Abstract
Cyanobacteria are a key constituent of biocrusts, communities dominated by lichens, mosses and associated microorganisms, which are prevalent in drylands worldwide and that largely determine their functioning. Despite their importance, there are large gaps in our knowledge of the composition and diversity of cyanobacteria associated with biocrusts, particularly in areas such as the Mediterranean Basin. We studied the diversity of these cyanobacteria in a gypsiferous grassland from Central Spain using both morphological identification after cultivation and genetic analyses with the 16S rRNA gene. Nine different morphotypes were observed, eight corresponding to filamentous, and one to unicellular cyanobacteria. We found cyanobacterial genera typical of biocrust communities, such as Microcoleus and Trichocoleus, and N-fixing cyanobacteria such as Scytonema and Nostoc. Genetic information allowed us to identify cultures belonging to recently described genera such as Roholtiella, Nodosilinea and Mojavia. We also describe two new phylotypes of Microcoleus and Scytonema, which are key genera contributing to ecosystem functioning in biocrust-dominated ecosystems worldwide.
Keywords: Soil cyanobacteria, Biocrust, Biological soil crust, Microcoleus, Cyanobacterial diversity, 16S rRNA
1. Introduction
Cyanobacteria are a key component of biocrusts, soil surface communities also formed by lichens, mosses, liverworts and other microorganisms that are a prevalent biotic feature of drylands worldwide (Büdel et al., 2016). Cyanobacteria are present in virtually all biocrust communities due to their capacity to adapt to a wide range of ecological conditions (Tamaru et al., 2005). Early successional biocrusts are dominated by filamentous pioneer cyanobacteria, which are the first colonizers of bare ground areas in drylands (Garcia-Pichel and Wojciechowski, 2009). These organisms secrete an exopolysaccharide (EPS) matrix that promotes soil stabilization and enhances microhabitat conditions for colonization of other cyanobacteria and the remaining biocrust constituents (Mager and Thomas, 2011). Cyanobacteria with heterocysts (heterocytes) are also important contributors to nitrogen fixation in oligotrophic ecosystems such as drylands (Belnap, 2002). Together with the other components of cryptogamic covers, heterocyst-forming cyanobacteria contribute to the fixation of nearly half of the total amount of biologically fixed nitrogen worldwide (Elbert et al., 2012).
Detailed studies of the composition and biogeography of cyanobacteria have been carried out in North America, Asia, Africa, Europe and Australia (e.g. Garcia-Pichel et al. 2013, Dojani et al. 2014, Hagemann et al. 2014, Kumar and Adhikary 2015, Williams et al. 2012, Williams et al. 2016). However, to date, few studies have analysed the cyanobacteria associated to biocrusts in gypsum habitats (Garcia-Pichel et al., 2001; Steven et al., 2013), even though they are hotspots of botanical diversity (Escudero et al., 2014) and harbour very conspicuous biocrust communities dominated by lichens (Castillo-Monroy et al., 2010; Martínez et al., 2006). We studied biocrusts in a gypsiferous semiarid site from Central Spain to advance our knowledge of cyanobacterial communities associated gypsum biocrusts. We used a combination of molecular and morphological information because this increases the number of identified sequences in molecular databases, which is a major concern in the study of cyanobacterial diversity nowadays (Thomazeau et al., 2010; Weber et al., 2016).
2. Materials and methods
2.1. Field site
This study was carried out in the Aranjuez Experimental Station, located in Central Spain (40°01'55.7"N - 3°32'48.3"W and 590 m above sea level). The climate is semiarid, with an intense summer drought lasting from June to September. Mean annual temperature is 15°C and annual precipitation is 349 mm. Soils are rich in gypsum, and are classified as Gypsiric Leptosols (IUSS Working Group WRB 2014; see Castillo-Monroy et al. 2010 for a physico-chemical characterization). The vegetation cover is sparse and dominated by herbaceous plants such as Macrochloa tenacissima (L.) Kunth. and shrubs such as Retama sphaerocarpa (L.) Boiss. and Helianthemum squamatum (L.) Dum. Cours. The soil in open areas located between plant patches is covered by a well-developed biocrust community dominated by the squamulous lichens Diplochistes diacapsis (Ach.) Lumbsch., Squamarina lentigera (Weber) Poelt. and Psora decipiens (Hedwig) Hoffm.; with patches of acrocarpous mosses Pleurochaete squarrosa (Brid.) Lindb. and Tortula revolvens (Schimp.) G. Roth. (see Maestre et al. 2013 for a full list of lichens and mosses found in the site).
2.2. Soil collection and morphological characterization of cyanobacteria
We randomly selected eight 50 x 50 cm plots in areas with a well-developed biocrust community in July 2013. At each plot, we collected five samples (0-1 cm depth), which were pooled and taken to the laboratory. Lichens and mosses were removed, and soil was sieved through a 2 mm sieve and kept dry in the dark.
Cyanobacterial strains were isolated using a modification of the procedure described in (Loza et al., 2013). Aliquots of ~1 g of soil were mixed with 1.5 ml of cyanobacterial culture media and distributed uniformly over different solid media (1.5% agar concentration). We used four common culture media for cyanobacteria: BG11, BG110 (Rippka et al., 1979), modified CHU 10, and modified CHU 10 without addition of N (Gómez et al., 2009). These media allowed the growth of cyanobacteria by providing a range of nutrient richness with and without N, which is important to isolate both N-fixing and non-N-fixing cyanobacteria. To avoid fungal contamination, we added cycloheximide (0.1 mg/ml). Cultures were incubated in a growth chamber at constant light and temperature (20-50 µmol photons m-2 s-1 and 28ºC) three to four weeks until colonies grew without overlapping. Cyanobacterial colonies were isolated under a dissecting microscope (Leica, Leica Microsystems, Wetzler, Germany) as described in Gómez et al. (2009). Cultures were kept in the same medium and conditions both in agar plates and in liquid medium to further promote their growth.
All colonies were characterized morphologically using a dissecting microscope and an Olympus BH2-RFCA (Olympus, Tokio, Japan) photomicroscope. Identification and morphological characterization of cyanobacteria were conducted considering the following attributes: colony morphology, trichome shape, presence of sheaths, details of cell morphology, number of trichomes per filament and end cell characteristics. Taxonomy was based on Geitler (1932), Anagnostidis and Komárek (1999), Komárek and Anagnostidis (2005) and Komárek (2013).
2.3. Genotypic characterization
DNA was extracted with the Ultraclean Microbial DNA Isolation Kit (Mobio, Carlsbad, CA, USA) following the manufacturer’s instructions. A prior step was added at the beginning of the procedure, as samples were homogenized and exposed to three cycles of thermal shock using alternating immersion in liquid N and heating to 60ºC to break the protective EPS that covers the surface of many cyanobacteria (Loza et al., 2013). PCR amplifications were performed using the bacterial 16S rRNA primers 27F and 1494R (Neilan et al., 1997). The PCR mixture (25μl) contained 2.5 μl Buffer 10X, 1.5 mM MgCl2, 50 μM dNTP, 10 pmol of each primer, BSA 1 mg/ml, 5 μl TaqMasterTM PCR Enhancer 5x (Eppendorf, Germany), 0.75 U Ultratools DNA polymerase (Biotools, Spain), miliQ H2O and 10 ng DNA. Amplification took place in a termocycler PCR Eppendorf Mastercycler (Eppendorf, Viena) with the reaction conditions described by Gkelis et al. (2005). Success in PCR was checked with agarose gel 1.5% using 1Kb Gene Ruler (MBL Biotools, Spain) and fluorescent DNA stain GelRed™. PCR products were purified with Real Clean Spin Kit (Real, Durviz, Spain) and sequenced at Centro Nacional de Investigaciones Oncológicas (Madrid, Spain). When sequences had low size (<200 bp) or quality (low confidence on % base assignation in sequence chromatograms), PCR products were cloned into pGEM-T vectors with the pGEM Easy Vector system (Promega, US) according to manufacturer recommendations and sequenced according to vector information and primers. Sequences were obtained for both strands independently.
We compared our results with sequences from the National Center for Biotechnology Information (NCBI) database to complement identifications. For phylogenetic analysis, sequences were aligned using ClustalW (Thompson et al., 1994) with the software Bioedit 7.2.5 (Ibis Biosiences, Carlsbad, CA). We obtained the most similar sequences and reference strains of the closest species from the NCBI database with BLAST (blast.ncbi.nlm.nih.gov) and then performed multiple alignment with all sequences (Altschul et al., 1990). Phylogenetic trees were generated with the MEGA7 software (Tamura et al., 2013) using the Maximum Likelihood, Neighbor Joining (Saitou and Nei, 1987), and Maximum Parsimony methods and the Tajima Nei matrix (Tajima and Nei, 1984) to calculate pairwise distances. The alignment was checked and corrected manually with the Bioedit software (Ibis Biosiences, Carlsbad, CA). All sequences obtained had an expected length ranging from 1046 to 1459 bp, except AR11, which had 532 bp. Independent phylogenetic trees made with and without AR11 produced similar clustering, therefore this sequence was included in the phylogenetic analysis. Phylogenetic trees were carried out using our nineteen 16S rRNA sequences together with 53 cyanobacterial sequences from the NCBI database. There were a total of 879 positions in the final dataset. Statistical significance was performed with the bootstrap test described in Felsenstein (1985), using 1000 and 500 replicates for the Neighbor Joining tree and both Maximum Parsimony and Maximum Likelihood trees, respectively. Sequence similarity between our sequences and those from the NCBI database was determined as 100*(1-P-distance), and was carried out using the MEGA7 software (Tamura et al., 2013).
Cultures were named after the site (Aranjuez, AR-) and were included in the culture collection of the Universidad Autónoma de Madrid (UAM). The nucleotide sequences obtained in this study were uploaded to the Genbank (NCBI) database (accession numbers: MF002044 - MF002062).
3. Results
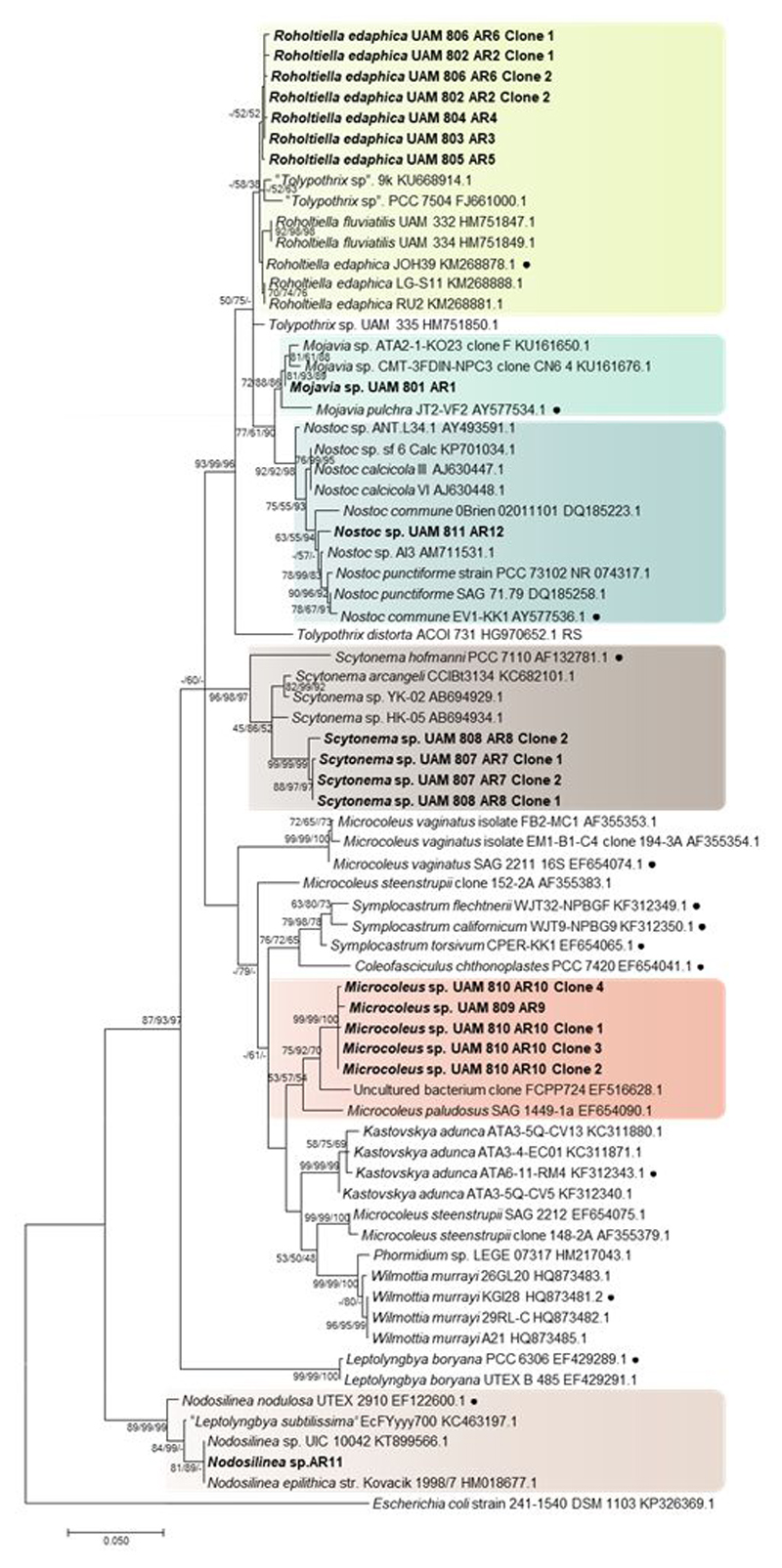
Macroscopic and microscopic evaluation of cultivated cyanobacteria yielded nine different cyanobacterial morphotypes and the successful isolation and sequencing of 12 strains. Three main types of morphologies were found: filamentous and heterocyst-forming cyanobacteria, filamentous cyanobacteria without heterocysts, and unicellular cyanobacteria (Figure 1; Table 1). The three methods used to obtain the phylogenetic tree (Maximum Likelihood, Neighbor Joining and Maximum Parsimony) produced similar clustering. Therefore, we show only the Maximum Likelihood tree, with the indication of the bootstrap values for all three approaches (Figure 2).
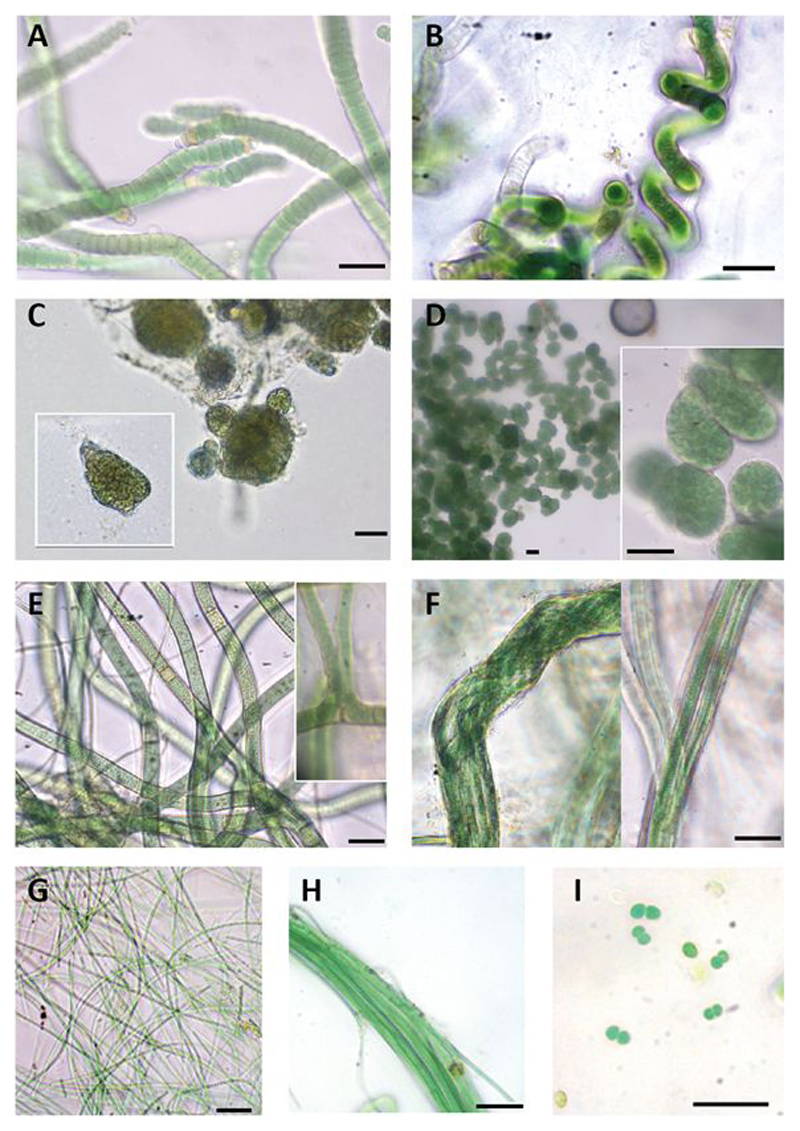
Figure 1.
Diversity of cyanobacterial morphotypes found in our study area. (A) Roholtiella edaphica. AR2; (B) Spirirestris sp; (C) Nostoc sp. AR12; (D) Mojavia sp. AR1; (E) Scytonema sp. In cultivation progress ; (F) Microcoleus sp. AR10; (G) Leptolyngbya sp. AR11, (H) Trichocoleus sp.; (I) Chroococcus sp. Scale bars equal 20 µm.
Table 1.
Morphological characteristics of the cyanobacterial strains observed in our study. B=breadth (µm); L= cell length (µm)
| Taxon | Culture-Strain | Morphological description | Dimensions (µm) | Figure |
|---|---|---|---|---|
| Roholtiella edaphica |
UAM 802- AR2 UAM 803 -AR3 UAM 804 -AR4 UAM 805 -AR5 UAM 806 -AR6 |
Heterocystous filamentous. Trichomes into a thin and transparent sheath. False branching occurs. Terminal and apical heterocysts. Rounded to flattened cells, sometimes attenuating towards the end of the trichome. | B=6.6-11.3 L=3.3-6 |
1 A |
| Spirirestris sp. | - | Thick filaments, that turn into spiralized and irregular shapes. Intermediate heterocysts. Rectangular cells, densely packed towards the end of the filament | B= 8-9.3 L=3.3-5.3 |
1 B |
| Nostoc sp. | UAM 812- AR12 | Heterocystous filamentous. Short chains of cells. Irregular to spherical colonies. Surrounded by a thin mucilage. Terminal or lateral heterocysts. Green to yellowish colour. Irregular cells, sometimes spherical. | B=5-7 L= 5.5-7.5 |
1 C |
| Mojavia sp. | UAM 801- AR1 | Heterocystous filamentous. Short chains of cells. Subspherical to ellipsoid, irregular colonies surrounded by a firm mucilaginous covering. Terminal or lateral heterocysts. Intensely colored green. Spherical to irregular cells, densely aggregated | B=4.3-6.4 L= 5.8-6.5 |
1 D |
| Scytonema sp. |
UAM 807 -AR7 UAM 808 -AR8 |
Heterocystous filamentous. Thick and cylindrical trichomes, yellow, blue-green and light green. Single and double false branching. Terminal and intermediate heterocysts. Necridia occurs in some filaments. Very thin and transparent sheath. Rectangular cells densely packed towards the end of the filament | B=7.5-9 L=3-5 |
1 E |
| Microcoleus sp. |
UAM 809 -AR9 UAM 810 -AR10 |
Ensheathed, bundle-forming filaments, surrounded by a very thick and transparent sheath. Sometimes 5-6 trichomes but also singular filaments. No heterocysts. | B=4.2-5 L=1.4-2.1 |
1 F |
| Nodosilinea sp. | UAM 811 -AR11 | Single, Long, thin and flexuous filaments. Colorless to Light green color. No heterocysts or akinetes. Cells isodiametric or longer than wide, cylindrical | B=2.2-3.2 L=3.5-5 |
1 G |
| Trichocoleus sp. | - | Ensheathed, bundle forming thick filaments, containing several trichomes. Sheaths fine and uncolored. Intense green and cylindrical trichomes, slightly attenuated at the end. Cylindrical cells, longer than wide. Apical cells are conical. | B=1.7-3.2 L=4.5-5.1 |
1 H |
| Chroococcus sp. | - | Single cells, aggregated into colonies usually with two cells surrounded by a transparent sheath. Light green | B=2-3.3 | 1 I |
Figure 2.
Phylogenetic tree based on 16S rRNA gene sequences obtained by the Maximum Likelihood method (log likelihood -17870.7546). The percentage of trees in which the associated taxa clustered together (Bootstrap) is shown next to the branches (>50% values are reported for Maximum Likelihood, Neigbor Joining and Maximum Parsimony analysis). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The 0.05 bar indicates substitutions per nucleotidic position. ● Reference strain. Newly sequenced strains are in bold.
Cluster I –Roholtiella. Sequences of isolated strains AR2 to AR6 were included in this cluster together with sequences of Roholtiella Bohuniká, Pietrasiak & Johansen., a recently described genus (Bohunická et al., 2015) from the Nostocaceae family. This genus includes some species formerly identified as Tolypothrix Kützing ex Bornet & Fahault, but phylogenetically distant to the Tolypothricaceae clade containing Tolypothrix sensu stricto. In addition, sequences of Tolypothrix not recognized as belonging to this genus (Hauer et al., 2014) were also included in this cluster. Our isolated strains shared morphological characteristics of the genus Roholtiella, such as single false branching, terminal and intercalary heterocysts, trichomes surrounded by a thin and transparent sheath and, in some cases, cells that attenuated their size towards the end of the trichome (Figure 1A). Strains AR2 to AR6 shared 99.43-99.89% of similarity between them, and were 99.32 to 99.66% similar to the reference strain of Roholtiella edaphica Bohunická & Lukesová JOH39. Therefore, these strains were assigned to this taxon.
Cluster II-Mojavia. This cluster contained sequences from Mojavia Řeháková & Johansen and our isolated strain AR1, which had 97.61-99.43 % similarity within the cluster. Mojavia is a new cyanobacterial genus that shows morphological resemblance to Nostoc Vaucher ex Bornet & Flahault (Řeháková et al., 2007). Isolated strain AR1 showed a high similarity (99.43%) with a sequence from the database corresponding to an isolated strain of Mojavia from the Atacama Desert, and a 98.37% similarity with the generitype Mojavia pulchra Řeháková & Johansen. Therefore, we assigned culture AR1 to this genus. Morphological analysis showed that this strain had short filaments, with terminal and intercalary heterocysts. Colonies were subsphaerical to irregular, densely aggregated and had an intense green color. A firm mucilaginous sheath covered thickly entwined trichomes (Figure 1C).
Cluster III-Nostoc. Cluster III included sequences corresponding to the Nostoc genus with a 97.75-100% similarity within the cluster. Isolated strain AR12 showed a similarity of 98.92% with Nostoc calcicola Brébisson ex Bornet & Flahault from this cluster, and a similarity of 98.47% with the generitype Nostoc commune Vaucher ex Bornet & Flahault. Cyanobacterial culture of strain AR12 had a thin mucilage and colonies with spherical shape, and a green to yellowish color (Figure 1D).
Cluster IV- Scytonema. This cluster harbored Scytonema Agardh ex Bornet & Fahault sequences that shared a 93.4-100 % similarity, and was strongly supported. However, cyanobacterial strains AR7 and AR8 were located in a cluster separated from the other Scytonema sequences, with only 97.6% of similarity to the closest relative Scytonema arcangeli Bornet & Flahault CCIBt3134. Our Scytonema cultures showed the typical characteristics of the genera, such as thick and cylindrical trichomes, filaments with double and single false branching, and terminal or intercalary heterocysts. Necridia appeared in some filaments, and their sheath was thin and transparent (Figure 1E).
Cluster V-Microcoleus. Isolated strains AR9 and AR10 correspond to the same species (99.49-100% similarity), and were grouped with Microcoleus paludosus Gomont and a sequence from an uncultured cyanobacterium with 96.45-100% similarity within the cluster. Filamentous strains AR9 and AR10 showed morphological features characteristic of Microcoleus Desmazierés ex Gomont: typical bundles or dense packages of trichomes (5-6 trichomes observed) surrounded by a very thick and transparent sheath, although single-trichomes in filaments were also observed (Figure 1F). Sequences of other bundle-forming cyanobacteria, such as Microcoleus vaginatus (Vaucher) Gomont, Microcoleus steenstrupii J.B.Petersen, Symplocastrum spp. (Gomont) Kirchner, Wilmottia spp. Strunecký, Elster & Komárek and Kastovskya adunca Mühlsteinova, Johansen & Pietrasiak, were located in other clusters and had low percentages of similarity to the sequences observed in our samples (all percentages of similarity below 95%).
Cluster VI -Nodosilinea. Cluster VI grouped sequences from Nodosilinea Perkenson & Casamatta, a new genus that resembles Leptolyngbya Anagnostidis & Komárek, but is morphologically and genetically distinct (Perkerson et al., 2011). This cluster exhibited a 97.42-100% sequence similarity and includes Nodosilinea generitype strain, as well as isolated strain AR11. Nevertheless, Leptolyngbya sensu stricto, with the generitype L. boryana (Gomont) Anagnostidis & Komárek, is placed in other cluster that exhibited only 86% sequence similarity (Figure 2). Culture AR11 showed single, long, thin and flexuous filaments that were green to transparent, and did not have akinetes or heterocysts (Figure 1G). The strain AR11 had a high similarity (99.6%) with Nodosilinea epilithica Kovacik, so it was assigned to this taxon.
In addition, three morphotypes were also observed in initial cultures, but could not be isolated and sequenced. They were identified as belonging to the genera Spirirestris Flechtner & Johansen, Trichocoleus Anagnostidis and Chroococcus Nägeli. Their morphological characteristics are described in Figure 1 and Table 1.
4. Discussion
4.1. Phylogenetic and morphological analysis of the isolated cyanobacteria
In this study, analysis of morphological features of isolated species in combination with phylogenetic analysis (polyphasic approach), allowed us to characterize the biocrust-forming cyanobacteria in a semiarid gypsiferous site from Central Spain. Biocrusts all over the world typically harbor genera reported in this study, such as bundle-forming filamentous Microcoleus, and heterocystous cyanobacteria from the genera Nostoc and Scytonema (Weber et al., 2016). We also found Nodosilinea, Trichocoleus, Roholtiella, Mojavia, Chroococcus and Spirirestris.
The genus Roholtiella contains cyanobacteria that formerly were included into the typical biocrust genus Tolypothrix which shares morphological characteristics, such as single and (less often) double false branching. However filaments of Tolypothrix are typically very long, cylindrical and do not attenuate towards the end, whereas Roholtiella shows tapering heteropolar trichomes during the first stages of its life cycle (Bohunická et al., 2015).
Strains AR1 and AR12 are morphologically similar to genus Nostoc, but could be separated on the basis of phylogenetic analysis. The sequence corresponding to strain AR12 was located into a pure Nostoc cluster, thus it clearly belongs to this genus. Strain AR1 was located in a cluster with all sequences from Mojavia. This genus was recently separated from the genus Nostoc based primarily on the distinctive secondary structure of the 16S-23S ITS region, and it is phylogenetically located as sister group to Nostoc sensu stricto (Řeháková et al., 2007), as found in our study (Figure 2). To our knowledge, our results provide the first record of Mojavia in Europe.
Regarding Scytonema, a wide representation of members of this genus has recently been analyzed (Komárek et al., 2013). The molecular evaluation of the species within this genus showed a separation of the traditional genus Scytonema in different clusters, which probably represented separate genera. Our cultures of Scytonema are clustered into the clade of Scytonema sequences in our phylogenetic tree. Nevertheless, the low similarity (97%) between our sequences with the closest relative Scytonema sp. HK-05, isolated from Japan, suggests that we found a novel biocrust-associated phylotype of Scytonema. A more comprehensive analysis, with a multi-locus evolutionary reconstruction and including a wider representation of Scytonemataceae, needs to be done in future studies.
Our phylogenetic analyses demonstrated the distinctiveness of strains of Microcoleus sp. AR9 and AR10 found in our samples. Their morphological evaluation showed that these strains had phenotypical characteristics of this genus, but genetically it is really different from other common Microcoleus found in biocrusts, such as the generitype Microcoleus vaginatus (only 91% similarity). The closer Microcoleus species was M. paludosus, which exhibited a similarity of 96.4-96-8% with our sequences. Microcoleus is a genus currently defined as polyphyletic, and is currently under taxonomic review (see Strunecký et al. (2013) and references therein). Based on molecular data, several bundle-forming cyanobacteria have been already separated from Microcoleus, such as Kastovskya Mühlsteinová, Johansen & Pietrasiak, Wilmottia, Coleofasciculus Siegesmund and Trichocoleus (Mühlsteinová et al., 2014) All of these genera are phylogenetically distant from the strains found in our study. The current state of cyanobacterial taxonomy makes sometimes difficult to compare results from different studies, particularly for non-taxonomic specialists. Therefore, and while awaiting further revision of the genus by studies using more bundle-forming representatives and combining information from several genes, we maintain this taxonomic assignation, particularly given its morphological and ecological closeness with typical Microcoleus from other biocrust types (see below).
Perkerson et al (2011) separated Nodosilinea from the polyphyletic genus Leptolyngbya on the basis of 16S rDNA sequences, the highly conserved 16S-23S ITS secondary structure, and morphology. Nodosilinea differs morphologically from Leptolyngbya by the presence of nodules when grown under low light conditions. However, we did not observe nodules in our AR11 strain, a response likely due the cultivation conditions employed. This character has never been reported from any other taxa in the Oscillatoriales (Perkerson et al., 2011).
4.2. Diversity of cyanobacteria from gypsum soils
Studies comparing different soil types have revealed that gypsum soils have distinct cyanobacterial communities. Using denaturalizing gradient gel electrophoresis (DGGE) and microscopy, cyanobacterial communities from gypsum soils in the Colorado Plateau (USA) were different to those from sandy, shale and silt soils (Garcia-Pichel et al., 2001). These authors also found that the common dominant filamentous cyanobacteria Microcoleus vaginatus appeared in all biocrusts except those from gypsiferous soils. In the same area, studies using high-throughput sequencing of the 16S rRNA gene have found that gypsum soils have a lower abundance and diversity of cyanobacteria, and a very low relative abundance of Microcoleus vaginatus (1-5%), when compared to other soil types (Steven et al., 2013). Biocrust-associated cyanobacteria have been rarely studied in Spain (Maestre et al., 2011). Using DGGE, Maestre et al. (2006) found 19 different phylotypes associated with biocrust-forming lichens from a calcareous site in south-eastern Spain. No heterocystous cyanobacteria were detected in that study, but common biocrust-forming genera, such as Leptolyngbya, Oscillatoria Vaucher ex Gomont or Phormidium Kützing ex Gomont, and the cosmopolitan Microcoleus steenstrupii were reported. We believe that the Microcoleus found at our study site could have the same ecological function as other Microcoleus because it showed the typical bundles contributing to soil stabilization. The lack of heterocystous cyanobacteria in Maestre et al. (2006) is possibly related to the molecular approach employed by these authors, and/or the greater difficulty in extracting DNA of these cyanobacteria due to their thick mucilage (Patzelt et al., 2014), which can underestimate this part of the cyanobacterial community that commonly appears in cultures (Garcia-Pichel et al., 2001).
4.3. Ecological significance
Biocrusts at our study site comprised a community of cyanobacteria dominated by filamentous forms (heterocystous and non-heterocystous), which enhance the ecological role of biocrusts. For example, the bundles of filaments of Microcoleus, surrounded by a sticky gelatinous sheath, form a net-like structure that binds soil particles together (Mager and Thomas, 2011). In addition, the thick EPS layer can remain over many years after the death of trichomes, contributing to biostabilization of soils, protection against erosion by wind and water, and enhancement of soil moisture (Büdel et al., 2016). The cohesion of soil particles by bundles of filaments allows the colonization by heterocystous cyanobacteria, such as Nostoc and Scytonema, and by lichens and mosses in later successional stages (Weber et al., 2016). Scytonema is able to grow and survive in habitats exposed to strong irradiation by producing the UV-protector pigment scytonemin (Sinha and Häder, 2008). The accumulation of this pigment substantially reduces soil albedo, with immediate consequences for the soil microbiome, as this induces the replacement of thermosensitive bacterial species with more thermotolerant forms (Couradeau et al., 2016). Dinitrogen fixation is the most important process in the N cycle of biocrusts, and it has been estimated to be responsible for about 30% of biologically fixed N in terrestrial ecosystems globally (Yeager et al., 2012). Most of this activity is carried out by heterocystous cyanobacteria such as Mojavia, Roholtiella, Scytonema or Nostoc, which play an important role in ecosystem N cycling within dryland soils (Belnap, 2002).
4.4. Concluding remarks
Given the important ecological roles they play, and the strong links between species composition and diversity and ecosystem functioning in biocrust communities (Bowker et al., 2013; Maestre et al., 2012; Yeager et al., 2012), understanding the composition and diversity of cyanobacterial communities can provide valuable information to assess ecosystem functioning and development in biocrust-dominated landscapes. This is particularly important in understudied habitats, such as gypsum outcrops from Mediterranean regions. We found cyanobacteria belonging to genera such as Microcoleus, Trichocoleus, Scytonema, Nostoc, Roholtiella, Nodosilinea, Chroococcus, Spirirestris and Mojavia. We also report two new phylotypes of Microcoleus and Scytonema, two of the most important cyanobacterial genera found in dryland soils worldwide. Our findings contribute to our understanding of biocrust-associated cyanobacteria from gypsum habitats, which are biodiversity hotspots and have a great conservation value.
Acknowledgements
This work was supported by grants from the Ministerio de Economía y Competitividad, Spain (CGL-2013-44870-R and CGL2013-44661-R). FTM and CCD acknowledge support from the European Research Council (ERC Grant Agreement 647038 [BIODESERT]).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anagnostidis K, Komárek J. Süsswasserflora von Mitteleuropa: Cyanoprokaryota; Teil 1: Chroococcales, Süsswasserflora von Mitteleuropa. Fischer; Jena: 1999. [Google Scholar]
- Belnap J. Nitrogen fixation in biological soil crusts from southeast Utah, USA. Biol Fertil Soils. 2002;35:128–135. doi: 10.1007/s00374-002-0452-x. [DOI] [Google Scholar]
- Bohunická M, Pietrasiak N, Johansen JR, Gómez EB, Hauer T, Gaysina LA, Lukešová A. Roholtiella, gen. nov. (Nostocales, Cyanobacteria)—A tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa. 2015;197:84–103. doi: 10.11646/phytotaxa.197.2.2. [DOI] [Google Scholar]
- Bowker MA, Maestre FT, Mau RL. Diversity and patch-size distributions of biological soil crusts regulate dryland ecosystem multifunctionality. Ecosystems. 2013;16:923–933. doi: 10.1007/s10021-013-9644-5. [DOI] [Google Scholar]
- Büdel B, Dulić T, Darienko T, Rybalka N, Friedl T. Cyanobacteria and algae of Biological Soil Crusts. In: Weber B, Büdel B, Belnap J, editors. Biological Soil Crusts: An organizing principle in drylands. Springer International Publishing; Cham: 2016. pp. 55–80. [DOI] [Google Scholar]
- Castillo-Monroy AP, Maestre FT, Delgado-Baquerizo M, Gallardo A. Biological soil crusts modulate nitrogen availability in semi-arid ecosystems: Insights from a Mediterranean grassland. Plant Soil. 2010;333:21–34. doi: 10.1007/s11104-009-0276-7. [DOI] [Google Scholar]
- Couradeau E, Karaoz U, Lim HC, Nunes da Rocha U, Northen T, Brodie E, Garcia-Pichel F. Bacteria increase arid-land soil surface temperature through the production of sunscreens. Nat Commun. 2016;7 doi: 10.1038/ncomms10373. 10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dojani S, Kauff F, Weber B, Büdel B. Genotypic and phenotypic diversity of cyanobacteria in biological soil crusts of the Succulent Karoo and Nama Karoo of Southern Africa. Microb Ecol. 2014;67:286–301. doi: 10.1007/s00248-013-0301-5. [DOI] [PubMed] [Google Scholar]
- Elbert W, Weber B, Burrows S, Steinkamp J, Büdel B, Andreae MO, Pöschl U. Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci. 2012;5:459–462. doi: 10.1038/ngeo1486. [DOI] [Google Scholar]
- Escudero A, Palacio S, Maestre FT, Luzuriaga AL. Plant life on gypsum: A review of its multiple facets. Biol Rev. 2014;90:1–18. doi: 10.1111/brv.12092. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 1985:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F, López-Cortés A, Nübel U. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol. 2001;67:1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science. 2013;340:1574–1577. doi: 10.1126/science.1236404. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F, Wojciechowski MF. The evolution of a capacity to build supra-cellular ropes enabled filamentous cyanobacteria to colonize highly erodible substrates. PLoS One. 2009;4:4–9. doi: 10.1371/journal.pone.0007801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitler L. Cyanophyceae. Johnson; New York: 1932. Cyanophyceae; p. 1196. [Google Scholar]
- Gkelis S, Rajaniemi P, Vardaka E, Moustaka-Gouni M, Lanaras T, Sivonen K. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from Lake Kastoria, Greece form a separate phylogenetic group. Microb Ecol. 2005;49:176–182. doi: 10.1007/s00248-003-2030-7. [DOI] [PubMed] [Google Scholar]
- Gómez N, Charles Donato J, Giorgi A, Guasch H, Mateo P, Sabater S. Conceptos y técnicas en Ecol Fluv. 2009. La biota de los ríos: los microorganismos autótrofos; pp. 219–247. [DOI] [Google Scholar]
- Hagemann M, Henneberg M, Felde VJMNL, Drahorad SL, Berkowicz SM, Felix-Henningsen P, Kaplan A. Cyanobacterial diversity in biological soil crusts along a precipitation gradient, Northwest Negev Desert, Israel. Microb Ecol. 2014:219–230. doi: 10.1007/s00248-014-0533-z. [DOI] [PubMed] [Google Scholar]
- Hauer T, Bohunická M, Johansen JR, Mareš J, Berrendero-Gomez E. Reassessment of the cyanobacterial family Microchaetaceae and establishment of new families Tolypothrichaceae and Godleyaceae. J Phycol. 2014;50:1089–1100. doi: 10.1111/jpy.12241. [DOI] [PubMed] [Google Scholar]
- IUSS Working Group WRB. World reference base for soil resources 2014. International soil classification system for naming soils and creating legends for soil maps, World Soil Resources Reports No. 106. 2014 doi: 10.1017/S0014479706394902. [DOI] [Google Scholar]
- Komárek J. Süsswasserflora von Mitteleuropa: Cyanoprokaryota; Teil 3: Heterocytous genera, Süsswasserflora von Mitteleuropa. Springer; Berlin: 2013. [Google Scholar]
- Komárek J, Anagnostidis K. Süsswasserflora von Mitteleuropa: Cyanoprokaryota; Teil 2: Oscillatoriales, Süsswasserflora von Mitteleuropa. Elsevier; München: 2005. [Google Scholar]
- Komárek J, Santanna CL, Bohunická M, Mareš J, Hentschke GS, Rigonato J, Fiore MF. Phenotype diversity and phylogeny of selected Scytonema-species (Cyanoprokaryota) from SE Brazil. Fottea. 2013;13:173–200. doi: 10.5507/fot.2013.015. [DOI] [Google Scholar]
- Kumar D, Adhikary SP. Diversity, molecular phylogeny, and metabolic activity of cyanobacteria in biological soil crusts from Santiniketan (India) J Appl Phycol. 2015;27:339–349. doi: 10.1007/s10811-014-0328-0. [DOI] [Google Scholar]
- Loza V, Berrendero E, Perona E, Mateo P. Polyphasic characterization of benthic cyanobacterial diversity from biofilms of the Guadarrama river (Spain): morphological, molecular, and ecological approaches1. J Phycol. 2013;49:282–297. doi: 10.1111/jpy.12036. [DOI] [PubMed] [Google Scholar]
- Maestre FT, Bowker MA, Cantón Y, Castillo-Monroy AP, Cortina J, Escolar C, Escudero A, Lázaro R, Martínez I. Ecology and functional roles of biological soil crusts in semi-arid ecosystems of Spain. J Arid Environ. 2011;75:1282–1291. doi: 10.1016/j.jaridenv.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Castillo-Monroy AP, Bowker MA, Ochoa-Hueso R. Species richness effects on ecosystem multifunctionality depend on evenness, composition and spatial pattern. J Ecol. 2012;100:317–330. doi: 10.1111/j.1365-2745.2011.01918.x. [DOI] [Google Scholar]
- Maestre FT, Escolar C, de Guevara ML, Quero JL, Lázaro R, Delgado-Baquerizo M, Ochoa V, Berdugo M, Gozalo B, Gallardo A. Changes in biocrust cover drive carbon cycle responses to climate change in drylands. Glob Chang Biol. 2013;19:3835–3847. doi: 10.1111/gcb.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Martín N, Díez B, López-Poma R, Santos F, Luque I, Cortina J. Watering, fertilization, and slurry inoculation promote recovery of biological crust function in degraded soils. Microb Ecol. 2006;52:365–377. doi: 10.1007/s00248-006-9017-0. [DOI] [PubMed] [Google Scholar]
- Mager DM, Thomas AD. Extracellular polysaccharides from cyanobacterial soil crusts: A review of their role in dryland soil processes. J Arid Environ. 2011;75:91–97. doi: 10.1016/j.jaridenv.2010.10.001. [DOI] [Google Scholar]
- Martínez I, Escudero A, Maestre FT, De La Cruz A, Guerrero C, Rubio A. Small-scale patterns of abundance of mosses and lichens forming biological soil crusts in two semi-arid gypsum environments. Aust J Bot. 2006;54:339–348. doi: 10.1071/BT05078. [DOI] [Google Scholar]
- Mühlsteinová R, Johansen JR, Pietrasiak N, Martin MP. Polyphasic characterization of Kastovskya adunca gen. nov et comb. nov. (Cyanobacteria: Oscillatoriales), from desert soils of the Atacama Desert, Chile. Phytotaxa. 2014;163:216. doi: 10.11646/phytotaxa.163.4.2. [DOI] [Google Scholar]
- Neilan BA, Jacobs D, Del Dot T, Blackall LL, Hawkins PR, Cox PT, Goodman AE, Therese DD, Blackall LL, Hawkins PR, Cox PT, et al. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int J Syst Bacteriol. 1997;47:693–697. doi: 10.1099/00207713-47-3-693. [DOI] [PubMed] [Google Scholar]
- Patzelt DJ, Hodač L, Friedl T, Pietrasiak N, Johansen JR. Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. J Phycol. 2014;50:698–710. doi: 10.1111/jpy.12196. [DOI] [PubMed] [Google Scholar]
- Perkerson RB, III, Johansen JR, Kovácik L, Brand J, Kaštovský J, Casamatta DA. A unique Pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen.nov. based on morphological and molecular data. J Phycol. 2011;47:1397–1412. doi: 10.1111/j.1529-8817.2011.01077.x. [DOI] [PubMed] [Google Scholar]
- Řeháková K, Johansen JR, Casamatta DA, Xuesong L, Vincent J. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia. 2007;46:481–502. doi: 10.2216/06-92.1. [DOI] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sinha RP, Häder DP. UV-protectants in cyanobacteria. Plant Sci. 2008;174:278–289. doi: 10.1016/j.plantsci.2007.12.004. [DOI] [Google Scholar]
- Steven B, Gallegos-Graves LV, Belnap J, Kuske CR. Dryland soil microbial communities display spatial biogeographic patterns associated with soil depth and soil parent material. FEMS Microbiol Ecol. 2013;86:101–113. doi: 10.1111/1574-6941.12143. [DOI] [PubMed] [Google Scholar]
- Strunecký O, Komárek J, Johansen J, Lukešová A, Elster J. Molecular and morphological criteria for revision of the genus Microcoleus (Oscillatoriales, Cyanobacteria) J Phycol. 2013;49:1167–1180. doi: 10.1111/jpy.12128. [DOI] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Tamaru Y, Takani Y, Yoshida T, Sakamoto T. Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Appl Environ Microbiol. 2005;71:7327–7333. doi: 10.1128/AEM.71.11.7327-7333.2005. doi:10.1128/AEM.71.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomazeau S, Houdan-Fourmont A, Coute A, Duval C, Couloux A, Rousseau F, Bernard C. The contribution of Sub-Saharian African strains to the phylogeny of Cyanobacteria: Focusing on the Nostocaceae (Nostocales, Cyanobacteria) J Phycol. 2010;46:564–579. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Belnap J, Büdel B. Synthesis on biological soil crust research. In: Weber B, Büdel B, Belnap J, editors. Biological Soil Crusts: An organizing principle in drylands. Springer International Publishing; Cham: 2016. pp. 527–534. [DOI] [Google Scholar]
- Williams L, Loewen-Schneider K, Maier S, Büdel B. Cyanobacterial diversity of western European biological soil crusts along a latitudinal gradient. FEMS Microbiol Ecol. 2016;92:fiw157. doi: 10.1093/femsec/fiw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager CM, Kuske CR, Carney TD, Johnson SL, Ticknor LO, Belnap J. Response of biological soil crust diazotrophs to season, altered summer precipitation, and year-round increased temperature in an arid grassland of the Colorado Plateau, USA. Front Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]