Metabolomics, the systematic analysis of small molecules present in a biologic specimen, is an increasingly useful tool for research in CKD.1 Ongoing methodologic improvements have expanded the capabilities of metabolomics platforms. Untargeted (or nontargeted) methods now simultaneously detect hundreds of known compounds from multiple pathways, including those involved in sugar, amino acid, organic acid, nucleotide, acylcarnitine, and lipid metabolism, as well as unknown compounds that are reproducibly found but yet to be identified.2 Initial metabolomic studies in CKD showed associations between several blood metabolites, eGFR, and future development of CKD.3,4 Continued technologic improvements, collaborative work across metabolomic platforms, and metabolite profiling in large cohort studies should yield new biomarkers, insights on CKD pathophysiology, and therapeutic opportunities.
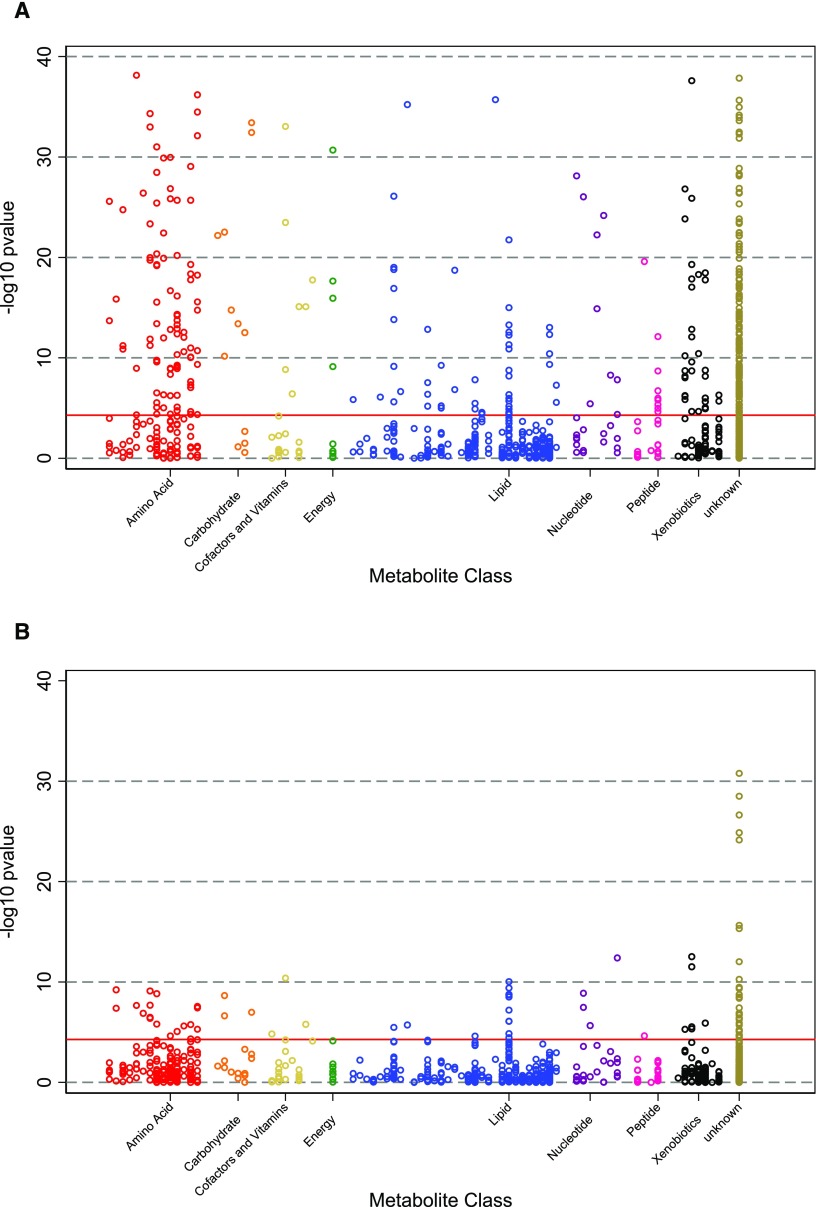
Statistical methods in metabolomics parallel those used in genetics, a field that has benefitted from widespread improvement in DNA sequencing methodologies over the past two decades. Figure 1A shows a traditional “Manhattan plot,” depicting P values from the associations of 953 serum metabolites with mortality risk among individuals with CKD and showing dozens of highly significant associations.5 This layout underscores the multiplicity of statistical comparisons and the need to adjust thresholds for statistical significance accordingly. Although there is no consensus approach, our view is that Bonferroni adjustment (i.e., adjusting for every metabolite) can be overly conservative, because many metabolites are highly intercorrelated, particularly within a given metabolite class or biochemical pathway. Alternative approaches include controlling for a false discovery rate, adjusting for the number of meaningful metabolite clusters determined by principal component analysis, or adopting a more lenient statistical threshold for discovery but requiring external validation.
Figure 1.
The number of statistically significant associations between metabolites and subsequent mortality is greatly reduced after adjustment for GFR. Figure depicts P-values of the demographic-adjusted associations between metabolites identified in an untargeted screen and future risk of mortality among 963 participants with CKD in the African American Study of Kidney Disease and Hypertension (A) before and (B) after adjustment for GFR measured by urinary clearance of iothalamate.
Although there are similarities between metabolomic and genetic analyses, important differences also exist. First, genetic data are relatively constant within an individual over time, whereas metabolites can and do vary according to a wide range of intrinsic and environmental factors, including diet, medications, and the microbiome (all of which contribute metabolites that can be detected in blood and other biologic specimens). This intraindividual variability can reduce power to detect true associations. Some metabolomic studies have required a degree of medium-term stability, evaluating only those metabolites with low intraindividual variation.6 Second, genetic studies can evaluate associations between genetic variants and phenotypes without regard to temporality given near certainty that the genetic variant was the antecedent event. In contrast, metabolite studies must consider longitudinal relationships, with careful accounting for baseline confounders of the relationship between metabolite and clinical outcomes.
One of the most important confounders in metabolomics studies of CKD is kidney function. Typically assessed as GFR, kidney function is related to approximately one third of detected metabolites in both general and CKD populations.7,8 Figure 1B shows the association between the same blood metabolites and risk of subsequent death depicted in Figure 1A after adjusting for baseline GFR, revealing a dramatic attenuation in the number of statistically significant metabolite associations. Thus, careful measurement and consideration of baseline GFR are critical. However, even perfect adjustment for GFR may not fully address the effect of kidney function on metabolite levels. Metabolites might vary because of differences in tubular secretion or resorption and disruptions in kidney catabolism or anabolism. Furthermore, there may be indirect effects of CKD, such as modifications to diet, the microbiome, and medications that may co-occur with CKD progression.
At present, even the best metabolomics platforms provide incomplete coverage of the human metabolome, detecting less than one quarter of the currently known endogenous and exogenous metabolites in a given biologic specimen. Different platforms provide different coverage of the metabolome, such that a metabolite detected on one platform may not be identified on another, complicating comparisons across studies. Within a platform, not all detected metabolites are measured equally well.9 Thus, it may be prudent to corroborate the assignments of promising metabolites with experiments that analyze samples spiked with the authentic standard.10 Development of dedicated targeted assays may also be necessary to improve precision and enable absolute quantitation, because untargeted assays typically report only relative results.
Taken together, metabolomics is emerging as a powerful tool for CKD research. However, it must be coupled with thoughtful study design, careful consideration of potential confounders, and rigorous quality control to yield meaningful results. Meta-analysis across large datasets significantly enhanced the power of genetic studies, and the ability to perform similar studies across different metabolomics platforms is a fundamental unmet need in the field. We view metabolomics studies as an important starting point for discovery and hypothesis generation, with subsequent efforts required for independent replication, individualized assay development, and mechanistic investigation.
Disclosures
None.
Acknowledgments
This work was supported by National Institutes of Health grants U01-DK-085689 (to M.E.G.), R01-DK-108803 (to M.E.G.), R01-HL-132372 (to T.S.), U01-DK-061022 (to T.S.), R01-DK-101500 (to T.S.), UH2-AG-056933 (to T.S.), and U01-DK-106981 (to E.P.R.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Hocher B, Adamski J: Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol 13: 269–284, 2017 [DOI] [PubMed] [Google Scholar]
- 2.Sévin DC, Kuehne A, Zamboni N, Sauer U: Biological insights through nontargeted metabolomics. Curr Opin Biotechnol 34: 1–8, 2015 [DOI] [PubMed] [Google Scholar]
- 3.Kalim S, Rhee EP: An overview of renal metabolomics. Kidney Int 91: 61–69, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, et al. : A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 24: 1330–1338, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu JR, Coresh J, Inker LA, Levey AS, Zheng Z, Rebholz CM, et al. : Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int 2018, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu B, Zheng Y, Nettleton JA, Alexander D, Coresh J, Boerwinkle E: Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 9: 1410–1417, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Römisch-Margl W, et al. : A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol 27: 1175–1188, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coresh J, Inker LA, Sang Y, Chen J, Shafi T, Post WS, et al. : Metabolomic profiling to improve glomerular filtration rate estimation: a proof-of-concept study. Nephrol Dial Transplant 2018, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, Yu B, Alexander D, Couper DJ, Boerwinkle E: Medium-term variability of the human serum metabolome in the Atherosclerosis Risk in Communities (ARIC) study. OMICS 18: 364–373, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tin AN, G, Evans AM, Winkler CA, Bottinger E, Rebholz CM, et al. : Lower serum 6-bromotryptophan identified as a risk factor for chronic kidney disease progression. J Am Soc Nephrol 2018, in press [DOI] [PMC free article] [PubMed] [Google Scholar]