Abstract
Canine Inflammatory Bowel Disease (IBD) is considered a multifactorial disease caused by complex interactions between the intestinal immune system, intestinal microbiota and environmental factors in genetically susceptible individuals. Although IBD can affect any breed, German shepherd dogs (GSD) in the UK are at increased risk of developing the disease. Based on previous evidence, the aim of the present study was to identify single nucleotide polymorphisms (SNPs), which may confer genetic susceptibility or resistance to IBD using a genome-wide association study (GWAS). Genomic DNA was extracted from EDTA blood or saliva samples of 96 cases and 98 controls. Genotyping of cases and controls was performed on the Canine Illumina HD SNP array and data generated was analyzed using PLINK. Several SNPs and regions on chromosomes 7,9,11 and 13 were detected to be associated with IBD using different SNP-by-SNP association methods and FST windows approach. Searching one Mb up-and down-stream of the most significant SNPs, as identified by single SNP analysis as well as 200Kb before and after the start and the end position of the associated regions identified by FST windows approach, we identified 63 genes. Using a combination of pathways analysis and a list of genes that have been reported to be involved in human IBD, we identified 16 candidate genes potentially associated with IBD in GSD.
1. Introduction
Inflammatory bowel disease (IBD) is a chronic debilitating disease that affects both humans and dogs. In dogs, it represents a group of common chronic enteropathies characterized by persistent or recurrent gastrointestinal signs (GI) and with histological evidence of inflammation (usually lymphoplasmacytic and/or eosinophilic) in the lamina propria of the small intestine, large intestine or both [1]. Despite recent progress and improvement in the diagnosis of IBD in dogs, the treatment of this condition remains challenging. IBD is not a curable disease, therefore the aim of current treatment protocols is to minimise the severity and frequency of the clinical signs including vomiting and diarrhoea. In general, treatment protocols include sequential dietary [2, 3], antibiotic [4,5] and corticosteroid [6] treatment trials. Currently, treatment with anti-inflammatory medications such as corticosteroid is the mainstay treatment for both human IBD and canine IBD patients [4]. However, the response to treatment is highly variable, and up to 50% of dogs with IBD that are initially managed with steroids will develop resistance and/or significant side effects, which ultimately leads to euthanasia in many dogs [6–8]. Recent advances in the field of molecular genetics in human IBD make it possible to use the patients’ genetic profile to predict the response to treatment [9, 10]. Similar to human, understanding the genes involved in canine IBD is predicted to reveal insights into disease pathogenesis in canine IBD. This could lead to the development of genetic screening tests for diagnosis and a “personalized medicine” approach, where dogs will be treated with specifically targeted therapeutics, taking their individual genetic makeup into account.
Although IBD can affect any dog, breed-specific associations and disease phenotypes have been reported in the literature [11, 12]. In the United Kingdom (UK), German shepherd dogs are at increased risk of developing the disease [13]. The exact aetiology of IBD in humans as well as dogs is not well understood, although it is considered to be a multifactorial immune-mediated disease in both species, resulting from a complex interaction between the intestinal innate and adaptive immune systems, the intestinal microbiome, and the genetic make-up of a potentially susceptible individual [14–17].
While recent genome-wide association studies (GWAS), investigating the complex genetic basis of IBD in humans have resulted in the identification of 163 susceptibility loci [18], the genetic factors contributing to susceptibility to canine IBD remain largely unknown. Genetic studies into canine IBD, using a candidate gene approach, have revealed a breed-independent association with non-synonymous single nucleotide polymorphisms (SNPs) in Toll-like receptor (TLR)-5, a gene encoding an important mucosal pattern recognition receptor (PRR). In addition, breed-specific SNPs in TLR-4 and nucleotide binding oligomerization domain protein 2 (NOD-2) were identified in GSD [19–21]. A recent study also reported association between SNPs in Major histocompatibility class (MHC) II haplotypes and a potentially increased resistance to IBD in GSD [22]. Despite of these findings, it is believed that similar to human IBD, canine IBD is a complex polygenic disorder with involvement of several more genetic factors that remain to be identified.
The development of high-throughput SNP genotyping technologies has facilitated the detection of genes responsible for complex diseases of the dog. Furthermore, long linkage disequilibrium within breeds resulting from genetic bottlenecks during domestication of dogs and breed formation, made the dog population structure suitable for association mapping techniques [23,24].
The aim of the current study was to detect additional genetic factors associated with IBD in a defined GSD population using GWAS approach. SNP profiles for a total of 190 GSDs were generated using the Canine Illumina HD SNP array. Using the resulting data set, GWAS was performed, which resulted in the subsequent identification of additional loci associated with IBD in GSD, some of them also having been described for human IBD.
2. Materials and methods
2.1. Ethics and welfare statement
All blood samples used in this study were collected in ethylenediaminetetraacetic acid (EDTA) and were residual material following completion of diagnostic testing, used for research with informed written owner consent. The use of residual EDTA blood and buccal swab samples was approved by the RVC Ethics and Welfare Committee (reference number 2013 1210).
2.2. Study population and genotyping
The case population consisted of 96 GSDs based in the UK, which were diagnosed with IBD at either the Royal Veterinary College (University of London, Uk), the Small Animal Teaching Hospital (University of Cambridge, UK), the Animal Health Trust (Newmarket, UK), or three small animal referral centres including Anderson Moores Veterinary Specialists (Hampshire, UK), Davies Veterinary Specialists (Hitchin, UK) and Hope Vets (Bovingdon, UK). The case population was identified using stringent inclusion criteria: all cases have been affected by chronic gastrointestinal signs (i.e. > 3 weeks duration), including diarrhoea and/or vomiting and/or anorexia and/or weight loss. Other causes of chronic gastrointestinal disease had been excluded, by performing routine haematology, serum biochemistry, faecal parasitology and bacteriology, abdominal ultrasonography, serum ACTH stimulation tests and measurement of serum canine trypsin‑like immunoreactivity concentration (TLI), as described in detail previously [22]. All dogs had undergone gastrointestinal endoscopic examination for the collection of mucosal intestinal biopsies. Histological examination of the biopsies had revealed lymphoplasmacytic and/or eosinophilic infiltration of the lamina propria. None of the cases suffered from concurrent diseases of known or suspected immune-mediated origin, dermatological conditions or diseases for which GSD are known to be predisposed for (i.e. anal furunculosis, atopic dermatitis or superficial keratitis).
The control population included 98 GSDs based in the UK. Animals in this group were only considered for inclusion in the study if they were older than 8 years of age, therefore reducing the likelihood to develop IBD later in life. Control dogs were retrospectively phenotyped, either by owner or veterinary surgeon telephone contact, to ensure they had neither current nor previous history of gastrointestinal signs for a duration of more than 5 consecutive days and did not suffer from conditions of known or suspected immune-mediated origin.
Residual blood samples stored in EDTA anticoagulant were available from the RVC sample archive for a total of 69 GSD diagnosed with IBD and 81 breed-matched geriatric controls. EDTA blood samples were also available for 10 cases that have been seen at the University of Cambridge (8 cases) and the Animal Health Trust (2 cases). In addition, buccal swab samples from the other cases and controls for which a residual blood sample was not available were provided by owners for inclusion in the control population.
Genomic DNA (gDNA) was extracted from blood using the GenElute™ Blood Genomic DNA Kit (Sigma-Aldrich®, Gillingham, UK) and from saliva samples using the Performagene® Kit (DNA Genotek Inc, Canada) according to manufacturer’s instructions. Following extraction, the amount of gDNA in all samples were quantified by nucleic acid fluorescent staining using a commercial kit and following the manufacturer’s instructions (Quant-iT™ PicoGreen® dsDNA Kit, Life Technologies, Paisley, UK), samples were normalised to a concentration of 20 ng μl-1 and stored at -20°C until use.
The genotyping of all cases and controls was performed using the Illumina 170K CanineHD® Beadchip (Illumina, San Diego, CA, USA). All SNPs with call rate <90%, minor allele frequency < 2%, monomorphic, unmapped to the CanFam3.1 reference sequence (GCF_000002285.3) and samples with call rate <80% were removed. After quality control, 94 cases, 96 controls and 117K SNPs remained for the final analysis.
To assess presence of population stratification, identity-by-decent and multidimensional scaling (MDS) analyses were performed using PLINK v1.07 software [25]. A scatter plot for the first 2 dimensions was produced. Population stratification analysis was also performed on the dataset with the Admixture software [26], with number of subpopulations (K) that ranging from 1 to 7. The cross-validation error generated at each K value was used to determine the best number of subpopulations across the dataset that is represented by the K with the lowest cross validation error.
2.3. Genome-wide association study
Association analyses were performed using different approaches: a basic case and control association (chi-square allelic test), a logistic regression and a logistic regression using the C1 and C2 components previously calculated with the MDS analysis as covariate in PLINK v1.07 software. P = -log10(5e-08) was considered as significant association, P = -log10(1e-05) considered as suggestive association. Genomic inflation factor (λ) and quantile-quantile (Q–Q) plots (S1 Fig) were obtained using the GenABEL software [27].
2.4. Fixation index (Fst) analysis
Association analysis was also performed using FST analysis SNP by SNP and using a sliding window of 1Mb, with 500Kb of overlapping. FST was calculated SNP by SNP, using the adapted formula below [23]
Where k is the SNP marker k, with frequency p1[k], p2[k]
The values can range from 0 (no differences) to 1 (complete different). For the window-based analysis, the mean FST was calculated across the SNPs included in the window.
The top four SNPs for single SNP and 5 windows for overlapping windows were considered as the most divergent between case and control groups.
2.5. Identification of potential candidate genes
Gene searching was conducted with a range of ±1 Mb for the SNPs based analyses and ±200Kb for the regions identified with the windows based analysis. To identify potential candidate genes associated with IBD in GSDs, pathway analysis was performed using the Enrichr tool, (http://amp.pharm.mssm.edu/Enrichr) to detect genes involved in pathway that could be related with the disease. For this analysis, the default statistical tests and corrections for multiple testing to maintain an overall p-values of 0.05 were applied. Moreover, all the genes identified were compared to a list of genes that has been already stablished for human IBD [18], to search for common genes. Genes that were detected with each of these two approaches were considered for the discussion.
3. Results
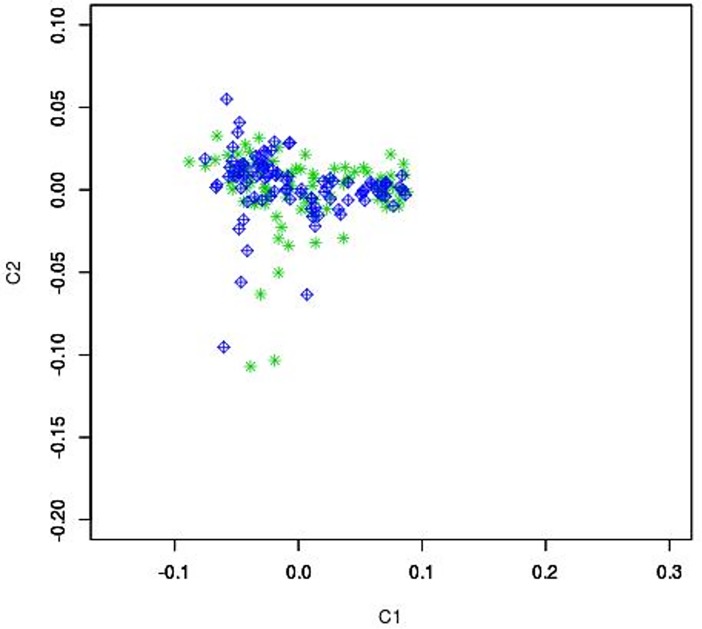
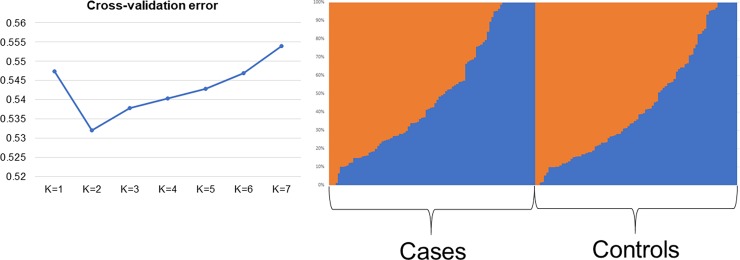
This study is the first GWAS on GSDs with the aim of investigating genetic factors associated with IBD. We performed genotyping of 170,000 SNP markers of the entire GSD cohort. The exclusion of non-informative markers, markers with low call rate and individuals with call rates <80% resulted in 200 individuals (94 cases, 96 controls) and 117K markers passing QC. Principal component analysis with PLINK (Fig 1) showed no evidence for population stratification within our GSD cohort (n = 190). The admixture analysis showed only two subpopulation that are balanced between cases and controls (Fig 2).
Fig 1. Scatter plot for the first 2 dimensions.
The x-axis is principal component 1 and y-axis is principal component 2. Green: case, Blue: control.
Fig 2.
Admixture plot of the case-control dataset (showed on the right) using the K with the lowest cross-validation error (showed on the left).
Results of the identity-by-descend analysis revealed three pairs of individuals with high Identity-by-descent proportion values (P_HAT> 0.90) in our study population. One sample of each pair was removed (two were cases and one control) prior to association analysis, resulting in 92 cases and 95 controls.
3.1. Association tests
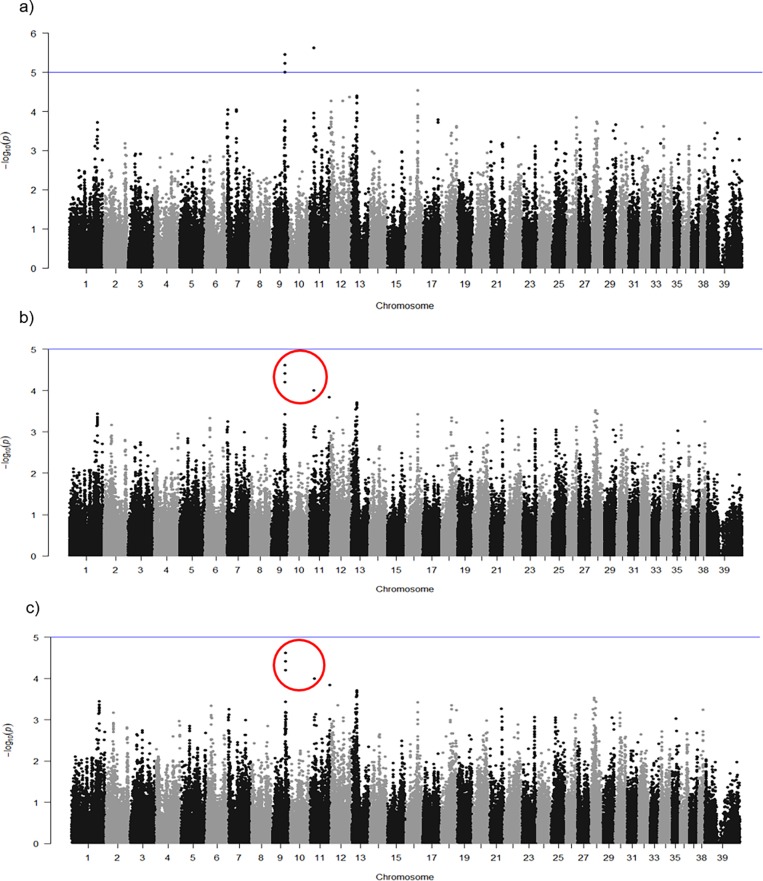
IBD in GSD appears to be associated with SNPs on Chromosomes 11. GWAS of 92 cases versus 95 controls (total of 187 GSDs), using different statistical approaches, identified a number of SNPs with high differences on chromosomes 7, 9, 11 and 13. Basic case/control association analysis (Fig 3A) revealed three SNPs on chromosome 9 and one SNP on chromosome 11 with suggestive association. The strongest association (P = 2.36 × 10−6) identified by basic case/control association analysis was for SNP BICF2S23033111 at position 20,056,580 on chromosome 11. A second peak was within a region on chromosome 9 spanning from position 51,531,181 to 51,544,743 bp. In this region, three SNPS (BICF2P812982, BICF2P436494 and BICF2P753594) were identified, Results are shown in Table 1.
Fig 3.
Manhattan plots of basic case-control association test (a), logistic regression without covariate (b) and logistic regression with covariate (c). Suggestive association threshold is indicated with the blue line. SNPs of the logistic analyses with the lowest p-value that overlapped the basic association study are within red circles.
Table 1. The most significant SNPs found by basic case/control association analysis.
| CHR | SNP | BP | A1 | F_A | F_U | A2 | CHISQ | P | OR |
|---|---|---|---|---|---|---|---|---|---|
| 9 | BICF2P753594 | 51531181 | C | 0,07609 | 0,2447 | G | 19,54 | 9,86E-06 | 0,2542 |
| 9 | BICF2P436494 | 51541093 | G | 0,07609 | 0,25 | A | 20,52 | 5,91E-06 | 0,2471 |
| 9 | BICF2P812982 | 51544743 | G | 0,07609 | 0,2553 | A | 21,51 | 3,52E-06 | 0,2402 |
| 11 | BICF2S23033111 | 20056580 | C | 0,212 | 0,04787 | A | 22,28 | 2,36E-06 | 5,349 |
CHR: Chromosome
SNP: SNP ID
BP: Physical position on CanFam3.1
A1: Minor allele name (based on whole sample)
F A: Frequency of this allele in cases
F U: Frequency of this allele in controls
A2: Major allele name
CHISQ: Basic allelic test chi-square
P: Asymptotic p-value for this test
OR:Estimated odds ratio (for A1, i.e. A2 is reference)
Logistic association approaches were also performed as our trait of interest was binary. These SNPs were found to have the lowest p-values by both logistic associations with no covariate (Fig 3B and S1 Table) and with covariate (Fig 3C and S2 Table), although they did not reach the significance threshold (Fig 3B and 3C and S1 and S2 Tables respectively).
Quantile–quantile plots for the three analyses (S1 Fig) showed the relationship between observed (y-axis) and expected (x-axis) test statistics and visualize the population substructure. The slight deviation in the upper right tail from the y = x line are suggestive of associations. Data generally falling on the y = x lines suggests no clear systemic bias. The degree of deviation from this line is measured by the 𝜆-statistic, where a value close to 1 indicates that the data follows the normal chi-squared distribution. The logistic with no covariate (Figure B in S1 Fig) showed the lowest 𝜆, which can be considered as most conservative model. The correction with population derived covariate (Figure C in S1 Fig) showed a difference in values. However, in both three cases, big differences were not observed, as the study population is fairly homogenous [28].
3.2. Fixation index (Fst) tests
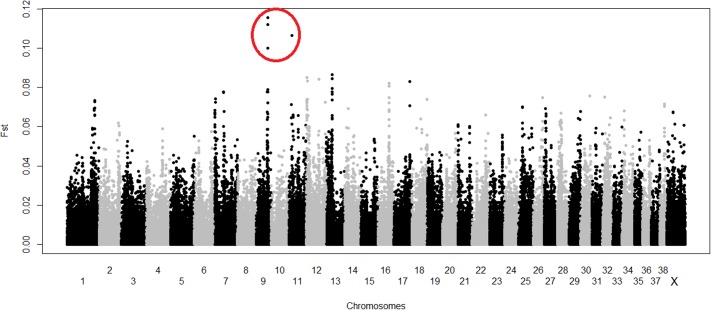
The FST analyses was carried out SNP by SNP, reported the top 5 most divergent SNPs on chromosomes 9, 11 and 12 (Fig 4 and S3 Table). Anyway, the to 4 SNPs, three on chromosome 9 and 1 on chromosome 11, overlapped those previously identified with the association analyses.
Fig 4. Single FST approach result.
SNPs with the highest FST value that overlap the basic case/control association test and logistic association are within red circles.
The results of the Fst window-based analysis indicated five main divergent regions between cases and controls: four on chromosome 7 and one on chromosome 13 with the highest m FST value (Table 2).
Table 2. Regions on chromosome 7 and 13, found by FST windows approach.
| chromosome | Start | End | N.Snps | mFST |
|---|---|---|---|---|
| 7 | 4000000 | 5000000 | 31 | 0.0362679 |
| 7 | 3500000 | 4500000 | 44 | 0.0384714 |
| 13 | 2500000 | 3500000 | 33 | 0.038702 |
| 7 | 2500000 | 3500000 | 17 | 0.0403834 |
| 7 | 3000000 | 4000000 | 39 | 0.0408544 |
Start: start position of the window. End: End position of the window. N.Snps: number of SNPs included in each window. mFST: mean FST value calculated for SNPs included in each window.
3.2 Candidate genes
The gene searching of the SNPs and windows-based analysis identified 80 genes, 44 detected with considering the consensus SNPs of the singles SNPs analyses (S4 Table), and 36 considering the regions detected with the windows-based Fst analysis (S5 Table). The combination of pathway association analysis and the literature search to look for genes already found to be involved in human IBD [18] identified a total of 16 candidate genes potentially associated with IBD in GSD (Table 3). Among these, a total of 11 genes have been previously reported to be associated with human IBD, eight of these genes are also involved in pathways such as Cytokines and Inflammatory Response and Glucocorticoid receptor regulations (Table 3 and S6 Table). Genes encoding for cytokines, including Th2 cytokine, IL-4 and IL-13, were amongst these genes, and were detected by both approaches.
Table 3. Enricher results.
Genes found to be enriched in biological processes and/or molecular components that are associated or directly/indirectly involved with human IBD.
| Chromosme region | gene symbol | gene name | gene start | gene end | database |
|---|---|---|---|---|---|
| 7 | PTPRC* | Protein tyrosine phosphatase, receptor type C | 4156687 | 4282147 | KEEG |
| 9 | TSC1 | Tuberous sclerosis 1 | 51419149 | 51454668 | KEEG |
| RALGDS | Ral guanine nucleotide dissociation stimulator | 51308323 | 51330270 | KEEG | |
| COL5A1 | Collagen type V alpha 1 chain | 50741552 | 50856744 | Wikipathways20016 | |
| RAPGEF1 | Rap guanine nucleotide exchange factor 1 | 52450597 | 52562778 | Wikipathways20016 | |
| 11 | IL4* | Interleukin-4 | 20972693 | 20981541 | KEEG; Wikipathways20016; Panther 2016; NCI-Nature 2016 |
| IL5* | Interleukin-5 | 20825469 | 20827269 | KEEG; Wikipathways20016; Panther 2016; NCI-Nature 2016 | |
| CSF2* | Granulocyte-macrophage colony-stimulating factor | 20344009 | 20346959 | KEEG; Wikipathways20016 | |
| IL13* | Interleukin-13 precursor | 20958464 | 20961391 | KEEG; Wikipathways20016; Panther 2016; NCI-Nature 2016 | |
| SLC22A4* | Solute carrier family 22 member 4 | 20598888 | 20644639 | KEEG | |
| SLC22A5* | Solute carrier family 22 member 5 | 20659221 | 20683073 | KEEG | |
| IRF1* | Interferon regulatory factor 1 | 20772643 | 20781207 | NCI-Nature 2016 | |
| ACSL6* | acyl-CoA synthetase long-chain family member 6 | 20223930 | 20285138 | - | |
| IL3* | Interleukin-3 | 20330480 | 20332668 | - | |
| PDLIM4* | PDZ and LIM domain 4 | 20570752 | 20587263 | - | |
| 13 | YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta | 2729503 | 2763613 | Wikipathways20016 |
*Genes found already to be associated with human IBD.
4. Discussion
In the present study, we identified candidate genes on several canine chromosomes that are potentially associated with IBD in GSD. These genes may have an impact on the immune-homeostasis in the canine gut, similar as described for the situation in human IBD.
A number of studies have highlighted the role of immunologic mechanisms and in particular, the involvement of specific cytokine subsets in the pathogenesis of canine IBD. Observations of increased numbers of immunoglobulin-containing cells and T cells in inflamed tissues [1, 29–31], upregulated mucosal and luminal expression of nitric oxide metabolites [32,33], and altered serum concentrations of select acute phase proteins, such as C-reactive protein, which is a marker of inflammation and tissue injuries, in IBD dogs [34,35], support the involvement of impaired immunoregulation in pathogenesis of canine IBD.
Our study results suggest a potential involvement of Th2 cytokines in the pathogenesis of IBD in GSDs, similar as described for ulcerative colitis (UC) in humans. According to the cytokine profile, CD4+ T cells can be divided into major subtypes, with Th1 cells producing mainly IL-2, IL-12, IFNg, TNFα, favoring the development of a cell-mediated immune response, whereas Th2 cells mainly produce IL-4, IL-5 and IL-13, which results in the induction of a humoral immune response [36].
In humans, IL-13, produced by specialized cells such as NK T-cells, was identified as an important effector cytokine in UC [37], and its release may impair epithelial barrier function by affecting epithelial apoptosis, tight junctions, and restitution velocity [38]. In addition, lamina propria mononuclear cells (LPMCs) from UC patients have been described to secrete significantly higher amounts of IL-13 and IL-5 than LPMCs from CD patients and healthy controls [37,38]. The IL-13 and IL-5 producing LPMCs bear the NK cell specific markers CD161 and detect CD1d, providing evidence that the release of these cytokines is due to activation of NK T-cells [37]. It was also shown that the NK T-cells isolated from UC patients are cytotoxic for HT-29 epithelial cells, and that this cytotoxicity was enhanced further by addition of IL-13 [37]. In addition, IL-13 signalling through the IL-13α2 receptor (IL-13Rα) induces TGF-β1 production and fibrosis [39]. Many functions of IL-13 are shared with the major Th2 cytokine IL-4. Results of studies performed in a murine model of IBD suggest that in T cell receptor α chain–deficient (TCRα-/-) mice, treatment with anti–IL-4 Ab resulted in a decrease of Th2 cytokines, and a concomitant increase of IFN-γ, suggesting that not only NK-T cells, but also CD4+ Th2 T cells play a major immunopathological role in the induction of IBD [40]. A similar finding of the potential involvement of IL-4 in canine IBD was confirmed by Jergens [41].
It is worth noticing however, that studies in canine IBD revealed contradictory results regarding the cytokine mRNA expression patterns in intestinal biopsies. German et al. [31] reported a mixed Th1-Th2 cytokine expression in GSDs with small intestinal enteropathy. However, this study included only four dogs diagnosed with IBD. In another study, up-regulation of IL-2 and TNF-α mRNA, two classic Th1 cytokines, in the colonic mucosa of dogs with lymphocytic-plasmacytic colitis (LPC) was reported in dogs with large or small intestinal IBD [42]. Furthermore, no differences in the mRNA expression of IL-2, IL-4, IL-5, IL-6, IL-10, IL-12, IL-18, IFN-γ, TNF-α and TGF-β was detected between healthy dogs or dogs with IBD in more recent studies using real-time q-RT-PCR [43,44]. Finally, Jergens et al. [41] demonstrated expression of a diverse range of pro-inflammatory, anti-inflammatory, and regulatory cytokines in the intestinal mucosa, which differed between dogs with IBD and clinically healthy dogs. Results of these studies suggested at the time, that a predominant Th1- or Th2 cytokine mRNA expression in the inflamed mucosa is not a feature of small- or large- intestinal IBD in dogs [41].
The variations in pattern of cytokine expression observed in these studies might be explained by different methods for mRNA quantification, including semi-quantitative RT-PCR [31,41,42] and real-time RT-PCR [43] techniques, the relatively small number of IBD dogs investigated in each study, and a potential over-representation of GSDs with enteropathies in most studies [31,42,43]. Furthermore, and in contrast to the present study, all of these studies involved a mixture of dog breeds, which may impact on the results obtained and could potentially suggest that different cytokine expression patterns are involved in the pathogenesis of IBD in different breeds. Therefore, to obtain a better insight into pathogenesis of the disease, the current study has focused on one breed and investigated relatively large number of dogs with IBD.
The GWAS data obtained in the present study on GSDs are in agreement with results obtained for UC in humans. Here, IBD is the result of disruption of intestinal immunological homeostasis, and particularly the alteration of the normal balance that the gut maintains between inflammatory and regulatory cytokines [45–47]. Interestingly, cytokine analysis in humans suggests a different pathology of the two main types of IBD. Inflammation associated with Crohn’s disease (CD) is believed to be driven by Th1 cells, producing IFN-ɣ and IL 2, and Th17 cells producing IL-17 and IL-22 [48–52]. In contrast, a Th2 cytokine pattern with IL-4 and IL-5 was found to be important in the pathogenesis of UC [50]. Furthermore, there is evidence that impaired regulation of immune responses by regulatory T cells (Treg) play a part in pathogenesis of human IBD [53]. Recently, these cells also have been characterized in dogs [54], and a decreased number of Treg cells was seen in the peripheral blood [55] and duodenal mucosa of dogs with IBD [56,57]. Whether this observation is a result of IBD, leading to the development of Treg cells that are unable to counteract the inflammatory process in the intestine of dogs with IBD, or a primary result of functional impairment of Tregs in dogs with IBD needs to be studied further.
In conclusion, based on the data provided within the present study, Th2 cytokines may have the potential of causing the epithelial cell damage in GSDs with IBD. Targeted re-sequencing of the genes identified in our study will help to identify causative SNPs and subsequent functional analysis of the causal SNPs may reveal insights into mechanisms involved in pathogenesis of canine IBD.
Supporting information
Quantile–quantile plots a, b and c demonstrate the relationship between observed (y-axis) and expected (x-axis) test statistics.
(DOCX)
(DOCX)
Note that results are the same as the S1 Table, but the genomic inflation factor is different between the two comparisons.
(DOCX)
Here, the top five most divergent SNPS are reported.
(DOCX)
The three consecutive SNP on chromosome 9 were considered as a cluster.
(DOCX)
(DOCX)
Using several databases (KEEG2016. WikiPathways 2016 and NCI-Nature 2016) with Pathway name. P values (P). Adjusted P values (Adj P) and list of genes detected.
(DOCX)
Acknowledgments
We are grateful to owners of GSD who gave permission for their dogs to participate in this study. We would like also to acknowledge Dr Mellora Sherman, Fabio Procoli, Julien Bazelle, Andrew Kent, and Poppy Clement for helping us with recruitments of IBD cases.
Data Availability
Data are all contained within the paper and/or Supporting Information files.
Funding Statement
We declare that the work described in this manuscript was funded through a BBSRC iCase PhD studentship stipend (BB/J01236X/1 to DW) and part-funded by Laboklin GmBH, Bad Kissingen, GE. Funding was also provided by the Ensminger fund, State of Iowa and Hatch funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jergens A.E. Inflammatory bowel disease—Current perspectives. Vet Clin North Am Small Anim Pract. 1999; 29(2): 501–521. [PubMed] [Google Scholar]
- 2.Luckschander N, Allenspach K, Hall J, Seibold F, Gröne A, Doherr MG, et al. Perinuclear antineutrophilic cytoplasmic antibody and response to treatment in diarrheic dogs with food responsive disease or inflammatory bowel disease. J Vet Intern Med. 2006;20(2): 221–227. [DOI] [PubMed] [Google Scholar]
- 3.Mandigers PJJ, Biourge V, van den Ingh TS, Ankringa N, German AJ. A Randomized, Open-Label, Positively-Controlled Field Trial of a Hydrolyzed Protein Diet in Dogs with Chronic Small Bowel Enteropathy. J Vet Intern Med. 2010; 24(6): 1350–1357. 10.1111/j.1939-1676.2010.0632.x [DOI] [PubMed] [Google Scholar]
- 4.German A J, Day MJ, Ruaux CG, Steiner JM, Williams DA, Hall EJ. Comparison of direct and indirect tests for small intestinal bacterial overgrowth and antibiotic-responsive diarrhea in dogs. J Vet Intern Med. 2003;17(1): 33–43. [DOI] [PubMed] [Google Scholar]
- 5.Westermarck E, Skrzypczak T, Harmoinen J, Steiner JM, Ruaux CG, Williams DA, et al. Tylosin-responsive chronic diarrhea in dogs. J Vet Intern Med. 2005; 19(2): 177–186. [DOI] [PubMed] [Google Scholar]
- 6.Allenspach K, Wieland B, Gröne A, Gaschen F. Chronic enteropathies in dogs: Evaluation of risk factors for negative outcome. J Vet Intern Med.2007; 21(4): 700–708. [DOI] [PubMed] [Google Scholar]
- 7.Allenspach K, Bergman PJ, Sauter S, Gröne A, Doherr MG, Gaschen F. P-glycoprotein expression in lamina propria lymphocytes of duodenal biopsy samples in dogs with chronic idiopathic enteropathies. J Comp Pathol. 2006;134:1–7. 10.1016/j.jcpa.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 8.Allenspach K, Rüfenacht S, Sauter S, Gröne A, Steffan J, Strehlau G, et al. Pharmacokinetics and clinical efficacy of cyclosporine treatment of dogs with steroid-refractory inflammatory bowel disease. J Vet Intern Med. 2006;20:239–244. [DOI] [PubMed] [Google Scholar]
- 9.Sandborn WJ. Pharmacogenomics and IBD—TPMT and thiopurines. Inflamm Bowel Dis. 2004;10: S35–S37. [DOI] [PubMed] [Google Scholar]
- 10.Roberts RL, Barclay ML. Current relevance of pharmacogenetics in immunomodulation treatment for Crohn's disease. J Gastroenterol Hepatol. 2012; 27(10): 1546–1554. 10.1111/j.1440-1746.2012.07220.x [DOI] [PubMed] [Google Scholar]
- 11.Kimmel SE, Waddell LS, Michek KE. Hypomagnesemia and hypocalcemia associated with protein-losing enteropathy in Yorkshire terriers: five cases (1992–1998). J Am Vet Med Assoc. 2000;217(5): 703–706. [DOI] [PubMed] [Google Scholar]
- 12.Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immun. 2006;74(8): 4778–4792. 10.1128/IAI.00067-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathrani A, Werling D, Allenspach K. Canine breeds at high risk of developing inflammatory bowel disease in the south-eastern UK. Vet Rec. 201;169(24): 635 10.1136/vr.d5380 [DOI] [PubMed] [Google Scholar]
- 14.Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–656. [DOI] [PubMed] [Google Scholar]
- 15.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006; 3(7): 390–407. 10.1038/ncpgasthep0528 [DOI] [PubMed] [Google Scholar]
- 16.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]
- 17.Fritz JH, Le Bourhis L, Magalhaes JG, Philpott DJ. Innate immune recognition at the epithelial barrier drives adaptive immunity: APCs take the back seat. Trends Immunol. 2008;29:41–49. 10.1016/j.it.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 18.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491(7422): 119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathrani A, House A, Catchpole B, Murphy A, German A, Werling D, et al. Polymorphisms in the Tlr4 and Tlr5 Gene Are Significantly Associated with Inflammatory Bowel Disease in German Shepherd Dogs. PLoS One. 2010;5(12): e15740 10.1371/journal.pone.0015740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathrani A, House A, Catchpole B, Murphy A, Werling D, Allenspach K. Breed-independent toll-like receptor 5 polymorphisms show association with canine inflammatory bowel disease. Tissue Antigens. 2011;78(2): 94–101.b 10.1111/j.1399-0039.2011.01707.x [DOI] [PubMed] [Google Scholar]
- 21.Kathrani A, Lee H, White C, Catchpole B, Murphy A, German A, et al. Association between nucleotide oligomerisation domain two (Nod2) gene polymorphisms and canine inflammatory bowel disease. Vet Immunol Immunopathol. 2014;161(1–2): 32–41. 10.1016/j.vetimm.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 22.Peiravan A, Allenspach K, Boag Al, Soutter F, Holder A,Catchpole B, et al. Single nucleotide polymorphisms in major histocompatibility class II haplotypes are associated with potential resistance to inflammatory bowel disease in German shepherd dogs. Vet immunol immunopathol. 2016; 182; 101–105. 10.1016/j.vetimm.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 23.Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N. et al. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. 10.1038/ng.2007.10 [DOI] [PubMed] [Google Scholar]
- 24.Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. 10.1126/science.1137045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007; 81(3): 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander D.H, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res.2009; 19(9), 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aulchenko Y.S, Ripke S, Isaacs A, Vanduijn C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23, 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Weedon M.N, Purcell S, Lettre G, Estrada K, Willer C.J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19, 807–812. 10.1038/ejhg.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jergens AE, Moore FM, Kaiser MS, Haynes JS, Kinyon JM. Morphometric evaluation of immunoglobulin A-containing and immunoglobulin G-containing cells and T cells in duodenal mucosa from healthy dogs and from dogs with inflammatory bowel disease or nonspecific gastroenteritis. Am J Vet Res. 1996;57:697–704. [PubMed] [Google Scholar]
- 30.Stonehewer J, Simpson JW, Else RW, Macintyre N. Evaluation of B and T lymphocytes and plasma cells in colonic mucosa from healthy dogs and dogs with inflammatory bowel disease. Res Vet Sci. 1998;65:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.German AJ, Helps CR, Hall EJ. Cytokine mRNA expression in mucosal biopsies from German shepherd dogs with small intestinal enteropathies. Dig Dis Sci. 2000;45(1):7–17. [DOI] [PubMed] [Google Scholar]
- 32.Gunawardana SC, Jergens AE, Ahrens FA, Niyo Y. Colonic nitrite and immunoglobulin G concentrations in dogs with inflammatory bowel disease. J Am Vet Med Assoc. 1997;211:318–321. [PubMed] [Google Scholar]
- 33.Jergens AE, Moore FM, Haynes JS, Miles KG. Idiopathic inflammatory bowel disease in dogs and cats: 84 cases (1987–1990). J Am Vet Med Assoc.1992; 201:1603–1608. [PubMed] [Google Scholar]
- 34.Jergens AE, Schreiner CA, Frank DE, Niyo Y, Ahrens FE, Eckersall PD, et al. A scoring index for disease activity in canine inflammatory bowel disease. J Vet Intern Med. 2003;17:291–297. [DOI] [PubMed] [Google Scholar]
- 35.Vermeire S, van Assche G, Rutgeerts P. C-reactive protein as a marker for inflammatory bowel disease. Inflamm Bowel Dis. 2004;10:661–665. [DOI] [PubMed] [Google Scholar]
- 36.O'Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets.J Immunol. 1998;161(2):547–551. [DOI] [PubMed] [Google Scholar]
- 37.Fuss IJF, Boirivant M, Leon F, Yoshida M, Fichtner-Feigl S, Yang Z, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490–1497. 10.1172/JCI19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heller F, Florian P, Bojarski C, Richter J, Christ M, Hillenbrand B, et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129 (2):550–564. 10.1016/j.gastro.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 39.Fichtner-Feigl S, Fuss IJ, Young CA, Watanab T, Geissler EK, Schlitt HJ, et al. Induction of IL-13 triggers TGF-beta1-dependent tissue fibrosis in chronic 2,4,6-trinitrobenzene sulfonic acid colitis. J Immunol. 2007;178:5859–5870. [DOI] [PubMed] [Google Scholar]
- 40.Iijima H, Takahashi I, Kishi D, Kim JK, Kawano S, Hori M, et al. Alteration of interleukin 4 production results in the inhibition of T helper type 2 cell-dominated inflammatory bowel disease in T cell receptor alpha chain-deficient mice. J Exp Med. 1999;190:607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jergens AE, Sonea IM, O'Connor AN, Kauffman LK, Grozdanic SD, Ackermann MR, et al. Intestinal Cytokine mRNA Expression in Canine Inflammatory Bowel Disease: A Meta-Analysis with Critical Appraisal. 2009. Comp med. 2009;59(2): 153–162. [PMC free article] [PubMed] [Google Scholar]
- 42.Ridyard AE, Nuttall TJ, Else RW, Simpson JW, Miller HR. Evaluation of Th1, Th2 and immunosuppressive cytokine mRNA expression within the colonic mucosa of dogs with idiopathic lymphocytic-plasmacytic colitis. Vet Immunol Immunopathol. 2002; 86: 205–214. [DOI] [PubMed] [Google Scholar]
- 43.Peters IR, Help CR, Calvert EL, Hall EJ, Day MJ. Cytokine mRNA quantification in duodenal mucosa from dogs with chronic enteropathies by real-time reverse transcriptase polymerase chain reaction. J Vet Intern Med. 2005; 19 (5): 644–653. [DOI] [PubMed] [Google Scholar]
- 44.De Majo, Pugliese M, Galia S, Mazzullo G, Camera E, Fera MT. Cytokine mRNA quantification in gastro-intestinal biopsies of dogs with idiopathic chronic enteropathies by Real Time RT-PCR: Preliminary results. Vet Res Commun. 2008;32 (1): S275–277. [DOI] [PubMed] [Google Scholar]
- 45.Garsid p. Cytokines in experimental colitis. Clin. Exp. Immunol. 1999;118 (3): 337–339. 10.1046/j.1365-2249.1999.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhan A, Mizoguchi E, Smith RN, Mizoguchi. Lessons for human inflammatory bowel disease from experimental models. Curr Opin Gastroenterol. 1999;15:285–90. [DOI] [PubMed] [Google Scholar]
- 47.Papadakis KA, Targan SR. Role of cytokines in pathogenesis of inflammatory bowel disease. Gastroenterol Clin North Am. 1999;28 (2): 283–296. [DOI] [PubMed] [Google Scholar]
- 48.Mullin GE, Lazenby AJ, Harris ML, Bayless TM, James SP. Increased interleukin-2 messenger RNA in the intestinal mucosal lesions of Crohn's disease but not ulcerative colitis. Gastroenterology. 1992;102(5):1620–1627. [DOI] [PubMed] [Google Scholar]
- 49.Plevy SE, Landers CJ, Prehn J, Carramanzana NM, Deem RL, Shealy D, et al. A role for TNF-alpha and mucosal T helper-1 cytokines in the pathogenesis of Crohn's disease. J Immunol. 1997;159(12):6276–82. [PubMed] [Google Scholar]
- 50.Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. J Immunol. 1996;157(3):1261–70. [PubMed] [Google Scholar]
- 51.Niessner M, Volk BA. Altered Th1/Th2 cytokine profiles in the intestinal mucosa of patients with inflammatory bowel disease as assessed by quantitative reversed transcribed polymerase chain reaction (RT-PCR). Clin Exp Immunol. 1995;101(3):428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290(4): G827–G838. 10.1152/ajpgi.00513.2005 [DOI] [PubMed] [Google Scholar]
- 53.Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, Stallmach A, et al. Peripheral and intestinal regulatory CD4+ CD25 (high) T cells in inflammatory bowel disease. Gastroenterology. 2005;128, 1868–1878. [DOI] [PubMed] [Google Scholar]
- 54.Pinheiro D, Singh Y, Grant CR, Appleton RC, Sacchini F, Walker KR, et al. Phenotypic and functional characterization of a CD4(+) CD25(high) FOXP3(high) regulatory T-cell population in the dog. Immunology. 2011; 132, 111–122.124. 10.1111/j.1365-2567.2010.03346.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkmann M, Hepworth MR, Ebner F. Frequencies of regulatory T cells in the peripheral blood of dogs with primary immune-mediated thrombocytopenia and chronic enteropathy: a pilot study. Vet J. 2014;202(3):630–633. 10.1016/j.tvjl.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 56.Maeda S, Ohno K, Fujiwara-Igarashi A, Uchida k, Tsujimoto H. Changes in Foxp3-Positive Regulatory T Cell Number in the Intestine of Dogs With Idiopathic Inflammatory Bowel Disease and Intestinal Lymphoma. Veterinary pathology. 53(1), 2015;102–112. 10.1177/0300985815591081 [DOI] [PubMed] [Google Scholar]
- 57.Junginger J, Schwittlick U, Lemensieck F. Immunohistochemical investigation of Foxp3 expression in the intestine in healthy and diseased dogs. Vet Res. 2012;43(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantile–quantile plots a, b and c demonstrate the relationship between observed (y-axis) and expected (x-axis) test statistics.
(DOCX)
(DOCX)
Note that results are the same as the S1 Table, but the genomic inflation factor is different between the two comparisons.
(DOCX)
Here, the top five most divergent SNPS are reported.
(DOCX)
The three consecutive SNP on chromosome 9 were considered as a cluster.
(DOCX)
(DOCX)
Using several databases (KEEG2016. WikiPathways 2016 and NCI-Nature 2016) with Pathway name. P values (P). Adjusted P values (Adj P) and list of genes detected.
(DOCX)
Data Availability Statement
Data are all contained within the paper and/or Supporting Information files.