Abstract
Microsporidiosis is an emerging and opportunistic disease, and Enterocytozoon bieneusi is the main cause of this disease in humans. Little information is available on prevalence and genotyping of E. bieneusi in minks. We collected 559 feces samples of minks from Heilongjiang and Jilin provinces in 2017, and studied E. bieneusi prevalence by nested PCR. A total of 23 out of 559 minks (4.1%) were detected as E. bieneusi-positive, and were raised in five of the seven investigated farms. Age was the only risk factor associated with E. bieneusi prevalence in investigated minks through logistic regression analysis. Sequence analysis of the ITS gene revealed that five E. bieneusi ITS genotypes, including Peru11, EbpC, and three novel genotypes (HLJM-1, HLJM-2 and JLM-1) were present, suggesting minks may be a potential source of human microsporidiosis.
Keywords: Enterocytozoon bieneusi, Epidemiology, Genotyping, Minks, China
Abstract
La microsporidiose est une maladie émergente et opportuniste, et Enterocytozoon bieneusi est la principale cause de la microsporidiose humaine. Peu d’informations sur la prévalence et le génotypage d’E. bieneusi chez les visons sont disponibles. 559 échantillons d’excréments de visons provenant des provinces de Heilongjiang et de Jilin ont été collectés en 2017 et la prévalence d’E. bieneusi a été étudiée par PCR nichée. Au total, 23 (4,1%) des 559 visons ont été détectés positifs pour E. bieneusi, et provenaient de cinq des sept fermes étudiées. L’âge était le seul facteur de risque, étudiés par analyse de régression logistique, associé à la prévalence d’E. bieneusi chez les visons. L’analyse de la séquence du gène ITS a révélé que cinq génotypes ITS d’E. bieneusi, y compris Peru11, EbpC, et trois nouveaux génotypes (HLJM-1, HLJM-2 et JLM-1), étaient présents, ce qui suggère que les visons pourraient être une source potentielle de microsporidiose humaine.
Introduction
Minks are a species of high economic importance: the animals are widely raised for their fur in Northern China, including in Heilongjiang and Jilin provinces. Minks can serve as reservoirs for many pathogens including influenza viruses [7], Aleutian mink disease virus [17], thrombocytopenia syndrome virus [21], Pentatrichomonas hominis [14], and Toxoplasma gondii [30]. Because minks are in close contact with their feeders, they can transmit many pathogens to humans, including Toxoplasma gondii [30]. Despite this, data regarding the prevalence and genotypes of Enterocytozoon bieneusi in minks are scarce.
The Microsporidia contains over 1300 named species, and has a worldwide distribution. Enterocytozoon bieneusi is the most frequent causative agent of human microsporidiosis [1, 11] and is responsible for more than 90% of human infections [3]. E. bieneusi can infect a variety of invertebrates and vertebrates [27, 28, 33], and can be transmitted through the anthroponotic, zoonotic, water-borne, and/or food-borne routes [4, 13, 19]. The symptoms of microsporidiosis caused by E. bieneusi are diarrhea and abdominal pain in immunodeficient individuals, while the infection appears asymptomatic in immunocompetent individuals who can shed spores into the environment and become a potential source of infection for other individuals [29].
More than 240 E. bieneusi genotypes have been defined based on the internal transcribed spacer (ITS) region of the rRNA gene [1]. All the genotypes can be grouped into 9 groups (named groups 1–9). The majority of human infections are caused by the zoonotic group 1 [6, 15]. However, some genotypes (such as I, J and BEB4) from the other zoonotic groups have also been found in humans [8].
In order to determine whether minks can be infected by E. bieneusi, and to assess the zoonotic risk of E. bieneusi between minks and humans, a total of 559 mink feces samples were collected from seven farms in Heilongjiang and Jilin provinces. The samples were tested to detect the prevalence of E. bieneusi and associated genotypes in minks by nested PCR amplification of the ITS region of E. bieneusi.
Materials and methods
Ethics statement
All animals were handled in strict accordance with good animal practices according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the study was approved by the Ethics Committee of Jilin Agricultural University.
Specimen collection
In all, 559 farmed mink fecal samples were randomly collected from Heilongjiang (43°26′~53°33′ N, 121°11′~135°05′ E) and Jilin (41°~46° N, 122°~131° E) provinces, northeastern China in 2017. More than 200 minks were bred at each farm, and the sampling percentage ranged from 5% to 10% on the different farms. Fresh dejections were immediately collected using a polyethylene glove, and were then stored in ice boxes and transported to the laboratory. The Farm ID, gender and age of minks were obtained from the owners.
DNA extraction and PCR amplification
The commercial E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) was used to extract genomic DNA, following the manufacturer’s instructions, and extracted DNA was stored at −20 °C. PCR targeting the ITS region was used to explore the prevalence and genotypes of E. bieneusi. All the PCR operations have been described in a previous study [31]. In each trial, positive and negative controls were present. The amplification products were observed using UV light after electrophoresis in a 1.5% agarose gel containing GoldViewTM (Solarbio, Beijing, China).
Sequence and phylogenetic analyses
Sangon Biotech Company (Shanghai, China) was contracted to sequence the PCR products. Sequence accuracy was evaluated by bidirectional sequencing. Replicates were made when new sequences were found (single nucleotide substitutions, insertions or deletions). ClustalX 1.83 was used to align the sequences. The neighbor-joining (NJ) method (Kimura 2-parameter model, 1000 replicates) was used to reconstruct the phylogenetic trees with Mega 5.0 software. Representative nucleotide sequences were deposited in GenBank under accession numbers MH052578–MH052582.
Statistical analysis
Data analysis of the prevalence of E. bieneusi infection in minks by age, gender, and different farms groups was performed by χ2 testing using SAS version 9.1 (SAS Institute, Cary, NC, USA) [16, 32]. When p < 0.05, the results were considered statistically significant. Odds ratios (ORs) and their 95% confidence intervals (95% CIs) were estimated to explore the strength of the association between E. bieneusi-positivity and the conditions investigated.
Results and discussion
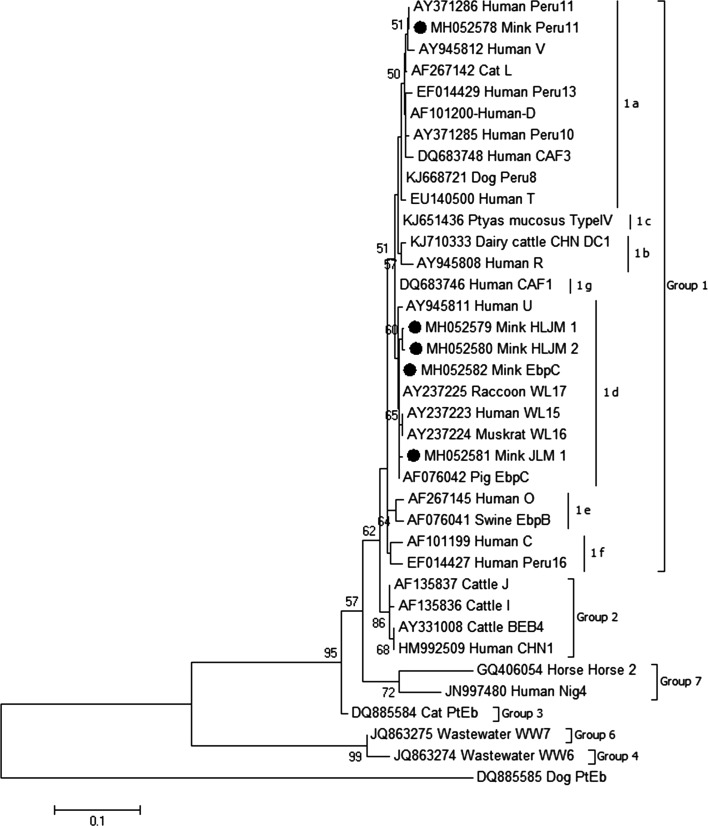
In the present study, 23 out of 559 minks (4.1%) were tested as E. bieneusi-positive. Female minks had a lower E. bieneusi infection rate than males (Table 1). The prevalence of E. bieneusi was 3.9% in minks from Heilongjiang Province, and 4.3% in minks from Jilin Province (Table 1). Minks aged more than three months had a significantly higher infection rate than those aged less than three months (Table 1). The E. bieneusi prevalence in the different investigated farms ranged from 0% to 7.5% (Table 2). Sequence analysis of the ITS region revealed that five E. bieneusi ITS genotypes (two known genotypes Peru11 and EbpC; three novel genotypes HLJM-1, HLJM-2 and JLM-1) were present in investigated minks (Fig. 1).
Table 1.
Prevalence of Enterocytozoon bieneusi in minks in Jilin and Heilongjiang provinces, China.
| Factor | Category | No. tested | No. positive | Prevalence (%) (95% CI) | p-value | OR (95% CI) |
|---|---|---|---|---|---|---|
| Region | Heilongjiang Province | 257 | 10 | 3.9% (1.5–6.3) | 0.81 | Reference |
| Jilin Province | 302 | 13 | 4.3% (2.0–6.6) | 1.1 (0.5–2.6) | ||
| Gender | Female | 279 | 9 | 3.2% (1.2–5.3) | 0.29 | 0.6 (0.3–1.5) |
| Male | 280 | 14 | 5.0% (2.4–7.6) | Reference | ||
| Age | ≤3 months | 244 | 5 | 2.0% (0.3–3.8) | 0.03 | Reference |
| >3 months | 315 | 18 | 5.7% (3.2–8.3) | 2.9 (1.1–7.9) | ||
| Total | 559 | 23 | 4.1% (2.5–15.8) |
Table 2.
Distribution of Enterocytozoon bieneusi genotypes on different farms.
| Farm ID | Sample size | Prevalence (%) | Genotype (No.) |
|---|---|---|---|
| 1 | 124 | 3.2 | HLJM-1 (1); Peru11 (3) |
| 2 | 133 | 4.5 | HLJM-1 (1); HLJM-2 (1); Peru11 (4) |
| 3 | 93 | 7.5 | Peru11 (4); EbpC (3) |
| 4 | 32 | 0 | – |
| 5 | 75 | 6.7 | Peru11 (2); EbpC (3) |
| 6 | 78 | 0 | – |
| 7 | 24 | 4.2 | JLM-1 (1) |
| Total | 559 | 4.1 | Peru11 (13); EbpC (6); HLJM-1 (2); HLJM-2 (1); JLM-1 (1) |
Fig. 1.
Phylogenetic analyses of Enterocytozoon bieneusi using the neighbor-joining (NJ) method (Kimura 2-parameter model). Bootstrap values below 50% from 1000 replicates are not shown. E. bieneusi isolates identified in the present study are pointed out by solid circles.
The overall infection rate of E. bieneusi in minks was 4.11%, which was higher than that in domestic rabbits (0.9%, 4/426) [32] in Jilin, pet chinchillas (3.6%, 5/140) [18] in Beijing, Zhengzhou, Anyang and Guiyang, and similar to the 4.1% infection rates in raccoon dogs (2/49) [29] in Liaoning, Heilongjiang and Jilin, and 4.6% prevalence in captive snakes (11/240) [10] in Guangxi. However, this rate was significantly lower than that in captive golden snub-nosed monkeys (46.2%, 74/160) [26] in Beijing, Shanghai, Anhui and Shanxi, and captive Asiatic black bears (27.4%, 29/106) [24] in Sichuan and Guizhou. The difference in E. bieneusi prevalence may be related to feeding conditions, sampling time, sample sizes, and animal husbandry practices, as well as variable susceptibility of different animals. Enterocytozoon bieneusi is a commonly enteric pathogen, and also exists extensively in the environment. Enterocytozoon bieneusi accumulation could occur throughout a minks lifetime. Therefore, minks aged more than 3 months (OR = 2.9, 95% CI 1.1–7.9, df = 1, p = 0.03) were at a 2.9 times higher risk of E. bieneusi infection compared to minks less than three months of age.
More than 50 E. bieneusi ITS genotypes have been found in captive animals in China [2, 5, 9, 10, 12, 18, 22–26, 29, 31, 32]. However, only five E. bieneusi ITS genes (two known genotypes, Peru11 and EbpC; and three novel genotypes, HLJM-1, HLJM-2 and JLM-1) were found in the present research (Table 2). Genotype Peru11 (distributed on four farms) was the most frequently found genotype of all four genotypes, and was responsible for 56.5% of all infections; genotype EbpC (n = 6, 26.1%) and HLJM-1 (n = 2, 8.7%) were found on two farms; HLJM-2 (n = 1, 4.3%) and JLM-1 (n = 1, 4.3%) were only identified on farm 2 and farm 7, respectively (Table 2). These findings suggest that the five genotypes were prevalent in investigated minks in Heilongjiang and Jilin, China. The Peru11 genotype was also found in captive non-human primates, laboratory macaques, and EbpC was prevalent in nonhuman primates and blue foxes in China [5, 9, 12, 18, 22–26, 29, 31, 32], which suggests that E. bieneusi may be transmitted among these captive animals.
The ITS sequence analysis demonstrated that the sequence of Accession Nos. MH052578 and MH052582 was identical to that of genotypes Peru11 (Accession No. KT922238) and EbpC (Accession No. KX905207), respectively. Moreover, all the five identified E. bieneusi genotypes were grouped into group 1 (Fig. 1). Peru11 was sub-classified into group 1a (Fig. 1); EbpC, HLJM-1, HLJM-2 and JLM-1 were sub-classified into group 1d (Fig. 1). More importantly, Peru11 and EbpC were also found in HIV-positive patients in Henan [20]. These findings suggest that minks may play an important role in human infections. Although no evidence of human microsporidiosis outbreaks originating from minks or other animals has been found, we should pay close attention to nosocomial transmission among humans, minks and other animals.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Project support was provided in part by the Natural Science Foundation of Shandong Province (Grant No. ZR2017PC004), and the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Grant No. SKLVEB2017KFKT007).
Cite this article as: Cong W, Qin S & Meng Q: Molecular characterization and new genotypes of Enterocytozoon bieneusi in minks (Neovison vison) in China. Parasite, 2018, 25, 34.
References
- 1. Deng L, Li W, Zhong Z, Chai Y, Yang L, Zheng H, Wang W, Fu H, He M, Huang X, Zuo Z, Wang Y, Cao S, Liu H, Ma X, Wu K, Peng G. 2018. Molecular characterization and new genotypes of Enterocytozoon bieneusi in pet chipmunks (Eutamias asiaticus) in Sichuan province, China. BMC Microbiology, 18, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deng L, Li W, Zhong Z, Gong C, Cao X, Song Y, Wang W, Huang X, Liu X, Hu Y, Fu H, He M, Wang Y, Zhang Y, Wu K, Peng G. 2017. Multi-locus genotypes of Enterocytozoon bieneusi in captive Asiatic black bears in southwestern China: High genetic diversity, broad host range, and zoonotic potential. PLoS One, 12, e0171772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desportes I, Le Charpentier Y, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R. 1985. Occurrence of a new microsporidan: Enterocytozoon bieneusi n.g., n. sp., in the enterocytes of a human patient with AIDS. Journal of Protozoology, 32, 250–254. [DOI] [PubMed] [Google Scholar]
- 4. Didier ES. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Tropica, 94(1), 61–76. [DOI] [PubMed] [Google Scholar]
- 5. Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, Wang RJ, Wang HY, Qi M, Yu SK. 2015. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling Mountains. Korean Journal of Parasitology, 53, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu Y, Feng Y, Huang C, Xiao L. 2014. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environmental Science & Technology, 48(24), 14219–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang W, Wang S, Zhang C, Li J, Hou G, Peng C, Chen J, Shan H. 2017. Characterization of H5N1 highly pathogenic mink influenza viruses in eastern China. Veterinary Microbiology, 201, 225–230. [DOI] [PubMed] [Google Scholar]
- 8. Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, Zhang S, Li W. 2015. Erratum for Jiang et al., 2015. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Applied and Environmental Microbiology, 81, 5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, Rume FI, Qi M, Jian F, Sun M, Yang G, Zou F, Ning C, Xiao L. 2014. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Applied and Environmental Microbiology, 80, 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karim MR, Yu F, Li J, Li J, Zhang L, Wang R, Rume FI, Jian F, Zhang S, Ning C. 2014. First molecular characterization of enteric protozoa and the human pathogenic microsporidian, Enterocytozoon bieneusi, in captive snakes in China. Parasitology Research, 113, 3041–3048. [DOI] [PubMed] [Google Scholar]
- 11. Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, Huang J, Wang R, Zhang L. 2016. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasites & Vectors, 9, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W, Deng L, Yu X, Zhong Z, Wang Q, Liu X, Niu L, Xie N, Deng J, Lei S, Wang L, Gong C, Zhou Z, Hu Y, Fu H, Xu H, Geng Y, Peng G. 2016. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasites & Vectors, 9, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li W, Song Y, Zhong Z, Huang X, Wang C, Li C, Yang H, Liu H, Ren Z, Lan J, Wu K, Peng G. 2017. Population genetics of Enterocytozoon bieneusi in captive giant pandas of China. Parasites & Vectors, 10(1), 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Li J, Zhang X, Yang Z, Yang J, Gong P. 2017. Prevalence of Pentatrichomonas hominis infections in six farmed wildlife species in Jilin, China. Veterinary Parasitology, 244, 160–163. [DOI] [PubMed] [Google Scholar]
- 15. Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, Guo Y, Wang L, Feng Y, Xiao L. 2015. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Veterinary Parasitology, 207, 220–227. [DOI] [PubMed] [Google Scholar]
- 16. Meng QF, Yao GZ, Qin SY, Wu J, Zhang XC, Bai YD, Cong W, Wang WL. 2017. Seroprevalence of and risk factors for Neospora caninum infection in yaks (Bos grunniens) in China. Veterinary Parasitology, 242, 22–23. [DOI] [PubMed] [Google Scholar]
- 17. Prieto A, Fernández-Antonio R, Díaz-Cao JM, López G, Díaz P, Alonso JM, Morrondo P, Fernández G. 2017. Distribution of Aleutian mink disease virus contamination in the environment of infected mink farms. Veterinary Microbiology, 204, 59–63. [DOI] [PubMed] [Google Scholar]
- 18. Qi M, Luo N, Wang H, Yu F, Wang R, Huang J, Zhang L. 2015. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitology International, 64, 339–341. [DOI] [PubMed] [Google Scholar]
- 19. Thellier M, Breton J. 2008. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite, 15, 349–358. [DOI] [PubMed] [Google Scholar]
- 20. Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, Liu L, Feng Y. 2013. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Neglected Tropical Diseases, 7, e2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang GS, Wang JB, Tian FL, Zhang HJ, Yin FF, Xu C, Xu D, Huang YT, Yu XJ. 2017. Severe fever with Thrombocytopenia Syndrome Virus infection in Minks in China. Vector Borne and Zoonotic Diseases, 17(8), 596–598. [DOI] [PubMed] [Google Scholar]
- 22. Xu C, Ma X, Zhang H, Zhang XX, Zhao JP, Ba HX, Rui Du, Xing XM, Wang QK, Zhao Q. 2016. Prevalence, risk factors and molecular characterization of Enterocytozoon bieneusi in raccoon dogs (Nyctereutes procyonoides) in five provinces of Northern China. Acta Tropica, 161, 68–72. [DOI] [PubMed] [Google Scholar]
- 23. Yang H, Lin Y, Li Y, Song M, Lu Y, Li W. 2017. Molecular characterization of Enterocytozoon bieneusi isolates in laboratory macaques in north China: zoonotic concerns. Parasitology Research, 116(10), 2877–2882. [DOI] [PubMed] [Google Scholar]
- 24. Yang Y, Lin Y, Li Q, Zhang S, Tao W, Wan Q, Jiang Y, Li W. 2015. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotype D in farmed foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in China: first identification and zoonotic concern. Parasitology Research, 114, 4341–4348. [DOI] [PubMed] [Google Scholar]
- 25. Yang Z, Zhao W, Shen Y, Zhang W, Shi Y, Ren G, Yang D, Ling H, Yang F, Liu A, Cao J. 2016. Subtyping of Cryptosporidium cuniculus and genotyping of Enterocytozoon bieneusi in rabbits in two farms in Heilongjiang Province, China. Parasite, 23, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu F, Wu Y, Li T, Cao J, Wang J, Hu S, Zhu H, Zhang S, Wang R, Ning C, Zhang L. 2017. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Veterinary Research, 13, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yue DM, Ma JG, Li FC, Hou JL, Zheng WB, Zhao Q, Zhang XX, Zhu XQ. 2017. Occurrence of Enterocytozoon bieneusi in donkeys (Equus asinus) in China: a public health concern. Frontiers in Microbiology, 8, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang XX, Cong W, Lou ZL, Ma JG, Zheng WB, Yao QX, Zhao Q, Zhu XQ. 2016. Prevalence, risk factors and multilocus genotyping of Enterocytozoon bieneusi in farmed foxes (Vulpes lagopus), Northern China. Parasites & Vectors, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao W, Zhang W, Yang Z, Liu A, Zhang L, Yang F, Wang R, Ling H. 2015. Genotyping of Enterocytozoon bieneusi in farmed blue foxes (Alopex lagopus) and raccoon dogs (Nyctereutes procyonoides) in China. PLoS One, 10, e0142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng WB, Zhang XX, Ma JG, Li FC, Zhao Q, Huang SY, Zhu XQ. 2016. Molecular detection and genetic characterization of Toxoplasma gondii in farmed minks (Neovison vison) in Northern China by PCR-RFLP. PLoS One, 11(11), e0165308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang XX, Cong W, Lou ZL, Ma JG, Zheng WB, Yao QX, Zhao Q, Zhu XQ. 2016. Prevalence, risk factors and multilocus genotyping of Enterocytozoon bieneusi in farmed foxes (Vulpes lagopus), Northern China. Parasites & Vectors, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang XX, Jiang J, Cai YN, Wang CF, Xu P, Yang GL, Zhao Q. 2016. Molecular characterization of Enterocytozoon bieneusi in domestic rabbits (Oryctolagus cuniculus) in Northeastern China. Korean Journal of Parasitology, 54, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang XX, Shi W, Zhang NZ, Shi K, Li JM, Xu P, Zhao Q, Du R. 2017. Prevalence and genetic characterization of Toxoplasma gondii in donkeys in northeastern China. Infection, Genetics and Evolution, 54, 455–457. [DOI] [PubMed] [Google Scholar]