Abstract
TrmD is an S-adenosyl methionine (AdoMet)-dependent methyl transferase that synthesizes the methylated m1G37 in tRNA. TrmD is specific to and essential for bacterial growth, and it is fundamentally distinct from its eukaryotic and archaeal counterpart Trm5. TrmD is unusual by using a topological protein knot to bind AdoMet. Despite its restricted mobility, the TrmD knot has complex dynamics necessary to transmit the signal of AdoMet binding to promote tRNA binding and methyl transfer. Mutations in the TrmD knot block this intramolecular signaling and decrease the synthesis of m1G37-tRNA, prompting ribosomes to +1-frameshifts and premature termination of protein synthesis. TrmD is unique among AdoMet-dependent methyl transferases in that it requires Mg2+ in the catalytic mechanism. This Mg2+ dependence is important for regulating Mg2+ transport to Salmonella for survival of the pathogen in the host cell. The strict conservation of TrmD among bacterial species suggests that a better characterization of its enzymology and biology will have a broad impact on our understanding of bacterial pathogenesis.
1. INTRODUCTION
All natural transfer RNA (tRNA) molecules contain posttranscriptional modifications to achieve their biological functions. More than 100 such posttranscriptional modifications have been identified to date. Those localized to the elbow region of the L-shaped tRNA tertiary structure generally stabilize the folding of the nucleic acid, while those localized at or near the anticodon ensure translational accuracy during protein synthesis on the ribosome. Each of these posttranscriptional modifications is synthesized in a pathway involving one or multiple enzymes. While the greatest majority of these enzymes are nonessential for life, acting for example as a chaperone to modulate tRNA activity [1], a very small number of these enzymes are absolutely required for cell growth and survival. TrmD is an example of one of these essential enzymes [2–8], responsible for methyl transfer from AdoMet to the N1 position of the G37 base to synthesize m1G37 on tRNA [9] (Fig. 1). The methylated m1G37 is on the 3′-side of the anticodon, and it is necessary for suppressing tRNA frameshifting during protein synthesis on the ribosome [10–12]. Unlike missense errors, frameshifting errors are almost always lethal, because they change the translational reading frame and introduce premature termination codons. TrmD is broadly conserved in sequence and structure among bacterial species, in both Gram (+) and Gram (−), but it is absent from the eukaryotic and archaeal domains [13,14]. Instead, eukaryotes such as humans and archaeal organisms use Trm5 to synthesize m1G37-tRNA. Intriguingly, while catalyzing the same chemical reaction, TrmD and Trm5 are fundamentally distinct from each other in nearly all aspects of the methyl transfer reaction, including the global architecture of the enzyme [15–19], the binding of AdoMet [20], the interaction with tRNA [21], the discrimination of the G37 base [22], the control of the rate-limiting step [23], and the catalytic mechanism [24–27]. TrmD and Trm5 are therefore considered as an analogous pair of enzymes that share no structural homology [28], but use the same chemical substrates and produce the same chemical products. As such, TrmD is ranked as a high-priority antimicrobial target [29], providing the attractive potential to isolate TrmD-specific drugs that would have no interaction with Trm5, thus limiting potential side effects. To fully explore this potential, a better understanding of TrmD structure and function and its biological roles in bacterial pathogenesis is necessary. Here, we highlight the most salient features of TrmD and provide an example illustrating how it regulates bacterial pathogenesis.
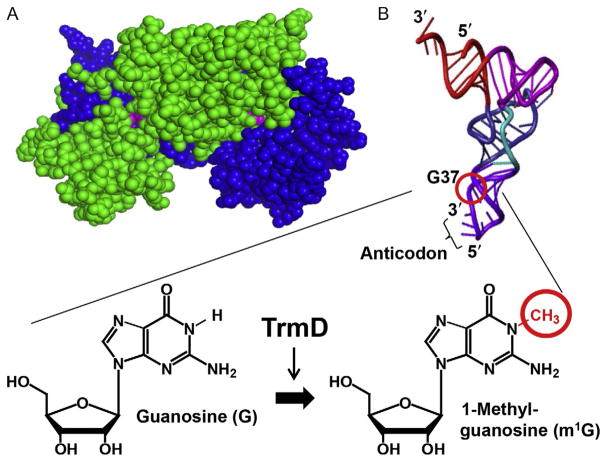
Fig. 1.
TrmD structure and activity. (A) The obligated dimer structure of TrmD (PDB: IUAK), showing chain A in green and chain B in blue and the two AdoMet molecules in pink at each dimer interface. (B) TrmD catalyzes methylation to the N1 position of G37 in tRNA, using AdoMet as the methyl donor and producing m1G37 on the 3′-side of the tRNA anticodon. The position of G37 and synthesis of m1G37 on the L-shaped tRNA tertiary structure is circled.
2. GLOBAL STRUCTURE OF TrmD
TrmD is an obligated homodimer that places each active site at the dimer interface (Fig. 1A) [15,16]. This global structure is different from the structure of Trm5, which is active as a monomer [14]. TrmD is strongly conserved in sequence among evolutionarily diverse bacterial species. High-resolution crystal structures of TrmD in a binary complex with AdoMet [15], or with the unreactive analog sinefungin [15], or with the reaction product AdoHcy (S-adenosyl homocysteine) [16] were known for more than 10 years before a ternary complex with sinefungin and a bound tRNA [17] became available. Most of these structures are based on Haemophilus influenzae TrmD (HiTrmD), which shares high sequence identify (>83%) and similarity (>93%) with Escherichia coli TrmD (EcTrmD), the best-characterized member of the TrmD family [20,23,30,31]. The close sequence conservation between these two enzymes permits exploration of the enzyme’s structure–function relationship in depth.
In all of the available structures of the TrmD dimer (e.g., Ref. [17]), each monomeric chain is made up of three distinct domains: an N-terminal domain (residues 1–160 in HiTrmD and EcTrmD) for binding AdoMet, a C-terminal domain for binding tRNA (residues 169–246), and a flexible linker in between (residues 161–168) (Fig. 2A). However, while both monomeric chains A and B are capable of binding AdoMet at the same time, only chain B is capable of binding tRNA by positioning the G37 base within its flexible linker [17]. This positioning organizes the otherwise disordered flexible linker into a helical structure. The resulting stoichiometry of one TrmD dimer binding with two AdoMet molecules but only one tRNA molecule is consistent between biochemical and structural studies (Fig. 2B) [23]. In this stoichiometry, the N-terminal domain of chain A is paired with the C-terminal domain of chain B to assemble the active site (Fig. 2C). Implicit in this stoichiometry is an asymmetry, in which only the AdoMet in chain A is active for methyl transfer, whereas the AdoMet in chain B is inactive. This half-of-the-sites asymmetry, which can alternate the active AdoMet between the two chains, provides one extreme example of negative cooperativity [32]. In contrast to positive cooperativity, where ligand binding to chain A activates ligand binding to chain B, negative cooperativity entails that the ligand bound to chain A prevents another ligand from binding to chain B. Model studies show that negative cooperativity can regulate ligand binding or enzyme activity between the two chains, depending on the substrate concentration [33]. When substrates are saturating, negative cooperativity modulates the two chains slowly, so that the inactivation of chain B by chain A is slow and gradual. Conversely, when substrates are limiting, the modulation becomes ultrasensitive, so that the inactivation of chain B by chain A is rapid and highly responsive to small fluctuations of substrate concentrations [33]. How TrmD responds to changes in substrate levels remains an important open question.
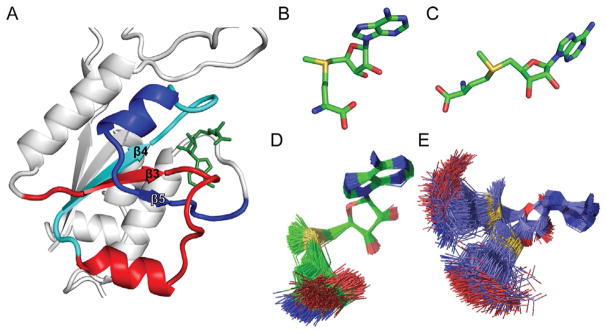
Fig. 2.
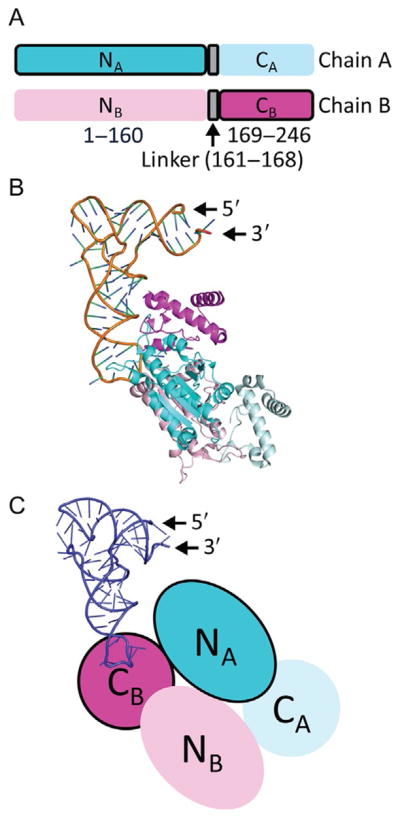
Domain structure of TrmD. (A) Each chain of the TrmD dimer has three distinct domains: an N-terminal domain (residues 1–160) for binding AdoMet, a flexible linker (residues 161–168) for binding G37 of tRNA, and a C-terminal domain (residues 169–246). In the dimer of TrmD, one active site is formed by the N-terminal domain of chain A (NA, dark blue with a black outline) and the C-terminal of chain B (CB, dark pink with a black outline). The other active site, which is not operational, is formed by the N-terminal domain of chain B (NB, light pink without an outline) with the C-terminal domain of chain A (CA, light blue without an outline). The sequence and length of each domain is highly conserved among TrmD enzymes across diverse organisms of the bacterial domain. (B) Each TrmD dimer (chain A in blue and chain B in pink) binds one tRNA molecule in the crystal structure of the ternary complex (PDB: 4YVI). The G37 base of the tRNA is inserted to the active site formed by the NA domain and the CB domain. (C) A cartoon diagram of the TrmD dimer, showing that one active site is assembled between the NA (in dark blue) and the CB (in dark pink), while the anticodon region of tRNA (in purple) is bound to the flexible linker of chain B. The darker color with a black outline indicates the active unit of the dimer structure.
3. THE TREFOIL KNOT OF TrmD AND MUTATIONS IN THE KNOT
In both monomeric chains of TrmD, AdoMet is bound in the N-terminal domain to the deep cleft of a trefoil knot fold [17], which is a topological knot that involves three crossings of the protein backbone through a loop (Fig. 3A). Proteins with a knotted fold are rare (~1.5% in the Protein Data Bank), but they occur across a wide range of families, containing either a trefoil (31), figure-of-eight (41), Gordian (52), or stevedore (61) knot in their structure [34]. While protein knots are generally proposed to enhance protein stability [35], the trefoil knot in TrmD was shown to be required for methyl transfer [31].
Fig. 3.
The trefoil knot structure of TrmD. (A) The trefoil knot is formed starting with the central β3 strand, which turns into β4 through a loop. The β4 makes a turn into β5, which makes a circular insertion through the loop to come out with another loop that binds the adenine base of AdoMet (PDB: 4YVI). The three β stands β3, β4, and β5, together with β1 and β2 (not labeled), form the central β sheet in the N-terminal domain of each monomeric chain in TrmD. (B) The bent conformation of AdoMet in the catalytically active monomer. (C) The open and extended conformation of AdoMet in the catalytically inactive monomer, which is similar to the structure observed in Trm5. (D) The bent conformation of AdoMet has rigid constraints and maintains the bent shape. (E) The open conformation of AdoMet has high flexibility and can extend from the bent shape to the open shape.
The TrmD trefoil knot consists of three β-strands at the central β-sheet (Fig. 3A). This knot starts with β3, which is followed by a loop that turns at the back of β3 and emerges into β4. The end of β4 is followed by another loop that turns into β5, which makes a circular insertion into the knot by crossing over β3 and coming out of the knot with a loop that binds the adenine ring of the methyl donor. In each TrmD trefoil knot, AdoMet is bound in an unusual bent shape [17], which constrains the adenosine and methionine moiety of the methyl donor to face each other (Fig. 3B). In contrast, the majority of methyl transferases (e.g., Trm5) bind AdoMet in the open space of a dinucleotide fold [18,19], which allows the two moieties to extend apart from each other (Fig. 3C). However, although both trefoil knots in TrmD adopt AdoMet in the bent shape in crystal structures, molecular simulation analysis indicates that the two knots have different dynamics and mobility [31]. The active knot is more constrained and binds AdoMet rigidly in the bent shape (Fig. 3D), whereas the inactive knot is more dynamic and binds AdoMet in a range of shapes from the bent to the extended open conformation (Fig. 3E). The different dynamics of the two knots reflects the asymmetry of their relationship.
Additional molecular simulation analysis shows that the trefoil knot is required for AdoMet to adopt the bent shape and that the bent shape is required for TrmD to catalyze methyl transfer [31]. Without the knot, as found in the crystal structure of Aquifex aeolicus TrmD [36], AdoMet cannot bend and can only exist in the open shape. Without being in the bent shape, AdoMet would be positioned in a spatial geometry incompatible with the position of the G37 base and unfavorable for methyl transfer. The importance of the trefoil knot is further manifested in its ability to transmit the signal of AdoMet binding in the bent shape, via intramolecular motions, to the tRNA site to stabilize the nucleic acid substrate binding. tRNA stabilization in the binding site then activates a signal transmitted to the active site to promote methyl transfer [31]. These intramolecular motions are not random, but are dedicated to harnessing the AdoMet binding energies to promote tRNA binding and to activate methyl transfer. Indeed, mutations that disrupt AdoMet binding also destabilize tRNA binding and decrease methyl transfer, while mutations that destabilize tRNA binding have little effect on AdoMet binding but directly affect methyl transfer [31]. Thus, the interaction of the trefoil knot with AdoMet in the bent shape promotes the initial step of the methyl transfer reaction. It is this interaction that determines the positioning of G37-tRNA on the enzyme and the assembly of the active site to catalyze methyl transfer.
Additional simulation analysis shows that the TrmD trefoil knot is important for asymmetric catalysis, because it mediates AdoMet signaling across the dimer interface [31]. This intermolecular signaling confers the asymmetry between the two active sites, such that one is catalytically active while the other is inactive. A mutation that disrupts the stability of the trefoil knot eliminates this asymmetry and equalizes the two active sites, making both AdoMet molecules mobile and both monomeric chains capable of binding one tRNA [31]. Intriguingly, despite enabling the two chains to methylate tRNA simultaneously, the mutant is compromised in both the affinity of tRNA binding and the rate of methyl transfer relative to the native and asymmetric enzyme (by 40- and 30-fold, respectively), resulting in a severe loss of catalytic efficiency (by 1200-fold) [31]. Thus, the asymmetry mediated by the trefoil knot at the dimer interface is important for producing the maximum efficiency of methyl transfer by TrmD.
The structural and molecular simulation analyses to date emphasize the notion that the trefoil knot of TrmD is required for the catalytic mechanism in three ways. It is the structure that enables AdoMet to achieve the bent shape necessary for methyl transfer. It is an organized protein fold within the enzyme that captures the free energy of AdoMet binding to stabilize and orient tRNA binding and to facilitate methyl transfer at the active site. It is also the mechanism for cross-chain communication between the two monomers of TrmD to coordinate with each other for the highest efficiency of methyl transfer. In addition, a mutation within the trefoil knot, which was isolated from a genetic analysis [37], renders the enzyme temperature sensitive and catalytically compromised [38]. This mutation, S88L, occurs at the beginning of the central β3 strand, which leads the way into the trefoil knot. The S88L mutation affects a position highly conserved among Gram (−), but not Gram (+), TrmD enzymes [38]. The temperature sensitivity induced by the mutation supports the general notion that a knot structure confers thermal stability to protein enzymes [35].
4. SYNTHESIS OF m1G37-tRNA BY TrmD
The synthesis of m1G37-tRNA by TrmD is a posttranscriptional event. The efficient methylation by the enzyme on transcripts of tRNA [23], lacking any posttranscriptional modifications, supports the notion that the enzyme catalyzes a primary reaction on tRNA without the requirement for any prior modifications. This is unlike secondary modification reactions on tRNA, where a prior modification is necessary. The synthesis of a complex modified base, such as cmo5U (5-carboxymethoxy uridine) or mcmo5 (5-methoxy-carbonyl methoxy uridine) at the tRNA wobble position, is initiated with the primary reaction, followed by a series of secondary reactions [39,40]. To understand the structural basis of how TrmD recognizes its tRNA substrate, a comprehensive analysis of individual domains of the nucleic acid was performed [21]. This analysis revealed that TrmD requires only a stem–loop structure, in contrast to the requirement for the complete tRNA structure by Trm5 [21]. Specifically, TrmD recognizes the anticodon stem–loop (ASL) of tRNA, which consists of 5bp of the stem and 7 nucleotides in the anticodon loop. However, this simple ASL is not sufficient, but an extension of the stem to 9bp is necessary [21]. The extended structure recapitulates the composite structure of the D stem (typically 4bp) coaxially stacking on the ASL in the native tRNA L-shape structure, which is the core structure of the vertical arm of the L-shape. This notion is intriguing, suggesting that TrmD recognizes the D-ASL vertical arm of the L in a model consistent with the ternary crystal structure of TrmD in complex with tRNA [17]. Indeed, TrmD makes contact only with the vertical arm of the L in the crystal structure, and the contact is mediated by interactions with the phosphodiester backbone of the nucleic acid in a mechanism known as “indirect readout” [41]. It is in contrast to a “direct readout” mechanism, where enzyme makes contact with nucleobases of the nucleic acid substrate.
Given that TrmD only requires the D-ASL structure for recognition of the tRNA substrate, the possibility exists that the m1G37 methylation may occur before the complete transcription of a tRNA. No data are available to date to address this possibility. The only available information is that the m1G37 methylation by TrmD does not need any other prior modification, aminoacylation, or even CCA addition to tRNA [21]. We do not yet know if the methylation can occur cotranscriptionally, or before tRNA processing at the 5′- or 3′-end. More research is necessary to understand the TrmD reaction relative to the biogenesis of tRNA processing and maturation.
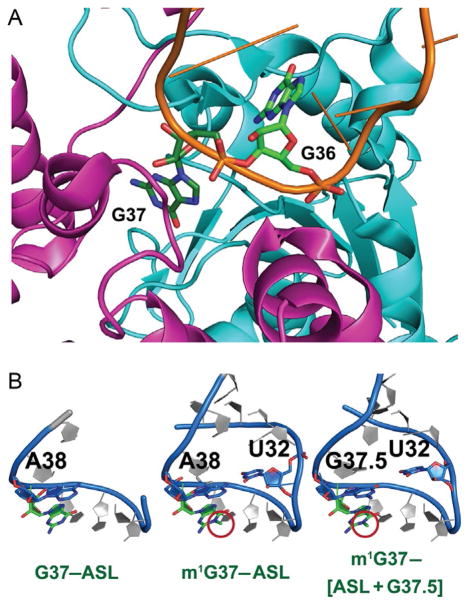
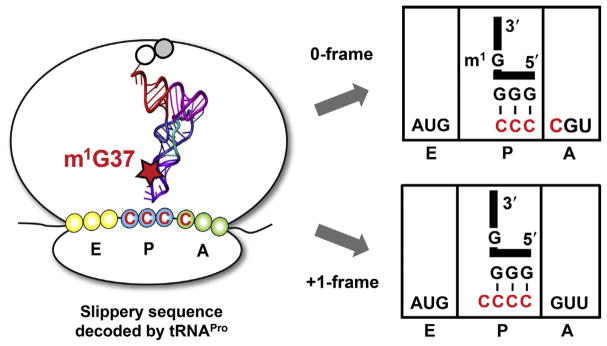
TrmD synthesizes the methylated m1G37 on bacterial tRNAs that contain both G37 and a preceding G36, the 3′-nucleotide of the anticodon [22]. Analysis of the TrmD–tRNA complex reveals that, after G37 is flipped out from the anticodon loop and is recognized by the flexible linker of chain B, the open space left between positions 35 and 38 is occupied by G36 in a stacked position stabilized by a pocket near the trefoil knot in chain A [17] (Fig. 4A). Thus, while G37 is the substrate for methylation, G36 provides additional interactions with the enzyme to induce further conformational changes that strengthen the positioning of G37. Importantly, the resulting m1G37 has the ability to remodel the structure of the tRNA anticodon loop relative to the unmethylated G37. In X-ray crystal structures of the ASL domain of tRNAPro/CGG (CGG: anticodon) in complex with a Thermus thermophilus ribosome [42], the unmethylated G37 prevents the ASL from establishing the typical hydrogen-bond interaction between the O2 atom of U32 and the N6 atom of A38, thus rendering the nucleotides from 30 to 32 on the 5′-side of the ASL disordered and invisible (Fig. 4B). In contrast, the methylated m1G37 is able to remodel the ASL structure and restore the U32–A38 interaction to the conformation consistent with a cognate codon–anticodon pair (Fig. 4B). The lack of structural ordering of the ASL as induced by the unmethylated G37 is striking and is also observed in a mutant structure of ASL containing an extra G37.5 nucleotide (the mutant denoted as [ASL+G37.5] = ASL with an insertion of G between G37 and A38) (Fig. 4B). This mutant [ASL+G37.5] structure is prone to +1-frameshifting [42], most likely because the G37.5 nucleotide forms an aberrant base pair with U32 by displacing A38 away from the normal position (Fig. 4B). Comparison of all three examples indicates that a signature for tRNA to shift to the +1-frame is the loss of the U32–A38 base pair in the ALS. By restoring this base pair, m1G37 provides a mechanism to suppress +1-frameshift.
Fig. 4.
G37 in TrmD and in the tRNA anticodon stem–loop structure. (A) G37 binding to TrmD organizes the flexible linker of chain B (magenta) and projects G36 to the trefoil knot of chain A (cyan) to stabilize the entire tRNA molecule bound to the enzyme (PDB: 4YVI). (B) The unmethylated G37 in the anticodon stem–loop (ASL) structure of tRNAPro/CGG (G37–ASL) prevents the interaction between U32 and A38 and renders the 5′-side of the ASL disordered and invisible (left, PDB: 4P70). The methylated m1G37 remodels the ASL structure (m1G37–ASL) to allow U32–A38 base pairing as in a canonical structure (middle, PDB: 4LT8). The insertion of G37.5 to the ASL, resulting in the structure of m1G37-[ASL + G37.5], disrupts U32–A38 pairing to form the aberrant U32–G37.5 pairing in a disordered structure (right, PDB: 4L47) similar to that in the G37–ASL structure.
5. RIBOSOME FRAMESHIFTING IN THE ABSENCE OF TrmD
The TrmD product m1G37-tRNA is an integral component of translational reading frame accuracy in bacteria [10–12]. In normal conditions, each bacterial ribosome decodes three mRNA nucleotides into a single amino acid at a rapid rate of ~20 residues per second, moving the associated tRNA from the A-site (the aminoacyl-tRNA site), to the P-site (the peptidyl-tRNA site), and through the E-site (the exit site). However, despite these complex and dynamic movements, an E. coli ribosome makes infrequent frameshifts in either the 5′ or 3′ direction. The shift by one nucleotide in the 5′ direction results in the ribosome moving backward with a −1 frameshift, while the shift by one nucleotide in the 3′ direction results in the ribosome moving forward with a +1 frameshift. Notably, bacteria do use frameshifts as a “programmed” mechanism to regulate gene expression. Examples include the programmed +1 frameshift for expression of the prfB gene (for expression of release factor RF2) [43,44] and the −1 frameshift for expression of the dnaX gene (for expression of DNA polymerase III) [45,46]. However, nonprogrammed frameshifts are considered translational errors, usually arising from shifting of a tRNA–ribosome complex on slippery mRNA sequences. Unlike missense errors, which replace one amino acid with another but still permit continued synthesis to the full-length protein, a frameshift error is deleterious, changing the reading frame and introducing premature termination codons. The frequency of nonprogrammed frameshifts is typically low, estimated to be less than one per 30,000 amino acids (or less than 0.003%) over all sequence contexts [47]. The methylated m1G37-tRNA is a key suppressor of +1 frameshift errors.
One of the most frameshift-prone mRNA sequences is CC[C/U]-[C/U], which codes for proline (Pro). In E. coli and throughout bacterial organisms, all three isoacceptors of tRNAPro contain m1G37 in the natural form (Fig. 5). The others that naturally contain m1G37 are the GAG and CAG isoacceptors of tRNALeu and the CCG isoacceptor of tRNAArg. The consistent association of m1G37 with tRNAPro emphasizes the importance of the methylation for translation of Pro codons. Of the three isoacceptors of tRNAPro, the UGG isoacceptor reads all four Pro codons (5′-CCN) through the use of cmo5U34 at the wobble position. This isoacceptor is essential for growth [48], and without m1G37, it is the most shift-prone among the three [11]. For example, in the absence of m1G37, the UGG isoacceptor can read the mRNA sequence CC[C/U]-N either in the 0-frame or the +1-frame with a similar free energy of stabilization, indicating a minimum energetic penalty for the tRNA to shift to the +1-frame. The next most shift-prone tRNA is the GGG isoacceptor [10], which can read the mRNA sequence CC[C/U]-[C/U] with identical stability in the 0- and +1-frame. Among the total sense codons in protein-coding genes, CC[C/U]-N occurs 17,000 times in the K12 genome of E. coli and 20,000 times in the LT2 genome of Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella). These high frequencies of occurrence pose a major challenge for translating ribosomes to suppress +1-frameshifting. Even the less frequent codon sequence CC[C/U]-[C/U] occurs 2300 times in the E. coli genome. In each natural occurrence, the CC[C/U]-N or CC[C/U]-[C/U] sequence can be directly next to the start codon AUG at the second position, or within a short distance from the start codon, or further downstream from the start codon.
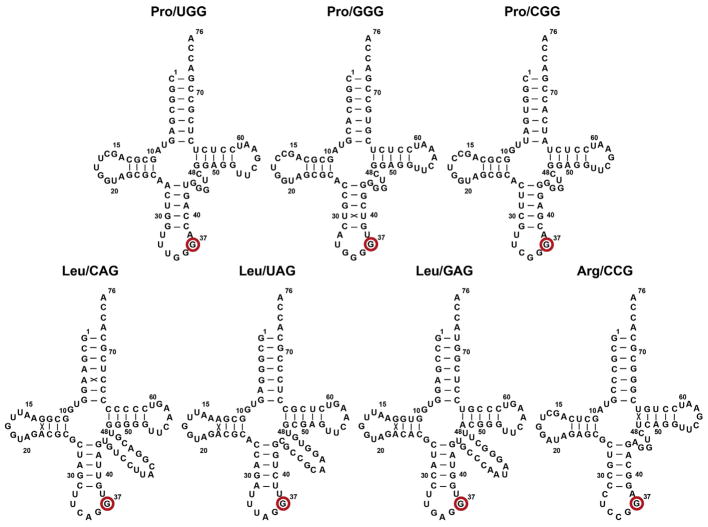
Fig. 5.
Sequence and cloverleaf structure of E. coli tRNA species that are substrates for TrmD, including the Leu/UAG isoacceptor that is most likely a substrate as well. The G37 base to be methylated to m1G37 is marked with a red circle. The numbering is based on the structure of yeast tRNAPhe [88].
Using the CC[C/U]-[C/U] sequence as an example, a cell-based reporter assay has examined its propensity of inducing ribosomal frameshifting at various positions throughout a reporter gene [10]. The results show that the sequence has the highest propensity of inducing frameshifts at the second codon position, when cells are lacking m1G37-tRNAPro [10]. At this position, the frequency of +1-frameshifting is ~1% when cells synthesize m1G37-tRNA, but the frequency is raised by almost 10-fold when cells lose the methylation. At any of the downstream positions, the effect of m1G37-tRNA is smaller (three- to four-fold). Thus, while m1G37-tRNA is important for suppressing +1-frameshifting throughout all sequence contexts, it has the strongest effect at the second codon. In a broader perspective, the CC[C/U]-N sequence is also slippery, and it is read by the UGG isoacceptor in the absence of m1G37. A genome-wide analysis of E. coli K12 bacteria identifies 48 protein-coding genes with the CC[C/U]-N sequence at the second codon, representing a frequency of 1.1% (out of 4289 genes). Some of these genes are essential themselves (e.g., lolB, a conserved outer membrane protein in Gram-negative bacteria). Maintenance of reading frame accuracy of these genes during translation on the ribosome is expected to depend on the presence of m1G37-tRNAPro (Fig. 6).
Fig. 6.
Suppression of ribosomal +1-frameshifting by m1G37-tRNA. On a slippery mRNA sequence AUG-CCC-C, the methylated m1G37-tRNAPro maintains the correct reading frame (0-frame), whereas the unmethylated G37-tRNAPro has a high propensity to shift to the +1-frame, which most frequently occurs during the tRNA sitting at the P-site next to an empty A-site. The tRNA is shown in the L-shape with the anticodon and G37 or m1G37 highlighted.
The second codon in bacterial mRNAs is unique. Its translation requires stable positioning of the initiator tRNA at the ribosomal P-site. This stabilization is enforced by EF-P, the translation factor that stimulates the first peptide bond formation [49]. Indeed, the cell-based reporter assay shows that EF-P also suppresses +1-frameshifting and that it is most effective at the second codon [10]. Notably, the suppression of +1-frameshifts by EF-P is an action on one Pro codon within the context of a slippery sequence and it is distinct from the other function of the factor, which is to release ribosome stalling when encountering poly-Pro codons [50,51].
The importance of m1G37 and EF-P for suppressing +1-frameshifts at the second codon position is further demonstrated in kinetic assays, which used purified ribosomes and protein factors to reconstitute an E. coli translational apparatus programmed by an mRNA [10,11]. These assays show that both m1G37 and EF-P execute suppression of +1-frameshifts most effectively at the second codon position relative to other positions. Among all natural posttranscriptional modifications in tRNAPro, m1G37 is the single determinant that suppresses +1-frameshifts [10]. However, while m1G37 alone is sufficient to completely arrest +1-frameshifts of the UGG isoacceptor, it is insufficient for the GGG isoacceptor and requires the assistance of EF-P [10]. The kinetic assays also reveal that +1-frameshifts can occur in one of two mechanisms. The slow mechanism is during tRNAPro sitting at the P-site next to an empty A-site (Fig. 6). This slow shift occurs on a timescale 103-fold slower relative to peptide bond formation [10], indicating that it is relevant only when cells are starved of nutrients and cannot synthesize sufficient amounts of Pro-tRNAPro to load up the A-site. In contrast, the fast mechanism is during tRNAPro translocation from the A-site to the P-site and it occurs on a timescale comparable to that of peptide bond formation [10], indicating that it has the potential to shift the reading frame even in normal cellular condition.
Both cell-based and kinetic assays have highlighted the propensity of the second codon position to promote +1-frameshifts. This codon position is associated with the first translocation after the first peptide bond is made, and the translating ribosome is ready to move through the second codon to position it at the P-site and open up the A-site for the third codon. The first translocation has a higher probability of shifting on the registry of the mRNA relative to later translocation events for several reasons. The first translocation lacks an E-site tRNA, which helps to maintain the reading frame [52,53], it retains residual interaction with the Shine–Dalgarno sequence [54], and it involves the structurally and dynamically unfavorable movements of transitioning the initiator tRNA from the P-site to the E-site [55]. The first translocation is also the defining moment for the translating ribosome to enter the elongation phase, and it is important for positioning the ribosome on the correct reading frame for downstream synthesis of full-length protein. Both TrmD and EF-P are strictly conserved in bacteria: while TrmD is essential for cell growth [10], EF-P is required for robust cell viability for most bacteria [56]. The dedicated action of TrmD and EF-P at the first translocation indicates an evolutionarily conserved process to take action as early as possible to safeguard the reading frame accuracy of protein synthesis.
6. Mg2+ DEPENDENCE FOR METHYL TRANSFER
Methyl transfer by TrmD requires Mg2+ in the catalytic mechanism [26]. This is highly unusual, because the methyl group of AdoMet is already positively charged and can be easily transferred without the need for metal ions. Indeed, the majority of AdoMet-dependent methyl transferases (e.g., Trm5) require no metal ion [26]. Some exceptions do exist, for instance, for methyl transferases that use Mg2+ to catalyze O-methyl transfer to catechols and flavonoids [57–59], and N6-methyl transfer to adenine in DNA [60,61]. In these exceptions, metal ions are used to modulate the selection and binding of the target substrates but not catalysis. The Mg2+ dependence of TrmD is also distinct from many nucleic acid metabolic enzymes (e.g., DNA and RNA polymerase, restriction enzymes, and catalytic ribozymes) [62,63]. While the latter enzymes use Mg2+ in their reaction, they use the metal ion to both stabilize the nucleic acid or nucleotide substrate and promote catalysis. Instead, TrmD uses Mg2+ exclusively to promote catalysis in the transition state of methyl transfer [26]. The chemical novelty of this transition state is that Mg2+ is a direct ligand to the nucleobase G37 in the tRNA, whereas DNA and RNA polymerases and related enzymes coordinate the metal ion with the phosphodiester backbone of the nucleic acid substrate.
In addition to Mg2+, TrmD can also use Ca2+ and Mn2+ as an active ion, but not Ni2+ or Co2+ [26]. Using EcTrmD as an example, analysis of the enzyme activity as a function of metal ion concentration shows that the Mg2+-dependent activity is maximal at the intracellular concentration of the metal ion in E. coli (2–3mM), but that the Ca2+- and Mn2+-dependent activity is below 1% (for Ca2+) or around 20% (for Mn2+) of the maximum at the intracellular concentration of the respective metal ion (0.1 and 80μM) [64,65]. Thus, Mg2+ is the only physiologically relevant metal ion, given that TrmD is essential for cell growth and a reduction of its activity to below 20% is sufficient to cause cell death [38]. Interestingly, the binding stoichiometry of Mg2+ to TrmD, as measured by the enzyme activity as a function of the metal ion, shows that it is one metal ion required for the activity of one dimer [26]. This is consistent with the stoichiometry of one methyl transfer per dimer [23].
Direct fluorescence-based binding assays with AdoMet or tRNA show that the single Mg2+ required for methyl transfer is not necessary for substrate binding [26]. Even inactive metal ions such as Co2+ or Ni2+, or even the metal-chelating EDTA, promote the same binding affinity. Instead, the single Mg2+ required for methyl transfer is involved in the abstraction of the N1 proton from G37-tRNA, which is likely the rate-limiting step of the TrmD-catalyzed methyl transfer [26]. The Mg2+ reaction has a solvent deuterium kinetic isotope effect (KIE) of 5–6, the Mn2+ reaction has a KIE of 1.8, and the Ca2+ reaction has a KIE of 3–4 [26]. The different magnitudes of KIE with the three metal ions reflect differences in their ability to stabilize a common species in the deprotonation of G37. The largest KIE for the Mg2+ reaction indicates that the reaction is most sensitive to perturbation of the proton abstraction, whereas the smaller KIEs for the Mn2+ and Ca2+ reactions suggest that other steps (e.g., rearrangement of catalytic residues) may have become partially rate limiting.
Metal rescue experiments show that Mg2+ stabilizes the developing negative charge on the O6 of G37 during proton abstraction [26]. The principle of metal rescue is that hard metal ions (e.g., Mg2+) prefer coordination with oxygen, whereas soft metal ions (e.g., Co2+) prefer coordination with sulfur [63]. Indeed, substitution of O6 of G37-tRNA with S6 reduces the rate of the Mg2+ reaction. In contrast, while Co2+ is inactive with O6 of G37-tRNA, it is fully active with S6 of G37-tRNA [63]. These results demonstrate a direct role of the metal ion in the coordination with the atom at the 6th position of G37. Further, in the fully active pair of Co2+-TrmD with the s6G37-tRNA substrate, the formation of a metal–thiolate bond is indicated by a unique UV–visible absorption spectral change for the charge transfer from the sulfur to the transition metal ion with d-shell electrons [63]. Further analysis shows that the R154A and D169A substitutions each abolish the charge transfer, indicating that the native residue of each is involved in catalysis [63]. This is supported by the tRNA-bound crystal structure of the enzyme [17], where the strictly conserved R154 provides a positive charge to stabilize the O6 of G37, whereas the negative charged D169 (or E169 in some bacterial species) provides the general base that abstracts the N1 proton from G37.
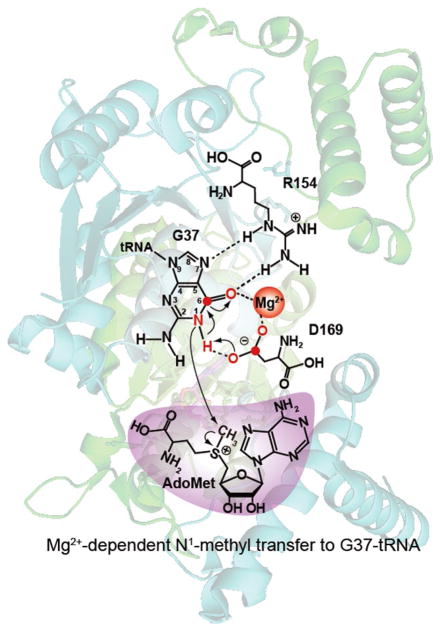
The catalytic role of Mg2+ to TrmD is transient, with a Kd (Mg2+) of 0.68±0.03mM for the E. coli enzyme [26] and a Kd (Mg2+) of 0.42±0.02mM for the Salmonella enzyme. None of the available crystal structures of TrmD contain a metal ion even though some complexes were soaked with 0.2M Ca2+ [20]. The transient nature of Mg2+ is consistent with the proposed catalytic mechanism involving G37-tRNA [26]. In this mechanism (Fig. 7), D169 is the general base to abstract the N1 proton from G37, while the deprotonation is accompanied by developing electron density on the O6 of G37. The developing negative charge on O6 of G37 is stabilized through coordination with Mg2+ and by hydrogen-bond interaction with the side chain of R154. The charge stabilization of O6 in turn facilitates Mg2+ to coordinate with the general base D169 and to help it to align more properly for proton abstraction. The activated N1 nucleophile is then poised for nucleophilic attack on the sulfonium center of AdoMet, resulting in synthesis of m1G37-tRNA and release of AdoHcy. The rate-limiting step is assigned to the action of D169, rather than to the protonation of the leaving group, due to the importance of D169 and the increase of activity as the proton concentration is lowered. The geometry of this catalytic step involves an eight-membered ring that may be nonplanar, such that Mg2+ can be the last member to join the transition state, providing a control for methyl transfer only when both G37-tRNA and AdoMet are appropriately bound.
Fig. 7.
The proposed Mg2+ in the catalytic mechanism of TrmD, involving an eight-membered ring that consists of the N1, C6, and O6 of the guanine ring, the hydrogen bond to the imino group of N1, the two coordination bonds of Mg2+, and the three atoms of the carboxylate of the general base D169. Atomic numbering is indicated for guanine.
7. REGULATION OF Mg2+ TRANSPORT IN SALMONELLA
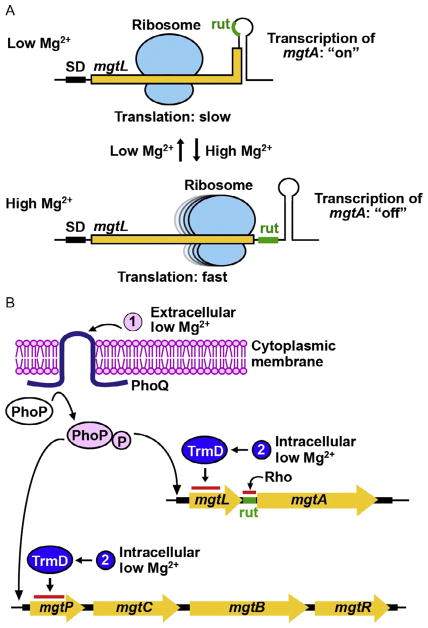
The biological function of the Mg2+ dependence of TrmD has been implicated in regulating metal ion transport in Salmonella [66]. This pathogen is the etiologic agent of human gastroenteritis, causing 1.2 million illnesses, 23,000 cases of hospitalization, and 450 deaths in the United States each year. Mg2+ transport is important for Salmonella survival, infection, and virulence in host cells. The host compartment for Salmonella is low in pH, low in Mg2+, and contains host antimicrobial peptides. This environment activates the PhoPQ two-component system to activate transcription of virulence genes, as well as transcription of the Mg2+ transporter mgtA gene and the mgtCBR virulence operon [67–71]. Transcription of mgtA and mgtCBR is then further regulated by translation of the 5′-leader of each mRNA in a mechanism that determines transcription attenuation of the downstream gene [72–77] (Fig. 8A). In this mechanism, the 5′-leader of mgtA is mgtL, coding for a small peptide (Fig. 8A). Rapid translation of the 5′-leader mgtL would expose a Rho-termination (rut: Rho-utilization) site, which terminates transcription ahead of the structural gene mgtA, whereas slow or stalled translation of mgtL would induce a conformational change of the 5′-leader mRNA and preclude the rut site, allowing transcription to continue through mgtA [76,78]. This translation-coupled transcription attenuation mechanism is reminiscent of the regulatory function of the short open reading frame in the trp, his, and other amino acid biosynthetic operons [79], in which the efficiency of translation of the leader peptide determines whether transcription is terminated at the rut site ahead of the structural gene or allowed to read-through the rut site and into the structural gene. Understanding how both transcription and translation regulate the expression of mgtA, the major Mg2+ transporter gene [80], is necessary to improve our management of Salmonella infection.
Fig. 8.
A proposed mechanism of Mg2+ sensing of Salmonella. (A) Regulation of mgtA transcription in response to Mg2+ level is mediated by the translation speed of the 5′-leader mRNA mgtL sequence, which codes for a short peptide. At low Mg2+, ribosomal translation of mgtL is slow, sequestering the rut sequence in a stem–loop structure and excluding the Rho-dependent transcription termination to permit transcription of mgtA in the “on” state. At high Mg2+, ribosomal translation of mgtL is fast, exposing the rut sequence and resulting in the Rho-dependent transcription termination of mgtA. (B) The two-component system PhoPQ provides a sensor for extracellular Mg2+. At low extracellular Mg2+, the activated PhoQ phosphorylates PhoP to turn on transcription of the Mg2+ transporter gene mgtA as well as the virulence operon mgtCBR, where mgtB is another Mg2+ transporter gene. Transcription of mgtA is then regulated by translation of the 5′-leader sequence mgtL, which encodes a small peptide. Rapid translation of mgtL exposes the Rho-utilization (rut) sequence and terminates transcription ahead of mgtA. In contrast, slow and incomplete translation of mgtL sequesters the rut sequence and allows transcription into the structural gene. The Mg2+-dependent methyl transferase activity of TrmD can act as an intracellular sensor that regulates the speed of translation of mgtL.
For mgtA expression, the 5′-leader mgtL contains multiple codons that require the TrmD product m1G37-tRNA for efficient and accurate translation (Fig. 8B). These include codons for Pro, Leu, and Arg [72]. The dominance of Pro codons in the short MgtL peptide (4 out of 17 residues) is striking. Indeed, the reduction of the level of Pro stalls translation of mgtL and activates transcription of mgtA [72]. However, the physiological connection between the abundance of Pro and the availability of Mg2+ is not obvious. Instead, recent data show that it is the efficiency of translation of the Pro codons in mgtL that is subject to Mg2+ regulation [66]. The efficiency of translation involves not just the level of Pro, but also the quality of translation when Pro-tRNAPro is reading a codon on the ribosome. In fact, the data suggest that the quality of translation of Pro-tRNAPro is worthy of more consideration. For example, different types of mutations in mgtL have the ability to influence the expression of mgtA [66]. A mutation in mgtL resulting in the sequence of three Pro codons in a row, which is a strong signal to stall the translating ribosome [50,51], activates transcription of mgtA. In contrast, substitution of three of the four natural Pro codons in mgtL reduces transcription of mgtA. Deletion of EF-P, which would promote +1-frameshifts and ribosome stalling, induces the expression of mgtA. This result of EF-P, which is related to the quality of translation of Pro codons, argues against the level of Pro in determining the response to Mg2+. Additionally, lesions in rpmA and rpmE, encoding ribosomal proteins L27 and L37, respectively, activate transcription of mgtA, possibly by reducing ribosomal protein synthesis. Most importantly, the trmD-S88L mutation, which reduces the synthesis of m1G37-tRNA [38], activates the transcription of mgtA to a constitutively high level.
Of all of the mutations that were examined recently [66], only the mutation in TrmD is associated with an Mg2+-dependent activity, suggesting a role for TrmD in Mg2+ sensing for regulation of the metal ion transport in Salmonella. This role is consistent with the observation that, while the substitution of Pro codons in mgtL abolishes the response to Pro, it does not abolish the response to Mg2+, but simply reduce the response [72]. The reduction of the response can be explained by the presence of the remaining Leu and Arg codons in mgtL that are still subject to the regulation by the Mg2+-dependent activity of TrmD. The data suggest that TrmD can be an intracellular sensor for Mg2+ (Fig. 8B). In high Mg2+, TrmD would be active and translation of the 5′-leader mgtL would be fast, leading to transcription termination ahead of the structural gene. In limiting Mg2+, however, TrmD would be inactivated and the deficiency in m1G37-tRNA would halt the translation of the 5′-leader mgtL, favoring transcription read-through into the mgtA gene.
The key advantage for the proposed mechanism is the ability of Salmonella to directly sense changes in the intracellular demand for Mg2+. Notably, when cell growth is rapid, intracellular Mg2+ is depleted by cell divisions, even though the metal ion is abundant outside. The coordination of TrmD sensing of intracellular Mg2+ with the PhoPQ sensing of the extracellular Mg2+ constitutes a dual-sensing model. The regulation of bacteria K+ homeostasis is an example of a dual-sensing model, where one single enzyme (KdpD) with two separate domains for sensing extra- and intracellular K+ is used to regulate the transcription of the main K+ transporter gene [81]. The potential synergy between TrmD sensing and PhoPQ sensing should confer the bacteria with robust homeostasis of Mg2+ for fitness in host cells. If this model is supported, it will have wide-reaching impact on bacterial pathogenesis, because the PhoPQ system [82,83], the 5′-leader of mgtA, and the TrmD enzyme are all broadly conserved among pathogenic species of Enterobacteriaceae.
8. CONCLUSION
TrmD is an important and critical enzyme for bacteria. It is ranked as a high-priority antimicrobial drug target [29], due to its essentiality for bacterial growth, strict conservation across a wide range of bacterial species, different structure and mechanism from its human counterpart Trm5, and the possession of an AdoMet binding site, where drug-like small molecules can bind to. However, while the pharmaceutical industry has attempted to target TrmD, progress has stalled [84], due to the lack of a TrmD-specific strategy. Insights into such a strategy are now beginning to emerge based on data from the extensive enzymology and cell-based studies as summarized here. The new focus should be to understand how TrmD differs from Trm5 and to determine the genes whose translation is most sensitive to targeting of TrmD. The unique trefoil knot of TrmD is a major difference from the active site of Trm5, and it is also distinct from the trefoil knot of other methyl transferases [26]. The trefoil knot of TrmD binds AdoMet in the unusual bent shape, and this binding is the basis to initiate substrate signaling to catalyze methyl transfer. The bent shape offers structural novelty and diversity from the conventional AdoMet analogs, and it should be explored for drug design. The Mg2+-dependent catalysis of TrmD is unique among known AdoMet-dependent methyl transferases. This Mg2+ dependence appears to have a role in regulating the transport and homeostasis of the metal ion when Salmonella infects its host. Given that successful drug targeting of HIV integrase has been achieved by disrupting the divalent ion interaction at the active site [85–87], drug targeting of the metal ion-binding site in TrmD would provide an attractive antimicrobial strategy. More efforts should be made to understand the Mg2+ dependence of TrmD and its biology in bacteria. The TrmD product m1G37-tRNA is the major suppressor of +1-frameshifts at Pro codons, but not all Pro codons share the same propensity of inducing frameshifting. Identifying Pro codons in bacterial genes that are most dependent on TrmD for translation would be a major step forward to develop a TrmD-specific strategy that will improve human health in the global population.
Acknowledgments
We thank the support of US National Institutes of Health grants GM108972 and GM114343 (to Y.-M.H.) and European Molecular Biology Organization (EMBO) installation grant 2057 and National Science Center grant Sonata BIS 2012/07/E/NZ1/01900 (to J.I.S). I.M. is a Japan Society for the Promotion of Science (JSPS) Overseas Research Fellow. The content is solely the responsibility of the authors and does not represent the official views of the granting agencies.
References
- 1.Keffer-Wilkes LC, Veerareddygari GR, Kothe U. RNA modification enzyme TruB is a tRNA chaperone. Proc Natl Acad Sci USA. 2016;113:14306–14311. doi: 10.1073/pnas.1607512113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, KGC, King P, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 5.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 7.Thanassi JA, Hartman-Neumann SL, Dougherty TJ, Dougherty BA, Pucci MJ. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 2002;30:3152–3162. doi: 10.1093/nar/gkf418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bystrom AS, Bjork GR. The structural gene (trmD) for the tRNA(m1G)methyltransferase is part of a four polypeptide operon in Escherichia coli K-12. Mol Gen Genet. 1982;188:447–454. doi: 10.1007/BF00330047. [DOI] [PubMed] [Google Scholar]
- 10.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat Commun. 2015;6:7226. doi: 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. The UGG isoacceptor of tRNAPro is naturally prone to frameshifts. Int J Mol Sci. 2015;16:14866–14883. doi: 10.3390/ijms160714866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 13.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian T, Evilia C, Williams S, Hou YM. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol. 2004;339:707–719. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 2003;22:2593–2603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Masuda I, Yoshida K, Goto-Ito S, Sekine S, Suh SW, Hou YM, Yokoyama S. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc Natl Acad Sci USA. 2015;112:E4197–E4205. doi: 10.1073/pnas.1422981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 19.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 20.Lahoud G, Goto-Ito S, Yoshida K, Ito T, Yokoyama S, Hou YM. Differentiating analogous tRNA methyltransferases by fragments of the methyl donor. RNA. 2011;17:1236–1246. doi: 10.1261/rna.2706011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol. 2007;373:623–632. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaguchi R, Giessing A, Dai Q, Lahoud G, Liutkeviciute Z, Klimasauskas S, Piccirilli J, Kirpekar F, Hou YM. Recognition of guanosine by dissimilar tRNA methyltransferases. RNA. 2012;18:1687–1701. doi: 10.1261/rna.032029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christian T, Lahoud G, Liu C, Hou YM. Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J Mol Biol. 2010;400:204–217. doi: 10.1016/j.jmb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christian T, Evilia C, Hou YM. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry. 2006;45:7463–7473. doi: 10.1021/bi0602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christian T, Lahoud G, Liu C, Hoffmann K, Perona JJ, Hou YM. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA. 2010;16:2484–2492. doi: 10.1261/rna.2376210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaguchi R, Lahoud G, Christian T, Gamper H, Hou YM. A divalent metal ion-dependent N(1)-methyl transfer to G37-tRNA. Chem Biol. 2014;21:1351–1360. doi: 10.1016/j.chembiol.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christian T, Gamper H, Hou YM. Conservation of structure and mechanism by Trm5 enzymes. RNA. 2013;19:1192–1199. doi: 10.1261/rna.039503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galperin MY, Walker DR, Koonin EV. Analogous enzymes: independent inventions in enzyme evolution. Genome Res. 1998;8:779–790. doi: 10.1101/gr.8.8.779. [DOI] [PubMed] [Google Scholar]
- 29.White TA, Kell DB. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp Funct Genomics. 2004;5:304–327. doi: 10.1002/cfg.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redlak M, Andraos-Selim C, Giege R, Florentz C, Holmes WM. Interaction of tRNA with tRNA (guanosine-1)methyltransferase: binding specificity determinants involve the dinucleotide G36pG37 and tertiary structure. Biochemistry. 1997;36:8699–8709. doi: 10.1021/bi9701538. [DOI] [PubMed] [Google Scholar]
- 31.Christian T, Sakaguchi R, Perlinska AP, Lahoud G, Ito T, Taylor EA, Yokoyama S, Sulkowska JI, Hou YM. Methyl transfer by substrate signaling from a knotted protein fold. Nat Struct Mol Biol. 2016;23:941–948. doi: 10.1038/nsmb.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauenstein SI, Hou YM, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283:21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha SH, Ferrell JE., Jr Thresholds and ultrasensitivity from negative cooperativity. Science. 2016;352:990–993. doi: 10.1126/science.aad5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulkowska JI, Rawdon EJ, Millett KC, Onuchic JN, Stasiak A. Conservation of complex knotting and slipknotting patterns in proteins. Proc Natl Acad Sci USA. 2012;109:E1715–E1723. doi: 10.1073/pnas.1205918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulkowska JI, Sulkowski P, Szymczak P, Cieplak M. Stabilizing effect of knots on proteins. Proc Natl Acad Sci USA. 2008;105:19714–19719. doi: 10.1073/pnas.0805468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Wang W, Shin DH, Yokota H, Kim R, Kim SH. Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold. Proteins. 2003;53:326–328. doi: 10.1002/prot.10479. [DOI] [PubMed] [Google Scholar]
- 37.Bjork GR, Nilsson K. 1-methylguanosine-deficient tRNA of Salmonella enterica serovar Typhimurium affects thiamine metabolism. J Bacteriol. 2003;185:750–759. doi: 10.1128/JB.185.3.750-759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda I, Sakaguchi R, Liu C, Gamper H, Hou YM. The temperature sensitivity of a mutation in the essential tRNA modification enzyme tRNA methyltransferase D (TrmD) J Biol Chem. 2013;288:28987–28996. doi: 10.1074/jbc.M113.485797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Xiao H, Bonanno JB, Kalyanaraman C, Brown S, Tang X, Al-Obaidi NF, Patskovsky Y, Babbitt PC, Jacobson MP, Lee YS, Almo SC. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature. 2013;498:123–126. doi: 10.1038/nature12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai Y, Miyauchi K, Kimura S, Suzuki T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016;44:509–523. doi: 10.1093/nar/gkv1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perona JJ, Hou YM. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007;46:10419–10432. doi: 10.1021/bi7014647. [DOI] [PubMed] [Google Scholar]
- 42.Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Natl Acad Sci USA. 2014;111:12740–12745. doi: 10.1073/pnas.1409436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 44.Craigen WJ, Cook RG, Tate WP, Caskey CT. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE, Puglisi JD. Dynamic pathways of −1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan S, Wen JD, Bustamante C, Tinoco I., Jr Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen F, Kurland CG. Processivity errors of gene expression in Escherichia coli. J Mol Biol. 1990;215:511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- 48.Nasvall SJ, Chen P, Bjork GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganoza MC, Aoki H. Peptide bond synthesis: function of the efp gene product. Biol Chem. 2000;381:553–559. doi: 10.1515/BC.2000.071. [DOI] [PubMed] [Google Scholar]
- 50.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 51.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 52.Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–265. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 55.Fei J, Richard AC, Bronson JE, Gonzalez RL., Jr Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol. 2011;18:1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. Loss of elongation factor P disrupts bacterial outer membrane integrity. J Bacteriol. 2012;194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferrer JL, Zubieta C, Dixon RA, Noel JP. Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 2005;137:1009–1017. doi: 10.1104/pp.104.048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kopycki JG, Rauh D, Chumanevich AA, Neumann P, Vogt T, Stubbs MT. Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J Mol Biol. 2008;378:154–164. doi: 10.1016/j.jmb.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 59.Lukacin R, Matern U, Specker S, Vogt T. Cations modulate the substrate specificity of bifunctional class I O-methyltransferase from Ammi majus. FEBS Lett. 2004;577:367–370. doi: 10.1016/j.febslet.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 60.Bist P, Rao DN. Identification and mutational analysis of Mg2+ binding site in EcoP15I DNA methyltransferase: involvement in target base eversion. J Biol Chem. 2003;278:41837–41848. doi: 10.1074/jbc.M307053200. [DOI] [PubMed] [Google Scholar]
- 61.Fedoreyeva LI, Vanyushin BF. N(6)-Adenine DNA-methyltransferase in wheat seedlings. FEBS Lett. 2002;514:305–308. doi: 10.1016/s0014-5793(02)02384-0. [DOI] [PubMed] [Google Scholar]
- 62.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 63.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 64.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262:12570–12574. [PubMed] [Google Scholar]
- 65.Hohle TH, O’Brian MR. The mntH gene encodes the major Mn(2+) transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol. 2009;72:399–409. doi: 10.1111/j.1365-2958.2009.06650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gall AR, Datsenko KA, Figueroa-Bossi N, Bossi L, Masuda I, Hou YM, Csonka LN. Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc Natl Acad Sci USA. 2016;113:15096–15101. doi: 10.1073/pnas.1612268113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groisman EA. The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays. 1998;20:96–101. doi: 10.1002/(SICI)1521-1878(199801)20:1<96::AID-BIES13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 68.Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol. 2008;10:576–582. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 69.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 72.Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 2011;30:1485–1496. doi: 10.1038/emboj.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee EJ, Groisman EA. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature. 2012;486:271–275. doi: 10.1038/nature11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee EJ, Choi J, Groisman EA. Control of a Salmonella virulence operon by proline-charged tRNA(Pro) Proc Natl Acad Sci USA. 2014;111:3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hollands K, Sevostiyanova A, Groisman EA. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc Natl Acad Sci USA. 2014;111:E1999–E2007. doi: 10.1073/pnas.1319193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 79.Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 80.Pontes MH, Yeom J, Groisman EA. Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol Cell. 2016;64:480–492. doi: 10.1016/j.molcel.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schramke H, Tostevin F, Heermann R, Gerland U, Jung K. A dual-sensing receptor confers robust cellular homeostasis. Cell Rep. 2016;16:213–221. doi: 10.1016/j.celrep.2016.05.081. [DOI] [PubMed] [Google Scholar]
- 82.Papp-Wallace KM, Maguire ME. Magnesium transport and magnesium homeostasis. EcoSal Plus. 2008;3 doi: 10.1128/ecosalplus.5.4.4.2. [DOI] [PubMed] [Google Scholar]
- 83.Groisman EA, Hollands K, Kriner MA, Lee EJ, Park SY, Pontes MH. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet. 2013;47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hill PJ, Abibi A, Albert R, Andrews B, Gagnon MM, Gao N, Grebe T, Hajec LI, Huang J, Livchak S, Lahiri SD, McKinney DC, et al. Selective inhibitors of bacterial t-RNA-(N(1)G37) methyltransferase (TrmD) that demonstrate novel ordering of the lid domain. J Med Chem. 2013;56:7278–7288. doi: 10.1021/jm400718n. [DOI] [PubMed] [Google Scholar]
- 85.Kawasuji T, Fuji M, Yoshinaga T, Sato A, Fujiwara T, Kiyama R. A platform for designing HIV integrase inhibitors. Part 2: a two-metal binding model as a potential mechanism of HIV integrase inhibitors. Bioorg Med Chem. 2006;14:8420–8429. doi: 10.1016/j.bmc.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 86.Long YQ, Jiang XH, Dayam R, Sanchez T, Shoemaker R, Sei S, Neamati N. Rational design and synthesis of novel dimeric diketoacid-containing inhibitors of HIV-1 integrase: implication for binding to two metal ions on the active site of integrase. J Med Chem. 2004;47:2561–2573. doi: 10.1021/jm030559k. [DOI] [PubMed] [Google Scholar]
- 87.Semenova EA, Johnson AA, Marchand C, Davis DA, Yarchoan R, Pommier Y. Preferential inhibition of the magnesium-dependent strand transfer reaction of HIV-1 integrase by alpha-hydroxytropolones. Mol Pharmacol. 2006;69:1454–1460. doi: 10.1124/mol.105.020321. [DOI] [PubMed] [Google Scholar]
- 88.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]