Abstract
Background
Atypical enteropathogenic Escherichia coli (aEPEC) is regarded as a globally emerging enteropathogen. aEPECs exhibit various level of resistance to a range of antibiotics, which is increasing alarmingly. The present study investigated the antimicrobial resistance of aEPEC isolates recovered from diarrheal patients, healthy carriers, animals, and raw meats.
Results
Among 267 aEPEC isolates, 146 (54.7%) were resistant to tetracycline, followed by ampicillin (49.4%), streptomycin (46.1%), and piperacillin (41.2%). Multidrug resistance (MDR) was detected in 128 (47.9%) isolates, and 40 MDR isolates were resistant to ≥ 10 antimicrobial agents. A total of 47 (17.6%) aEPEC isolates were identified as extended-spectrum β-lactamase (ESBL)-producers. The blaCTX-M-14 and blaCTX-M-15 genes were predominant among ESBL-producing isolates.
Conclusions
This investigation depicted the occurrence of multidrug-resistant and ESBL-producing aEPEC isolates in China. The results suggested that it is necessary to continuously monitor the emergence and spread of MDR aEPEC.
Electronic supplementary material
The online version of this article (10.1186/s13099-018-0234-0) contains supplementary material, which is available to authorized users.
Keywords: Enteropathogenic E. coli, Antimicrobial resistance, Multidrug resistance, ESBL
Background
Escherichia coli remains one of the most common etiological agents of diarrheal illness among children under 5 years old in developing countries [1, 2]. Six major diarrheagenic E. coli are well-characterized: enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), and diffusely adherent E. coli (DAEC) [3]. EPEC are the primary cause of summer diarrhea in infants in developing countries [4]. It was estimated that about 79,000 deaths every year are linked with EPEC, which was the first to be identified and is the most prevalent pathotype of diarrheagenic E. coli [5].
EPEC isolates carry the locus of enterocyte effacement (LEE) island, which can induce the hallmark histopathology on the surfaces of intestinal epithelial cells, known as the attaching and effacing (A/E) lesion. A/E results in electrolyte disruption and eventual diarrhea [3, 6, 7]. Some EPEC isolates possess the adherence factor (EAF) plasmid, which carries the bundle-forming pilus genes, the plasmid-encoded regulator genes, and other virulence-related factors [3]. Depending on the presence or absence of the EAF plasmid, EPEC strains are divided into two subgroups: typical EPEC (tEPEC) and atypical EPEC (aEPEC) [8]. In developing countries, tEPEC was considered to be the main cause of infantile diarrhea for decades [6]. However, further studies have shown an apparent increase in the involvement of aEPEC strains in endemic childhood diarrhea and outbreaks in adults in recent years [9–14]. Thus, aEPEC strains have been regarded as emerging enteropathogens and have caused a number of infections [15–17]. Humans and animals, including food-production animals and pet animals, can be reservoirs of aEPEC, while the major reservoirs of tEPEC are humans [6].
Multidrug resistance (MDR), which was designated as resistance to one agent in three or more antibiotic classes [18], has been increasing alarmingly in E. coli (http://www.ecdc.europa.eu/en/healthtopics/antimicrobial_resistance/database/Pages/map_reports.aspx) [19]. The establishment of MDR is mediated by many diverse and interactive mechanisms, e.g., drug efflux, enzymatic inactivation, and target protection [20]. The determinants responsible for MDR are widely distributed among E. coli isolates, irrespective of their resources [20]. Production of extended-spectrum β-lactamase (ESBL) is one of the main mechanisms conferring the spread of MDR [21], because most ESBL-producing isolates show extensive resistance to other antimicrobial agents [22]. The genes encoding ESBLs are usually located on plasmids and different types of ESBLs have been identified globally [23]. According to their amino acid sequences, ESBLs are classified into several types, such as TEM, SHV, CTX, OXA, PER, and GES [24]. Currently, the most frequently detected genetic type of ESBL is CTX-M [25]. There are five major sublineages of CTX-M: 1, 2, 8, 9, and 25 [26].
The spread of antibiotic resistance among pathogens has become an emerging public health concern [21]. In China, aEPEC appeared to be one of the most common pathogens associated with infectious diarrhea [27]. However, there are few data available regarding the resistance of aEPEC. The present study aimed to determine the overall antimicrobial resistance profiles, the current prevalence of MDR, the ESBL genotype distribution, and the determinants of resistance in aEPEC isolates recovered from diarrheal patients, healthy carriers, animals, and raw meat in China. The results will fill in large knowledge gaps concerning this pathogen in China, and provide further information and guidance for the application antimicrobials in farm animals and in clinical treatment.
Methods
Isolation and identification of aEPEC isolates
Samples from different sources (diarrheal patients, healthy carriers, animals, and raw meat) were collected during 2006–2015 in ten geographical regions (Henan, Shanxi, Heilongjiang, Beijing, Qinghai, Guangdong, Sichuan, Shanghai, Guizhou, and Anhui) of China. Fecal samples of diarrheal patients were collected when patients were admitted to sentinel hospitals; stools from healthy carriers were sampled during routine physical examinations; while stool samples of animals and raw meat samples were collected during routine surveys.
The samples were processed as previously described [28]. In brief, the overnight enrichment culture of each sample was centrifuged and the cells were lysed in lysis buffer (10 mM Tris–HCl [pH 8.3], 100 mM NaCl, 1 mM EDTA [pH 9.0], 1% Triton X-100). The released DNA was then examined for eae gene by polymerase chain reaction (PCR) assays. The enrichment culture with eae+ were streaked on CHROMagar™ ECC plate (CHROMagar Co., Paris, France) and incubated at 37 °C for 18–24 h. Ten E. coli-like colonies from each culture were selected to detect the presence of the eae gene. The eae+ colonies were then subcultured on Luria–Bertani (LB) (Oxoid, Basingstoke, UK) plates, incubated for another 18–24 h, and subjected to PCR assays for the eae, stx1, stx2, and bfpA genes. Isolates that were eae positive, but bfpA and stx1/stx2 negative, were defined as aEPEC [6].
A total of 267 aEPEC isolates were identified and included in this study (Additional file 1). Among them, 151, 32, and 51 isolates were recovered from the stools of diarrheal patients, healthy carriers, and animals (cattle, pig, chicken, bird, pika, and marmot), respectively. The remaining 33 strains were isolated from raw meat (beef, pork, mutton, and chicken meat).
Phenotypic antimicrobial susceptibility testing
Susceptibility to a panel of 23 drugs belonging to 12 classes was determined using the disc diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) (2017) [29]: penicillins: ampicillin (AM, 10 μg), piperacillin (PRL, 100 μg); β-lactam/β-lactamase inhibitor combinations: amoxicillin–clavulanic acid (AMC, 20/10 μg), ampicillin–sulbactam (SAM, 10/10 μg); cephems: cefepime (FEP, 30 μg), cefotaxime (CTX, 30 μg), ceftriaxone (CRO, 30 μg), cefuroxime (CXM, 30 μg), ceftazidime (CAZ, 30 μg); monobactams: aztreonam (ATM, 30 μg); carbapenems: imipenem (IPM, 10 μg), meropenem (MEM, 10 μg); aminoglycosides: gentamicin (CN, 10 μg), kanamycin (K, 30 μg), streptomycin (S, 10 μg); tetracyclines: tetracycline (TE, 30 μg); quinolones: nalidixic acid (NA, 30 μg); fluoroquinolones: ciprofloxacin (CIP, 5 μg), norfloxacin (NOR, 10 μg), levofloxacin (LEV, 5 μg); folate pathway inhibitors: trimethoprim–sulfamethoxazole (SXT, 1.25/23.75 μg); phenicols: chloramphenicol (C, 30 μg); and nitrofurans: nitrofurantoin (F, 300 μg) (Oxoid). E. coli ATCC® 25922 served as the control. Strains were resuspended at a concentration of 0.5 McFarland standards in saline solution (0.85% NaCl) (BioMerieux, Marcyl’Etoile, France) and plated on Muller-Hinton agar plate (Thermo Fisher Scientific, Waltham, MA, USA) and grown at 37 °C for 16–18 h. Using a Scan 1200 (Interscience, Saint Nom, France), the diameters of the zone of inhibition were measured to the nearest 0.1 mm and recorded. Each isolate was determined as susceptible (S), intermediate (I), or resistant (R) according to the CLSI standards (2017). Isolates exhibiting resistance to at least one agent in three or more antimicrobial classes were defined as MDR strains [18].
Screening and confirmation of ESBL producing isolates
ESBL production was screened phenotypically using cefotaxime (30 μg). The presumptive isolates were confirmed by combination disk tests with cefotaxime and ceftazidime (30 μg), with and without clavulanic acid (10 μg), as described by the CLSI guidelines [29]. A ≥ 5 mm increase in the zone diameter for cefotaxime or ceftazidime in combination with clavulanic acid versus the zone diameter of the corresponding antimicrobial agent alone defined an ESBL producer [29]. Klebsiella pneumoniae ATCC® 700603 was used as a positive control.
Identification of β-lactamase genes
DNA templates were prepared by crude extraction, as previously described [30]. All isolates were screened for the presence of the blaCTX-M [26], blaTEM, and blaSHV [31] gene using PCR. Four sets of group-specific primers were further used to identify five subgroups (blaCTX-M-1, blaCTX-M-2, blaCTX-M-8/25/26, and blaCTX-M-9) of blaCTX-M [26]. The PCR products were resolved on a 1% agarose gel and then subjected to sequencing using an ABI 3730 Automated DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The resulting sequences were compared against the sequences in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Whole genome sequencing and identification of antimicrobial resistance genes
Based on their serotypes, pulse-field gel electrophoresis (PFGE) patterns and multi-locus sequence typing (MLST), 96 isolates (69 from diarrheal patients, 16 from healthy carriers, and 11 from raw meat) were selected from among the 267 aEPEC strains for whole genome sequencing. Bacterial genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega Co., Madison, WI, USA) according to the manufacturer’s instructions. Genomic DNA was sequenced using an Illumina HiSeq 2500 PE125 instrument (Illumina, Santiago, CA, USA) with 500-bp libraries at the Beijing Novogene Bioinformatics Technology Co., Ltd. Coverage greater than 100× was obtained. The sequence read data was filtered by quality control using the Illumina data pipeline. High-quality filtered reads were assembled into contigs and scaffolds using SOAP de novo (http://soap.genomics.org.cn/soapdenovo.html). Based on the N90, N50, minimum contig size, maximum contig size, and number of contigs, the optimum genome assembly was chosen. Contigs with length > 500 bp were used for further analysis. Assembled draft genomes of all 96 isolates were then used to predict coding genes using the GeneMarkS program [32]. tRNAs and rRNAs were identified using tRNAscan-SE [33] and the rRNAmmer [34], respectively. Seven databases (Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups, Non-Redundant Protein Database, Transporter Classification Database, Swiss-Prot, and TrEMBL) were used to predict gene functions. The Antibiotic Resistance Genes Database (http://ardb.cbcb.umd.edu/) was used to search for antimicrobial resistance genes [35]. The raw data of these genomes have been submitted to the GenBank under accession numbers listed in Additional file 2.
Statistical analysis
Differences in the antimicrobial resistance patterns among aEPEC origins were assessed by a two-tailed Chi square test or Fisher’s exact test, with a level of significance of P < 0.05. All statistical analyses were performed using Epi Info software, version 3.5.3 [36].
Results
Antimicrobial resistance of aEPEC isolates
Of the 267 aEPEC isolates tested, the highest levels of resistance were to tetracycline (54.7%), followed by ampicillin (49.4%), streptomycin (46.1%), and piperacillin (41.2%). Resistances against other antibiotics were as follows: trimethoprim–sulfamethoxazole (39.3%), nalidixic acid (35.2%), gentamicin (28.8%), kanamycin (14.6%), cefuroxime (19.5%), cefotaxime (18.4%), ceftriaxone (18.0%), and chloramphenicol (10.5%). However, most isolates were sensitive to cephalosporins (93.6% for cefepime and 97.0% for ceftazidime), fluoroquinolones (95.1% for ciprofloxacin, 96.6% for norfloxacin, and 95.5% for levofloxacin), and nitrofurantoin (98.5%). All isolates were susceptible to carbapenems (imipenem and meropenem) (Table 1, Additional file 1).
Table 1.
Antimicrobial susceptibility profiles of 267 aEPEC strains isolated from different sources
| Class/antimicrobial | No. of resistant isolates from different sources (%) | Total | P value | |||
|---|---|---|---|---|---|---|
| Diarrheal patient (151) | Healthy carrier (32) | Animal (51) | Raw meat (33) | |||
| Penicillins | ||||||
| Ampicillin | 84 (55.6) | 15 (46.9) | 20 (39.2) | 13 (39.4) | 132 (49.4) | 0.1185 |
| Piperacillin | 71 (47.0) | 13 (40.6) | 19 (37.3) | 7 (21.2) | 110 (41.2) | 0.0484 |
| β-Lactam/β-lactamase inhibitor combinations | ||||||
| Amoxicillin–clavulanic acid | 14 (9.3) | 0 | 10 (19.6) | 3 (9.1) | 27 (10.1) | 0.0319 |
| Ampicillin–sulbactam | 24 (15.9) | 0 | 11 (21.6) | 9 (27.3) | 44 (16.5) | 0.0177 |
| Cephems | ||||||
| Cefepime | 14 (9.3) | 3 (9.4) | 0 | 0 | 17 (6.4) | 0.0396 |
| Cefotaxime | 39 (25.8) | 5 (15.6) | 1 (2.0) | 4 (12.1) | 49 (18.4) | 0.0013 |
| Ceftriaxone | 38 (25.2) | 5 (15.6) | 2 (3.9) | 3 (9.1) | 48 (18.0) | 0.0029 |
| Ceftazidime | 7 (4.6) | 1 (3.1) | 0 | 0 | 8 (3.0) | 0.2622 |
| Cefuroxime | 39 (25.8) | 5 (15.6) | 2 (3.9) | 6 (18.2) | 52 (19.5) | 0.0071 |
| Monobactams | ||||||
| Aztreonam | 20 (12.6) | 3 (9.4) | 0 | 1 (3.0) | 23 (8.6) | 0.0202 |
| Carbapenems | ||||||
| Imipenem | 0 | 0 | 0 | 0 | 0 | – |
| Meropenem | 0 | 0 | 0 | 0 | 0 | – |
| Aminoglycosides | ||||||
| Gentamicin | 57 (37.7) | 3 (9.4) | 11 (21.6) | 6 (18.2) | 77 (28.8) | 0.0019 |
| Kanamycin | 22 (14.6) | 1 (3.1) | 9 (17.6) | 7 (21.2) | 39 (14.6) | 0.1782 |
| Streptomycin | 78 (51.7) | 9 (28.1) | 20 (39.2) | 16 (48.5) | 123 (46.1) | 0.0692 |
| Tetracyclines | ||||||
| Tetracycline | 89 (58.9) | 15 (46.9) | 21 (41.2) | 21 (63.6) | 146 (54.7) | 0.0816 |
| Quinolones | ||||||
| Nalidixic acid | 62 (41.1) | 7 (21.9) | 17 (33.3) | 8 (24.2) | 94 (35.2) | 0.0866 |
| Fluoroquinolones | ||||||
| Ciprofloxacin | 8 (5.3) | 0 | 4 (7.8) | 1 (3.0) | 13 (4.9) | 0.4053 |
| Norfloxacin | 5 (3.3) | 0 | 4 (7.8) | 0 | 9 (3.4) | 0.1447 |
| Levofloxacin | 8 (5.3) | 0 | 4 (7.8) | 0 | 12 (4.5) | 0.2020 |
| Folate pathway inhibitors | ||||||
| Trimethoprim–sulfamethoxazole | 72 (47.7) | 6 (18.8) | 18 (35.3) | 9 (27.3) | 105 (39.3) | 0.0060 |
| Phenicols | ||||||
| Chloramphenicol | 11 (7.3) | 0 | 9 (17.6) | 8 (24.2) | 28 (10.5) | 0.0020 |
| Nitrofurans | ||||||
| Nitrofurantoin | 3 (2.0) | 0 | 1 (2.0) | 0 | 4 (1.5) | 0.7275 |
Although the isolates from different sources showed the highest resistance to tetracycline, the resistance rate of other antibiotics was different among isolates from diarrheal patients, healthy carriers, animals, and raw meat (Table 1). Of the 151 aEPEC strains isolated from diarrheal patients, 89 (58.9%) showed resistance to tetracycline, followed by ampicillin (55.6%), streptomycin (51.7%), trimethoprim–sulfamethoxazole (47.7%), piperacillin (47.0%), and nalidixic acid (41.1%).
Among the 32 strains isolated from healthy-carrier, resistances against tetracycline, ampicillin, and piperacillin were observed in 15 (46.9%), 15 (46.9%) and 13 (40.6%) isolates, respectively. In contrast, all isolates from healthy carriers were susceptible to β-lactam/β-lactamase inhibitor combinations (amoxicillin–clavulanic acid and ampicillin–sulbactam), fluoroquinolones (ciprofloxacin, norfloxacin and levofloxacin), chloramphenicol, and nitrofurantoin.
Of the 51 animal-originated strains, resistance to tetracycline was dominant (41.2%), followed by ampicillin (39.2%), streptomycin (39.2%), piperacillin (37.3%), and trimethoprim–sulfamethoxazole (35.3%). However, all 51 isolates were susceptible to cefepime, ceftazidime, and aztreonam.
Isolates from raw meat displayed the highest level of resistance to tetracycline (63.6%), followed by streptomycin (48.5%) and ampicillin (39.4%). However, all 33 isolates were susceptible to cefepime, ceftazidime, norfloxacin, levofloxacin, and nitrofurantoin.
MDR aEPEC isolates
MDR was detected in 128 (47.9%) isolates. The prevalence of MDR was 55.6% (84/151), 31.3% (10/32), 37.3% (19/51), and 45.5% (15/33) among aEPEC isolates from diarrheal patients, healthy carriers, animals, and raw meat, respectively. Significant differences were observed in the overall distribution of MDR isolates among the four sources (χ2 = 9.563, P = 0.023). The prevalence of MDR in isolates from diarrheal patients was significantly higher than that from healthy carriers (χ2 = 6.282, P = 0.012) and animals (χ2 = 5.150, P = 0.023) (Table 2). Forty (31.3%) MDR isolates were resistant to ≥ 10 antimicrobial agents tested in the study. It was noteworthy that two patient isolates were resistant to 17 and 19 antibiotics, respectively.
Table 2.
The distribution of multidrug resistance (MDR) strains among 267 aEPEC isolates
| No. of antimicrobial group | No. of resistant isolates from different sources (%) | Total | |||
|---|---|---|---|---|---|
| Diarrheal patient | Healthy carrier | Animal | Raw meat | ||
| 0 | 38 (25.2) | 10 (31.3) | 24 (47.1) | 7 (21.2) | 79 (29.6) |
| 1–2 | 29 (19.2) | 12 (37.5) | 7 (13.7) | 11 (33.3) | 59 (22.1) |
| ≥ 3 | 84 (55.6) | 10 (31.3) | 19 (37.3) | 15 (45.5) | 128 (47.9) |
| Total | 151 (100) | 32 (100) | 51 (100) | 33 (100) | 267 (100) |
ESBL producing aEPEC isolates
A total of 47 (17.6%) ESBL-producing isolates were identified among 267 aEPEC isolates. The isolates from diarrheal patients showed the highest rate of ESBL-producing (38/151, 25.2%), compared to those from healthy carrier isolates (5/32, 15.6%), raw meat (3/33, 9.1%), and animals (1/51, 2.0%) (Table 3). Most (83.0%) ESBL-producing isolates were MDR strains. Compared with the non-ESBL producing isolates, ESBL producers displayed significantly higher rates of resistance to ampicillin, piperacillin, amoxicillin–clavulanic acid, ampicillin–sulbactam, cefepime, cefotaxime, ceftriaxone, ceftazidime, cefuroxime, aztreonam, gentamicin, kanamycin, streptomycin, tetracycline, nalidixic acid, trimethoprim–sulfamethoxazole, and nitrofurantoin (Fig. 1).
Table 3.
Characteristics of 47 ESBL-producing aEPEC isolates
| Origin (no. of isolates) | Isolates | Antimicrobial resistance pattern | bla CTX-M | bla TEM | |
|---|---|---|---|---|---|
| CTX-M-1 group | CTX-M-9 group | ||||
| Diarrheal patients (38) | EP004 | AM, PRL, SAM, FEP, CTX, CRO, CXM, CAZ, ATM, CN, K, S, TE, NA, SXT | CTX-M-15 | CTX-M-14 | TEM-1 |
| EP008 | AM, PRL, SAM, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP012 | AM, PRL, SAM, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP013 | AM, PRL, SAM, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP014 | AM, PRL, SAM, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP017 | AM, PRL, AMC, SAM, CTX, CRO, CXM, CN, K, S, TE, NA, SXT, C | CTX-M-14 | TEM-1 | ||
| EP028 | AM, PRL, FEP, CTX, CRO, CXM, CAZ, ATM, CN, TE, NA, SXT | CTX-M-15 | TEM-1 | ||
| EP033 | AM, PRL, CTX, CRO, CXM, CN, S, TE | CTX-M-14 | TEM-1 | ||
| EP041 | AM, PRL, CTX, CRO, CXM, CN, S, SXT | CTX-M-3 | TEM-1 | ||
| EP043 | AM, PRL, CTX, CRO, CXM, ATM | CTX-M-55 | TEM-1 | ||
| EP064 | AM, PRL, CTX, CRO, CXM, CN, NA, SXT | CTX-M-3 | TEM-1 | ||
| EP074 | AM, PRL, AMC, SAM, FEP, CTX, CRO, CXM, CAZ, ATM, CN, S, TE, NA, SXT | CTX-M-15 | |||
| EP079 | AM, PRL, AMC, SAM, FEP, CTX, CRO, CXM, CAZ, ATM, CN, K, S, TE, NA, SXT | CTX-M-15 | TEM-1 | ||
| EP088 | AM, PRL, FEP, CTX, CRO, CXM, CN, S, TE, SXT | CTX-M-3 | |||
| EP103 | AM, PRL, SAM, FEP, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, CIP, LEV, C, F | CTX-M-55 | CTX-M-14 | TEM-1 | |
| EP105 | AM, PRL, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, CIP, LEV, SXT, F | CTX-M-65 | |||
| EP109 | AM, PRL, AMC, CTX, CRO, CXM, ATM, CN, S, NA, SXT | CTX-M-15 | TEM-1 | ||
| EP112 | AM, PRL, SAM, CTX, CRO, CXM, CN, S, TE, NA, SXT | CTX-M-14 | |||
| EP115 | AM, PRL, AMC, SAM, FEP, CTX, CRO, CXM, ATM, CN, K, S, TE, NA, CIP, LEV, SXT, C, F | CTX-M-15 | TEM-1 | ||
| EP116 | AM, PRL, AMC, FEP, CTX, CRO, CXM, CN, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP136 | AM, PRL, CTX, CRO, CXM, ATM | CTX-M-15 | |||
| EP155 | AM, PRL, CTX, CRO, CXM, CN, S, TE, SXT | CTX-M-14 | |||
| EP163 | AM, PRL, CTX, CRO, CXM, S, SXT | CTX-M-14 | |||
| EP166 | AM, PRL, CTX, CRO, CXM, TE, SXT | CTX-M-14 | TEM-1 | ||
| EP171 | AM, PRL, FEP, CTX, CRO, CXM, CAZ, ATM, CN, TE, SXT | CTX-M-55 | TEM-1 | ||
| EP176 | AM, PRL, FEP, CTX, CRO, CXM, CN, K, S, TE, NA | CTX-M-14 | TEM-1 | ||
| EP179 | AM, PRL, SAM, CTX, CRO, CXM, CN, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP180 | AM, PRL, FEP, CTX, CRO, CXM, CAZ, ATM, S, TE, NA | CTX-M-15 | CTX-M-14 | ||
| EP182 | AM, PRL, SAM, CTX, CRO, CXM, CN, K, S, TE, NA | CTX-M-14 | |||
| EP186 | AM, PRL, CTX, CRO, CXM, CN, K, S, TE, NA, CIP, NOR, LEV, SXT, C | CTX-M-14 | TEM-1 | ||
| EP187 | AM, PRL, AMC, FEP, CTX, CRO, CXM, CN, S, TE, SXT | CTX-M-3 | TEM-1 | ||
| EP191 | AM, PRL, SAM, CTX, CRO, CXM, CN, K, S, TE, SXT | CTX-M-14 | TEM-1 | ||
| EP193 | AM, PRL, SAM, FEP, CTX, CRO, CXM, CAZ, ATM, CN, K, S | CTX-M-55 | TEM-1 | ||
| EP239 | AM, PRL, AMC, SAM, CTX, CRO, CXM, CN, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP370 | AM, PRL, FEP, CTX, CRO, CXM, CN, TE, NA, SXT | CTX-M-14 | |||
| EP408 | AM, PRL, SAM, CTX, CRO, CXM, ATM, S, TE, NA, SXT | CTX-M-14 | TEM-1 | ||
| EP410 | AM, PRL, CTX, CRO, CXM, ATM, CN, NA | CTX-M-15 | |||
| EP412 | AM, PRL, CTX, CRO, CXM, TE | CTX-M-13 | |||
| Healthy carriers (5) | EP318 | AM, PRL, FEP, CTX, CRO, CXM, CAZ, ATM | CTX-M-15 | ||
| EP361 | AM, PRL, FEP, CTX, CRO, CXM, K, S, TE, NA | CTX-M-14 | |||
| EP 402 | AM, PRL, FEP, CTX, CRO, CXM, ATM, TE | CTX-M-3 | |||
| EP404 | AM, PRL, CTX, CRO, CXM, S | CTX-M-14 | |||
| EP415 | AM, PRL, CTX, CRO, CXM, ATM, TE | CTX-M-15 | |||
| Animal (1) | EP298 | AM, PRL, CTX, CRO, CXM, K, S, TE, NA, SXT, C | CTX-M-14 | ||
| Raw meat (3) | EP244 | AM, PRL, AMC, SAM, CTX, CRO, CXM, ATM, S, TE, NA, SXT, C | CTX-M-14 | ||
| EP299 | AM, PRL, CTX, CRO, CXM, K, S, TE, NA, SXT, C | CTX-M-14 | |||
| EP344 | AM, PRL, CTX, CRO, CXM, K, S, TE, C | CTX-M-14 | |||
Fig. 1.
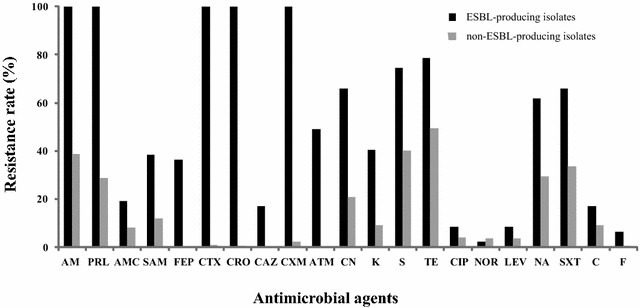
Comparison of antimicrobial susceptibility profiles of ESBL-producing and non-ESBL-producing aEPEC isolates. Proportions (%) of isolates (y-axis) resistant to antimicrobial agents (x-axis) among ESBL-producing and non-ESBL-producing isolates
Molecular characteristics of ESBL genes
The presence of blaCTX-M, blaTEM, and blaSHV genes in 47 ESBL-producing isolates was screened using PCR. The blaCTX-M-1 subgroup was identified in 20 (42.6%) ESBL-producing isolates, with 17 from diarrheal patients and three from healthy carriers. The blaCTX-M-9 subgroup was found in 30 (63.8%) isolates, with 24 from diarrheal patients, three from raw meat, two from healthy carriers, and one from animals. A total of 26 isolates recovered from diarrheal patients possessed the blaTEM subgroup (Table 3). None of the 47 isolates examined in this study was positive for the genes belonging to subgroups blaCTX-M-2, blaCTX-M-8/25/26, or blaSHV.
DNA sequencing showed that blaCTX-M-14 gene was the most prevalent, and was present in 28 (59.6%) ESBL-producing isolates, with 22 from diarrheal patients, three from raw meat, two from healthy carriers, and one from animal. The blaCTX-M-15 gene was identified in 11 (23.4%) isolates, with nine from diarrheal patients and two from healthy carriers. The blaCTX-M-55 and blaCTX-M-3 genes were found in four and five isolates, respectively. The two genes, blaCTX-M-13 and blaCTX-M-65, belonging to the subgroup blaCTX-M-9, were only found in two separate diarrheal patient-derived isolates. In addition, all of the 26 blaTEM genes were identified as blaTEM-1. The coexistence of subgroup blaCTX-M-1 and blaCTX-M-9 genes was identified in three diarrheal patient isolates, including one that harbored blaCTX-M-14 and blaCTX-M-55, and two that harbored blaCTX-M-14 and blaCTX-M-15 (Table 3).
Distribution of antimicrobial resistance determinants
Among the 96 genome-sequenced aEPEC isolates, 50 were resistant to ampicillin and possessed β-lactamase-related genes, including blaTEM-1 (48.0%), blaCTX (16.0%), blaOXA (6.0%), blaTEM-1 + blaCTX (16.0%), blaTEM-1 + blaLEN (2.0%), blaCTX + blaLEN (4.0%), and blaCTX + blaOXA (2.0%) (Table 4, Additional file 2). There was a significant association (χ2 = 84.715, P = 0.000) between the presence of these genes and resistance to ampicillin. Fifty-one isolates resistant to tetracycline harbored resistance associated determinants, including tetA (52.9%), tetB (3.9%), tetC (2.0%), tetA + tetC (17.6%), and tetB + tetC (10.0%). A significant association was observed between resistance to tetracycline and the occurrence of tetA (χ2 = 47.172, P = 0.000) and tetB (P = 0.062), but not with tetC (χ2 = 1.129, P = 0.288). Three and five chloramphenicol-resistant isolates harbored cat and cml genes, respectively. The sul1 + dfra12/17 (37.5%) and sul1 + sul2 + dfra5/12/17 (35.0%) were the predominant resistance genes among the 40 isolates that were resistant to trimethoprim–sulfamethoxazole. The combination of sul and dfra was detected more frequently in resistant strains than in sensitive strains (χ2 = 72.432, P = 0.000). The most frequent resistance gene observed in 33 phenotypically gentamicin-resistant isolates was aac3iia (69.7%). Four different genes or gene combinations, i.e., ant3ia, aph33ib, aph33ib + aph6id, and aph33ib + aph6id + ant3ia, were found in four (9.1%), two (4.5%), 24 (54.5%), and one (2.3%) of the 44 streptomycin-resistant isolates, respectively (Table 4, Additional file 2). Significant associations between the presence of these genes and streptomycin resistance were also observed (χ2 = 57.281, P = 0.000).
Table 4.
Resistance-related genes among 96 genome sequenced aEPEC isolates
| Phenotype of resistance (no. of isolates) | Resistance genes | No. of isolates (%) |
|---|---|---|
| Ampicillin (50) | bla TEM-1 | 24 (48.0) |
| bla CTX | 8 (16.0) | |
| bla OXA | 3 (6.0) | |
| blaTEM-1 + blaCTX | 8 (16.0) | |
| blaTEM-1 + blaLEN | 1 (2.0) | |
| blaCTX + blaLEN | 2 (4.0) | |
| blaCTX + blaOXA | 1 (2.0) | |
| Tetracycline (51) | tetA | 27 (52.9) |
| tetB | 2 (3.9) | |
| tetC | 1 (2.0) | |
| tetA + tetC | 9 (17.6) | |
| tetB + tetC | 5 (10.0) | |
| Chloramphenicol (8) | cat | 3 (37.5) |
| cml | 5 (62.5) | |
| Trimethoprim–sulfamethoxazole (40) | sul1 + dfra12/17 | 15 (37.5) |
| sul2 + dfra14/17 | 6 (15.0) | |
| sul3 + dfra12 | 3 (7.5) | |
| sul1 + sul2 + dfra5/12/17 | 14 (35.0) | |
| dfra1 | 2 (5.0) | |
| dfra17 | 1 (2.5) | |
| Gentamicin (33) | aac3iia | 23 (69.7) |
| aac3iia + ant2ia | 3 (9.1) | |
| aac3iia + aph3ia | 2 (6.1) | |
| aac3iia + ant2ia + aph3ia | 1 (3.0) | |
| Streptomycin (44) | ant3ia | 4 (9.1) |
| aph33ib | 2 (4.5) | |
| aph33ib + aph6id | 24 (54.5) | |
| aph33ib + aph6id + ant3ia | 1 (2.3) | |
| Kanamycin (12) | ant2ia | 1 (8.3) |
| aph3ia | 3 (25.0) | |
| ant2ia + aph3ia | 2 (16.7) |
Discussion
Globally, EPECs displaying different levels of resistance to a range of antibiotics are increasing alarmingly [37]. The antimicrobial resistance of EPEC has been reported in many countries, including Brazil [38, 39], India [40], Iran [41], Ireland [42], the United Kingdom [43], and Singapore [44]. In China, only two studies are available: one characterizing 39 EPEC isolates in ready-to-eat foods [45] and another examining 58 EPEC isolates recovered from pediatric diarrheal patients [46]. These EPEC strains were either restricted to being from foods or were regionally restricted. In the present study, the 267 aEPEC isolates were recovered from different sources (diarrheal patients, healthy carriers, animals and raw meat) from ten provinces/cities of China. This was the first study to reveal the comprehensive antimicrobial resistance of aEPEC in China and to provide further insight into the current situation of this specific diarrheagenic E. coli.
Of the 151 diarrheal patient-derived aEPEC isolates, the highest resistance rate was to tetracycline, followed by ampicillin and streptomycin, which was different from reports in Iran [47], Brazil [39], and India [40]. Physicians in China should pay attention to the antimicrobial resistance of clinical aEPEC isolates, because EPEC is still one of the most common pathogens associated with infectious diarrhea. Domestic animals, such as sheep, cattle, poultry, and pigs, have been considered as the main reservoirs of aEPEC [14]. In Europe, the predominant antimicrobial agents administered to animals are sulphonamides and/or trimethoprim, tetracyclines and β-lactams [48]. However, there is little antimicrobials consumption data available in this field in China. It was reported that high doses and multiple types of veterinary antimicrobial products were used routinely in livestock husbandry [49]. The agents mentioned above are also included in the antimicrobials that can be used in the treatment and prevention of animal diseases. The high prevalence of antimicrobial-resistant aEPEC in raw meat and animals could be explained by the possible overuse and misuse of tetracyclines, ampicillin, and trimethoprim/sulphonamides in veterinary practice and agriculture. Poor sanitary conditions or practices might also play a role in the spread of resistant aEPEC.
The emergence of multidrug resistance, especially among Enterobacteriaceae, i.e., E. coli, has become a critical public concern [18]. In this study, nearly half of the 267 aEPEC strains were multidrug resistant. These MDR strains showed high resistance to tetracycline (92.2%) and ampicillin (89.8%), and 31.3% of that showed resistance to ≥ 10 antimicrobial agents. In addition, in this study, significantly more aEPEC strains from diarrheal patients showed multidrug resistance than did strains from healthy carriers and animals. Thus, diarrheal patients may be the main source of MDR aEPEC strains in China and clinicians should be careful when using antibiotics as therapy for EPEC infections. A recent study showed that wild birds could also act as carriers of MDR EPEC [50]. Consistent with this, we found that 19 (37.3%) aEPEC strains from animals, including birds, pika, and marmot, were MDR. In this sense, MDR aEPEC could emerge in the natural environment and then pose potential risk to public health.
Most multidrug resistances in Enterobacteriaceae are associated with ESBLs [51]. E. coli has become one of main producers of ESBL and has posed a major challenge in the treatment of bacterial infection [19]. A previous study showed that occurrence of ESBL-producing E. coli in patients in China varied from 30.2 to 57.0% [52]. In our study, 47 (17.6%) aEPEC isolates were identified as ESBL-producing strains, with 38 the isolates coming from diarrheal patients. Most ESBL-producing isolates showed co-resistance to other antimicrobial agents, such as aminoglycosides, tetracyclines, and sulfonamides, and even to fluoroquinolones [22]. The present results showed that ESBL-producing aEPEC isolates displayed co-resistance to aminoglycosides, tetracycline, nalidixic acid, trimethoprim–sulfamethoxazole, and nitrofurantoin, but not to fluoroquinolones. It is worth noting that MDR E. coli usually implies significant increase of resistance and pathogenic potential, such as the emergence of ESBL-producing clone ST131 [53] and another clinically relevant ESBL-producing clone ST410 [54]. The multi-locus sequence typing (MLST) analysis in our previous study indicated that these aEPEC isolates showed high clonal diversity, but none of them were identified as ST131 or ST410 [28].
TEM, SHV, and CTX-M are the three main genetic types of ESBLs [19]. Currently, the CTX-M-type ESBLs have dramatically increased and largely outnumber other types of ESBLs [25]. However, there are extensive geographical variations in the distribution of dominant CTX-M types across different countries, such as CTX-M-2 in Japan, CTX-M-1 in Italy, and CTX-M-2 and CTX-M-15 in Brazil. By contrast, CTX-M-15 widespread throughout the world [22, 55, 56]. In the present study, all 47 ESBL-producing aEPEC isolates possessed CTX-M genes. No TEM or SHV type ESBL genes were detected. The most prevalent gene was blaCTX-M-14, followed by blaCTX-M-15, with majority being from diarrheal patients. These findings revealed that CTX-M-14 and CTX-M-15 were predominant among aEPEC isolates in China. This is consistent with previous reports that CTX-M-14 was the most abundant CTX-M type among E. coli strains from animals [57] and clinical patients in China [52]. CTX-M-55 was observed only in four aEPEC strains from diarrheal patients, although it was demonstrated to be widespread in E. coli isolates from food-producing animals and environmental samples in China [58, 59]. These findings suggested that humans might acquire these strains from animals, as well as from the food chain.
High levels of resistance to tetracycline, ampicillin, and streptomycin were identified among 96 genome sequenced aEPEC isolates. More than half of the ampicillin resistant strains harbored the blaTEM-1 gene in this study. It has been reported that blaTEM was the most frequent β-lactamase gene involved in ampicillin resistance in E. coli [60]. Of the known tetracycline resistance genes, only tetA, tetB, and tetC (alone or in combination) were detected, indicating that the major mechanism involved in tetracycline resistance in aEPEC isolates is active efflux. This is consistent with the investigation of EPEC from diarrheic rabbits in Portugal [60]. Among the aEPEC resistant to aminoglycosides, 69.7% of the isolates resistant to gentamicin carried aac3iia; 54.5% isolates resistant to streptomycin possessed genes aph33ib and aph6id; and most isolates resistant to kanamycin harbored aph3ia. These results suggested that aminoglycoside acetyltransferases are the main mechanism of resistance to gentamicin, while aminoglycoside phosphotransferases are the predominant mechanism mediating streptomycin and kanamycin resistance. With respect to determinants responsible for resistance to trimethoprim–sulfamethoxazole, our results demonstrated that sul1, sul2, dfra12, and/or dfra17 were the predominant genes, as revealed by a previous study [60].
Some limitations exist in this study. Compared with the number of strains from diarrheal patients, fewer isolates from healthy carriers, animals, and raw meat were included. Further investigations are needed to clarify the association between virulence and antimicrobial resistance.
In conclusion, our investigation revealed the occurrence of multidrug-resistant and ESBL-producing aEPEC isolates in China. These results suggest that it is necessary to continuously monitor the emergence and spread of MDR aEPEC to guide the application of antimicrobials in farm animals and in clinical treatment.
Additional files
Additional file 1. Antimicrobial susceptibility of 267 aEPEC strains tested in the study.
Additional file 2. Antimicrobial susceptibility profiles and resistance-related genes of 96 genome-sequenced aEPEC strains.
Authors’ contributions
YX designed the project. YXu, HS, XB, SF and RF performed the experiments. YXu analyzed the data. YXu and YX drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional files).
Consent for publication
All authors gave the consent for publication.
Ethics approval and consent to participate
The present study was approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, China CDC (Approval No. ICDC2014003).
Funding
This work was supported by the National Natural Science Foundation of China [Grant Numbers 81772152, 81701977], the State Key Laboratory of Infectious Disease Prevention and Control [Grant Number 2015SKLID504], and the National Basic Research Priorities Program of China [Grant Number 2015CB554201].
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- A/E
attaching and effacing
- AM
ampicillin
- AMC
amoxicillin–clavulanic acid
- ATM
aztreonam
- aEPEC
atypical EPEC
- C
chloramphenicol
- F
nitrofurantoin
- CAZ
ceftazidime
- CIP
ciprofloxacin
- CN
gentamicin
- CRO
ceftriaxone
- CTX
cefotaxime
- CXM
cefuroxime
- EPEC
enteropathogenic Escherichia coli
- ESBL
extended-spectrum β-lactamase
- FEP
cefepime
- K
kanamycin
- LEE
locus of enterocyte effacement
- LEV
levofloxacin
- MDR
multidrug resistance
- MEM
meropenem
- NA
nalidixic acid
- NOR
norfloxacin
- IPM
imipenem
- PRL
piperacillin
- S
streptomycin
- SAM
ampicillin–sulbactam
- SXT
trimethoprim–sulfamethoxazole
- TE
tetracycline
- tEPEC
typical EPEC
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13099-018-0234-0) contains supplementary material, which is available to authorized users.
Contributor Information
Yanmei Xu, Email: xyanmei2006@126.com.
Hui Sun, Email: sunhui@icdc.cn.
Xiangning Bai, Email: baixiangning@icdc.cn.
Shanshan Fu, Email: 543195534@qq.com.
Ruyue Fan, Email: fryforever@163.com.
Yanwen Xiong, Phone: +8610 58900749, Email: xiongyanwen@icdc.cn.
References
- 1.Walker CL, Aryee MJ, Boschi-Pinto C, Black RE. Estimating diarrhea mortality among young children in low and middle income countries. PLoS ONE. 2012;7:e29151. doi: 10.1371/journal.pone.0029151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379:2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 3.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HD, Frankel G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev. 2005;29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE, et al. Global causes of diarrheal disease mortality in children < 5 years of age: a systematic review. PLoS ONE. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Torres AG. Enteropathogenic Escherichia coli: foe or innocent bystander? Clin Microbiol Infect. 2015;21:729–734. doi: 10.1016/j.cmi.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke SC, Haigh RD, Freestone PP, Williams PH. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin Microbiol Rev. 2003;16:365–378. doi: 10.1128/CMR.16.3.365-378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalita A, Hu J, Torres AG. Recent advances in adherence and invasion of pathogenic Escherichia coli. Curr Opin Infect Dis. 2014;27:459–464. doi: 10.1097/QCO.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afset JE, Bevanger L, Romundstad P, Bergh K. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J Med Microbiol. 2004;53:1137–1144. doi: 10.1099/jmm.0.45719-0. [DOI] [PubMed] [Google Scholar]
- 10.Robins-Browne RM, Bordun AM, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, et al. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg Infect Dis. 2004;10:1797–1805. doi: 10.3201/eid1010.031086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen MB, Nataro JP, Bernstein DI, Hawkins J, Roberts N, Staat MA. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J Pediatr. 2005;146:54–61. doi: 10.1016/j.jpeds.2004.08.059. [DOI] [PubMed] [Google Scholar]
- 12.Franzolin MR, Alves RC, Keller R, Gomes TA, Beutin L, Barreto ML, et al. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem Inst Oswaldo Cruz. 2005;100:359–363. doi: 10.1590/S0074-02762005000400004. [DOI] [PubMed] [Google Scholar]
- 13.Estrada-Garcia T, Lopez-Saucedo C, Thompson-Bonilla R, Abonce M, Lopez-Hernandez D, Santos JI, et al. Association of diarrheagenic Escherichia coli pathotypes with infection and diarrhea among Mexican children and association of atypical Enteropathogenic E. coli with acute diarrhea. J Clin Microbiol. 2009;47:93–98. doi: 10.1128/JCM.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandes RT, Elias WP, Vieira MA, Gomes TA. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett. 2009;297:137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 15.Sakkejha H, Byrne L, Lawson AJ, Jenkins C. An update on the microbiology and epidemiology of enteropathogenic Escherichia coli in England 2010–2012. J Med Microbiol. 2013;62:1531–1534. doi: 10.1099/jmm.0.062380-0. [DOI] [PubMed] [Google Scholar]
- 16.Ochoa TJ, Barletta F, Contreras C, Mercado E. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans R Soc Trop Med Hyg. 2008;102:852–856. doi: 10.1016/j.trstmh.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staples M, Doyle CJ, Graham RM, Jennison AV. Molecular epidemiological typing of enteropathogenic Escherichia coli strains from Australian patients. Diagn Microbiol Infect Dis. 2013;75:320–324. doi: 10.1016/j.diagmicrobio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2011;11:1499–1504. doi: 10.1016/j.meegid.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe CF, Katz K, McGeer AJ, Muller MP. Efficacy of admission screening for extended-spectrum beta-lactamase producing Enterobacteriaceae. PLoS ONE. 2013;8:e62678. doi: 10.1371/journal.pone.0062678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Moxon CA, Paulus S. Beta-lactamases in Enterobacteriaceae infections in children. J Infect. 2016;72(Suppl):S41–S49. doi: 10.1016/j.jinf.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22:90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Ensor V, Gossain S, Nye K, Hawkey P. Rapid and simple detection of blaCTX-M genes by multiplex PCR assay. J Med Microbiol. 2005;54:1183–1187. doi: 10.1099/jmm.0.46160-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhang ZK, Lai SJ, Yu JX, Yang WQ, Wang X, Jing HQ, et al. Epidemiological characteristics of diarrheagenic Escherichia coli among diarrhea outpatients in China, 2012–2015. Zhonghua liu xing bing xue za zhi. 2017;38:419–423. doi: 10.3760/cma.j.issn.0254-6450.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Bai X, Jin Y, Hu B, Wang H, Sun H, et al. High prevalence of virulence genes in specific genotypes of atypical enteropathogenic Escherichia coli. Front Cell Infect Microbiol. 2017;7:109. doi: 10.3389/fcimb.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CLSI. Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100S. Wayne: CLSI; 2017.
- 30.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 31.Lee WC, Yeh KS. Characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolated from fecal samples of piglets with diarrhea in central and southern Taiwan in 2015. BMC Vet Res. 2017;13:66. doi: 10.1186/s12917-017-0986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldonado NA, Munera MI, Lopez JA, Sierra P, Robledo C, Robledo J, et al. Trends in antibiotic resistance in Medellin and municipalities of the metropolitan area between 2007 and 2012: results of six years of surveillance. Biomedica. 2014;34:433–446. doi: 10.7705/biomedica.v34i3.1658. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian K, Selvakkumar C, Vinaykumar KS, Goswami N, Meenakshisundaram S, Balakrishnan A, et al. Tackling multiple antibiotic resistance in enteropathogenic Escherichia coli (EPEC) clinical isolates: a diarylheptanoid from Alpinia officinarum shows promising antibacterial and immunomodulatory activity against EPEC and its lipopolysaccharide-induced inflammation. Int J Antimicrob Agents. 2009;33:244–250. doi: 10.1016/j.ijantimicag.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 38.Scaletsky IC, Souza TB, Aranda KR, Okeke IN. Genetic elements associated with antimicrobial resistance in enteropathogenic Escherichia coli (EPEC) from Brazil. BMC Microbiol. 2010;10:25. doi: 10.1186/1471-2180-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitondo-Silva A, Nakazato G, Falcao JP, Irino K, Martinez R, Darini AL, et al. Phenotypic and genetic features of enteropathogenic Escherichia coli isolates from diarrheal children in the Ribeirao Preto metropolitan area, Sao Paulo State, Brazil. APMIS. 2015;123:128–135. doi: 10.1111/apm.12314. [DOI] [PubMed] [Google Scholar]
- 40.Malvi S, Appannanavar S, Mohan B, Kaur H, Gautam N, Bharti B, et al. Comparative analysis of virulence determinants, antibiotic susceptibility patterns and serogrouping of atypical enteropathogenic Escherichia coli versus typical enteropathogenic E. coli in India. J Med Microbiol. 2015;64:1208–1215. doi: 10.1099/jmm.0.000131. [DOI] [PubMed] [Google Scholar]
- 41.Bakhshi B, Fallahzad S, Pourshafie MR. The occurrence of atypical enteropathogenic Escherichia coli strains among children with diarrhea in Iran. J Infect Chemother. 2013;19:615–620. doi: 10.1007/s10156-012-0526-0. [DOI] [PubMed] [Google Scholar]
- 42.Bolton DJ, Ennis C, McDowell D. Occurrence, virulence genes and antibiotic resistance of enteropathogenic Escherichia coli (EPEC) from twelve bovine farms in the north-east of Ireland. Zoonoses Public Health. 2014;61:149–156. doi: 10.1111/zph.12058. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins C, Smith HR, Lawson AJ, Willshaw GA, Cheasty T, Wheeler JG, et al. Serotypes, intimin subtypes, and antimicrobial resistance patterns of atypical enteropathogenic Escherichia coli isolated in England from 1993 to 1996. Eur J Clin Microbiol Infect Dis. 2006;25:19–24. doi: 10.1007/s10096-005-0075-x. [DOI] [PubMed] [Google Scholar]
- 44.Lim YS, Ngan CC, Tay L. Enteropathogenic Escherichia coli as a cause of diarrhoea among children in Singapore. J Trop Med Hyg. 1992;95:339–342. [PubMed] [Google Scholar]
- 45.Zhang S, Wu Q, Zhang J, Zhu X. Occurrence and characterization of enteropathogenic Escherichia coli (EPEC) in retail ready-to-eat foods in China. Foodborne Pathog Dis. 2016;13:49–55. doi: 10.1089/fpd.2015.2020. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Shan XF, Deng H, Huang YJ, Mu XP, Huang AL, et al. Epidemiology, antimicrobial resistance and beta-lactamase genotypic features of enteropathogenic Escherichia coli isolated from children with diarrhea in Southern China. Jpn J Infect Dis. 2015;68:239–243. doi: 10.7883/yoken.JJID.2014.120. [DOI] [PubMed] [Google Scholar]
- 47.Mahmoudi-Aznaveh A, Bakhshi B, Najar-Peerayeh S. The trend of enteropathogenic Escherichia coli towards atypical multidrug resistant genotypes. J Chemother. 2017;29:1–7. doi: 10.1080/1120009X.2016.1154683. [DOI] [PubMed] [Google Scholar]
- 48.Grave K, Torren-Edo J, Mackay D. Comparison of the sales of veterinary antibacterial agents between 10 European countries. J Antimicrob Chemother. 2010;65:2037–2040. doi: 10.1093/jac/dkq247. [DOI] [PubMed] [Google Scholar]
- 49.Hu Y, Cheng H. Use of veterinary antimicrobials in China and efforts to improve their rational use. J Glob Antimicrob Resist. 2015;3:144–146. doi: 10.1016/j.jgar.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Borges CA, Cardozo MV, Beraldo LG, Oliveira ES, Maluta RP, Barboza KB, et al. Wild birds and urban pigeons as reservoirs for diarrheagenic Escherichia coli with zoonotic potential. J Microbiol. 2017;55:344–348. doi: 10.1007/s12275-017-6523-3. [DOI] [PubMed] [Google Scholar]
- 51.Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum beta-lactamases in Gram-negative rods. Infect Genet Evol. 2012;12:883–893. doi: 10.1016/j.meegid.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. 2014;14:659. doi: 10.1186/s12879-014-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaufler K, Semmler T, Wieler LH, Wöhrmann M, Baddam R, Ahmed N, et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410—another successful pandemic clone? FEMS Microbiol Ecol. 2015;92:fiv155. doi: 10.1093/femsec/fiv155. [DOI] [PubMed] [Google Scholar]
- 55.Ranjan A, Shaik S, Hussain A, Nandanwar N, Semmler T, Jadhav S, et al. Genomic and functional portrait of a highly virulent, CTX-M-15-producing H30-Rx subclone of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2015;59:6087–6095. doi: 10.1128/AAC.01447-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocha FR, Pinto VP, Barbosa FC. The spread of CTX-M-type extended-spectrum beta-lactamases in Brazil: a systematic review. Microb Drug Resist. 2016;22:301–311. doi: 10.1089/mdr.2015.0180. [DOI] [PubMed] [Google Scholar]
- 57.Xu G, An W, Wang H, Zhang X. Prevalence and characteristics of extended-spectrum beta-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front Microbiol. 2015;6:1103. doi: 10.3389/fmicb.2015.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng H, Zeng Z, Chen S, Liu Y, Yao Q, Deng Y, et al. Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J Antimicrob Agents. 2012;39:305–310. doi: 10.1016/j.ijantimicag.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Ma J, Liu JH, Lv L, Zong Z, Sun Y, Zheng H, et al. Characterization of extended-spectrum beta-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl Environ Microbiol. 2012;78:3668–3673. doi: 10.1128/AEM.07507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poeta P, Radhouani H, Goncalves A, Figueiredo N, Carvalho C, Rodrigues J, et al. Genetic characterization of antibiotic resistance in enteropathogenic Escherichia coli carrying extended-spectrum beta-lactamases recovered from diarrhoeic rabbits. Zoonoses Public Health. 2010;57:162–170. doi: 10.1111/j.1863-2378.2008.01221.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Antimicrobial susceptibility of 267 aEPEC strains tested in the study.
Additional file 2. Antimicrobial susceptibility profiles and resistance-related genes of 96 genome-sequenced aEPEC strains.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its additional files).


