Abstract
Background
Epigenetic disturbances are crucial in cancer initiation, potentially with pleiotropic effects, and may be influenced by the genetic background.
Methods
In a subsets (ASSET) meta-analytic approach, we investigated associations of genetic variants related to epigenetic mechanisms with risks of breast, lung, colorectal, ovarian and prostate carcinomas using 51,724 cases and 52,001 controls. False-discovery-rate corrected p-values (q-values < 0.05) were considered statistically significant.
Results
Among 162,887 imputed or genotyped variants in 555 candidate genes, SNPs in eight genes were associated with risk of more than one cancer type. For example, variants in BABAM1 were confirmed as a susceptibility locus for squamous cell lung, overall breast, ER-negative breast, overall prostate, overall and serous ovarian cancer; the most significant variant was rs4808076 (odds ratio (OR)=1.14, 95% confidence interval (CI)=1.10–1.19, q=6.87*10−5). DPF1 rs12611084 was inversely associated with ER-negative breast, endometrioid ovarian, overall and aggressive prostate cancer risk (OR=0.93, 95% CI=0.91–0.96, q=0.005). Variants in L3MBTL3 were associated with colorectal, overall breast, estrogen receptor (ER)-negative breast, clear cell ovarian, and overall and aggressive prostate cancer risk (e.g. rs9388766: OR=1.06, 95% CI=1.03–1.08, q= 0.02). Variants in TET2 were significantly associated with overall breast, overall prostate, overall ovarian and endometrioid ovarian cancer risk, rs62331150 showing bidirectional effects. Analyses of sub-pathways did not reveal gene subsets that contributed disproportionately to susceptibility.
Conclusion
Functional and correlative studies are now needed to elucidate the potential links between germline genotype, epigenetic function, and cancer etiology.
Impact
This approach provides novel insight into possible pleiotropic effects of genes involved in epigenetic processes.
Keywords: Cancer, Single Nucleotide Polymorphism, Pleiotropy, Meta-Analysis
INTRODUCTION
Genetic and epigenetic alterations are hallmarks of cancer initiation and progression and can influence each other to work cooperatively (1). Dysfunction of epigenetic processes, such as DNA methylation, chromatin remodeling and covalent histone modifications can be as important in carcinogenesis as the change of the genetic material itself (2). Since the first studies that described the global hypomethylation of cancer genomes and the hypermethylation of the promoter sequence of mainly tumor suppressor genes, several “pan-cancer” DNA methylation patterns (patterns across multiple cancer types) have been identified (reviewed in (3)). The CpG island methylator phenotype (CIMP) was first described in colorectal cancer (4) and later similar patterns were observed in several other tumor types. Highlighting the interplay between genetic and epigenetic changes, CIMP subtypes usually present with characteristic genetic alterations. CIMP-H colorectal cancers are frequently characterized by BRAF mutations, while CIMP-L tumors tend to harbor KRAS mutations (5). Non-CIMP colorectal cancer, the B-CIMP-negative breast cancer and the low methylated tumor group of serous ovarian cancers frequently acquire TP53 mutations (5–8).
Furthermore, somatic mutations in epigenetic regulatory genes that are either carcinogenic driver or passenger mutations are known to exist. Important mutations have been shown for example in DNMTs, IDH1, IDH2 and TETs (as important players of DNA methylation), in EZH2 and KDM1A (involved in histone modifications) and in ARID1A (participant of chromatin remodeling) (reviewed in (2)). In addition, inherited genetic variants related to epigenetic regulatory processes were described in association with multiple cancers (9, 10). Given the fundamentality of epigenetic processes, germline variants in genes related to epigenetic pathways presumably have pleiotropic effects on the initiation of different cancers.
As part of the U.S. National Cancer Institute’s Genetic Associations and Mechanisms in Oncology (GAME-ON) Network (http://epi.grants.cancer.gov/gameon/), we have previously shown the value of cross-cancer analyses in inflammation pathways (11).
An additional value is that our datasets include large numbers of cancer subtypes that were not studied in The Cancer Genome Atlas (TCGA). The present study was focused and approved by the GAME-ON consortium for the overall analyses of pleiotropy, where we aimed to identify cross-cancer associations of epigenetically related polymorphisms that advance our understanding of the role of epigenetics in cancer development. Given the central role of epigenetic processes in carcinogenesis, germline variants in genes related to epigenetic pathways show pleiotropic effects on the initiation of different cancers. Consequently, we investigated whether common polymorphisms in epigenetic genes are associated with risk of multiple cancer types (breast, colorectal, lung, ovarian and prostate cancer) and their subtypes.
METHODS
Study population
Within the GAME-ON Network, 32 studies from North America and Europe participated in this investigation (12–21). Studies included frequency matched cases and controls on at least age, and all subjects were of European descent based on ancestry analyses. The study characteristics are summarized in Table 1. In total, 51,724 cancer patients (breast, colorectal, lung, ovarian and prostate with respective subtypes) and 52,001 controls were included in the analysis.
Table 1.
Overview of cancer types and studies participating in the original meta-analyses
| Cancer site Studies |
Genotyping platforms | Subtype | Covariates | No. of Studies | N cases | N controls |
|---|---|---|---|---|---|---|
|
Colorectal MECC, CFRa, KY, ACS, Australia, NF |
Affymetrix Axiom | All | Age, sex, PCs | 6 | 5100 | 4831 |
|
| ||||||
|
Breast ABCFS, HEBCS, UK2, SASBAC, MARIE, CPSII, EPIC, MEC, NHS2, PBCS, PLCO |
Illumina arrays (317K-1.2M) | All | Age, PCs (vary by studies) | 11 | 15569 | 18204 |
| ESR1 (ER)-negative | Age, PCs (vary by studies) | 8 | 4760 | 13248 | ||
|
| ||||||
|
Lung UK, MDACC, IARC, NCI, SLRI, HGF |
Illumina arrays (317K-610K) | All | Age, sex, PCs | 6 | 12527 | 17285 |
| Adenocarcinoma | Age, sex, PCs | 6 | 3804 | 16289 | ||
| Squamous cell | Age, sex, PCs | 6 | 3546 | 16434 | ||
|
| ||||||
|
Ovary UKGWAS, USGWAS, U19GWAS |
Illumina arrays (317K-2.5M) | All | Study, PCs | 3 | 4368 | 9123 |
| Endometrioid | Study, PCs | 3 | 2553 | 9123 | ||
| Serous | Study, PCs | 3 | 715 | 9123 | ||
| Clear cell | Study, PCs | 3 | 355 | 9123 | ||
|
| ||||||
|
Prostate BPC3, CRUK1, CRUK2, CAPS |
Illumina and Affymetrix arrays | All | Age, study | 6 | 14160 | 12712 |
| Aggressive subtype | Age, study, PCs | 6 | 4446 | 12724 | ||
Colon-CFR: 1660 cases, 1393 controls
ER = estrogen receptor; PCs = principal components representing residual European ancestry.
Gene and variant selection, pathway assignment
Genes (n=634) involved in epigenetic processes were identified using GO and GeneCards databases by searching for the following keywords: DNA methylation, DNA demethylation, histone acetylation, deacetylation, methylation, demethylation, and other histone modification, chromatin remodeling, chromatin modification and histones. The recent literature was also reviewed. After excluding genes on sex chromosomes and those not covered in all cancer sites, 555 genes were included in the analysis, which were categorized into one or more of epigenetic sub-pathways (Supplementary Table S1).
We analyzed all single nucleotide polymorphisms (SNPs) residing within 50 kb of the largest transcript for each gene (Databases see in Supplementary Table S2). Overall, 162,887 polymorphisms were included in the final analysis. In the combined dataset, the major alleles (according dbSNP) were used as reference alleles.
Statistical analysis
Cancer sites were further divided into subtypes and for each cancer type and subtype, a fixed effect meta-analysis was conducted to combine results from individual studies (Table 1). This method used log-additive models adjusted for age, European principal components, and sex (where appropriate).
The beta values and standard errors for each cancer or cancer subtype were then combined using the association analysis based on a subsets (ASSET) meta-analytic approach, which allows for disease heterogeneity and potential opposite directions of the same genetic variant on different cancer types (22). It searches for the most parsimonious grouping based on the test statistics using any of the five cancers or cancer subtypes simultaneously as the outcome variables. Overlapping subjects amongst cancer subtypes (e.g. overlapping cases and controls between overall lung cancer and its subtypes) and across cancer types (e.g. UK ovary and UK breast GWAS both used controls from Welcome Trust Case Control Consortium, WTCCC) were accounted for in the covariance matrix when estimating the standard errors (11). The resulting p-values were adjusted using false-discovery rate (FDR) correction. Results with FDR q<0.05 were considered statistically significant (Supplementary Table S3). All association analyses were performed in R (3.2.5).
Functional annotation
The overall approach of the functional annotation is summarized in Supplementary Fig. S1. For each gene with more than five significant SNPs (FDR q<0.05 in the ASSET meta-analysis), we selected tagSNPs to represent these regions in subsequent analysis. Specifically, a linkage disequilibrium (LD) map was prepared using the Haploview 4.2 software and tagSNPs were identified with the tagger algorithm of Haploview using 1000Genomes data (release 20130502). Variants with more than two alleles based on 1000Genomes were excluded from LD mapping. As a result we were able to investigate SNPs that were not covered in the original meta-analysis but potentially have functional effect on the genes in the region of interest.
To assess if any of the epigenetic sub-pathways shown in Supplementary Table S1 were enriched with genes containing significant associations with cancer types or subtypes, pathway analyses were conducted using the ALIGATOR algorithm of the SNPath R package.
The possible functional annotation of the tagSNPs and the region-representative SNPs (functional follow-up (FFU) SNPs) were then assigned using the FunciSNP R/Bioconductor package (23). Using the package, we identified all the corresponding SNPs of our tagSNPs using 50 kb searching window and r2>=0.8 as a linkage threshold. In the next step, FunciSNP package checks if the corresponding SNPs or the tagSNPs show overlap with DNA segments with predicted functional importance. To annotate these biofeatures, we used the combined genome segmentation assessed by the ENCODE Project Consortium. These results represent ChIP-seq data for eight chromatin marks (H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac, H3K27me3, H3K36me3, H4K20me1), RNA Polymerase II and the CTCF transcription factor, as well as DNase-seq and FAIRE-seq data. This data is processed with ChromHMM and Segway software which segments the genome into seven disjoint segments based on their predicted functional role (24). Since the goal of the study was to identify those polymorphisms that change the function of the epigenetic related genes, we interpreted polymorphisms that overlap with a predicted transcribed region only, if they were in the gene of interest. We used the data available on Huvec, H1hesc and Gm12878 cell lines. Unfortunately, comprehensive information for the genome segmentation track was not available for all cell lines of the respective cancer types. We thus decided to use data from normal cell lines. Additionally, an ENCODE Uniform transcription factor binding site (TFBS) track was used, that encompasses data for 161 transcription factors from 91 cell types. Supplementary Table S4 summarizes the functional annotation of all SNPs based on FunciSNP (SNPs that were annotated as not functional are not listed). Furthermore, the functionality of the ASSET-identified SNPs as well as their corresponding SNPs were annotated using RegulomeDB, version 1.1.
All software packages and databases that were used are listed in Supplementary Table S2.
RESULTS
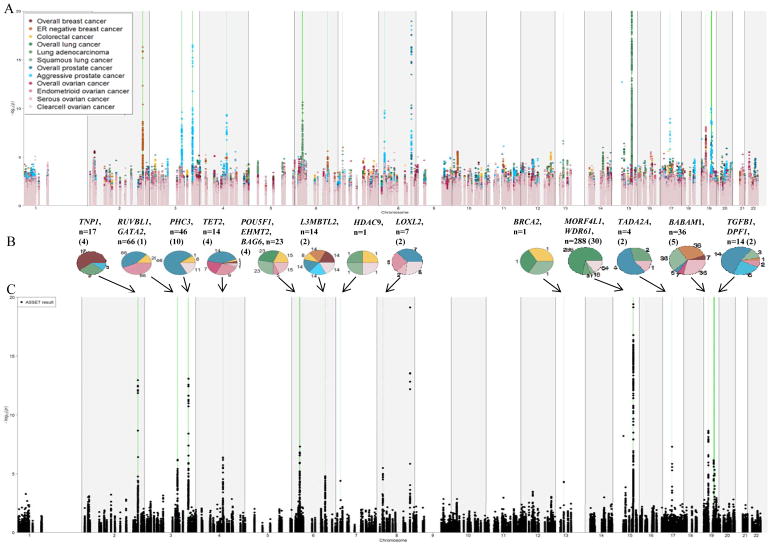
The results of the original (individual study based) meta-analyses and the ASSET-based risk associations are summarized in Figure 1. Ovarian cancer was associated with the largest number of variants (98), followed by prostate (70), lung (50), breast (46) and colorectal (10) cancers. Interestingly, all of the endometrioid ovarian cancer specific SNPs also showed an association with overall prostate cancer risk. These polymorphisms were mainly located in the RUVBL1 gene regions. Variants in the flanking region of MORF4L1 on 15q25 were mainly associated with lung and ovarian cancer. Due to the proximity of MORF4L1 to CHRNA5, CHRNA3 and CHRNB4 genes and their well-known association with lung cancer, we excluded this region from further analysis (25–27). The number of remaining SNPs that were associated with lung cancer risk was 35 and with ovarian cancer was 83. Furthermore, variants in PHC3 (3q26) were solely associated with risk of overall prostate cancer and will not further be discussed.
Figure 1.
Manhattan plot showing the original meta-analyses (A) and the results of the ASSET-based meta-analysis (C) on the selected SNPs available for all studies. Variants with –log10 (p values) higher than 20 are not shown. Regions showing significant pleiotropic association in the ASSET analysis are marked in green.
Pie charts (B) show the number of variants that were significant in the ASSET analysis. Numbers in brackets depict the number of independent risk loci. Each diagram represents a gene region and the numbers of SNPs associated with a specific cancer type (in the same colors as indicated in the Manhattan plot (A)) are shown. SNPs associated with multiple cancer types are counted in each of the respective cancer sections. Overlap is not visualized.
When combining genes into epigenetic sub-pathways (see above), we observed no significant risk association with more than one cancer type or subtype (p values>0.05) indicating that all pathways were similarly important for cancer risk.
Overall 99 SNPs in 8 genes (excluding MORF4L1: 84 SNPs in 7 genes) showed significant associations (FDR q<0.05) with risk of more than one cancer type (Supplementary Fig. S2. A and B). Genes with associated SNPs were: RUVBL1 (3q21), TET2 (4q24), L3MBTL3 (6q23), HDAC9 (7p21), BRCA2 (13q12), MORF4L1 (15q25), BABAM1 (19p13) and DPF1 (19q13) (Table 2, Supplementary Fig. S3). Previous GWAS-identified cancer risk associations in these and other genes located in these regions are listed in Supplementary Table S5.
Table 2.
Summary of gene regions significantly associated with more than one cancer.
| Candidate genes | Select. region | Nr. of SNPs/region | Nr. of associated SNPsa | Associated cancers | Strongest association | Previously in GWAS associated cancersb |
|---|---|---|---|---|---|---|
| RUVBL1 | 3: 127733628-127922757 | 346 | 27 (1) | Endometrioid ovarian cancer, overall prostate cancer, colorectal cancer | Endometrioid ovarian cancer, Overall prostate cancer: rs144609957 - OR: 1.13; 95% CI: 1.08–1.19; p: 3.44*10−7 | Prostate cancer |
| TET2 | 4: 106017842-106250960 | 573 | 9 (3) | Overall prostate cancer, overall ovarian cancer, endometrioid ovarian cancer, overall breast cancer, clear cell ovarian cancer, colorectal cancer | Overall prostate cancer, endometrioid ovarian cancer: rs6839705 - OR: 1.11; 95% CI: 1.07–1.16; p: 3.32*10−7 | Prostate cancer, breast cancer |
| L3MBTL3 | 6: 130289728-130512594 | 590 | 11 (2) | Colorectal cancer, overall breast cancer, ER-neg breast cancer, clear cell ovarian cancer, overall prostate cancer, aggressive prostate cancer | Colorectal cancer, overall breast cancer, ESR1 (ER)-negative breast cancer, clear cell ovarian cancer, overall prostate cancer, aggressive prostate cancer: rs9388766 - OR 1.06; 95% CI: 1.03–1.08; p: 1.07*10−6 | None |
| HDAC9 | 7: 18076572-18758466 | 1307 | 1 (1) | Lung adenocarcinoma, squamous cell lung cancer, colorectal cancer, clear cell ovarian cancer | Lung adenocarcinoma, squamous cell lung cancer, colorectal cancer, clear cell ovarian cancer: rs190505819 - OR: 1.88; 95% CI: 1.44–2.45; p: 2.95*10−6 | None |
| BRCA2 | 13: 32839617-33023809 | 196 | 1 (1) | Overall lung cancer, squamous cell lung cancer, colorectal cancer | Overall lung cancer, squamous cell lung cancer, colorectal cancer: rs56404467 - OR: 1.30; 95% CI: 1.15–1.48; p: 3.26*10−5 | Breast cancer, lung cancer |
| MORF4L1 | 15: 79115123-79240081 | 327 | 15 (2) | Overall lung cancer, lung adenocarcinoma, squamous cell lung cancer, clear cell ovarian cancer | Overall lung cancer, lung adenocarcinoma, clear cell ovarian cancer: rs7179953 - OR: 0.93; 95% CI: 0.91–0.96; p: 5.34*10−7 | Lung cancer |
| BABAM1 | 19: 17328232-17443811 | 404 | 33 (5) | Squamous cell lung cancer, ER-neg breast cancer, serous ovarian cancer, overall breast cancer, overall ovarian cancer, overall prostate cancer, | Squamous cell lung cancer, ESR1 (ER)-negative breast cancer, serous ovarian cancer: rs4808076 - OR: 1.14; 95% CI: 1.10–1.19; p: 1.77*10−10 | Breast cancer, ovarian cancer |
| DPF1 | 19: 38651649-38770317 | 284 | 2 (1) | Overall prostate cancer, ER-neg breast cancer, endometrioid ovarian cancer, aggressive prostate cancer | Overall prostate cancer, ER-neg breast cancer, endometrioid ovarian cancer, aggressive prostate cancer: rs12611084 - OR 0.93; 95% CI: 0.91–0.96; p: 8.40*10−8 | Prostate cancer |
Nr=Number; SNP=Single nucleotide polymorphism; GWAS=Genome-wide association study; OR=Odds ratio; CI=Confidence interval; ER=Estrogen receptor
Independent associations
associated with breast, colorectal, lung, ovarian or prostate cancer
The most pleiotropic genes were TET2, BABAM1, DPF1and especially L3MBTL3 (Figure 1, Table 2). Eleven variants in L3MBTL3 were associated with cancer risk, all with pleiotropic effects. The highest OR (odds ratio) in this region was 1.06 (rs9388766, 95% CI (confidence interval) =1.03–1.08, FDR q= 0.02), which was associated with risk of colorectal, overall breast, ESR1 (ER)-negative breast, clear cell ovarian, overall and aggressive prostate cancer. L3MBTL3 is a member of the putative Polycomb group (PcG) proteins. Two SNPs, rs9375694 and rs6569648, were previously identified as eQTLs (expression quantitative trait locus) for L3MBTL3 (RegulomeDB score: 1d and 1f, respectively) (28). The variant allele of rs6899976 may also be functionally important, since it overlaps with CTCF enriched regions in all cell lines as well as a transcription factor binding site. However, this variant has a RegulomeDB score of only 4.
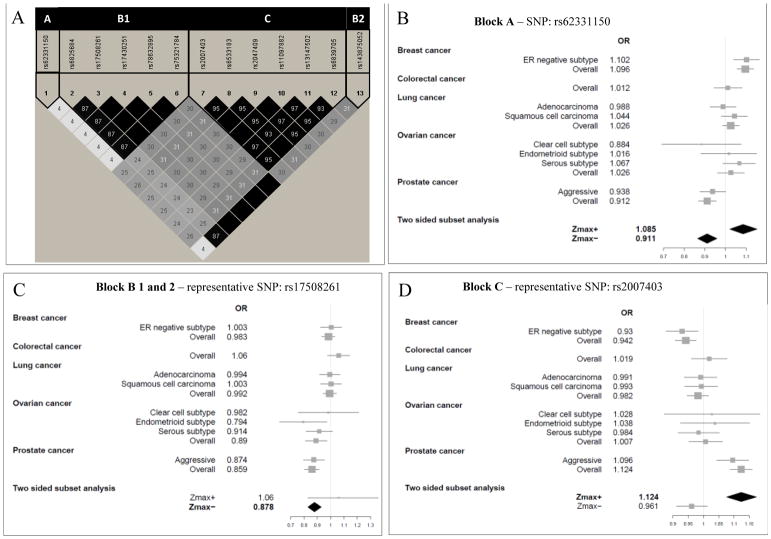
TET2 (tet methylcytosine dioxygenase 2) at 4q24 encodes a protein catalyzing the conversion of methylcytosine to 5-hydroxymethylcytosine. Nine variants at this locus were significantly associated with risk of at least two cancer types, of which one variant (rs6825684) was associated with decreased risk of four cancers or subtypes: colorectal, overall prostate, overall and endometrioid ovarian cancer (OR=0.89, 95% CI=0.85–0.93, FDR q=0.02) and one polymorphism (rs62331150) showed a bidirectional effect. The variant allele of rs62331150 increased the risk of overall breast and serous ovarian cancer (OR:1.09, 95% CI: 1.02–1.15, p-value=0.009) and decreased the risk of clear cell ovarian and prostate cancer (OR=0.91, 95% CI= 0.87–0.96, p-value=0.0004) with a combined q-value of 0.04 (Figure 2). Most of the variants were positioned within TET2. The non-synonymous rs34402524 was predicted to be deleterious (SIFT) and possibly damaging (PolyPhen). Among the ASSET identified and FFU SNPs, further functional annotation singled out polymorphisms with a possible functional role. Rs62331150, (RegulomeDB score =2b), overlaps with a transcription start site, a transcription factor binding site and an enhancer region.
Figure 2.
(A) Linkage disequilibrium (LD) plot encompassing the significant SNPs in the TET2 region. Selected SNPs representing each LD block with respective forest plots are shown for (B) rs62331150 representing the single-variant block A; (C) rs17508261representing block B1 and B2; and (D) rs2007403 representing block C.
33 variants, all pleiotropic, showed an association with cancer susceptibility in the region containing BABAM1, a known ovarian and breast cancer locus. The strongest association was observed for rs4808076, which conferred 14% increased risk of ESR1 (ER)-negative breast, serous ovarian and squamous cell lung cancer (OR=1.14, 95% CI=1.10–1.19, FDR q=6.87*10−5). Five variants decreased the risk of six cancer types and subtypes; overall prostate, overall breast, ESR1 (ER)-negative breast, squamous cell lung, overall and serous ovarian cancer risk (strongest signal for rs8100241: OR=0.95, 95% CI=0.93–0.97, FDR q=1.78*10−3). Besides BABAM1, the captured region (19p13) additionally contains ANKLE1, ABHD8 and USHBP (Supplementary Table S5). BABAM1 was selected for its involvement in chromatin modifications, namely ubiquitination as part of the BRCA1 A complex. The ASSET identified SNPs in this region were in LD with several variants that may play an important role in regulatory processes. The most important ones are shown in Table 3. Apart from the variants in regulatory regions, five SNPs were in coding sequences. Important features of these variants, as well as their SIFT (29) and PolyPhen (30) scores are shown in Table 4.
Table 3.
Variants in the region 19p13 with a putative functional effect using ENCODE combined genome segmentation assessed in Huvec, H1hesc and Gm12878 cell lines
| SNP ID | SNP position | RegulomeDB score | TFBSa | Promoter flanking | Enhancer | Weak enhancer | Transcription start site (TSS) | CTCF rich |
|---|---|---|---|---|---|---|---|---|
| rs10406920 | 17389648 | 3a | + | |||||
| rs113299211 | 17400765 | 5 | Gm12878 | |||||
| rs11540855 | 17403361 | 2a | + | Huvec, H1hesc, Gm12878 | ||||
| rs11667661 | 17390579 | 2b | ||||||
| rs11669059 | 17400453 | 2b | + | Huvec | ||||
| rs12982178 | 17371568 | 3a | H1hesc | Huvec | ||||
| rs2363956 | 17394124 | 5 | Huvec | |||||
| rs34084277 | 17387176 | 1f | ||||||
| rs35686037 | 17359535 | 2b | + | H1hesc | ||||
| rs4808616 | 17403033 | 2b | H1hesc | Gm12878 | ||||
| rs55924783 | 17404072 | 5 | H1hesc, Gm12878 | |||||
| rs56069439 | 17393925 | 4 | + | H1hesc | H1hesc | |||
| rs66753001 | 17394839 | 5 | H1hesc | |||||
| rs73509996 | 17393449 | 4 | + | Huvec | Huvec, H1hesc | |||
| rs8100241 | 17392894 | 4 | + | Huvec, H1hesc | Gm12878 | |||
| rs8108174 | 17393530 | 2b | + | Huvec, H1hesc, Gm12878 | ||||
| rs8170 | 17389704 | 4 | + |
Indicated with + when overlapping with the ENCODE Uniform transcription factor binding site (TFBS) track
Table 4.
Pleiotropic polymorphisms in the BABAM1 region that are located in exons and their predicted effect on the proteins
| SSNP | Position | Gene | Nucleotide change | Amino acid change | EUR MAF | SIFT | PolyPhen-2 |
|---|---|---|---|---|---|---|---|
| rs8170 | 19:17389636 | BABAM1 | AAG ⇒ AAA | K [Lys] ⇒ K [Lys] | A:0.16 | n.a. | n.a. |
| rs10425939 | 19:17389155 | ANKLE1 | GGC ⇒ GGT | G [Gly] ⇒ G [Gly] | T:0.16 | n.a. | n.a. |
| rs10425939 | 19:17389155 | ANKLE1 | GCT ⇒ GTT | A [Ala] ⇒ V [Val] | T:0.16 | n.a. | n.a. |
| rs8100241 | 19:17392893 | ANKLE1 | GCG ⇒ ACG | A [Ala] ⇒ T [Thr] | A:0.58 | deleterious(0) | probably_damaging (0.998) |
| rs8108174 | 19:17393529 | ANKLE1 | CTG ⇒ CAG | L [Leu] ⇒ Q [Gln] | A:0.58 | deleterious(0) | probably_damaging (1) |
| rs2363956 | 19:17394123 | ANKLE1 | TTG ⇒ TGG | L [Leu] ⇒ W [Trp] | G:0.57 | deleterious(0.03) | probably_damaging (0.999) |
SNP= Single nucleotide polymorphism; EUR MAF= Minor allele frequency in European population
DPF1 is part of the neuron-specific chromatin remodeling complex (nBAF complex). One variant (rs12611084) was significantly associated with endometrioid ovarian, ESR1 (ER)-negative breast, overall and aggressive prostate cancer risk (OR=0.93, 95% CI=0.91–0.96, FDR q=0.005) and one variant (rs8100395) additionally with lung adenocarcinoma (OR=0.93, 95% CI=0.90–0.96, FDR q=7.2*10−3). Both variants were located upstream of DPF1, some were overlapping with other genes in this region, PPP1R14A and SPINT2, and were captured by one tagSNP in the FunciSNP analysis. Seven FFU SNPs showed a possible functional role, among them rs7250689, which was previously reported to be an eQTL for PPP1R14A (28). Based on RegulomeDB, rs8100395 and rs12611084 (both significant in the ASSET analysis) likely affect binding, and additionally overlap with enhancer regions as well as transcription factor binding sites and, in the case of rs8100395, overlaps with a CTCF enriched region.
Overall, 27 polymorphisms in RUVBL1 were associated with risk of prostate and endometrioid ovarian cancer, while one SNP was additionally associated with colorectal cancer risk. The strongest association was observed for rs144609957 with increased risk of prostate and endometrioid ovarian cancer (OR=1.13, 95% CI=1.08–1.19, FDR q=0.01). None of the SNPs had reached genome-wide significance in the original meta-analysis. RUVBL1 plays a role in chromatin organization. All associated SNPs belonged to the same LD block and were captured by one tagging SNP. Further, FunciSNP analysis revealed seven variants that overlapped with multiple biofeatures (transcription factor binding site, weak enhancer region and promoter flanking region). These variants also had low RegulomeDB scores, the lowest being 2b for rs9879865 and rs9879866, variants that likely affect binding.
DISCUSSION
We performed the first large-scale association study of variants in epigenetic-related genes and cancer risk utilizing the extensive genomic data on 51,724 cancer patients and 52,001 controls and identified eight epigenetic-related genes with pleiotropic effects on cancer risk.
Epigenetic disturbances are common drivers of carcinogenesis, yet, effects of germline variants and their potential pleiotropic mechanisms are not well understood. Thus, we investigated the risk association of SNPs related to epigenetic processes with multiple cancers. Using a subset-based meta-analysis, we were able to account for different subsets of cancer types and subtypes even with contrasting risk associations.
The L3MBTL3 gene on 6q26 is a member of the putative Polycomb group (PcG). It contains a methyl-lysine reader Malignant Brain Tumor (MBT) domain that is responsible for the recognition of the mono- and di-methylated lysines of H3 and H4 histone tails. MBT domain proteins are associated with gene expression repression and their dysregulation has been shown to contribute to different diseases (31). In our analysis, two variants (rs9375694 and rs6569648), which were previously identified as eQTLs, were significantly associated with risk of prostate and breast cancer (and their subtypes), and to a lesser extent with risk of clear cell ovarian and colorectal cancer (28). Interestingly, previous GWAS identified an association of rs6569648 and rs6899976, both hits in our analysis, with height (32) and height is associated with risk of several cancers including breast, ovarian, prostate and colorectal cancer (33). Our findings suggest the link between height and cancer risk may be vis-a-vis altered epigenetic processes, but this requires further investigations.
Several SNPs located in and around TET2 showed significant associations with risk of overall prostate, overall ovarian, endometrioid ovarian, overall breast and colorectal cancer. Previous studies reported significant associations of variants at the TET2 locus with risk of cancer including ovarian and breast cancer (9, 21, 34). A large number of functional variants were identified in this region forming multiple pleiotropic linkage blocks that support the role of TET2 and its germline variants in the development of multiple cancer types. Furthermore, an association between rs62331150 and TET2 gene expression in breast normal and tumor tissue was recently shown (9). The bidirectional association of the rs62331150 variant allele implies that the effect of TET2 genetic variation may be of a different nature for distinct cancers, increasing the risk of breast cancer, but decreasing the risk of prostate cancer. Similar associations were observed for a group of highly linked polymorphisms, namely rs2007403, rs2047409, rs6533183, rs6839705, rs11097882 and rs13147502 confirming previous studies (21, 35); however, with only one statistically significant risk direction.
Several functional variants were found at 19p13 with significant associations observed for risk of ESR1 (ER)-negative breast cancer, serous ovarian cancer and squamous cell lung cancer, but also with overall ovarian, breast and prostate cancer. BABAM1 is involved in chromatin modifications (ubiquitination), as part of the BRCA1 complex and regulates the retention of BRCA1 at double strand DNA breaks to maintain stability of this complex at the sites of DNA damage (36). Previous GWAS associated this region with breast (37) and ovarian cancer (10), with some of the SNPs showing triple-negative breast cancer specificity (38). However, to our knowledge we are the first to describe an association with squamous cell lung cancer or overall prostate cancer risk. Demonstrating a limitation of our selective candidate gene approach, new evidence suggests that nearby 19p13 genes ANKLE1 and/or ABHD8, rather than BABAM1, may be the functional drivers in breast and ovarian cancer [Lawrenson et al, Nature Communications in press]. The complexity of this region requires detailed functional follow up to disentangle the combined effect of individual variants and to understand their role in carcinogenesis.
DPF1 is part of the mSWI/SNF (also called BAF) chromatin remodeling complex with a central role in carcinogenesis (39). Mutations in DPF1 were seen in solid tumors (7). Furthermore, significant overexpression of DPF1 was observed in breast and squamous cell lung cancers (40). Our results also support a pleiotropic effect of DFP1 during carcinogenesis through potentially functional polymorphisms in this gene. However, as in each region of interest, we cannot exclude the potential relevance of the other genes in this region (PPP1R14A, SPINT2).
Polymorphisms in 3q21 were previously only observed in association with prostate cancer risk (41); however, our analysis has detected additional associations with endometrioid ovarian and colorectal cancer risk. RUVBL1 is a member of the INO80 family protein remodeling complex. It interacts with MYC and CTNNB1 (β-catenin), participates in many signal transduction pathways and is overexpressed in many cancer types (42). We have identified several polymorphisms with seemingly strong functional impacts. Interestingly, a proportion of endometrioid ovarian and colorectal cancers arise from common etiologies associated with hereditary non-polyposis cancer (HNPCC) or Lynch syndrome (43) and also show de novo promoter methylation silencing of DNA mismatch repair genes (44) and altered β-catenin signaling (45). RUVBL1 may represent novel susceptibility genes that further unify endometrioid ovarian and colorectal cancer development.
The major strength of this study is the large sample size of more than 100,000 subjects across five cancer types and their subtypes, some of which were not studied in TCGA. In addition, by searching the most parsimonious grouping based on the test statistics using any of the five cancers or cancer subtypes simultaneously as the outcome variables, the ASSET-subset-based meta-analysis (1) increased the power to detect associations, which may not have been detected in the individual analyses of the five cancer types, (2) allowed estimation of associations with opposing effects, and (3) provided new insights into pleiotropy that were not observed in the original analyses (22). Further, the overlapping subjects (cases and controls) are accounted for during the analysis (11). Finally, our focused approached reduced the genome-wide multiple testing burden and allowed for examination of functionally grouped subsets of epigenetic-related genes (i.e., sub-pathways). We were thus able to confirm established and identify new risk genes, including TET2 and L3MBTL3.
Although the odds ratios that are discovered as pleiotropic across cancer types may be considered modest, there is potential clinical significance. First, the ORs for individual cancers may be higher than the summary OR. Second, the combination of several SNPs with low ORs may become relevant through creation of a risk score, and third, the association of SNPs with disease may be modified and, in some instances, strengthened by environmental factors.
While our approach provides interesting insights into the pleiotropic effects of selected regions, it is limited with respect to the assignment of the identified predisposing variants to genes by chromosomal position rather than the actual cancer-initiating processes. Of note, several of the identified pleiotropic associations cannot clearly be linked to the selected epigenetic genes, as some of the regions additionally contain genes that were previously described for their effect on carcinogenesis.
Further investigations are required to elucidate the functional link between the identified pleiotropic variants and their impact on epigenetic processes such as the potential effect of TET2 polymorphisms on DNA methylation. Indeed, our pathway-based selection of epigenetic-related genes overlooked the subtleties of complex gene networks, and most genes are involved in multiple biological processes. Finally, this dataset did not allow for the investigation of interactions with other genetic or environmental factors, which are undoubtedly of great importance.
In summary, using a unique, large dataset, we identified novel pleiotropic variants in epigenetic-related genes that are associated with susceptibility to multiple cancer types and subtypes. This study provides the basis for future studies investigating the impact of these variants, their causal relationship to epigenetic processes, and the mechanisms leading to carcinogenic pleiotropy.
Supplementary Material
Acknowledgments
Financial support:
This work was supported by TRICL (Transdisciplinary Research for Cancer of Lung) and International Lung Cancer Consortium (ILCCO): National Institute of Health U19 CA148127-01 (PI: Amos), Canadian Cancer Society Research Institute (no. 020214, PI: Hung). DRIVE (Discovery, Biology, and Risk of Inherited Variants in Breast Cancer): National Institute of Health U19 CA148065. CORECT (ColoRectal Transdisciplinary Study): National Institute of Health U19 CA148107; R01 CA81488, P30 CA014089. ELLIPSE (ELLIPSE, Elucidating Loci in Prostate Cancer Susceptibility): This work was support by the GAME-ON U19 initiative for prostate cancer (ELLIPSE), U19 CA148537. CRUK GWAS: This work was supported by the Canadian Institutes of Health Research, European Commission’s Seventh Framework Programme grant agreement n∘ 223175 (HEALTH-F2-2009-223175), Cancer Research UK Grants C5047/A7357, C1287/A10118, C5047/A3354, C5047/A10692, C16913/A6135, and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. The Prostate Cancer Program of Cancer Council Victoria also acknowledge grant support from The National Health and Medical Research Council, Australia (126402, 209057, 251533, 396414, 450104, 504700, 504702, 504715, 623204, 940394, 614296,), VicHealth, Cancer Council Victoria, The Prostate Cancer Foundation of Australia, The Whitten Foundation, PricewaterhouseCoopers, and Tattersall’s. EAO, DMK, and EMK acknowledge the Intramural Program of the National Human Genome Research Institute for their support. CAPS GWAS study was supported by the Swedish Cancer Foundation (grant no 09-0677, 11-484, 12-823), the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linneus Centre (Contract ID 70867902) financed by the Swedish Research Council, Swedish Research Council (grant no K2010-70X-20430-04-3, 2014-2269). The BPC3 was supported by the U.S. National Institutes of Health, National Cancer Institute (cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to S.M.G., U01-CA98216 to E.R., and U01-CA98758 to B.E.H., and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics).
FOCI (Transdisciplinary Cancer Genetic Association and Interacting Studies): National Institutes of Health U19 CA148112- 01 (PI: T.A. Sellers), R01-CA122443, P50-CA136393, P30-CA15083 (PI: E.L. Goode), Cancer Research UK (C490/A8339, C490/A16561, C490/A10119, C490/A10124 [PI: P. Pharoah]). NHS by the National Institutes of Health (P01 CA087969, UM1 CA186107, R01 CA151993, R35 CA197735 and P50 CA127003). The Colon CFR data collection was supported by grant UM1 CA167551 and the Colon CFR Illumina GWAS was supported by grants U01 CA122839 and R01 CA143237 to G. Casey from the National Cancer Institute, National Institutes of Health. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CCFR. PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. Additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Prostate Cancer GWAS (Yeager M, et al. Nat Genet. 2007;39(5):645–649), Colon CGEMS pancreatic cancer scan (PanScan) (Amundadottir L, et al. Nat Genet. 2009;41(9):986–990 and Petersen GM, et al. Nat Genet. 2010;42(3):224–228), and the Lung Cancer and Smoking study. The prostate and PanScan study datasets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207.v1.p1 and phs000206.v3.p2, respectively, and the lung datasets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093.v2.p2. Funding for the Lung Cancer and Smoking study was provided by National Institutes of Health (NIH), Genes, Environment, and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446, and NIH GEI U01 HG-004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping. PMH: National Institutes of Health (R01 CA076366 to P.A. Newcomb).
NHS: We would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping for NHS under the supervision of Dr. Immaculata De Vivo and Dr. David Hunter, Qin (Carolyn) Guo and Lixue Zhu who assisted in programming for NHS. We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. PLCO: The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff or the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc., and Drs. Bill Kopp, Wen Shao, and staff, SAIC Frederick. Most importantly, we acknowledge the study participants for their contributions to making this study possible. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI. DRIVE: The DRIVE GAME-ON consortium (http://epi.grants.cancer.gov/gameon/) would like to thank the following key investigators (institution and location): Muriel Adank (VU University Medical Center, Amsterdam, the Netherlands), Habibul Ahsan (University of Chicago, Chicago, IL), Irene Andrulis (Mount Sinai Hospital, Toronto, Canada), Kristiina Aittomäki (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Lars Beckman (Institute for Quality and Efficieny in Health Care, Cologne, Germany), Carl Blomquist (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Federico Canzian (German Cancer Research Center, Heidelberg, Germany), Jenny Chang-Claude (Deutsches Krebsforschungszentrum [DKFZ], Heidelberg, Germany), Laura Crisponi (Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche, Cagliari, Italy), Kamila Czene (Karolinska Institut, Stockholm, Sweden), Norbert Dahmen (University of Mainz, Mainz, Germany), Isabel dos Santos Silva (London School of Hygiene and Tropical Medicine, London, UK), Olivia Fletcher (The Institute of Cancer Research, London, UK), Lorna Gibson (London School of Hygiene and Tropical Medicine, London, UK), Per Hall (Karolinska Institut, Stockholm, Sweden), HEBON (Hereditary Breast and Ovarian Cancer Research Group Netherlands), Rebecca Hein (Deutsches Krebsforschungszentrum, Heidelberg, Germany, and University of Cologne, Cologne, Germany), Albert Hofman (Erasmus Medical Center, Rotterdam, the Netherlands), John L. Hopper (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Astrid Irwanto (Genome Institute of Singapore, Singapore), Rudolf Kaaks (German Cancer Research Center, Heidelberg, Germany), Muhammad G. Kibriya (University of Chicago, Chicago, IL), Peter Lichtner (German Research Center for Environmental Health, Neuherberg, Germany), Jianjun Liu (Genome Institute of Singapore, Singapore), Enes Makalic (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Alfons Meindl (Technische Universität München, Munich, Germany), Hanne Meijers-Heijboer (VU University Medical Center, Amsterdam, the Netherlands), Bertram Müller-Myhsok (Max Planck Institute of Psychiatry, Munich, Germany), Taru A. Muranen (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Heli Nevanlinna (Univesity of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Julian Peto (London School of Hygiene and Tropical Medicine, London, UK), Ross L. Prentice (Fred Hutchinson Cancer Research Center, Seattle, WA), Nazneen Rahman (Institute of Cancer Research, Sutton, UK), Daniel F. Schmidt (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Rita K. Schmutzler (University of Cologne, Cologne, Germany), Melissa C. Southey (The University of Melbourne, Melbourne, Victoria, Australia), Clare Turnbull (Institute of Cancer Research, Sutton, UK), Andre G. Uitterlinden (Erasmus Medical Center, Rotterdam, the Netherlands), Rob B. van der Luijt (University Medical Center Utrecht, Utrecht, the Netherlands), Quinten Waisfisz (VU University Medical Center, Amsterdam, the Netherlands), Alice S. Whittemore (Stanford University, Stanford, CA), and Wei Zheng (Vanderbilt University, Nashville, TN). FOCI: FOCI consortium would like to thank the following investigators for their contribution: Mike Birrer, Ann Chen, Julie Cunningham, Ed Iversen, John McLaughlin, Steven Narod, Harvey Risch, Jenny Permuth-Wey, Paul Pharoah, Simon Gayther, and Susan Ramus. UK ovarian cancer GWAS: We thank all the individuals who took part in this study. We thank all the researchers, clinicians, and administrative staffs who have enabled the many studies contributing to this work. This study made use of data generated by the Wellcome Trust Case Control consortium with its project funding provided by the Wellcome Trust under award 076113. We thank the support of the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge and University College Hospital.
Footnotes
Rosalind A. Eeles1,2, Zsofia Kote-Jarai1, Kenneth Muir3,4, Ali Amin Ala Olama5,6, Sara Benlloch5,1, David E Neal7,8, Jenny L Donovan9, Freddie C Hamdy10, Graham G Giles11,12, Fredrik Wiklund13, Henrik Gronberg13, Christopher A Haiman14, Fredrick Schumacher15, David J Hunter17, Peter Kraft18, Ruth C Travis19, Elio Riboli20, Stephen J Chanock21, Sonja I Berndt22, Demetrius Albanes21
The Institute of Cancer Research
Royal Marsden NHS Foundation Trust,
Institute of Population Health, University of Manchester
Warwick Medical School, University of Warwick
Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge
University of Cambridge, Department of Clinical Neurosciences
University of Cambridge, Department of Oncology,
Cancer Research UK Cambridge Research Institute, Li Ka Shing Centre
School of Social and Community Medicine, University of Bristol,
Nuffield Department of Surgical Sciences, University of Oxford, Oxford, UK, Faculty of Medical Science, University of Oxford,
Cancer Epidemiology Centre, The Cancer Council Victoria
Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne
Department of Medical Epidemiology and Biostatistics, Karolinska Institute
Department of Preventive Medicine, Keck School of Medicine, University of Southern California/Norris Comprehensive Cancer Center
Case Western Reserve University, School of Medicine
Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard School of Public Health
Program in Molecular and Genetic Epidemiology, Department of Epidemiology, Harvard School of Public Health
Cancer Epidemiology, Nuffield Department of Population Health University of Oxford
Department of Epidemiology and Biostatistics, School of Public Health
Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH
Epidemiology and Genetics, National Cancer Institute, NIH
Conflict of interest:
The authors disclose no potential conflicts of interest
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–80. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 3.Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome Med. 2014;6:66. doi: 10.1186/s13073-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–82. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X, Long J, Zeng C, Michailidou K, Ghoussaini M, Bolla MK, et al. Fine-Scale Mapping of the 4q24 Locus Identifies Two Independent Loci Associated with Breast Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2015;24:1680–91. doi: 10.1158/1055-9965.EPI-15-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung RJ, Ulrich CM, Goode EL, Brhane Y, Muir K, Chan AT, et al. Cross Cancer Genomic Investigation of Inflammation Pathway for Five Common Cancers: Lung, Ovary, Prostate, Breast, and Colorectal Cancer. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amin Al Olama A, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22:408–15. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–8. 8e1–2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nature genetics. 2010;42:874–9. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Human genetics. 2012;131:217–34. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature genetics. 2013;45:362–70. 70e1–2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Human Molecular Genetics. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nature genetics. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Human Molecular Genetics. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. 61e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. American Journal of Human Genetics. 2012;90:821–35. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012;40:e139. doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–41. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 27.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2010;21:221–30. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiren S, Haggstrom C, Ulmer H, Manjer J, Bjorge T, Nagel G, et al. Pooled cohort study on height and risk of cancer and cancer death. Cancer Causes Control. 2014;25:151–9. doi: 10.1007/s10552-013-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song F, Amos CI, Lee JE, Lian CG, Fang S, Liu H, et al. Identification of a melanoma susceptibility locus and somatic mutation in TET2. Carcinogenesis. 2014;35:2097–101. doi: 10.1093/carcin/bgu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–28. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnad F, Doll S, Manning G, Arnott D, Zhang Z. Bioinformatics analysis of thousands of TCGA tumors to determine the involvement of epigenetic regulators in human cancer. BMC Genomics. 2015;16(Suppl 8):S5. doi: 10.1186/1471-2164-16-S8-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenbaum J, Baek SH, Dutta A, Houry WA, Huber O, Hupp TR, et al. The emergence of the conserved AAA+ ATPases Pontin and Reptin on the signaling landscape. Sci Signal. 2013;6:mr1. doi: 10.1126/scisignal.2003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch HT, Casey MJ, Snyder CL, Bewtra C, Lynch JF, Butts M, et al. Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management. Mol Oncol. 2009;3:97–137. doi: 10.1016/j.molonc.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Albarracin CT, Chang KH, Thompson-Lanza JA, Zheng W, Gershenson DM, et al. Microsatellite instability and expression of hMLH1 and hMSH2 proteins in ovarian endometrioid cancer. Mod Pathol. 2004;17:75–80. doi: 10.1038/modpathol.3800017. [DOI] [PubMed] [Google Scholar]
- 45.Wu R, Zhai Y, Fearon ER, Cho KR. Diverse mechanisms of beta-catenin deregulation in ovarian endometrioid adenocarcinomas. Cancer Res. 2001;61:8247–55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.