Summary
Molecular chaperones are responsible for managing protein folding from translation through degradation. These crucial machines ensure that protein homeostasis is optimally maintained for cell health. However, “too much of a good thing” can be deadly, and the excess of chaperones can be toxic under certain cellular conditions. For example, overexpression of Ssa1, a yeast Hsp70, is toxic to cells in folding-challenged states such as [PSI+]. We discovered that overexpression of the nucleotide exchange factor Sse1 can partially alleviate this toxicity. We further argue that the basis of the toxicity is related to the availability of Hsp70 cofactors, such as Hsp40 J-proteins and nucleotide exchange factors. Ultimately, our work informs future studies about functional chaperone balance and cautions against therapeutic chaperone modifications without a thorough examination of cofactor relationships.
Keywords: Molecular chaperone, Hsp70, nucleotide exchange factor, prion, yeast
Graphical abstract
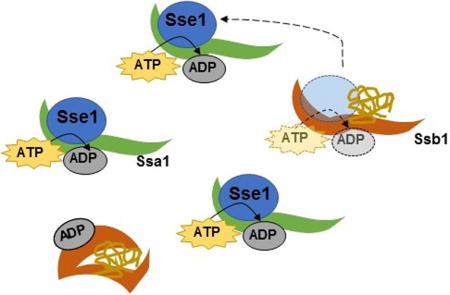
Introduction
Correct protein folding is achieved through a network of chaperones, co-chaperones, and nucleotide exchange factors that collaborate to configure a protein into an ordered state (Hartl et al., 2011). When left unchecked, improper protein folding can lead to disrupted protein homeostasis and cell death, and, in mammalian systems, it can cause diseases such as Alzheimer’s disease and ALS (Labbadia and Morimoto, 2015). Thus, extensive networks of molecular chaperones have evolved to protect organisms from these adverse events.
Saccharomyces cerevisiae have more than 60 molecular chaperones that act on misfolded proteins, and hundreds of cofactors that assist these activities (Gong et al., 2009). Hsp70s are one class of chaperones that rely upon other proteins in order to facilitate ATP cycling and binding to clients (Kampinga and Craig, 2010; Mayer, 2013). Hsp70s work in concert with other cochaperones to change the conformation of misfolded proteins to promote proper folding. Hsp40 J-proteins are responsible for both delivering substrates and stimulating the ATPase domain of Hsp70s (Craig et al., 2006; Sharma and Masison, 2009). Nucleotide exchange factors remove ADP and allow Hsp70s to return to a “ready” state (Dragovic et al., 2006). The repeated binding and release of clients with exposed hydrophobic patches allows the misfolded proteins to regain their correct conformations (Mayer, 2013).
Prion aggregation is one consequence of protein misfolding in yeast. Many prions exist in yeast, and they regulate diverse cellular functions including transcription, translation termination, and nitrogen metabolism (Liebman and Chernoff, 2012). Prion propagation occurs via specific misfolding of prion proteins into beta sheet-rich structures that promote additional monomers to convert and join the prion template. Yeast prions act as epigenetic mechanisms of inheritance as they are transmitted from mother to daughter cell (Liebman and Chernoff, 2012). One well-characterized yeast prion, [PSI+], is caused by the misfolding and aggregation of the translation termination factor Sup35 (Chernoff et al., 1993; Paushkin et al., 1996; Patino et al., 1996). Many chaperones act upon Sup35 aggregates, including members of the Hsp70 and Hsp40 families (Liebman and Chernoff, 2012). The activities of these chaperones can either prevent or promote de novo prion formation, depending on expression level (Reidy and Masison, 2011).
Given the thermodynamic and cytosolic pressures that can oppose proper protein folding, one may hypothesize that “more is better” when it comes to intracellular chaperones. It stands to reason that higher levels of chaperone expression could benefit cells by providing more opportunities for misfolded proteins to be captured and refolded. Interestingly, this is not always the case.
We previously reported that the levels of Hsp70s, namely Ssa1 and Ssb1, are at a fine balance in the yeast Saccharomyces cerevisiae (Keefer and True, 2016). When more Ssa1 is available relative to Ssb1, [PSI+] yeast experience exacerbated toxicity when Sup35 is concurrently overproduced. This phenomenon did not occur with overexpression of other chaperones, such as Ssb1. The toxicity associated with excess Sup35 is known to be related to the reduced availability of both Sup35 and Sup45, another yeast translation termination factor (Dagkesamanskaya and Ter-Avanesyan, 1991; Chernoff et al., 1992; Derkatch et al., 1997; Vishveshwara et al., 2009). This toxicity can be rescued by overexpressing Sup45 or the non-prion C-terminal region of Sup35 (Vishveshwara et al., 2009). The reasons for increased toxicity associated with increased Ssa1 are not clear, but could include direct effects upon the [PSI+] prion, the titration of other important chaperone cofactors, or the sequestration of key client proteins.
We wondered what characteristics of Ssa1 influenced its toxic phenotypes. Here, we demonstrate that modification of [PSI+] is unlikely to be a major contributor to Ssa1-related toxicity. We demonstrate that Hsp70 cofactor availability plays a role in this toxicity, especially when those cofactors are shared between Ssa1 and Ssb1.
Results
Ssa1 overexpression is toxic to yeast containing aggregated Sup35
Yeast containing the strong [PSI+] prion show toxicity when Sup35, the constituent of [PSI+], is overexpressed (Dagkesamanskaya and Ter-Avanesyan, 1991; Chernoff et al., 1992; Vishveshwara et al., 2009) (Figure 1A). When these yeast are subject to a concurrent overexpression of the chaperone Ssa1, the toxicity phenotype is exacerbated (Figure 1B). We previously reported this phenomenon in the context of chaperone balance, and concluded that the relative availability of the Hsp70s is a factor that influences toxicity (Keefer and True, 2016). However, we further demonstrated that imbalances can occur without changes in chaperone expression level. Other factors, such as localization or cofactor availability, can create these functional imbalances. Indeed, Ssa1 overexpression toxicity is not rescued by simultaneous overexpression of Ssb1 from the same promotor (Figure 1C). This indicated that imbalanced Hsp70 levels are not the cause of the enhanced toxicity, as Ssb1 and Ssa1 are known to have opposing activity upon [PSI+] (Allen et al., 2005). This result further suggested that chaperone imbalance-related toxicity is related to chaperone cofactors, and not the chaperones exclusively. We hypothesized that Ssa1-dependent toxicity may be due to a change of activity of its cofactors.
Figure 1. Overexpression of Sse1 rescues toxicity associated with excess Ssa1.
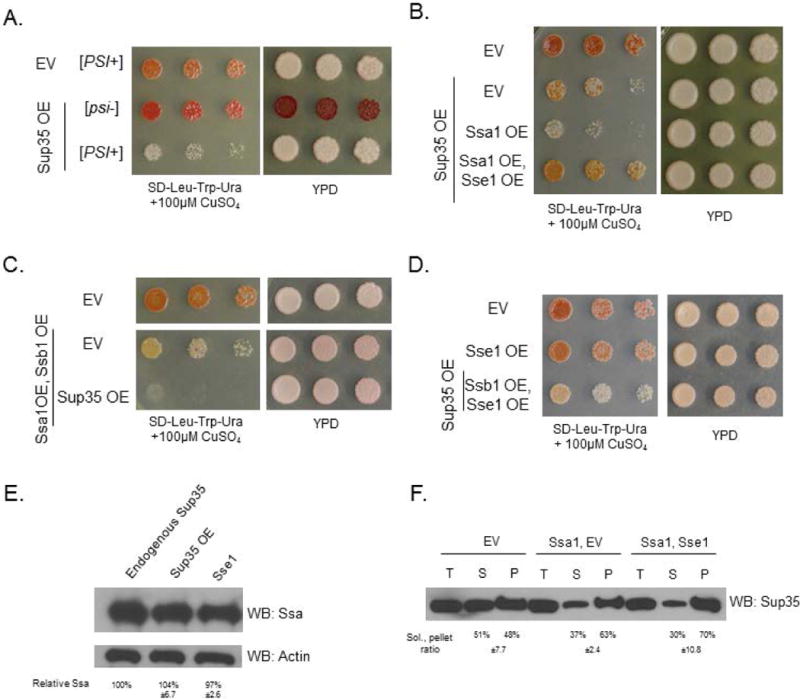
Throughout Figure 1, the top row empty vector (EV) control strains contain three vectors (LEU2-, TRP1-, and URA3-marked) with no open reading frames (Table S1). (A) Sup35 overexpression is toxic to strong [PSI+] yeast. (B) Ssa1 overexpression is toxic in strong [PSI+] yeast at low levels of Sup35 overexpression. toxicity is rescued by the overexpression of nucleotide exchange factor Sse1. (C) Overexpressing Ssb1 does not ameliorate the toxicity of Ssa1 overexpression. Though excess Ssb1 is generally beneficial to cells, the overabundance of Ssa1 continues to compete with Ssb1 for cofactors. Horizontal white bars separate spottings from the same plate. (D) Sse1 overexpression does not cause enhanced growth of cells relative to empty vector controls. Middle row: the Sse1 OE strain contains LEU2- and TRP1-marked empty vectors. (E) The overexpression of Sup35 or Sse1 does not change the levels of Ssa1 expression in yeast. Blots were quantified with ImageJ and normalized to actin. Quantifications are the means of three replicates. (F) A solubility assay was performed to assess the amount of total (T), soluble (S), and pellet/insoluble (P) Sup35. There were no differences in the solubility of Sup35 in the Ssa1 overexpression strain relative to the Ssa1/Sse1 overexpression strain, indicating that the Sse1 toxicity rescue is not due to modification of Sup35 aggregates. As expected, Ssa1 overexpression decreased the soluble pool of Sup35 relative to the EV control, as expected (Newnam et al., 1999). Blots were quantified with ImageJ, quantifications are the means of three replicates.
We therefore examined the role of Sse1 on Ssa1 activity. Sse1 is a nucleotide exchange factor (NEF) that is specific to the Hsp70 family of chaperones (Shaner et al., 2005; Andréasson et al., 2008). We chose Sse1 because it interacts with both the Ssa and Ssb protein families, perhaps exclusively (Bukau et al., 2006), and its overexpression is able to cure weak variants of [PSI+] (Kryndushkin and Wickner, 2007; Fan et al., 2007). It is known that Hsp70 cofactors, including NEFs, are typically present in sub-stoichiometric amounts in healthy cells (Kampinga and Craig, 2010). Thus, we hypothesized that Ssa1-related toxicity could be due to insufficient NEF activity relative to Ssa1 activity or abundance.
To test this, we overexpressed Ssa1 and Sse1 concurrently in [PSI+] cells overexpressing Sup35. We found that the expression of Sse1 strikingly rescued the toxicity associated with Ssa1 overexpression (Figure 1B). We tested Sse1 overexpression alone and in conjunction with Ssb1 overexpression (Figure 1D), and observed no change in growth relative to empty vector controls. This indicated that Sse1 overexpression does not globally enhance growth of yeast.
We assessed the effects of Sse1 overexpression on levels of Ssa1 and upon Sup35 solubility. We observed no changes in Ssa1 expression levels in response to the overexpression of Sup35 or Sse1 (Figure 1E). The overexpression of Sse1 together with Ssa1 did not change the amounts of soluble or insoluble Sup35 relative to overexpressing Ssa1 alone (Figure 1F; controls shown in Figure S1), indicating that the toxicity rescue effect is not due to resolubilization of Sup35 aggregates. Thus, we investigated other mechanisms by which Sse1 could be improving viability in conjunction with Ssa1 overexpression.
Ssa1 overexpression could trap clients or titrate cofactors
Sse1 is known to interact with both Ssa1 and Ssb1 (Shaner et al., 2005). We verified the role of Ssb by assessing Sup35 toxicity in an ssb1Δssb2Δ strain. We observed that overexpression of Sup35 was quite toxic (Figure 2). Overexpression of Sse1 did not rescue the toxicity (Figure 2), indicating that its role in rescuing Ssa-related toxicity is due to an Ssb-dependent effect.
Figure 2. Sse1 rescues toxicity via a mechanism that is dependent upon Ssb.
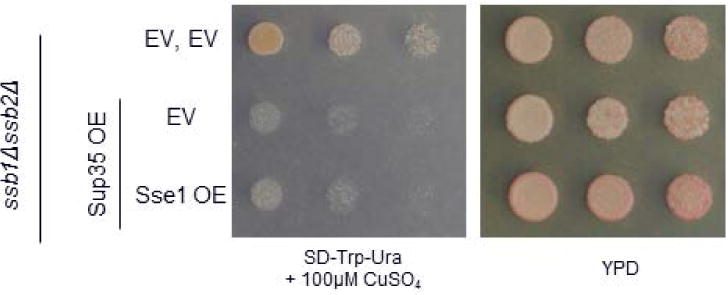
The ssb1Δssb2Δ strain experiences severe toxicity when subjected to Sup35 overexpression. This toxicity is not rescued by the overexpression of Sse1, indicating that the Sse1 rescue of Ssa1-induced toxicity occurs via an interaction with Ssb.
Thus, we generated two theories about why Sse1 overexpression rescued the toxicity associated with Ssa1 overexpression (Figure 3). The first model suggests that the Ssa1 toxicity is due to a “trapping” of substrates caused by an insufficient amount of nucleotide exchange. In other words, overexpression of Ssa1 may overload the cell’s nucleotide exchange factors and create a stoichiometric mismatch. After a client protein binds to the substrate-binding domain (SBD), the SBD closes and traps the client until the ADP dissociates and a new ATP binds to open the Hsp70 (Bukau et al., 2006). With too few NEFs available to remove bound ADPs, clients would be sequestered, potentially impacting cell viability. Overexpressing Sse1 would partially relieve the imbalance and allow client proteins to be folded efficiently.
Figure 3. Model for cause of Ssa1 toxicity.
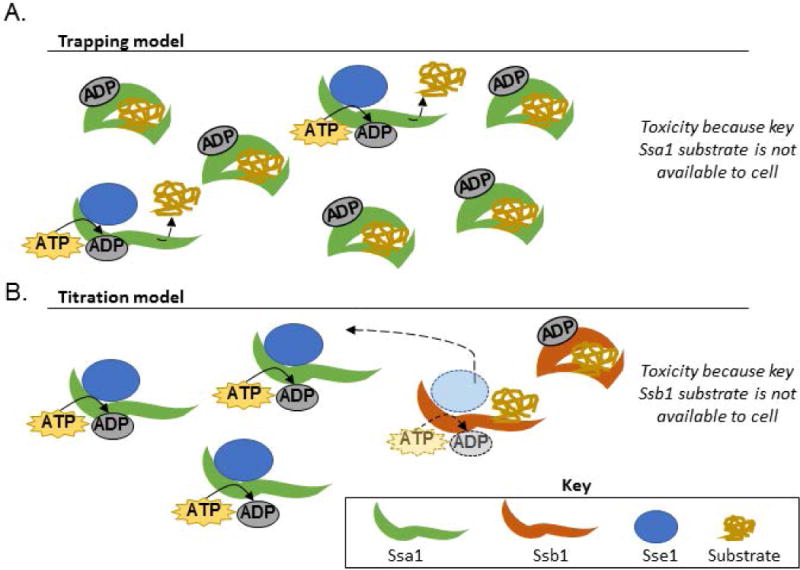
(A) The “trapping” model suggests that overexpressing Ssa1 is toxic because crucial proteins are not made available to the cell. Ssa1 overexpression leads to a relative lack of available nucleotide exchange factors, so bound substrates are not efficiently cycled through the folding process. Overexpressing Sse1 partially relieves the mismatch between available Ssa1 and available NEFs. (B) The “titration” model suggests that Ssa1 overexpression causes cofactors to interact less frequently with Ssb1 due to stoichiometric imbalances. Titrating cofactors, such as the Sse1, could lead to decreased Ssb1 activity, causing reduced growth. Overexpressing Sse1 would restore the activity of both chaperones by increasing cofactor availability.
The second model proposes that overexpressing Ssa1 titrates certain nucleotide exchange factors away from other Hsp70s, such as Ssb1. Sse1 is a cofactor for both Ssa1 and Ssb1. An increase in Ssa1 activity could reduce the amount of contact between Sse1 and Ssb1. Overexpressing Sse1 would allow both classes of Hsp70s to resume normal function. The major distinction between the Ssa1 overexpression models lies in which Hsp70 remains trapped in the ADP-bound state: in the “trapping” model, Ssa1 retains substrates, while the “titration” model suggests that Ssa1 cycles substrates while Ssb1 remains ADP-bound.
To distinguish between these models, we first decided to examine the Hsp70 domains. Wild type Hsp70s contain three domains: a 40kDa N-terminal nucleotide binding domain, a 20kDa substrate binding domain, and a 10kDa variable domain (Figure 4A) (Sharma and Masison, 2009; Kampinga and Craig, 2010). We utilized chimeric Hsp70 proteins in order to probe the effects of each domain of Ssa1 and Ssb1 (James et al., 1997). In total, we employed six chimeras representing all combinatorial groupings of these domains (Figure 4B), along with the wild type proteins in identical vectors. Each construct is referred to by its N- to C-terminal identity. For example, “ABA” refers to the chimera that contains the Ssa1 nucleotide binding domain, the Ssb1 substrate binding domain, and the Ssa1 variable domain.
Figure 4. The nucleotide binding domain of Ssa1 is responsible for toxicity.
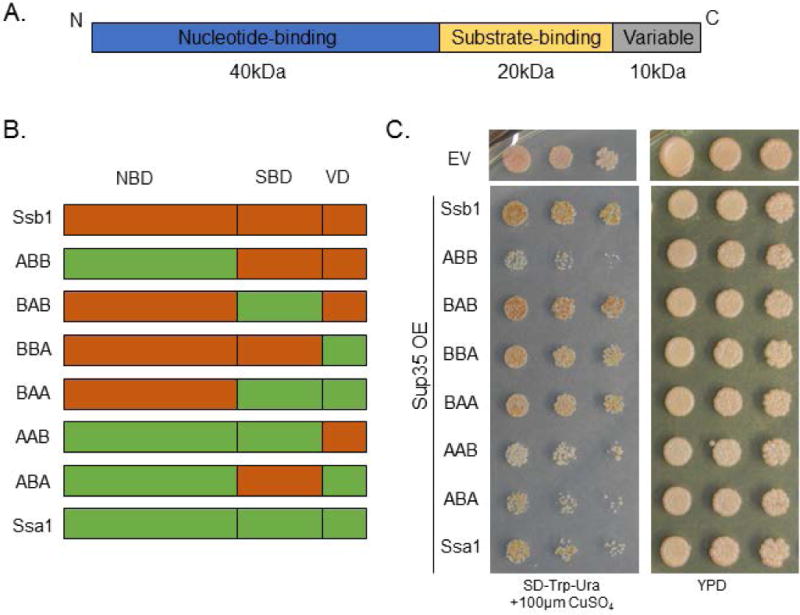
(A) The Saccharomyces cerevisiae Hsp70s have a common domain architecture consisting of an N-terminal nucleotide binding domain, an intermediate substrate-binding domain, and a C-terminal variable domain. (B) We utilized chimeric proteins to investigate the effect of each domain of Ssa1 and Ssb1. All combinatorial possibilities were tested. Chimeras are named by their domains, e.g. “ABB” contains the Ssa1 nucleotide binding domain, the Ssb1 substrate binding domain, and the Ssb1 variable domain. (C) Expressing the Hsp70 chimeras demonstrates that the ABB and ABA proteins are toxic in strong [PSI+] yeast in conjunction with Sup35 overexpression. Horizontal white bar separates spottings from different plates, but performed at the same time on identical media. The empty vector (EV) control strain contains two vectors (TRP1-, and URA3-marked) with no open reading frames (Table S1).
We expressed these fusion proteins in conjunction with Sup35 overexpression and observed that the nucleotide binding domain of Ssa1 was responsible for the most toxic phenotypes (Figure 4C). Conversely, all chimeras containing the Ssb1 nucleotide binding domain grew strongly when challenged with Sup35 overexpression. Surprisingly, one of the most toxic combinations was ABB, indicating that the Ssa1 nucleotide binding domain alone can be harmful to cell growth. The ABA chimera was also quite toxic, and yeast containing AAB experienced toxicity similar to WT Ssa1. We considered the possibility that a mismatch between nucleotide binding domains and substrate binding domains may be innately toxic; however, the BAB and BAA chimeras both grew robustly.
This result supported the “titration” model; that is, the NBD of Ssa1 draws nucleotide exchange factors away from Ssb1. We arrived at this interpretation by examination of the AAB and ABA chimeras. If excess Ssa1 was retaining key substrates (the “trapping” model), then the AAB chimera would exhibit similar toxicity to Ssa1, but ABB would grow strongly because of its lack of Ssa1 substrate binding domain. However, ABB was even more toxic than Ssa1. We hypothesized that the extreme toxicity of the ABB and ABA chimeras were due to two factors. First, the presence of the Ssa1 nucleotide binding domain titrated NEFs away from Ssb1. Second, the presence of the Ssb1 substrate binding domain attracted client proteins away from the likely more-functional endogenous Ssb1, further harming cellular viability.
Sse1 overexpression confirms the importance of the Ssa1 nucleotide binding domain
To confirm the role of the Ssa1 nucleotide binding domain in toxicity, we again analyzed the effect of Sse1 overexpression. We co-expressed Sse1, Sup35, and each Hsp70 chimera and assessed yeast viability (Figure 5). Again, we found that Sse1 overexpression partially rescued the toxicity associated with ABB, as well as other chimeras containing the Ssa1 nucleotide binding domain.
Figure 5. Sse1 overexpression rescues Hsp70 chimera toxicity.
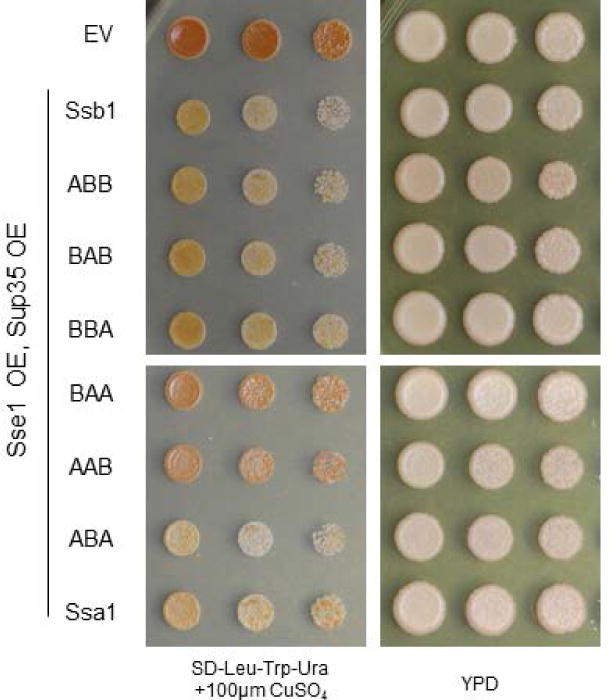
Introducing excess Sse1 into yeast containing Hsp70 chimeras rescued the toxicity associated with the Ssa1 nucleotide binding domain. Horizontal white bars separate spottings from the same plate. The empty vector (EV) control strain contains three vectors (LEU2-, TRP1-, and URA3-marked) with no open reading frames (Table S1).
A functional chaperone and cofactor balance is essential to survival
To further validate the titration model, we challenged the Ssa1 nucleotide exchange cycle. We hypothesized that a more active Ssa1 ATPase domain would exacerbate the toxicity associated with Ssa1 overexpression, as more NEFs would be titrated from Ssb1 in order to exchange ADP. Therefore, we examined the effect of Ydj1 on Ssa1-related toxicity. Ydj1 is an Hsp40 J-protein that stimulates Ssa1 ATPase activity, though not Ssb1 ATPase activity, and delivers client proteins to Ssa1 (Cyr and Douglas, 1994; Shorter and Lindquist, 2008). We hypothesized that increased levels of Ydj1 could speed the overall nucleotide cycle via enhancing ATPase activity or delivering more substrates to Ssa1 (likely a combination of the two), which would increase the need for NEFs. Indeed, we observed that Ydj1 overexpression was highly toxic, even more than Ssa1 overexpression alone (Figure 6, compare to Figure 1A).
Figure 6. Functional chaperone balance is not restored by overexpressing Ydj1 or Ssb1.
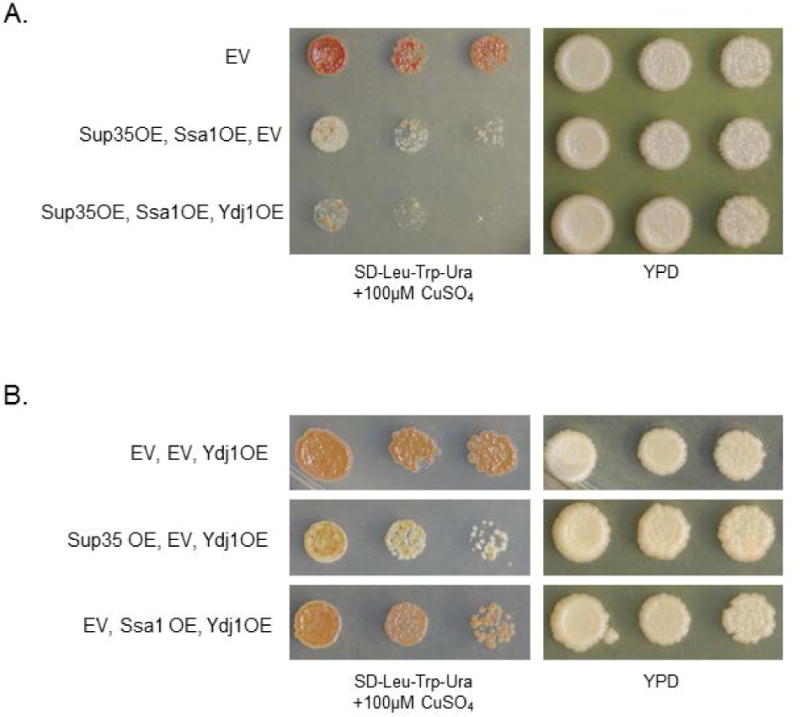
(A) Ydj1 is an Hsp40 J-protein that enhances Ssa1 ATPase activity. Overexpressing Ydj1 with Sup35 and Ssa1 is more toxic than Sup35 and Ssa1 overexpression alone. The EV strain contains LEU2-, TRP1-, and URA3-marked empty vectors. (B) Ydj1 overexpression alone is not toxic. The co-overexpression of Ssa1 and Ydj1 is not toxic without Sup35 overexpression.
Discussion
Here, we have shown that chaperone-related toxicity is caused in part by stoichiometric imbalances of cofactors. In particular, we demonstrated that the prion-dependent toxicity of Ssa1 overexpression is influenced by changes to the relative availability of the nucleotide exchange factor Sse1. We have provided evidence to support a “titration model,” in which Ssa1 overexpression titrates cofactors away from Ssb1, reducing its functionality and its ability to cycle through substrates. Further, we have demonstrated that this effect is dependent upon Ssb (Figure 2) and is not a result of modification of Sup35 aggregates (Figure 1F).
Though we limited this investigation to Ssa1, Ssb1, and Sse1, there are undoubtedly other consequences of chaperone imbalance that are not described herein. Changes in chaperone levels or folding ability can lead to enhanced genetic variation, prion modification, and changes in cytoskeletal organization (Rutherford and Lindquist, 1998; Newnam et al., 1999; Johnson et al., 2015). These effects are not limited to yeast, as Hsp70 overproduction in Drosophila can be deleterious to growth (Feder et al., 1992). Future investigation into the downstream effects of chaperone imbalance will shed light on the mechanisms by which they contribute to cells monitoring and reacting to protein folding challenges.
In our lab strains, Ssa1 overexpression was not toxic in [PSI+] cells without the concurrent overproduction of the prion-forming protein Sup35 (Keefer and True, 2016). The Sup35 overexpression toxicity is known to be caused by the sequestration of translation termination factors (Vishveshwara et al., 2009); however, the exacerbation of the toxicity by Ssa1 was a surprising result. We theorize that Ssa1 enhances the toxicity due to the greater folding burden that is assumed by cells when Sup35 is overexpressed in a [PSI+] phenotypic state. Ssa1 overproduction, and subsequent Ssb1 under-function, may be tolerable under endogenous conditions, but becomes toxic when widespread protein misfolding challenges the chaperone network. This is in line with the hypothesis that cells contain excess free chaperones in order to protect against unforeseen folding needs (Morimoto, 2008). It is reasonable to believe that other methods of inducing protein misfolding could also lead to Ssa1-related toxicity. Though Ssa1 and Sse1 overproduction do not affect Sup35 and Sup45 sequestration, they do demonstrate that chaperone and NEF interventions can exacerbate or ameliorate toxicity associated with misfolded proteins.
In our previous study, we investigated a system that mimicked Ssa1 overexpression, via increased availability of Ssa1 relative to Ssb1, without any change in expression of either Hsp70 (Keefer and True, 2016). Others have also reported that expression levels are less important than the interaction, or competition, between the Hsp70s (Kiktev et al., 2015). Practically, this means that different genetic backgrounds or cellular stressors could make an organism more susceptible to chaperone-related toxicity. Cofactors are typically present in sub-stoichiometric amounts relative to their Hsp70s, a fine balance that is perturbed by either over- or under-expression of J-proteins and NEFs (Kampinga and Craig, 2010). Similar conclusions have been reached for Hsp90s and their cofactors (Johnson et al., 2014). These studies indicate that chaperone networks are finely-tuned and that there are perils associated with altering Hsp availability relative to their cofactors.
To that end, modification of chaperones has been suggested in the treatment of various human diseases, ranging from neurodegenerative conditions to cancer (Mahalingam et al., 2009; Ebrahimi-Fakhari et al., 2011; Rappa et al., 2012; Lindberg et al., 2015). Our work informs important considerations for these future research avenues; namely, each chaperone must be considered as part of an interconnected network. Modulating the activity of one chaperone can have unforeseen consequences upon other chaperones, in a manner that may be unpredictable under conditions of cellular stress.
Experimental Procedures
Yeast cultures and transformation
Yeast were cultured using standard techniques (Guthrie and Fink, 1991) Plasmid transformations were performed by the PEG/LiOAC method (Guthrie and Fink, 1991). All yeast are WT 74D-694 and are strong [PSI+] (Derkatch et al., 1996). A complete list of plasmids is available in Table S1.
Yeast spotting
Yeast cultures were grown overnight in selective media. Cultures were pelleted, washed, and resuspended in water to an optical density of 1.0. The normalized yeast solutions were pipetted into a 96-well plate, and serial dilutions (1:5) were made using a multichannel pipette. Yeast were spotted onto plates using an ethanol-sterilized 48-pin replicator that was placed in the appropriate wells and then onto an agar plate. Throughout the paper, YPD plates are used as controls to show the total number of cells plated, as even the slight amount of copper in synthetic media is enough to induce toxicity in yeast that contain p314CUP1-Sup35. For all experiments, the amount of CuSO4 in media was titrated to compare growth within each experiment. CuSO4 levels were altered to reveal relative effects of protein overexpression.
Protein expression quantification
Cultures of yeast were grown to mid-log phase and lysed via mechanical disruption in lysis buffer containing 20mM Tris, 0.5M NaCl, 5mM imidazole, 2M urea, 0.1% IGEPAL, 50mM NEM, 3mM PMSF, and 1 tablet Roche Protease Inhibitor Cocktail #4693159001. Lysates were normalized by total protein and boiled in SDS-PAGE sample buffer (200mM Tris-HCl pH 6.8, 4% SDS, 0.4% bromophenol blue, 40% glycerol) prior to loading on a 10% SDS-PAGE gel. Western blotting was performed by standard protocols. Blots were quantified with ImageJ.
Sup35 Solubility
The Sup35 solubility assay was adapted from Geiler-Samerotte et al., 2011. Liquid cultures of yeast were pelleted, washed, and resuspended in 25μl Soluble Protein Buffer (50mM Tris-HCL, 500mM NaCl, protease inhibitors) and transferred to curve-bottomed 2mL microcentrifuge tubes (United Laboratory Plastics) that each contained one 5mM diameter steel ball (Retsch). Tubes were frozen in liquid N2. Cells were lysed by vortexing the tubes for 4x90s intervals, returning to liquid N2 between rounds. Following standardization via Bradford assay, a “total” fraction was taken and the remaining lysate was spun at 100,000xg for 20 minutes at 4°C in a Beckman Optima TLX Ultracentrifuge (TLA100). The supernatant was retained as the “soluble” fraction, and the pellet was resuspended in wash buffer (50mM Tris-HcL, 150mM NaCl, protease inhibitors) and spun at 100,000xg for 20 minutes. The remaining pelleted material was resuspended in 200μl insoluble protein buffer (50mM Tris-HCl, 150mM NaCl, 8M Urea, 2% SDS, protease inhibitors) and retained as the “insoluble” fraction. All fractions were boiled in SDS-PAGE sample buffer (final concentration 50mM Tris-Cl pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100mM DTT) for 5 minutes before standard SDS-PAGE (10% polyacrylamide) and Western blotting.
Supplementary Material
Acknowledgments
We thank Drs. E. Craig and S. Lindquist for reagents. We are especially grateful to Dr. E. Craig for the use of the Ssa/Ssb chimera constructs and the anti-Ssa antibody. We are appreciative of members of the True lab for helpful discussions and comments on the manuscript and to Dr. Ankan Bhadra for technical assistance. The authors have no conflict of interest to declare.
Grant Number: R01 GM072778
Footnotes
Author Contributions
K.K. and H.T. designed experiments. K.K. performed experiments. K.K. and H.T. wrote the manuscript. H.T. secured funding.
References
- Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 Is a Nucleotide-activated Exchange Factor for Hsp70. J Biol Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular Chaperones and Protein Quality Control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Inge-Vechtomov SG, Derkach IL, Ptyushkina MV, Tarunina OV, Dagkesamanskaya AR, Ter-Avanesyan MD. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 1992;8:489–499. doi: 10.1002/yea.320080702. [DOI] [PubMed] [Google Scholar]
- Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Dagkesamanskaya AR, Ter-Avanesyan MD. Interaction of the yeast omnipotent suppressors SUP1(SUP45) and SUP2(SUP35) with non-mendelian factors. Genetics. 1991;128:513–520. doi: 10.1093/genetics/128.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Molecular Chaperones in Parkinson’s Disease – Present and Future. J Park Dis. 2011;1:299–320. [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA. An atlas of chaperone–protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Academic Press Inc; San Diego, CA: 1991. [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Johnson CR, Weems AD, Brewer JM, Thorner J, McMurray MA. Cytosolic chaperones mediate quality control of higher-order septin assembly in budding yeast. Mol Biol Cell. 2015;26:1323–1344. doi: 10.1091/mbc.E14-11-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Zuehlke AD, Tenge VR, Langworthy JC. Mutation of essential Hsp90 co-chaperones SGT1 or CNS1 renders yeast hypersensitive to overexpression of other co-chaperones. Curr Genet. 2014;60:265–276. doi: 10.1007/s00294-014-0432-3. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer KM, True HL. Prion-Associated Toxicity is Rescued by Elimination of Cotranslational Chaperones. PLOS Genet. 2016;12:e1006431. doi: 10.1371/journal.pgen.1006431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Melomed MM, Lu CD, Newnam GP, Chernoff YO. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol Microbiol. 2015;96:621–632. doi: 10.1111/mmi.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. The Biology of Proteostasis in Aging and Disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prions in Yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg I, Shorter J, Wiseman RL, Chiti F, Dickey CA, McLean PJ. Chaperones in Neurodegeneration. J Neurosci. 2015;35:13853–13859. doi: 10.1523/JNEUROSCI.2600-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Targeting HSP90 for cancer therapy. Br J Cancer. 2009;100:1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic Interactions between Yeast Chaperones Hsp104 and Hsp70 in Prion Curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Rappa F, Farina F, Zummo G, David S, Campanella C, Carini F, et al. HSP-molecular chaperones in cancer biogenesis and tumor therapy: an overview. Anticancer Res. 2012;32:5139–5150. [PubMed] [Google Scholar]
- Reidy M, Masison DC. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion. 2011;5:245–249. doi: 10.4161/pri.5.4.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Sharma D, Masison DC. Hsp70 Structure, Function, Regulation and Influence on Yeast Prions. Protein Pept Lett. 2009;16:571–581. doi: 10.2174/092986609788490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


