SUMMARY
Cell proliferation can be dependent on the non-essential amino acid serine, and dietary restriction of serine inhibits tumor growth, but the underlying mechanisms remain incompletely understood. Using a metabolomics approach, we found that serine deprivation most predominantly impacts cellular acylcarnitine levels, a signature of altered mitochondrial function. Fuel utilization from fatty acid, glucose, and glutamine is affected by serine deprivation, as are mitochondrial morphological dynamics leading to increased fragmentation. Interestingly, these changes can occur independently of nucleotide and redox metabolism, two known major functions of serine. A lipidomics analysis revealed an overall decrease in ceramide levels. Importantly, supplementation of the lipid component of bovine serum or C16:0-ceramide could partially restore defects in cell proliferation and mitochondrial fragmentation induced by serine deprivation. Together, these data define a role for serine in supporting mitochondrial function and cell proliferation through ceramide metabolism.
In Brief
Cell proliferation can be serine dependent, but the underlying mechanisms remain incompletely understood. Using metabolomic and lipidomic approaches, Gao et al. find that, in addition to their functional role in nucleotide and redox metabolism, serine-derived lipids, ceramides, are essential for mitochondrial function and cell proliferation.
INTRODUCTION
Serine, a non-essential amino acid, participates in a metabolic network that interlinks the folate cycle and methionine cycle to support cell proliferation (Ducker and Rabinowitz, 2017; Locasale, 2013; Mattaini et al., 2016; Yang and Vousden, 2016). Modern cancer chemotherapy is built upon antagonizing folate metabolism, and recent studies have uncovered newfound roles of one-carbon metabolism in cancer pathology (Locasale, 2013; Newman and Maddocks, 2017; Yang and Vousden, 2016). Serine derives from both nutritional uptake and the serine synthesis pathway that branches from glycolysis. Changes in serine metabolism have long been noted in cancer pathogenesis (Davis et al., 1970; de Koning et al., 2003; Snell et al., 1988; Snell and Weber, 1986). Recent findings have reported on amplification of and a pro-oncogenic role for phosphoglycerate dehydrogenase, the enzyme that initiates de novo serine synthesis (Locasale et al., 2011; Possemato et al., 2011). Dietary restriction of serine and glycine also shows therapeutic effects on tumor growth (Maddocks et al., 2013, 2017).
Serine acts as a central node in the metabolic network for many aspects of cell survival and proliferation (Locasale, 2013; Mehrmohamadi et al., 2014; Nilsson et al., 2014; Tedeschi et al., 2013; Yang and Vousden, 2016). It is a precursor of glycine and cysteine that constitutes components of the cellular antioxidant glutathione (GSH). It donates carbon units via the folate cycle for synthesis of thymidine and purines, and methionine, providing the methyl donor for histones and DNA (Kottakis et al., 2016; Maddocks et al., 2016; Mehrmohamadi et al., 2016; Mentch et al., 2015). Recent studies have also revealed that serine can be a major source of the cellular reducing agent nicotinamide adenine dinucleotide phosphate (NADPH) (Ducker et al., 2016; Fan et al., 2014; Lewis et al., 2014; Tedeschi et al., 2013; Ye et al., 2012). It is also the head group of the phospholipid phosphatidylserine and provides a substrate for sphingolipid synthesis, but whether this component of serine metabolism participates in cellular physiology is largely unknown.
Serine-derived folate metabolism is partitioned in mitochondrial and nucleocytoplasmic compartments, where it contributes to nucleotide and redox metabolism in both compartments (Ducker and Rabinowitz, 2017; Tibbetts and Appling, 2010; Yang and Vousden, 2016). The complete oxidation of folate carrier generates formate and produces NADH, a reducing equivalent used in the electron transport chain. Recently it has been suggested that overflow of serine-derived formate could possibly contribute to oxidative phosphorylation in the mitochondria (Meiser et al., 2016). Furthermore, genetic defects in mitochondrial function result in retrograde responses that alter one-carbon metabolism (Bao et al., 2016; Celardo et al., 2017; Nikkanen et al., 2016). Thus, serine is known to connect to metabolism in the mitochondria through folate; however, it is poorly understood whether other serine-derived metabolic pathways partake in mitochondrial function.
In this study, we first used a comparative metabolomics approach (Liu et al., 2015; Ser et al., 2016) to investigate the metabolic processes affected by serine. This analysis led to the discovery that mitochondrial metabolism appears to be the predominantly affected metabolic function responsive to serine availability. Surprisingly, however, this maintenance of mitochondrial function occurs through a mechanism independent of redox and nucleotide metabolism, which are the two main known functions of serine in the mitochondria. Instead, we define a mechanism whereby lipid metabolism, particularly ceramide synthesis, is essential for cell proliferation that together expands the scope of the role of serine metabolism in mediating cellular physiology.
RESULTS
Comparative Metabolomics Reveals that Serine Deprivation Compromises Mitochondrial Fatty Acid Metabolism
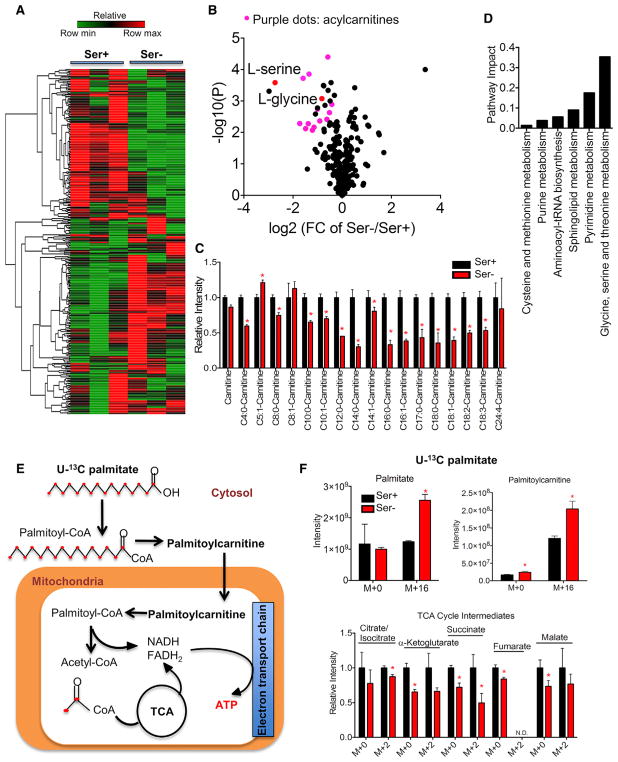
Serine deprivation reduces cell proliferation in HCT116 cells (Labuschagne et al., 2014; Maddocks et al., 2013, 2016, 2017). To study the global effects of metabolism in this context, we considered a comparative metabolomics approach that generates a profile of over 300 metabolites from diverse chemical classes. We observed a distinct metabolite profile in HCT116 cells cultured in serine-depleted media for 24 hr compared with cells cultured in basal conditions (Figure 1A). Serine deprivation altered cellular amino acid levels, with, as expected, the most dramatic reduction observed in the levels of serine and glycine (Figures 1B and S1). Prominently, a pattern of acylcarnitine-related metabolites appeared most affected as shown in the volcano plot (Figure 1B). A closer inspection of the acylcarnitine species revealed an overall reduction in both medium- and long-chain acylcarnitines (Figure 1C), likely reflecting altered mitochondrial fatty acid oxidation (FAO) (Koves et al., 2008). A pathway analysis of significantly altered metabolites (fold change [FC] > 1.5, p < 0.05, Student’s t test) identified glycine, serine, and threonine metabolism, pyrimidine metabolism, and sphingolipid metabolism as the three most affected metabolic pathways (Figure 1D). Consistent with what is known about serine, purine metabolism and cysteine and methionine metabolism were among the top six pathways affected (Figure 1D).
Figure 1. Serine Deprivation Compromises Mitochondrial Fatty Acid Metabolism.
(A) Heatmap of over 300 metabolites from HCT116 cultured with RPMI media containing serine (Ser+, 0.285 mM) or no serine (Ser−) with addition of 10% FBS for 24 hr.
(B) Volcano plots of metabolite in (A): acylcarnitines are highlighted in purple.
(C) Relative ion intensity of acylcarnitines in (A).
(D) Pathway analysis of significantly changed (FC > 1.5, p < 0.05 by Student’s t test) metabolites in (A).
(E) Schematic of U-13C palmitate tracing in HCT116 cells.
(F) HCT116 cells cultured in RPMI media containing 0.8 mM (Ser+) or no serine (Ser−) plus 10% dialyzed FBS was traced with 100 μM U-13C palmitate for 24 hr. Ion intensity of ATP, M+0 and M+16 palmitate, M+0 and M+16 palmitoylcarnitine, and relative ion intensity of M+0 and M+2 TCA cycle intermediates (citrate/isocitrate, α-ketoglutarate, fumarate, succinate, and malate) from (E).
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test (n = 3 per group). N.D., not detected. See also Figure S1.
To examine the hypothesis that reduced acylcarnitine levels corresponded to changes in FAO, we fed cells with U-13C palmitate that is channeled into the mitochondria by a carrier palmitoylcarnitine for FAO (Lopaschuk et al., 2010) (Figure 1E). Both labeled M+16 palmitate and M+16 palmitoylcarnitine were elevated under serine deprivation (Figure 1F). The tricarboxylic acid (TCA) cycle intermediates showed an overall reduction in M+0 and M+2 isotopomers. Moreover, acetylcarnitine, a surrogate of acetyl-CoA, was decreased by serine deprivation as well (Figure S1B). A 45% reduction in ATP levels that was observed upon pre-loading of palmitate (Figure S1B) further suggests a reduction of mitochondrial FAO. Together, these findings indicate serine deprivation results in a compromised mitochondrial FAO.
Serine Deprivation Alters Mitochondrial Glucose and Glutamine Metabolism
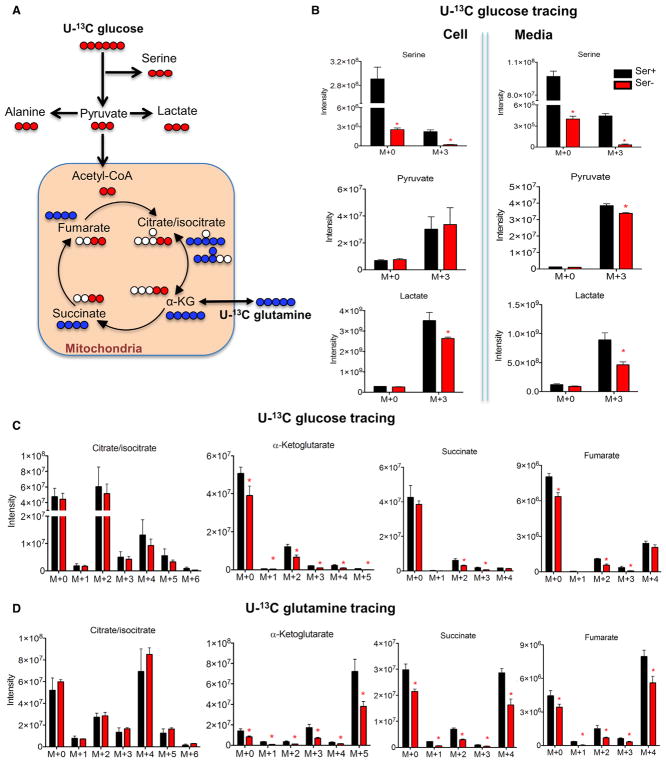
Glucose and glutamine are two major anaplerotic sources for citric acid cycle metabolism in the mitochondria (DeNicola and Cantley, 2015; Pavlova and Thompson, 2016). Thus, we explored further the effect of serine deprivation on metabolism of glucose and glutamine using U-13C glucose and U-13C glutamine tracers (Figure 2A). Previous studies have reported that serine deprivation reduces glycolytic flux to lactate (Gravel et al., 2014; Ye et al., 2012). Within 6 hr of tracing, cells under serine deprivation had lower levels of M+3 pyruvate and M+3 lactate secreted, indicating reduced glycolytic flow to lactate (Figure 2B). Unexpectedly in these conditions, serine deprivation also led to a reduction of M+3 serine in both cells and culture media (Figure 2B), indicating a relative lower amount of glucose diverted to serine synthesis. This was in contrast to an observed increase in M+3 alanine (Figure S2A), suggesting an increase of glucose-derived pyruvate toward alanine synthesis. Thus, serine deprivation reduces glycolytic flow to lactate and its branching toward serine synthesis in the cytosol.
Figure 2. Serine Deprivation Alters Mitochondrial Glucose and Glutamine Metabolism.
(A) Schematic of U-13C glucose (11 mM) and U-13C glutamine (2 mM) tracing in HCT116 cells cultured in RPMI media containing serine (Ser+, 0.8 mM) or no serine (Ser−) plus 10% dialyzed FBS for 24 hr.
(B) Ion intensity of pyruvate, lactate, serine, alanine, and their M+3 13C-isotopmers in the cell and in the culture media from study in (A).
(C and D) Ion intensity of tricarboxylic acid cycle metabolites citrate/isocitrate, α-ketoglutarate, succinate, and fumarate, and their 13C-isotopmers from U-13C glucose tracing study (C) and U-13C glutamine tracing study (D) in (A).
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test (n = 3 per group). See also Figure S2.
Examining the isotopomer distribution of TCA cycle intermediates, we found a significant reduction in M+2 α-ketoglutarate, M+2 succinate, and M+2 fumarate derived from U-13C glucose (Figure 2C). This was in addition to an overall reduction of α-ketoglutarate, succinate, and fumarate isotopomers under serine deprivation (Figure S2B). Similarly, serine deprivation decreased U-13C glutamine-derived TCA cycle intermediates M+5 α-ketoglutarate, M+4 succinate, and M+4 fumarate (Figure 2D), as well as the overall levels of α-ketoglutarate, succinate, and fumarate (Figure S2C). The distribution of citrate/isocitrate again remained the same, with a larger portion of M+4 citrate/isocitrate versus M+5 citrate/isocitrate (Figure 2D). Therefore, in addition to a compromised mitochondrial FAO, serine deprivation also leads to reduction of glucose and glutamine catabolism in the mitochondria that is altogether consistent with an imbalance in mitochondrial fuel utilization.
Serine Deprivation Leads to Mitochondrial Fragmentation
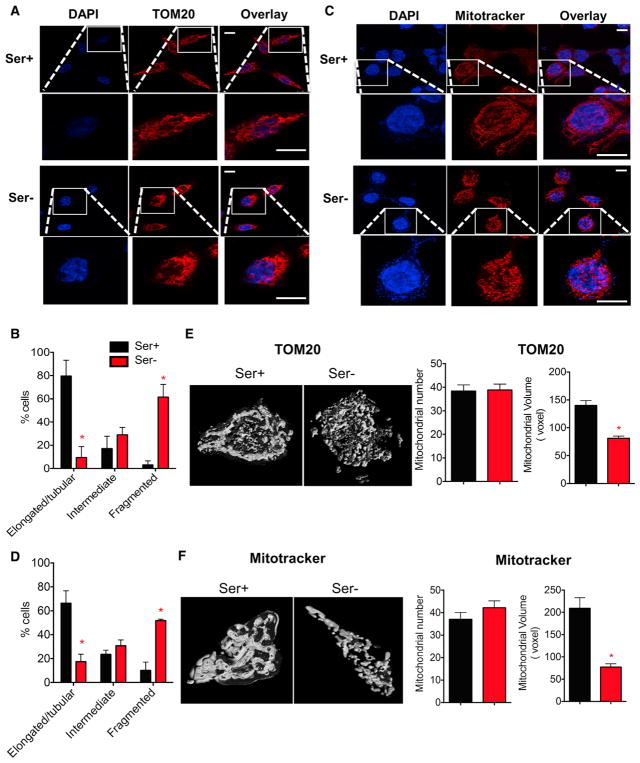
Given the observed alterations in mitochondrial substrate utilization, we sought to look for possible sources of these effects. Immunofluorescence staining with the mitochondrial outer membrane protein TOM20 demonstrated that fragmented mitochondria were present in a larger proportion of cells under serine deprivation (Figures 3A and 3B). A similar observation was also obtained by visualizing mitochondria with a MitoTracker dye, which labels mitochondria through its mildly thiol-reactive chloromethyl moiety (Figures 3C and 3D). Image analysis revealed a reduced mitochondrial volume but a constant mitochondrial number in serine-deprived cells (Figures 3E and 3F), further supporting fragmentation of the mitochondria. Furthermore, mitochondrial membrane potential was altered by serine deprivation (Figure S3). Together, in addition to affecting substrate utilization, serine deprivation also changes mitochondrial dynamics that result in increased mitochondrial fragmentation.
Figure 3. Serine Deprivation Leads to Mitochondrial Fragmentation.
HCT116 cells were cultured in RPMI media containing serine (Ser+, 0.8 mM) or no serine (Ser−) plus 10% FBS for 24 hr.
(A and B) Representative of confocal imaging of cells probed with mitochondrial marker TOM20 in (A). Scale bars, 10 μm. (B) Quantification of images from (A).
(C) Representative of confocal imaging of cells probed with MitoTracker. Scale bars, 10 μm.
(D) Quantification of images from (C). In (B) and (D), the relative percentages of cells were scored in three categories by shapes of mitochondria: tubular, fragmented, and intermediate. Over 150 cells were counted from three independent studies.
(E and F) Representative image of mitochondrial reconstruction using ImageJ in a single cell stained with TOM20 (E) or MitoTracker (F), and quantification of the mitochondria from 20 cells from three independent experiments.
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test. See also Figure S3.
Serine Deprivation Can Disrupt Mitochondrial Metabolism while Maintaining Overall Cellular Redox Balance
Serine is known to be involved in many pathways that can affect mitochondrial metabolism (Ducker and Rabinowitz, 2017; Mattaini et al., 2016; Newman and Maddocks, 2017; Yang and Vousden, 2016). It maintains cellular redox balance through production of NADPH in the mitochondria and GSH. It also donates one-carbon units for synthesis of nuclear and mitochondrial DNA and RNA. Although lesser explored, it also provides substrate for synthesis of lipids, sphingolipids, and glycerolipids. Therefore, we next investigated the relative contribution of each of these aspects of serine metabolism to mitochondrial function.
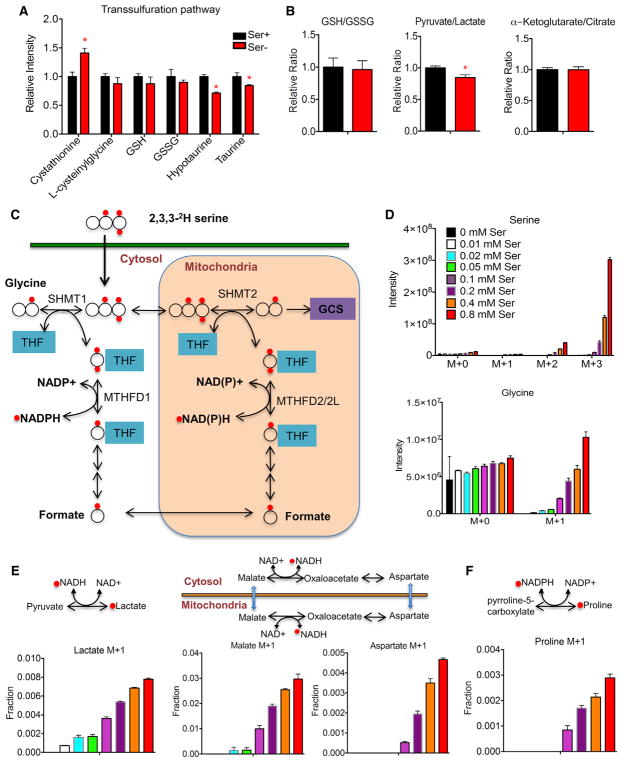
The metabolite profile under serine deprivation revealed no change in the oxidized or reduced forms of GSH, or in the levels of the precursor cysteinylglycine (Figure 4A). Both hypotaurine and taurine, the end products of the transsulfuration pathway, were reduced by serine deprivation (Figure 4A). The ratio of GSH to oxidized glutathione (GSSG) and α-ketoglutarate/citrate, a surrogate of the NADH/NAD+ ratio, also remained the same (Figure 4B). There was a small reduction in the ratio of pyruvate/lactate, the interconversion of which depends on the cytosolic NADH/NAD+ ratio (Figure 4B), consistent with the earlier observation with reduction in glycolytic flow toward lactate but an increase of alanine production (Figure 2A). Thus, serine deprivation seems to have a minimal impact on cellular redox balance by 24 hr.
Figure 4. Serine Deprivation Can Disrupt Mitochondrial Metabolism while Maintaining Overall Cellular Redox Balance.
(A) Relative ion intensity of metabolites in the transsulfuration pathway in HCT116 cells cultured in RPMI media containing serine (Ser+, 0.285 mM) or no serine (Ser−) plus 10% FBS for 24 hr.
(B) Relative intensity ratio of GSH/GSSG, pyruvate/lactate, and α-ketoglutarate/citrate.
(C) Schematic of 2,3,3-2H serine tracing study.
(D–F) HCT116 cells cultured in RPMI media containing 0.8 mM (Ser+) or no serine (Ser−) plus 10% dialyzed FBS were traced with 2,3,3-2H serine for 24 hr. (D) Ion intensity of serine and glycine isotopomers. (E) Fraction of M+1 lactate, M+1 malate, and M+1 aspartate, and schematic reactions involved in NADH/NAD+.
(F) Fraction of M+1 proline, and schematic reaction involved in NADPH/NADP+.
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test (n = 3 per group).
Serine metabolism is compartmentalized in the cytosol and mitochondria (Ducker and Rabinowitz, 2017; Tibbetts and Appling, 2010). To distinguish serine metabolism in these two compartments, we traced cells in a dose-dependent manner with 2,3,3-2H serine (Figure 4C), as has been previously employed (Ducker et al., 2016). The enrichment of M+3 serine and M+1 glycine showed as expected dose dependence (Figure 4D), indicating successful incorporation of labeling (Figure 4E). In the cytosol, production of lactate from pyruvate requires the presence of NADH. Cytosolic NADH is interchangeable with mitochondrial NADH through malate-aspartate shuttle. In the mitochondria, malate is oxidized by malate dehydrogenase to generate oxaloacetate, converting NAD+ to NADH. Oxaloacetate is then used for aspartate production via aspartate aminotransferase. M+1 malate, M+1 aspartate, and M+1 lactate were all serine dose dependent, and the labeling, after the appropriate natural abundance corrections, consisted of 3%, 0.5%, and 0.8% of their respective total metabolite abundance in the presence of 0.8 mM 2,3,3-2H serine (Figure 4E). These findings confirm that the vast majority (>97%) of the NADH generated in the TCA cycle is not from serine.
Mitochondrial serine metabolism is thought to account for a substantial portion of cellular NADPH production (Ducker et al., 2016; Fan et al., 2014; Lewis et al., 2014; Tedeschi et al., 2013). Proline, whose biosynthesis is driven by NADPH in the initial step catalyzed by pyrroline-5-carboxylate synthase, was labeled in a serine dose-dependent manner (Figure 4F). In the presence of 0.8 mM 2,3,3-2H serine, only 0.3% of proline was 2H-labeled, supporting a minor contribution of serine-derived NADPH in our conditions in HCT116 cells. Together with the earlier metabolomics profiling of the overall redox state, our data show that although TCA cycle activity is affected, electrons derived directly from serine make a small contribution to redox balance in our settings.
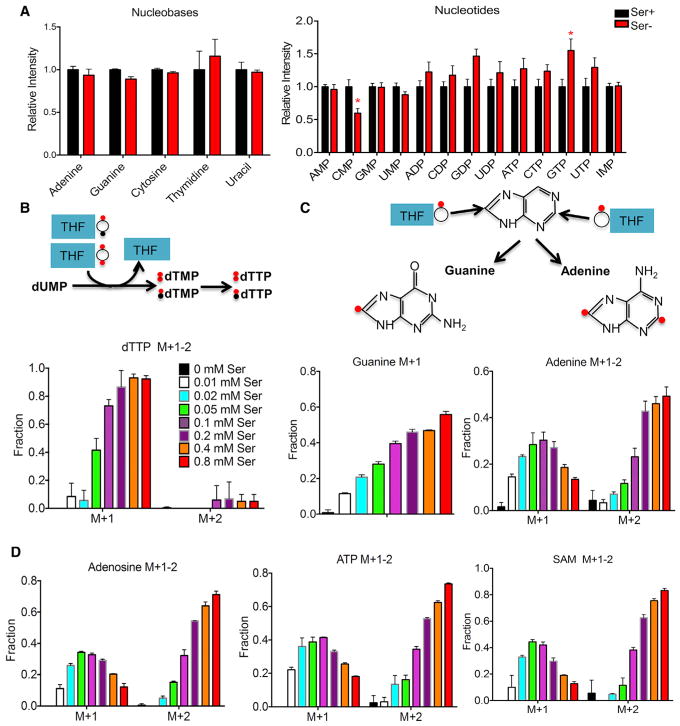
Serine Donates One-Carbon Units for Nucleotide Biosynthesis, but Nucleotide Levels Are Maintained
Next, we examined the effect of serine deprivation on nucleotide synthesis. Serine deprivation for 24 hr had a minimal effect on the levels of cellular nucleobases and nucleotides (Figure 5A). A recent report showed that serine supports methionine cycle and production of S-adenosylmethionine (SAM), a universal methyl-donor (Maddocks et al., 2016). Surprisingly this is achieved through de novo ATP production, but not by directly donating the one-carbon unit to homocysteine remethylation into methionine (Maddocks et al., 2016). Consistent with this finding, we observed a reduction in cellular SAM and MTA (5′-methyladenosine), an intermediate in the methionine salvage pathway. Serine deprivation also elevated cellular S-adenosylhomocysteine (SAH), but not homocysteine (Figure S4). Together, serine deprivation leads to a reduction in SAM/SAH, but not SAM/MTA (Figures S4A and S4B). Notably by 24 hr, ATP levels were not significantly changed (Figure 5A). Thus, serine deprivation for 24 hr reduces cellular SAM levels with levels of ATP maintained.
Figure 5. Serine Donates One-Carbon Units for Nucleotide Biosynthesis, but Nucleotide Levels Are Maintained.
(A) Relative ion intensity of nucleotides and nucleobases in HCT116 cells cultured in RPMI media containing serine (Ser+, 0.285 mM) or no serine (Ser−) plus 10% FBS for 24 hr.
(B) Schematic reaction from dUMP to dTTP and fraction of M+1 and M+2 dTTP from 2,3,3-2H serine tracing study in Figure 4C.
(C) Schematic presents the one-carbon unit from tetrahydrofolate (THF) contributes to adenine and guanine synthesis, and fraction of M+1 adenine and guanine isotopomers (M+1 and M+2) from 2,3,3-2H serine tracing study in Figure 4C.
(D) Fraction of adenosine, ATP, and S-adenosylmethionine (SAM) isotopomers (M+1 and M+2) from 2,3,3-2H serine tracing study in Figure 4C.
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test (n = 3 per group). See also Figure S4.
To evaluate the contribution of serine to nucleotide synthesis, we used a 2,3,3-2H serine tracer to probe the contribution of serine to different pathways. Production of thymidine (the methylation of deoxyuridine monophosphate [dUMP] to deoxythymidine monophosphate [dTMP]) depends on cytosolic CH2-THF (5,10-methylenetetrahydrofolate). CH2-THF from cytosolic serine catabolism by SHMT (serine hydroxymethyl transferase)1 contributes to M+2 deoxythymidine trisphosphate (dTTP). CH2-THF from mitochondrial serine metabolism enters the cytosol through exchange of formate and the interchange between CH2-THF and CHO-THF (10-formyl-THF) (Figure 4C), leading to M+1 dTTP production. M+1 dTTP was serine dose dependent, and accounting again for natural abundance, 93% of dTTP with one deuterium label from 0.8 mM 2,3,3-2H serine, whereas minimal labeling was observed in M+2 dTTP (Figure 5B), suggesting in the presence of serine, much of CH2-THF in thymidine synthesis is from mitochondrial serine metabolism. Serine also donates one-carbon units in the form of CHO-THF for guanine and adenine (purine) synthesis. Indeed, both M+1 guanine and labeled (M+1 and M+2) adenine showed dose-dependent labeling from serine. Labeling with 0.8 mM 2,3,3-2H serine for 24 hr, we observed that 56% of guanine and 63% of adenine possessed 2H label (Figure 5C). Additionally, adenosine, ATP, and SAM all showed a similar labeling pattern as adenine, with label enrichment each reaching the saturation limit for quantitation because of natural abundance containing 2H from 2,3,3-2H serine (Figure 5D). Therefore, in the presence of replete serine, serine is a predominant one-carbon donor for both thymidine and purine synthesis. However, under serine deprivation, the minimal effect on cellular nucleotide levels at 24 hr indicates an adaptive mechanism over time with one-carbon units derived from other sources.
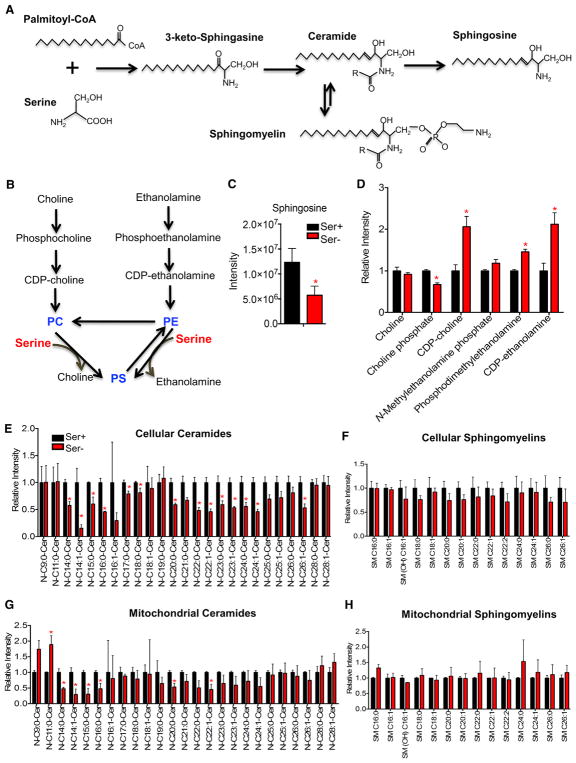
Serine Deprivation Alters Metabolism of Glycerophospholipids and Sphingolipids
Serine is also the precursor for sphingolipid and glycerophospholipid synthesis (Figures 6A and 6B), and this motivated us to examine metabolites in these less studied pathways. Sphingosine levels were 53% reduced by serine deprivation (Figure 6C). Intermediates in glycerophospholipid metabolism were also altered: a reduction in choline phosphate, whereas a significant elevation in CDP-choline, CDP-ethanolamine, and phosphodimethylethanolamine (Figure 6D), and an intermediate in phosphatidylcholine (PC) synthesis from methylation of phosphatidylethanolamine (PE). The above observations suggest a possible effect of serine deprivation in sphingolipid and glycerophospholipid metabolism.
Figure 6. Serine Deprivation Alters Metabolism of Glycerophospholipids and Sphingolipids.
(A) Schematic of sphingolipid metabolism.
(B) Schematic of phospholipid metabolism: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in mammalian cells.
(C) Ion intensity of sphingosine in HCT116 cells cultured in RPMI media containing serine (Ser+, 0.285 mM) or no serine (Ser−) plus 10% FBS for 24 hr.
(D) Relative ion intensity of metabolites in phospholipid metabolism from HCT116 cells cultured in RPMI media containing serine (Ser+, 0.285 mM) or no serine (Ser−) plus 10% FBS for 24 hr.
(E–H) HCT116 cells were cultured in RPMI media containing 0.8 mM (Ser+) or no serine (Ser−) with addition of 10% charcoal-stripped FBS for 24 hr. (E and F) Relative ion intensity of cellular ceramides (E) and sphingomyelins (F). (G and H) Relative ion intensity of ceramides (G) and sphingomyelins (H) in isolated mitochondrial fractions.
Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test (n = 2–3 per group). See also Figure S5.
To more closely examine the effect of serine on sphingolipid and glycerophospholipid metabolism, we developed a lipidomics method to profile multiple classes of lipids. To eliminate the interference of preexisting lipids in FBS (fetal bovine serum), we incubated cells with media containing 0.8 or 0 mM serine plus 10% charcoal-stripped (delipidated) FBS for 24 hr. The lipidomics analysis revealed an overall reduction in ceramides (C14:0-, C14:1-, C15:0-, C16:0-, C17:0-, C18:0-, C20:0-, C22:0-, C22:1-, C23:0-, C23:1-, C24:0-, C24:1-, and C26: 1-ceramides), yet sphingomyelins remained unchanged (Figures 6E and 6F). Serine deprivation showed minimal to no impact on phospholipids, PC, PE, or phosphatidylserine (PS) (Figure S5A). The mitochondria are one major site for certain lipid synthesis such as cardiolipin and PE (Tatsuta et al., 2014). Thus, serine deprivation could initially alter mitochondrial lipid biosynthesis, which then triggers a series of functional effects. To test this, we isolated mitochondrial fractions (Figure S5B) and performed a lipidomics analysis on isolated mitochondria. Mitochondrial PC and PE were largely unaffected, whereas certain PS species (C34:1-, C34:2-, C40:1-, C40:2-, C40:7-, C42:1-, and C42:2-PS species) were elevated (Figure S5C). Mitochondrial ceramides, but not sphingomyelins, showed an overall reduction under serine deprivation (Figures 6G and 6H). Together, the data reveal a prominent role for serine in ceramide metabolism.
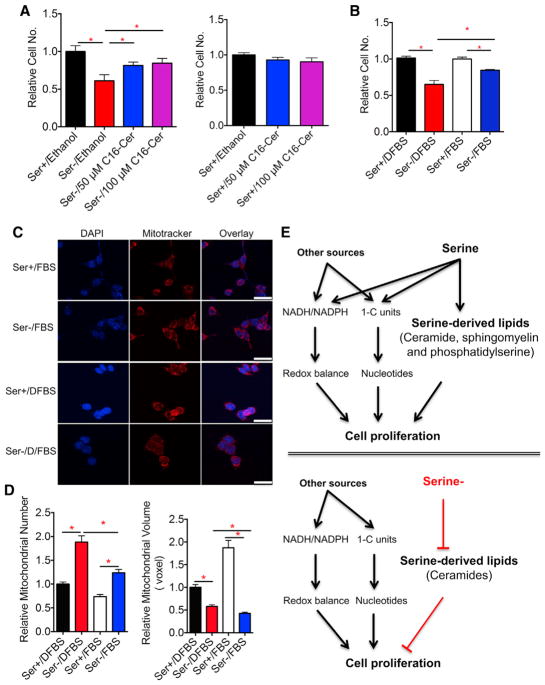
Lipid Supplementation Partially Restores Defects in Mitochondrial Morphology and Cell Proliferation upon Serine Deprivation
Serine deprivation affects cell proliferation, and dietary serine and glycine deprivation also delays tumor growth (Labuschagne et al., 2014; Maddocks et al., 2013; Maddocks et al., 2017). Thus, we tested whether replenishment of ceramides or a full set of lipids could rescue the defective cell proliferation caused by serine deprivation. Supplementation of 50 or 100 μM C16:0-ceramide, which was decreased both overall and in the mitochondrial compartment under serine deprivation, partially restored the cell number after 72 hr (Figure 7A). C2-ceramide, which is cell membrane permeable, failed to reverse the delayed cell growth and appeared cytotoxic at 25 μM as previously reported (Autelli et al., 2009) (Figure S6A). Furthermore, supplementing delipidated serum with a full complement of lipids also rescued cell proliferation in part (Figure 7B). Thus, serine-derived lipid metabolism contributes to cell proliferation.
Figure 7. Lipid Supplementation Partially Restores Defects in Mitochondrial Morphology and Cell Proliferation upon Serine Deprivation.
(A) Relative numbers of HCT116 cells cultured in RPMI media plus 10% of charcoal-stripped FBS (DFBS) containing serine (Ser+, 0.8 mM), or no serine (Ser−) with addition of 50 or 100 μM C16-ceramide (dissolved in ethanol) for 72 hr, and all conditions contain a final 1% ethanol.
(B) Relative numbers of HCT116 cells cultured in RPMI media with 0.8 mM Ser or without serine (Ser−) plus 10% of DFBS or FBS for 72 hr.
(C) HCT116 cells were cultured in RPMI media with 0.8 mM Ser or without serine (Ser−) plus 10% of DFBS or 10% of FBS for 24 hr. Representative confocal imaging of cells probed with MitoTracker. Scale bars, 25 μm.
(D) Quantification of images from 20 cells in (C). Data are mean ± SD. *p < 0.05 by two-tailed Student’s t test.
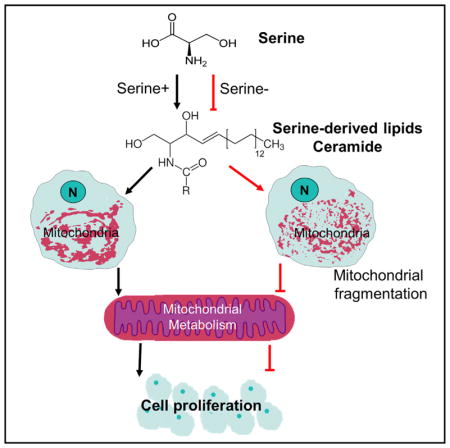
(E) Model: serine supports cell growth through production of NADH and NADPH that counterbalances cellular redox, generation of one-carbon (1-C) units that supports nucleotide synthesis, and provides substrate for serine-derived lipid synthesis. In the absence of serine, other factors come into play to minimize the effect on cellular redox and nucleotides caused by serine deprivation. Meanwhile, serine also supports the synthesis of serine-derived lipids, and ceramide synthesis is sensitive to serine deprivation; the decrease of ceramides accounts for the defective cell growth at least in part.
See also Figure S6.
We then evaluated whether supplementing a full set of lipids or C16:0-ceramide could rescue the extent of mitochondrial fragmentation caused by serine deprivation. Mitochondrial visualization with a MitoTracker dye showed that supplementation of a full set of lipids (FBS) or C16:0-ceramide alleviated mitochondrial fragmentation, mainly by increasing mitochondrial number, but not mitochondrial volume (Figures 7C, 7D, S6B, S6C). This finding is consistent with the partial rescue of serine-deprivation-induced delay of cell proliferation.
Furthermore, we have also evaluated our major findings in other cell lines including human colorectal adenocarcinoma cell line HT29, human breast cancer cell line MDA-MB-231, and human pulmonary adenocarcinoma cell line Calu-6. In all these cell lines, serine deprivation led to inhibition of cell proliferation (Figure S6D). Such growth defect was at least partially rescued by addition of the full spectrum of serum (FBS) in all these cell lines (Figure S6D). Addition of C16:0-ceramide rescued partially the inhibition of cell proliferation in HT29, but not in MDA-MB-231 or Calu-6 (Figure S6D), suggesting that the impact of serine deprivation on ceramide metabolism is also cell type dependent. Together, serine-derived lipid metabolism determines mitochondrial morphology that at least partially contributes to cell proliferation (Figures 7E).
DISCUSSION
Recent work has highlighted the importance of serine in nucleotide synthesis and redox balance (Ducker et al., 2016; Fan et al., 2014; Labuschagne et al., 2014; Mehrmohamadi et al., 2014). Yet, it largely has remained unknown as to the contribution of serine-derived lipid metabolism to cell physiology. Here using a metabolomics approach, unexpectedly we found serine plays a major role in maintaining mitochondrial metabolism by affecting lipid metabolism. This is exemplified by decreases in acylcarnitine levels, compromised mitochondrial metabolism of fatty acid, glucose, and glutamine, and altered mitochondrial morphology. This occurs largely independent of two of the main functions of serine in the mitochondria involving nucleotide and redox metabolism but requires serine-derived lipid metabolism.
The metabolomics analysis reveals an alteration of sphingolipid metabolism induced by serine limitation. Further lipidomics analysis uncovers a significant reduction in a variety of ceramide species that is most prominent in the mitochondrial subcellular compartment. Increased ceramide levels are known to perturb mitochondrial membrane structure, increase membrane permeability, and trigger cell apoptosis (Sentelle et al., 2012; Siskind, 2005). Ceramides can also cause autophagic cell death, independent of the apoptosis mechanism (Jiang and Ogretmen, 2013; Sentelle et al., 2012). In our setting we expand on these known functions of ceramide by showing that deficiencies in ceramide levels, which do not lead to massive apoptosis, also alter mitochondrial function through structural mechanisms. Defective cell proliferation by serine deprivation is, in part, rescued by C16:0-ceramide alone or complementing media with the lipid component of serum. Together, these data highlight a previously undefined mechanism for serine-derived lipids in mitochondrial biology and cell proliferation. In a very recent finding, using a yeast system, Hwang et al., (2017) independently reported that the serine-derived sphingolipids are also important for the proliferation of aneuploid cells. Supporting this conclusion, in human subjects, over 50% of consumed serine has not been accounted for in certain studies (Davis et al., 2004). Thus, it is possible that serine-derived lipid products constitute much of this unaccounted portion.
Mitochondrial morphology is highly dynamic and regulated by mitochondrial fusion and fission (Westermann, 2010). In parallel with its role in oxidative metabolism, we show that serine also maintains mitochondrial morphology and membrane potential (Figures 3 and S3). Fragmentation that occurs during limitations of serine availability is accompanied with a reduction in mitochondrial volume with the number of mitochondria maintained. Because phospholipid levels are largely maintained, we speculate that such mitochondrial fragmentation and reduction of mitochondrial membrane potential are likely attributable to the reduction of mitochondrial ceramides. Dysfunctional mitochondria caused by deletion of mitochondrial DNA or lesions in the electron transport chain also are known to impair one-carbon metabolism (Bao et al., 2016). Our data thus define a reciprocal relationship showing that serine (one-carbon) metabolism also impacts mitochondrial function and is consistent with recent reports showing serine metabolism can synergize with agents such as biguanides, which target the mitochondria (Gravel et al., 2014).
EXPERIMENTAL PROCEDURES
Reagents
FBS, penicillin, and streptomycin were purchased from HyClone Laboratories. RPMI 1640 medium and charcoal-stripped FBS were purchased from Thermo Fisher. Dialyzed FBS was from Life Technologies. Optima ammonium acetate, ammonium hydroxide, Optima liquid chromatography–mass spectrometry (LC-MS) grade, acetonitrile, methanol, and water were purchased from Fisher Scientific. U-13C glucose, U-13C glutamine, and 2,3,3-2H serine were obtained from Cambridge Isotope Laboratories. U-13C palmitate, C16-ceramide, and C2-ceramide were purchased from Sigma-Aldrich.
Cell Culture
MDA-MB-231 and Calu-6 cell lines were gifts from Dr. Donald McDonnell and Dr. Kris Wood, respectively. HCT116 and HT29 cell lines were gifts from Dr. Lewis Cantley’s laboratory. HCT116, HT29, MDA-MB-231, and Calu-6 cells were maintained in RPMI 1640 supplemented with 10% FBS and 100,000 U/L penicillin and 100 mg/L streptomycin. Cells were grown at 37°C with 5% CO2.
Cell Proliferation
Cell growth rescue by ceramides or FBS was determined by cell counting using Moxi Z Mini Automated Cell Counter (ORFLO Technologies). In brief, cells were seeded at a density of 5,000–10,000 cells per well in 24-well plates. After overnight incubation, media were replaced with conditional RPMI medium containing 0.8 or 0 mM serine plus 10% charcoal-stripped FBS for an additional 72 hr. For rescue with C16-ceramide or C2-ceramide, conditional RPMI medium also contained vehicle (1% ethanol) or C16-ceramide (50 or 100 μM, in ethanol) or C2-ceramide (5 or 25 μM, in ethanol). For rescue with FBS, media were replaced with conditional RPMI medium containing 0 μM serine plus 10% FBS for 72 hr.
Immunofluorescence
Cells were cultured in conditional media containing 0.8 or 0 mM serine with an addition of 10% FBS on glass coverslips (VWR) in six-well dishes for 24 hr. Next, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After one wash in 1× PBS, cells were permeabilized in 0.15% Triton X-100 for 15 min at room temperature. Cells were then blocked in 10% BSA for 20 min at room temperature, followed by incubation in primary antibody solution (1:500 dilution in 10% BSA) for overnight at 4°C. After three washes using 1× PBS, cells were incubated in secondary antibody solution in 10% BSA and followed by another three times of wash. Cells were then mounted onto glass slides using Fluoromount-G mounting solution (Southern Biotech) for imaging acquisition on a spinning-disk confocal microscope (Leica) using a × 100/1.4NA oil or a ×40/1.25NA oil (Leica Plan Apochromat) objective. Images were acquired and processed using the Leica LAS AF program software.
Analysis of Mitochondrial Morphology
To access changes of mitochondrial morphology, we labeled mitochondria with a MitoTracker dye, MitoTracker Red CMXRos (final concentration 100 nM; Invitrogen, Molecular Probes), for 20 min in a CO2 incubator or stained with anti-TOM20 antibody TOM20 (sc-11415; Santa Cruz) as previously described (Norris et al., 2015). Cells were divided as previously described (Karbowski et al., 2006) into three categories: “Normal” with >90% of mitochondria forming elongated interconnected networks, “Intermediate” with mixed tubular and short mitochondria, and “Fragmented” with >90% short punctiform mitochondria. The percentage of cells with elongated, intermediate, or fragmented mitochondria was plotted, and 150 cells per condition were quantified in samples from three independent experiments. Mitochondrial number and volume were quantified using ImageJ (NIH) as before (Pennanen et al., 2014). Images were first deconvolved and then volume-reconstituted using the VolumeJ plug-in. The volume and number of mitochondria in each cell were quantified using the ImageJ-3D Object Counter plug-in. Mitochondria from 17–21 cells were counted per condition.
Analysis of Mitochondrial Membrane Potential
HCT116 cells were seeded in six-well dishes for overnight. After overnight incubation, cells were washed once and cultured in conditional media containing 0.8 or 0 mM serine plus 10% FBS for 24 hr. Cells were then stained with 100 nM TMRE (tetramethylrhodamine, ethyl ester) for 20 min at 37°C. Fluorescence intensity of cells was evaluated by fluorescence-activated cell sorting (excitation 488 nm, emission 575 nm). As the negative control, cells were pre-incubated with 20 μM fluorocarbonyl cyanide phenylhydrazone (FCCP) for 10 min before TMRE staining.
Mitochondrial Fractionation
HCT116 cells were seeded in 10-cm dishes at a density of 2.0 × 105 cells/mL × 10 mL/dish. After overnight incubation, cells were washed with PBS once and cultured for an additional 24 hr with 10 mL of conditional RPMI medium containing 0.8 or 0 mM serine plus 10% charcoal-stripped FBS. 2.0 × 107 cells per condition were used for mitochondrial fractionation using a commercially available mitochondria isolation kit from Thermo Scientific (89874), following the manufacturing protocol.
Immunoblotting
The purity of mitochondrial fractions was determined by immunoblotting mitochondrial fractions and whole-cell lysate with antibodies against mitochondrial markers VDAC (4866; Cell Signaling) and TOM20 (sc-11415; Santa Cruz), plasma marker Na+/K+-ATPase (3010; Cell Signaling), cytosol marker β-ACTIN (3700S; Cell Signaling), Golgi marker GM130 (610823; BD Biosciences), and lysosomal marker LAMP1 (Developmental studies Hybridoma Bank). The protein concentration was determined by Bradford assay. Immunoblotting signals were detected by ChemiDoc MP Imaging System with Image Lab Software (Bio-Rad).
Metabolite Profiling and Isotope Tracing
HCT116 cells were seeded in six-well plates at a density of 2.0 × 105 cells per well. For overall polar metabolite profile, cells were washed once with PBS after overnight incubation and cultured for an additional 24 hr with 2 mL of conditional RPMI medium containing 0.8 or 0 mM serine plus 10% FBS. For 13C isotope tracing, after overnight incubation and one-time wash with PBS, cells were first incubated in 2 mL of conditional RPMI medium containing 0.8 or 0 mM serine plus 10% dialyzed FBS for 16 hr, and then medium was replaced with fresh conditional RPMI medium (0.8 or 0 mM serine) containing tracer U-13C glucose, U-13C glutamine, or U-13C palmitate plus 10% dialyzed FBS. Cells were traced with 11 mM U-13C glucose or 2 mM U-13C glutamine for 6 hr, or with 100 μM U-13C palmitate for 24 hr. For 2,3,3-2H serine tracing, after overnight culture, cells were washed once with PBS and cultured for an additional 24 hr with 2 mL of conditional RPMI medium containing 10% dialyzed FBS and 0 μM serine with an addition of 2,3,3-2H serine at different concentrations (0, 0.01, 0.02, 0.05, 0.1, 0.2, 0.4, 0.8 mM). For lipidomics analysis, after overnight incubation and one-time wash with PBS, cells were treated with 2 mL of conditional RPMI medium containing 0.8 or 0 mM serine plus 10% charcoal-stripped FBS for an additional 24 hr. Media and/or cellular metabolites were extracted at the end of tracing.
Metabolite Extraction
Polar metabolite extraction has been described previously (Liu et al., 2015). In brief, cells cultured in six-well plates were placed on top of dry ice right after medium removal. 1 mL of ice-cold extraction solvent (80% methanol/water) was added to each well and the extraction plate was quenched at −80°C for 10 min. Cells were then scraped off the plate into an Eppendorf tube. Samples were vortexed and centrifuged at 20,000 × g for 10 min at 4°C. For media metabolite extraction, 20 μL of spent media was added to 80 μL of ice-cold water in Eppendorf tube on ice, followed by the addition of 400 μL of ice-cold methanol. Samples were vortexed at the highest speed for 1 min before centrifugation at 20,000 ×g for 10 min at 4°C. The supernatant was transferred to a new Eppendorf tube and dried in a vacuum concentrator. The dry pellets were stored at −80°C for LC-HRMS analysis. Samples were reconstituted into 30 μL of sample solvent (water:methanol:acetonitrile, 2:1:1, v/v/v) and were centrifuged at 20,000 × g at 4°C for 3 min. The supernatant was transferred to LC vials, and 3 μL was injected for hydrophilic interaction chromatography method (HILIC) chromatography.
For lipid extraction, 50 μg of mitochondrial fraction or 150 μg of whole-cell lysate were extracted in final extraction solution methanol/MTBE (methyl tert-butyl ether)/water (1:3:1, v/v/v). Samples were vortexed at the highest speed for 1 min, followed by centrifugation at 3,500 g × 10 min. The top layer was collected and dried in a vacuum concentrator. The dry pellets were stored at −80°C for LC-HRMS analysis. Samples were reconstituted into 30 μL of sample solvent (Isopropanol) and centrifuged at 20,000 × g at 4°C for 10 min. The supernatant was transferred to LC vials and the injection volume was 3 μL (equivalent to 2.5 μg of mitochondrial protein or 7.5 μg of whole-cell lysate) for HILIC chromatography.
High-Performance Liquid Chromatography
Ultimate 3000 UHPLC (Dionex) was coupled to Q Exactive-Mass spectrometer (QE-MS; Thermo Scientific) for metabolite separation and detection.
For additional polar metabolite analysis, a HILIC with an Xbridge amide column (100 × 2.1 mm internal diamenter [i.d.], 3.5 μm; Waters) was used for compound separation at room temperature. The mobile phase and gradient information were described previously (Liu et al., 2015).
For lipid analysis, a reverse-phase liquid chromatography method with an Xbridge BEH C18 column (100 × 2.1 mm i.d., 2.5 μm; Waters) at 40°C was used for compound separation. The column was employed with mobile phase A: acetonitrile:water (60:40, v/v) with 0.1% formic acid and 10 mM ammonium formate, and mobile phase B: Isopropanol:acetonitrile (90:10, v/v) with 0.1% formic acid and 10 mM ammonium formate. Linear gradient was: 0 min, 40% B; 1.5 min, 40% B; 5 min, 85% B; 12 min, 97% B; 16 min, 97% B; 16.5 min, 40% B, and 20.5 min, 40% B. Flow rate is 0.2 mL/min.
Mass Spectrometry
The Q Exactive mass spectrometer (QE-MS) is equipped with a heated electrospray ionization (HESI) probe. For polar metabolites, the relevant parameters were as listed: heater temperature, 120°C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 3.0 kV for positive (pos) mode and 2.5 kV for negative (neg) mode. Capillary temperature was set at 320°C, and S-lens was 55. A full scan range was set at 70–900 (mass to charge [m/z]) for positive mode (1.31–12.5 min) and negative mode (1.31–6.6 min) and 100–1,000 for negative mode (6.61–12.5 min). The resolution was set at 70,000 (at m/z 200). The maximum injection time (max IT) was 200 ms. Automated gain control (AGC) was targeted at 3 × 106 ions. Customized mass calibration was performed before data acquisition.
For lipids, the relevant parameters are as listed: heater temperature, 120°C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 4.0 kV for both pos mode and neg modes. Capillary temperature was set at 320°C, and S-lens was 65. A full scan range from 200–1,500 (m/z) was used. The resolution was set at 70,000. The max IT was 200 ms with typical injection times around 50 ms. The above settings resulted in a duty cycle of around 550 ms to carry out scans in both the pos and neg modes. AGC was targeted at 3 × 106 ions. Standard mass calibration was performed before sample analysis.
Data Analysis
LC-MS peak extraction and integration were analyzed with commercially available software Sieve 2.0 (Thermo Scientific). The integrated peak intensity was used for further data analysis. For tracing studies using U-13C glucose, U-13C glutamine, or U-13C palmitate, 13C natural abundance was corrected as previously described (Yuan et al., 2008). For 2H tracing studies using 2,3,3-2H serine, 2H natural abundance was corrected. Based on our Orbitrap mass resolution of 70,000 (at m/z 200), 2H peaks were separable from other isotopes by the mass spectrometer. When 2H peaks could not be directly resolvable, they were also corrected for 13C, 18O, and 15N natural abundance (correction was only required for the unresolvable combination of one or two 13C, 18O, and 15N atoms).
Statistical Analysis and Bioinformatics
Pathway analysis of metabolites was carried out with software Metaboanalyst (http://www.metaboanalyst.ca/MetaboAnalyst/faces/home.xhtml) using the KEGG pathway database (http://www.genome.jp/kegg/). All data were represented as mean ± SD, unless otherwise indicated. The p values were calculated by a two-tailed Student’s t test unless otherwise noted.
Data and Software Availability
The metabolomics and lipidomic data reported in this study were deposited to Mendeley Data (https://doi.org/10.17632/62v47m8y35.1) and are available at https://data.mendeley.com/datasets/62v47m8y35/draft?a=496e4cf9-5e32-4423-ad48-7dea4ebc87a1.
Supplementary Material
Highlights.
Serine maintains mitochondrial metabolism
Serine deficiency causes mitochondrial fragmentation
Serine metabolism regulates ceramide and sphingolipid metabolism
Lipid supplementation partially restores serine-deprivation-induced phenotypes
Acknowledgments
Support from the NIH (grants R01CA193256 and P30CA014236 to J.W.L.) and the Canadian Institutes of Health Research (CIHR) (funding reference number 146818 to X.G.) are gratefully acknowledged. We thank Kris Wood and Tso-Pang Yao for sharing reagents and Grace Anderson, Jin Yan, and members of the Locasale lab for helpful discussions on mitochondrial imaging and quantification.
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.017.
AUTHOR CONTRIBUTIONS
X.G. and J.W.L. designed the study and wrote the manuscript. X.G., K.L., M.A.R., S.M.S., C.Q., and S.L. performed experiments. J.L. developed methods for lipidomics.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Autelli R, Ullio C, Prigione E, Crepaldi S, Schiavone N, Brunk UT, Capaccioli S, Baccino FM, Bonelli G. Divergent pathways for TNF and C(2)-ceramide toxicity in HTC hepatoma cells. Biochim Biophys Acta. 2009;1793:1182–1190. doi: 10.1016/j.bbamcr.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016;5:e10575. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celardo I, Lehmann S, Costa AC, Loh SH, Miguel Martins L. dATF4 regulation of mitochondrial folate-mediated one-carbon metabolism is neuroprotective. Cell Death Differ. 2017;24:638–648. doi: 10.1038/cdd.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JL, Fallon HJ, Morris HP. Two enzymes of serine metabolism in rat liver and hepatomas. Cancer Res. 1970;30:2917–2920. [PubMed] [Google Scholar]
- Davis SR, Stacpoole PW, Williamson J, Kick LS, Quinlivan EP, Coats BS, Shane B, Bailey LB, Gregory JF., 3rd Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab. 2004;286:E272–E279. doi: 10.1152/ajpendo.00351.2003. [DOI] [PubMed] [Google Scholar]
- de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R. L-serine in disease and development. Biochem J. 2003;371:653–661. doi: 10.1042/BJ20021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Cantley LC. Cancer’s fuel choice: new flavors for a picky eater. Mol Cell. 2015;60:514–523. doi: 10.1016/j.molcel.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M, Zhao Y, Topisirovic I, St-Pierre J, Pollak M. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res. 2014;74:7521–7533. doi: 10.1158/0008-5472.CAN-14-2643-T. [DOI] [PubMed] [Google Scholar]
- Hwang S, Gustafsson HT, O’Sullivan C, Bisceglia G, Huang X, Klose C, Schevchenko A, Dickson RC, Cavaliere P, Dephoure N, Torres EM. Serine-Dependent Sphingolipid Synthesis Is a Metabolic Liability of Aneuploid Cells. Cell Rep. 2017;21:3807–3818. doi: 10.1016/j.celrep.2017.11.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Ogretmen B. Ceramide stress in survival versus lethal autophagy paradox: ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Autophagy. 2013;9:258–259. doi: 10.4161/auto.22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539:390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sadhukhan S, Sun S, Wagner GR, Hirschey MD, Qi L, Lin H, Locasale JW. High-resolution metabolomics with acyl-CoA profiling reveals widespread remodeling in response to diet. Mol Cell Proteomics. 2015;14:1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–257. doi: 10.1083/jcb.201604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrmohamadi M, Liu X, Shestov AA, Locasale JW. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014;9:1507–1519. doi: 10.1016/j.celrep.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrmohamadi M, Mentch LK, Clark AG, Locasale JW. Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism. Nat Commun. 2016;7:13666. doi: 10.1038/ncomms13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, Mackay GM, Gottlieb E, Blyth K, Vousden K, et al. Serine one-carbon catabolism with formate overflow. Sci Adv. 2016;2:e1601273. doi: 10.1126/sciadv.1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AC, Maddocks ODK. Serine and Functional Metabolites in Cancer. Trends Cell Biol. 2017;27:645–657. doi: 10.1016/j.tcb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Nikkanen J, Forsström S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamäki J, Roivainen A, Marjamäki P, et al. Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. Cell Metab. 2016;23:635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris KL, Hao R, Chen LF, Lai CH, Kapur M, Shaughnessy PJ, Chou D, Yan J, Taylor JP, Engelender S, et al. Convergence of Parkin, PINK1, and α-Synuclein on Stress-induced Mitochondrial Morphological Remodeling. J Biol Chem. 2015;290:13862–13874. doi: 10.1074/jbc.M114.634063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen C, Parra V, López-Crisosto C, Morales PE, Del Campo A, Gutierrez T, Rivera-Mejías P, Kuzmicic J, Chiong M, Zorzano A, et al. Mitochondrial fission is required for cardiomyocyte hypertrophy mediated by a Ca2+-calcineurin signaling pathway. J Cell Sci. 2014;127:2659–2671. doi: 10.1242/jcs.139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser Z, Gao X, Johnson C, Mehrmohamadi M, Liu X, Li S, Locasale JW. Targeting One Carbon Metabolism with an Antimetabolite Disrupts Pyrimidine Homeostasis and Induces Nucleotide Overflow. Cell Rep. 2016;15:2367–2376. doi: 10.1016/j.celrep.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siskind LJ. Mitochondrial ceramide and the induction of apoptosis. J Bioenerg Biomembr. 2005;37:143–153. doi: 10.1007/s10863-005-6567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Weber G. Enzymic imbalance in serine metabolism in rat hepatomas. Biochem J. 1986;233:617–620. doi: 10.1042/bj2330617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Scharwey M, Langer T. Mitochondrial lipid trafficking. Trends Cell Biol. 2014;24:44–52. doi: 10.1016/j.tcb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Tedeschi PM, Markert EK, Gounder M, Lin H, Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, DiPaola RS, Hirshfield KM, et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4:e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci USA. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.