Abstract
Traditional 2D cell cultures do not accurately recapitulate tumor heterogeneity, and insufficient human cell lines are available. Patient-derived xenograft (PDX) models more closely mimic clinical tumor heterogeneity, but are not useful for high-throughput drug screening. Recently, patient-derived organoid cultures have emerged as a novel technique to fill this critical need. Organoids maintain tumor tissue heterogeneity and drug-resistance responses, and thus are useful for high-throughput drug screening. Among various biological tissues used to produce organoid cultures, circulating tumor cells (CTCs) are promising, due to relative ease of ascertainment. CTC-derived organoids could help to acquire relevant genetic and epigenetic information about tumors in real time, and screen and test promising drugs. This could reduce the need for tissue biopsies, which are painful and may be difficult depending on the tumor location. In this review, we have focused on advances in CTC isolation and organoid culture methods, and their potential applications in disease modeling and precision medicine.
Keywords: Cancer, Organoids, Circulating tumor cells, Precision medicine
1. Introduction
Circulating tumor cells (CTCs) are shed from primary tumor and/or metastatic lesions into the vasculature and initiate metastatic lesions at distant sites. In 1869, Thomas Ashworth, an Australian physician, first identified cells similar to cancer cells in blood drawn from the saphenous vein [1]. In 1889, Stephen Paget proposed a ‘seed and soil’ hypothesis, in which certain tumor cells (“seeds”) have a specific affinity for the environment of specific organs (‘soil’), and compatibility between the seed and soil leads to metastasis [2]. CTCs are considered as ‘seeds’ distributed by primary tumors for potential initiation of metastatic growth at distant organ sites. CTCs reflect tumor heterogeneity, and could be genotyped and functionally characterized to study and target the evolving mutational landscape of primary and/or metastatic tumors [3–5].
CTCs have opened a new avenue towards combating cancer by acting as important indicators of metastatic disease and prognostic biomarkers. Several studies in different solid cancers such as breast [6,7], lung [8], prostate [9], and esophageal squamous cell carcinoma [10] have suggested that effective chemotherapy or hormonal therapy are associated with decreased CTCs. Correlations between numbers of CTCs, and survival time – both progression-free survival (PFS) and overall survival (OS) before and after surgery – are considered important pharmacodynamics data in treatment response studies to determine clinical outcomes and risk of relapse [11,12]. Meta-analyses of patients with ovarian [13] and lung cancer [14] showed a strong link between number of CTCs and cancer progression and treatment response, determined as shorter PFS and OS [15]. Normanno et al. (2013) noted that after the first cycle of chemotherapy, CTCs decreased in patients with small cell lung cancer [16]. In gastrointestinal cancers (e.g. pancreatic, gastric, and colorectal cancers), CTCs are being used to predict distant metastasis and patient survival, and are helpful in tumor staging during chemotherapy and/or radiotherapy [17,18]. Higher expression of multidrug-resistance-related proteins (MRPs) and aldehyde dehydrogenase 1 (ALDH1) in CTCs are associated with shorter PFS and predicts response to chemotherapy in breast cancer [19]. Blassl et al. (2016) identified therapeutic resistance in ovarian cancer cells through gene expression profiling of CTCs [20]. In short, CTCs have now emerged as a ‘liquid biopsy’, offering a safe, low-cost and repeatable tissue source that is an alternative to invasive biopsies, and can be immensely useful in the diagnosis and prognosis of various cancers.
Immense efforts have been made to isolate live CTCs and culture them for genetic and epigenetic characterization of tumors. Further, CTCs in culture could be useful to screen promising drugs and making important treatment decisions in emerging precision medicine and targeted treatment regimens. This review focuses on the current status of efforts to isolate and culture CTCs, to grow three-dimensional organoids for potential applications in cancer research and drug development.
2. Current methodologies for isolation and characterization of CTCs
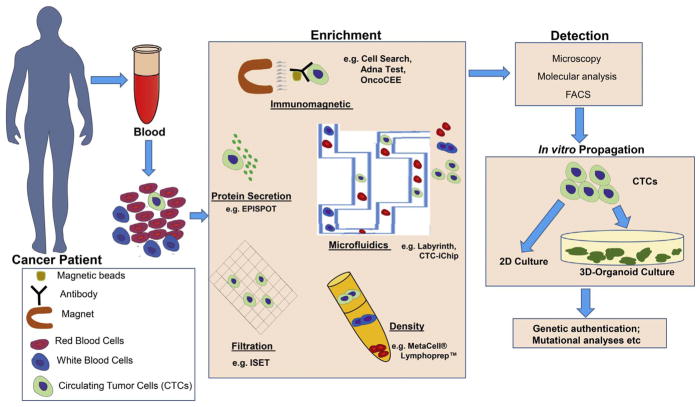
For organoid cultures, it is important to isolate sufficient numbers of viable CTCs from blood. In the last few decades, several methods have been developed for isolation of CTCs using biological characteristics (such as surface marker expression) and physical properties (e.g. density, size and electrical charge) (see summary in Table 1 and Fig. 1).
Table 1.
Comparative analysis of different isolation and enrichment techniques for CTCs.
| Sl no. | Technique | Methods of separation | Detection marker | Cancer | Key feature | Limitation | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Cell Search® | Immunomagnetic | EpCAM | Prostate, colorectal, breast | FDA-approved method; clinically relevant automated system; quantitative and highly reproducible | Detection depends on EpCAM-positivity only; expensive and subjective image evaluation | [22–25] |
| 2 | AdnaTest | Immunomagnetic | EpCAM | Breast, ovarian, prostate, colorectal | Enrichment through anti-EpCAM-coated magnetic particles followed by RT-PCR analysis of cancer-specific transcripts; highly sensitive; cost-effective; requires less sample | Detection depends on EpCAM and MUC1 positivity; preprocessing of blood leads to loss of CTCs | [26–28] |
| 3 | OncoCEE | Immunomagnetic | antibody cocktail | Breast, colorectal | Capture CTCs through EpCAM, detection through cocktail of antibodies (TROP2, MUC1, HER2, EGFR and N-cadherin); high sensitivity | EpCAM based capture; blood needs to be preprocessed | [31,32] |
| 4 | MagSweeper | Immunomagnetic | EpCAM | Breast, prostate, colorectal | No preprocessing of blood; CTCs could be separated through magnetic rod labelled with anti EpCAM antibody; genetic analysis can be performed in CTCs | Detection depends on EpCAM-positivity only | [38–42] |
| 5 | CTC-Chip | Micropost Array | EpCAM, size | Prostate, breast, pancreatic, colon, lung | Isolation of 99% of viable CTCs from blood; visual confirmation; CTCs could be harvested for genetic analyses | Detection depends on EpCAM-positivity only; potential subjectivity in analysis | [43] |
| 6 | Geometrically enhanced differential immunocapture (GEDI) device | Microfluidic-based enrichment | PSMA/HER2, size | Prostate, breast, gastric | Uses a combine approach of antibody-coated microposts (positive enrichment) and hydrodynamic chromatography (size-based separation) | Nontransparent in nature; larger-scale production is difficult. | [44,45] |
| 7 | Herringbone (HB) chip, Graphene oxide (GO) chip and Geometrically enhanced mixing (GEM) chip | Surface-capture microfluidic devices | Surface-coated with EpCAM in the microfluidic device | Prostate, pancreatic, breast, lung | Transparent surface-coated devices useful in high-resolution imaging of CTCs; suited for large-scale production | Requires larger volume of blood; immobilization of captured CTCs on the surface makes it harder to retrieve | [46–48] |
| 8 | Magnetic sifters | Microfluidic chip | Size, EpCAM | Lung | Magnetic pores arranged in a honeycomb pattern and uses a flow-through fluidic array configuration; high-throughput capture of CTCs followed by efficient release | Detection depends on EpCAM positivity only | [49] |
| 9 | Ephesia chip | Microfluidic and immunomagnetic-based | EpCAM-based sieving method | Prostate and breast | CTCs are captured through antibody coated magnetic beads self-assembled in the microchip | Detection depends on EpCAM-positivity only | [50,51] |
| 10 | Liquid Biopsy ® | Microfluidic device | EpCAM-based positive selection | Breast | Authenticated platform that captures CTCs labelled with magnetic nanoparticle from blood sample. Useful for Next Generation Sequencing Studies | Detection depends on EpCAM-positivity only | [52] |
| 11 | IsoFlux | Microfluidic platform | Uses flow control and immune-magnetic capture | Prostate | Larger-diameter magnetic beads (4.5 μm) cause larger magnetic moment compared to nanoscale particles employed by the CellSearch System; maximum recovery of low EpCAM-expressing cells with fewer beads; enables molecular characterization of intact viable CTCs | Limited flow rate | [53] |
| 12 | CTC-iChip | Microfluidic immunomagnetic-based | EpCAM, CD45/CD15, CD66b | Melanoma, prostate, breast | Allows sequential separation of different blood components through micropillar array, hydrodynamic size-based sorting and magnetophoresis | Samples not suitable for DNA sequencing | [54,55] |
| 13 | Spiral microfluidics | Microfluidic device | Size and deformability | Breast, lung | Label-free, tumor antigen–independent; optically transparent microfluidic device; enumerate CTCs in suspension; high specificity and high purity of sample; greater accuracy | Cannot isolate CTCs < 12 μm in size | [57,58] |
| 14 | Labyrinth | Microfluidic device (size-based) | Combination of long loops and sharp corners | Breast, Pancreatic | Label-free and surface expression-independent microfluidic device with high-throughput isolation of CTCs | Needs to be tested further in other cancer models | [59] |
| 15 | MetaCell® and Lymphoprep™ | Size, density | Density gradient centrifugation | Colorectal cancer | RT-PCR analysis to detect CTCs | Low specificity | [60,61] |
| 16 | Ficoll-Paque | Density | Density gradient centrifugation | Colorectal | Detection of both EpCAM-positive and negative CTCs; easy to handle and inexpensive | Chance of cross contamination between different layers; low specificity | [62] |
| 17 | OncoQuick® plus | Density, size | Density gradient centrifugation | Breast | Detection of both EpCAM-positive and negative CTCs; easy to handle; inexpensive: no chance of cross contamination among different layers | Low specificity, preprocessing leads to CTC loss | [63] |
| 18 | Epithelial ImmunoSPOT Assay (EPISPOT) | Protein secretion | Cytokeratin-19, mucin-1 and PSA | Breast, Prostate and Colon | Discriminates between viable and apoptotic CTCs using protein secretion; limited number of markers | Proteins must be actively secreted | [64,65] |
| 19 | ISET technology | Size | Filtration | Breast, hepatomas, prostate, lung | Isolation of intact CTCs, without a previous immune-based selection; easy and rapid; can isolate EpCAM-negative CTCs; CTCs can be further analyzed for multiplexed imaging and genetic analysis | Retention of larger CTCs (size) of leukocyte range only; lower specificity | [67,68] |
| 20 | ScreenCell® | Size | Filtration | Prostate | Isolates live CTCs within minutes with a high recovery rate; immunocytochemistry and FISH assays can be performed directly on the filter; more chances of getting high-quality genetic material | Single use; variations in cell size within a single population lead to CTC loss | [69,70] |
Fig. 1.
Development and characterization of 3D organoids from CTCs. CTCs are isolated from a patient’s blood, followed by enrichment and detection as shown. Thereafter, CTCs are cultured in 2D to generate cell lines, or 3D on Matrigel® in defined media conditions to generate organoids.
Immunoaffinity-based enrichment techniques are the first reported and most widely used techniques for CTC enrichment [21]. The Cell-Search® system (Veridex LLC) is an FDA-approved technique used for detection of CTCs in samples from patients with prostate [22,23], colorectal [24], and breast cancer [25]. It employs immunomagnetic separation (ferrofluid nanoparticles functionalized with an EpCAM antibody) combined with fluorescence imaging technology using antibodies to identify CTCs (with the criteria of EpCAM+, DAPI+, cytokeratins 8, 18+, and/or 19+, and CD45−) from white blood cells (with the criteria of cytokeratins-, CD45+, and DAPI+).
To further improve efficiency and detection speed, several new methods have been developed. These include AdnaTest (Adnagen AG), a commercial platform [26] that detects CTCs through an optimized cocktail of antibodies and a combination of tumor-associated markers. In this method, CTCs are first enriched through antibody-coated magnetic particles (including EpCAM), followed by reverse transcription-polymerase chain reaction (RT-PCR) analysis of tumor-associated genes (e.g. CA15-3, GA 733-2, and Her2) [26–28]. OncoCEE (commercialized by Biocept) uses an anti-EpCAM antibody for capture and a cocktail of TROP-2, MUC-1, HER2, EGFR and N-cadherin antibodies for detection of CTCs [29]; this approach had high detection efficiency in samples from patients with metastatic breast cancer [30].
However, two major drawbacks exist with these types of detection systems. First, the epithelial cell surface marker EpCAM is used to isolate CTCs, although EpCAM is not expressed in various sub-types of the same cancer [31]. EpCAM-based detection of CTCs did not recognize breast cancer cells of a normal cell-like subtype characterized by aggressive behavior [32]. Furthermore, downregulation of epithelial markers during epithelial–mesenchymal transition (EMT) is common in CTCs [33,34]. These problems have been overcome by using additional surface marker/s frequently expressed on cells lacking EpCAM, such as N-cadherin, vimentin, EGFR, CD133, and O-cadherin [35–37].
Preprocessing of blood using centrifugation or cell lysis can cause significant loss of CTCs. To resolve this issue, new enrichment techniques have been developed, such as MagSweeper, which used a magnetic rod to separate CTCs (magnetically labelled) from non-magnetically labelled cells [38]. This technique was used for genetic profiling studies in breast cancer [39,40] and prostate cancer [41], and for analyses of stem cells in colorectal cancer [42]. Cell-antibody interaction is the key for efficient capture of CTCs, which can be controlled by sample flow velocity and direction. In 2007, a microfluidic device called CTC-Chip was developed. It consists of a collection of microposts coated with anti-EpCAM antibody specifically designed for CTC enrichment [43]. Precise control of fluid flow promoted isolation of viable CTCs from blood of patients with metastatic prostate, breast, pancreatic, colon, and lung cancer [43]. This lead to further microfluidic-based enrichment techniques, such as a geometrically enhanced differential immunocapture (GEDI) device, which uses a combination of antibody-coated microposts (positive enrichment) and hydrodynamic chromatography (size-based margination) to reduce non-specific leukocyte adhesion. This technique was used to isolate CTCs in samples from patients with castration-resistant prostate cancer (PSMA+/CD45− cells) [44], as well as breast and gastric cancer [45].
Although promising, these micropost devices have limitations, such as their nontransparent nature combined with post structures (difficult for high-resolution imaging) and complexity of production at a larger scale. As a result, new surface-capture microfluidic devices have been developed, including a herringbone (HB) chip [46], a graphene oxide (GO) chip [47], and a geometrically enhanced mixing (GEM) chip [48]. By using surface-coated antibodies, these transparent devices allow high-resolution imaging of CTCs suited for large-scale production. However, trypsinization is required to retrieve CTCs, which are immobilized on these devices [47,48]; this step could cleave major surface receptors on CTCs important for their characterization. To address this problem, magnetic sifters (small microfluidic chips) were developed with a dense array of magnetic pores arranged in a honeycomb pattern. These chips sieve samples by vertical flow centrifugation and reportedly allow efficient capture of CTCs followed by high-throughput release [49]. Another device called the Ephesia chip contains antibody-coated magnetic beads self-assembled in the microchip; these were reported to capture CTCs in samples from patients with prostate and breast cancer [50,51].
Currently, a few automated commercial hybrid CTC enrichment platforms are also available based on both microfluidic and immunomagnetic principles (Table 1). These include Liquid Biopsy® (Cyvenio), an automated authenticated platform that captures CTCs labelled with magnetic nanoparticles from blood samples, and can be used in next-generation sequencing studies [52]. IsoFlux (Fluxion Biosciences) is a system using three interconnected fluidic reservoirs to separate cells labelled with anti-EpCAM coated magnetic beads from unbound cells, under the influence of a high magnetic field. This method was more sensitive than CellSearch in detecting CTCs in prostate cancer samples [53]. The CTC-iChip (Janssen Diagnostics), a microfluidic immunomagnetic-based CTC enrichment technique [54], allows sequential separation of different blood components through a micropillar array, hydrodynamic size-based sorting, and magnetophoresis. CTC-iChip uses inertial fluidics to focus all the nucleated cells and then use a positive or negative selection using magnetic beads to pull CTCs or WBCs [54,55].
Another class of microfluidics based separation technologies uses inertial hydrodynamic forces in microfluidic channels to separate the cells based on size, hence named “label free technologies” [56]. The advantage of these technologies is their high throughput. Using inertial forces either in linear or curved channels, cells can be focused and diverted into different streamlines based on their size. Spiral Microfluidics (Clearbridge Biomedics), a straightforward spiral microfluidic device with a trapezoidal cross-section for label-free enrichment of CTCs, leads to greater accuracy in genome sequencing and mapping, as well as in single-cell analysis [57,58]. More recently, a microfluidic labyrinth was developed to isolate CTCs at a 2.5 mL/min flow rate entirely based on size, with no positive or negative selection [59].
Other currently used methods are MetaCell®, a size-based enrichment technique for viable CTC enrichment from peripheral blood [60]; and Lymphoprep™ by Nycomed Pharm AS [61]; and Ficoll-Paque by GE-Amersham Biosciences, a density-based approach followed by RT-PCR analysis which has been used to detect CTCs in colorectal cancer [62]. OncoQuick® Plus by Greiner Bio One uses a combined approach of density gradient centrifugation and filtration to capture CTCs, and its use in samples from patients with metastatic breast cancer has been reported [63]. RosetteSep CTC Enrichment Cocktail (STEMCELL Technologies) is designed to enrich CTCs by negative selection, and unwanted cells are removed using tetrameric antibody complexes recognizing CD2, CD16, CD19, CD36, CD38, CD45, CD66b and glycophorin A, followed by centrifugation over a density gradient medium. Epithelial ImmunoSPOT Assay (EPISPOT), a secreted protein-based approach, has been used to capture CTCs in samples from patients with several different cancers [64,65].
Other commercial kits/systems based on filter-based size exclusion technologies use both molecular and cytopathologic approaches [66]. These include Isolation by Size of Epithelial Tumor cells (ISET technology) by Rare Cell Diagnostics; this technology was used to isolate CTCs as intact cells, without a previous immune-based selection, from a variety of cancer types [67,68]. ScreenCell® is a small filter-based device used to isolate and characterize CTCs [69,70]. These new technologies could overcome the drawbacks of existing technologies and facilitate long-term culture of CTCs in 2D and 3D organoid models.
3. CTC applications in cell culture models
3.1. 2D and 3D cell culture models
Cancer-related mortality rates remain high, in part, because of a high rate of failure in drug development due to the lack of sufficient clinically relevant preclinical models. Currently available cancer cell lines fall short of expectations as an effective clinically relevant model because of issues related to genotypic drift, cross-contamination with other cell lines, difficulty in establishment of permanent cell lines from primary tumors, loss of tumor heterogeneity, and adaptation to in vitro growth [71]. Furthermore, for several cancer types, sufficient numbers of clinically relevant cell lines are not available. For example, although prostate cancer is among the most common malignancies, hardly any cell lines for primary prostate cancer are available in public repositories. Further, cancer cell lines representing different races are also not available. These limitations demand novel measures to develop cancer cell culture models more representative of clinical situations. As CTCs are mostly derived from primary tumors, CTCs in culture could be a potential source of information about molecular drivers of cancer progression that could inform treatment decisions.
Due to the rareness of CTCs (i.e. approx. 1 in 1,000,000 of circulating cells), it has been challenging to establish cell culture and permanent cell lines in vitro or grow them in xenografts (in vivo) to perform functional analyses. However, significant advances have been made towards isolation and culture of CTCs. Alix-Panabieres et al. (2005) introduced the concept of short-term culture of CTCs in vitro on a membrane coated with antibodies to capture the secreted proteins and detect CTCs [64].
In 2013, Zhang et al. established primary cultures from CTCs (an ex-vivo expansion) obtained from patients with advanced stage breast cancer [72]. After isolating EpCAM (−) CTCs with the brain metastasis-selected markers (BMSMs) HER2(+)/EGFR(+)/HPSE(+)/Notch1(+), this group studied the invasiveness of these CTC lines [72]. CTC lines with BMSMs had high invasiveness and led to development of brain and lung metastasis in nude mice [72].
One year later, Yu et al. (2014) established six CTC lines using samples from ER+ breast cancer patients, through CTC-iChip technology [73]. They maintained the CTCs in serum-free media for>6 months, with basic fibroblast growth factor (FGF) and epidermal growth factor (EGF), under hypoxic conditions (4% O2) [73]. Genome sequencing confirmed the mutational status of PIK3CA gene, estrogen receptor gene (ESR1) and fibroblast growth factor receptor gene (FGFR2) in these CTC-derived cell lines. Moreover, they confirmed the cytological similarity between cultured CTCs and primary CTCs (isolated from a patient) and tumorigenic properties of these cell lines in mice [73]. In another study, Zhang et al. (2014) established a three-dimensional (3D) co-culture model for better in-situ capture and culture of CTCs [74]. After isolating CTCs in samples from patients with lung cancer, they cultured CTCs on microfluidic chips along with tumor-associated fibroblasts and extracellular matrix proteins to construct a tumor microenvironment favorable for growth of CTCs. Matched mutations were detected between expanded CTCs and primary tumors [74]. In another study, this 3D co-culture model [74] was used to capture and expand CTCs from multiple blood draws through the treatment cycle of a patient with lung adenocarcinoma [75]. This study demonstrated the usefulness of CTCs to assess ALK rearrangement as well as serial genetic alterations, matching similar observations in tumor biopsies [75].
3.2. Organoid culture model
Organoids are miniscule models of tissues grown in a 3D semisolid extracellular matrix with specific growth factor–supplemented medium [76,77]. Single epithelial cells can form organoids in 7–10 days; these can be dissociated into single cells to reinitiate organoid formation. A major achievement in organoid culture emerged in 2009, when Sato et al. established the mini gut culture system from mouse small intestinal crypts with defined media conditions for better growth [77]. The technology was subsequently adapted for other digestive epithelial tissues, such as the epithelium of stomach, colon, pancreatic ducts, and liver bile ducts, as well as various cancer types [76,78–80].
In organoid culture systems, isolated single cells are grown in Matrigel® (as a substitute for basal lamina), a 3D laminin and collagen-rich matrix along with optimal niche factors to form organoids. The niche factors (briefly summarized in Table 2) include B27, N2, R-spondin1, noggin, N-acetylcysteine, recombinant EGF, nicotinamide, recombinant FGF, recombinant FGF10, and SB-431542. R-spondin-1 (Wnt signal-enhancer) is considered responsible for long-term expansion of intestinal epithelial organoids [77] and for prostate development, including luminal cell differentiation [81]. Noggin is a known inhibitor of bone morphogenetic protein (BMP) signaling [82] and considered essential for proliferation of epithelial cells and prostate budding. Therefore, it is used to promote organoid formation and expansion. EGF, FGF2, FGF10, and prostaglandin E2 (PGE2) support epithelial cell proliferation and are important factors in human small intestinal cultures [83,84]. SB202190 (p38 inhibitor) and nicotinamide also are considered essential for human small intestinal cultures [76] and SB202190 is considered important for keratinization [85]. A83–01 (Alk3/4/5 inhibitor) is included to prevent the proliferative block in prostate cells via inhibition of the TGF-β signaling cascade [86]. The Rho/ROCK kinase inhibitor Y-27632 is necessary for long-term expansion of primary prostate epithelial and stroma-derived feeder cells [87]. Dihydrotestosterone (DHT) in media significantly enhances the efficiency of prostate organoid formation [85]. Differentiation can be achieved by withdrawing growth factors and simultaneously blocking Notch signaling (dibenzazepine, a γ-secretase inhibitor) [76,83]. In contrast to normal human tissues, several niche factors could be dispensable for the growth of organoids derived from cancerous tissue.
Table 2.
Organoid media composition.
| Organoids | Culture conditions | Function | Reference |
|---|---|---|---|
| Intestine | WNT3A and FGF4 | Differentiation (hindgut specification and morphogenesis) | [76,112] |
| R-spondin 1, Noggin, EGF, FGF4, WNT, L-glutamine, HEPES, N2 supplement, B27 supplement | Maturation | ||
| Colon | EGF, R-spondin 1, Noggin, WNT3A, Nicotinamide, Gastrin, TGFβ inhibitor (A-83-01), p38 inhibitor (SB202190) | Establishment | [76] |
| Without WNT3A, p38 MAP kinase inhibitor and nicotinamide | Differentiation | ||
| Gastric | EGF, R-spondin 1, Noggin, FGF10, WNT, Gastrin, Nicotinamide, A-83-01, RHOK (Y-27632), insulin-like growth factor (IGF), SB202190, (GSK)3β inhibitor (CHIR99021); prostaglandin E (PGE)2, retinoic acid | Organoid formation | [113,114] |
| IGF, p38 inhibitor, GSK3b inhibitor, and A-83-01 | Induced budding structures | ||
| Liver | Noggin, WNT, ROCK inhibitor | Establishment | [115] |
| N2 supplement, B27 supplement, N-Acetylcysteine, Gastrin, EGF, R-spondin 1, FGF10, Hepatocyte growth factor, Nicotinamide, A83-01, Forskolin | Differentiation | ||
| Pancreas | A83-01, Noggin, R-spondin 1, WNT3A, EGF, FGF10, Nicotinamide, PGE2 | Establishment | [99] |
| Prostate | EGF, R-spondin 1, Noggin, A83-01, SB202190, FGF10, FGF2, PGE2, Nicotinamide and Dihydrotestosterone (DHT) | Establishment | [85,89,116] |
| Lung | Wnt, FGF, cAMP and Glucocorticoids | Establishment | [117] |
| Brain (cerebral organoid) | N2 supplement, Glutamax, Non-essential aminoacid (NEAA) and heparin | Formation of neuroepithelial tissues | [118] |
| N2 supplement, B27 supplement without vitamin A, Glutamax, NEAA, 2-mercaptoethanol and insulin | Maturation | ||
| B27 supplement with Vitamin A, Retinoic acid | Differentiation | ||
| Kidney | GSK3α inhibitor (CHIR99021) | Differentiation (Nephrogenesis) | [119] |
| FGF9, Heparin | Organoid formation |
3D organoids have been successfully developed from primary tumors, metastatic lesions, and CTCs [80,88,89]. Patient-derived organoids are easy to initiate, propagate and store; represent and maintain tumor heterogeneity; are biologically stable; can easily be used for genetic manipulations, and are suitable for high-throughput screening assays [88]. Gao et al. (2014) established seven 3D organoid cultures from biopsies of metastatic prostate cancer and CTCs from blood samples of patients with prostate cancer [89]. For isolation of CTCs, they used a simple Ficoll-Paque technique with a CD45 depletion cocktail; and used germline and organoid DNA to characterize the organoid lines at molecular level including mutational status (by whole exome sequencing), copy number alterations (by array comparative genomic hybridization), and identification of the fusion gene and transcriptional landscape (using paired-end RNA sequencing) [89]. These organoid lines showed histologic features highly reminiscent of the parent prostate tumor [89]. Importantly, these organoid lines harbor genetic alterations quite similar to the parent prostate cancer, including PTEN loss, TMPRSS2-ERG interstitial deletion, SPOP and FOXA1 mutations, and CHD1 loss [89]. These genomic alterations remained even after months of subsequent culturing, confirming the maintenance of tumor heterogeneity in organoids [89]. Furthermore, the organoid lines presented the phenotypic diversity of castration-resistant prostate cancer (CRPC), including androgen receptor (AR)-dependent adenocarcinoma, AR-negative adenocarcinoma, neuroendocrine carcinoma, and squamous differentiation [89]. Although, it is easier to develop organoids from metastatic tumor tissues, it is not always practical or possible to obtain a metastatic tumor biopsy for organoid culture. Therefore, CTCs are the most preferred biological tissue for organoid culture.
3.3. Patient-derived xenograft (PDX) versus organoid model
In PDX models, freshly resected tumor pieces are subcutaneously or orthotopically implanted into immunocompromised mice [90]. These models mimic the original tumor conditions more closely than in vitro conditions and show less genetic divergence when compared to cancer cell lines [91]. In addition, several sub-clones grow in parallel and partially conserve parental tumor heterogeneity. These benefits make PDX models valid for preclinical research and allow assays to test drug efficacy and develop predictive biomarkers for standard and novel anticancer drugs based studies [92]. However, PDX models have several caveats. These include the delay between murine engraftment and patient treatment, and lymphomagenesis of human tumors in mice. Most importantly, this model is expensive, time-consuming, labor-intensive, not amenable to high-throughput drug screening, and could be ethically problematic [93]. Further, it is quite challenging to develop PDX models in slow-growing cancers such as prostate cancer.
On the other hand, organoids have architectural and physiological similarities to native organ systems, and are superior to traditional two-dimensional homogeneous cell lines [94]. Additionally, organoids have self-organizing ability, are easy to handle, are accessible to genetic engineering, can be used for large-scale drug screening within a shorter time span, and are cheaper than PDX models [80]. The success rate of establishment of organoids from early-stage tumors is much higher compared to cancer cell lines or PDX models [80]. Organoids fall between purely clonal cancer cell lines and PDX models in terms of tumor heterogeneity. They can also be used as tools for genetic relatedness, identification of biomarkers, screening of drugs, and preclinical evaluation of precision medicine strategies [95,96]. With these advantages, organoids may be poised to become a model that fits research needs between simple cancer cell lines and complex PDX models.
4. Potential applications of CTC-derived organoid cultures
4.1. Disease modeling
Carcinogenesis is a complicated process, and it is challenging to link specific genetic events with different stages in carcinogenesis, such as angiogenesis, metastasis, and drug-resistance development. Organoids could be easily manipulated using retroviruses, inhibitors, and/or CRISPR/Cas9 approaches; therefore, organoids are useful in cancer modeling and in identifying key “driver mutations” involved in cancer development [97–99]. Drost et al. (2015) used a human intestinal organoid model to identify specific genetic alterations involved in colorectal cancer (CRC) growth and progression [97]. They used a CRISPR/Cas 9 genomic editing tool to generate organoids (small intestine and colon) with specific mutations (APCKO, TP53KO, KRASG12D and SMAD4KO); they reported that these organoids could grow in vitro in the absence of all stem-cell-niche factors [97]. In vivo, organoids with triple mutations (APCKO, TP53KO, and KRASG12D) showed slower growth resembling ‘adenoma’ when injected subcutaneously in immunodeficient mice; however, organoids with quadruple mutations (APCKO, TP53KO, KRASG12D and SMAD4KO) showed highly proliferative and invasive growth similar to ‘invasive carcinoma’ [97]. Using a similar human colon organoid model, Fumagalli et al. (2017) showed that sequential accumulation of oncogenic mutations (APCKO, TP53KO, KRASG12D and SMAD4KO) facilitates primary tumor growth, migration and metastasis following orthotopic transplantation of organoids [98]. Boj et al. (2015) reported the utility of an organoid model to better understand the development of pancreatic ductal adenocarcinoma (PDA) [99]. Following transplantation, organoids derived from murine and human PDA generated lesions reminiscent of pancreatic intraepithelial neoplasia, which then progressed to invasive PDA [99]. Further, they demonstrated the utility of organoids to identify molecular pathways correlated with PDA progression, offering novel therapeutic and diagnostic opportunities [99]. CTC-derived organoids could be immensely useful to model metastatic progression and drug-induced selection, by establishing multiple organoid lines from the same patient over a period of time (e.g. early, progressing, and metastasized cancers; pre-treatment and posttreatment) or patients treated in parallel. In future, biobanking of organoids derived from diseased tissues will be useful in better understanding disease pathogenesis and the development of new diagnostic tools (Fig. 2).
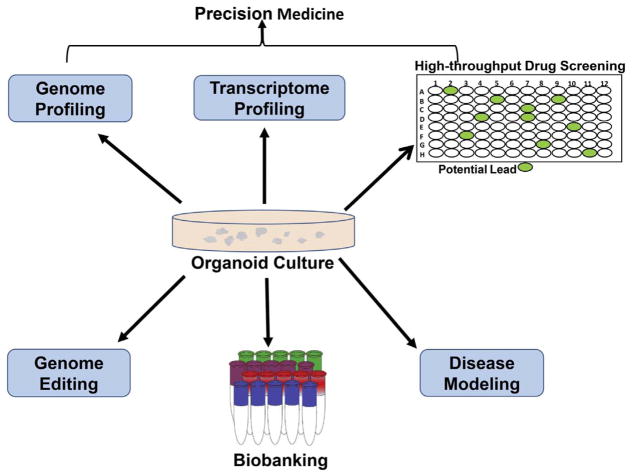
Fig. 2.
Application of CTC-derived 3D organoids in precision medicine and biomedical research. CTC-derived 3D organoids could be useful in genome and transcriptome profiling, high-throughput drug screening, disease modeling, biobanking, and genome editing.
4.2. Genetic instability
Genetic instability is considered an absolute requirement for the generation of multiple mutations that underlie cancer. Organoid models have proven useful in characterizing the importance of genetic instability during cancer development and progression [97,100]. Matano et al. (2015) showed that besides driver mutations, genetic instability is required for metastatic progression of CRC [100]. Using a CRISPR/Cas9 genome editing system, genetic alterations (deletion of APC, TP53, and SMAD4; and point mutations in KRASG12V and PIK3CAE545K) were introduced into organoids derived from normal human intestinal epithelium [100]. Importantly, even with these driver mutations, the engineered organoids were largely devoid of aneuploidy or copy number alterations, indicative of genomic stability [100]. Importantly, when xenotransplanted in NOG mice, these organoids did not metastasize. However, metastases were produced when driver mutations were introduced into colorectal adenoma organoid lines with proven chromosomal instability [100]. Drost et al. (2015), using a human intestinal organoid model, showed that loss of both APC and p53 promote chromosomal instability and aneuploidy and render cells sensitive of further accumulation of genetic alterations [97]. Van de Wetering et al. (2015) showed the presence of common genomic alterations and microsatellite instabilities of CRCs in organoid culture [101]. Webber et al. (2015) reported similar finding in human CRCs with identical somatic mutations and DNA copy number between organoids and tumor biopsy of the same patient [88]. Zhang et al. (2017) used 3D cultures of CTCs isolated from the blood of a patient with lung adenocarcinoma to detect ALK rearrangement (EML4-ALK fusion) [75], suggesting the usefulness of CTCs-derived organoids in analyzing genetic instability.
4.3. Drug discovery
Patient-derived organoid models are a reliable, robust, and biomimetic screening platform that could bridge the gap between primary 2D cell-based drug screening and PDX animal models. Organoids could be useful for testing drug efficacy, drug toxicity studies in liver organoids, or drug bioavailability studies in intestinal organoids [102,103]. In particular, CTC-derived models with relevant pathologies of patients could be a key link to screening specific drug/s [75,89,104]. Gao et al. (2014) reported the usefulness of prostate cancer patient-derived organoid lines, including CTCs-derived organoids for testing the second-generation androgen receptor antagonist (enzalutamide) and PI3K-kinase pathway inhibitors (everolimus and BKM-120) [89]. Hodgkinson et al. (2014) reported that CTCs derived from patients with small-cell lung carcinoma were tumorigenic in immune-compromised mice, and mirrored the donor patient’s response to platinum and etoposide treatment [105]. This study suggested that CTC-derived explants could be useful to monitor the changing patterns of a tumor’s drug susceptibility and to identify potential new therapeutic targets [105]. Boehnke et al. (2016) demonstrated the usefulness of patient-derived CRC organoids for high-throughput screening and drug discovery [106]. Van de Wetering et al. (2015) also demonstrated the successful application of human CRC organoids in a systematic and unbiased high-throughput screening to identify clinically relevant biomarkers [101]. Therefore, all these studies suggest the possibility and efficacy of organoid technology in unraveling the molecular basis of drug response.
4.4. Precision medicine
Precision medicine is an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person [107]. Next-generation sequencing and mutational analyses of tumor tissues have allowed identification of molecular biomarkers predicting success or resistance to specific therapies [108]. In this regard, organoids could be useful both in ‘bottom-up’ functional oncogene validation in wild-type tissue organoids to identify ‘driver mutations’, as well as in ‘top-down’ target validation and drug screening for precision therapy [109]. As mentioned above, several studies have characterized the function of driver mutations in carcinogenesis using organoid models [97–99]. Similarly, Van de Wetering et al. (2015) reported that tumor organoids could be useful to study various CRC molecular subtypes as well as to perform gene-drug association studies [101]. This study also showed that porcupine (a small molecule inhibitor of Wnt secretion) was effective only against a patient-derived organoid line carrying a mutation in the Wnt feedback regulator RNF43 [101], suggesting the usefulness of this inhibitor in a subset of CRC patients carrying the RNF43 mutation.
Bartucci et al. (2016) described an interdisciplinary approach to develop patient-derived organoids by using adaptive T cell and chimeric antigen receptor immunotherapy [110]. Recent studies have also indicated that CTC-derived models could be useful in longitudinal genetic profiling to monitor the evolving mutational landscape and drug sensitivity patterns and customize therapies for individual patients [75,105,111]. For example, CTCs accurately predicted ALK rearrangement as well as ALK mutations over time in a patient with lung adenocarcinoma; and CTC in vitro culture also predicted the treatment response to specific ALK inhibitors (ceritinib and crizotinib) [75]. Therefore, CTC-derived organoids offer a useful tool to screen drugs in the pipeline, based on the most recent genetic profiling as patients develop resistance or do not respond to specific treatment (Fig. 2).
5. Challenges and future directions
Because CTCs are extremely rare in the blood, the biggest challenge at present is rapid enrichment and isolation of viable CTCs from patient blood samples. Currently, the CellSearch® system is the only FDA-approved method for CTC detection and enumeration. However, there is increasing interest in developing novel tools and technologies for quick isolation and characterization of CTCs. Sequential analyses of CTCs and/or CTC-derived organoids could answer several important clinical questions, such as: How do metastatic progression and treatment relapse develop over the course of disease in real time? What factors are responsible for cancer dormancy? Answers to such questions could help in targeting metastatic progression, micrometastasis, and disease relapse. Further, CTC-derived organoids offer more predictive drug screening platforms and could play an important role in developing patient-specific treatments (Fig. 2).
However, CTC-derived organoid cultures still have some inherent limitations. These models lack the complexity of the in vivo immune system, vascularization, or fibroblasts and cannot determine the ratelimiting organ toxicity of drugs. Therefore, the efficacy of single organoid models to recapitulate interactions at the tissue level in the human body is still limiting. It is also not clear if CTC-derived organoids capture the complete heterogeneity of the tumor. Further investigations are required to establish sophisticated co-culture organoid models (e.g. with cancer-associated fibroblasts, endothelial cells, or immune cells) as reproducible and standardized tools for translational research and drug discovery. By further improving our understanding of the impact of the microenvironment on tumor progression, we may be able to generate predictive data from more biologically relevant organoid models that incorporate multicellular constituents and physical properties of a tumor.
Supplementary Material
Acknowledgments
Dr. Deep acknowledges grant support from NCI (R21CA199628-03S1), Dr. Nagrath acknowledges grant support from NIH (5-R33-CA-202867-02 & 1-R01-CA-208335-01-A1). We acknowledge the editorial assistance of Karen Klein, MA, in the Wake Forest Clinical and Translational Science Institute (UL1 TR001420; PI: McClain).
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.TA A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869:14. [Google Scholar]
- 2.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 3.Mishima Y, Paiva B, Shi J, Park J, Manier S, Takagi S, Massoud M, Perilla-Glen A, Aljawai Y, Huynh D, Roccaro AM, Sacco A, Capelletti M, Detappe A, Alignani D, Anderson KC, Munshi NC, Prosper F, Lohr JG, Ha G, Freeman SS, Van Allen EM, Adalsteinsson VA, Michor F, San Miguel JF, Ghobrial IM. The mutational landscape of circulating tumor cells in multiple myeloma. Cell Rep. 2017;19:218–224. doi: 10.1016/j.celrep.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorges TM, Kuske A, Rock K, Mauermann O, Muller V, Peine S, Verpoort K, Novosadova V, Kubista M, Riethdorf S, Pantel K. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin Chem. 2016;62:1504–1515. doi: 10.1373/clinchem.2016.260299. [DOI] [PubMed] [Google Scholar]
- 5.Lyberopoulou A, Aravantinos G, Efstathopoulos EP, Nikiteas N, Bouziotis P, Isaakidou A, Papalois A, Marinos E, Gazouli M. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One. 2015;10:e0123902. doi: 10.1371/journal.pone.0123902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giuliano M, Giordano A, Jackson S, Hess KR, De Giorgi U, Mego M, Handy BC, Ueno NT, Alvarez RH, De Laurentiis M, De Placido S, Valero V, Hortobagyi GN, Reuben JM, Cristofanilli M. Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving firstline systemic treatment. Breast Cancer Res. 2011;13:R67. doi: 10.1186/bcr2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierga JY, Hajage D, Bachelot T, Delaloge S, Brain E, Campone M, Dieras V, Rolland E, Mignot L, Mathiot C, Bidard FC. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol. 2012;23:618–624. doi: 10.1093/annonc/mdr263. [DOI] [PubMed] [Google Scholar]
- 8.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao Y, Li J, Shi C, Wang W, Qu X, Xiong M, Sun Y, Li D, Zhao X, Zhang D. Prognostic value of circulating tumor cells in the peripheral blood of patients with esophageal squamous cell carcinoma. OncoTargets Ther. 2017;10:1363–1373. doi: 10.2147/OTT.S129004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LW. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 12.Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, Rao SB, Eng-Wong J, Seillier-Moiseiwitsch F, Noone AM, Isaacs C. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L, Kwong J, Wang CC. Prognostic value of circulating tumor cells and disseminated tumor cells in patients with ovarian cancer: a systematic review and meta-analysis. J Ovarian Res. 2015;8:38. doi: 10.1186/s13048-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma XL, Xiao ZL, Liu L, Liu XX, Nie W, Li P, Chen NY, Wei YQ. Metaanalysis of circulating tumor cells as a prognostic marker in lung cancer. Asian Pac J Cancer Prev. 2012;13:1137–1144. doi: 10.7314/apjcp.2012.13.4.1137. [DOI] [PubMed] [Google Scholar]
- 15.Hou JM, Greystoke A, Lancashire L, Cummings J, Ward T, Board R, Amir E, Hughes S, Krebs M, Hughes A, Ranson M, Lorigan P, Dive C, Blackhall FH. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am J Pathol. 2009;175:808–816. doi: 10.2353/ajpath.2009.090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Normanno N, Rossi A, Morabito A, Signoriello S, Bevilacqua S, Di Maio M, Costanzo R, De Luca A, Montanino A, Gridelli C, Rocco G, Perrone F, Gallo C. Lung Cancer. Vol. 85. Amsterdam, Netherlands: 2014. Prognostic value of circulating tumor cells’ reduction in patients with extensive small-cell lung cancer; pp. 314–319. [DOI] [PubMed] [Google Scholar]
- 17.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 18.Hiraiwa K, Takeuchi H, Hasegawa H, Saikawa Y, Suda K, Ando T, Kumagai K, Irino T, Yoshikawa T, Matsuda S, Kitajima M, Kitagawa Y. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann Surg Oncol. 2008;15:3092–3100. doi: 10.1245/s10434-008-0122-9. [DOI] [PubMed] [Google Scholar]
- 19.Gradilone A, Naso G, Raimondi C, Cortesi E, Gandini O, Vincenzi B, Saltarelli R, Chiapparino E, Spremberg F, Cristofanilli M, Frati L, Agliano AM, Gazzaniga P. Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): prognosis, drug resistance and phenotypic characterization. Ann Oncol. 2011;22:86–92. doi: 10.1093/annonc/mdq323. [DOI] [PubMed] [Google Scholar]
- 20.Blassl C, Kuhlmann JD, Webers A, Wimberger P, Fehm T, Neubauer H. Gene expression profiling of single circulating tumor cells in ovarian cancer - establishment of a multi-marker gene panel. Mol Oncol. 2016;10:1030–1042. doi: 10.1016/j.molonc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LW, Uhr JW. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 23.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Gao P, Song Y, Sun J, Chen X, Zhao J, Xu H, Wang Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch system in colorectal cancer. BMC Cancer. 2015;15:202. doi: 10.1186/s12885-015-1218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 26.Andreopoulou E, Yang LY, Rangel KM, Reuben JM, Hsu L, Krishnamurthy S, Valero V, Fritsche HA, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 27.Gorges TM, Stein A, Quidde J, Hauch S, Rock K, Riethdorf S, Joosse SA, Pantel K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the CellSearch(R) system and the AdnaTest(R) PLoS One. 2016;11:e0155126. doi: 10.1371/journal.pone.0155126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capoun O, Mikulova V, Jancikova M, Honova H, Kolostova K, Sobotka R, Michael P, Zima T, Hanus T, Soukup V. Prognosis of castration-resistant prostate cancer patients - use of the AdnaTest(R) system for detection of circulating tumor cells. Anticancer Res. 2016;36:2019–2026. [PubMed] [Google Scholar]
- 29.Mikolajczyk SD, Millar LS, Tsinberg P, Coutts SM, Zomorrodi M, Pham T, Bischoff FZ, Pircher TJ. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol. 2011;2011:252361. doi: 10.1155/2011/252361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalinsky K, Mayer JA, Xu X, Pham T, Wong KL, Villarin E, Pircher TJ, Brown M, Maurer MA, Bischoff FZ. Correlation of hormone receptor status between circulating tumor cells, primary tumor, and metastasis in breast cancer patients. Clin Transl Oncol. 2015;17:539–546. doi: 10.1007/s12094-015-1275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Packeisen J, Kaup-Franzen C, Knieriem HJ. Detection of surface antigen 17-1A in breast and colorectal cancer. Hybridoma. 1999;18:37–40. doi: 10.1089/hyb.1999.18.37. [DOI] [PubMed] [Google Scholar]
- 32.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, Garcia-Blanco MA. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mostert B, Kraan J, Bolt-de Vries J, van der Spoel P, Sieuwerts AM, Schutte M, Timmermans AM, Foekens R, Martens JW, Gratama JW, Foekens JA, Sleijfer S. Detection of circulating tumor cells in breast cancer may improve through enrichment with anti-CD146. Breast Cancer Res Treat. 2011;127:33–41. doi: 10.1007/s10549-010-0879-y. [DOI] [PubMed] [Google Scholar]
- 36.Bitting RL, Boominathan R, Rao C, Kemeny G, Foulk B, Garcia-Blanco MA, Connelly M, Armstrong AJ. Development of a method to isolate circulating tumor cells using mesenchymal-based capture. Methods. 2013;64:129–136. doi: 10.1016/j.ymeth.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murlidhar V, Reddy RM, Fouladdel S, Zhao L, Ishikawa MK, Grabauskiene S, Zhang Z, Lin J, Chang AC, Carrott P, Lynch WR, Orringer MB, Kumar-Sinha C, Palanisamy N, Beer DG, Wicha MS, Ramnath N, Azizi E, Nagrath S. Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. Cancer Res. 2017;77:5194–5206. doi: 10.1158/0008-5472.CAN-16-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng G, Krishnakumar S, Powell AA, Zhang H, Mindrinos MN, Telli ML, Davis RW, Jeffrey SS. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GS, Li T, Chabot MS, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim ST, Sohn I, Do IG, Jang J, Kim SH, Jung IH, Park JO, Park YS, Talasaz A, Lee J, Kim HC. Transcriptome analysis of CD133-positive stem cells and prognostic value of survivin in colorectal cancer. Cancer Genomics Proteomics. 2014;11:259–266. [PubMed] [Google Scholar]
- 43.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP, Navarro VN, Tagawa ST, Bander NH, Nanus DM, Giannakakou P. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS One. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galletti G, Sung MS, Vahdat LT, Shah MA, Santana SM, Altavilla G, Kirby BJ, Giannakakou P. Isolation of breast cancer and gastric cancer circulating tumor cells by use of an anti HER2-based microfluidic device. Lab Chip. 2014;14:147–156. doi: 10.1039/c3lc51039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C, Fan ZH. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip. 2014;14:89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, Schwartz EJ, Weissman IL, Jeffrey SS, Neal JW, Rohatgi R, Wakelee HA, Wang SX. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14:78–88. doi: 10.1039/c3lc50580d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Autebert J, Coudert B, Champ J, Saias L, Guneri ET, Lebofsky R, Bidard FC, Pierga JY, Farace F, Descroix S, Malaquin L, Viovy JL. High purity microfluidic sorting and analysis of circulating tumor cells: towards routine mutation detection. Lab Chip. 2015;15:2090–2101. doi: 10.1039/c5lc00104h. [DOI] [PubMed] [Google Scholar]
- 51.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci U S A. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winer-Jones JP, Vahidi B, Arquilevich N, Fang C, Ferguson S, Harkins D, Hill C, Klem E, Pagano PC, Peasley C, Romero J, Shartle R, Vasko RC, Strauss WM, Dempsey PW. Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell. PLoS One. 2014;9:e86717. doi: 10.1371/journal.pone.0086717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M, Ionescu-Zanetti C. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl Oncol. 2013;6:528–538. doi: 10.1593/tlo.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Microfluidic marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murlidhar V, Rivera-Baez L, Nagrath S. Small. Vol. 12. Weinheim an der Bergstrasse; Germany: 2016. Affinity versus label-free isolation of circulating tumor cells: who wins? pp. 4450–4463. [DOI] [PubMed] [Google Scholar]
- 57.Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AA, Chaudhuri PK, Tan DS, Lim WT, Lee SC, Chen PC, Lim CT, Han J. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14:128–137. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 58.Warkiani ME, Khoo BL, Wu L, Tay AK, Bhagat AA, Han J, Lim CT. Ultrafast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc. 2016;11:134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 59.Lin E, Rivera-Baez L, Fouladdel S, Yoon HJ, Guthrie S, Wieger J, Deol Y, Keller E, Sahai V, Simeone DM, Burness ML, Azizi E, Wicha MS, Nagrath S. High-throughput microfluidic labyrinth for the label-free isolation of circulating tumor cells. Cell Syst. 2017;5:295–304. e294. doi: 10.1016/j.cels.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 60.Eliasova P, Pinkas M, Kolostova K, Gurlich R, Bobek V. Circulating tumor cells in different stages of colorectal cancer. Folia Histochem Cytobiol. 2017;55:1–5. doi: 10.5603/FHC.a2017.0005. [DOI] [PubMed] [Google Scholar]
- 61.Weitz J, Kienle P, Lacroix J, Willeke F, Benner A, Lehnert T, Herfarth C, von Knebel Doeberitz M. Dissemination of tumor cells in patients undergoing surgery for colorectal cancer. Clin Cancer Res. 1998;4:343–348. [PubMed] [Google Scholar]
- 62.Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, Thorban S, Nekarda H, Siewert JR. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 63.Konigsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, de Santis M, Zeillinger R, Hudec M, Dittrich C. Acta Oncol. Vol. 50. Stockholm, Sweden: 2011. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients; pp. 700–710. [DOI] [PubMed] [Google Scholar]
- 64.Alix-Panabieres C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients, Recent results in cancer research, Fortschritte der Krebsforschung. Progres Dans les Recherches sur le Cancer. 2012;195:69–76. doi: 10.1007/978-3-642-28160-0_6. [DOI] [PubMed] [Google Scholar]
- 65.Alix-Panabieres C, Pantel K. Liquid biopsy in cancer patients: advances in capturing viable CTCs for functional studies using the EPISPOT assay. Expert Rev Mol Diagn. 2015;15:1411–1417. doi: 10.1586/14737159.2015.1091729. [DOI] [PubMed] [Google Scholar]
- 66.Alama A, Truini A, Coco S, Genova C, Grossi F. Prognostic and predictive relevance of circulating tumor cells in patients with non-small-cell lung cancer. Drug Discov Today. 2014;19:1671–1676. doi: 10.1016/j.drudis.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Hofman V, Long E, Ilie M, Bonnetaud C, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, Selva E, Marquette CH, Poudenx M, Sibon S, Kelhef S, Venissac N, Jais JP, Mouroux J, Molina TJ, Vielh P, Hofman P. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology. 2012;23:30–38. doi: 10.1111/j.1365-2303.2010.00835.x. [DOI] [PubMed] [Google Scholar]
- 68.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, Yanagita M, Wang L, Berkowitz JA, Distel RJ, Cayre YE. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 70.Chen CL, Mahalingam D, Osmulski P, Jadhav RR, Wang CM, Leach RJ, Chang TC, Weitman SD, Kumar AP, Sun L, Gaczynska ME, Thompson IM, Huang TH. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate. 2013;73:813–826. doi: 10.1002/pros.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Wittner BS, Stojanov P, Brachtel E, Sgroi D, Kapur R, Shioda T, Ting DT, Ramaswamy S, Getz G, Iafrate AJ, Benes C, Toner M, Maheswaran S, Haber DA. therapy. Cancer, Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, Fouladdel S, Chang AC, Lin L, Jiang H, Waghray M, Luker G, Simeone DM, Wicha MS, Beer DG, Ramnath N, Nagrath S. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014;5:12383–12397. doi: 10.18632/oncotarget.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z, Shiratsuchi H, Palanisamy N, Nagrath S, Ramnath N. Expanded circulating tumor cells from a patient with ALK-positive lung cancer present with EML4-ALK rearrangement along with resistance mutation and enable drug sensitivity testing: a case study. J Thorac Oncol. 2017;12:397–402. doi: 10.1016/j.jtho.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 77.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 78.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 79.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 81.Francis JC, Thomsen MK, Taketo MM, Swain A. beta-Catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS Genet. 2013;9:e1003180. doi: 10.1371/journal.pgen.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cook C, Vezina CM, Allgeier SH, Shaw A, Yu M, Peterson RE, Bushman W. Noggin is required for normal lobe patterning and ductal budding in the mouse prostate. Dev Biol. 2007;312:217–230. doi: 10.1016/j.ydbio.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 84.Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, Witte ON. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RG, Cuppen E, Chen Y, Sawyers CL, Clevers HC. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qin J, Wu SP, Creighton CJ, Dai F, Xie X, Cheng CM, Frolov A, Ayala G, Lin X, Feng XH, Ittmann MM, Tsai SJ, Tsai MJ, Tsai SY. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, Haddad BR, Rhim JS, Dritschilo A, Riegel A, McBride A, Schlegel R. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van der Velden DL, Peeper DS, Cuppen EP, Vries RG, Clevers H, Voest EE. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112:13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin K, Teng L, Shen Y, He K, Xu Z, Li G. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin Transl Oncol. 2010;12:473–480. doi: 10.1007/s12094-010-0540-6. [DOI] [PubMed] [Google Scholar]
- 91.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, Watkins DN. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernards R. A missing link in genotype-directed cancer therapy. Cell. 2012;151:465–468. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 94.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 97.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 98.Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ, Begthel H, Beerling E, Tan EH, Sansom OJ, Cuppen E, Clevers H, van Rheenen J. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A. 2017;114:E2357–e2364. doi: 10.1073/pnas.1701219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 101.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao J, Zeng Z, Sun J, Zhang Y, Li D, Zhang X, Liu M, Wang X. A novel model of P-glycoprotein inhibitor screening using human small intestinal organoids. Basic Clin Pharmacol Toxicol. 2017;120:250–255. doi: 10.1111/bcpt.12680. [DOI] [PubMed] [Google Scholar]
- 103.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, JNM IJ, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ranga A, Gjorevski N, Lutolf MP. Drug discovery through stem cell-based organoid models. Adv Drug Deliv Rev 69–. 2014;70:19–28. doi: 10.1016/j.addr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 105.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, Polanski R, Burt DJ, Simpson KL, Morris K, Pepper SD, Nonaka D, Greystoke A, Kelly P, Bola B, Krebs MG, Antonello J, Ayub M, Faulkner S, Priest L, Carter L, Tate C, Miller CJ, Blackhall F, Brady G, Dive C. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 106.Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, Haybaeck J, Liebs S, Lange M, Schafer R, Regenbrecht CR, Reinhard C, Velasco JA. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J Biomol Screen. 2016;21:931–941. doi: 10.1177/1087057116650965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nabipour IAM. Precision medicine, an approach for development of the future medicine technologies. ISMJ. 2016;19:167–184. [Google Scholar]
- 108.Roberts PJ, Stinchcombe TE, Der CJ, Socinski MA. Personalized medicine in non-small-cell lung cancer: is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 109.Cantrell MA, Kuo CJ. Organoid modeling for cancer precision medicine. Genome Med. 2015;7:32. doi: 10.1186/s13073-015-0158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bartucci M, Ferrari AC, Kim IY, Ploss A, Yarmush M, Sabaawy HE. Personalized medicine approaches in prostate cancer employing patient derived 3D organoids and humanized mice. Front Cell Dev Biol. 2016;4:64. doi: 10.3389/fcell.2016.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tran NH, Cavalcante LL, Lubner SJ, Mulkerin DL, LoConte NK, Clipson L, Matkowskyj KA, Deming DA. Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther Adv Med Oncol. 2015;7:252–262. doi: 10.1177/1758834015591952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136. e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR. In vitro generation of human pluripotent stem cell derived lung organoids. elife. 2015:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Takasato M, Er PX, Chiu HS, Little MH. Generation of kidney organoids from human pluripotent stem cells. Nat Protoc. 2016;11:1681–1692. doi: 10.1038/nprot.2016.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.