Abstract
Background Nephronophthisis (NPH) is the most prevalent genetic cause for ESRD in children. However, little is known about the prevalence of NPH in adult-onset ESRD. Homozygous full gene deletions of the NPHP1 gene encoding nephrocystin-1 are a prominent cause of NPH. We determined the prevalence of NPH in adults by assessing homozygous NPHP1 full gene deletions in adult-onset ESRD.
Methods Adult renal transplant recipients from five cohorts of the International Genetics and Translational Research in Transplantation Network (iGeneTRAiN) underwent single-nucleotide polymorphism genotyping. After quality control, we determined autosomal copy number variants (such as deletions) on the basis of median log2 ratios and B-allele frequency patterns. The findings were independently validated in one cohort. Patients were included in the analysis if they had adult-onset ESRD, defined as start of RRT at ≥18 years old.
Results We included 5606 patients with adult-onset ESRD; 26 (0.5%) showed homozygous NPHP1 deletions. No donor controls showed homozygosity for this deletion. Median age at ESRD onset was 30 (range, 18–61) years old for patients with NPH, with 54% of patients age ≥30 years old. Notably, only three (12%) patients were phenotypically classified as having NPH, whereas most patients were defined as having CKD with unknown etiology (n=11; 42%).
Conclusions Considering that other mutation types in NPHP1 or mutations in other NPH-causing genes were not analyzed, NPH is a relatively frequent monogenic cause of adult-onset ESRD. Because 88% of patients had not been clinically diagnosed with NPH, wider application of genetic testing in adult-onset ESRD may be warranted.
Keywords: genetic renal disease, transplantation, end-stage renal disease, human genetics, cystic kidney
Nephronophthisis (NPH) is a Mendelian genetic disease and a classic pediatric kidney disease. Although NPH is considered a rare disorder (incidence of 0.1–0.2 per 10,000 live births), NPH is the most prevalent genetic cause for ESRD in children, with a frequency of 15%.1,2 The most common variant is NPH type 1 (OMIM 256100), in which patients generally present at age 13 years old with ESRD.3
The etiology of NPH lies in the primary cilium, which functions as a sensory organelle in the renal cell.3,4 Mutations in genes coding for proteins essential to the primary cilium lead to structurally or functionally aberrant cilia, making NPH a ciliopathy.3,4 To date, causative mutations in many genes encoding these proteins have been reported, but the largest proportion of NPH (20%–25%) is caused by homozygous full gene deletions of the NPHP1 gene (OMIM 607100).5,6 These full gene deletions are recurrent in the general population, a result of recurrent complex rearrangements at this locus due to flanking low copy repeats (elements with highly similar sequence identities).7–9 Mutations in NPHP1 are completely penetrant; thus, the presence of the mutation always leads to an NPH phenotype.5,6 For the other NPH genes, no such recurrent full gene deletion is known.5,6
Clinically, NPH generally starts around age 6 years old with nonspecific and mild symptoms due to an impaired ability to concentrate urine and retain water.10 This leads to early symptoms, such as polyuria, polydipsia, and secondary enuresis.10 If performed, renal ultrasound may show echogenicity with loss of corticomedullary differentiation.11 The disease always progresses to ESRD, in general around age 13 years old, with a need for RRT (dialysis or renal transplantation).3 In some patients with NPH, renal ultrasound shows corticomedullary cysts, although the majority display small atrophic kidneys at the ESRD stage.11 Some patients with NPH (15%) also display additional extrarenal abnormalities, such as neurologic anomalies (Joubert syndrome) or ophthalmologic dysplasias (Senior–Løken syndrome), which can guide diagnosis.12 However, isolated NPH may prove difficult to diagnose clinically, because the phenotype is generally nonspecific, can be variable, and often only becomes clinically apparent in the ESRD stage.3 Therefore, genetic testing is the sole method to diagnose NPH with certainty.6
Because it almost always presents in midchildhood, NPH is primarily considered a pediatric kidney disease. Nevertheless, anecdotal cases of patients with adult-onset ESRD have been reported.13–15 Although these patients suggest that the diagnosis of NPH is rare in adults, little is known about the overall prevalence of NPH in the adult-onset ESRD population.13–18
We set out to investigate the prevalence of NPH in adults by analyzing, as a proxy, the prevalence of homozygous NPHP1 full gene deletions in patients with adult-onset ESRD. We proposed to do this by assessing autosomal copy number variants (CNVs; e.g., large deletions) in genomic data generated for a genome-wide association study in multiple cohorts of renal transplant recipients and (corresponding donor) controls from the International Genetics and Translational Research in Transplantation Network (iGeneTRAiN) Consortium.19
Methods
CNV Analyses
We genotyped five iGeneTRAiN Consortium cohorts, all consisting of renal transplant recipients and (donor) controls.19 The DeKAF Genomics, Gen03, TransplantLines-Genetics, and Vienna cohorts were genotyped with the custom Affymetrix Axiom Tx GWAS Array (Affymetrix Inc., Santa Clara, CA).20 This array, designed for the iGeneTRAiN Consortium, contains single-nucleotide variants, single-nucleotide polymorphisms (SNPs), and monomorphic markers for approximately 782,000 positions across the whole genome (on the basis of human reference genome build GRCh37).20 Of these, approximately 22,000 variants were used to cover approximately 2200 manually curated CNV regions.20 The GoCAR cohort was genotyped with the Infinium HumanOmniExpressExome-8 v1B and OmniExpressExome-8 v1.1A (Illumina, San Diego, CA), providing whole-genome coverage with markers for approximately 960,000 SNPs.19
Quality control was performed by excluding all low-quality SNPs, leaving only high-quality SNPs (call rate >0.99; Hardy–Weinberg equilibrium P>0.001; minor allele frequency <0.1) that were linkage disequilibrium pruned to leave no pairs with r2>0.2. Also, we removed SNPs in regions with known long stretches of linkage disequilibrium and nonautosomal SNPs. Only data on the 5606 patients with adult-onset ESRD (start of first RRT at 18 years old or older) were analyzed: n=3192 from the DeKAF Genomics and Gen03 combined cohorts, n=1230 from the TransplantLines-Genetics cohort, n=500 from the GoCAR cohort, and n=684 from the Vienna cohort.
The data generated were used to determine autosomal CNVs (i.e., deletions and duplications). CNV calling was performed using the default settings of two well published algorithms: the BRLMM-P algorithm designed by Affymetrix Inc. (DeKAF Genomics, Gen03, and TransplantLines-Genetics) or PennCNV (GoCAR and Vienna).21,22 All calls with logR ratio SD >0.3, B-allele frequency drift >0.01, waviness factor >0.05, or waviness SD >0.15 were excluded. Loci where no definite copy number call could be made were also excluded from further analysis.
To assess NPHP1 full gene deletions, all samples containing a CNV with an overlap of at least one nucleotide with the NPHP1 region (chromosome 2: 110879888–110962643 on the basis of GRCh37) were identified using R Studio (version 1.0.153 for Windows; RStudio Inc., Boston, MA). All patients with a copy number call of zero in this entire region (i.e., homozygous full gene deletions) were selected for further analysis. We additionally assessed the number of recipients and donors with a copy number call of one (i.e., heterozygous full gene deletion carriers). The regions of the other known NPH genes were also assessed for CNVs.6
All NPHP1 deletions called by the algorithms were manually inspected to ensure that only true calls were made. Furthermore, the dense SNP arrays applied to the cohorts are well established and validated, and the NPHP1 region is large, leading to a near zero chance of false findings.19–22 To further validate this, the TransplantLines-Genetics samples that displayed a homozygous deletion copy number call for NPHP1 (n=11) were independently validated with multiplex ligation–dependent probe amplification, showing 100% concordance.5,9,15
Demographic and Phenotypic Statistics
Demographic and phenotypic information was retrieved per cohort for all patients with adult-onset ESRD, which was defined as start of first RRT at 18 years old or older. For patients with adult-onset NPH, we studied various phenotypical characteristics, namely the age at ESRD onset (defined as age at first RRT [dialysis or transplantation]), ethnicity, and primary renal disease diagnosis. If patient consent (discussed in Ethical Constraints below) allowed for data retrieval from the patient file, data on family history, hypertension, polyuria, proteinuria, and extrarenal NPH-associated anomalies were retrieved.
Ethnicity was determined by principal component analysis using 1000 Genomes Phase 3 data (1092 samples and 14 different ancestries).23,24 Populations were determined by visual inspection of the first two principal components (by J.v.S.). Descriptive statistics were generated for the demographic and primary renal diagnosis variables using SPSS (version 23 for Windows; IBM Corp., Armonk, NY). Additionally, we applied a two-sided Fisher exact test to assess these binomial variables. All samples were also analyzed on identity by state patterns to identify patients who were related up to and including to the third degree (identity by state <10%).
Ethical Constraints
The enrollment of participants for all iGeneTRAiN Consortium cohorts was approved by the institutional review boards of the hospitals at which participants were included. All participants signed informed consent for genomic data analysis via SNP array. For the GoCAR cohort, TransplantLines-Genetics, and Vienna cohorts, the principal consent form included retrieval of relevant information from their patient file. Participants enrolled in the DeKAF Genomics and Gen03 cohorts had to additionally consent for data retrieval from their patient file.
Furthermore, participants were not asked to consent to return of genomic study results on an individual level. Thus, we could not notify NPHP1 homozygous or heterozygous gene deletion carriers of this finding.
Results
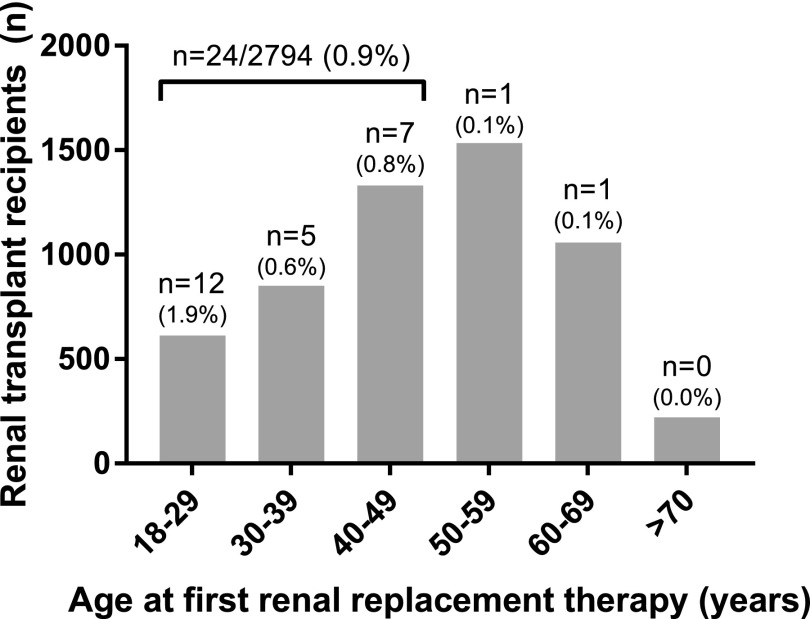
In total, 5606 renal transplant recipients with start of first RRT at any age ≥18 years old were included. Overall descriptive characteristics of the five cohorts were reported previously by the iGeneTRAiN Consortium.19 Of these patients with adult ESRD onset, 26 (0.5%) patients (Figure 1) displayed the same approximately 96,389-base pair deletion (size on the basis of SNP array) (example in Figure 2) on both alleles, including all NPHP1 gene exons. The 26 patients were not related up to and including to the third degree.
Figure 1.
Homozygous NPHP1 full gene deletions have an overall prevalence of 0.5% in adult onset ESRD patients, while the prevalence is even 0.9% in patients aged 18–50 at ESRD onset. The number of renal transplant recipients (n) in the five combined cohorts is displayed by age decade of start of RRT. The number of homozygous NPHP1 full gene deletion carriers (n) and the percentage per specific age category are shown above each bar.
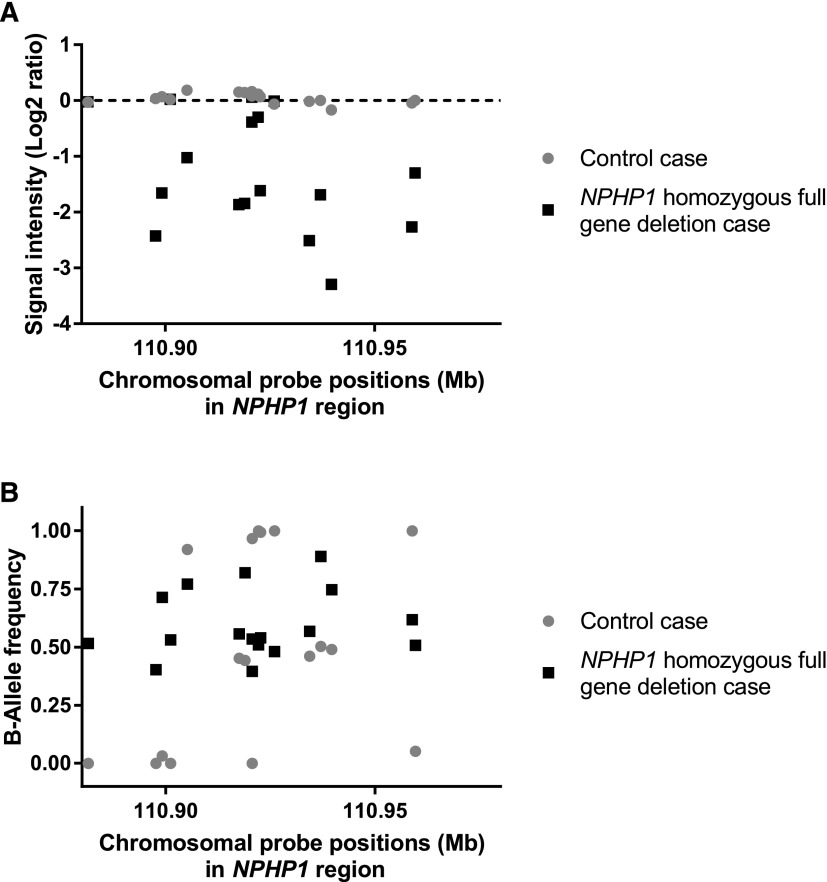
Figure 2.
The graphical representation of (A) log2 ratio and (B) B-allele frequency of all single-nucleotide polymorphism markers in the NPHP1 gene region clearly distinguishes between patients with a NPHP1 homozygous full gene deletion and healthy controls. Displayed here are an example homozygous NPHP1 full gene deletion patient (patient 19, Table 1), shown with black squares, and a healthy control sample, shown with gray circles. (A) The log2 ratio represents the normalized signal intensity for a specific marker; thus, if there is signal for that marker, the ratio is zero, whereas any value below zero indicates genomic deletion. An average log2 ratio of −2 specifically indicates a homozygous deletion. (B) The B-allele frequency displays if the particular marker is present in a homozygous state (either B-allele frequency of approximately 0.00 or 1.00) or a heterozygous state (B-allele frequency of approximately 0.50), such as is clear in the control patient shown here. In patients with a homozygous deletion, the algorithm cannot accurately determine the B-allele frequency, because no markers are present at all. This leads to a ”waterfall“ configuration, with markers being assigned B-allele frequencies anywhere between 0.00 and 1.00 at random.30
None of the 3311 (donor) controls displayed this homozygous deletion. None of the 5606 recipients showed a deletion of any of the other 19 known NPH genes.6 Markedly, we detected a higher number of heterozygous NPHP1 deletions in transplant recipients (n=36) compared with the transplant (donor) controls (n=10; P<0.001). Although these patients were not additionally assessed with next generation sequencing, this finding points to the possibility of compound heterozygosity (a full gene deletion on one allele and a different pathogenic mutation on the other allele) in the recipients.
When addressing the phenotype of the patients with NPHP1 deletions (Figure 1 and Table 1), the median age at start of RRT was 30 years old (range, 18–61), with 14 patients (54%) ages 30 years old or older. Interestingly, the prevalence of homozygous NPHP1 deletions was 0.9% in recipients between 18 and 50 years old at the start of first RRT (n=24 of 2794) and even higher (2.1%) in recipients ages 18–29 years old (Figure 1).
Table 1.
Phenotypical information about the 26 patients with NPHP1 homozygous gene deletion, including age at presentation, age at first RRT, and the presence of various renal and extrarenal symptoms of nephronophthisis
| Patient No. | Sex | Ancestry (Geographic Origin) | Clinical Diagnosis | Age at Presentation, yr | Age at First RRT, yr | Positive Family History of Renal Disease | Hypertension, Age in yr | Polyuria, Age in yr | Proteinuria, Age in yr | Bone Disease | Ophtamologic Anomalies Associated with NPH | Neurologic Anomalies Associated with NPH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Woman | Caucasian (United States) | Tubular and interstitial disease | Not retrievable | 18 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 2 | Woman | Caucasian (Europe) | NPH | 19 | 19 | NR | — | — | +, Age NR | — | — | — |
| 3 | Man | Caucasian (United States) | Chronic renal failure with unknown etiology | Not retrievable | 21 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 4 | Woman | Caucasian (United States) | Chronic renal failure with unknown etiology | Not retrievable | 21 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 5 | Man | Caucasian (Europe) | Glomerular disease | 22 | 22 | — | 22 | NR | 22 | — | — | — |
| 6 | Woman | Caucasian (Europe) | Chronic renal failure with unknown etiology | 21 | 23 | — | 21 | — | — | — | — | — |
| 7 | Woman | Caucasian (United States) | Chronic renal failure with unknown etiology | 23 | 23 | — | 23 | NR | NR | NR | — | — |
| 8 | Woman | Caucasian (United States) | Chronic renal failure with unknown etiology | 23 | 23 | + | 23 | NR | NR | + | — | — |
| 9 | Man | African (United States) | Hypertensive nephrosclerosis | 25 | 25 | — | 25 | NR | NR | NR | — | — |
| 10 | Woman | Caucasian (Europe) | Sporadic primary reflux nephropathy | 27 | 28 | — | 27 | — | NR | — | — | — |
| 11 | Man | Caucasian (Europe) | GN, histologically examined | 28 | 28 | + | — | — | NR | — | — | Severe developmental delay |
| 12 | Woman | Caucasian (Europe) | NPH | 29 | 29 | — | — | — | NR | + | — | — |
| 13 | Woman | Caucasian (United States) | Chronic renal failure with unknown etiology | Not retrievable | 30 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 14 | Man | Caucasian (Europe) | NPH | 27 | 30 | NR | 27 | — | — | — | — | — |
| 15 | Man | Caucasian (Europe) | Vascular nephropathy | NR | 30 | — | +, Age NR | NR | NR | NR | Congenital nystagmus | Tremor |
| 16 | Man | Caucasian (United States) | Medullary cystic disease | NR | 31 | NR | — | NR | — | NR | NR | NR |
| 17 | Man | Caucasian (Europe) | Chronic renal failure with unknown etiology | 30 | 34 | + | 30 | — | — | — | — | — |
| 18 | Woman | Caucasian (Europe) | Chronic renal failure with unknown etiology | 9 | 40 | + | 9 | — | — | — | — | — |
| 19 | Man | Caucasian (Europe) | Urate nephropathy | 28 | 40 | — | — | — | — | — | — | — |
| 20 | Woman | Caucasian (Europe) | Chronic renal failure with unknown etiology | 39 | 42 | — | 34 | — | NR | — | — | — |
| 21 | Woman | Caucasian (Europe) | Medullary cystic disease | NR | 42 | NR | — | NR | — | NR | NR | NR |
| 22 | Man | Caucasian (United States) | Chronic renal failure with unknown etiology | Not retrievable | 43 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 23 | Man | Caucasian (United States) | Chronic renal failure with unknown etiology | Not retrievable | 44 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 24 | Man | Caucasian (United States) | Tubular and interstitial disease | Not retrievable | 46 | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable | Not retrievable |
| 25 | Woman | Caucasian (Europe) | Autosomal dominant polycystic kidney disease | 42 | 52 | + | 50 | — | — | — | — | — |
| 26 | Woman | Caucasian (Europe) | Hypertensive nephrosclerosis | NR | 61 | — | +, Age NR | NR | NR | NR | Congenital blindness | — |
Patients are sorted by age at first RRT (youngest to oldest). For seven patients, not all data were retrievable due to the patients not consenting to additional data retrieval from the patient file (information noted as “not retrievable”). Additional information is discussed in Ethical Constraints in the Methods section. NPH, nephronophthisis; NR, not reported in patient file; —, symptom not present; . +, symptom present.
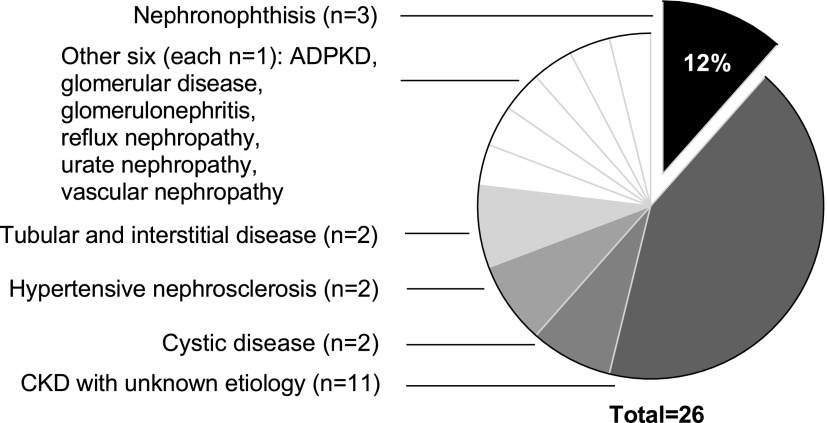
Regarding the clinical primary renal disease diagnosis, only three patients (12%), of whom two were younger than 30 years old at the start of first RRT, were classified as having NPH (Figure 3). The other patients (88%) were diagnosed with CKD with unknown etiology (n=11), cystic disease (n=2), hypertensive nephrosclerosis (n=2), tubular and interstitial disease (n=2), glomerular disease (n=1), GN (histologically examined; n=1), sporadic primary reflux nephropathy (n=1), vascular nephropathy (n=1), urate nephropathy (n=1), and autosomal dominant polycystic kidney disease (n=1; no mutation in PKD1 or PKD2).
Figure 3.
Only 12% of homozygous NPHP1 full gene deletion cases were correctly identified as having nephronophthisis, while 88% were clinically diagnosed as having something other than nephronophthisis.
Two patients displayed extrarenal anomalies associated with NPH, namely congenital blindness (possibly Senior–Løken syndrome type 125) and severe neurodevelopmental delay (possibly Joubert syndrome type 426). Both of these patients had not been clinically diagnosed with NPH but with hypertensive nephrosclerosis and GN (histologically examined), respectively (Table 1).
Discussion
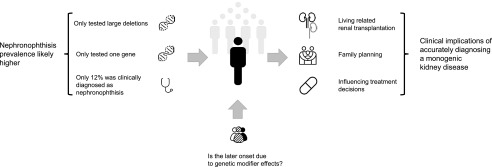
Our data indicate that, with a 0.5% prevalence of homozygous NPHP1 full gene deletions, the frequency of NPH in adult-onset ESRD is considerably higher than previously reported.6,16 When interpreting our results, one should note that, inherent to the method applied in our study (which only analyzes larger deletions and duplications), the overall prevalence of NPH in adult-onset ESRD that we report here is very likely to be an underestimation (Figure 4). First, with this method, any combination of two smaller NPHP1 mutations, such as (homozygous) pathogenic intragenic deletions up to approximately 700 base pairs and single-nucleotide variants, could not be analyzed.22 Because this is outside the scope of this study, compound heterozygosity and homozygosity for other mutations were not assessed. The assumption that NPHP1-related disease is probably more frequent in adult-onset ESRD is supported by our recipients showing a significantly higher frequency of heterozygous deletion carriers than donors, suggesting that a subset of these deletion carriers likely also carries a second mutation on the other allele. Second, the causative mutations in other NPH genes generally are not recurrent full gene deletions.5,6 This is underscored by the fact that our analysis showed no large deletions in these genes.6 We have not performed any (additional) next generation sequencing to assess any other types of mutation. In the light of these two considerations, our findings point to an underestimation of the number of causative NPH mutations in the overall cohort.
Figure 4.
At least one in 200 (0.5%) patients has all adult-onset ESRD due to nephronophthisis, but the overall prevalence is likely higher. The late presentation of nephronophthisis might be due to genetic modifier effects. Accurately diagnosing a monogenic disease such as nephronophthisis can have wide-ranging clinical implications.
With regard to age at onset of ESRD, over one half of the patients with NPHP1 in our study were 30 years old or older at first onset of ESRD. Therefore, we postulate that NPH is not merely a pediatric disease entity. In the literature so far, only six patients across four families have been described with NPHP1 mutations and ESRD onset after 30 years of age, with the oldest being 56 years old.13–15 Our study extends this age of onset to 61 years old. In the subpopulation with onset between 18 and 50 years old, we observed an NPHP1 deletion prevalence of 0.9%. The mechanism leading to phenotypic variance in age of ESRD onset remains unclear. It has been hypothesized previously that this nonpediatric onset of ESRD, especially after 30 years old, is due to the influence of yet unknown modifier genes.14,15 Because of the ancestry composition of our cohorts (the majority being Caucasian; data not shown), we could not analyze the presence of distinct ethnicity-specific modifier effects.6,8 Nevertheless, modifier effects, whether ethnicity specific or not, could play a role in the discrepancy in age at onset between the patients in our report and those in older literature.
In assessing the clinical diagnoses of the patients with NPHP1 deletions, we show that there is likely an underdiagnosis of NPHP1-related disease in adult-onset ESRD in clinical practice. The underdiagnosis, which was universally present among five cohorts originating from several countries in Europe as well as the United States, might, at least in part, be due to the nonspecific phenotype, because it can be especially difficult to recognize in adults presenting with advanced renal failure.14 This is underscored by our finding that 88% of patients with NPHP1 had received a clinical diagnosis other than NPH, even in patients with NPH-associated extrarenal anomalies. Notwithstanding, it is important to accurately define the etiology of patients with ESRD not only to prevent misdiagnosis and useless treatment but also, because a monogenic disease can have a variety of clinical implications.27 For instance, it can affect decisions related to living related kidney donation, which is contraindicated in siblings at risk.28 It might also influence choices regarding family planning, especially in consanguineous relationships.29
In summary, we are the first to show in a large cohort that NPH due to NPHP1 homozygous full gene deletions has a prevalence of one in 200 patients (0.5%) in all adult-onset ESRD. Although the incidence was clearly higher in patients with an ESRD onset between 18 and 50 years old (prevalence of 0.9%), NPH can have an onset at up to 61 years of age. Because the method that we used underestimates the total number of causal mutations, we conclude that NPH is a relatively frequent monogenic cause of adult-onset ESRD that is likely underdiagnosed in current daily practice. Given the potential clinical implications of having a genetic diagnosis, our data warrant wider application of genetic testing in adult-onset ESRD.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank Affymetrix Inc. (Santa Clara, CA) for performing part of the copy number variant calling using their BRLMM-P algorithm.21 All images in Figure 4 were purchased from thenounproject.com under a royalty-free license. For two images, the color was changed from black to gray, and one image was cropped to ensure adequate fit.
The DeKAF-Genomics cohort and the GEN03 cohort were supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grants 5U19-AI070119 and 5U01-AI058013. R.S., N.V.A.M.K., and A.M.v.E. are supported by Dutch Kidney Foundation grants KSTP12.010, 15OP14, and Kouncil CP11.18, and A.M.v.E. is supported by NutsOhra Foundation grant 070-1303. Z.Z. is partially supported by the Translational Collaborative Research Initiative Grant from the Icahn School of Medicine at Mount Sinai.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “The Underestimated Burden of Monogenic Diseases in Adult-Onset ESRD,” on pages 1583–1584.
References
- 1.Levy M, Feingold J: Estimating prevalence in single-gene kidney diseases progressing to renal failure. Kidney Int 58: 925–943, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Hildebrandt F: Genetic kidney diseases. Lancet 375: 1287–1295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf MTF, Hildebrandt F: Nephronophthisis. Pediatr Nephrol 26: 181–194, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaats GG, Lilien MR, Giles RH: Nephronophthisis: Should we target cysts or fibrosis? Pediatr Nephrol 31: 545–554, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, et al.; GPN Study Group : Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 132: 865–884, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun DA, Hildebrandt F: Ciliopathies. Cold Spring Harb Perspect Biol 9: pii: a028191, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunier S, Calado J, Benessy F, Silbermann F, Heilig R, Weissenbach J, et al.: Characterization of the NPHP1 locus: Mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet 66: 778–789, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan B, Liu P, Gupta A, Beck CR, Tejomurtula A, Campbell IM, et al.: Comparative genomic analyses of the human NPHP1 locus reveal complex genomic architecture and its regional evolution in primates. PLoS Genet 11: e1005686, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konrad M, Saunier S, Heidet L, Silbermann F, Benessy F, Calado J, et al.: Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet 5: 367–371, 1996 [DOI] [PubMed] [Google Scholar]
- 10.Ala-Mello S, Koskimies O, Rapola J, Kääriäinen H: Nephronophthisis in Finland: Epidemiology and comparison of genetically classified subgroups. Eur J Hum Genet 7: 205–211, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Blowey DL, Querfeld U, Geary D, Warady BA, Alon U: Ultrasound findings in juvenile nephronophthisis. Pediatr Nephrol 10: 22–24, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Wolf MTF: Nephronophthisis and related syndromes. Curr Opin Pediatr 27: 201–211, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georges B, Cosyns JP, Dahan K, Snyers B, Carlier B, Loute G, et al.: Late-onset renal failure in Senior-Loken syndrome. Am J Kidney Dis 36: 1271–1275, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Hoefele J, Nayir A, Chaki M, Imm A, Allen SJ, Otto EA, et al.: Pseudodominant inheritance of nephronophthisis caused by a homozygous NPHP1 deletion. Pediatr Nephrol 26: 967–971, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haghighi A, Savaj S, Haghighi-Kakhki H, Benoit V, Grisart B, Dahan K: Identification of an NPHP1 deletion causing adult form of nephronophthisis. Ir J Med Sci 185: 589–595, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Bollée G, Fakhouri F, Karras A, Noël L-H, Salomon R, Servais A, et al.: Nephronophthisis related to homozygous NPHP1 gene deletion as a cause of chronic renal failure in adults. Nephrol Dial Transplant 21: 2660–2663, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Chaari I, Trabelsi M, Goucha R, Elaribi Y, Kharrat M, Guarguah T, et al.: Prevalence and incidence estimation of large NPHP1 homozygous deletion in Tunisian population. Pathol Biol (Paris) 60: e84–e86, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Omran H, Fernandez C, Jung M, Häffner K, Fargier B, Villaquiran A, et al.: Identification of a new gene locus for adolescent nephronophthisis, on chromosome 3q22 in a large Venezuelan pedigree. Am J Hum Genet 66: 118–127, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Genetics & Translational Research in Transplantation Network (iGeneTRAiN) : Design and implementation of the international genetics and translational research in transplantation network. Transplantation 99: 2401–2412, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YR, van Setten J, Verma SS, Lu Y, Holmes MV, Gao H, et al.: Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med 7: 90, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The P, just P, finally P: BRLMM-P: A genotype calling method for the SNP 5.0 Array. In: ReVision, 2007, pp 1–16 [Google Scholar]
- 22.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al.: PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17: 1665–1674, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolliffe IT, Cadima J: Principal component analysis: A review and recent developments. Philos Trans A Math Phys Eng Sci 374: 20150202, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. : A global reference for human genetic variation. Nature 526: 68–74, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loken AC, Hanssen O, Halvorsen S, Jolster NJ: Hereditary renal dysplasia and blindness. Acta Paediatr 50: 177–184, 1961 [DOI] [PubMed] [Google Scholar]
- 26.Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DWW, et al.: The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet 75: 82–91, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakash S, Gharavi AG: Diagnosing kidney disease in the genetic era. Curr Opin Nephrol Hypertens 24: 380–387, 2015 [DOI] [PubMed] [Google Scholar]
- 28.Huang E, Samaniego-Picota M, McCune T, Melancon JK, Montgomery RA, Ugarte R, et al.: DNA testing for live kidney donors at risk for autosomal dominant polycystic kidney disease. Transplantation 87: 133–137, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Eerde AM, Krediet CTP, Rookmaaker MB, van Reekum FE, Knoers NVAM, Lely AT: Pre-pregnancy advice in chronic kidney disease: Do not forget genetic counseling. Kidney Int 90: 905–906, 2016 [DOI] [PubMed] [Google Scholar]
- 30.Illumina: Interpreting infinium assay data for whole-genome structural variation. In: Analysis, 2010, pp 1–9 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.