015. Salt stress sensing and early signalling events in plant roots: current knowledge and hypothesis. Plant Science 241, 109–119. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. 2000. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences, USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. 2003. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology 21, 81–85. [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. 2002. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. The Plant Cell 14, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P, Gerós H. 2009. Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signaling and Behavior 4, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, et al. 2005. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. The Plant Journal 44, 928–938. [DOI] [PubMed] [Google Scholar]
- Thomson SJ, Hansen A, Sanguinetti MC. 2015. Identification of the intracellular Na+ sensor in Slo2.1 potassium channels. Journal of Biological Chemistry 290, 14528–14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJ, Furch AC, Will T, Buxa SV, Musetti R, Hafke JB. 2014. Spread the news: systemic dissemination and local impact of Ca2+ signals along the phloem pathway. Journal of Experimental Botany 65, 1761–1787. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Paepe A, Speleman F. 2002. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT–PCR. Analytical Biochemistry 303, 95–98. [DOI] [PubMed] [Google Scholar]
- Verdoy D, Lucas MM, Manrique E, Covarrrubias AA, De Felipe MR, Pueyo JJ. 2004. Differential organ-specific response to salt stress and water deficit in nodulated bean (Phaseolus vulgaris). Plant, Cell and Environment 27, 757−767. [Google Scholar]
- Wang P, Li Z, Wei J, Zhao Z, Sun D, Cui S. 2012. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. Journal of Biological Chemistry 287, 44062–44070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shabala L, Liu X, et al. 2015. a Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Frontiers in Plant Science 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shabala L, Zhou M, Stefano G, Pandolfi C, Mancuso S, Shabala S. 2015b Developing and validating a high-throughput assay for salinity tissue tolerance in wheat and barley. Planta 242, 847–857. [DOI] [PubMed] [Google Scholar]
- Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM. 2004. Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Science 167, 849–859. [Google Scholar]
- Yatime L, Laursen M, Morth JP, Esmann M, Nissen P, Fedosova NU. 2011. Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. Journal of Structural Biology 174, 296–306. [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E. 2001. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nature Biotechnology 19, 765–768. [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E. 2001. Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proceedings of the National Academy of Sciences, USA 98, 12832–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Johnson P, Ryan PR, Delhaize E, Zhou M. 2012. Quantitative trait loci for salinity tolerance in barley (Hordeum vulgare L.). Molecular Breeding 29, 427–436. [Google Scholar]
- Zhu JK. 2003. Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology 6, 441–445. [DOI] [PubMed] [Google Scholar]
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S. 2016. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. Journal of Experimental Botany 67, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.