Abstract
A wide variety of bacterial pathogens express phase-variable DNA methyltransferases that control expression of multiple genes via epigenetic mechanisms. These randomly switching regulons – phasevarions – regulate genes involved in pathogenesis, host-adaptation and antibiotic resistance. Individual phase-variable genes can be identified in silico as they contain easily recognised features such as simple sequence repeats (SSR) or inverted repeats (IR) that mediate the random switching of expression. Conversely, phasevarion-controlled genes do not contain any easily identifiable features. The study of DNA methyltransferase specificity using Single-Molecule, Real-Time (SMRT) sequencing and methylome analysis has rapidly advanced the analysis of phasevarions by allowing methylomics to be combined with whole transcriptome/proteome analysis to comprehensively characterise these systems in a number of important bacterial pathogens.
Keywords: phasevarion, DNA methyltransferase, SMRT sequencing, methylome analysis, phase-variation
Bacterial epigenetics, phase-variation, and ‘phasevarions’
Epigenetics is the study of heritable changes in gene expression that occur without changes in DNA sequence [1]. Many mechanisms exist by which these changes are mediated, including DNA methylation, histone modification, and genomic imprinting [1]. DNA methylation is one of the best-studied epigenetic mechanisms, and several well-characterised systems exist within bacteria whereby DNA-methylation leads to changes in gene expression. For example, variable expression of the Pap pilus and antigen 43 in Escherichia coli is mediated by Dam (DNA adenine methyltransferase) methylation of sites in the gene’s promoter region, which alters the ability of the LRP and OxyR regulatory proteins to bind [2]. Loss of Dam leads to decreased virulence in a number of human pathogens, such as Salmonella enterica and Haemophilus influenzae [3]. As another example of methyltransferase action regulating phenotype, the cell cycle of Caulobacter crescentus is regulated by the methyltransferase CcrM (Cell cycle regulated methyltransferase). The functions of solitary DNA methyltransferases, such as Dam and CcrM, have been reviewed in detail previously [2–4].
Many bacterial DNA methyltransferases (see Glossary) exist as part of a restriction-modification (R-M) systems. Four main classes of R-M system exist - types I, II, III & IV, which differ in their subunit composition, cofactor requirements, and DNA cleavage position and sequence specificity [5] (BOX 1). R-M systems are classically considered to confer protection to the bacterial cell against bacteriophages and other horizontal DNA transfers [6], with the cognate methyltransferase protecting ‘self’ DNA from ‘non-self’ foreign DNA, which is degraded by the restriction enzyme component. However, diverse roles of R-M systems have been described, such as genomic island stabilization, genome evolution, and co-factor utilization [7], in addition to their role in epigenetic regulation of gene expression [8].
Box 1. Restriction-Modification (R-M) systems.
Four major classes of restriction-modification (R-M) systems have been characterized in bacteria, differing in their subunit composition, cleavage position, sequence specificity, and cofactor requirements [5]. R-M systems consist of restriction enzymes, which cleave DNA in a sequence specific manner, and cognate methyltransferase enzymes, which methylate the sequences recognized by the restriction components, protecting DNA from cleavage.
Type I systems consist of co-transcribed hsdM, hsdR and hsdS genes, encoding Methyltransferase (M), Restriction (R) and Specificity (S) subunits, respectively [62]. The M and R subunits are highly conserved, whereas the S subunits are highly variable. Each specificity protein is made up of two separate target recognition domains (TRDs). These domains recognize two separate 3 or 4bp sequences separated by a central spanning domain. Shuffling of the DNA sequences encoding the separate TRDs generates variability within the resulting S subunits, leading to restriction/methylation at different sequences [62]. The M2S trimers are active, stand-alone methyltransferases; the R2M2S pentamer is required for DNA cleavage.
Type II systems consist of two independent enzymes: a methyltransferase (Mod) and a restriction endonuclease (Res). Recognition sequences are typically 4–8nt long and palindromic. Eleven distinct subtypes of type II class Res exist, characterised by their behavior and cleavage properties [63]. Mod and Res are both active, stand-alone enzymes.
Type III systems consist of a methyltransferase (encoded by mod) and a restriction endonuclease (encoded by res). These genes are transcribed together and form a two-subunit complex [64]. Mod catalyses the methylation of a single strand of DNA at a specific 4–6 bp asymmetrical recognition sequence, independently of the Res subunit [65]. Mod (M2) is active as a methyltransferase alone, whereas Res requires a complex with the Mod subunit in an R2M2 stoichiometry in order to cleave DNA, usually 25–27 bp downstream of the sequence recognized by Mod [66]. REBASE contains over 1500 type III systems [40], although only ~60 of these have a defined specificity [65].
Type IV systems are methylation dependent restriction systems, and although they are useful tools for epigenetic research [67] they are not associated with cognate methyltransferases [5, 67].
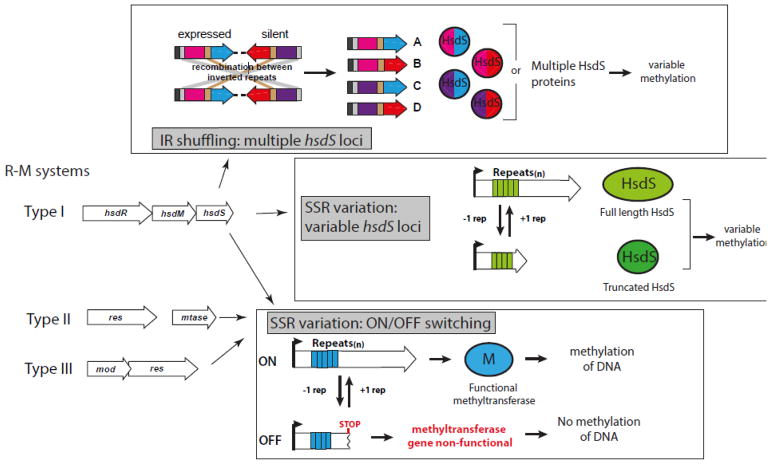
Intriguingly, the genomes of many bacterial pathogens contain DNA methyltransferase genes, associated with R-M systems, that are phase-variable [9–16]. Phase-variation is the random and reversible switching of gene expression (BOX 2), and is typically associated with bacterial surface structures [17]. Phase-variation of DNA methyltransferases can occur due to either (1) hypermutation of simple sequence repeats (SSRs) in the open reading frame (ORF), resulting in ON-OFF switching of gene expression, or (2) through genetic ‘shuffling’ of expressed and silent genes via inverted repeats (IRs), which results in multiple allelic variants of a single protein (BOX 2). Phase-variation of DNA methyltransferase expression results in differential DNA methylation throughout the genome, leading to variable expression of multiple genes via epigenetic mechanisms (Figure 1; Key Figure). These systems are called phasevarions (phase-variable regulons) [8, 18]. The concept of the phasevarion was first described in H. influenzae strain Rd [16]. In this system, phase-variable ON/OFF switching of a type III DNA methyltransferase gene, modA1, occurs as a result of reversible changes in the number of simple sequence repeats located in the modA1 open reading frame. Comparison of the modA1 ON vs. OFF variants revealed that fifteen genes are differentially expressed, including heat-shock proteins dnaK and dnaJ, and outer-membrane opacity protein opa [16]. Since this initial characterization, phase-variable DNA methyltransferases have been identified and shown to control phasevarions in many human-adapted pathogens including the pathogenic Neisseria [14], Helicobacter pylori [15] Moraxella catarrhalis [10] and Streptococcus pneumoniae [11]. All these phasevarions regulate expression of genes that are involved in host colonisation, survival, and pathogenesis, and many regulate expression of putative vaccine candidates.
Box 2. Phase-variation.
Phase-variation is the random and reversible switching of gene expression. It is traditionally associated with genes encoding bacterial surface features, such as adhesins [68], pili [69], iron acquisition proteins [70, 71], and lipo-oligosaccharide (LOS) [72, 73]. Phase-variation allows a population of organisms to generate a phenotypically diverse population. These mixed populations may contain individuals that are, for example, primed to evade an immune response, or better equipped to colonise certain host niches. This random switching of expression means that proteins encoded by phase-variable genes are not ideal vaccine candidates, as their expression is not stable. Phase-variable genes contain sequence features that are easily identified in silico, meaning the proteins they encode can be discounted from development as vaccine candidates. These easily identifiable features are inverted repeats (IRs), and simple-sequence repeats (SSRs) (Figure 1) [8, 17]. Recombination between homologous IRs results in gene shuffling between expressed and silent variants of particular loci. Therefore, the protein encoded by a gene containing IRs is always expressed, but shuffles between a number of allelic variants. SSR tracts are unstable, and vary in length through DNA polymerase slippage during replication. Depending on the number of SSRs present in the tract, genes containing SSRs are in-frame, and expressed (ON), or are out-of-frame, resulting in a premature stop codon, and not expressed (OFF).
Figure 1. An illustration of the ways methyltransferase loci (R-M) phase-vary.
Type I R-M systems can produce multiple methyltransferase variants via shuffling of variable, duplicated hsdS loci through the presence of inverted repeats (IRs) in the ORF of these hsdS genes. Type I loci can also generate multiple HsdS proteins through variation in simple sequence repeats (SSR). Variation in SSR length leads to ON/OFF switching of all three types of R-M loci.
This review aims to detail the current state of phasevarion research, and highlight the role of phase-variable DNA methyltransferases in several major human pathogens.
Detection of DNA methylation and the advent of SMRT sequencing/methylome analysis
The epigenetic nature of phasevarions complicates the in silico identification of stably expressed proteins, as the regulated genes do not contain any identifiable features [8]. The only way to identify genes in a phasevarion is by detailed study of the organisms containing these systems, using gene and/or protein expression analysis techniques. Although epigenetic gene regulation has been studied for many years, the actual characterization of the DNA methyltransferases themselves, in particular the sequences methylated and their genomic context, has been difficult and time consuming. This is especially true for bacteria, in which adenine methylation is the most common form of DNA methylation [19]. Many methods have been developed for eukaryotic CpG methylation, which is important in a variety of processes, including X-chromosome inactivation, carcinogenesis, and chromatin structure [20]. Specific methods to study CpG methylation, such as bisulphite sequencing [21], are not applicable to other forms of methylation. Other methods based on bisulphite sequencing require knowledge of sequence context within which methylations occur, such as methylation specific PCR [22] or methylation specific co-immunoprecipitation [23]. Methods for monitoring adenine methylation are rare, with those developed requiring extensive experimentation, such as chemical modification and bond formation using modified oligonucleotides and chemical crosslinking [24], or the use of radio-labelled AdoMet [25]. Restriction-inhibition assays using methylation sensitive restriction enzymes can be used [14, 26], but these are time consuming and not guaranteed to be successful as restriction enzyme sites may not overlap the particular DNA sequence that is methylated. Mass spectrometry can be used to detect the methyl group itself, but this technique gives no information on the actual sequence context. These limitations therefore made study of adenine methyltransferases particularly difficult, as no high-throughput method was available to rapidly detect the motifs methylated by these systems. Knowledge of DNA methyltransferase sequence context facilitates the identification and study of genes controlled by differential methylation. Recently, Oxford Nanopore MinION DNA sequencing technology has been used to map methylated adenine and cytosine residues using bacterial genomic DNA [27], and of methylated cytosine residues using human genomic DNA [28]. However, this technology has not yet been used to discover the specificity of uncharacterized methyltransferases, although these recent advances are an excellent development for the field of methylomics.
Single Molecule Real Time (SMRT) sequencing was developed as a new DNA sequencing technology in 2010 by Pacific Biosciences (PacBio), and was applied to the study of genome wide methylation patterns [29, 30]. During SMRT sequencing, analysis of the kinetics of DNA synthesis allow a sequence to be generated and the position of modifications such as methylation to be identified [29, 31] (BOX 3). SMRT sequencing/methylome analysis therefore provides a complete, closed genome sequence, and reveals the position of every DNA modification in that genome [32]. A thorough review of SMRT methylome analysis has been published previously [33] and provides significant detail about this technique.
Box 3. Single-Molecule, Real-Time (SMRT) DNA sequencing and methylome analysis.
SMRT sequencing technology uses fluorescently labeled nucleotides, and directly synthesizes DNA from the input template in order to generate a sequence by monitoring the pulse from each nucleotide as it is incorporated into the nascent polynucleotide chain [29, 31]. It is possible to monitor the time between pluses using this system, with the time between the incorporation of two adjacent bases known as the inter-pulse duration (IPD). When bases are modified on the template strand, e.g., a methyl group is present on an adenine residue, the IPD is increased, as this modification delays incorporation of the complementary thymidine base into the nascent daughter strand. Through thousands of reads, the average IPD of every position can be calculated, and that base called as modified or unmodified based on the average IPD for that context in known unmodified samples [29, 30]. Therefore, SMRT sequencing coupled to whole genome methylome analysis not only gives complete, closed genomes for the organism under study, but also shows exactly which residues are modified, and their position in the genome.
Over the last ~5 years, SMRT sequencing/methylome analysis has been used to verify existing DNA methyltransferase specificities [30] and to identify new, previously uncharacterized methyltransferases in a variety of bacterial species [30, 34]. SMRT sequencing/methylome analysis has also been used to characterize the complete methylomes of a number of important bacterial pathogens, including Campylobacter jejuni strains 11168 and 81–176 [34], E. coli ST131 [35] and several strains of H. pylori [36, 37]. Knowledge of the methylome will be invaluable in further understanding the pathobiology of these organisms. Methylome studies provide the opportunity to investigate the roles of DNA methyltransferases in bacterial physiology; for example, the role of DNA methyltransferases during the cell cycle has been characterized in C. crescentus using SMRT sequencing/methylome analysis [38]. The power of SMRT sequencing/methylome analysis has also been demonstrated while analyzing phase-variable DNA methyltransferases that control phasevarions. Comparison of genomic DNA from a pair of isolates containing a phase-variable DNA methyltransferase gene, with the gene expressed in one sample (i.e., phase-varied ON) and not-expressed in the other (i.e., phase-varied OFF, or a knock-out mutant; Figure 2), has been used in several different bacteria to identify the exact sequence recognized and methylated by that particular DNA methyltransferase [9, 10, 39]. In addition, knowledge of methylation differences can be correlated with gene expression profiles, facilitating studies of the exact mechanism responsible for differential gene expression. There are currently over 4,000 PacBio SMRT records in the restriction enzyme database REBASE [40], with almost half of the identified methylation motifs assigned to known methyltransferases.
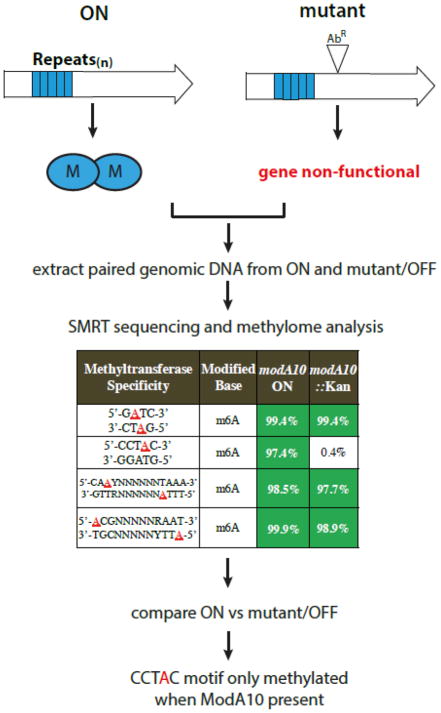
Figure 2. Determination of phase-variable methyltransferase specificity using SMRT sequencing/methylome analysis.
By using genomic DNA preparations from paired isolates that express the methyltransferase (ON variant) or do not express the methyltransferase (OFF variant or a knock-out mutant), it is possible to determine the specificity of the methyltransferase under investigation. Paired genomic DNA samples are subjected to SMRT sequencing, the resulting methylomes are compared, and the phase-variable methyltransferase specificity is identified as the motif that is absent from the OFF variant/knock-out. This example is actual SMRT data generated to decipher ModA10 specificity in NTHi [9].
Phase-variable type III mod genes are the most well-studied phasevarion-controlling methyltransferases
Since the first description of a type III mod gene in H. influenzae strain Rd that controlled a phasevarion [16], a number of phase-variable type III mod genes encoding a phase-variable methyltransferase have been identified in human-adapted bacterial pathogens. In every case studied to date, phase-variable ON/OFF switching of the type III DNA methyltransferase, mediated by SSRs, results in differential regulation of multiple genes. Currently, well characterized phasevarions and mod genes include modA in non-typeable Haemophilus influenzae (NTHi), Neisseria meningitidis and Neisseria gonorrhoeae; modB in N. meningitidis and N. gonorrhoeae [14, 41]; modD in N. meningitidis [13]; modH in H. pylori [15]; and modM in Moraxella catarrhalis [12].
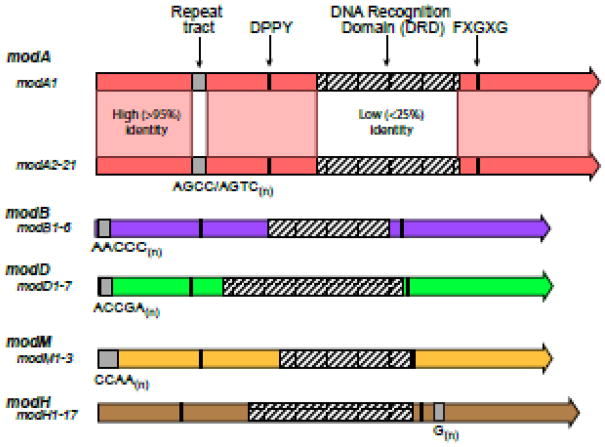
The mod family is highly variable, with SSR tract unit and length, and tract location varying considerably between mod genes (Figure 3). There is very little sequence identity between mod genes [42]. Thus phase-variation of mod genes, and consequently phasevarions, appear to have evolved independently several times in different bacterial species. This implies that the phenotypic diversity resulting from mod phase-variation provides a considerable advantage. Each individual mod gene (modA, modB, etc) is highly conserved in the N- and C- terminal regions, with only the central DNA recognition domain (DRD) showing significant allelic variation [14, 15, 42] (Figure 3). The DRD dictates the specificity of the enzyme. Therefore, different Mod alleles with distinct DRDs methylate different DNA sequences, and therefore regulate expression of different genes. For modA, different alleles evolve from shuffling and transfer of different modA sub-sequences, leading to new DRDs, and therefore new alleles with different methylation specificities [42]. This process has led to the evolution of twenty-one different modA alleles in H. influenzae and Neisseria spp. [9, 42–44]. A recent study describing the phasevarions controlled by the five most prevalent modA alleles (modA2, 4, 5, 9, 10) in NTHi isolated from children with middle ear infection used SMRT sequencing and methylome analysis to rapidly define the methylation specificities of these alleles, as well as generate closed, annotated genomes for prototypical strains containing each of these alleles [9]. This analysis also showed differential regulation of the putative NTHi vaccine candidates HMW, OMP P5, and OMP P6 by modA phase variation in NTHi [9]. Multiple allelic variants of other mod genes have also evolved: six modB and seven modD alleles have been identified in the pathogenic Neisseria [39, 44]; H. pylori strains contain one of seventeen different modH alleles [15]; and at least three modM alleles have been identified in M. catarrhalis [10, 12]. The study of modM in M. catarrhalis used SMRT sequencing/methylome analysis to define the methylation specificity of the most prevalent modM allele, modM2 [10], and was also used recently to determine the methylation specificity of the modM3 allele [45].
Figure 3. Type III mod genes have evolved phase-variable expression multiple times.
There are currently five well characterized phase-variable type III mod genes, all of which control phasevarions – modA, modB, modD, modH, and modM. Each of these mod genes is distinct, with low sequence homology between each class of mod gene. Within each class of mod gene there are multiple allelic variants, which are highly conserved except for their central DNA recognition domain (DRD). Diversity of the DRD means that each mod allele methylates a different DNA sequence, and therefore controls a different phasevarion.
Some strains of N. meningitidis can contain up to three separate phase-variable mod genes - modA, modB and modD. Each individual mod gene has a different methyltransferase specificity, and all have been shown to control individual phasevarions [13, 14, 25, 39, 44]. Even though the pathogenic Neisseria contain multiple mod genes, SMRT sequencing/methylome analysis allowed the specificity of the most common mod alleles present in these organisms to be rapidly identified [39, 44]: modA11 (5′-CGYm6AG-3′), modA12 (5′-ACm6ACC-3′) and modD1 (5′-CCm6AGC-3′). SMRT sequencing technology was particularly powerful in determining the ModA11 recognition sequence, which has a highly relaxed specificity around the core recognition motif of CGYm6AG. The level of methylation was also dependent on the bases flanking this core regions, ranging from 4.6% methylation at GCGCm6AGG sites, to 100% methylation at ACGTm6AGG sites (core sequence underlined) [39]. Determination of this specificity would have been almost impossible without the power of SMRT sequencing/methylome analysis.
There is significant evidence that particular mod alleles, meaning particular phasevarions, are associated with virulence and pathogenesis, and this was reviewed in detail recently [8]. The selection for NTHi containing modA2 ON has been demonstrated to occur in the middle ear during experimental otitis media [9], and the switch from the modA2 OFF to ON state within the middle ear is associated with increased disease severity [46]. Phase-variation of modA2 also leads to differential responses to oxidative stress and neutrophil killing [47]. Phase variation of the modA10 allele in NTHi leads to increased cellular adhesion and invasion, and in increased host death when it is switched OFF compared to ON [48]. A preference for the modM3 allele in M. catarrhalis has been suggested during middle ear infection, with a significant number of middle ear isolates containing this allele when compared to strains isolated from the nasopharynx [10]. A recently identified phasevarion in the paediatric pathogen Kingella kingae modulates the host immune response, and increased bacterial toxin production is seen when the modK1 allele is ON, relative to modK1 OFF [49].
The most prevalent modH allele in H. pylori, modH5, has been shown recently to control expression of the flagellum of this organism [50]. SMRT sequencing and methylome analysis revealed that ModH5 methylates the sequence 5′-Gm6ACC-3′. This sequence was found to be over-represented in a number of virulence associated genes, including the major flagellar component, flaA [50]. Determination of the recognition sequence of ModH5 by SMRT sequencing and methylome analysis subsequently allowed the demonstration that differential methylation of a 5′-GACC-3′ motif in the promoter of flaA leads directly to expression differences in this gene. This is the first demonstration of methyltransferase phase-variation directly controlling the gene expression of a member of a phasevarion [50], with rapid elucidation of the methylation specificity of ModH5 key to this demonstration.
Phase-variable type I R–M systems switch their expression and specificity through a variety of methods
Type I R-M systems have been shown to phase-vary through changes in length of SSRs, and by genetic shuffling of sequences via IRs (BOX 2). Changes in the length of SSRs leads to ON/OFF switching of DNA methyltransferase activity, akin to the type III mod systems, and also changes the specificity of some type I systems. Shuffling of sequences leads to multiple methyltransferase activities by producing a variety of HsdS proteins.
Variation in SSR length can occur in both hsdM and hsdS subunits. For example, ON/OFF switching of a type I hsdM gene in H. influenzae occurs due to changes in length of a pentanucleotide GACGA(n) SSR located in the hsdM open reading frame. This ON/OFF switching results in differences in resistance to phage infection [51]. Phase-variation of an hsdM gene in the bovine pathogen Mannheimia haemolytica results in variable production of the leukotoxin produced by this organism [52]. This hsdM gene also contains a pentanucleotide SSR, but the repeating unit is CAGCA(n).
N. gonorrhoeae contains an unusual phase-variable type I system, with the hsdS gene of this loci split into two different open reading frames (hsdSNgoAV1 and hsdSNgoAV2), due to changes in length of a poly-guanidine tract in the 3′ end of the gene. This hsdS gene produces a truncated (hsdSNgoAV1 only) or full length (hsdSNgoAV1 and hsdSNgoAV2 fused as a single polypeptide) HsdS specificity protein, dependent on the length of this poly-guanidine tract [26]. The truncated HsdS protein designated NgoAV, results in methylation of the sequence 5′-GCm6A(N8)TGC-3′/3′-GC m6A(N8)TGC-5′. whereas the full length HsdS protein, NgoAVΔ, results in methylation of 5′-GCm6A(N7)GTCA-3′/3′-TGm6AC(N7)TGC-5′ and 5′-GCA(N7)CTCA-3′/3′-TGm6AG(N7)TGC-5′, although the latter sequence is methylated only on the complementary strand [26]. Thus, rather than variation in SSRs leading to ON/OFF methyltransferase switching, two distinct methyltransferase activities result from SSR changes in this gene. Since this phase variation would result in distinct genomic methylation patterns, distinct phasevarions may be controlled, although this remains to be investigated. The specificity of these enzymes was determined by time-consuming restriction-inhibition assays [26], which are dependent on restriction enzymes that cut at the same site as the methyltransferase acts and are inhibited by this methylation.
A third type of phase variable type I system has been identified that contains multiple variable hsdS genes, which recombine through shuffling of different hsdS genes to produce methyltransferases with distinct specificities. An excellent review has recently been published describing the role and variety of these ‘locus inverting’ phase-variable type I methyltransferases [53]. Rearrangements occur between distinct IRs located in the hsdS loci, and may be facilitated by a locus-associated recombinase. The first example of a ‘locus inverting’ phase-variable type I system controlling phasevarions was described in the human pathogen Streptococcus pneumoniae [11, 54]. Shuffling between the variable hsdS genes in the SpnD39III locus results in six different HsdS proteins, termed SpnIIID39A-F, that have six different methylation specificities, and result in six distinct gene expression patterns [11]. Several genes involved in capsule biosynthesis are downregulated when the SpnD39III-B variant (specificity of 5′-CRAm6AN9TTC-3′/3′-GYTTN9m6AAG-5′) is expressed, adding a further layer to the complexity of capsule regulation in the pneumococcus. A distinct phasevarion was shown to be regulated when the SpnD39III-A variant is expressed (specificity of 5′-CRAm6AN8CTG-3′/3′-GYTTN8m6GAC-5′), with genes involved in the stress response (dnaK, gpx) and nutrient acquisition (psaABC, fucA, K, U) differentially regulated compared to other SpnD39III alleles. The six DNA methyltransferase activities where rapidly defined by PacBio SMRT sequencing/methylome analysis by using strains where each methyltransferase was ‘locked’ into a single hsdS allele and unable to switch [11]. The unusual motifs recognized and methylated by type I enzymes would have made elucidation extremely difficult and time-consumingusing conventional methods.
A similar system with duplicated, variable hsdS loci (hsdS and hsdS’) containing IRs has been identified in the zoonotic pathogen Streptococcus suis [55]. This system is associated with human invasive disease [56], although the methylation specificity and demonstration of methyltransferase phase-variation has yet to be demonstrated.
Phase-variable type II methyltransferases have been identified in closely related gastric pathogens
In H. pylori, the genes encoding DNA methyltransferases associated with several type II R–M systems contain SSRs [57], and many are associated with colonization and virulence [58, 59]. For example, a survey of the gene encoding the M.HpyAIV methyltransferase in clinical isolates of H. pylori found changes in a poly-adenine tract correlated with ON/OFF switching of this methyltransferase [60]. Methylation by M.HpyAIV, at 5′-Gm6ANTC-3′ sites, was also shown to influence expression of catalase (katA), and was demonstrated to induce a more robust host response in mice, suggesting it controls a phasevarion [60]. In another DNA methyltransferase of H. pylori, Hpy99XXII, changes in the length of a poly-guanidine tract located in the gene resulted in expression of this methyltransferase from an inactive form with a different poly-guanidine tract length [36]. SMRT sequencing/methylome analysis showed that the recognition sequence of this methyltransferase is 5′-TCm6AN6TRG-3′. Analysis of the genome sequences of multiple strains of H. pylori show variation in the length of the poly-guanidine tract found in the gene encoding this methyltransferase [36], indicating phase-variable expression; however, control of a phasevarion needs to be experimentally confirmed.
C. jejuni contains a phase-variable type II restriction modification system, Cj0031 [61]. ON/OFF switching of this R-M system resulted from variation in the length of a poly-guanidine tract located in the open reading frame of cj0031. A number of clinically significant phenotypes such as biofilm formation and cellular invasion were significantly altered by ON/OFF switching of this methyltransferase [61]. Genes such as the autotransporter capA, the adhesin cadF, and the periplasmic binding protein peb1A were all regulated by phase-variable ON/OFF switching of the cj0031 gene. The specificity of the Cj0031 methyltransferase enzyme was determined to be 5′-CCYGm6A-3′ using SMRT sequencing/methylome analysis [61]. The variability in this site would have been almost impossible to determine using standard restriction-inhibition assays, providing another example of the power of SMRT sequencing/methylome analysis to determine the specificity of previously uncharacterised systems.
To our knowledge, no other phase-variable type II R-M systems have been identified to date, which leads to the intriguing possibility that they have only evolved in closely related pathogens that cause disease in a specific niche, i.e., the human digestive tract.
Concluding remarks and future perspectives
The list of phase-variable DNA methyltransferases controlling phasevarions is ever expanding, with many new systems characterised within the last five years. The identification of a variety of phase-variable methyltransferases, that switch their expression via distinct mechanisms, implies that phasevarions have evolved independently in different species and suggests that this type of variable epigenetic regulation provides a strong selective advantage. The random and reversible switching of phase-variable DNA methyltransferases leads to multiple distinct phenotypes in a population that are subject to periodic selection and counter-selection in different environments. Improvements in DNA sequencing technology has identified many new R-M systems in a variety of bacterial species, with SMRT sequencing/methylome analysis allowing the specificity of these newly identified systems to be rapidly identified. Although extensive effort is still required to elucidate the genes regulated by phase-variable methyltransferase expression, ongoing advances in transcriptomic and proteomic technologies have made this process much easier. Identification of the sites of methyltransferase activity by SMRT sequencing/methylome analysis allows coupling of phenotypic analysis with gene expression studies to comprehensively identify all members of a phasevarion
A thorough understanding of phase-variable methyltransferases, and the phasevarions they control, is required for a better understanding of bacterial pathogenesis as well as the development of new and novel vaccines and treatments (see Outstanding Questions). Due to the complexity and variability of epigenetic regulation in bacterial pathogens, in silico methods to determine if a gene is phase-variable may no longer be adequate; even targets that contain no identifiable features associated with phase-variation may be subject to variable expression as they can be regulated in a phasevarion. Therefore, rapid methods to identify the sequences modified by phase-variable DNA methyltransferases (such as SMRT sequencing/methylome analysis) and to determine the genes within the phasevarions (such as RNA-Seq and/or SWATH proteomics), should be widely employed during bacterial studies. A comprehensive characterization of phasevarions is a necessity to direct and inform future vaccine development and treatments for the ever-growing list of pathogenic bacteria containing these systems.
OUTSTANDING QUESTIONS.
-
The exact molecular mechanism by which differential methylation affects gene expression needs to be investigated, and is a major area of work currently underway in this field.
Does differential methylation directly affect gene expression as a result of methylation in promoter regions that affects binding by regulators/transcription factors/RNA polymerase, etc?
Does methylation indirectly affect gene expression due to regulation of an unlinked locus, for example an activator of the gene in the phasevarion?
Most phasevarion characterization to date has been carried out in vitro. While this provides invaluable information about the genes controlled by each phasevarion, many genes may not be identified as they may be regulated only during in vivo conditions. Therefore, what are expression profiles from bacteria isolated in vivo?
What is the combined effect of multiple phase-variable methyltransferases in single strains? For example, pathogenic Neisseria can contain up to three phase-variable mod genes (modA, modB and modD), all switching their expression independently, and all controlling different phasevarions.
Several phase-variable methyltransferases have been highlighted in this review, but do they control phasevarions?
TRENDS BOX.
Phase variable DNA methyltransferases mediate epigenetic regulation in many human pathogens
Phase variable regulons, phasevarions, play important roles in bacterial virulence and pathobiology
In all characterised phasevarions, methyltransferase phase variation controls genes involved in pathobiology, and contain current and putative vaccine candidates
SMRT DNA sequencing and methylome analysis has revolutionised the field of bacterial epigenetics
Understanding phasevarions is key to the development of effective treatments and vaccines
Acknowledgments
This study was funded by Australian National Health and Medical Research Council (NHMRC) (Project Grant 1021631 and Career Development Fellowship to KLS; Project Grant 1099279 to KLS and JMA; Program Grant 1071659 to MPJ); Garnett Passe & Rodney Williams Memorial Foundation (GPRWMF) Research Training Fellowship to AT, and National Institute of Health (NIH) USA grant R01 DC015688 to LOB and MPJ.
GLOSSARY
- Inverted repeat; IR
a short sequence, typically ~10–80 bases long, that is duplicated and inverted a number of base pairs downstream allowing recombination, or shuffling, of the DNA between the two repeated sequences
- Modification enzyme/Methyltransferase
enzymes that add a methyl (CH3) group to a specific base in DNA, usually in a sequence specific manner. They can protect ‘self’ DNA from degradation by a cognate restriction enzyme
- Phase-variation
the rapid and reversible switching of gene expression
- Phasevarion
phase-variable regulon. The suite of genes regulated by phase-variation of a single methyltransferase
- Simple sequence repeat; SSR
a short simple genetic sequence (e.g., G(n), TA(n), AGCC(n)) repeated a number of times within or associated with an open reading frame
- Restriction enzyme
bacterial enzymes that degrade DNA in a sequence specific manner
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dupont C, et al. Epigenetics: Definition, Mechanisms and Clinical Perspective. Semin Reprod Med. 2009;27:351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadesús J, Low DA. Programmed Heterogeneity: Epigenetic Mechanisms in Bacteria. J Biol Chem. 2013;288:13929–13935. doi: 10.1074/jbc.R113.472274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadesús J, Low D. Epigenetic Gene Regulation in the Bacterial World. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev. 2016;40:575–591. doi: 10.1093/femsre/fuw023. [DOI] [PubMed] [Google Scholar]
- 5.Roberts RJ, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bickle TA, et al. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978;75:3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasu K, Nagaraja V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol Mol Biol Rev. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan A, et al. The Capricious Nature of Bacterial Pathogens: Phasevarions and Vaccine Development. Front Immunol. 2016;7:586. doi: 10.3389/fimmu.2016.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atack JM, et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun. 2015;6 doi: 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blakeway LV, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 11.Manso AS, et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 2014;5 doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seib KL, et al. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Mic. 2002;32:159–165. doi: 10.1111/j.1574-695X.2002.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 13.Seib KL, et al. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J. 2011;25:3622–3633. doi: 10.1096/fj.11-183590. [DOI] [PubMed] [Google Scholar]
- 14.Srikhanta YN, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009;5:e1000400. doi: 10.1371/journal.ppat.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srikhanta YN, et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One. 2011;6:e27569. doi: 10.1371/journal.pone.0027569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikhanta YN, et al. The phasevarion: A genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A. 2005;102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moxon R, et al. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Ann Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 18.Srikhanta YN, et al. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Micro. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 19.Blow MJ, et al. The Epigenomic Landscape of Prokaryotes. PLoS Genet. 2016;12:e1005854. doi: 10.1371/journal.pgen.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 21.Krueger F, et al. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 24.Dohno C, et al. Discrimination of N6-methyl adenine in a specific DNA sequence. Chem Commun. 2010:46. doi: 10.1039/c0cc00172d. [DOI] [PubMed] [Google Scholar]
- 25.Adamczyk-Poplawska M, et al. Characterization of the NgoAXP: phase-variable type III restriction-modification system in Neisseria gonorrhoeae. FEMS Microbiol Lett. 2009;300:25–35. doi: 10.1111/j.1574-6968.2009.01760.x. [DOI] [PubMed] [Google Scholar]
- 26.Adamczyk-Poplawska M, et al. Deletion of One Nucleotide within the Homonucleotide Tract Present in the hsdS Gene Alters the DNA Sequence Specificity of Type I Restriction-Modification System NgoAV. J Bacteriol. 2011;193:6750–6759. doi: 10.1128/JB.05672-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rand AC, et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods. 2017;14:411–413. doi: 10.1038/nmeth.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson JT, et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 29.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark TA, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korlach J, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 32.Beaulaurier J, et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat Commun. 2015;6 doi: 10.1038/ncomms8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis BM, et al. Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr Opin Microbiol. 2013;16:192–198. doi: 10.1016/j.mib.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray IA, et al. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forde BM, et al. Lineage-Specific Methyltransferases Define the Methylome of the Globally Disseminated Escherichia coli ST131 Clone. mBio. 2015;6:e01602–01615. doi: 10.1128/mBio.01602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krebes J, et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 2014;42:2415–2432. doi: 10.1093/nar/gkt1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorrell R, Kwok T. The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence. In: Tegtmeyer N, Backert S, editors. Molecular Pathogenesis and Signal Transduction by Helicobacter pylori. Springer International Publishing; 2017. pp. 105–127. [DOI] [PubMed] [Google Scholar]
- 38.Kozdon JB, et al. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc Natl Acad Sci U S A. 2013;110:E4658–E4667. doi: 10.1073/pnas.1319315110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seib KL, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015;43:4150–4162. doi: 10.1093/nar/gkv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts RJ, et al. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwiatek A, et al. Type III methyltransferase M.NgoAX from Neisseria gonorrhoeae FA1090 regulates biofilm formation and human cell invasion. Frontiers in microbiology. 2015;6 doi: 10.3389/fmicb.2015.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gawthorne JA, et al. Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae. PLoS One. 2012;7:e32337. doi: 10.1371/journal.pone.0032337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox KL, et al. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–5252. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan A, et al. Distribution of the type III DNA methyltransferases modA, modB and modD among Neisseria meningitidis genotypes: implications for gene regulation and virulence. Sci Rep. 2016;6 doi: 10.1038/srep21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan A, et al. Complete Genome Sequence of Moraxella catarrhalis Strain CCRI-195ME, Isolated from the Middle Ear. Genome Announc. 2017;5:e00384–00317. doi: 10.1128/genomeA.00384-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brockman KL, et al. ModA2 Phasevarion Switching in Nontypeable Haemophilus influenzae Increases the Severity of Experimental Otitis Media. J Infect Dis. 2016;214:817–824. doi: 10.1093/infdis/jiw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brockman KL, et al. The ModA2 Phasevarion of nontypeable Haemophilus influenzae Regulates Resistance to Oxidative Stress and Killing by Human Neutrophils. Sci Rep. 2017;7:3161. doi: 10.1038/s41598-017-03552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.VanWagoner TM, et al. The modA10 phasevarion of nontypeable Haemophilus influenzae R2866 regulates multiple virulence-associated traits. Microb Pathogenesis. 2016;92:60–67. doi: 10.1016/j.micpath.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srikhanta YN, et al. Phasevarion regulated virulence in the emerging paediatric pathogen Kingella kingae. Infect Immun. 2017;85:e00319–00317. doi: 10.1128/IAI.00319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srikhanta Y, et al. Methylomic and phenotypic analysis of the ModH5 phasevarion of Helicobacter pylori. Sci Rep. 2017;7:16140. doi: 10.1038/s41598-017-15721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaleski P, et al. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology. 2005;151:3361–3369. doi: 10.1099/mic.0.28184-0. [DOI] [PubMed] [Google Scholar]
- 52.Highlander SK, Garza O. The restriction-modification system of Pasteurella haemolytica is a member of a new family of type I enzymes. Gene. 1996;178:89–96. doi: 10.1016/0378-1119(96)00340-x. [DOI] [PubMed] [Google Scholar]
- 53.De Ste Croix M, et al. Phase-variable methylation and epigenetic regulation by type I restriction–modification systems. FEMS Microbiol Rev. 2017;41:S3–S15. doi: 10.1093/femsre/fux025. [DOI] [PubMed] [Google Scholar]
- 54.Li J, et al. Epigenetic Switch Driven by DNA Inversions Dictates Phase Variation in Streptococcus pneumoniae. PLoS Pathog. 2016;12:e1005762. doi: 10.1371/journal.ppat.1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willemse N, Schultsz C. Distribution of Type I Restriction–Modification Systems in Streptococcus suis: An Outlook. Pathogens (Basel, Switzerland) 2016;5:62. doi: 10.3390/pathogens5040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willemse N, et al. An emerging zoonotic clone in the Netherlands provides clues to virulence and zoonotic potential of Streptococcus suis. Sci Rep. 2016;6:28984. doi: 10.1038/srep28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin LF, et al. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc Natl Acad Sci U S A. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ando T, et al. Restriction-modification systems may be associated with Helicobacter pylori virulence. J Gastroenterol Hepatol. 2010;25(Suppl 1):S95–98. doi: 10.1111/j.1440-1746.2009.06211.x. [DOI] [PubMed] [Google Scholar]
- 59.Gauntlett JC, et al. Phase-variable restriction/modification systems are required for Helicobacter pylori colonization. Gut Pathog. 2014;6:35. doi: 10.1186/s13099-014-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skoglund A, et al. Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori. J Bacteriol. 2007;189:8914–8921. doi: 10.1128/JB.00108-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anjum A, et al. Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168. Nucleic Acids Res. 2016;44:4581–4594. doi: 10.1093/nar/gkw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loenen WAM, et al. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pingoud A, et al. Type II restriction endonucleases--a historical perspective and more. Nucleic Acids Res. 2014;42:7489–7527. doi: 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 65.Rao DN, et al. Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res. 2014;42:45–55. doi: 10.1093/nar/gkt616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raghavendra NK, et al. Mechanistic insights into type III restriction enzymes. Front Biosci (Landmark Ed) 2012;17:1094–1107. doi: 10.2741/3975. [DOI] [PubMed] [Google Scholar]
- 67.Loenen WA, Raleigh EA. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014;42:56–69. doi: 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atack JM, et al. Selection and counter-selection of Hia expression reveals a key role for phase-variable expression of this adhesin in infection caused by non-typeable Haemophilus influenzae. J Infect Dis. 2015;212:645–653. doi: 10.1093/infdis/jiv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blyn LB, et al. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richardson AR, Stojiljkovic I. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ren Z, et al. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J Bacteriol. 1999;181:5865–5870. doi: 10.1128/jb.181.18.5865-5870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fox KL, et al. Selection for phase variation of LOS biosynthetic genes frequently occurs in progression of non-typeable Haemophilus influenzae infection from the nasopharynx to the middle ear of human patients. PLoS One. 2014;9:e90505. doi: 10.1371/journal.pone.0090505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poole J, et al. Analysis of nontypeable Haemophilus influenzae phase variable genes during experimental human nasopharyngeal colonization. J Infect Dis. 2013;208:720–727. doi: 10.1093/infdis/jit240. [DOI] [PMC free article] [PubMed] [Google Scholar]