Abstract
Many enveloped RNA viruses recruit host cell proteins during assembly as a mechanism to limit antiviral effects of complement. Using viruses which incorporated CD46 alone, CD55 alone or both CD46 and CD55, we addressed the role of these two host cell regulators in limiting complement-mediated neutralization of Parainfluenza virus 5 (PIV5). PIV5 incorporated functional forms of both CD55 and CD46 into virions. PIV5 containing CD55 was highly resistant to Complement-mediated neutralization, whereas CD46-containing PIV5 was as sensitive to neutralization as virus lacking both regulators. PIV5 infected cells had increased levels of cell surface CD55, which was further upregulated by exogenous treatment with tumor necrosis factor alpha. PIV5 derived from cells with higher CD55 levels was more resistant to complement-mediated neutralization in vitro than virus from control cells. We propose a role for virus induction of host cell complement inhibitors in defining virus growth and tissue tropism.
Keywords: complement, parainfluenza virus
Introduction
The complement system constitutes an integral part of host defense that most animal viruses must face during natural infections. Complement consists of a complex group of both soluble and cell-associated proteins that together can serve to link innate and adaptive immunity through a large number of activities, including recognition of viruses, direct neutralization of infectivity, recruitment and stimulation of leukocytes, opsonization by immune cells, and activation of T and B cell responses (Blue et al, 2004; Carroll, 2004; Kemper and Polack, 2007). Complement activation and the ability of viruses to counteract complement can play important roles in viral pathogenesis (e.g., Mehlhop et al, 2006; Morrison et al, 2007; Delgado and Polack 2004; Stoermer and Morrison, 2001), but can also be harnessed to improve the effectiveness of vaccines and therapeutic vectors (Johnson et al., 2011; Bergmann-Leitner, 2006; Schauber-Plewa et al., 2005). The overall goal of the work described here was to determine the relative role of two important host regulators of complement in limiting neutralization of Parainfluenza virus 5 (PIV5).
Complement is activated by virus structures that can trigger the classical, lectin or alternative pathways (Markiewski and Lambis, 2007; Roozendaal and Carroll, 2006). For example, we have shown that a single point mutation in the PIV5 Fusion protein (F) can alter the virion structure such that complement activation shifts from the alternative to the classical pathway (Johnson et al, 2013). Once activated, these pathways converge on a central component C3, which is cleaved into C3a and C3b. C3a serves as an anaphylatoxin to promote inflammation. C3b can bind covalently to viral components to aid in opsonization and phagocytosis. In addition, C3b can associate with other factors such as cleavage products from Factor B to form the C3 convertase (e.g. C3bBb), and this functions to amplify the initially deposited C3b by further cleavage of C3 molecules in a feedback loop (Kerr, 1980). The association of C3b with further downstream components such as C5 through C9 can lead to formation of the membrane attack complex (MAC) which is capable of lysing virus particles or infected cells. Thus, C3 is a key molecule for complement activities.
The complement system is highly regulated in order to prevent inappropriate damage to normal cells and healthy tissues (e.g., Atkinson et al., 2005; Pangburn et al., 1983). This regulation involves a series of host cell proteins called regulators of complement activation (RCA), many of which target the C3 convertase (Liszewski et al., 2008; Kim and Song. 2006). Two key membrane-bound RCA proteins include CD46 and CD55, both of which act to inhibit formation and stability of the C3 convertase through two major mechanisms. The cofactor CD46 is a glycosylated integral membrane RCA protein expressed on a wide range of tissues and cell types (Liszewski et al., 1991; Russell et al., 1992). CD46 combines with the protease Factor I to mediate inactivation of C3b into iC3b, rendering it incapable of functioning as a convertase. CD55 (or decay accelerating factor, DAF) is membrane-associated through a glycosyl-phosphatidylinositol (GPI) linkage (Lublin and Atkinson, 1989). The mechanism by which CD55 blocks propagation of the complement pathway is distinct from that of CD46, since CD55 lacks cofactor activity (Pangburn et al, 1983; Medof et al, 1984). Instead, CD55 functions to disrupt complement pathways by rapidly promoting the decay of previously formed convertases.
Many large DNA viruses encode analogs which directly inhibit complement pathways, or act as mimics of host cell RCAs (reviewed in Ahmad et al, 2007; Bernet et al., 2003; Lambris et al., 2008). By contrast, the small coding capacity of most RNA virus genomes is thought to preclude dedication of viral proteins solely to complement inhibition. Thus, most RNA viruses can associate with soluble or membrane-bound host cell RCAs as a mechanism to limit complement neutralization (reviewed in Cummings et al., 2007; Stoermer and Morrison, 2001). Examples include human immunodeficiency virus type 1 (HIV 1) which incorporates CD55, CD59 and CD46 into progeny virions (Saifuddin et al., 1997), hepatitis C virus which assembles with CD55 (Mazumdar et al., 2013) and the paramyxoviruses which utilize CD55 and CD46 (Biswas et al., 2014; Johnson et al., 2009).
For a number of negative strand RNA viruses, both CD46 and CD55 have been shown to be incorporated into the membrane of progeny virions, where they function to inhibit complement pathways. This includes studies with PIV5, Mumps Virus (MuV), Vesicular Stomatitis Virus (VSV), and Newcastle Disease Virus (Johnson et al., 2009; 2012; Biswas et al., 2014). For MuV and VSV, we have previously shown that both of these RCAs can contribute to complement inhibition, but virion-associated CD55 is much more potent than CD46 in delaying complement-mediated neutralization in vitro (Johnson et al., 2012). In the present study, we demonstrate for the prototype paramyxovirus PIV5 that virion-associated CD55 plays a much more potent role in complement evasion than does CD46. PIV5-associated CD46 showed relatively low in vitro activity, and this was consistent with low level incorporation of this cofactor into PIV5 virions.
Given the critical role of virion-associated CD55 in the delay of neutralization of many negative strand viruses, we tested the hypothesis that virus derived from cells expressing higher levels of CD55 would be more resistant to neutralization. Here we show that infection with PIV5 as well as other RNA viruses results in increased levels of CD55 on the cell surface. PIV5 derived from cells with higher cell surface CD55 was more resistant to neutralization by normal human serum (NHS) in vitro. We propose a model for the role of virus induction of host cell RCAs in defining virus growth and tissue tropism.
MATERIALS AND METHODS
Cells and Viruses
CV-1, A549 and MRC-9 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat inactivated fetal calf serum (Hyclone, Logan, UT). Chinese hamster ovary (CHO) cells that overexpress the CYT2 isoform of human CD46 (CHO-CD46) and the control drug-resistant CHO-K1 cells were kindly provided by Dr. Denis Gerlier (Gerlier et al., 1994). CHO-CD55 and CHO-CD46/55 cells were generated as described previously (Johnson et al, 2012). All CHO cells were maintained in DMEM supplemented with 4.5 g/l glucose, 10 mM HEPES pH 7.2, 10 ug/ml Gentamycin, L-glutamine, 1% non essential amino acid and 6% heat inactivated FBS.
PIV5 expressing GFP and the PIV5 leader mutant Le-(U5C, A14G) have been described previously (He et al., 1997; Manuse and Parks, 2009). MuV, Respiratory Syncytial Virus (RSV) were grown in CV-1 and Hep2 cells, respectively. Zika virus MR766 strain (ATCC) and La Crosse virus (kindly provided by Dr. Pekosz, Johns Hopkins) were grown and titered in Vero cells. PIV5 and VSV were purified by sucrose gradient centrifugation as detailed previously (Johnson et al., 2012). For neutralization assays, 100 PFU of PIV5 was treated at 37°C for varying times or concentrations of normal human serum (NHS) or heat inactivated (HI) NHS. After incubation, remaining infectivity was determined by plaque assays. Reported results were the average of three to six reactions, with the significance of data points calculated using the student’s t-test.
Complement reagents, Factor I cofactor activity and decay acceleration activity assays
NHS was collected from healthy donors, processed and divided into small aliquots before freezing at −80°C as published previously (Johnson et al., 2008). Purified complement proteins were purchased from Complement Technologies (Tyler, TX). Recombinant CD55 (rCD55) and rCD46 were from R&D science (Minneapolis, MN) and Sino Biologicals (China), respectively. Sheep and rabbit red blood cells (RBCs) were from Lampire Biologicals (Pipersville, PA).
Factor I-mediated cleavage of C3b was analyzed as described earlier (Johnson et al, 2012) using purified virus as a source of cofactor activity. Briefly, various concentrations of purified PIV5 or VSV was incubated with 3 ug of C3b and 100 ng of Factor I for different time periods at 37°C in a total volume of 20 ul of PBS. Reactions were stopped by adding sample buffer containing SDS and beta-mercaptoethanol and boiled. Samples were analyzed on 9% SDS-PAGE gels, and protein bands were visualized with Gelcode Blue Stain reagent (Thermo Scientific, IL).
Alternative pathway C3 convertase activity associated with purified virions was measured by forming the enzyme C3bBb on rabbit red blood cells (ER) as detailed earlier (Johnson et al, 2012). Classical pathway C3 convertase (C4b2a) activity was measured on antibody sensitized sheep erythrocytes as described previously (Johnson et al, 2012). Convertase activity is expressed as percent lysis by absorbance at 405 nm normalized against 100% lysis in assay without inhibitor
Flow Cytometric Analysis
Virus-infected or mock infected cells were trypsinized, suspended in DMEM supplemented with 10% FBS and washed once with PBS. Cell surface CD55 was detected by indirect immunofluorescence staining using mouse anti-human CD55 primary antibody at a 1:100 dilution (EMD Millipore) and Alexa Fluor 633-conjugated goat anti-mouse secondary antibody at a 1:500 dilution (Invitrogen). Flow cytometric analysis was performed using the FACS Canto (BD Biosciences) recording at least 10,000 independent events. Results were analyzed using Flowlogic software.
Reverse Transcription and Real Time PCR
Total RNA was purified from 105 cells using TRIzol (Invitrogen) and 1 ug of RNA was used to generate cDNA using the TaqMan® Reverse Transcription Reagents (Applied Biosystems) as described by the manufacturer. Quantitative real-time PCR was carried out using Fast SYBR® FAST Green Master Mix (Applied Biosystems) and Bio-Red CFX Connect Real-Time. Primers used include: CD55 (Hs00892618_m1; ThermoFisher Scientific), β-actin forward 5′-GATCATTGCTCCTCCTGAGC-3′, and β-actin reverse 5′-ACTCCTGCTTGCTGATCCAC-3′. Relative CD55 replication was analyzed using the web-based service (RT2 Profiler PCR array analysis, SABiosciences) as described previously (Li et al., 2013).
Western blotting
Cell lysates or gradient purified virions were analyzed for levels of CD46 and CD55 by SDS-PAGE followed by Western blot analysis using antibodies against either CD46 or CD55 (Santa Cruz Biotechnology) at a dilution of 1:500. Polyclonal rabbit serum for the PIV5 P protein was used to normalize for virion amounts. Blots were visualized by horseradish peroxidase-conjugated antibodies and enhanced chemiluminescence substrate (Pierce Chemicals).
Electron microscopy
Electron microscopy (EM) analysis of sucrose gradient-purified PIV5 was carried as previously described (Johnson et al, 2012). Briefly, viruses were placed on carbon-coated 200-mesh gold grids (EM Sciences, PA) and incubated in a humidified chamber for 5 min before blocking with a solution of 1% BSA in PBS. Samples were probed with a mouse monoclonal anti-CD46 antibody (1ug/10ul; R&D) or mouse anti-CD55, (1ug/10ul; Millipore). Samples were then incubated with 10 ul of 1:10 dilution of anti-mouse 6 nm gold-labeled antibody (for anti-CD46) and/or anti-mouse 12 nm gold labeled antibody for (anti-CD55; Jackson Immuno Research, CA). Samples were negatively stained with 2% phospho-tungstic acid and analyzed with a Technai transmission electron microscope.
RESULTS
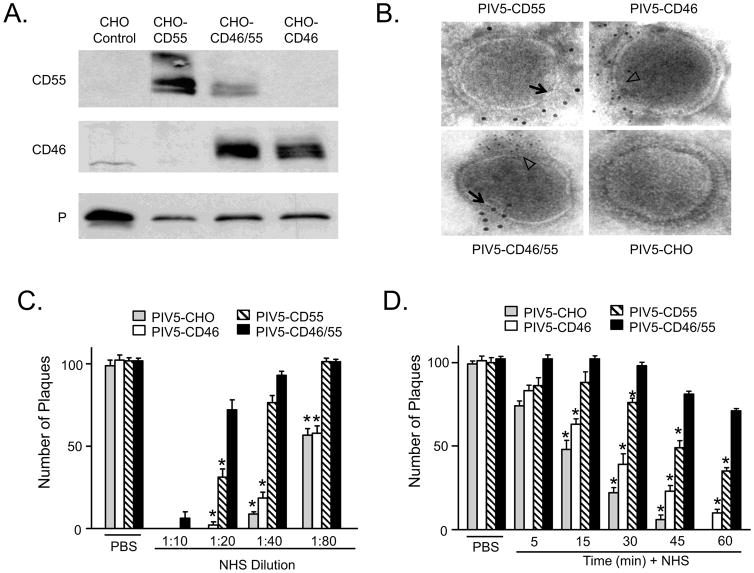
PIV5 harboring CD55 is more resistant to in vitro neutralization by NHS than virus harboring CD46 To generate PIV5 containing CD46 and/or CD55, virus was grown in control CHO-K1 cells which lack regulators or CHO cells expressing CD55 alone, CD46 alone or both regulators (CHO-CD46/55). Virus was purified by gradient centrifugation and analyzed by western blotting. As shown in Fig. 1A, virus derived from CHO-CD46 and CHO-CD46/55 cells contained approximately the same levels of CD46. Virus derived from CHO-CD55 cells contained slightly more CD55 (0.1 ug/ug virus) than that found for virus from the CHO-CD46/55 cells (0.03 ug/ug virus). Levels of CD46 in viruses from CHO-CD46 cells (0.03 ug/ug virus) or from the CHO-CD46/55 cells (0.02 ug/ug virus) were very similar. To confirm association with virions, PIV5 particles grown in the different CHO cell lines were probed with antibodies to CD46 or CD55 and gold-labeled secondary antibody before visualizing by electron microscopy (EM). As shown in Fig. 1B, virus particles had clear immunostaining for CD55 or CD46 that was clustered along one face of the virus particle (arrows and white arrowheads). In the case of virus grown in the CD46/CD55 cells, staining for these two regulators clustered on opposite faces of the virions (Fig. 1B), as described previously for our work with VSV and MuV (Johnson et al., 2012).
Figure 1.
PIV5 containing CD55 is more resistant to in vitro complement-mediated neutralization than virus containing CD46 alone. A) Western blotting. PIV5 derived from the indicated cell lines was purified by gradient centrifugation and 10 ug was analyzed by western blotting for the presence of CD46, CD55 or P protein as a loading control. B) EM analysis. Purified virus derived from CHO cells expressing CD46, CD55 or both CD46 and CD55 was treated with the indicated antibodies, followed by 6 nm (CD46) or 12 nm (CD55) colloidal gold goat anti-mouse antibody. Samples were analyzed by EM at a magnification of 55,000X. Black arrows and white arrow heads indicate the location of staining for CD55 and CD46, respectively. C and D) One hundred PFU of PIV5 derived from indicated cell lines was incubated with PBS as a control, with the indicated dilutions of NHS for one h (panel C), or with a 1:20 dilution of NHS for the indicated time points (panel D). Remaining infectivity was determined by plaque assay. Data are from six independent reactions and error bars represent standard deviation. *, p<0.001 when comparing CHO-derived, or CD46-containing virus to PIV5 containing both CD46 and CD55.
To determine the effect of virion-associated CD46 and CD55 on complement-mediated neutralization, a dilutions of NHS were incubated with one hundred PFU of PIV5 containing these regulators alone or in combination. Remaining infectivity was determined by plaque assay. As shown in Fig 1C, the contribution of virion-associated CD46 alone to a delay in in vitro neutralization of PIV5 was minimal. This is evident by the finding that at all NHS dilutions tested PIV5 containing CD46 alone (white bars) was neutralized to nearly the same extent as seen with the control virus lacking regulators (gray bars). By contrast, virus with CD55 alone (hatched bars) was more resistant than virus with CD46 alone. The effect of CD55 (hatched bars) alone was very similar to CD46 plus CD55 (black bars) at dilutions of NHS which were greater than at least 1:40.
Timecourses of neutralization were carried out to determine the role of CD55 and CD46 in the kinetics of neutralization by NHS. As shown in Fig 1D, PIV5 from control CHO cells lacking regulators was neutralized very rapidly (gray bars), with almost no infectivity remaining by 45 min. PIV5 containing CD46 alone (white bars) was only slightly more resistant to neutralization than control virus. By contrast, PIV5 with CD55 alone (hatched bars) was much more resistant to neutralization (e.g., 30 min timepoint). PIV5 containing both CD46 and CD55 (black bars) showed the slowest rate of neutralization, with slightly more resistance to neutralization than PIV5 with CD55 alone. These data indicate that PIV5-associated CD55 has a much greater impact on delaying neutralization than does CD46.
In vitro functional analysis of PIV5-associated CD55 activity
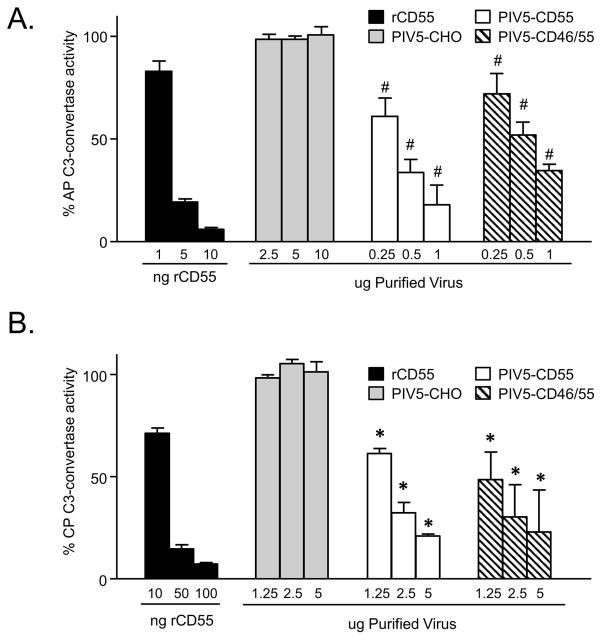
To determine the DAF activity associated with virions containing CD55, rabbit RBCs were used to assemble the C3bBb convertase from purified C3, Factor B and Factor D as described in Materials and Methods. When exposed to complement, these convertase-containing cells are lysed due to formation of the MAC (Mullick et al, 2005). Virion-associated DAF activity was determined as a function of the ability of CD55 to inhibit convertase activity and thus prevent cell lysis which was set at 100% for control samples lacking CD55.
As shown in Fig 2A, purified recombinant rCD55 had potent dose-dependent DAF activity against the alternative pathway (AP) convertase, with ~75% of the C3 convertase activity inhibited using 5 ng of recombinant protein (black bars). As expected, PIV5 derived from control CHO-K1 cells showed little DAF activity (gray bars), and the convertase-containing RBCs were efficiently lysed by complement (100% convertase activity). PIV5 derived from either the CHO-CD55 cells (white bars) or the CHO-CD46/55 cells (hatched bars) showed a significant dose-dependent inhibition of C3 convertase activity, but this was less potent than that seen with soluble rCD55. For example, one ug of PIV5 from CHO-CD55 cells had DAF activity that was close to that seen with 5 ng of rCD55 protein.
Figure 2.
Alternative and classical pathway DAF activity associated with CD55-containing PIV5 particles. Purified PIV5 derived from the indicated cells was assayed for decay accelerating activity against the alternative pathway (AP, panel A) or classical pathway (CP, panel B) C3-convertase as detailed in Materials and Methods. C3-mediated hemolytic activity is expressed as a percentage of the lysed control sample set at 100%. Data are from triplicate samples and error bars represent standard deviation. For panels A and B: #, p<0.001 and *, p<0.01 when comparing results of each concentration of PIV5-CD55 and PIV5-CD46/55 with PIV5-CHO control virus.
Virus particles were also tested for DAF activity against the classical pathway (CP) C3 convertase. Here, the C4bC2a complex was assembled on antibody-sensitized sheep RBCs by sequential addition of purified C1, C2, and C4, and then these RBCs were used in assays with purified PIV5 particles as a source of CD55. Inhibition of C3 convertase activity was assayed by decreases in hemolysis. As shown in Fig. 2B, higher levels of rCD55 were required to achieve 50% inhibition of the CP convertase (10 to 50 ng, panel B) compared to the AP convertase (1–10 ng, panel A), consistent with the known specificity of CD55 (Mullick et al, 2005). Most importantly, PIV5 derived from CHO-CD55 and CHO-CD46/55 cells showed strong convertase activity (50% at ~1 ug). These data indicate that PIV5-associated CD55 has strong activity against both the CP and AP convertases.
PIV5-associated CD46 has low cofactor activity against C3b
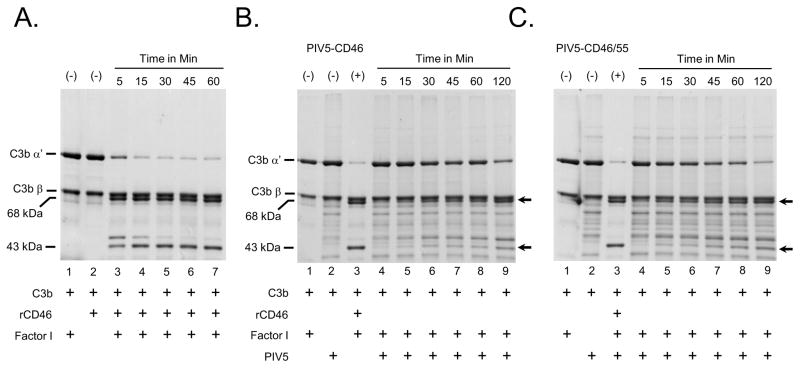
The above finding that CD46 contributed relatively little to blocking PIV5 neutralization raised the hypothesis that virion-associated CD46 had low functional activity. To test the potency of virion-associated CD46, Factor I-mediated cleavage of C3b into iC3b was reconstituted in vitro using purified virions and purified components as described previously (Johnson et al, 2012). In this assay, the disappearance of the C3b α′ chain and appearance of the 68 kDa and 43 kDa iC3b cleavage products was monitored by SDS-PAGE and coomassie blue staining. As shown in Fig. 3A for the positive control soluble recombinant CD46 (rCD46), the C3b α′ chain was rapidly cleaved in as little as 5 min incubation with Factor I protease. By contrast, when PIV5 containing CD46 alone (Fig. 3B) or CD46 plus CD55 (panel C) was used with Factor I, there was only very slow cleavage of the α′ chain. For example, by 45 min of incubation less than half of the C3b α′ chain had been cleaved and little of the 43 kDa product was detected. These data indicate that PIV5-associated CD46 has relatively weak cofactor activity against C3b compared to recombinant CD46. While there appear to be differences in activity between recombinant CD46 and that found in virions, this could reflect differences in soluble versus membrane-tethered native form of the protein or reduced activity when incorporated into the virus particle.
Figure 3.
PIV5 containing CD46 has low cofactor activity. A) Factor I-mediated C3b cleavage with rCD46. Twenty ng of rCD46 was incubated for the indicated times with purified C3b and Factor I as detailed in Materials and Methods. The cleavage of the α′ chain of C3b was monitored by SDS-PAGE and coomassie blue staining. Arrows indicate the positions of the α′ and β chains of C3b, and the 68 and 43/46 kDa cleavage products. Lanes 1 and 2 are samples showing negative controls lacking rCD46 or Factor I, respectively. B and C) Five ug of purified PIV5-CD46 or PIV5-CD46/55 was incubated for the indicated times with purified C3b and Factor I and analyzed by SDS-PAGE as detailed for panel A. (+) lanes are positive controls of C3b incubated with Factor I and rCD46 to demonstrate near complete cleavage of the α ′ chain into the 68 and 43/46 kDa cleavage products.
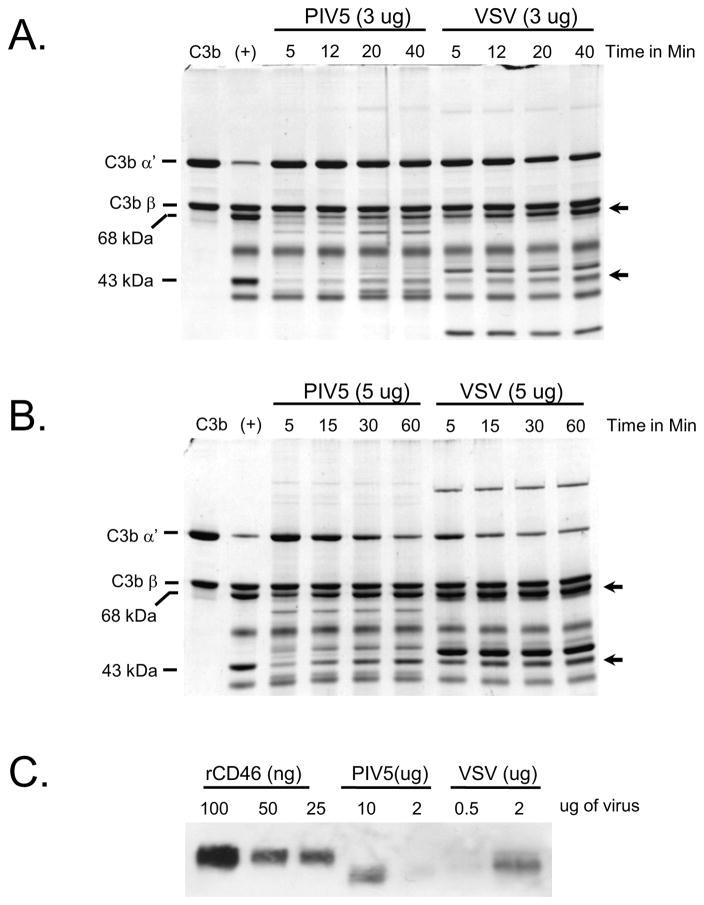
Low CD46 activity could be due to low incorporation into virion particles. CD46 cofactor activity was compared using CHO-CD46 derived PIV5 or VSV, the latter of which we have shown to have relatively high CD46 cofactor activity (Johnson et al, 2012). As shown in Fig. 4, timecourses of C3b cleavage with 3 ug (panel A) or 5 ug (panel B) of purified viruses showed more rapid Factor I-mediated cleavage with VSV compared to PIV5 (e.g., compare 5 and 15 min appearance of 43 kDa cleavage product in Fig. 4B). Western blotting (Fig. 4C) to quantitate virion-associated CD46 showed that VSV contained ~5–10 times more CD46 than PIV5 (e.g., compare 2 ug VSV and 10 ug PIV5). Thus, low cofactor activity of PIV5-associated CD46 and low impact of virion-associated CD46 on neutralization can be explained by relatively low incorporation of this cofactor into PIV5 virions.
Figure 4.
Comparison of virion-associated CD46 cofactor activity from purified PIV5-CD46 versus VSV-CD46. A and B) C3b cleavage assays were reconstituted in vitro using purified C3b, Factor I and either 3 ug (panel A) or 5 ug (panel B) of purified PIV5 and VSV derived from CD46-expressing CHO cells. After the indicated times at 37°C, samples were analyzed by SDS-PAGE and coomassie blue staining. The positions of the α′ and β chains of C3b are indicated, and arrows show the position of the 68 and 43 kDa cleavage products. C) Levels of PIV5- and VSV-associated CD46 from purified virus preparations were analyzed by western blotting along with the indicated amounts of purified rCD46.
Upregulation of cell surface CD55 confers greater resistance of virions to complement-mediated neutralization
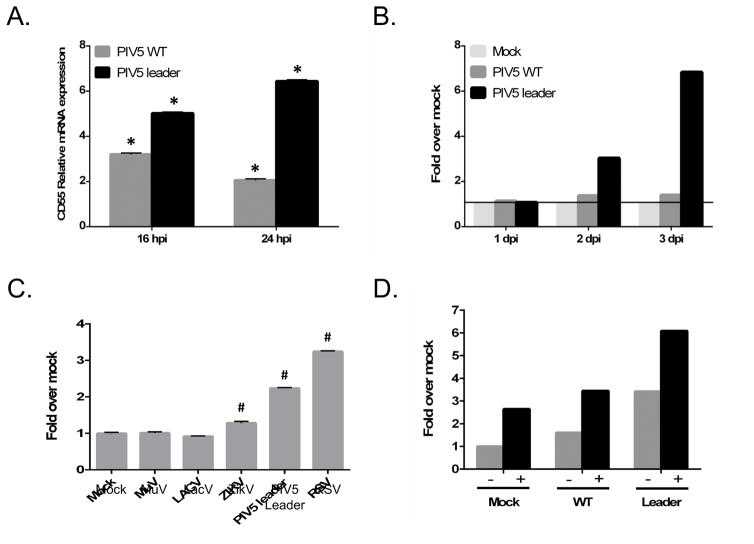
Our result that CD55 was important in evading complement led to the hypothesis that virus infection would increase CD55 surface expression and lead to virions that are more resistant to neutralization. To test this hypothesis, levels of CD55 were measured in infected normal human lung fibroblast MRC-9 cells. For these studies, cells were infected with WT PIV5 and with PIV5 Le-(U5C,A14G), a mutant with two point mutations in the leader promoter which we have shown previously to induce a strong host cell antiviral response despite the presence of a functional V protein (Manuse and Parks, 2009). As shown in Fig. 5A, cells infected with both WT and the Leader mutant PIV5 increased the expression of CD55 mRNA relative to mock infected cells. As shown in Fig. 5B, this increase in mRNA was also reflected in an increase in cell surface CD55 as detected by flow cytometric analysis, with the most dramatic increase being seen in the case of cells infected with the Le-(U5C, A14G) mutant. Cell surface CD55 expression was also tested in A549 lung epithelial cells infected with a range of RNA viruses (Fig. 5C). In this case, CD55 cell surface expression was increased by infection with respiratory syncytial virus (RSV), the Le-(U5C, A14G) mutant, and Zika virus (Zikv), but not by La Crosse virus (LacV) or MuV.
Figure 5.
Cell surface CD55 upregulation following infection with various RNA viruses. A and B) MRC-5 cells were mock infected or infected at an moi of 10 with WT PIV5 or the Le-(U5C, A14G) mutant. At the indicated hpi, either total RNA was isolated for RT-PCR quantitation of relative levels of CD55 mRNA (panel A) or MFI levels of cell surface CD55 were quantitated by flow cytometry relative to mock infected cells (panel B). C) A549 cells were mock infected or infected at an moi of 10 with the indicated viruses, and cell surface CD55 was quantitated by flow cytometry relative to mock infected cells at 2 days pi. Data are from triplicate samples and error bars represent standard deviation. D) TNF-α upregulation of cell surface CD55. A549 cells were mock infected or infected at an moi of 10 with WT PIV5 or the Le-(U5C, A14G) mutant. Cells were left untreated (−) or treated with 100 ng/ml TNF-α (+), and cell surface CD55 was determined by flow cytometry at 2 days pi. Results are an example of three separate experiments. For panels A and C: *, p<0.01 and #, p<0.001
CD55 cell surface expression has been shown to be induced by some proinflammatory cytokines such as TNF-α (Ahmad et al., 2003). As shown in Fig. 5D, TNF-α treatment of mock infected and PIV5 infected A549 cells gave a substantial increase in cell surface CD55. The largest increase was seen with TNF-α treatment of cells infected with the Le-(U5C, A14G) mutant.
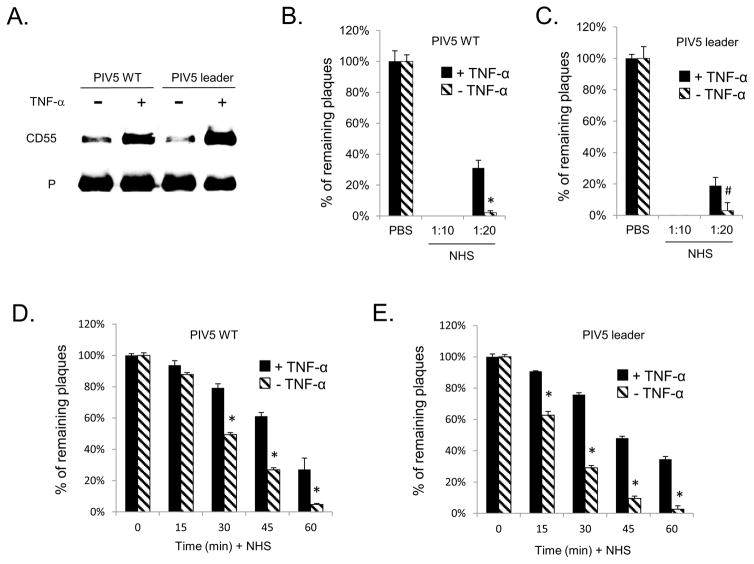
WT and Leader mutant PIV5 was grown in cells that had been treated with or without TNF-α. Virus was purified and analyzed for levels of CD55 and viral P protein as a load control. As shown in Fig. 6A, WT and Leader mutant PIV5 from TNF-α treated cells contained on average 2.3 and 3.3 fold more CD55 compared to virus from untreated control cells, respectively. We tested the hypothesis that virus derived from cells with increased CD55 surface expression would be more resistant to complement-mediated neutralization. WT PIV5 or the Le-(U5C, A14G) mutant were grown in control untreated A549 cells or in TNF-α treated cells. Neutralization assays were then carried out on the resulting progeny virus using varying concentrations of NHS and times of incubation. As shown in prior work (Johnson et al, 2011; 2013), WT PIV5 and the Le-(U5C, A14G) mutant derived from TNF-α treated A549 cells was sensitive to neutralization by NHS but not by heat inactivated NHS (data not shown), indicating a high dependence on intact complement pathways. As shown in Fig. 6, both WT PIV5 (panel B) and the Le-(U5C, A14G) mutant (panel C) derived from TNF-α-treated cells showed significantly more resistance to neutralization than virus derived from control untreated cells. This increased resistance was more evident in experiments that varied the time of incubation with NHS. As shown in Fig. 6, both WT PIV5 (panel D) and the Le-(U5C, A14G) mutant (panel E) virus that was derived from TNF-α treated cells showed significant delays in neutralization when compared to virus from untreated control cells (e.g., see 30 and 45 min timepoint). Together, these data support the contention that paramyxovirus infection can induce the upregulation of CD55 expression at the cell surface, resulting in progeny virions with enhanced ability to evade complement-mediated neutralization.
Figure 6.
Viruses from cells with upregulated CD55 expression have higher levels of CD55 and higher resistance to complement-mediated neutralization. A) WT PIV5 and Le-(U5C, A14G) mutant were grown in A549 cells treated with (+) or without TNF-α treatment. Purified virus was analyzed by western blotting for levels of CD55 and viral P protein. Panels B–D) WT PIV5 (panel B and D) or the Le-(U5C, A14G) mutant (panel C and E) were grown in untreated (−, cross hatched bars) or TNF-α treated (+, black bars) A549 cells. One hundred PFU of each virus was then incubated with PBS as a control, with the indicated dilutions of NHS for one h (panels A and B) or with a 1:30 dilution of NHS for the indicated time points (panels C and D). Remaining infectivity was determined by plaque assay. Data are from six independent reactions and error bars represent standard deviation. *, p<0.001; *, and #, p<0.01 when comparing results with TNF-α treated cells to control cells.
DISCUSSION
Using a nonhuman primate model, we have previously shown that the neutralizing potency of antibodies elicited during a primary respiratory tract infection with PIV5 is highly dependent on the presence of complement (Mayer et al., 2014). In the vast majority of cases, in vitro neutralization of PIV5 by NHS is dependent on active complement pathways (Johnson et al., 2012; 2013). Likewise, there are examples of antibodies elicited by virus infection that are highly dependent on complement for specific effector functions (e.g. Vogt et al, 2011; Mehlhop and Diamond, 2009). These examples highlight the importance of understanding mechanisms employed by RNA viruses to block complement pathways, since evasion of this innate immune response not only affects virus neutralization but also the potency of adaptive immune responses.
The overall goals of this work were to: 1) determine the extent to which PIV5-associated CD46 and CD55 contribute individually or in combination to complement evasion and 2) test the hypothesis that increased expression of a cell surface RCA would confer decreased sensitivity of progeny virions to complement-mediated neutralization. Our results show that CD55 is a much more powerful inhibitor of complement-mediated neutralization of PIV5 than CD46. We found that cells infected with WT and mutant PIV5, as well as RSV, displayed upregulated CD55 cell surface expression which was further enhanced by treatment with TNF-α. Finally, cells with upregulated CD55 produced progeny PIV5 with enhanced capacity to evade complement neutralization. Together, these data support the contention that paramyxoviruses induce an increase in cell surface expression of key RCAs as a mechanism to evade complement neutralization.
Why does viron-associated CD46 contribute little to the resistance of PIV5 to complement inactivation? Our data indicate that the levels of CD46 incorporation into PIV5 particles is low relative to the related virus VSV (~5–10 fold lower) and this correlates with lower cofactor activity against C3b and a lower contribution of CD46 to complement resistance. Thus, a simple explanation is that limitations on CD46 incorporation into budding PIV5 can account for a low contribution of this RCA to Complement inactivation. Other factors such as steric hindrance, interactions with the PIV5 glycoproteins HN and F may also contribute to low cofactor activity. It is noteworthy that PIV5-associated CD46 had an increased electrophoretic mobility consistent with removal of sialic acid residues due to HN neuraminidase activity. However, treatment of recombinant CD46 with sialidase did not alter in vitro activity against C3b (data not shown), suggesting that viral neuraminidase does not alter CD46 activity.
The signals and mechanisms that control incorporation or exclusion of host cell proteins into the envelope of budding negative strand RNA viruses are incompletely understood. Evidence supports a model whereby membrane microdomains containing either viral or host cell membrane proteins cluster along with internal proteins such as matrix protein and the nucleocapsid at the site of virion assembly (reviewed in Lyles, 2013). Thus, one possibility is that the membrane microdomains which drive PIV5 assembly and budding may not include high levels of CD46. In this regard, we were able to show by immune-gold microscopy that CD55 and CD46 map to opposing faces of the PIV5, MuV and VSV virions (Johnson et al., 2012), consistent with their incorporation into virions by partitioning into distinct membrane microdomains.
Similar to other negative strand viruses (e.g., VSV, MuV, NDV), we have shown that CD55 is a very important contributor to PIV5 evasion of complement neutralization. The exact mechanism whereby virion-associated CD55 is more important than CD46 for complement evasion is not known. CD55 is a GPI-linked molecule (Lublin and Atkinson, 1989) compared to CD46 which is a type I transmembrane protein (Liszewski et al., 1991), raising the possibility that differences in their structure within the confines of the virion envelope could contribute to individual activities against complement. An additional consideration is that CD46 functions in a trimeric complex along with C3b and Factor I, resulting in the cleavage of C3b into iC3b fragments. Thus, it is conceivable that steric hindrance due to tethering of CD46 in the virion particle and the need to have both C3b and Factor I colocalize for activity may limit effectiveness compared to CD55.
CD55 was found to be upregulated by infection with a number of RNA viruses, including RSV, WT and mutant PIV5, and Zika virus. It has also been shown to be upregulated by infection with other RNA viruses such as hepatitis C virus (Mazumdar et al 2013). CD55 induction is thought to involve PI3-kinase, p38 MAPK and NFkB signaling pathways (Ahmed et al., 2003). Prior work has shown that PIV5 does not block NFkB signaling pathways when induced by extracellular stimuli (Arimilli et al, 2009), and that PI3K pathways are not activated by PIV5 infection (Sun et al., 2008). Work is in progress to determine the pathways that PIV5 uses to increase cell surface CD55 and the contributions of p38 MAPK and NFkB to this induction. It is noteworthy that the Le-(U5C, A14G) mutant which contains alterations to the leader promoter increased CD55 expression to a greater extent than WT PIV5, and that this higher induction of a host cell protein correlates with the much higher-than-WT level of viral gene expression seen for this mutant (Manuse et al, 2009). These correlations suggest that specific viral gene products generated during an infection such as the viral glycoproteins or dsRNA are inducers of CD55 expression pathways.
TNF-α treatment of PIV5-infected cells led to increased CD55 cell surface expression above that seen with virus infection alone. Thus, PIV5 fails to block TNF-α signaling in infected A549 cells, but instead can take advantage of TNF-α signaling pathways for further complement evasion. CD55 expression can be enhanced through treatment of cells with cytokines derived from immune cells, including interferon-gamma, IL1-β or TNF-α (Halme et al, 2009). This raises the interesting possibility that paramyxoviruses exploit the production of extracellular cytokines and their ability to induce RCAs during in vivo infections. In this model, cytokines are produced in vivo by infected cells, neighboring uninfected cells or by lymphocytes that are recruited to the site of infection. This results in upregulated RCA expression on virus infected cells, incorporation of higher levels of RCA into progeny virions and enhanced evasion of complement neutralization.
Our results have implications for the understanding of host cell determinants of viral tropism and dissemination of virus through specific tissues of an infected animal. It is known that many tissues and cell types can vary widely in their expression of both complement factors and RCAs (e.g. Bolger et al. 2007; Veerhuis et al., 2011). Thus, the ability of a virus to enhance cell surface RCA concentration following infection and the factors that dictate assembly of specific complement regulators into virions could be important factors in tissue tropism, spread through an organ, and dissemination from the site of initial infection.
Highlights.
Parainfluenza virus 5 infection results in upregulation of cell surface CD55.
Increased cell surface CD55 results in more incorporation of this complement regulator into budded virions.
Virus derived from cells with upregulated CD55 are more resistant to complement-mediated neutralization in vitro
Acknowledgments
We thank members of the Parks lab for input and Namita Varudkar for excellent technical assistance. This work was supported by NIH grants AI083253 (GDP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad M, Pyaram K, Mullick J, Sahu A. Viral complement regulators: the expert mimicking swindlers. Indian J Biochem Biophys. 2007;44:331–343. [PubMed] [Google Scholar]
- Ahmad SR, Lidington EA, Ohta R, Okada N, Robson MG, Davies KA, Leitges M, Harris CL, Haskard DO, Mason JC. Decay-accelerating factor induction by tumour necrosis factor-alpha, through a phosphatidylinositol-3 kinase and protein kinase C-dependent pathway, protects murine vascular endothelial cells against complement deposition. Immunology. 2003;110:258–68. doi: 10.1046/j.1365-2567.2003.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimilli S, Johnson JB, Alexander-Miller MA, Parks GD. TLR-4 and -6 agonists reverse apoptosis and promote maturation of Simian Virus 5 infected human dendritic cells through NFkB-dependent pathways. Virology. 2007;365:144–156. doi: 10.1016/j.virol.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson JP, Liszewski MK, Richards A, Kavanagh D, Moulton EA. Hemolytic uremic syndrome: an example of insufficient complement regulation on self tissue. Ann NY Acad Sci. 2005;1056:144–152. doi: 10.1196/annals.1352.032. [DOI] [PubMed] [Google Scholar]
- Bergmann-Leitner ES, Leitner WW, Tsokos GC. Complement 3d: from molecular adjuvant to target of immune escape mechanisms. Clin Immunol. 2006;121:177–185. doi: 10.1016/j.clim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Bernet J, Mullick J, Singh AK, Sahu A. Viral mimicry of the complement system. J Biosci. 2003;28:249–64. doi: 10.1007/BF02970145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M, Kumar S, Johnson J, Parks GD, Subbiah E. Incorporation of host complement regulatory proteins into Newcastle Disease virus enhances complement evasion. J Virol. 2012;86:12708–12716. doi: 10.1128/JVI.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blue CE, Spiller OB, Blackbourn DJ. The relevance of complement to virus biology. Virology. 2004;319:176–184. doi: 10.1016/j.virol.2003.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger MS, Ross DS, Jiang H, Frank MM, Ghio AJ, Schwartz DA, Wright JR. Complement levels and activity in the normal and LPS-injured lung. Am J Physiol Lung Cell Mol Physiol. 2007;292:L748–L759. doi: 10.1152/ajplung.00127.2006. [DOI] [PubMed] [Google Scholar]
- Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5:981–6. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory factor H. Proc Natl Acad Sci. 2006;103:19111. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KL, Waggoner SN, Tacke R, Hahn YS. Role of complement in immune regulation and its exploitation by virus. Viral Immunology. 2007;20:505. doi: 10.1089/vim.2007.0061. [DOI] [PubMed] [Google Scholar]
- Delgado MF, Polack FP. Involvement of antibody, complement and cellular immunity in the pathogenesis of enhanced respiratory syncytial virus disease. Expert Rev Vaccines. 2004;3:693–700. doi: 10.1586/14760584.3.6.693. [DOI] [PubMed] [Google Scholar]
- Gerlier D, Loveland B, Varior-Krishnan G, Thorley B, McKenzie IFC, Rabourdin-Combe C. Measles virus receptor properties are shared by several CD46 isoforms differing in extracellular regions and cytoplasmic tails. J Gen Virol. 1994;75:2163–71. doi: 10.1099/0022-1317-75-9-2163. [DOI] [PubMed] [Google Scholar]
- He B, Paterson RG, Ward CD, Lamb RA. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- Johnson JB, Grant K, Parks GD. The paramyxoviruses simian virus 5 and mumps virus recruit host cell CD46 to evade complement-mediated neutralization. J Virol. 2009;83:7602. doi: 10.1128/JVI.00713-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JB, Aguilar H, Lee B, Parks GD. Interactions of human complement with virus particles containing the Nipah virus glycoproteins. J Virol. 2011;85:5940–5948. doi: 10.1128/JVI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JB, Lyles DS, Alexander-Miller MA, Parks GD. Virion-associated CD55 is more potent than CD46 in mediating resistance of mumps virus and VSV to neutralization. J Virol. 2012;86:9929–9940. doi: 10.1128/JVI.01154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JB, Schmitt AP, Parks GD. Point mutations in the paramyxovirus F protein that enhance fusion activity shift the mechanism of complement-mediated virus neutralization. J Virol. 2013;87:9250–9259. doi: 10.1128/JVI.01111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MA. The human complement system: assembly of the classical pathway C3 convertase. Biochem J. 1980;189:173–181. doi: 10.1042/bj1890173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Atkinson JP. T-cell regulation: with complements from innate immunity. Nat Rev Immunol. 2007;7:9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- Kim DD, Song WC. Membrane complement regulatory proteins. Clin Immunol. 2006;118:127–36. doi: 10.1016/j.clim.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion of human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kakinami C, Li Q, Yang B, Li H. Human Apolipoprotein A-I Is Associated with Dengue Virus and Enhances Virus Infection through SR-BI. PLoS ONE. 2013;8(7):e70390. doi: 10.1371/journal.pone.0070390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Fang CJ, Atkinson JP. Inhibiting complement activation on cells at the step of C3 cleavage. Vaccine. 2008;26s:122–127. doi: 10.1016/j.vaccine.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin DM, Atkinson JP. Decay-accelerating factor: Biochemistry, Molecular biology and function. Ann Rev Immunol. 1989;7:35–58. doi: 10.1146/annurev.iy.07.040189.000343. [DOI] [PubMed] [Google Scholar]
- Lyles DS. Assembly and budding of negative-strand RNA viruses. Adv Virus Res. 2013;85:57–90. doi: 10.1016/B978-0-12-408116-1.00003-3. [DOI] [PubMed] [Google Scholar]
- Manuse MJ, Parks GD. A role for the paramyxovirus genomic promoter in limiting host cell antiviral responses and cell killing. J Virol. 2009;83:9057. doi: 10.1128/JVI.01055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathology. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar B, Kim H, Meyer K, Bose SK, Di Bisceglie AM, Ray RB, Diamond MS, Atkinson JP, Ray R. Hepatitis C virus infection upregulates CD55 expression on the hepatocyte surface and promotes association with virus particles. J Virol. 2013;87:7902–10. doi: 10.1128/JVI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhop E, Nelson S, Jost CA, Gorlatov S, Johnson S, Fremont DH, Diamond MS, Pierson TC. Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nilevirus. Cell Host Microbe. 2009;6:381–91. doi: 10.1016/j.chom.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhop E, Diamond MS. Protective immune responses against West Nile virus are primed by distinct complement activation pathways. J Exp Med. 2006;203:1371–81. doi: 10.1084/jem.20052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Fraser RJ, Smith PN, Mahalingam S, Heise MT. Complement contributes to inflammatory tissue destruction in a mouse model of Ross River virus-induced disease. J Virol. 2007;81:5132–5143. doi: 10.1128/JVI.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick J, Bernet J, Panse Y, Hallihosur S, Singh AK, Sahu A. Identification of complement regulatory domains in vaccinia virus complement control protein. J Virol. 2005;79:12382–12393. doi: 10.1128/JVI.79.19.12382-12393.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Deficiency of an erythrocyte membrane protein with complement regulatory activity in paroxysmal nocturnal hemoglobinuria. Proc Natl Acad Sci USA. 1983;80:5430–34. doi: 10.1073/pnas.80.17.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal R, Carroll MC. Emerging patterns in complement-mediated pathogen recognition. Cell. 2006;125:29–32. doi: 10.1016/j.cell.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Russell SR, Sparrow RL, McKenzie IFC, Purcell DFJ. Tissue-specific and allelic expression of the complement regulator CD46 is controlled by alternative splicing. Eur J Immunol. 1992;22:1513–1518. doi: 10.1002/eji.1830220625. [DOI] [PubMed] [Google Scholar]
- Saifuddin M, Hedayati T, Atkinson JP, Holguin MH, Parker CJ, Spear GT. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidyinositor-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement mediated destruction. J Gen Virol. 1997;78:1907–1911. doi: 10.1099/0022-1317-78-8-1907. [DOI] [PubMed] [Google Scholar]
- Schauber-Plewa C, Simmons A, Tuerk MJ, Pacheco CD, Veres G. Complement regulatory proteins are incorporated into lentiviral vectors and protect particles against complement inactivation. Gene Therapy. 2005;12:238–245. doi: 10.1038/sj.gt.3302399. [DOI] [PubMed] [Google Scholar]
- Stoermer KA, Morrison TE. Complement and viral pathogenesis. Virology. 2011;411:362–73. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Fuentes SM, Timani K, Sun D, Murphy C, Lin Y, August A, Teng MN, He B. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J Virol. 2008;82:105–14. doi: 10.1128/JVI.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerhuis R, Nielsen HM, Tenner AJ. Complement in the Brain. Mol Immunol. 2011;48:1592–603. doi: 10.1016/j.molimm.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt MR, Dowd KA, Engle M, Tesh RB, Johnson S, Pierson TC, Diamond MS. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms. J Virol. 2011;85:11567–80. doi: 10.1128/JVI.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]