Abstract
The TLO genes are a family of subtelomeric ORFs in the fungal pathogens Candida albicans and C. dubliniensis encoding a subunit of the Mediator complex homologous to Med2. The more virulent pathogen C. albicans has 15 copies of the gene whereas the less pathogenic species C. dubliniensis has only two. To investigate if expansion of the TLO repertoire in C. dubliniensis has an effect on phenotype and virulence we expressed three representative C. albicans TLO genes (TLOβ2, TLOγ11 and TLOα12) in a wild type C. dubliniensis background, under the control of either their native or the ACT1 promoter. Expression of TLOβ2 resulted in a hyperfilamentous phenotype, while overexpression of TLOγ11 and TLOα12 resulted in enhanced resistance to oxidative stress. Expression of all three TLO genes from the ACT1 promoter resulted in increased virulence in the Galleria infection model. In order to further investigate if individual TLO genes exhibit differences in function we expressed six representative C. albicans TLO genes in a C. dubliniensis Δtlo1/Δtlo2 double mutant. Differences were observed in the ability of the expressed CaTLOs to complement the various phenotypes of the mutant. All TLO genes with the exception of TLOγ7 could restore filamentation, however only TLOα9, γ11 and α12 could restore chlamydospore formation. Differences in the ability of CaTLO genes to restore growth in the presence of H2O2, calcofluor white, Congo red and at 42°C were observed. Only TLOα3 restored wild-type levels of virulence in the Galleria infection model. These data show that expansion of the TLO gene family in C. dubliniensis results in gain of function and that there is functional diversity amongst members of the gene family. We propose that this expansion of the TLO family contributes to the success of C. albicans as a commensal and opportunistic pathogen.
Introduction
Candida species (spp.) are an important component of the human microbiota. They are found in a wide range of anatomic niches, particularly in the gastrointestinal and vaginal tracts and if host conditions provide an opportunity they can evade immune responses and cause a spectrum of diseases, ranging from superficial infections of the mucosae to life threatening systemic infections in severely immunocompromised patients. In particular, Candida spp. have been cited as the fourth most common cause of nosocomial bloodstream infections [1].
The most pathogenic Candida species are C. albicans, C. parapsilosis, C. tropicalis and C. glabrata. [2]. C. albicans is by far the most commonly identified cause of candidiasis and is often regarded as the most pathogenic fungal species in humans. C. albicans is a highly versatile microorganism that has the ability to activate rapid transcriptional responses in order to adapt to changing environmental conditions, potentially allowing it to colonise and infect multiple anatomic sites [3].
One of the reasons why C. albicans is more pathogenic than other Candida spp. is that, apart from Candida dubliniensis, it is the only truly dimorphic Candida species, having the ability to switch between yeast and filamentous forms of growth [4]. Hyphal cells and hypha-specific proteins are well documented contributors to virulence and facilitate adherence to the host, penetration of tissues and the formation of biofilms [5–7].
Candida dubliniensis, which was first identified in 1995 [8] is very closely related to C. albicans. The genomes of both species are highly similar (i.e. 98% synteny [9]) and they share many phenotypic traits, including the capacity to form hyphae. Despite their very close relationship C. dubliniensis has been shown to be far less virulent than C. albicans and is only rarely found to be the cause of systemic infections [10]. Comparison of the genomes to identify the underlying genetic differences for the disparity in virulence found gene family size as the primary differentiating feature. Some of these differences were found in gene families known to contribute to virulence (e.g. the agglutinin-like sequence (ALS) and secretory aspartyl proteinase (SAP) gene families), however, one of the greatest differences was observed in the composition of the TLO (TeLOmere-associated) family in each species. The C. albicans SC5314 genome contains 15 TLO genes compared with just two in C. dubliniensis [9]. In C. albicans, as the name suggests, the TLO genes are situated close to the telomeres of each chromosome. The C. albicans TLO genes can be divided into four distinct clades based on the structure of their genes (Fig 1A). These include the relatively highly expressed α clade containing six members, a single β clade gene, the γ clade containing seven members, and the ψ clade containing a single pseudogene member [11–13]. TLO copy number varies between strains (10–15), with variation in the number of α and γ clade genes [14]. However, a single β clade gene is present in all genomes analysed to date [14]. The N-terminus of the C. albicans TLO genes encodes a conserved Med2 domain and these genes are now known to encode the Med2 component of the Mediator complex [11,15]. Mediator is a large multi-subunit protein complex which is conserved throughout eukaryotes and mediates interaction between RNA polymerase II and the machinery used in the initiation of transcription at target gene promoters [16,17]. Uwamahoro et al. (2012) demonstrated that C. albicans Mediator has a role in the expression of genes related to virulence traits [17] and it has also recently been demonstrated to play a role in resistance to antifungal drugs [18,19]. Tlo/Med2 forms a part of the Tail module of Mediator, along with Med3 and Med15 [15]. Different Tlos are found at different levels in C. albicans and C. dubliniensis [15] suggesting that there are pools of Mediator in each species with a different Med2 component. Given the size of the TLO family in C. albicans it has also been proposed that there is a substantial pool excess of “Mediator-free” Tlo in this species [11].
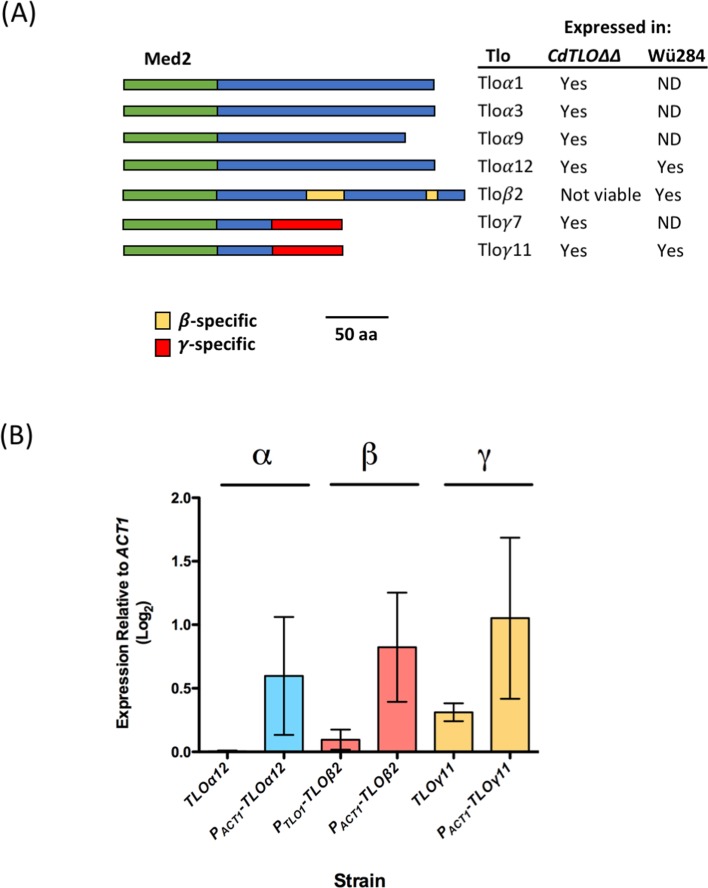
Fig 1. Structure and expression of C. albicans TLO genes.
(A) Diagram comparing the structure of the Tlo proteins analysed in this study, based on the models of Anderson et al. [11]. The green box represents the conserved Med2-like domain. The blue box represents the clade-specific c-terminus. The γ- and β-specific regions are indicated by yellow and red boxes, respectively. The table on the right indicates which genes have been expressed in wild-type C. dubliniensis Wü284 and the TLO null derivative ΔΔtlo. ND = not done. (B) RT-PCR expression data of C. albicans TLOs expressed in the C. dubliniensis WT Wü284 strain. RT-PCR expression graphs represent the results of three independent experiments.
Deletion of the two TLO genes in C. dubliniensis resulted in defects in activation of transcriptional responses associated with a number of virulence traits including tolerance of oxidative stress and hypha formation [12], while overexpression of CdTLO2 (and creation of a pool of “free” Tlo), but not CdTLO1, in C. dubliniensis results in hyperfilamentation [20]. As well as confirming the role of Med2 in virulence, the data from C. dubliniensis suggest differences in functionality amongst the two Tlo proteins expressed in that species.
We have previously proposed that the increased virulence of C. albicans compared to other Candida species may be due to an increased transcriptional flexibility due to its expanded family of Tlo proteins which may have differences in functionality [12]. Evidence that individual Tlo proteins have specific function(s) in C. albicans has recently been provided by Dunn et al. [21] who investigated the phenotypic effect of controlling the expression of individual TLO genes using a Tet-ON system. In order to investigate our hypothesis, we have added to the repertoire of TLO genes in C. dubliniensis by heterologously expressing representative C. albicans TLO genes in a wild-type C. dubliniensis strain. The rationale for selecting C. dubliniensis as the host species was due to the low copy number of native TLO genes in this species as we reasoned that it could be difficult to determine a phenotypic effect in a C. albicans background of 15 TLO genes. In addition, we also attempted to investigate the functional diversity within the C. albicans TLO gene family, by expressing TLO representative CaTLO genes in a C. dubliniensis ΔΔtlo double mutant and identified the phenotypes conferred by each gene. We propose that our data demonstrate possible evolutionary advantages associated with TLO gene duplication and diversification.
Materials & methods
Candida strains & culture conditions
The strains of Candida spp. used in this study and their genotypes are listed in S1 Table. All Candida strains were routinely grown on Yeast Extract Peptone Dextrose (YEPD) agar at 37°C. Nourseothricin-resistant transformants were cultured on YEPD agar containing nourseothricin [100 μg/ml (NAT100)]. Lee’s Medium [22] and Spider medium [23] were used to induce filamentation. Cornmeal agar supplemented with 1% Tween® 80 was used for chlamydospore formation.
For spot plate assay experiments, a suspension of 2 x 106 cells/ml was prepared from overnight cultures and 7 μl from serial dilutions (100 to 10−4) were spotted onto YEPD agar plates containing the indicated agents. The plates were incubated at 37°C for 48 h in a static incubator. Growth was recorded using a Flash n’ Go plate visualizer (IUL Instruments). Each experiment was carried out on three separate occasions.
For liquid culture, YEPD broth was used in an orbital incubator at 200 r.p.m. at the indicated temperature. In order to determine the doubling times of strains, the optical densities of cultures were measured at 600nm during the exponential growth phase and plotted using Prism GraphPad (GraphPad, CA, USA). Doubling times were calculated from 3 replicate growth curves. Galactose (2% w/v) was substituted for glucose where indicated. Induction of filamentation in liquid cultures was carried out with cells from overnight YEPD broth cultures grown at 30°C, which were washed twice with sterile Milli-Q water (Millipore Ireland B.V., Co. Cork, Ireland) and added to hyphal-induction medium (10% v/v foetal bovine serum in dH2O) to a density of 2 x 105 cells/ml in a six-well tissue culture plate at 37°C. The numbers of true hyphal cells were quantified using a Nikon E600 microscope and a Nikon TMS-F inverted light microscope (Nikon U.K., Surrey, U.K.). Experiments were carried out on three separate occasions.
Heterologous expression of C. albicans TLO genes in C. dubliniensis
C. albicans TLO genes β2, γ11 and α12 were heterologously expressed in the C. dubliniensis wild type strain Wü284. TLOγ11 and α12 were expressed under the control of their native promoters, however the TLOβ2 promoter sequence is incomplete in the SC5314 genome sequence so a fusion gene with the TLOα1 promoter (S1 File) was synthesised by GeneWiz (Essex, UK) and inserted in the XhoI and HindIII restriction endonuclease sites of pCDRI [24]. TLOγ11 and α12 were amplified from SC5314 using gene-specific primers (S2 Table) containing recognition sequences for XhoI and HindIII restriction endonucleases. Digested amplimers were ligated to XhoI/HindIII cut pCDRI plasmid using T4 DNA ligase (Promega, Wisconsin, USA) and transformed into E. coli XL10 competent cells (Sigma-Aldrich, Missouri, USA) using standard protocols. Transformants were selected on pre-warmed Lysogeny (L) agar supplemented with 100 μg/ml ampicillin. Plasmid pCDRI and its derivatives were linearised with Eco47III and transformed in C. dubliniensis as described by Staib et al. [25]. Transformants were selected on YEPD agar containing 100 μg/ml nourseothricin. Additional plasmid constructs containing TLOs β2, γ11 and α12 were also generated in pGM161, which is a derivative of pCDRI allowing expression from the ACT1 promoter, using the same cloning strategy [24].
C. albicans TLO genes α1, α3, γ7, α9, γ11 and α12 were heterologously expressed in the C. dubliniensis ΔΔtlo double mutant under the control of their native promoters. Each gene was PCR amplified and cloned in pCDRI and introduced in the ΔΔtlo double mutant as described above.
cDNA synthesis and qualitative real-time PCR
RNA was extracted and used to generate cDNA as described by Flanagan et al. [26]. qRT-PCR was carried out on the Applied Biosystem 7500 Fast Real Time PCR System as described by Flanagan et al. [26]. Plates were set up in triplicate with the endogenous control, ACT1, run alongside each target. Results were exported into Microsoft Excel and the delta Ct values calculated for each sample. These were ultimately graphed using GraphPad Prism version 6 (San Diego, California, USA, www.graphpad.com).
Biofilm induction assays
Biofilm mass was determined using crystal violet to quantify biomass. Cells were grown in YEPD at 37°C overnight with shaking at 200 rpm. Following overnight incubation, 100 ml was removed and transferred to YNB with 100 mM glucose and incubated overnight at 37°C with shaking at 200 rpm. Following the second night of incubation, cells were washed in 1X PBS and resuspended in 1 ml of YNB with 100 mM glucose at a cell density of 2 x 106 cells/ml. 100 μl of each strain was placed in triplicate into a 96 well plate and incubated at 37°C for 90 min. Following incubation, the medium was aspirated, and cells washed twice with 150 ml 1X PBS. A 100 μl volume of YNB containing 100 mM glucose was placed onto the washed cells and the plates were incubated at 37°C for 24 and 48 hr. Following incubation, the wells were washed three times with 200 μl sterile 1X PBS to remove non-adherent cells and 110 μl of 0.4% (v/v) crystal violet was added to each well and stained at room temperature for 45 min. The crystal violet was removed and each well washed with 200 μl of dH2O three times. The wells were de-stained with 200 μl of 95% (v/v) ethanol for 45 min. A 100 μl aliquot of each suspension was transferred to a new 96-well plate and the absorbance measured at an OD540 using a Tecan Plate Reader system (Tecan). Results were analysed using GraphPad Prism version 6.
In vivo infection model
Candidal virulence was assessed using the wax moth larva Galleria mellonella obtained from Live Foods Direct (Sheffield, England). Larvae were stored at 15°C in wood shavings in the dark prior to use and those that weighed between 0.20 to 0.30 g were used within 2 weeks of receipt. For each infection experiment, 10 larvae were placed into sterile 9 cm petri dishes lined with Whatman filter paper and wood shavings. Candida strains were assigned a random code prior to each experiment to facilitate blind assessment of virulence. For infection, 1 X 106 yeast cells in 20 μl were injected into the haemocoel via the last left pro-leg with a 30G insulin U-100 Micro-Fine syringe (BD New Jersey, USA) as described by Cotter et al. (2000) [27]. The inoculated larvae were incubated at 30°C and larval mortality was assessed at 24 h intervals, as described by Cotter et al. (2000) [27]. The results were analysed using GraphPad Prism version 6.
Results
Expression of Candida albicans TLO genes in wild type C. dubliniensis
In order to investigate the effect on phenotype of expanding the repertoire of TLO genes in C. dubliniensis, we expressed the C. albicans TLOβ2, TLOγ11 and TLOα12 genes (representing each of the three CaTLO clades) in the C. dubliniensis WT Wü284 background under the control of a native TLO promoter and that of the constitutively expressed ACT1 gene. In the case of TLOβ2 for which no promoter sequence was available, we used the TLOα1 promoter as a proxy native promoter as both TLOβ2 and TLOα1 exhibit similar mRNA expression levels [11].
Quantitative Real Time PCR was used to determine the level of expression of each TLO under the expression of their native promotor and that of the ACT1 gene (Fig 1B). The level of each TLO expressed under their native promotor was lower compared with that of the ACT1 gene. TLOβ2, under the expression of the native TLOα1 promoter, was expressed at 0.1 relative to ACT1. When placed under the expression of the ACT1 gene, the expression increased to 1.31 relative to ACT1, a fold-change of 13.1. TLOγ11 under the control of its native and the ACT1 promoters was expressed at 0.45 and 1.05, respectively, relative to ACT1, a fold-change of 2.3. Similarly, TLOα12 under the control of the native promoter and ACT1 gene showed expression levels of 0.005 and 0.5 relative to ACT1, a fold-change of 118.
Once the level of expression of each gene had been determined, a range of phenotypic tests was then performed to determine whether the expression of additional C. albicans TLO genes had the ability to affect the phenotype of the host strain.
TLOβ2 expression in wild type C. dubliniensis results in hyperfilamentous growth
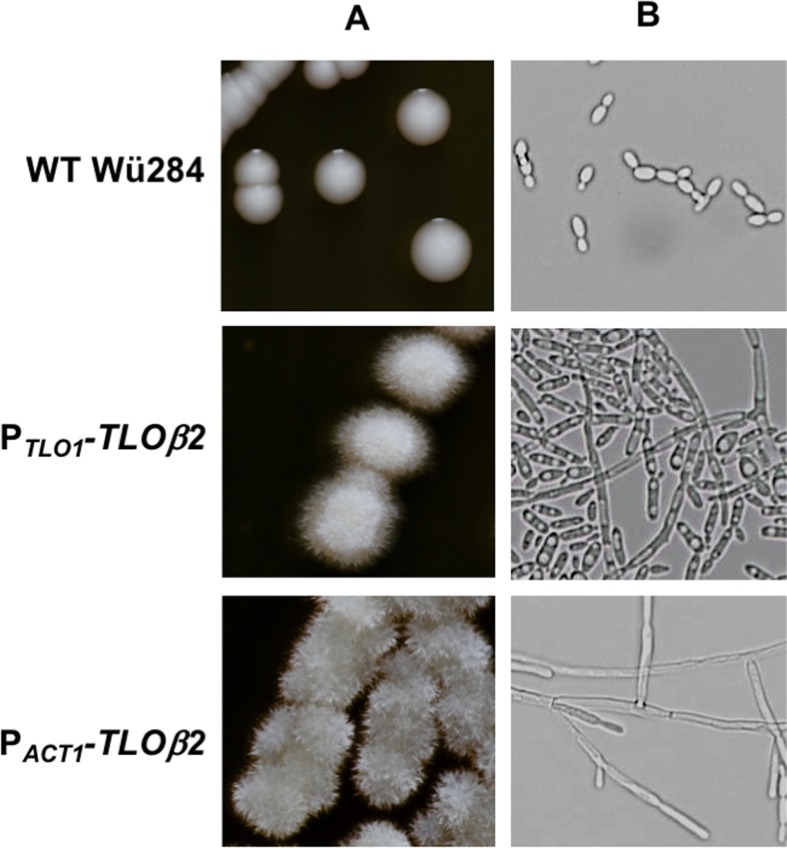
In wild type C. dubliniensis Wü284, expression of TLOγ11 and TLOα12 under the native or ACT1 promoter did not affect the colony morphology of the strain on YEPD agar. However, TLOβ2 whether expressed under the comparatively weak TLOα1 promoter or the ACT1 promoter in wild-type C. dubliniensis resulted in wrinkled colonies on YEPD agar and hypha formation in YEPD broth (Fig 2). This phenotype was affected by expression levels of TLOβ2, with the TLOα1 promoter variant exhibiting a predominantly pseudohyphal mode of growth in YEPD broth, and the ACT1 variant producing longer filaments with evidence of true hyphal growth (Fig 2B).
Fig 2. C. albicans TLOβ2 confers filamentous growth in C. dubliniensis.
Colony (A) and cellular (B) morphology of C. dubliniensis WT Wü284 and derivatives harboring TLOβ2 expressed from the TLO1 promoter (PTLO1-TLOβ2) and the ACT1 promoter (PACT1-TLOβ2). Colonies were grown for 48 h on solid YEPD agar. The morphology of the cells in representative colonies of each derivative was visualised using a x40 objective lens.
TLO expansion in wild type C. dubliniensis affects growth rate
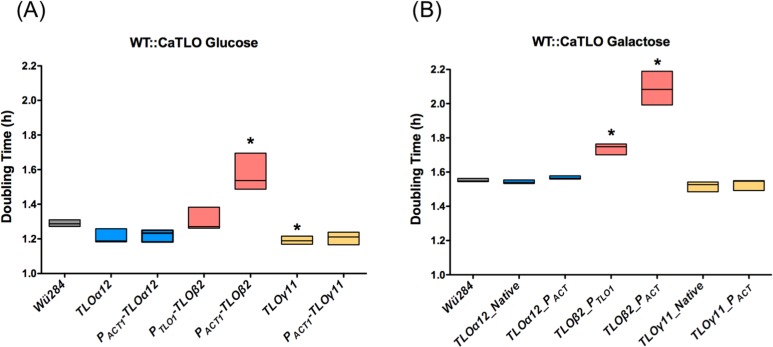
It has previously been shown that deletion of the two TLO genes present in the C. dubliniensis genome leads to reduced growth rate in YEPD and also results in greatly increased doubling times when galactose is the sole source of carbon [12]. To investigate the ability of an expanded TLO gene repertoire to affect growth rate we cultured all strains in YEP-Glucose and YEP-Galactose over an 8 h time course. In WT C. dubliniensis Wü284, expression of TLOα12 under the control of its native or the ACT1 promoter did not affect doubling times in YEP-Glucose or YEP-Galactose (Fig 3). Expression of TLOγ11 under its native promoter in WT Wü284 reduced the doubling time in YEP-Glucose by approximately 10 min (Fig 3A). Expression of TLOβ2 had the effect of greatly reducing growth in both media, and this was most significant in strain PACT1-TLOβ2 compared to PTLO1-TLOβ2 (Fig 3B). This effect on growth rate is most likely due to the filamentous morphology exhibited by these strains (Fig 2).
Fig 3. The effect of C. albicans TLO genes on growth rates in YEP-Glucose and -Galactose broth.
Doubling times of WT Wü284 and derivatives expressing the indicated C. albicans TLO genes in YEP-Glucose (A) and -Galactose (B). Asterisks indicate significant difference from Wü284. Data were generated in three replicate experiments.
TLO expansion in wild type C. dubliniensis enhances H2O2 resistance
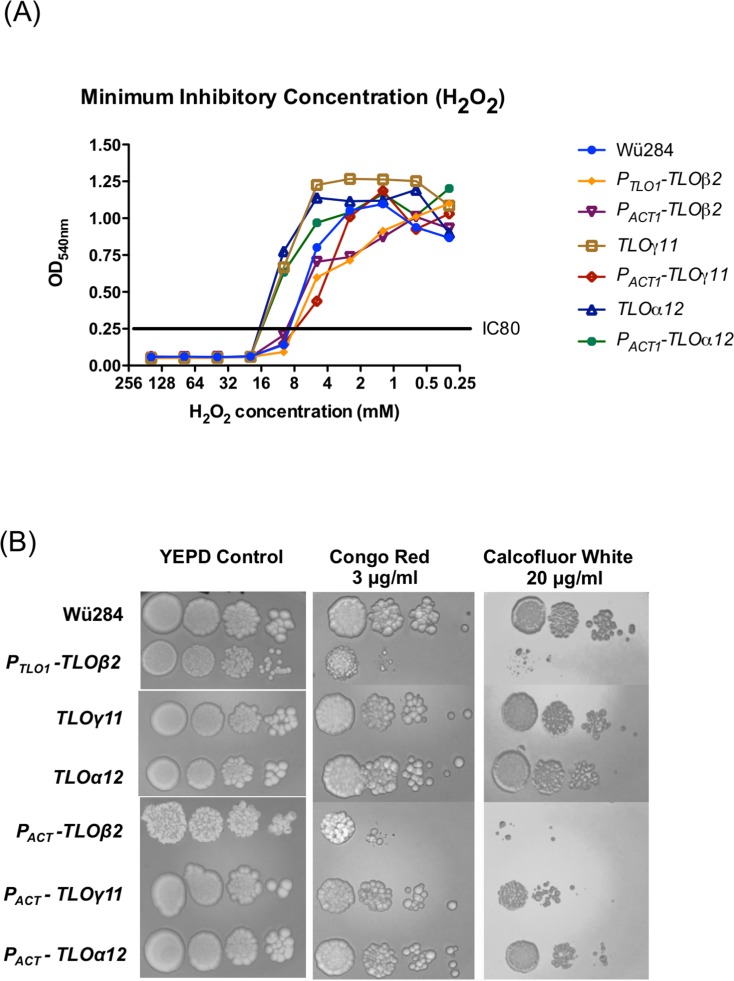
Using a broth dilution MIC test for H2O2 we showed that expression of TLOγ11 and TLOα12 in WT C. dubliniensis Wü284 under their native promoters led to a doubling of MIC80 from 10mM to 20mM. TLOα12 expressed using the ACT1 promoter also led to a similar increase in MIC80. Using this assay, expression of TLOβ2 under the control of the TLOα1 or ACT1 promoters did not affect susceptibility to H2O2 (Fig 4A).
Fig 4. The effect of C. albicans TLO genes on susceptibility to H2O2 and cell wall damaging agents.
(A) Minimum inhibitory concentration of H2O2 was determined by broth dilution. The IC80 is indicated and shows the concentration of H2O2 that reduced growth of the derivatives tested below 80% of the inhibitor-free control. (B) Ten-fold serial dilutions (left to right) of 2 x 104 cells were spotted on to plates containing 3 μg/ml Congo Red and 20 μg/ml Calcofluor White.
TLOβ2 increases susceptibility to cell wall damaging agents
In order to determine if the C. albicans TLO genes differ in their effects on cell wall stress responses we compared the effect of these genes on growth on media containing the β-1,3-glucan-binding dye Congo Red and the chitin-binding dye Calcofluor White. Expression of TLOγ11 and TLOα12 in WT C. dubliniensis Wü284 under their native promoters did not affect susceptibility to Congo Red or Calcofluor White, while expression of TLOβ2 under the control of the TLOα1 or ACT1 promoters in wild type C. dubliniensis resulted in increased susceptibility to both agents (Fig 4B).
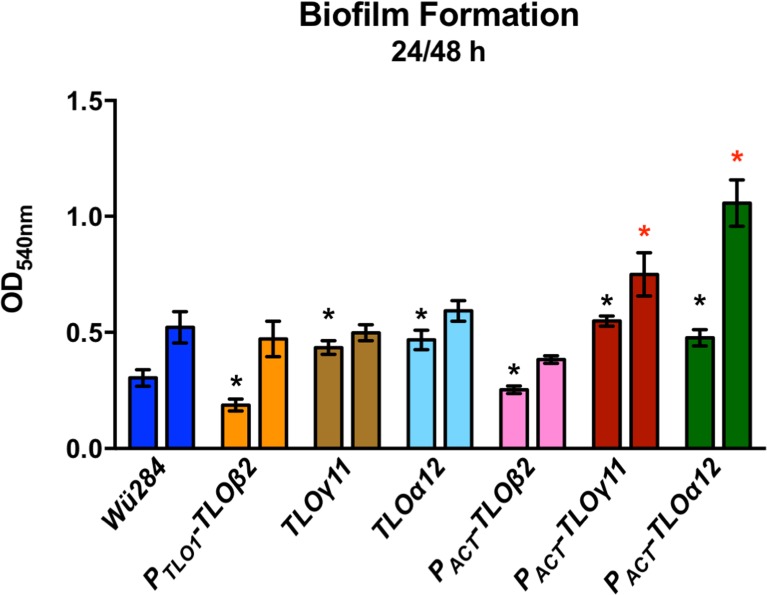
Biofilm formation
The ability to form biofilm on plastic surfaces following 24 h and 48 h incubation was assessed using a Crystal Violet staining assay (Fig 5). Expression of TLOβ2 in strain Wü284 resulted in a significant decrease in biofilm formation at 24 h (Fig 5). Expression of TLOγ11 and TLOα12 resulted in a higher degree of biofilm formation at the 24 h timepoint (Fig 5). The greatest increase in biofilm formation relative to Wü284 was observed in the PACT1-TLOγ11 and PACT1-TLO α12 expressing strains, which exhibited increased biofilm at 24 h and 48 h (Fig 5).
Fig 5. The effect of C. albicans TLO genes on biofilm formation on plastic surfaces.
Each strain was grown in Spider medium in a 96-well plate for 48 h and biomass measured using a Crystal Violet assay. An asterisk indicates significant differences with * at 24 h and red * at 48 h.
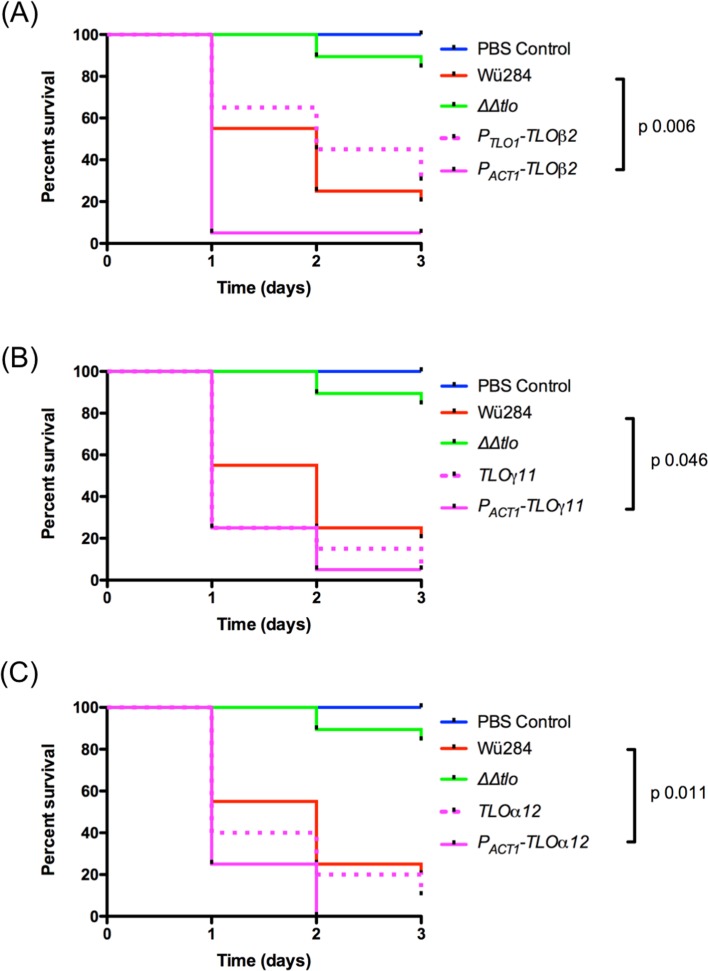
TLO expansion in wild type C. dubliniensis enhances virulence in the Galleria mellonella infection model
Given the differential effects of specific TLO genes on various virulence attributes, such as morphology, stress tolerance and cell wall integrity, we decided to investigate if differences in virulence could be detected using an in vivo infection model. Virulence of WT C. dubliniensis Wü284 expressing TLOβ2, TLOγ11 and TLOα12 from a native TLO promoter or the ACT1 promoter was investigated using the insect larval G. mellonella model. Although genes expressed from native TLO promoters did not confer significant increases in virulence, expression of these C. albicans TLO genes in WT C. dubliniensis under the control of the ACT1 promoter significantly enhanced virulence in this larval infection model (Fig 6). This effect was most significant in the case of the PACT1- TLOβ2 expressing strain.
Fig 6. The effect of C. albicans TLO genes on the virulence of wild-type Wü284.
Ten G. mellonella larvae were inoculated with 1 X 106 cells of each indicated strain (performed blind) and viability was monitored over 3 days. Results presented represent three independent infection experiments. P values indicate results of a Log-Rank (Mantel-Cox) test against the wild type Wü284 survival curve.
Expression of Candida albicans TLO genes in the C. dubliniensis Δtlo1/Δtlo2 (ΔΔtlo) double mutant
In order to further investigate the range of phenotypes regulated by C. albicans Tlo proteins, we also expressed representative members of the C. albicans TLO gene family in the C. dubliniensis Δtlo1/Δtlo2 (ΔΔtlo) double mutant under the control of their native upstream regulatory elements. Several attempts were made to generate stable transformants expressing TLOβ2, however, no viable transformants were recovered in these experiments. Quantitative Real Time PCR was used to determine the level of expression of the C. albicans TLOs in the C. dubliniensis ΔΔtlo backgrounds (S1 Fig). TLO 1, 3, 9 and 12, all of which belong to the αclade, show similar expression levels of expression relative to ACT1 (0.016 to 0.088, Fig 1B). Interestingly, the ɣclade genes TLOγ7 (0.38 relative to ACT1) and TLOγ11 (0.006 relative to ACT1) differed greatly in expression levels compared with one another (S1 Fig).
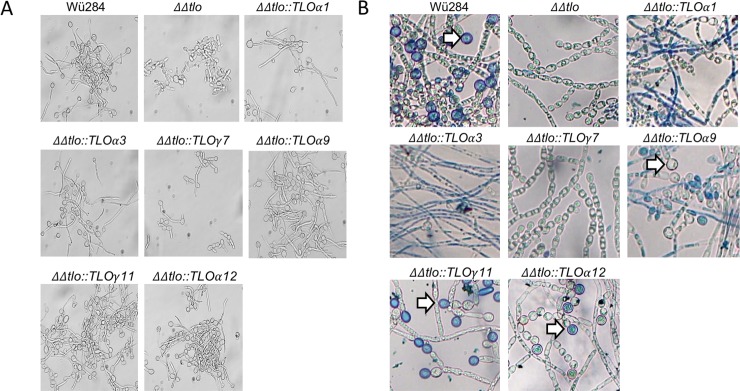
All TLO genes tested, with the exception of TLOγ7, restored filamentous growth in the C. dubliniensis ΔΔtlo mutant, which is normally not capable of forming true hyphae (Fig 7A). TLOα9, γ11 and α12 restored the ability to produce chlamydospores in the deletion mutant, while TLOα1, α3 and γ7 were unable to do so (Fig 7B).
Fig 7. Morphology of C. dubliniensis ΔΔtlo expressing CaTLO genes.
A. Photomicrographs of C. dubliniensis ΔΔtlo and derivatives harboring the indicated C. albicans TLO genes following 4 h growth in water supplemented with 10% (v/v) foetal bovine serum at 37°C. B. Chlamydospore formation of C. dubliniensis ΔΔtlo and derivatives harboring the indicated C. albicans TLO genes on cornmeal agar supplemented with tween. Chlamydospores are indicated by arrows. Identical results were observed in replicate experiments.
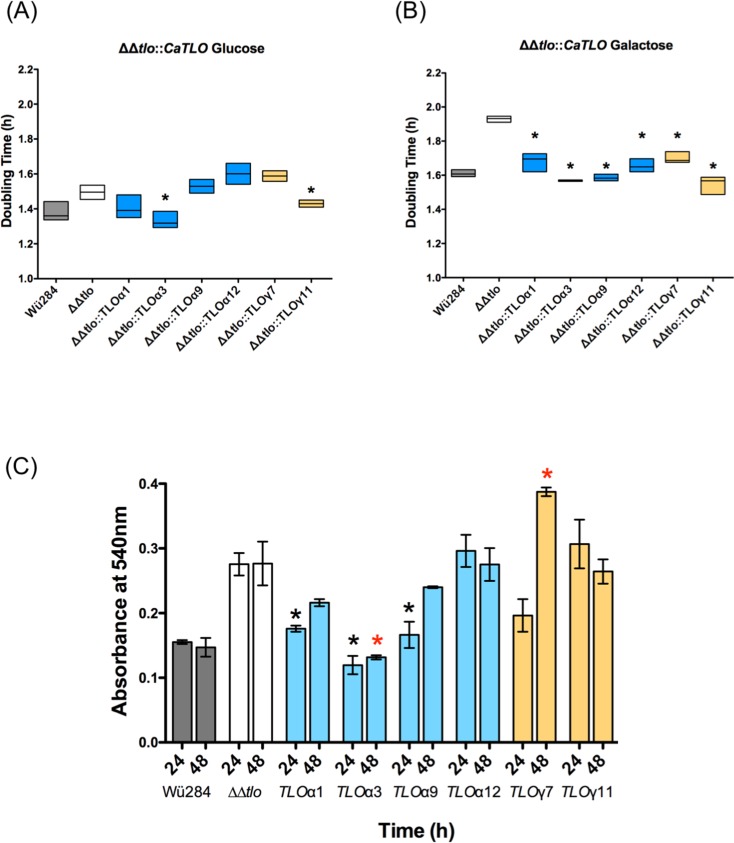
All of the TLO genes complemented the defective growth of the ΔΔtlo mutant in YEP-Galactose (Fig 8A and 8B). The ΔΔtlo C. dubliniensis mutant has previously been shown to produce excess levels of biofilm on plastic surfaces relative to wild type [12]. Following 24 h growth under biofilm forming conditions, the ΔΔtlo::TLOα1, α3, and α9 strains exhibited reduced biofilm formation relative to the ΔΔtlo double mutant and comparable to that observed with WT Wü284. The remaining genes tested either resulted in similar or greater (e.g. TLOγ7) levels of biofilm than the ΔΔtlo double mutant (Fig 8C).
Fig 8. Growth of C. dubliniensis ΔΔtlo and derivatives harboring C. albicans TLO genes.
A and B show doubling times of WT Wü284, the ΔΔtlo double mutant and derivatives expressing indicated C. albicans TLO genes in YEP-Glucose (A) and -Galactose (B). Stars indicate strains exhibiting doubling times significantly different from ΔΔtlo (p ≤ 0.05). Panel C shows biofilm formation on plastic surfaces. Each ΔΔtlo::TLO strain was grown in the presence of YEPD in a 96-well plate for 48 h. Biomass was measured using a crystal violet assay in three replicate experiments. Asterisks indicate significant differences from ΔΔtlo at 24 h (*) and 48 h (red *), respectively. Data are the result of three independent experiments.
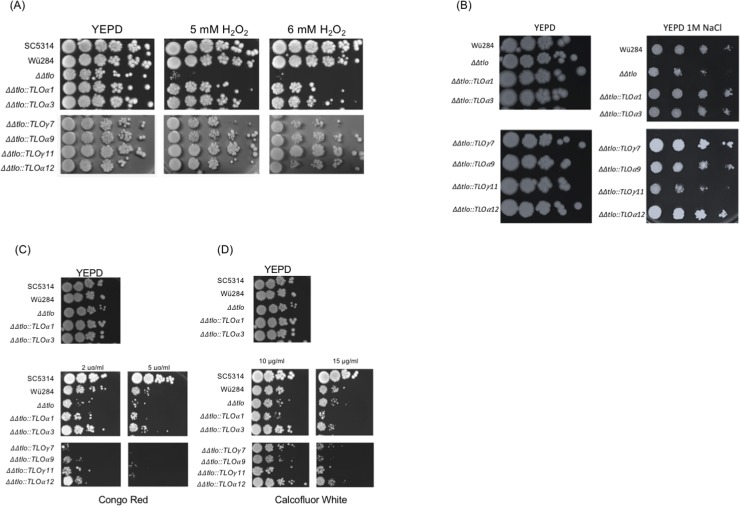
TLOγ11 also conferred increased resistance to oxidative stress. At a concentration of 6 mM H2O2 ΔΔtlo::TLOγ11 had the greatest effect on enhancing tolerance of oxidative stress, with the remaining genes conferring tolerance, but to a lesser extent (Fig 9A). When incubated on solid YEPD supplemented with 1 M NaCl, all CaTLO genes tested, with the exception CaTLOγ11, resulted in increased growth compared to the ΔΔtlo mutant strain (Fig 9B). In the presence of the cell wall perturbing compounds Congo Red (2 μg/ml) and Calcofluor White (10 μg/ml) TLOα3 consistently restored growth of the ΔΔtlo mutant to wild-type levels. ΔΔtlo::TLOα12 also exhibited enhanced levels of growth on Calcofluor White (10 μg/ml) compared to the ΔΔtlo mutant (Fig 9C and 9D).
Fig 9. C. albicans TLO genes differentially affect tolerance of environmental stress conditions.
Growth of each ΔΔtlo::CaTLO strain in the presence (A) H2O2, (B) NaCl, (C) Congo Red and (D) Calcofluor White. Ten-fold serial dilutions (left to right) of 2 x 104 cells were spotted onto YEPD agar and YEPD agar containing the indicated agents. Plates were incubated for 48 h at 37°C.
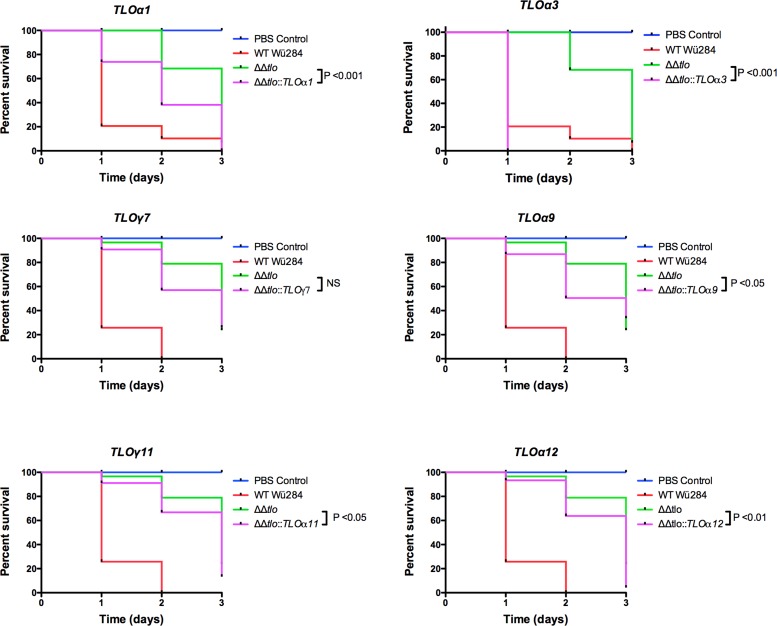
Finally, examination of the virulence of the CaTLO expressing strains in the Galleria mellonella model showed that ΔΔtlo::TLOα3 enhanced virulence to the greatest extent, with survival rates significantly less than the parental ΔΔtlo mutant and similar to the WT strain (Fig 10). The remaining TLO genes tested were shown to result in a restoration of virulence in the infection model with mortality rates greater than that of the ΔΔtlo double mutant but less than the WT Wü284.
Fig 10. The effect of C. albicans TLO genes on the virulence of the ΔΔtlo mutant.
Ten G. mellonella larvae were inoculated with 1 X 106 cells of each indicated strain (performed blind) and viability was monitored over 3 days. Results presented represent three independent infection experiments. P values indicate results of a Log-Rank (Mantel-Cox) test against the ΔΔtlo mutant survival curve.
Discussion
One of the largest gene families in C. albicans is the TLO family, which consists of up to 15 members, each encoding a protein orthologous to the Med2 subunit of the transcriptional regulator complex Mediator [28]. This expansion is unique to C. albicans and there is significant variation in the copy number of genes in this family between different strains [14]. There are only two TLO genes encoded in the genome of C. dubliniensis, the species most closely related species to C. albicans. Deletion of the two TLO genes in C. dubliniensis resulted in significant transcriptional and phenotypic defects, including an inability to produce hyphae and reduced tolerance of oxidative stress. Reintroduction of each of the C. dubliniensis TLO genes into the double mutant background indicated that the CdTLO1 and CdTLO2 genes differ in their ability to complement the mutant phenotypes, suggesting they may have distinct functions in gene control [12]. The purpose of the current study was to investigate if expansion of this two-membered family in C. dubliniensis affects phenotypes associated with virulence. Such a finding would lend support to our hypothesis that expansion of the C. albicans TLO gene family played a role in the evolution of the enhanced virulence of this species in comparison with other related species.
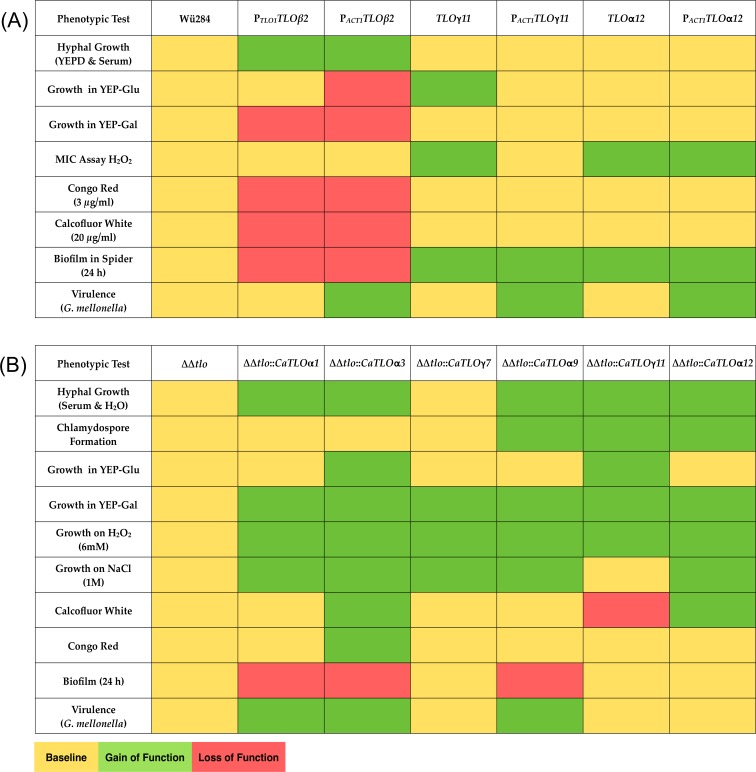
We expanded the repertoire of Tlo proteins in C. dubliniensis Wü284 using representatives of the α, β, and γ TLO families, namely TLOβ2, TLOγ11 and TLOα12 and a summary of the effects of this expansion is shown in Fig 11A. Expression of TLOβ2 had the most dramatic effect on morphology, resulting in the production of wrinkled colonies containing cells with hyphal morphologies. Interestingly, the extent of this phenotype was influenced by the expression level of TLOβ2, with the ACT1-promoter driven gene resulting in more highly-wrinkled colonies with a higher proportion of true hyphae. The more pronounced phenotype in the highly expressed construct indicates that expression level influences phenotype. This affect may be exerted by displacing endogenous CdTlo1 and CdTlo2 from the Mediator complex, therefore promoting Tloβ2 regulated functions. Alternatively, the higher expression levels may create a pool of Tlo in excess of Mediator. A similar phenotype was recently described following overexpression of CdTlo2 to create a Mediator excess population of Tlo in C. dubliniensis [20]. Unexpectedly, TLOβ2 could not be expressed in the C. dubliniensis ΔΔtlo double mutant, suggesting that a Mediator complex exclusively containing Tloβ2 is lethal to the cell.
Fig 11. Heat maps summarizing phenotypic effects of CaTLO genes.
The phenotypic effects of expressing each CaTLO gene in the C. dubliniensis wild-type (panel A) and in C. dubliniensis ΔΔtlo (panel B) are colour coded; yellow indicates the same phenotype as the mutant, green indicates a gain of function and red indicates a loss of function.
Although wild-type Wü284 expressing TLOγ11 and TLOα12 were not affected in morphology, these strains exhibited a specific enhanced resistance to H2O2, whereas the TLOβ2 expressing strains which were filamentous were highly susceptible to oxidative and cell wall stress, suggesting that the morphology of the cells may contribute to their ability to tolerate stress. Expression of TLOβ2, TLOγ11 and TLOα12 in wild type Wü284 had varying effects on growth rates in vitro. TLOβ2 generally increased doubling times in YEP-Glucose and -Galactose and this might be related to the hyper-filamentous, polarised growth pattern exhibited by this strain. In contrast, TLOγ11 transformants exhibited a small (approximately 10 min) but significant reduction in doubling time in YEP-Glucose.
Infection of G. mellonella larvae with these strains showed that expression of the heterologous genes at low level using native TLO promoters had limited effects on larval survival, however expression under the control of the stronger ACT1 promoter resulted in significantly reduced larval survival (similar to C. albicans SC5314). These data for the first time provide experimental evidence supporting enhanced fitness as a result of TLO copy number expansion in Candida species. It is interesting to note that the phenotypic effect was most significant when the genes were expressed under the control of the stronger ACT1 promoter, indicating that a critical level of Tlo is required for this gain of function. This may partly explain why C. albicans has expanded the TLO family to such a significant extent.
In the second part of our study, to better understand the diversity of functions regulated by TLOs, we expressed a range of C. albicans TLO genes in a ΔΔtlo C. dubliniensis background. In general, heterologous expression of the C. albicans TLO genes in the ΔΔtlo C. dubliniensis background could restore all phenotypes examined, including some subtle and some major differences in the ability of individual paralogs to complement the phenotypes. A summary of these phenoptypes is shown in Fig 11B. It is clear from this heatmap that individual CaTLOs differ in their ability to affect specific phenotypes and in the magnitude of their restorative capability. For example, TLOα1, α3 and γ7 do not have the ability to restore chlamydospore production in the mutant, while TLOγ11 and α12 did not suppress biofilm formation (despite being active inducers of stress responses). The exception to this was TLOγ7, which despite having the highest expression level of all of the C. albicans TLOs (0.38 relative to ACT1) tested in the ΔΔtlo C. dubliniensis background, had the least effect on restoring the phenotypes in the mutant. TLOγ7 failed to restore the ability to form hyphae and chlamydospores or growth in media containing cell perturbing compounds. However, TLOγ7 did restore growth in YEP-Gal and tolerance of sodium chloride and H2O2, indicating that the gene possesses some functionality. In order to investigate if there are differences in the effects of specific CaTLO genes on virulence we tested the virulence of strains in the Galleria mellonella larval infection model. TLOα3 was found to restore virulence in the C. dubliniensis ΔΔtlo mutant to a greater extent than the other TLO genes tested, suggesting a clear disparity in the activity of specific TLO genes.
These data complement the findings of Dunn et al. [21] who used a ‘Tet-ON’ missexpression system to probe individual TLO genes in C. albicans SC5314. The authors concluded that TLOs controlled multiple phenotypes and that single phenotypes were often regulated by multiple TLOs, including virulence in G. mellonella. However, it is difficult to directly compare the results of individual phenotypic tests in both studies due the different nature of the host strains (C. albicans and C. dubliniensis) and the phenotypic tests used.
In summary, the C. albicans TLO gene family is comprised of fifteen genes, mainly situated in the subtelomeric region of the chromosomes. These regions have been shown in other organisms to undergo rapid evolution and gene families in the subtelomeres have been demonstrated to expand rapidly and undergo functional divergence [29]. Our data support the hypothesis that there is functional diversity in the C. albicans TLO gene family and also indicate that the high copy number of TLO genes in C. albicans may have evolved to increase gene dosage, which in our larval infection model has a significant effect on virulence. Studies are now underway to confirm these hypotheses by attempting to deplete the TLO gene family in C. albicans using CRISPR-Cas9 mutagenesis.
Supporting information
RT-PCR expression data of respresentative C. albicans TLO genes expressed in the C. dubliniensis ΔΔtlo mutant strain. RT-PCR expression graphs represent three independent experiments.
(TIF)
(DOCX)
(DOCX)
(TXT)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The project was supported by the Dublin Dental University Hospital and Science Foundation Ireland (11/RFP.1/GEN/3042). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang YL. Virulence factors of Candida species. J Microbiol Immunol Infect. 2003;36: 223–228. [PubMed] [Google Scholar]
- 2.Mendes Giannini MJS, Bernardi T, Scorzoni L, Fusco-Almeida AM, Sardi JCO. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62:10–24. 10.1099/jmm.0.045054-0 [DOI] [PubMed] [Google Scholar]
- 3.Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr Opin Microbiol. 2004;7: 336–341. 10.1016/j.mib.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 4.Gow NAR, Brown AJP, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5: 366–371. [DOI] [PubMed] [Google Scholar]
- 5.Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, De Koster CG, et al. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology. 2011;157: 2297–2307. 10.1099/mic.0.049395-0 [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Su C, Liu H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014;22: 707–714. 10.1016/j.tim.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69: 71–92. 10.1146/annurev-micro-091014-104330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141: 1507–1521. 10.1099/13500872-141-7-1507 [DOI] [PubMed] [Google Scholar]
- 9.Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, et al. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 2009;19: 2231–2244. 10.1101/gr.097501.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran GP, Coleman DC, Sullivan DJ. Candida albicans versus Candida dubliniensis: Why Is C. albicans more pathogenic? Int J Microbiol. 2012;2012: 205921–205927. 10.1155/2012/205921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson MZ, Baller JA, Dulmage K, Wigen L, Berman J. The three clades of the telomere-associated TLO gene family of Candida albicans have different splicing, localization, and expression features. Eukaryotic Cell. American Society for Microbiology; 2012;11: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haran J, Boyle H, Hokamp K, Yeomans T, Liu Z, Church M, et al. Telomeric ORFs (TLOs) in Candida spp. encode mediator subunits that regulate distinct virulence traits. PLoS Genet. 2014;10: e1004658 10.1371/journal.pgen.1004658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson MZ, Wigen LJ, Burrack LS, Berman J. Real-Time evolution of a subtelomeric gene family in Candida albicans. Genetics; 2015;200: 907–919. 10.1534/genetics.115.177451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015;25: 413–425. 10.1101/gr.174623.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A, Petrov KO, Hyun ER, Liu Z, Gerber SA, Myers LC. The Tlo proteins are stoichiometric components of Candida albicans mediator anchored via the Med3 subunit. Eukaryot Cell. 2012;11: 874–884. 10.1128/EC.00095-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conaway RC, Conaway JW. Function and regulation of the mediator complex. Curr Opin Genet Devel. 2011;21: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uwamahoro N, Qu Y, Jelicic B, Lo TL, Beaurepaire C, Bantun F, et al. The functions of mediator in Candida albicans support a role in shaping species-specific gene expression. PLoS Genet. 2012;8: e1002613 10.1371/journal.pgen.1002613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa JL, Boeszoermenyi A, Vale-Silva LA, Torelli R, Posteraro B, Sohn Y- J, et al. Inhibiting fungal multidrug resistance by disrupting an activator-mediator interaction. Nature. 2016;530: 485–489. 10.1038/nature16963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Myers LC. Mediator tail module is required for Tac1-activated CDR1 expression and azole resistance in Candida albicans. Antimicrob Agents Chemother. 2017;61: e01342–17. 10.1128/AAC.01342-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Moran GP, Sullivan DJ, Maccallum DM, Myers LC. Amplification of TLO mediator subunit genes facilitate filamentous growth in Candida spp. PLoS Genet. 2016;12: e1006373 10.1371/journal.pgen.1006373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn MJ, Kinney GM, Washington PM, Berman J, Anderson MZ. Functional diversification accompanies gene family expansion of MED2 homologs in Candida albicans. PLoS Genet. 2018;14: e1007326 10.1371/journal.pgen.1007326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycellal and yeast forms of Candida albicans. Sabouraudia: J Med Vet Mycol. 2009;13: 148–153 [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Köhler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994; 266: 1723–1726. [DOI] [PubMed] [Google Scholar]
- 24.Moran GP, Maccallum DM, Spiering MJ, Coleman DC, Sullivan DJ. Differential regulation of the transcriptional repressor NRG1 accounts for altered host‐cell interactions in Candida albicans and Candida dubliniensis. Mol Microbiol. 2007;66: 915–929. 10.1111/j.1365-2958.2007.05965.x [DOI] [PubMed] [Google Scholar]
- 25.Staib P, Moran GP, Sullivan DJ, Coleman DC, Morschhauser J. Isogenic strain construction and gene targeting in Candida dubliniensis. J Bacteriol. 2001;183: 2859–2865. 10.1128/JB.183.9.2859-2865.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flanagan PR, Liu N-N, Fitzpatrick DJ, Hokamp K, Köhler JR, Moran GP, et al. The Candida albicans TOR-activating GTPases Gtr1 and Rhb1 coregulate starvation responses and biofilm formation. mSphere. 2017;2: e00477–17. 10.1128/mSphere.00477-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol. 2000;27: 163–169. [DOI] [PubMed] [Google Scholar]
- 28.Van Het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8: R52 10.1186/gb-2007-8-4-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. Elsevier; 2010;20: 895–903. 10.1016/j.cub.2010.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR expression data of respresentative C. albicans TLO genes expressed in the C. dubliniensis ΔΔtlo mutant strain. RT-PCR expression graphs represent three independent experiments.
(TIF)
(DOCX)
(DOCX)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.