Abstract
Complex biological processes, such as organogenesis and homeostasis, are stringently regulated by genetic programs that are fine-tuned by epigenetic factors to establish cell fates and/or to respond to the microenvironment. Gene regulatory networks that guide cell differentiation and function are modulated and stabilized by modifications to DNA, RNA and proteins. In this review, we focus on two key epigenetic changes – DNA methylation and histone modifications – and discuss their contribution to retinal development, aging and disease, especially in the context of age-related macular degeneration (AMD) and diabetic retinopathy. We highlight less-studied roles of DNA methylation and provide the RNA expression profiles of epigenetic enzymes in human and mouse retina in comparison to other tissues. We also review computational tools and emergent technologies to profile, analyze and integrate epigenetic information. We suggest implementation of editing tools and single-cell technologies to trace and perturb the epigenome for delineating its role in transcriptional regulation. Finally, we present our thoughts on exciting avenues for exploring epigenome in retinal metabolism, disease modeling, and regeneration.
Keywords: Chromatin, DNA methylation, Histone modification, Next Generation Sequencing, Neuronal Differentiation, Photoreceptor, Retina neurodegeneration
1. Introduction
Higher order brain functions in humans, e.g., emotions, learning and behavior, are largely dictated by the visual world. The retina is the most approachable structure of the central nervous system with five major types of neurons and one type of glia (called Müller glia) organized in intricate highly-stratified circuits that are involved in the processing and transmission of visual information to the brain. Vision begins with hyperpolarization of rod and cone photoreceptors in response to light. Chemical signals from photoreceptors are sent to retinal ganglion cells (RGCs) after amplification, integration and refinement by bipolar, horizontal and amacrine cells. Axons of the RGCs form the optic nerve and carry visual information in the form of electrical signals to the brain. Numerous distinct cell types within bipolar and amacrine neurons and in RGCs are necessary to accurately process visual images (Masland, 2012). This extensive neural diversity, together with an easier access to cells, have inspired in-depth investigations into molecular mechanisms associated with retinal cell-fate specification, morphogenesis and physiology.
Early during retinal development, at embryonic day (E)8 in mice, expression of a group of genes (including Pax6, LhX2, Rax and Six3) in the anterior neural plate defines the eye field, which eventually gives rise to the optic cup (Agathocleous and Harris, 2009; Gregory-Evans et al., 2013; Heavner and Pevny, 2012). Distinct pools of retinal progenitor cells (RPCs) acquire specific competence states, achieved by the combinatorial action of intrinsic regulatory factors at various development time points, and produce different types of neurons and Müller glia. Overlapping waves of differentiation begin as early as E11 and no proliferating cell is detectable by postnatal day (P)10 (Young, 1985). RGCs, horizontal and amacrine cells, cones and ~15% of the rods are generated prenatally in the mouse and rat retina, whereas most rods, bipolar cells and Müller glia are born postnatally (Carter-Dawson and LaVail, 1979). Genetic programs are the primary drivers of retinal development, coordinating proliferation and cell cycle exit, deciding cell fates, controlling cell numbers and guiding cell maturation (Reese and Keeley, 2016). Transcription factors (TFs) and signaling pathways that specify cell-types and orchestrate functional maturation in specific lineages have been discussed in several excellent reviews and will not be elaborated here (Bassett and Wallace, 2012; Brzezinski and Reh, 2015; Cepko, 2014; Sanes and Zipursky, 2010; Swaroop et al., 2010).
In addition to genetic control, cell-cell interactions, soluble factors and signals from the microenvironment are necessary for developing a tightly-controlled cellular organization and subsequent maintenance of a functional retina. A plethora of neurons within the major cell types that emerge from similar competence-restricted RPC pools makes it imperative to utilize distinctive epigenetic mechanisms for fine-tuning gene expression patterns and to achieve specific morphology and function. The overall contribution of the epigenome for generating such diversity is expected to involve a variety of molecular machineries, participation of non-coding RNAs, as well as diverse DNA and histone modifications.
The concept of epigenetics has evolved since the term was first introduced (Waddington, 1942; Waddington, 2012). Waddington combined the embryologic model of epigenesis that describes how specialized structures develop from general precursors in gradual steps and the role of genetics during development, thereby integrating the fields of embryology and genetics. Waddington’s model of an epigenetic landscape represented cell differentiation in terms of a hillside with a slope showing epigenesis, where the fate-choices of a cell travelling downhill will become increasingly restrictive until it reaches a delta of valleys, which in turn epitomized cell-fates formed as a consequence of genetics (Waddington, 1940). Subsequently, this hypothesis was expanded to suggest that epigenetic mechanisms determine gene expression and are responsible for cellular memory (Nanney, 1958).
DNA methylation was proposed as a mechanism for cellular memory (maintained through cell division), which could explain the phenomenon of X-inactivation and heritable changes in gene expression (Holliday and Pugh, 1975; Riggs, 1975). Subsequent studies correlated DNA methylation with transcriptional outputs creating a new definition of epigenetics that puts emphasis on molecular mechanisms affecting chromosomal features maintained during cell division. However, the discovery of short-lived chromatin modifiers allowed for an even broader view of epigenetics that referred to all molecular signatures in chromosomes generated in response to stimuli; more specifically, “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” (Bird, 2007). Another current definition of epigenetics still includes the concept of heritability as “the inheritance of variation (genetic) above and beyond (epi) changes in DNA sequence” (Bonasio et al., 2010). In this review, we will refer to epigenetics as the study of the epigenome, specifically the chromatin regulatory modifications that relate to the memory of past stimuli without necessarily implying mitotic or meiotic inheritance, in accordance to Bird’s definition.
Chromatin is composed of stable interactions of DNA with histones and non-histone components, allowing a tight packaging of the nearly two-meter-long DNA within a small nucleus, which further imposes many constraints for regulatory factors to access DNA. During evolution, the compact organization within the nucleus likely led to disparate molecular interactions, intermediary between the genetic material and the transcriptional output, thereby establishing stringent controls on gene expression. Epigenetic signals are diverse and include DNA and histone modifications, histone variants and positioning, higher order chromatin structure, and non-histone factors associating with chromatin including RNA. Chemical modifications of DNA and histones exert a strong influence on the accessibility of genomic regulatory sequences to distinct molecular complexes that control transcription and splicing. Here, we provide a comprehensive overview of the accumulated knowledge on epigenetic mechanisms, focusing on DNA methylations and histone modifications, involved in retinal development and homeostasis. We have highlighted mouse and human studies in health and disease, as well as articulated our thoughts on exciting avenues for future exploration. We recommend the following excellent reviews on related topics, such as nucleosome remodeling, 3D chromatin organization and RNA-mediated gene regulation (Becker and Workman, 2013; Holoch and Moazed, 2015; Soshnev et al., 2016; Zelinger, 2018).
2. DNA methylation
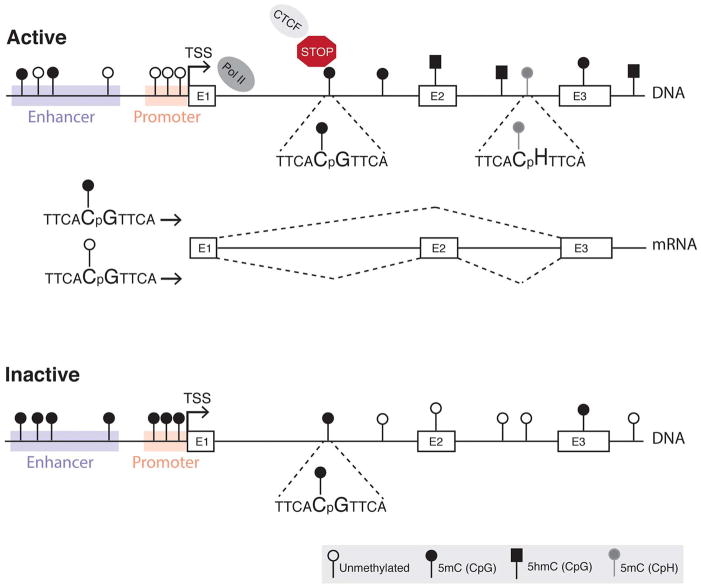
Covalent addition of methyl groups to DNA can influence transcription and provide genomic stability (Smith and Meissner, 2013). The importance of DNA methylation in the retina is evidenced by studies reporting genomewide changes of this mark during retinogenesis underscoring its key role in regulating gene expression. The retina seems to harbor unique methylation forms, previously shown to be functional only in stem cells and brain (Fig. 1) (Mo et al., 2016; Perera et al., 2015). Here, we will describe general features of DNA methylation and the main molecular players involved in its regulation before giving specific examples of its role in the retina (Section 4).
Fig. 1.
Context-dependent roles of DNA methylation. In active genes, DNA methylation is usually absent at the promoter, is low at enhancer regions and variable at gene bodies (Jones, 2012). Neuronal genes may harbor 5hmC and 5mC in the CpG context and/or 5mC in the CpH context (H = A, T or C) at their gene bodies (Lister et al., 2013). CTCF links DNA methylation to RNA splicing by binding to DNA near an alternative exon (E2), halting RNA polymerase and allowing the incorporation of E2 into the transcript (Shukla et al., 2011). DNA methylation can prevent CTCF binding, and thereby the incorporation of alternative exons. Inactive genes usually are methylated at their promoter and enhancer regions, and contain variable methylation at gene bodies with no 5hmC or 5mC in the CpH context.
A methyl group is commonly added on cytosine at position C5, and C5-methylcytosine (5mC) was the first discovered epigenetic mark (Hotchkiss, 1948). Other known DNA modifications include methylation on adenine at position N6 (6mA) (Wu et al., 2016) and oxidative derivatives of 5mC including C5-hydroxymethylation (5hmC) (Tahiliani et al., 2009), C5-formylcytosine (5fc) and C5-carboxylcytosine (5caC) (Ito et al., 2011). We will limit our discussion to 5mC and 5hmC because of their key roles in neuronal development and homeostasis.
DNA methylation was initially recognized as a repressive mark associated with X chromosome inactivation, imprinting, and silencing of repetitive genomic elements (Jones, 2012). However, DNA methylation is now demonstrated to impart context-dependent functions and regulate diverse aspects of mammalian biology. The function of DNA methylation is dependent on CpG dinucleotide density and their precise location within a gene (Jones, 2012; Zemach et al., 2010). In addition, DNA methylation may change with cellular activities (Guo et al., 2011a; Martinowich et al., 2003). Therefore, the targeting and function of DNA methylation is tightly controlled and may involve multiple regulatory mechanisms.
DNA methylation impedes gene expression if present at or near the transcription start site (TSS), particularly within a CpG island (CpGI, a region of high CpG content) (Jones, 2012) (Fig. 1). The mammalian genome possesses less CpG content than would be expected by random chance since deamination of methylated cytosine results in thymine, which happens to be the most frequent genetic variation in humans (Rideout et al., 1990). Although CpGI methylation usually leads to gene repression, most CpGIs remain unmethylated and instead harbor the repressive histone modification H3K27me3, established by the polycomb complex (Reddington et al., 2013). Less than 10% of the CpGIs are methylated in the mammalian genome and usually located in regions of long-term repression, including imprinted genes and the X-chromosome (Illingworth and Bird, 2009).
DNA methylation in the gene body is positively correlated with enhanced transcription (Ball et al., 2009) but can also influence RNA splicing (Shukla et al., 2011) or alternative promoter usage (Maunakea et al., 2010) (Fig. 1). We have limited understanding of the functional significance of methylation in enhancers and insulators (Stadler et al., 2011; Ziller et al., 2013). Methylation in intergenic regions is reported to antagonize polycomb repression, permitting the expression of neurogenic genes (Wu et al., 2010). Conversely, early in development, unmethylated enhancer regions could serve as permissive marks for later activation of cell-type specific genes (Wiench et al., 2011). DNA methylation can also exert an inhibitory effect on insulators by preventing CTCF binding to DNA (Bell and Felsenfeld, 2000), thus allowing interactions between adjacent regions of the genome that could be insulated from each other. Together, these findings indicate that the role of intergenic methylation depends on the cellular and/or genomic context.
DNA methylation can occur in the CpH context (H = A, T, C) as well. Patterns of 5mC in CpH regions were first observed in embryonic stem cells (ESCs) and appear to represent ~ 25% of the DNA methylation (Lister et al., 2009). The mammalian brain contains high levels of CpH methylation, which increases during synaptogenesis (Lister et al., 2013). Notably, mouse cones and rods also exhibit DNA methylation in the CpH context although at lower levels than cortical neurons (Mo et al., 2016). Other forms of methylation (such as 5hmC) are abundant in neurons (Kriaucionis and Heintz, 2009) and accumulate during neuronal maturation in the brain (Lister et al., 2013) and the retina (Perera et al., 2015), demonstrating its potential functions in activity-dependent gene regulation.
2.1. Writers and erasers
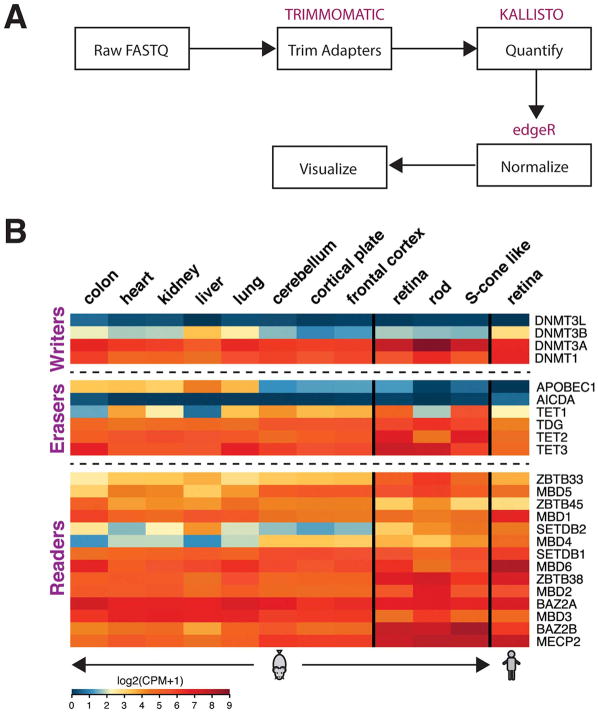
Addition and removal of a methyl group at position C5 of cytosines is catalyzed enzymatically by DNA methyltransferases (DNMTs) (called writers) and Tet-eleven translocation (TET) enzymes (called erasers), respectively (Wu and Zhang, 2014). DNMT1 copies methylation patterns during DNA replication and preferentially modifies hemimethylated DNA, whereas the de novo methyltransferases DNMT3a and DNMT3b establish novel patterns of methylation during development and adulthood (Goll and Bestor, 2005). In the retina, DNMT1 and DNMT3 have an important role in photoreceptor and retinal pigment epithelium (RPE) development (Nasonkin et al., 2013; Rhee et al., 2012; Singh et al., 2017) (see Section 4). DNMT2 mediates tRNA methylation (Goll et al., 2006) and its role in DNA methylation is unclear (Jeltsch et al., 2016). DNA methylation can be passively lost through cellular divisions or actively removed by TET enzymes, which generate 5hmC, 5fc and 5caC as part of a stepwise oxidation process (Ito et al., 2011; Tahiliani et al., 2009). Notably, Tet1 is expressed at low levels in purified adult rods but shows high expression in the whole retina (Fig. 2B), suggesting specific functions in other retinal neurons. The removal of a methyl group can also be catalyzed by AID (activation-induced cytidine deaminase) and APOBECs (apolipoprotein B mRNA-editing enzyme catalytic polypeptides), which convert 5hmC to 5-hydroxymethyluracil (5hmU) (Guo et al., 2011b). TDG (thymine DNA glycosylase) mediates base excision repair to remove 5caC as a last step in methyl group removal (Kohli and Zhang, 2013). Additionally, Gadd45b (growth arrest and DNA damage-inducible 45b) mediates activity-dependent demethylation of promoters in neurogenic genes (Ma et al., 2009). Expression profiles of DNMTs and TET enzymes in the mouse and human retina compared to other tissues are presented in Fig. 2.
Fig. 2.
Expression of writers, erasers and readers of DNA methylation. (A) Schematic of expression analysis. Raw RNA-seq reads from adult mouse tissues and human retina were initially filtered for low quality and adapter contamination using Trimmomatic (v0.36). Transcript level quantification was performed using the kallisto (v0.43) utility. All data were analyzed as single end sequences. Gene level quantification was computed using the tximport R package on Ensembl (version 84 and 82 for mouse and human data sets, respectively). Normalization was performed using trimmed mean of M-values (TMM) and counts per million (CPM) values were computed using the edgeR package. RNA-seq data were obtained from the following sources: sorted rods and S-cone-like cells (Kim et al., 2016b), mouse retina (Hoshino et al., 2017), unpublished human retina (available at https://neicommons.nei.nih.gov/#/), and non-retinal tissues (Pervouchine et al., 2015). (B) Heatmaps of expression data from indicated adult tissues. We included only genes having a one-to-one mapping between human and mouse annotations. The retina expression data are more similar to neural tissues. Scale of expression values is shown as log2 of Counts per million (CPM) +1, with blue representing low and red high expression values.
2.2. Readers
Some proteins, such as pioneer TFs, are methylation insensitive and help other regulatory factors to bind DNA by inducing demethylation (Domcke et al., 2015), whereas others can recognize DNA in a methylation-dependent manner and are called “readers” (Ludwig et al., 2016). The methylated CpG dinucleotide can serve as a signal to directly recruit TFs (Zhu et al., 2016) or chromatin-modifying regulators (Kinde et al., 2015; Ludwig et al., 2016).
The CpG dinucleotide can be recognized by proteins containing a methyl-CpG binding domain (MBD), which is comprised of 75–80 amino acid residues and binds symmetrically to methylated CpG dinucleotides (Ohki et al., 2001). At least 11 MBD proteins have been identified so far; these include MBD1-6, MeCP2 (methyl-CpG-binding protein 2) (Hendrich and Bird, 1998), the histone methyltransferases SETDB1 and SETDB2 (KMT1E and KMTF, respectively), and BAZ2A and BAZ2B (Hendrich and Tweedie, 2003). We noted that expression of SETDB2 in the adult mouse liver and retina was higher compared to other tissues (Fig. 2). Other protein domains can bind methylated CpG as well. These include transcription repressors ZBTB33, ZBTB4 and ZBTB38 containing C2H2 zinc fingers (Filion et al., 2006), basic leucine zipper (bZIP) factor C/EBPa (CCAAT/enhancer-binding protein-a) (Rishi et al., 2010), zinc-finger protein ZFP57, and KAP1 (KRAB-associated protein 1) (Quenneville et al., 2011). The analysis of methylation levels within immunoprecipitated chromatin followed by bisulfite sequencing (ChIP-BS-seq) suggests that many transcriptional regulatory proteins can bind to methylated genomic motifs in vivo (Zhu et al., 2016). Screenings aimed at finding proteins that bind to methylated nucleotides and their oxidized derivatives have been performed using microarrays or DNA pull-down assays and identified numerous 5mC- and 5hmC- binding proteins (Hu et al., 2013; Spruijt et al., 2013).
Unmethylated CpGs are preferentially recognized by CXXC-domain containing proteins, which can prevent methylation at these sites (Song et al., 2011). In concordance, the loss of the CXXC-containing histone demethylase FBXL10 (KDM2B) in polycomb complex-bound promoters causes de novo methylation and transcriptional silencing (Boulard et al., 2015).
The repertoire of proteins that can read DNA methylation is very diverse and extensive, and we are just beginning to understand the complex interplay of regulatory proteins and modifications that allow and maintain a tight control of cell-type specific gene expression. Most of the proteins that regulate DNA methylation, including methyl-CpG binding proteins, are expressed in the mammalian retina (Fig. 2).
3. Histone modifications
Histones contribute to the compaction of DNA in the nucleus forming macromolecular structures, called nucleosomes, which typically include a 147-bp DNA fragment wrapped about 1.7 times around an octamer of two copies of each of the core histones H2A, H2B, H3 and H4 (Campos and Reinberg, 2009). Nucleosomes are separated by a short linker DNA fragment and form a “beads-on-a-string” fiber where DNA can be made accessible to other proteins (euchromatin). This fiber can be further compacted with the help of the linker histone H1, making DNA largely inaccessible (heterochromatin). Two categories of heterochromatin are possible: facultative heterochromatin is a dynamic structure that can be decondensed when genes are turned on, while constitutive heterochromatin is more (but not completely) rigid and locks the telomeric, centromeric and pericentric regions of the chromosomes (Wang et al., 2016).
Due to their impact on chromatin structure and DNA accessibility, histones are key regulators of all major chromatin-related processes, including DNA transcription, replication and repair (Zentner and Henikoff, 2013). Histone post-translational modifications, particularly acetylation and methylation, constitute the most studied epigenetic marks in the retina. As in DNA methylation, genomewide changes of histone marks during retinal development have revealed their key role in regulating gene expression. The importance of histone modifications in the retina has also been established by pharmacological or genetic inactivation of enzymes that participate in this process. In addition, changes in histone marks are observed during aging and age-related retinal diseases suggesting their involvement in disease pathogenesis. In this section, we will summarize the basic mechanisms involved in histone modifications, with emphasis on acetylation and methylation, before providing specific examples of their role in retinal development, aging and disease later in Sections 4 and 5.
3.1. A combinatorial interplay of histone modifications to interpret the genome
All four core histones have a flexible amino-terminal (Nter) tail that protrudes away from the nucleosome and undergo an array of post-translational modifications, such as methylation, acetylation, ubiquitylation and phosphorylation (Kouzarides, 2007). Likewise, the carboxy-terminal end of histones H2A and H2B and the globular domain of the four core histones, as well as the complete histone H1, are subjected to many post-translational modifications (Zhao and Garcia, 2015). The combinatorial interplay of histone modifications participates in a plethora of gene regulatory mechanisms that are now referred as the histone code (Jenuwein and Allis, 2001). Whether these modifications are instructive or primarily stabilize nucleosomes is still poorly understood (Henikoff and Shilatifard, 2011; Rando, 2012).
Furthermore, histone variants and histone tail clipping provide two additional layers of regulation (Azad and Tomar, 2014; Henikoff and Smith, 2015). Here, we will only introduce the best-characterized histone marks, namely acetylation and methylation of lysine (K) residues of histone H3 and H4 Nter tails. Histone lysine residues can accept one, two or three methyl groups (me1, me2, me3) but only one acetyl (ac) moiety. Some modifications are mutually exclusive on a single histone tail but can coexist on adjacent histones within a nucleosome. Thus, the two H3 histones in a given nucleosome may carry distinct combinations of modifications (= asymmetry) (Voigt et al., 2012).
Histone acetylation is associated with euchromatin and active transcription (Roh et al., 2005). Acetylation destabilizes nucleosomes, decreases DNA wrapping and establishes loading docks for readers (Struhl, 1998). Highly acetylated promoters are protected from DNA methylation (Mutskov et al., 2002). Many lysine residues at the Nter tails of histones H3 and H4 can be acetylated (Fig. 4) and the accumulation of the modification rather than its position likely affects the transcriptional outcome (Dion et al., 2005). In mouse retinal neurons, acetylated histones are localized in the euchromatin, except H3K4ac that spans both euchromatin and facultative heterochromatin compartments (Eberhart et al., 2013) (Fig. 3). In mouse rod photoreceptors, some acetylated histones are particularly detected in the facultative heterochromatin compartment close to the euchromatin interface (Kizilyaprak et al., 2010) (Fig. 3).
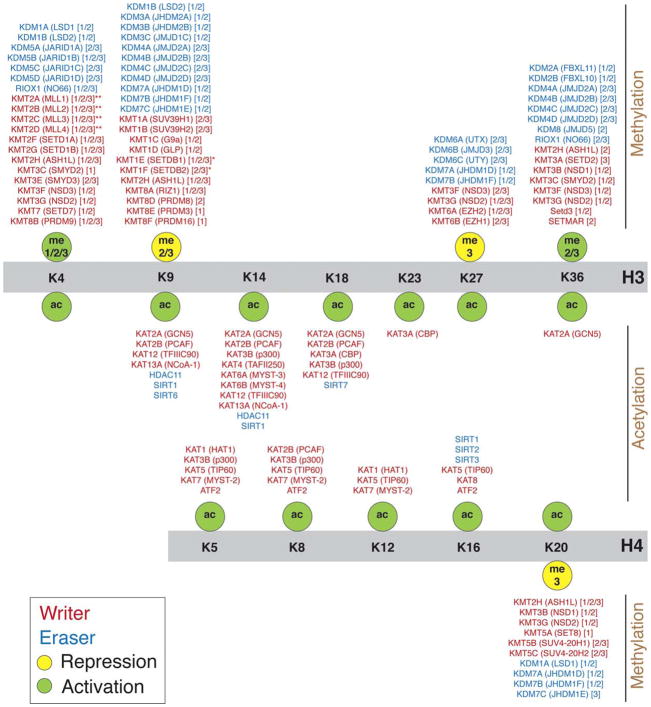
Fig. 4.
Targets of (de)acetylating and (de)methylating enzymes on lysine (K) residues of histone H3 and H4 Nter tails. A diverse group of enzymes can add (writers, shown in red) or remove (erasers, shown in blue) acetyl or methyl groups to/from histone tails. Enzyme names are according to the nomenclature established in (Allis et al., 2007) with common names indicated in parenthesis. The degree of methylation targeted by the methyltransferases and demethylases are shown in square brackets. The key hallmarks associated with gene activation (shown in green) and repression (shown in yellow) are explained in Section 3.1. *Methyltransferases containing the methyl-CpG binding domain (MBD). **Methyltransferases containing the CXXC domain recognizing unmethylated cytosines. Additional details pertaining to these enzymes are provided in Tables 1 and 2.
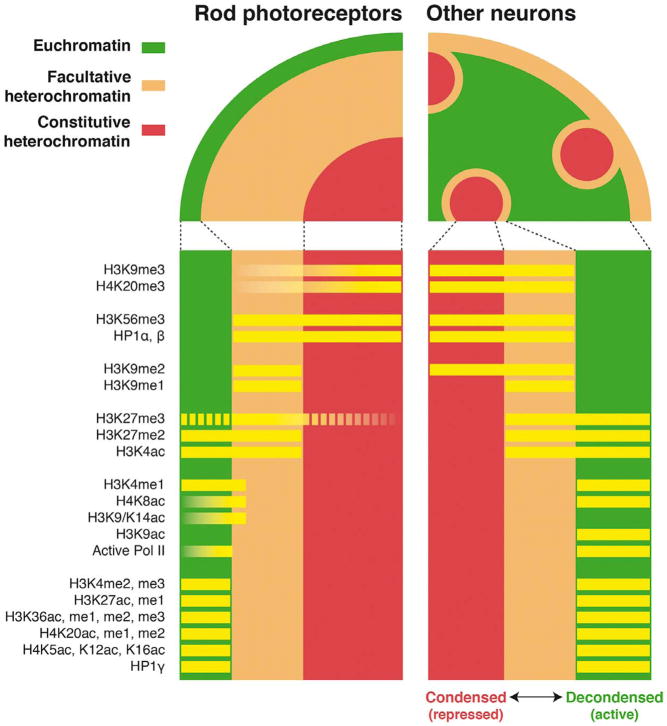
Fig. 3.
Distribution of histone modifications and associated proteins along distinct chromatin domains in retinal neurons. All neurons in the mouse retina, except rods, have conventional nuclei comprising of central euchromatin and peripheral clusters of heterochromatin; however, rod photoreceptors have central domains of heterochromatin with euchromatin localized at the periphery (Eberhart et al., 2013; Solovei et al., 2009). H3K9me2 is the only mark exhibiting a differential distribution between rod photoreceptors and other retinal neurons, and the presence of H3K27me3 in euchromatin and constitutive heterochromatin is discordant between studies (represented by dashed lines). The data was compiled from (Eberhart et al., 2013) and (Kizilyaprak et al., 2010).
Histone methylation is well characterized at lysine residues 4, 9, 27 and 36 of histone H3 and at lysine 20 of histone H4 Nter tails (Fig. 4). The outcome of these modifications on gene expression is based on the species/cell type, location (promoter/enhancer/gene body), degree of modification (me1/2/3), and neighboring epigenetic marks (Hyun et al., 2017). The constitutive heterochromatin is marked by H3K9me3 and H4K20me3 (Schotta et al., 2004), whereas the facultative heterochromatin generally harbors H3K9me2 and H3K27me3 (Rosenfeld et al., 2009; Wen et al., 2009). However, most of these marks can also be associated with active genes in certain contexts, especially when combined with other marks or at specific genes or chromatin positions (Vakoc et al., 2005; Young et al., 2011). The heterochromatin compartments of mouse retinal neurons exhibit these respective hallmarks but one mark diverges: H3K9me2 mark is present only in the facultative heterochromatin of rod nuclei but in both heterochromatin compartments of other nuclei (Eberhart et al., 2013) (Fig. 3).
Active genes are marked at their TSS by H3K4me3 which facilitates histone acetylation and favors transcription initiation, but it is not a sign of successful elongation (Guenther et al., 2007; Wang et al., 2009). H3K4me1 and H3K4me2 are detected downstream of active TSSs (Schneider et al., 2004) and seem to be better markers of successful transcription than H3K4me3. The TSS of poised genes exhibit a bivalent mark with coexisting H3K4me3 and H3K27me3 (Bernstein et al., 2006; Mikkelsen et al., 2007). This repressed state permits a rapid activation by the removal of H3K27me3 in response to specific signals and is observed especially during development before cells commit to a particular fate (Voigt et al., 2013). The status of enhancers is determined by similar combinations of histone signatures: H3K4me1 is associated with H3K27ac in active enhancers but with H3K27me3 in poised enhancers (Shlyueva et al., 2014). Histone marks localized in gene bodies exert distinct functions according to their precise location. H3K36me2 and me3 are commonly present in the gene bodies of actively transcribed genes, especially toward their 3′ end (Bannister et al., 2005). H3K36me3 at the 5′ end of an exon is a mark of exon expression and correlates with alternative splicing (Hon et al., 2009). All the marks mentioned in this paragraph are localized in the euchromatin of mouse retinal neurons, with di- and trimethylated H3K27 being also present in the facultative heterochromatin (Eberhart et al., 2013) (Fig. 3).
3.2. Histone writers, erasers and readers
We will briefly describe the enzymes that modify or simply recognize the marks mentioned previously (Fig. 4). The enzymes capable of modifying histones fall into two categories: the writers that add post-translational modifications and the erasers that remove them. The proteins that can recognize specific histone modification(s) but do not edit them are named readers. Many of the histone writers, erasers and readers are not exclusive and can also target non-histone proteins, but we will focus only on the histone targets.
3.2.1. Writers and erasers of histone lysine acetylation
Acetylation of lysine residues on histones is catalyzed by histone acetyltransferases (HATs), also called lysine acetyltransferases (KATs). All KATs share a common core binding motif for the acetyl donor but different domains for substrate recognition, catalysis and autoregulation, and thus show varying specificity toward histones (Table 1A and Fig. 4) (Marmorstein and Zhou, 2014). However, this latter characteristic is still poorly described for several of them and challenged by their participation in different complexes (Carrozza et al., 2003).
Table 1.
Histone acetyltransferases (A) and histone deacetylases (B).
| A | ||
|---|---|---|
| Protein | Gene | Substrates |
| HAT4 | NAA60 | |
| KAT1 (HAT1) | HAT1 | H4K5,12 |
| KAT2A (GCN5) | KAT2A | H3K9,14,18,36 |
| KAT2B (PCAF) | KAT2B | H3K9,14,18; H4K8 |
| KAT3A (CBP) | CREBBP | H3K18,23 |
| KAT3B (p300) | EP300 | H3K14,18; H4K5,8 |
| KAT4 (TAFII250) | TAF1 | H3K14 |
| KAT5 (TIP60) | KAT5 | H4K5,8,12,16 |
| KAT6A (MYST-3) | KAT6A | H3K14 |
| KAT6B (MYST-4) | KAT6B | H3K14 |
| KAT7 (MYST-2) | KAT7 | H4K5,8,12 |
| KAT8 | KAT8 | H4K16 |
| KAT9 (ELP3) | ELP3 | H3 |
| KAT12 (TFIIIC90) | GTF3C4 | H3K9,14,18 |
| KAT13A (NCoA-1) | NCOA1 | H3K9,14 |
| KAT13B (NCoA-3) | NCOA3 | H3, H4 |
| KAT13C (NCoA-2) | NCOA2 | H3, H4 |
| KAT13D (CLOCK) | CLOCK | H3, H4 |
| (ATF-2) | ATF2 | H4K5,8,16 |
| B | ||
|---|---|---|
| Class | Gene/Protein | Targets |
| Class I | HDAC1 | Broad spectrum |
| HDAC2 | Broad spectrum | |
| HDAC3 | Unclear | |
| HDAC8 | Broad spectrum | |
|
| ||
| Class IIa | HDAC4 | Broad spectrum |
| HDAC5 | Broad spectrum | |
| HDAC7 | ||
| HDAC9 | ||
|
| ||
| Class IIb | HDAC6 | Broad spectrum |
| HDAC10 | ||
|
| ||
| Class III | SIRT1 | H3K9,14; H4K16 |
| SIRT2 | H4K16 | |
| SIRT3 | H4K16 | |
| SIRT6 | H3K9 | |
| SIRT7 | H3K18 | |
|
| ||
| Class IV | HDAC11 | H3K9,14 |
Targets of histone acetyltransferases (A) and histone deacetylases (B) were compiled from (Allis et al., 2007; Seto and Yoshida, 2014; Zhao and Garcia, 2015). Only targets on histone H3 and H4 Nter tails are listed, though many of these enzymes have additional targets (on histone and/or non-histone proteins). Sirtuins 4 and 5 are mitochondrial and do not have histones among their targets. Histone acetyltransferase names are given according to the nomenclature established in (Allis et al., 2007) with one common name in brackets. Histone deacetylases have matching gene and protein names; gene names match human nomenclature.
Deacetylation of histone lysines is carried out by histone deacetylases (HDACs) and sirtuins (SIRTs) that are distributed into 4 classes: Class I includes HDACs 1, 2, 3 and 8; Class II includes HDACs 4, 5, 9 and 10 (Class IIa) as well as HDACs 6 and 10 (Class IIb); Class III comprises SIRTs 1 to 7 (SIRTs 4 and 5 are exclusively mitochondrial and do not target histones); and Class IV is formed by HDAC11 only (Seto and Yoshida, 2014). SIRTs target well-defined histone residues (Table 1B and Fig. 4), contrary to HDACs whose specificities are still unclear, notably because many of them possess low activity on their own and exert their influence as part of multiprotein complexes. Most HDACs are suggested to cover a broad spectrum of histone lysine residues with varying efficiency, but this remains poorly characterized (Seto and Yoshida, 2014).
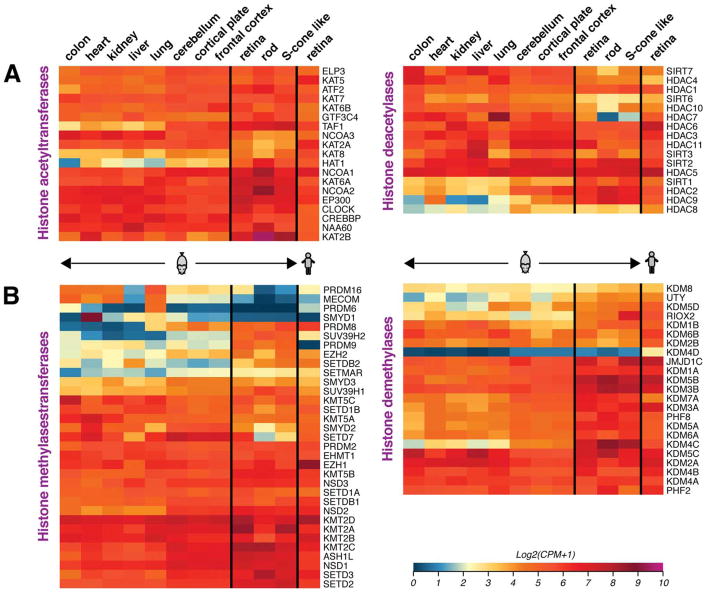
In the human and mouse retina, all KATS are highly expressed, whereas SIRTs and HDACs have more variable expression levels (Fig. 5A). Several of the histone acetyltransferases and deacetylases interact with specific transcription factors as part of co-activator and co-repressor complexes (see Section 4.5 and Fig. 10). SIRT1 is reported to exhibit decreased expression with aging and in age-related diseases (see Section 5), and maintaining its activity seems to protect retina and RPE cells against light or oxidative damage (Mimura et al., 2013). Changes in histone acetylation through HDACs is suggested to be involved in early stages of apoptosis in the retina (Pelzel et al., 2010).
Fig. 5.
Heatmaps showing gene expression of enzymes that catalyze (de)acetylation and (de)methylation of lysine residues in histone H3 and H4 Nter tails. Expression data was extracted from RNA-seq profiles of indicated tissues and cell types, as elaborated in the legend of Fig. 2. While a majority of genes are widely expressed, the retina displays unique patterns of expression. High expression of Setd7, Hdac7 and Prdm16 in the retina, but not in photoreceptors, suggests their potential functions in the inner retina. Scale of expression values is shown as log2 of Counts per million (CPM) +1, with blue representing low and red high expression values.
Fig. 10.
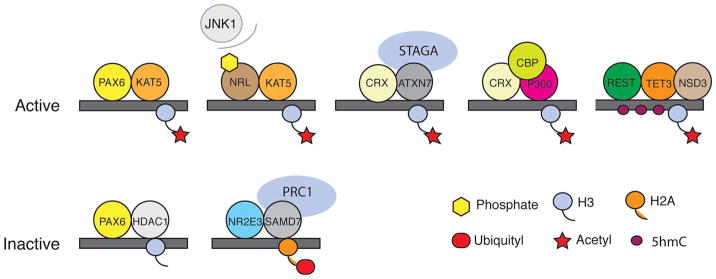
Interaction of transcription factors with epigenetic modifiers in the mammalian retina. Binding of transcription factors to distinct epigenetic modifiers helps in establishing cell-type specific epigenetic signatures and transcriptional profiles. A few examples are given here. PAX6, which is expressed in RPCs and interneurons, can interact with both histone acetyltransferase KAT5 (Kim et al., 2012a) and deacetylase HDAC1 (Kim et al., 2017), respectively regulating activation and repression of its target genes. Similarly, NRL can be phosphorylated by JNK1 and interacts with the histone acetyltransferase KAT5 to drive the expression of its target genes in rod photoreceptors (Kim et al., 2012b). CRX can interact with various histone-modifying complexes (ATXN7 and P300/CBP) that contain histone deacetylases to activate its target genes (Palhan et al., 2005; Peng and Chen, 2007). NR2E3 is shown to interact with SMAD7 to repress gene expression in rod photoreceptors through the PRC1 complex (Omori et al., 2017). REST interacts with TET3 to increase 5hmC in the gene bodies of neuronal-specific genes, and TET3, in turn, recruits the histone methyltransferase NSD3, which methylates H3K36 in those regions (Perera et al., 2015).
3.2.2. Writers and erasers of histone lysine methylation
Methyl moieties are deposited on lysine residues of histone H3 and H4 Nter tails by lysine methyltransferases (KMTs) and removed by lysine demethylases (KDMs). KMTs exert their catalytic activity through a common catalytic SET domain (Black et al., 2012), whereas KDMs segregate into 2 categories. The first group contains only two H3K4 demethylases, LSD1 (KDM1A) and LSD2 (KDM1B), which cannot work on trimethylated residues. All other KDMs form the second category and possess a JmjC (Jumonji C) catalytic domain able to handle trimethylation (Kooistra and Helin, 2012). Targets of all KMTs have not been characterized; this is especially true for the PRDM family (Hyun et al., 2017). KMTs and KDMs generally target a specific histone residue, add/remove a limited number of methyl groups and work sequentially (Table 2 and Fig. 4) (Black et al., 2012; Hyun et al., 2017; Kooistra and Helin, 2012). For instance, many KMTs can add the first two methyl moieties to H3K36 (such as the NSD group), but only SETD2 (KMT3A) catalyzes the addition of the third moiety. Similarly, SET8 (KMT5A) attaches the first methyl group to H4K20, and then SUV4-20H1 (KMT5B) and SUV4-20H2 (KMT5C) add the other two. H3K9 methyltransferases are segregated into constitutive and facultative heterochromatin-specific enzymes: SETDB1 (KMT1E), SUV39H1 (KMT1A) and SUV39H2 (KMT1B) are involved in constitutive heterochromatin methylation in a sequential manner, whereas G9a (KMT1C) and GLP (KMT1D) form heterodimers to suppress gene expression in euchromatin (Hyun et al., 2017). As is the case with modifiers of histone acetylation, KMTs and KDMs are part of multiprotein complexes. For example, the H3K27 methyltransferases EZH1 and EZH2 (KMT6B and 6A, respectively), belong to the polycomb repressive complex 2 (PRC2) and are not active on their own, requiring other PRC2 subunits to bind to histone H3 Nter tail and even to the trimethylated H3K27 for spreading methylation to adjacent targets (Hyun et al., 2017).
Table 2.
Histone methyltransferases (A) and histone demethylases (B) targeting lysine residues 4, 9, 27 and 36 at histone H3, and lysine 20 at histone H4.
| A | ||
|---|---|---|
| Protein | Gene | Products |
| KMT1A (SUV39H1) | SUV39H1 | H3K9me2/3 |
| KMT1B (SUV39H2) | SUV39H2 | H3K9me2/3 |
| KMT1C (G9a, EHMT2) | EHMT2 | H3K9me1/2 |
| KMT1D (GLP, EHMT1) | EHMT1 | H3K9me1/2 |
| KMT1E (SETDB1) | SETDB1 | H3K9me1/2/3 |
| KMT1F (SETDB2) | SETDB2 | H3K9me2/3 |
|
| ||
| KMT2A (MLL1) | KMT2A | H3K4me1/2/3 |
| KMT2B (MLL2) | KMT2B | H3K4me1/2/3 |
| KMT2C (MLL3) | KMT2C | H3K4me1/2/3 |
| KMT2D (MLL4) | KMT2D | H3K4me1/2/3 |
| KMT2F (SETD1A) | SETD1A | H3K4me1/2/3 |
| KMT2G (SETD1B) | SETD1B | H3K4me1/2/3 |
| KMT2H (ASH1L) | ASH1L | H3K4me1/2/3, H3K9me1/2/3, H3K36me2, H4K20me1/2/3 |
|
| ||
| KMT3A (SETD2) | SETD2 | H3K36me3 |
| KMT3B (NSD1) | NSD1 | H3K36me1/2, H4K20me1/2 |
| KMT3C (SMYD2) | SMYD2 | H3K36me1/2, H3K4me1 |
| KMT3D (SMYD1) | SMYD1 | |
| KMT3E (SMYD3) | SMYD3 | H3K4me2/3 |
| KMT3F (NSD3) | NSD3 | H3K4me1/2, H3K27me2/3, H3K36me1/2 |
| KMT3G (NSD2) | NSD2 | H3K4me1/2, H3K27me1/2/3, H3K36me1/2, H4K20me1/2/3 |
|
| ||
| KMT5A (SET8) | KMT5A | H4K20me1 |
| KMT5B (SUV4-20H1) | KMT5B | H4K20me2/3 |
| KMT5C (SUV4-20H2) | KMT5C | H4K20me2/3 |
|
| ||
| KMT6A (EZH2) | EZH2 | H3K27me1/2/3 |
| KMT6B (EZH1) | EZH1 | H3K27me2/3 |
|
| ||
| KMT7 (SETD7) | SETD7 | H3K4me1/2 |
|
| ||
| KMT8A (RIZ1) | PRDM2 | H3K9me1/2/3 |
| KMT8B (PRDM9) | PRDM9 | H3K4me1/2/3 |
| KMT8C (PRDM6) | PRDM6 | |
| KMT8D (PRDM8) | PRDM8 | H3K9me2 |
| KMT8E (PRDM3) | MECOM | H3K9me1 |
| KMT8F (PRDM16) | PRDM16 | H3K9me1 |
|
| ||
| (Setd3) | SETD3 | H3K36me1/2 |
| (SETMAR) | SETMAR | H3K36me2 |
| B | ||
|---|---|---|
| Protein | Gene | Substrates |
| KDM1A (LSD1) | KDM1A | H3K4me1/2, H3K9me1/2 |
| KDM1B (LSD2) | KDM1B | H3K4me1/2, H3K9me1/2 |
|
| ||
| KDM2A (FBXL11) | KDM2A | H3K36me1/2 |
| KDM2B (FBXL10) | KDM2B | H3K36me1/2, H3K4me3 |
|
| ||
| KDM3A (JHDM2A) | KDM3A | H3K9me1/2 |
| KDM3B (JHDM2B) | KDM3B | H3K9me1/2 |
| KDM3C (JHDM2C) | JMJD1C | H3K9me1/2 |
|
| ||
| KDM4A (JMJD2A) | KDM4A | H3K9me2/3, K36me2/3 |
| KDM4B (JMJD2B) | KDM4B | H3K9me2/3, K36me2/3 |
| KDM4C (JMJD2C) | KDM4C | H3K9me2/3, K36me2/3 |
| KDM4D (JMJD2D) | KDM4D | H3K9me2/3, K36me2/3 |
| KDM4E | KDM4E | H3K9me2/3, K36me2/3 |
|
| ||
| KDM5A (JARID1A) | KDM5A | H3K4me2/3 |
| KDM5B (JARID1B) | KDM5B | H3K4me1/2/3 |
| KDM5C (JARID1C) | KDM5C | H3K4me2/3 |
| KDM5D (JARID1D) | KDM5D | H3K4me2/3 |
|
| ||
| KDM6A (UTX) | KDM6A | H3K27me2/3 |
| KDM6B (JMJD3) | KDM6B | H3K27me2/3 |
| KDM6C (UTY) | UTY | H3K27me2/3 |
|
| ||
| KDM7A (JHDM1D) | KDM7A | H3K9me1/2, H3K27me1/2, H4K20me1/2 |
| KDM7B (PHF8) | PHF8 | H3K9me1/2, H3K27me1/2, H4K20me1/2 |
| KDM7C (PHF2) | PHF2 | H3K9me2, H4K20me3 |
|
| ||
| KDM8 (JMJD5) | KDM8 | H3K36me2 |
|
| ||
| (RIOX1, NO66) | RIOX1 | H3K4me1/2/3, H3K36me2/3 |
| (RIOX2, MINA) | RIOX2 | |
Lists of the histone methyltransferases (A) and histone demethylases (B) targeting lysine residues 4, 9, 27 and 36 at histone H3 and lysine 20 at histone H4, compiled from (Allis et al., 2007; Black et al., 2012; Hyun et al., 2017; Kooistra and Helin, 2012; Zhao and Garcia, 2015). Many of these enzymes have additional targets (on histone and/or non-histone proteins). Protein names are given according to the nomenclature established in (Allis et al., 2007) with common names in brackets; gene names match human nomenclature.
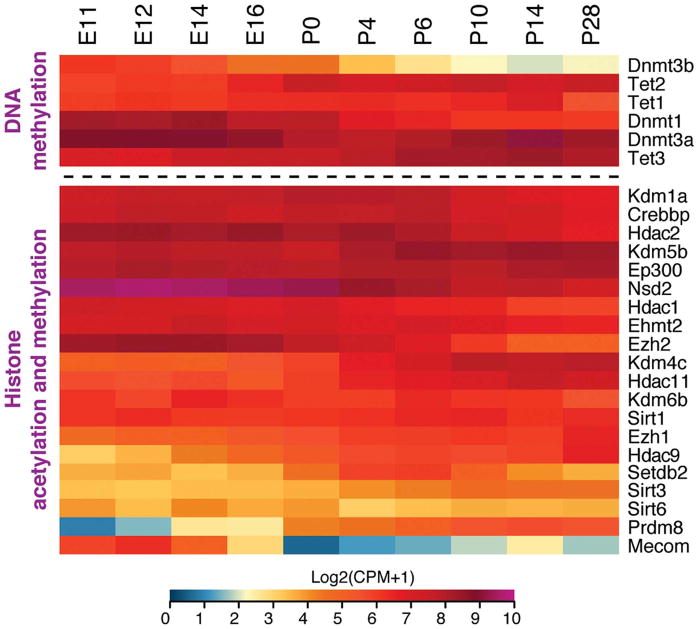
EZH2 plays a central role in retinal development (see Section 4.4), but its expression decreases in the adult retina; however, the expression of EZH1 increases during development (Fig. 9). Setd7 (encoding KMT7) and Prdm16 (encoding KMT8F) are expressed at lower levels in mature mouse photoreceptors compared to the adult retina (Fig. 5B), suggesting their function in non-photoreceptor cells. Notably, Prdm16 (KMT8F) seems to be specifically expressed in the RPE and in a subset of RGCs in the mouse retina (Groman-Lupa et al., 2017).
Fig. 9.
Heatmaps showing gene expression of epigenetic enzymes during mouse retinal development. Gene-level analysis of RNA-seq data during mouse retinogenesis shows variable patterns of expression of epigenetic modifiers involved in DNA methylation (upper panel) and histone modifications (lower panel). This temporal and spatial distribution contributes to the regulation of distinct biological processes and cellular diversity during retinal development. The figure shows only enzymes whose function has been studied in mice or that are differentially expressed during development. The RNA-seq data was obtained from (Hoshino et al., 2017).
3.2.3. Readers of histone lysine acetylation and methylation
Bromodomain is the key structural motif that binds to acetylated lysine residues in histones (Dhalluin et al., 1999). This domain is part of transcription-associated factors (such as TAF1 and BET proteins), chromatin remodelers (e.g., SMARCA4), as well as some HATs (such as KATs 2A, 2B, 3A and 3B), which can act as transcription coactivators (Marmorstein and Zhou, 2014; Sanchez and Zhou, 2009).
The methylated state of lysine residues in histones can be recognized by an array of proteins carrying distinct domains (Fischle, 2012): (1) the superfamily Royal that includes the chromo, chromobarrel, Tudor, MBT and PWWD domains; (2) the zinc-binding domain group including the PHD finger, ADD and CW modules; (3) WD40, ankyrin and HEAT repeat domains; and (4) the BAH domain. The PHD fingers and chromo and PWWD domains primarily favor binding to me2 and me3, whereas the MBT domain and HEAT and ankyrin repeats bind only to me1 and me2. The chromobarrel, Tudor, ADD and CW domains and the WD40 repeat are reportedly less selective.
Many transcriptional regulatory proteins as well as histone modifiers contain one or more reader domains or a combination of domains that recognize specific arrangement of modifications within the same histone tail (cis) or in trans (Ruthenburg et al., 2011), accounting for a complex combinatorial interplay among histone modifications. Interaction among multiple factors within chromatin complexes can impart additional avenues for crosstalk (Torres and Fujimori, 2015; Zhang et al., 2015c).
3.3. Crosstalk between DNA methylation and histone modifications
Histone modifications and DNA methylation are closely intertwined in regulating gene expression (Bintu et al., 2016). DNA methylation influences histone mark deposition, such as H3K27me3 and H3K27ac at promoters and enhancers (King et al., 2016), and some histone modifying enzymes are reported to be sensitive to the methylation status of DNA (Bartke et al., 2010; Blackledge et al., 2010). Conversely, DNMTs can read histone marks (Dhayalan et al., 2010; Ooi et al., 2007; Zhang et al., 2010) and target sites exhibiting specific histone modifications (Choi et al., 2011).
DNMT1, DNMT3a, DNMT3b and DNMT3L can interact with HDAC2 and/or HDAC1 and exert a co-repressive function beyond their methyltransferase activity (Aapola et al., 2002; Fuks et al., 2000; Fuks et al., 2001; Geiman et al., 2004; Rountree et al., 2000). DNMTs can also associate with the H3K9 methyltransferases SUV39H1 and SUV39H2 (KMT1A and KMT1B, respectively) and with HP1α and HP1β to reinforce constitutive heterochromatin condensation (Fuks et al., 2003; Lehnertz et al., 2003). Similarly, DNMT3a and DNMT3b can recruit HP1 and the H3K9 methyltransferase G9a (KMT1C) to allow facultative heterochromatin establishment in ESCs (Epsztejn-Litman et al., 2008). We recommend the following reviews about these crosstalk mechanisms (Cheng, 2014; Du et al., 2015).
4. Epigenetic regulation of retinal development
The epigenetic landscape in the retina is shaped by DNA and histone modifications that together with the limitations imposed by the nuclear architecture allow appropriate levels of gene expression during development. In this section, we will highlight perturbation assays and genomewide studies of the epigenome that have provided insights into retinal development. We have summarized common methodologies in Box 1 and associated analytical tools in Boxes 2 and 3. Changes in the epigenome appear to be conserved in the developing retina of rodents and humans, making translation to human disease possible. Additionally, combined analysis of Hi-C data from mouse cortex (Dixon et al., 2012), to map topologically associated domains, with mouse retina epigenomic data sets has uncovered previously-unrecognized conserved enhancer and super-enhancer regions (Aldiri et al., 2017). Epigenomic studies could also be used for designing therapies since sequence variants associated with common multifactorial diseases are often located in non-coding and distal gene regulatory regions (Schaub et al., 2012).
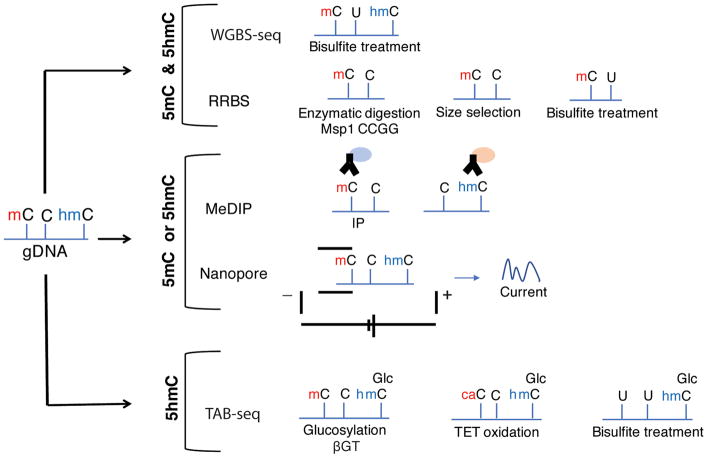
BOX 1. Investigating DNA and histone modifications – emerging technologies.
DNA methylation
DNA methylation can be determined globally or in targeted genomic regions using a variety of methods, with BS-seq being the most widely-used protocol (Fig. 6). Affinity-based 5mC pull-down (Mohn et al., 2009) and restriction enzymes (Brunner et al., 2009) are often used in combination with BS-seq, PCR (polymerase chain reaction) or microarrays. Reduced representation bisulfite sequencing (RRBS) that is based on size selection of restriction fragments and enriches for regions of high CG content (Meissner et al., 2005), as well as DNA microarrays that cover most RefSeq genes and CpGIs (Horvath, 2013), have been used widely to map a subset of the genome with high coverage and low cost. In addition, different assays have been developed to study 5hmC using specific antibodies or enzymatic modifications (Olkhov-Mitsel and Bapat, 2012). New sequencing technologies, such as Single-Molecule Real-Time (SMRT) (Lluch-Senar et al., 2013) or Mycobacterium smegmatis porin A (Msp)-based nanopore sequencing (Laszlo et al., 2013), can directly read modified bases and even map other modifications such as 5hmC (Laszlo et al., 2013).
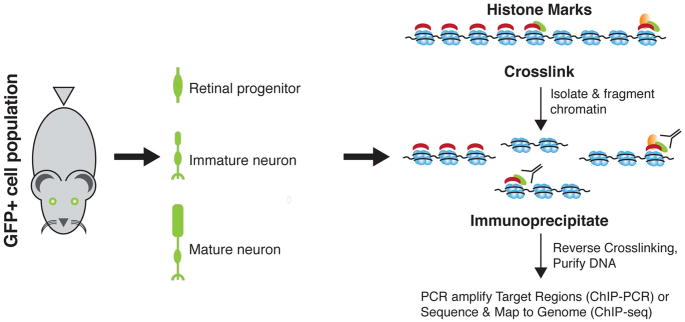
Histone modifications
Identification of histone modifications, or their regulatory machinery, can be achieved genome-wide or at specific loci by combining immunoprecipitation using antibodies with next-generation sequencing (ChIP-seq) or with PCR (Fig. 7) (Chaitankar et al., 2016; Park, 2009). Several variations of these methods have been developed to lower the input material and/or increase resolution; these include the addition of an exonuclease to remove DNA sequences not involved in interaction with target proteins (Rhee and Pugh, 2011), the use of native chromatin to decrease artefactual interactions resulting from crosslinking (Orsi et al., 2015), and in situ DNA cleavage to obtain information on long-range chromatin contacts without chromatin solubilization and fixation (Skene and Henikoff, 2017).
Epigenetic modifications are context- and cell type- dependent. Within a specific tissue (such as retina), distinct cell types or even individual cells can possess unique patterns of DNA methylation and histone modifications to sustain functional homeostasis at a given time and/or spatial location. While most of the epigenetic studies pertaining to the retina have been performed using whole tissue (Aldiri et al., 2017; Perera et al., 2015; Popova et al., 2012; Rapaport et al., 2004), the significance of investigating the epigenome of specific retinal cells is increasingly being recognized by using purified cells (Aldiri et al., 2017; Kim et al., 2016b; Mo et al., 2016).
Single-cell studies
Mapping of DNA methylation has been performed at the single cell resolution using bisulfite converted DNA (Farlik et al., 2015; Guo et al., 2015a; Smallwood et al., 2014), a fluorescent reporter system (Stelzer et al., 2015), or restriction enzymes coupled with single-cell PCR (Lorthongpanich et al., 2013); however, these studies did not achieve full coverage of the genome. Chromatin accessible sites have been identified with single cell resolution by integrating the assay for transposase accessible chromatin followed by high-throughput sequencing (ATAC-seq) with combinatorial cellular indexing (Cusanovich et al., 2015) or microfluidic systems (Buenrostro et al., 2015). Importantly, three-dimensional chromosomal architecture studies at the single-cell level have revealed that chromosome territories are highly dynamic yet preserve some local and global organizational features (Nagano et al., 2013). Single cell techniques still require a survey of larger number of cells due to cell-to-cell variations and probability of false-positives from low-sequencing depth or enrichment level (Schwartzman and Tanay, 2015). New sequencing platforms capable of directly reading modified nucleotides (Laszlo et al., 2013) are being developed and represent promising tools to survey the epigenome at the single-cell level.
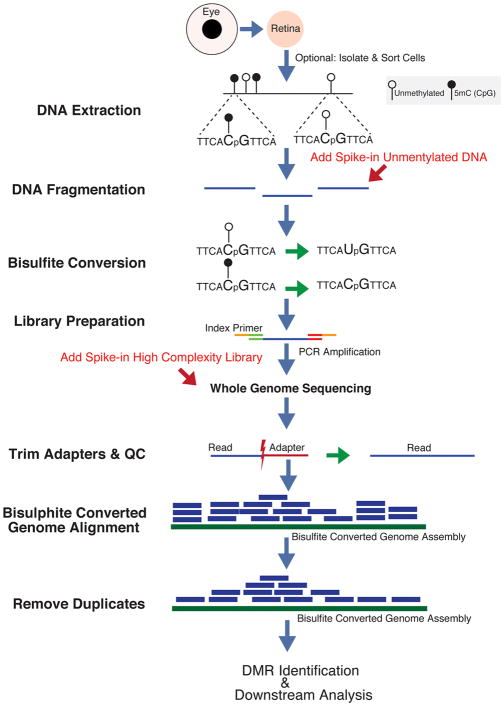
BOX 2. Computational analysis of whole genome bisulfite sequencing data.
Primary Processing
Quality control of raw sequencing reads
A workflow for whole-genome bisulfite sequencing is presented in Fig. 8. Raw sequencing data obtained from a next-generation sequencer are initially pruned for low-quality bases and adapter contamination, which can otherwise lead to inference of false C-T conversions. Removing duplicates for data sets originating from larger genomes is highly recommended; however, deduplication of the data should be ignored for RRBS or array-based target enrichment methods (Bock, 2012). FastQC is currently the standard software that performs a quality check on raw sequencing reads, and TrimGalore is the benchmark tool for clipping adapters.
Alignment
After performing quality control of sequencing reads, the next step is to align these reads to a reference genome. Unlike alignment to the standard reference genome as in other next-generation sequencing (NGS) methods, many BS-seq specific aligners (MethylCoder (Pedersen et al., 2011), BS-Seeker2 (Guo et al., 2013), BRAT-BW (Harris et al., 2012), Bismark (Krueger and Andrews, 2011) and Bison (Ryan and Ehninger, 2014)) generate a transformed reference genome by computationally converting C to T. An alternative strategy used by a second class of aligners (BSMAP (Lee et al., 2015), LAST (Frith et al., 2012)) considers T nucleotides as C and allows certain level of C-T mismatches. Though several popular BS-seq aligners exist, there is no gold-standard aligner at this stage. Comparative studies have demonstrated a comparable performance of different BS-seq aligners (Chatterjee et al., 2012).
Additional quality check considerations
Incomplete conversions that result when an unmethylated C nucleotide is not converted to T can lead to false methylation calls. A generalized approach to measure the conversion rate is to add spike-in sequences with unmethylated C nucleotides (Fig. 8). An important post-alignment consideration is to restrict any further downstream analysis to unique regions of the reference genome. A read may uniquely map to a repetitive region of the genome since most aligners allow a certain number of mismatches to handle genetic variations or sequencing errors. In BS-seq data sets, the conversion of C nucleotides to T can almost double the number of repetitive regions (from 18.7% to 33.5%) in the human reference genome (Karimzadeh, 2016), thereby creating computational challenges and resulting in biases that could be hard to interpret and investigate (Treangen and Salzberg, 2012). Several tools and programs exist for identifying uniquely mappable regions on the reference genome (Derrien et al., 2012; Storvall et al., 2013). As of July 2017, only software that evaluates unique mappability of the methylome (i.e., bisulfite-converted genome) is Bismap (http://bismap.hoffmanlab.org) (Karimzadeh, 2016).
Identification of differentially methylated regions
Downstream analysis of BS-seq targets identifies the conversion of unmethylated C to T, referred as Methylation Calling. Global profiling of methylation calls is followed by inferring differentially methylated regions (DMRs), which can correspond to a single base or an entire gene locus (Bock, 2012). For BS-seq data, DMR identification is performed on methylation calls as opposed to alignment data. Standard statistical methods, such as t-test or chi-square test, compare methylation calls for each C between the two groups. More advanced approaches for defining statistically significant DMRs include mixed model (Wang, 2011), Shannon entropy (Zhang et al., 2011), feature selection (Zhuang et al., 2012), and statistical correction for copy-number aberrations (Huang et al., 2014; Robinson et al., 2012). Given that the number of methylation calls in DMR analysis can span several million bases, multiple hypothesis correction should be performed by controlling false discovery rate (FDR). Complementary statistical approaches can be implemented for increased power in case of weak differences in methylation calls (Bock, 2012; Pirrotta, 2016).
BOX 3. Computational analysis of ChIP-seq data.
Genome Alignment
After a fastqc analysis and a trimming step similar to the BS-seq primary analysis procedure (see section 4.2.1), mapping of the sequence reads to the reference genome is the first critical step in ChIP-seq analysis. BWA (Li and Durbin, 2010) and Bowtie (Langmead, 2010; Meissner et al., 2005) are two widely-used open source aligners. BWA includes three different algorithms for alignment, and one of these algorithms is chosen based on sequencing-read-lengths. BWA-Backtrack algorithm is preferred over BWA-MEM for < 70 bp reads. Before downstream analysis, we recommend that only reads uniquely aligned to the genome are selected and duplicate reads are purged to mitigate PCR amplification bias.
Peak identification
Peak calling refers to identification of genomic regions that are significantly enriched for aligned reads (i.e., histone or TF ChIP-seq signal) compared to the control (e.g., ChIP-seq with IgG). For single-end sequencing experiments, a bimodal distribution of aligned reads is observed on positive and negative strands around the true binding site (Shin et al., 2013). For this reason, many peak identification algorithms, such as MACS2 (Zhang et al., 2008), extend the reads on both strands by predicted fragment length prior to peak estimation. Statistical significance (p-value) and peak quality metrics (FDR), estimated by peak-calling algorithms, help users identify the high confidence peaks. An alternative to FDR is the fold-change metric that is defined as the ratio of aligned reads in peak regions between sample and control experiments.
Quality control and replicate concordance
A ratio of > 0.5 for uniquely-mapped reads over the total number of reads is an indicator of successful library preparation. In general, the number of duplicate reads that can potentially arise due to PCR amplification bias must not be greater than the number of uniquely mapped reads. Five percent FDR with a fold-change of 5 compared to the control, and a majority of peaks showing fold-change ≥ 20, are considered good cut-offs for high-quality ChIP-seq enrichment regions. At least 50% overlap of ChIP-seq peaks in two independent biological replicates is a good measure of concordance. A more robust measure for replicate concordance is the irreproducible discovery rate (IDR) (Landt et al., 2012) that uses a specific number of overlapping regions across consistent peaks and correlation between peak ranks based on their quality. If the replicate concordance test is successful, the reads originating from replicates must be merged and divided in random subsets to produce pseudo-replicates. Then, common peaks originating from pseudo-replicates should be used for further analysis.
Chromatin-state discovery and characterization
A single chromatin mark seldom plays a functional role in isolation, and a combinatorial interplay between distinct histone marks (histone code) eventually defines the coding and/or non-coding genomic regions. Genome Segmentation based on the histone code can detect regulatory regions and elucidate molecular events that can contribute to decoding complex biological processes, such as cell fate specification. An unsupervised approach to identify combinatorial marks is to segregate the genome based on gene annotation and check for co-occurrence of marks on these regions. Using every gene as an anchor, five regions including gene body, 5′ untranslated region (UTR), 3′ UTR, 1 kb upstream (promoter), and 1 kb downstream were chosen to reconstruct regulatory network of Drosophila melanogaster (Marbach et al., 2012). Another efficient, and probably more accurate machine-learning-based software to partition the genome is chromatin hidden Markov modeling (ChromHMM) (Ernst and Kellis, 2012). This tool implements a multivariate hidden Markov model-based approach to identify de novo combinatorial interactions between histone marks. Unlike gene annotation-based segmentation, ChromHMM by default analyzes the chromatin marks across 200 bp intervals that roughly approximate to nucleosome size. Epigenetic states were annotated during retinogenesis for human and mouse genomes using an 11-state model (Aldiri et al., 2017).
4.1. Chromatin organization and gene expression- insights from mouse photoreceptors
A common pattern of chromatin organization in the nuclei of most eukaryotic cells, including retinal neurons, reveals central euchromatin and peripheral clusters of heterochromatin (Eberhart et al., 2013; Solovei et al., 2009) (Fig. 3). However, rod photoreceptors are characterized by a unique pattern of chromatin organization acquired gradually during their differentiation (between P0 and P28) and characterized by distinct heterochromatin domains that relax progressively, generating concentric patterns with euchromatin localized at the outermost exterior ring. This inverted pattern is commonly observed in rods of nocturnal mammals and proposed to facilitate low-light vision (Solovei et al., 2009). Upregulation of the histone variant H1c contributes to the high chromatin compaction and inverted pattern in rods, and the loss of Hist1h1c, Hist1h1e and H1f0 (h1c/H1e/H10) results in larger nuclei and a reduction of heterochromatin (Kizilyaprak et al., 2011; Popova et al., 2013). Thus, rod photoreceptors in rodents can unravel novel insights into the relationship between chromatin architecture, chromatin states and gene expression.
The distribution of most histone modifications in rods is similar to other cells (Fig. 3 and Section 3). One key distinction is that the H3K9me2 mark is not present in the constitutive heterochromatin compartment in rods as it is in other cell types (Eberhart et al., 2013) (Fig. 3). RNA Polymerase II has been observed only in the highly acetylated region of the euchromatin that is close to the facultative heterochromatin interface (Kizilyaprak et al., 2010) (Fig. 3). Notably, the subnuclear localization of repressed genes is not always correlated with epigenetic states that are predicted from combinations of chromatin modifications using the chromHMM model (see Box 3) (Aldiri et al., 2017). However, this gene localization appears consistent among different rod cells, indicating a tight regulation of 3D chromatin organization within their nucleus (Aldiri et al., 2017).
ATAC-seq (assay for transposase accessible chromatin followed by high-throughput sequencing) analysis, a method to map open chromatin loci, has demonstrated that chromatin is more condensed in rods than in cones (Hughes et al., 2017), probably through the rod differentiation factor NRL (Mears et al., 2001) or one of its targets [such as NR2E3 (Oh et al., 2008) or the histone deacetylase KDM5B (Hao et al., 2012)]. Indeed, the methylome of rd7 mouse retina (having loss of NR2E3 function) represents an intermediate state between rods and cones (Mo et al., 2016), consistent with gene expression status (Cheng et al., 2011; Corbo and Cepko, 2005). Interestingly, rods appear to have a larger fraction of hypomethylated regions that do not overlap ATAC-seq peaks compared to other cell types, and that could represent vestigial enhancers (putative enhancers in fetal neural tissue). Hypomethylation in these regions is proposed to be the result of limited access of DNMTs to the highly compacted heterochromatin domains of rods (Mo et al., 2016).
Importantly, analysis of open chromatin has also provided insights into photoreceptor-specific DNA regulatory regions. Based on the ATAC-seq peaks, rod-specific gene expression may be regulated by proximal regulatory sequences compared to cones where distal enhancers may play a larger role in gene regulation (Hughes et al., 2017; Mo et al., 2016). Rods and cones share accessible chromatin regions around photoreceptor genes (Mo et al., 2016), consistent with a similar ontogeny of the two photoreceptors (Kim et al., 2016b; Ng et al., 2011; Swaroop et al., 2010). However, rods and cones ATAC-seq peaks seem to be enriched for different sets of TF motifs. For example, CTCF motifs seem to be present in enhancers enriched for general TFs but are absent in regions with photoreceptor-specific motifs (Hughes et al., 2017). Moreover, cone open chromatin is enriched for Q50, bHLH, paired NR and bZIP motifs (Hughes et al., 2017), and a de novo motif with unknown biological significance was identified in rod open chromatin (Mo et al., 2016). Further studies on the 3D architecture of photoreceptors, in addition to combinatorial codes of transcription factors and epigenetic modifiers, will provide insights into the basic principles that regulate the interplay of chromatin structure and gene expression in these cell types.
4.2. DNA methylation and retinogenesis
High expression of DNA methylation writers, erasers and readers in the mammalian retina (see Fig. 2) suggests an important role of DNA methylation(s) in regulating gene expression relevant to neuronal diversity. In the brain, the expression of DNMTs is cell-type or developmental-stage specific. For example, Dnmt3a is expressed in neural stem cells of the sub-ventricular zone (SVZ) as well as in differentiated neurons throughout life whereas Dnmt3b is expressed in the SVZ only until E15.5 (Feng et al., 2005). Mutations in Dnmt1, 3a and/or 3b cause developmental abnormalities and embryonic/postnatal death (Okano et al., 1999). Selective loss-of-function of these enzymes in the neural tissue has unraveled their cell-type and gene-specific functions during neurogenesis. Dnmt1−/− mice exhibit aberrant glial differentiation because of increased JAK-STAT signaling (Fan et al., 2005). Mice selectively deficient in Dnmt3a in neural precursors show defects in the neuromuscular system with reduction in motor neurons and abnormalities in neuromuscular junctions (Nguyen et al., 2007). DNMT3B-knockdown in human ESCs results in precocious neuronal differentiation and maturation, concurrent with the reduction of H3K27me3 and of the H3K27 methyltransferase EZH2 on neuronal promoters (Martins-Taylor et al., 2012).
In the retina, Dnmt1, 3a and 3b are expressed in RPCs and their expression persists in differentiated cell types to a varying degree (Nasonkin et al., 2011) (Figs. 2 and 9). The expression of Dnmt1 becomes restricted to differentiated cells at E15.5, whereas Dnmt3a and b exhibit a dynamic pattern with high expression in early developing retina, low expression at E15.5, and again high expression in early-born neurons (Nasonkin et al., 2011). However, RNA-seq analysis of postnatally-born rods shows continued expression of Dnmt3a (Kim et al., 2016b). Mutation analysis of Dnmts has revealed a crucial role of methylation in photoreceptor and RPE maturation (Table 3). Conditional knockout of Dnmt1 in the mouse retina using a Chx10-Cre line resulted in cell-cycle defects in RPCs and aberrant photoreceptor differentiation without altering cell fates (Rhee et al., 2012). Similarly, Dnmt1-conditional knockout in the retina generated by using the Rx-Cre line showed correct specification of all cell types, with the exception of S-cones, and defects in photoreceptor outer segment morphogenesis (Nasonkin et al., 2013). This latter phenotype was the consequence of abnormalities in RPE, which exhibited improper morphology and polarization (Nasonkin et al., 2013). A hypomorph model of Dnmt1/3a/3b triple-mutations supports an important role of methylation in photoreceptor maturation and function with aberrant expression of synaptic and phototransduction genes (Singh et al., 2017). In the zebrafish, dnmt3 knockdown resulted in defects in the brain and retinal development but not in other tissues, and dnmt1 overexpression could not rescue dnmt3 defects (Rai et al., 2010). In addition, dnmt3a cooperated with the H3K9 methyltransferase G9a in this model in controlling the expression of lef1, a regulator of neurogenesis. Notably, Dnmt2 has not been studied in the mammalian retina, but it is reported to methylate tRNA in the cytoplasm to regulate retinal development in the zebrafish (Rai et al., 2007).
Table 3.
Phenotypes associated with loss of function of epigenome modifier genes in mice.
| Gene | Type | Target | Phenotype | Reference |
|---|---|---|---|---|
| Dnmt1 | Rx-Cre | RPC | Thinner retinal layers / Absence of outer segments Lack of S-cones / Aberrant morphology and polarity of RPE |
(Nasonkin et al., 2013) |
| Dnmt1 | Chx10-Cre | RPC | RPC cell-cycle defects / Degeneration Aberrant photoreceptor differentiation |
(Rhee et al., 2012) |
| Dnmt1/3a/3b | Rx-Cre | RPC | Global hypomethylation / Disorganization of synaptic layer Aberrant photoreceptor differentiation/Lack of S-cones |
(Singh et al., 2017) |
| Ep300/Crebbp | Opsin-Cre | PR | Disrupted PR cell morphology and function Loss of chromatin structure |
(Hennig et al., 2013) |
| Sirt1 | ESCs | Germline | Thin retinas with rosette-like structures | (Cheng et al., 2003) |
| Sirt6 | ESCs | Germline | Disrupted ERG / Dysregulation of metabolic genes | (Silberman et al., 2014) |
| Kdm5b (Jarid1b) | shRNA | P0 retina | Aberrant rod morphology / Dysregulation of gene expression | (Hao et al., 2012) |
| Kdm6b (Jmjd3) | shRNA | E17 retina | Failed differentiation of rod bipolar cells | (Iida et al., 2014) |
| Bmi1 | ESCs | Germline | Decreased proliferation of peripheral RPCs | (Chatoo et al., 2010) |
| Bmi1 | ESCs | Germline | Degeneration of cone bipolar cells and cone photoreceptors | (Barabino et al., 2016) |
| Samd7 | ESCs | Germline | De-repression of non-rod genes in rod photoreceptors | (Omori et al., 2017) |
| Ehmt2 (G9a) | Dkk-Cre | RPC | Loss of photoreceptor / Rosette formation Persistent proliferation |
(Katoh et al., 2012) |
| Ehmt2 (G9a) | Crx-Cre | PR | No overt phenotype | (Katoh et al., 2012) |
| Ezh2 | Chx10-Cre | RPC | Postnatal photoreceptor degeneration Dysregulation of photoreceptor genes |
(Yan et al., 2016) |
| Ezh2 | Math5-Cre | RGC | No overt phenotype | (Yan et al., 2016) |
| Ezh2 | Pax6-Cre or Six-Cre | RPC | Reduction of postnatal precursor proliferation Aberrant lamination Increased differentiation of late-born neurons Upregulation of non-retinal genes |
(Zhang et al., 2015a) |
| Ezh2 | Dkk-Cre | RPC | Microphthalmia / Impaired postnatal proliferation Increased proportion of ON-bipolar cells Accelerated differentiation of rods and Müller glia |
(Iida et al., 2015) |
| Ezh2 | Dkk-Cre | RPC | Upregulation of rod-specific genes in rods | (Ueno et al., 2016) |
| Ezh2 | Rx-Cre | RPC | Increased differentiation of Müller glia | (Fujimura et al., 2018) |
| Eed | Rx-Cre | RPC | Reduced RPC proliferation / Increased proportion of amacrine cells / Postnatal cell-death Aberrant bipolar | (Fujimura et al., 2018) |
| Prdm8 | ESCs | Germline | cell differentiation and survival | (Jung et al., 2015) |
ESC, embryonic stem cell; PR, Photoreceptor; RGC, retinal ganglion cell; RPC, retinal progenitor cell.
The relationship between DNA methylation and gene expression has been explored by examining the methylation status of the promoters of selected genes. In this scenario, photoreceptor-specific genes exhibit cell-type specific DNA methylation patterns, and DNA methylation in the promoter generally anti-correlates with gene repression. For example, Rho and other photoreceptor-specific genes are unmethylated around the TSS in rod photoreceptors but methylated in other cell-types (Merbs et al., 2012). Similar negative correlations have been observed in a genomewide study that compared global transcriptome with patterns of DNA methylation in developing rod photoreceptors (Kim et al., 2016a). Moreover, an identical negative correlation is detected in other cell types, such as in RGCs at the promoter of EphA5, an ephrin receptor (Petkova et al., 2011). More importantly, methylation appears to directly regulate gene expression and is not merely a correlative mark. As an example, changes in DNA methylation preceded the changes in Irbp gene activation during retinal development (Liou et al., 1994), and exogenous methylation of CpG sites in the Irbp promoter diminished protein binding to DNA and promoter activity (Boatright et al., 2000). In concordance, genomewide analysis of DNA methylation in purified rods and cones showed that the level of methylation around the TSS and the gene body were lower in cell type-specific genes. Rods purified from the rd7 mouse (loss of Nr2e3 function) possessed methylation in cone-specific genes with the exception of NR2E3-regulated genes (Mo et al., 2016). Similarly, MBD2b/MBD3L1 enrichment for methylated DNA in the mouse retina suggested the same anti-correlation of DNA methylation with gene expression (Oliver et al., 2013b).
However, a genomewide analysis of DNA methylation using the whole retina has revealed low correlation of methylation and transcriptional changes as retinogenesis proceeded (Aldiri et al., 2017). These findings could be partially explained by signals emerging from multiple retinal populations, which might lead to an inaccurate estimation of the epigenetic changes related to cell-type-specific gene expression. In fact, some of the genes harboring repressive marks appeared to be upregulated in the whole retina but were downregulated when studied in a purified rod population (Aldiri et al., 2017). Furthermore, as the analysis of DNA methylation in this study did not take into account the genomic context, the average methylation levels might have obscured specific correlations of gene expression with methylation at distinct locations on the genome.
4.2.1. Multiple roles of DNA methylation in the retina
Diverse roles of DNA methylation have been suggested in the brain and the retina depending on the context (e.g., CpG vs CpH, genomic location) and the degree of oxidation of the methyl group (see Section 2 and Fig. 1). Brain neurons utilize methylation extensively as a dynamic mark that changes upon neuronal activation (Guo et al., 2011a). For example, hydroxymethylation increases during neuronal differentiation and is accompanied by the loss of H3K27me3 in neural-specific genes (Hahn et al., 2013). Moreover, methylation in the CpH context appears to be relevant for the maturation of neurons because of its accumulation during synapse formation (Lister et al., 2013). The role of methylation in the CpH context remains to be established during neuronal maturation in the retina; however, previous genomewide studies have reported about 0.1% of DNA methylation in the CpH context in adult rods (Mo et al., 2016). Notably, the rd7 retina exhibits an intermediate level of CpH methylation between rods and cones; given that Nr2e3 is expressed after terminal mitosis (Oh et al., 2008), mCpH is likely increased post-mitotically and may be important for cell maturation as it is in the brain (Mo et al., 2016). Furthermore, mass spectrometry analysis has revealed the presence of 5hmC and its accumulation during terminal maturation of the rodent retina (Perera et al., 2015). An antibody against 5hmC identified over 5,000 intragenic regions, which were enriched for neuronal genes. In this study, TET3, the highest expressed Tet gene in the developing retina, was reported to interact with the transcriptional repressor REST, which directed TET3 to its specific target genes. Importantly, 5hmC co-localized with H3K36me3, a mark of actively transcribed genes, suggesting that 5hmC induces chromatin remodeling of its targets. Overexpression of TET3 resulted in enhanced expression of neuronal genes and deposition of H3K36me3 in their gene bodies (Perera et al., 2015). In concordance, 5hmC has been detected mainly in regions of euchromatin in mouse rods (Singh et al., 2017). Taken together, these studies show that DNA methylation has diverse roles in regulating gene expression in the retina and that the type of methylation and the context should be taken into account when correlating DNA methylation and transcription.
By comparing the pattern of DNA methylation in brain and retina, many tissue-specific differentially methylated regions (T-DMR) (see Box 2) have been detected within exons and introns. Interestingly, a subset (14%) of alternatively-spliced genes are associated with T-DMRs in gene bodies (Wan et al., 2013). A correlation of genomewide expression levels with 2498 T-DMRs, obtained by comparing brain and retina, has revealed that, as predicted, a majority of promoter T-DMRs correlate negatively with gene expression (Wan et al., 2015). Interestingly, a substantial number of genes also show a positive correlation, suggesting that DNA methylation at the promoters has divergent functions that may include augmented transcription in specific contexts (Wan et al., 2015). Notably, CRX (cone-rod homeobox), an important transcriptional regulator of photoreceptor development, is shown to bind to a methylated DNA motif (CGTAATTAGGAAGGTAAATC) that is different from its canonical motif, in a microarray assay (Hu et al., 2013). Correlation with ChIP-seq peaks and further confirmation of its binding to methylated regions would be required to establish whether methylation increases the repertoire of CRX binding sites and/or if it provides CRX with “pioneer” properties that enable chromatin accessibility.
4.3. Histone modifications and retinogenesis
Human and mouse retina express most of the genes encoding the histone-modifying enzymes that have been described so far (Fig. 5). Importantly, some of them (e.g., Setd7, Hdac7 and Prdm16) demonstrate high expression in the adult mouse retina but show minimal or no expression in mature photoreceptors (Fig. 5), underscoring their importance in cell-type specific functions of the inner retina. Temporal and spatial expression of enzymes during development allows implementation of regulatory programs essential for cell-type specification and maturation (Fig. 9). Interestingly, genes encoding histone methyltransferases such as Ezh1, Ezh2, Mecom and Prdm8 have dynamic expression patterns through development, suggesting their importance in retinogenesis; a fact that is supported by loss of function studies of Ezh2 and Prdm8 in mice (see Section 4.3.1).
During cell-type specification, repression of pluripotent genes is generally associated with the deposition of H3K9me3, H3K27me3 and DNA methylation on their promoters, whereas lineage-specific genes usually harbor bivalent marks (e.g., both H3K27me3 and H3K4me3) that become monovalent after induction of differentiation (Mikkelsen et al., 2007). Active genes then acquire the H3K4me3 mark at their promoters and H3K4me1 and H3K27ac marks at their enhancers. In contrast, silenced genes associate with H3K27me3 and exhibit DNA methylation at their promoter regions. Many studies in the mammalian retina corroborate this signature, whereas a few suggest alternative patterns.
4.3.1. Histone methylation during retinogenesis
Several groups have explored the relationship between transcription of specific genes during retinal development and corresponding deposition of H3K4me2/3 (associated with activation) and H3K27me3 (associated with repression). For example, transcriptional activation of the Ath5 gene, encoding a key TF of the bHLH family, during retinal development has been correlated with H3K4me2 deposition at its promoter (Skowronska-Krawczyk et al., 2004). Similarly, the expression levels of Sox4 and Sox11 are positively correlated with H3 acetylation and H3K4me3 and negatively correlated with H3K27me3 (Usui et al., 2013). The analysis of Hes1 promoter revealed progressive acquisition of the H3K27me3 repressive mark in all cell types during retinal development, with the exception of Müller glia where Hes1 is normally expressed. Accordingly, Hes1 was upregulated in the H3K27 methyltransferase Ezh2-knockout retina (Ueno et al., 2017). Purified developing rods also revealed a progressive loss of H3K4me3 and accumulation of H3K27me3 at the promoter of Opn1sw (Kim et al., 2016b).
Genomewide studies of histone modifications using whole retina have demonstrated low correlation between gene expression and epigenetic marks. Genes with similar expression level harbored different patterns of H3K4me2 and H3K27me3; however, some groups of genes showed consistent signatures (Popova et al., 2012). For example, many genes that did not harbor H3K27me3 at any time during retinal development were never expressed in the retina, and rod-specific genes that correspond to visual perception shared a similar histone signature with lack of H3K27me3 during development and de novo accumulation of H3K4me2 at the TSS and in the gene body (Popova et al., 2012). In concordance, Nrl and Rho promoters never showed H3K27me3 marks and their expression was correlated to acquisition of the H3K4me3 signature (Kim et al., 2016b). In another study, some downregulated genes (e.g., Uhrf1) never acquired histone repressive marks, whereas others (e.g., Ascl1) gained such modifications during retinal development (Aldiri et al., 2017). A few of the genes showing upregulation during development, such as Stx3, were initially poised and then lost their repressive marks before activation of transcription. Interestingly, derepression of cell-type-specific genes was more prominent than gain of repression-related marks in progenitor genes. This is in agreement with the lack of H3K27me3 observed in non-retinal genes (Popova et al., 2012) and lower expression of Ezh2 in rod photoreceptors (Iida et al., 2015) (Fig. 5B). Other chromatin modifications may therefore be necessary to stabilize gene repression in the mature retina.
Perturbation studies are key to identifying the specific role of each epigenetic modification during development (Table 3). Targeted knockout of the H3K9 methyltransferase-encoding gene Ehmt2 (G9a) severely affected retinal development, with loss of photoreceptor cells, rosette formation, and persistent cell proliferation (Katoh et al., 2012). G9a (KMT1C) appears to function in RPCs as its loss in photoreceptors using Crx promoter did not reveal an overt phenotype (Katoh et al., 2012). The histone methyltransferase PRDM8 (KMT8D) forms a complex with bHLHe22 (bHLHb5) to direct the assembly of neural circuits (Ross et al., 2012) and is necessary for the survival of rod bipolar and certain OFF-cone bipolar cells (Jung et al., 2015). However, the role of PRDM8 in histone modifications was not investigated in these studies and remains poorly understood. Similarly, loss of Kdm6b (encoding the H3K27 demethylase JMJD3) resulted in a cell type-specific phenotype, with reduction of Bhlhb4 expression and consequently impaired differentiation of rod bipolar cells (Iida et al., 2014). Knockdown of Kdm5b, a histone H3K4 demethylase that is highly expressed in rods (Fig. 5B) and regulated by NRL, impaired rod homeostasis (Hao et al., 2012). Pharmacological inhibition of the histone demethylase LSD1 (KDM1A) resulted in aberrant expression of progenitor genes and increased levels of H3K4me2 in their promoters and gene bodies, preventing rod development (Popova et al., 2016).
4.3.2. Histone acetylation during retinogenesis
An appropriate balance of histone acetylation is crucial for cell survival and regulation of RPC proliferation and differentiation. Treatment with Trichostatin A (TSA), a general inhibitor of HDACs, resulted in augmented transcription of Apaf-1 and Casp3 and apoptosis in P5 and P15 explant retina but not in mature retina (P60) (Wallace et al., 2006). Addition of TSA to newborn mouse explant cultures resulted in downregulation of genes associated with cell cycle regulation and rod development, thereby affecting RPC proliferation and rod differentiation (Chen and Cepko, 2007). Remarkably, the proportion of different cell types was not equally affected; these cultures had increased number of bipolar cells and were devoid of photoreceptors and Müller glia. Similarly, pharmacological inhibition of HDAC1 in mouse explant retina and concomitant increase in H3K9ac and H4K12ac, but not H3K27ac, prevented rod differentiation and maintained the expression of progenitor genes, such as Hes1 and Vsx2 (Ferreira et al., 2017). In concordance, the loss of HDAC1 in zebrafish resulted in continued RPC proliferation through the activation of Wnt and Notch/Hes pathways (Yamaguchi et al., 2005).
A challenge when studying histone-modifying enzymes is that some of their effects may be mediated through histone-independent mechanisms. For example, Hdac4 knockdown in the mouse retina resulted in an excess of bipolar cells, likely due to impaired cell death pathways, and HDAC4 overexpression reduced natural cell death and rescued degeneration in rd1 mutant mice; however, these effects of HDAC4 are mediated through its role in the cytoplasm and not on histone modifications (Chen and Cepko, 2009). HDAC4 overexpression and increased stability is reported to preserve photoreceptors and restore visual function in a model of retinitis pigmentosa (Guo et al., 2015b). HDAC4 interacts with MEF2 factors, which target it to the nucleus (Wang and Yang, 2001). Notably, MEF2C is regulated by NRL (Hao et al., 2011) and MEF2D interacts with CRX (Andzelm et al., 2015); these two proteins control gene expression in photoreceptors. Whether these MEFs cooperate with HDAC4 in regulating photoreceptor development remains to be determined.
Mice deficient in Sirt1, encoding a deacetylase, exhibit eye defects as early as E12.5 and display small and irregularly shaped eyes with thin retina and rosette-like structures (Cheng et al., 2003). In contrast, knockout of Sirt6, another deacetylase, did not show any overt retinal defects but instead demonstrated electrophysiological alterations and aberrant expression of glycolytic genes (Silberman et al., 2014). Thus, Sirt6 seems to affect neuronal function rather than RPC proliferation or differentiation.
4.4. The Polycomb and the SWI/SNF complexes during retinal development
The Polycomb and SWI/SNF are large and evolutionary conserved multi-subunit complexes that often function antagonistically in controlling mammalian development (Ho and Crabtree, 2010; Simon and Kingston, 2013). The polycomb complexes comprise of enzymes that modify histone tails to repress gene expression, while the SWI/SNF complexes alter chromatin structure to facilitate transcription. Polycomb proteins are grouped into two complexes: the repressive complex 1 (PRC1) and 2 (PRC2). PRC1 adds a ubiquityl moiety on H2A lysine 119 (H2AK119ub1) (Simon and Kingston, 2013). Mutations in two components of the PRC1 complex, BMI1 and SAMD7, have profound effects on retinal development. BMI1 is required for self-renewal of immature RPCs, and its deficiency affects proliferation and postnatal maintenance of peripheral RPCs (Chatoo et al., 2010). BMI1 is also expressed in differentiated neurons, and its knockout results in postnatal loss of cone bipolar cells and cone photoreceptors (Barabino et al., 2016). SAMD7 is specifically expressed in developing rod photoreceptors, and its loss causes global transcriptional changes in the retina with upregulation and downregulation of cone and rod genes, respectively. SAMD7 loss also resulted in the reduction of H2AK119ub in upregulated genes, in agreement with its role within the PRC1 complex (Omori et al., 2017). Thus, SAMD7 is an example of a cell type-specific epigenetic modifier that contributes to the repression of non-cell type-specific genes. Importantly, mutations in Samd7 regulatory regions have been identified in patients with autosomal recessive retinitis pigmentosa (Van Schil et al., 2016).
PRC2 catalyzes the addition of methyl groups to H3K27 and is involved in maintaining a balance between proliferation and differentiation during development (Margueron and Reinberg, 2011). In Xenopus, PRC2 regulates RPC proliferation and knockdown of ezh2 (a core member of the PRC2 complex) impairs the differentiation of neural retina cell types while promoting the Müller-glial fate (Aldiri et al., 2013). Similarly, targeted knockout of Ezh2 in early retinal progenitors in mice has revealed a crucial role of H3K27 methylation in controlling the timing of postnatal differentiation and cell survival (Table 3) (Fujimura et al., 2018; Iida et al., 2015; Zhang et al., 2015a). In contrast, loss of Ezh2 later in development resulted in normal retina in the perinatal period but postnatal loss of differentiated cells (Yan et al., 2016). However, no overt retinal phenotype was observed when Ezh2 was deleted in RPCs expressing Math5, which is critical for RGC differentiation (Yan et al., 2016). The conditional deletion of Eed (another core component of PRC2) in retinal progenitors resulted in decreased cell proliferation and increased proportions of amacrine cells as well as cell death at postnatal stages (Fujimura et al., 2018).
The SWI/SNF chromatin remodeling complexes regulate gene expression by disrupting the interaction of histones and DNA in an ATP-dependent manner (Ho and Crabtree, 2010). These complexes are divided into two sub-classes: the BAF (BRG/hBRM associated factors) complexes, and the PBAF (polybromo associated BAF) complexes (Ho and Crabtree, 2010). In the retina, the ATPase subunit BRG1 (encoded by Smarca4) directly controls genes involved in cell proliferation, adhesion, polarity and photoreceptor differentiation (Aldiri et al., 2015). Knockdown of Smarca2 (BRM), another ATPase subunit homologous to Smarcd4, perturbs the differentiation of RGCs in the embryonic retina by reducing the expression of Pou4f2 (Brn3b) and inhibiting Notch signaling (Das et al., 2007). Smarcd3 (BAF60C), also a member of the SWI/SNF complex, is expressed in RPCs and contributes to their proliferation by interacting with the Notch pathway; it’s expression decreases during differentiation but is re-established in Müller glia that reenter the cell cycle after neurotoxic damage (Lamba et al., 2008).
4.5. Targeting of epigenetic modifiers by transcription factors to regulate gene expression
Temporal and cell type- or tissue-specific expression of epigenetic modifiers (Figs. 2, 5 and 9) and their selective interaction with TFs (Fig. 10) ensure stringent regulation of gene expression during development. PAX6, a key eye-field transcription factor, can bind HDAC1 through its paired transactivation domains to mediate transcriptional repression during retinal development (Kim et al., 2017). PAX6 can also interact with the histone acetyltransferase KAT5 (TIP60), which enhances its transcriptional activity in postnatal mouse retina (Kim et al., 2012a). Similarly, phosphorylation of NRL at Ser50 by JNK1 (c-Jun N-terminal kinase 1) is reported to result in the recruitment of KAT5, which in turn can stimulate H3/H4 acetylation and Rhodopsin promoter activity (Kim et al., 2012b). Mutations at Ser50 and Pro51 that alter NRL phosphorylation lead to retinal degeneration (Bessant et al., 1999; Kanda et al., 2007). CRX binds to the histone acetyltransferases CBP (CREB-binding protein) and p300, which mediate H3/H4 acetylation, leading to H3 acetylation at the promoters of photoreceptor genes and subsequent recruitment of other TFs that potentiate transcription (Peng and Chen, 2007). Conditional knockout of both Ep300 and Crebbp resulted in chromatin decondensation and redistribution of repressive and active marks in rods (Hennig et al., 2013). CRX also interacts with the STAGA (SPT3-TAF9-ADA-GCN5 acetyltransferase) transcription coactivator complex through Ataxin-7 (ATXN7, SCA7), and ChIP studies revealed an association of CRX, STAGA and H3ac in CRX target genes (Palhan et al., 2005). A rod-specific mutation in Atxn7 resulted in the aberrant recruitment of STAGA complex to large genomic regions, inducing histone H3 hyperacetylation, chromatin decondensation and transcriptional dysregulation of rod genes (Helmlinger et al., 2006; Kizilyaprak et al., 2011). In Drosophila, loss of Ataxin-7, results in neural and retinal degeneration and decreased H2B ubiquitylation and H3K9 acetylation (Mohan et al., 2014). Less H2B ubiquitylation is also observed in human cells in the absence of Ataxin-7 (Mohan et al., 2014). Additional investigations are needed to define the role of histone H2B ubiquitylation/deubiquitylation in modulating retinal gene expression. Furthermore, NR2E3 was shown to interact with the PRC1 subunit SAMD7 to repress non-rod genes in rod photoreceptors (Omori et al., 2017). Finally, the TF REST was shown to recruit the DNA demethylase TET3 to increase 5hmC in the gene bodies of neuronal genes and thus their transcriptional output (Perera et al., 2015) (Fig. 10).
5. Epigenetic changes during retinal aging and diseases
5.1. Aging
In humans, a gradual deterioration is experienced in many psychophysical parameters of the visual function with advanced age; these include contrast sensitivity and dark adaptation, with rod-mediated scotopic vision being the most affected (Owsley, 2016). Structural and cellular changes in the aging retina are associated with dysfunction or loss of neuronal and non-neuronal cells, even in the absence of any ocular pathology (Cavallotti et al., 2004). Rods are more affected than cones, especially in the central retina where cones are stable whereas the number of rods decrease (Bonnel et al., 2003). Neural retina and RPE are extensively exposed to light and oxidative stress, leading to cellular and molecular alterations including mitochondrial DNA damage, impaired lysosomal and mitochondrial functions, accumulation of lipofuscin in RPE (Bonnel et al., 2003), local chronic inflammation and drusen accumulation in the Bruch’s membrane (Booij et al., 2010).
The availability of array-based methods has permitted large-scale profiling of expression changes, and the first such study of the aging human retina uncovered differential expression of genes associated with energy metabolism and stress response (Yoshida et al., 2002). Another analysis of the human retina revealed differential aging-related changes in gene expression between macula and peripheral retina and possible dysregulation of Wnt-pathway genes (Cai et al., 2012). Microarray profiling of purified mouse rod photoreceptors identified significant alterations with advanced age in genes that belong to lipid/retinoid metabolism, angiogenesis, oxidative phosphorylation, and stress and immune response (Parapuram et al., 2010). Additionally, the analysis of the transcriptome of aged and young rat retinas by RNA-seq identified 160 differentially-expressed genes that were primarily related to extracellular matrix organization and immune response (Kozhevnikova et al., 2013).
Cellular aging is accompanied by an altered epigenetic landscape (Fig. 11), resulting in chromatin reorganization and aberrant gene expression (Benayoun et al., 2015; Pal and Tyler, 2016). Accumulation of epigenetic changes is highly heterogeneous within a tissue, leading to an epigenetic drift that generates transcriptional noise and makes the tissue less responsive/adaptive to its environment. Such a differential response to the environment is exemplified by the presence of distinct epigenetic signatures of astrocytes following ischemia in middle-aged rats compared to young adults (Chisholm et al., 2015).
Fig. 11.
Aging-associated epigenetic alterations. An aging cell undergoes changes in its epigenetic landscape with varying alterations in histone modifications, global loss of core histones, histone variant exchange (e.g., incorporation of H3.3 or macroH2A), global reduction with local increase in DNA methylation levels, and redistribution and loss of heterochromatin (Benayoun et al., 2015; Pal and Tyler, 2016).
The epigenetic age of a cell or tissue is reflected by the level of methylation of discrete CpG sites that constitute an “epigenetic clock” (Horvath, 2013; Stubbs et al., 2017). This epigenetic clock is considered as a biomarker of aging (Christiansen et al., 2016; Levine et al., 2015) and may even be part of a programmed cellular aging process (Mitteldorf, 2015). The epigenetic clock is based on DNA methylation and does not include the status of histone marks. The trend is a loss of repressive and gain of activating histone marks (Benayoun et al., 2015; Pal and Tyler, 2016), and studies focused on mammalian neuronal systems tend to confirm this tendency (Table 4). Lack of consensus about global changes in histone marks during aging could be the consequence of using mixed cell populations and thus diluting the signal from specific cell-types. Indeed, studies emphasize that some histone modifications are altered depending on the brain and chromatin regions studied (Ryu et al., 2011; Snigdha et al., 2016), and are modulated by environmental factors (Gong et al., 2015; Morse et al., 2015; Peleg et al., 2010).
Table 4.
Global changes of histone post-translational modifications during aging of the mammalian nervous system.
| Mark | Change | Species | Tissues / Cell types | Age | Reference |
|---|---|---|---|---|---|
| H3K4me2 | + | Macaque | Prefrontal cortex | 0.4, 9, 22, 26Y | (Han et al., 2012) |
| H3K4me3 | + | Human | Prefrontal cortex neurons | 0.5 to 69Y | (Cheung et al., 2010) |
| + | Human | Prefrontal cortex neurons | 20–28Y vs 38–55Y | (Han et al., 2012) | |
| + | Mouse | Hippocampus | 3–4M vs 18–20M | (Snigdha et al., 2016) | |
| = | Mouse | Cerebellum | |||
| + | Rat | Hippocampus | 3M vs 19–22M | (Morse et al., 2015) | |
| H3K9ac | − | Mouse | Cochlea | 8W vs 132W | (Watanabe and Bloch, 2013) |
| = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) | |
| + | Mouse | Cortical neurons | 4–8W vs 55–65W | (Rodrigues et al., 2014) | |
| H3K9me(all) | − | Rat | Hippocampus | 3M vs 20M | (Elsner et al., 2013) |
| H3K9me2 | + | Mouse | Cochlea | 8W vs 132W | (Watanabe and Bloch, 2013) |
| H3K9me3 | − | Mouse | Cortical neurons | 4–8W vs 55–65W | (Rodrigues et al., 2014) |
| H3K14ac | = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) |
| H3K20me2 | − | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H3K27ac | = | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H3K27me3 | − | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H3K56ac | = | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H3K79me3 | − | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H3R2me2 | − | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
| H4K5ac | = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) |
| H4K8ac | = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) |
| H4K12ac | = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) |
| H4K16ac | = | Mouse | Hippocampus | 3M vs 16M | (Peleg et al., 2010) |
| = | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) | |
| H4R3me2 | = | Mouse | Brain | 3M vs 22M | (Gong et al., 2015) |
Data for retinal aging are not available. + and − represent higher and lower expression in aging, respectively. =, no change. Not all changes were normalized to the total levels of the respective histone. Ages are expressed in years (Y), months (M), or weeks (W).
Little is understood about epigenetic changes during healthy aging of the retina. Age-related changes have been reported in cytosine methylation at the promoters of the Major Histocompatibility Complex I components H2-D1 and H2-K1 in the retina as well as in other mouse brain regions (Mangold et al., 2017). Expression and activity of the histone deacetylase SIRT1 seem to be reduced with aging in the rat and human retina and RPE (Lamoke et al., 2015; Peng et al., 2011). With age, retinal cells undergo a higher rate of apoptosis and resveratrol, a Sirtuin activator, is shown to reduce this rate, increase SIRT1 expression and improve the ERG signal in rats (Zeng and Yang, 2015). Aged RPE cells also become more prone to apoptosis, and SIRT1 is involved in protecting RPE cells against apoptosis (Bhattacharya et al., 2012) and against premature senescence induced by oxidative stress (Zhuge et al., 2014). However, both studies reported the modulation of p53 acetylation as a mediator of SIRT1 effects. Indeed, SIRT1 has several non-histone targets, including NF-κB and p53, and much remains to be done for distinguishing the epigenetic from the non-epigenetic functions of SIRT1 in healthy aging as well as in disease. Finally, resveratrol, was able to increase the viability and proliferation of cultured retinal stem cell-like cells that were obtained from old rats (Peng et al., 2010).
5.2. Epigenetics of age-related retinal diseases
Neurodegenerative diseases are associated with epigenetic age acceleration (Horvath et al., 2016). Several physiological features are common to pathological states of the retina and normal aging (Chader and Taylor, 2013; Jackson et al., 2002; Swaroop et al., 2009), differing in their extent/severity (Owsley et al., 2014); thus, accelerated aging is predicted to contribute to age-related disease pathogenesis (Ardeljan and Chan, 2013; Lamoke et al., 2015; Mortuza et al., 2013). Notably, genes associated with an acceleration of the cerebellum epigenetic clock and with brain aging exhibit significant overlap with those associated with age-related macular degeneration (AMD) and type 2 diabetes (Lu et al., 2017). We will review DNA methylation and histone modification changes in the retina in two age-related diseases, AMD and diabetic retinopathy (DR).
5.2.1. Epigenetic changes associated with age-related macular degeneration
AMD is a multifactorial disease of advanced age with varying pathology affecting the macula and consequently leading to loss of central vision. Complex, and as yet poorly understood, interaction of genetic susceptibility and environmental risk factors can impair RPE, Bruch’s membrane and choroidal functions, eventually leading to photoreceptor cell death (Fritsche et al., 2014). Genomewide association studies have identified at least 52 independent variants at 34 genetic loci that are associated with risk of both geographic atrophy and choroidal neovascularization in advanced stage of AMD (Fritsche et al., 2016). The presence of reported, and perhaps additional unknown, genetic variants can greatly enhance the susceptibility to AMD; however, clinical manifestations of the disease are highly heterogeneous and likely guided by environmental factors (such as smoking and diet) through epigenetic mechanisms (Gemenetzi and Lotery, 2014; Sobrin and Seddon, 2014).
To date, no genomewide methylation profiles have been reported for AMD retina and/or RPE. The analysis of human RPE/choroid and neural retina has revealed the hypermethylation of GSTM1 and GSTM5 in AMD, in concordance with their decreased expression levels (Hunter et al., 2012). Hypermethylation has also been reported in the promoter region of PRSS50 in the retina and whole blood of AMD patients (Oliver et al., 2015). Methylation differences have been observed in peripheral blood mononuclear cell DNA of AMD patients, specifically at the IL17RC locus (Wei et al., 2012); however, the latter finding was not reproducible in leukocytes, retina and RPE of other cohorts (Oliver et al., 2013a), and no genomewide significant differences were detected in global methylation profiles of whole blood of AMD patients and controls (Oliver et al., 2015; Pinna et al., 2016). Weak evidence of differential allele-specific methylation pattern was reported in whole blood of AMD cases and controls, including monozygotic twins discordant for AMD stage, in genomic regions spanning the AMD susceptibility genes – CFH, C2 and CFB (Hutchinson et al., 2014). Notably, the J haplogroup of mtDNA, which is associated with a higher risk for AMD, may influence global DNA methylation levels in RPE, and especially the expression of several epigenetic enzymes (including HAT1, HDAC1, DNMT1, DNMT3A and DNMT3B) and nuclear genes (CFH, EFEMP1, VEGFA and NFkB2), compared to the AMD-protective H haplogroup, and this effect may likely be due to haplotype-specific SNPs (Atilano et al., 2015).
No genomewide information is currently available about histone modifications in AMD; nevertheless, we list a few related reports. Reduced expression of HDACs 1/2, 5, and 6 was observed in the AMD retina compared to age-matched controls (Anderson et al., 2015). SIRT1 expression was also decreased in the AMD retina and RPE (Peng et al., 2011) as well as in RPE cells differentiated from induced pluripotent stem cells (iPSCs) of AMD patients (Golestaneh et al., 2016). Such changes in histone modifiers can potentially have a broad impact on the epigenetic landscape of the cells associated with AMD pathology. Inhibition of HDACs by Trichostatin A is reported to exert an anti-angiogenic effect in human RPE and vascular endothelial cells as well as in a mouse model of choroidal neovascularization (Chan et al., 2015). One case study has investigated the effects of a resveratrol-based drug in three AMD patients and reported an improvement in visual function, mirrored by anatomical restoration (Richer et al., 2013), as well as long-term beneficial effects (Richer et al., 2014).
5.2.2. Role of epigenetic changes in the metabolic memory of diabetic retinopathy
DR is a common complication of diabetes with varying clinical features, including macular edema, angiogenesis, microvascular damage and proliferative retinopathy (Wong et al., 2016). Changes in retinal gene expression have been observed during the progression of retinopathy in a mouse model of diabetes (Kandpal et al., 2012). Microarray profiling of donor retina from diabetic patients revealed differential expression of mitochondria-associated genes that are involved in angiogenesis, anti-oxidant defense and energy production (Govindarajan et al., 2017). Genetic susceptibility variants linked to DR exert much smaller impact on disease compared to AMD-associated variants though some commonalities seem to exist (Gangwani et al., 2014). Of interest for epigenetics is the reported association of a polymorphism in the SUV39H2 gene, encoding the H3K9 histone methyltransferase KMT1B, with DR (Syreeni et al., 2011).
Hyperglycemia is the driver of diabetes, and the notion of “metabolic memory” or “hyperglycemic memory” has been propagated since the disease develops after long, and even interrupted, exposure to a persistent hyperglycemic state, suggesting an epigenetic imprint (Kowluru, 2017). The epigenetic imbalance of the diabetic retina was first suggested by studies showing global though contradictory changes in histone acetylation levels and altered expression/activity of HDACs and HATs that persisted after the termination of hyperglycemia in streptozotocin-induced diabetic rats (Kadiyala et al., 2012; Zhong and Kowluru, 2010). A mass spectrometry analysis demonstrated that 67 out of 135 identified histone marks (involving all canonical and some non-canonical histones) are significantly altered in the retina of streptozotocin-induced diabetic rats compared to non-diabetic animals (Wang et al., 2017). Some of these changes appear to be modified by the presence of marks harbored by nearby residues, underlying a complex combinatorial interplay that remains to be explored.
Numerous studies have linked the dysregulation of histone and DNA modifying enzymes to perturbations of the epigenetic environment and expression of genes involved in the progression of DR. As this role of epigenetics in DR has been reviewed recently (Kowluru, 2017; Kowluru et al., 2015), we will only highlight a few findings. Changes in the epigenetic landscape of MMP9 and SOD2 promoters are likely due to altered activity of several epigenetic writers and erasers, in concordance with the proposed role of oxidative stress and mitochondrial dysfunction in DR. In addition, increase histone and DNA methylation at other promoters such as of Keap1, and POLG, respectively, as well as hypermethylation of mitochondrial DNA are reported to impact the expression of respective genes in hyperglycemia or DR models. Interestingly, evidence of these epigenetic changes in the retina from patients with DR was also shown (Kowluru et al., 2015).
Upregulation of the histone acetyltransferase gene Ep300 in the diabetic retina and of two histone arginine methyltransferases, PRMT1 and PRMT4, in the diabetic RPE have been proposed to impact the expression of endothelial and extracellular matrix factors and to aggravate oxidative stress and apoptosis of RPE cell via H3R17 dimethylation, respectively (Cao et al., 2014; Kaur et al., 2006; Kim et al., 2015; Kim et al., 2014). Conversely, downregulation of Sirtuins, and especially of SIRT1, has been reported in models of DR (Duarte et al., 2015; Zhao et al., 2016) and associated with retinal vasculature defects (Lamoke et al., 2015), which could be reversed by over-expressing SIRT1 (Mortuza et al., 2015). An early decrease of SIRT6 and associated increase of H3K9 and H3K56 acetylation have been observed in the retina of diabetic mice, and proposed to be a driver of BDNF (brain-derived neurotrophic factor) decrease and VEGF (vascular endothelial growth factor) increase (Zorrilla-Zubilete et al., 2018). Another study reported an increased expression of major components of the PRC2 complex in animal and cell models of DR, and linked this to a subsequent increased repression of miR-200b that led to higher VEGF production in vitro (Ruiz et al., 2015).
The possibility of treating DR has also been explored by modulating epigenetic enzymes in animal and in vitro models. A glucagon-like peptide 1 analog, extendin-4, stimulated the expression SIRT1 and SIRT3 in the diabetic retina and could mitigate deleterious effects of diabetes (Zeng et al., 2016), probably by inducing SOD3 expression through enhanced H3 acetylation at its promoter (Yasuda et al., 2016). Likewise, fenofibrate, an anti-inflammatory agent, could reverse the effects of high glucose-induced stress via the upregulation of Sirt1 expression (Zhao et al., 2015). Taken together, these and other studies (Xie et al., 2014) suggest that epigenetic enzymes could be good candidates for therapies of DR.
6. Challenges and Opportunities
Identification of epigenetic contributions to gene regulation in the retina will be invaluable for delineating mechanisms of cell fate acquisition during development, impact of genetic variations on pathways associated with retinal disease, and influence of the environment on phenotypes. Despite the progress in elucidating epigenetic patterns during development, we have limited understanding of how chromatin architecture and epigenome affect transcriptional machineries during aging and disease, especially in retinal cells. As an example, the catalog of histone posttranslational modifications at different residues is growing fast and now includes phosphorylation, sumoylation and ubiquitylation among others; however, most of these have not been studied in the retina. Similarly, we have little understanding of precise roles of most epigenetic enzymes that display dynamic expression patterns during retinal development.
The advent of next generation sequencing technology has allowed the generation of mRNA and epigenetic reference maps for the developing and mature retina and for some isolated cell-populations (Aldiri et al., 2017; Hoshino et al., 2017; Kim et al., 2016a; Macosko et al., 2015; Siegert et al., 2012); however, more comprehensive temporal genomewide maps must be produced for multiple epigenetic modifications, including DNA methylation, core histone modifications and eventually non-coding RNAs, for integrated gene regulatory networks that control development and disease (Chaitankar et al., 2016; Yang et al., 2015). Implementation of single cell technologies (or at least the analysis of a purified cell type) will be crucial to define the temporal and unique cellular states that are obscured when averaging epigenetic profiles (Schwartzman and Tanay, 2015) (see Box 1). So far, these studies are few in the retina due to the low abundance of most retinal cell types and lack of appropriate markers for their isolation. Moreover, most of the phenotypes related to global acetylation and deacetylation, reported to date, appear to be observed in rod photoreceptors, and the high abundance of this cell type obscures changes present in other cell populations.
We wanted to point out that phenotypes related to global perturbations in the epigenome, especially in histone modifications, are complex and affect both cell cycle and survival of progenitors, and thus invariably influence relative cell number proportions. We suggest cell type-specific loss-of-function studies of individual epigenome-associated enzymes for assessing primary targets of chromatin modifiers. Equally important would be improvements in analytical algorithms that permit integration of all chromatin modifications for defining their significance in an individual cell, a specific cell type, and/or within the context of the retina as a whole. We suggest that gene regulation analysis should also take into consideration a plethora of chromatin modifications, including DNA methylation and histone modifications, that depend on genomic context and may influence transcription or associated processes (such as alternative splicing) when present in gene bodies. Finally, newly-discovered epigenome editing tools (Liu et al., 2016) should allow us to clarify the causal role of epigenetic modifications in controlling gene expression and consequently dysregulation of cellular pathways during aging and disease.
6.1. Retinal metabolism and epigenetic regulation
Epigenetic modifications provide a link between the environment, the cell’s metabolic state and gene expression (Barres and Zierath, 2011). Notably, many intermediary factors of the cell metabolism are cofactors of histone- and DNA-modifying enzymes. Sirtuin histone deacetylases use NAD+ as a cofactor, and histone demethylases utilize either FAD or alpha-ketoglutarate. Similarly, acetyltransferases and methyltransferases use acetyl-coenzyme A and S-adenosylmethionine as acetyl and methyl donors, respectively (Berger and Sassone-Corsi, 2016). Conversely, metabolic derivatives can inhibit histone methytransferases and demethylases (Black et al., 2012). Fluctuations in the levels of metabolites may contribute to temporal control of gene expression and the activity of chromatin-modifying enzymes (Kaelin and McKnight, 2013). Diet also influences the epigenome; for example, deficiency in methyl donors affects DNA methylation in various model systems (Barres and Zierath, 2011). Caloric restriction, a condition known to increase lifespan and modulate age-related epigenetic, transcriptomic and cellular changes (Li et al., 2011), can slow down age-related changes in histone post-translational modifications (Gong et al., 2015) and DNA methylation (Hahn et al., 2017; Maegawa et al., 2017) and increase the activity of histone deacetylases (Guarente, 2013). In the retina, caloric restriction has shown protective effects in pathologic contexts as well as during normal aging (Kawashima et al., 2013; Kim et al., 2004; Kong et al., 2012). Therefore, the metabolic state of cells appears to be a critical regulator of the epigenetic landscape during development and disease pathogenesis.
The retina is a highly metabolically active tissue, and the photoreceptors have delicately-balanced energy homeostasis with high anaerobic glycolysis as well as low reserve capacity during mitochondrial oxygen consumption (Chinchore et al., 2017; Hurley et al., 2015; Kooragayala et al., 2015). The retina also uses fatty acids as a source of energy and performs lipid β-oxidation for energy needs. Whether the levels of acetyl-coenzyme A or other metabolites correlate with global acetylation changes during retinal development would require further investigations. Photoreceptors exhibit a high metabolic rate, pronounced metabolic changes in response to light and circadian rhythms, accessibility to drug delivery and susceptibility to degeneration, thereby representing an excellent system to investigate metabolic regulation of the epigenome and its impact on functional homeostasis.
6.2. In vitro differentiation of the retina
Three-dimensional organoids, derived in vitro, from iPSCs can recapitulate early retinal development in vivo (Chen, 2016; Clevers, 2016; Sasai, 2013; Volkner et al., 2016); however, epigenetic memory during the derivation of iPSC lines from somatic cell types (Bar-Nur et al., 2011; Kim et al., 2010) is reported to influence the outcomes during cell differentiation (Roessler et al., 2014). The retina generated from rod-derived iPSCs displayed higher efficiency of producing differentiated cell types in organoid culture (Hiler et al., 2015). Profiling DNA and histone modifications of retinal cell types and organoids during in vitro reprogramming and their comparison with in vivo development (as elucidated for transcriptomes of photoreceptors in organoids (Chen, 2016; Kaewkhaw et al., 2015)) can generate enhancer profiles and “barcodes of neural identity” (Telese et al., 2013) that would be useful potentially for improving the quality and efficiency of culture system and provide avenues to dissect complex biological processes, such as morphogenesis, lamination, synapse formation, and neurotransmission. Manipulating the epigenetic landscape through chromatin-modifying enzymes, either by ectopic expression (Akiyama et al., 2016; Albini et al., 2013) or small molecule treatments (Zhang et al., 2015b) can therefore be a convenient and effective strategy for reproducible differentiation of discrete cell types and of organoids in vitro for investigations on retinal development, disease mechanisms and treatment design.
6.3. Retinal regeneration
Müller glia can function as a stem cell in teleost fish retina to replenish neurons after injury (Ramachandran et al., 2010; Raymond et al., 2006). Even in the mammalian and chicken retina, Müller glia exhibit stem cell-like features but have limited capacity for regeneration (Das et al., 2006; Fischer and Reh, 2001), and extensive efforts are being made to understand molecular mechanisms underlying such differences. Notably, the transcriptome of purified Müller glia is similar to that of retinal progenitors cells (Roesch et al., 2008; Sifuentes et al., 2016; Ueno et al., 2017), and the epigenome seems to be “ready” for activation of dedifferentiation pathways as promoters of pluripotency factors are hypomethylated in quiescent Müller glia in both zebrafish and mouse (Powell et al., 2013). Considerable reconfiguration of the epigenome occurs in Müller glia cells transitioning from quiescence to a progenitor state in zebrafish. An initial global hypomethylation wave is followed by de novo methylation, which correlates with changes in gene expression, and the inhibition of dnmt enzymes with 5-aza-2′-deoxycytidine (5-dAza) showed reduced progenitor proliferation and interfered with reprograming (Powell et al., 2013). Similarly, knockdown of the cytidine deaminases-encoding genes apobec2a and apobec2b inhibited the induction of ascl1, a proneural transcription factor necessary for regeneration in teleost fish, and decreased Müller glia proliferation as well as optic nerve regeneration (Powell et al., 2012). Remarkably, pharmacological induction of histone H3K27 acetylation in mice is shown to promote Müller glia production of neurons after retinal injury. The histone deacetylase inhibitor Trichostatin A stimulated transdifferentiation of Müller glia into bipolar or amacrine cells after glutamate toxicity in mice overexpressing Ascl1 (Jorstad et al., 2017). Thus, the epigenome of mammalian Müller glia seems to be permissive to reprogramming, and identifying specific epigenetic changes needed to drive transdifferentiation into different cell types may allow the design of novel therapeutic paradigms for retinal repair.
7. Concluding Remarks
Epigenetic studies have begun to unravel valuable insights into retina and cell type-specific gene regulation and biological processes associated with development and homeostasis. Given the clinical variability of age-related retinal diseases, one can also anticipate a significant role of epigenetics in conjunction with non-coding genomic variations and/or environmental influence in disease pathogenesis. Currently, we have limited, and somewhat descriptive, understanding of epigenetic modifications and modifiers associated with retinal development, aging and disease pathogenesis. Efforts are being made to integrate global epigenetic profiles from whole retina and of distinct cell types within to corresponding transcriptome data sets, generated over a dynamic temporal period. In conjunction with lineage-specific loss-of-function phenotyping and epigenome editing, future investigation will permit construction of comprehensive gene regulatory networks that can then be manipulated for designing knowledge-based therapies of degenerative blinding diseases.
Supplementary Material
Fig. 6.
Survey of DNA methylation. Methods to identify 5-methyl-cytosine (5mC) and 5-methyl-hydroxy-cytosine (5hmC) are frequently used in combination with polymerase chain reaction (PCR), next-generation sequencing, or DNA microarrays. Some of these methods, such as genomewide bisulfite sequencing (WGBS-seq) and reduced representation bisulfite sequencing (RRBS), cannot distinguish between 5mC and 5hmC. In bisulfite sequencing, a bisulfite salt converts all unmethylated cytosines to uracyl. In RRBS, specific methylation-insensitive enzymes are used to digest DNA and, after size selection, only a fraction of the genome biased towards CpG rich regions is surveyed (Meissner et al., 2005). To distinguish 5mC from 5hmC, specific antibodies are used in Methylated DNA immunoprecipitation (MeDIP) method (Thu et al., 2009). Nanopore sequencing can detect the electrolytic current signals of base modifications by moving the single-strand DNA molecule through a protein pore (Laszlo et al., 2013). In Tet-assisted bisulfite sequencing (TAB-seq) (Yu et al., 2012), 5hmC is glucosylated by β-glucosyltransferase (βGT) to protect 5hmC from the action of TET enzymes, which convert the 5mC into 5caC. Bisulfite treatment then converts 5caC into U leaving the glucosylated 5hmC intact.
Fig. 7.
Chromatin immunoprecipitation of specific retinal cell types to investigate histone modifications. Various retinal cell types can be purified by: immunopanning (Wang et al., 2007), immunomagnetic purification (Sajgo et al., 2017), laser capture microdissection (Kim et al., 2006), isolation of nuclei tagged in specific cell types (INTACT) (Mo et al., 2016), or fluorescence-activated cell sorting (FACS) (Kim et al., 2016b). FACS and INTACT have been used for ChIP of histone modifications in the retina (Kim et al., 2016b; Mo et al., 2016). Several fluorescent reporter lines could be used to purify different retinal populations (Siegert et al., 2009). Whole retina or purified cell populations are crosslinked with paraformaldehyde (Park, 2009). The nuclear fraction is isolated, followed by sonication to shear chromatin into small fragments. Antibodies against specific histone modifications can pulldown accompanying genomic DNA regions. Reverse-crosslinking and DNA purification are then followed by PCR and/or next generation sequencing. Mapping to the genome and subsequent comparison with transcriptome datasets permit correlations of histone modifications with transcription (Chaitankar et al., 2016; Yang et al., 2015).
Fig. 8.
Whole-genome bisulfite sequencing workflow and analysis. Genomic DNA from cells or tissues is fragmented and treated with sodium bisulfite to convert unmethylated cytosine residues to uracyl (Lizardi et al., 2017). Unmethylated DNA from lambda phage is often added to control for bisulfite conversion efficiency. Library preparation includes adapter ligation and PCR amplification. Addition of a high-complexity library is necessary to obtain high quality sequencing reads. Raw FASTQ sequencing reads are validated for quality and adapter contamination. After trimming low quality reads and adapters, the reads are aligned to a bisulfite-converted reference genome, duplicate alignments are removed and differentially methylated regions (DMRs) are identified. Further downstream analysis includes annotation of DMRs and their correlation to the transcriptome.
Acknowledgments
We are grateful to Hyunjin Yang and Jung-woong Kim for discussions, and to Colin Barnstable, Valerie Wallace, Tiziana Cogliati, and Freekje van Asten for constructive comments on the manuscript. We acknowledge the contribution of Adrien Gontier with two of the illustrations. Our research is supported by the Intramural Research program (EY000450, EY000546) of the National Eye Institute, National Institutes of health, Bethesda, MD, USA.
Abbreviations
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- AMD
Age-related macular degeneration
- ATAC-seq
Assay for transposase accessible chromatin followed by high-throughput sequencing
- BS-seq
Bisulfite sequencing
- ChIP
Chromatin immunoprecipitation
- CpGI
CpG Island
- CRX
Cone-rod homeobox protein
- DMR
Differentially methylated region
- DNMT
DNA methyltransferases
- DR
Diabetic retinopathy
- ESC
Embryonic stem cell
- HDAC
Histone deacetylases
- KAT
Lysine acetyltransferase
- KDM
Lysine demethylases
- KMT
Lysine methyltransferase
- MBD
Methyl-CpG binding domain
- NRL
Neural retina leucine zipper
- PRC
Polycomb repressive complex
- RGC
Retinal ganglion cell
- RPC
Retinal progenitor cell
- RPE
Retinal pigment epithelium
- SIRT
Sirtuin
- TET
Tet-eleven translocation
- TF
Transcription factor
- TSS
Transcription start site
Glossary
- Adapters
Synthetic oligonucleotide sequences ligated to DNA for cloning or PCR amplification
- Alignment
The computational process of assigning sequencing reads to a location on the genome
- Chromatin
Genomic DNA tightly-packaged by histone and other protein complexes and primarily structured in nucleosomes
- CpG
A DNA region where a cytosine is followed by a guanine nucleotide in 5′ to 3′ direction
- DNA methylation
An epigenetic mark involving methylation of cytosine or adenine
- Enhancer
A distance- and orientation-independent regulatory region
- Epigenetics
The study of adaptive chromatin modifications that do not involve alterations in DNA sequence
- Eraser
An enzyme that removes a covalent modification from DNA or histones
- Euchromatin
A decondensed form of chromatin that is transcriptionally accessible (active)
- FASTA
A file format for storing sequencing data
- Hemimethylation
A state of DNA in which a cytosine nucleotide is methylated in only one strand
- Heterochromatin
A condensed form of chromatin that is transcriptionally inactive and generally possesses low gene content
- Histone
A family of basic and highly conserved proteins that condense the DNA to form chromatin
- Histone code
The combinatorial interplay of histone modifications leading to chromatin regulatory mechanisms
- Histone modifications
Covalently-ligated chemical groups on histones that can modulate chromatin structure and/or gene expression
- Next Generation Sequencing (NGS)
High throughput and massively parallel sequencing of DNA without the need for cloning
- Peak
An identified genomic region with enriched signal originating from NGS experiments, e.g., ChIP-seq
- Phototransduction
The process of converting a photon into electrochemical signals by the retinal photoreceptors
- Promoter
A regulatory region upstream or adjacent to the transcription start site of a gene
- Reader
A protein that recognizes chromatin modifications
- Retinogenesis
The process of retinal development
- RNA splicing
The removal of intronic sequences to generate mature mRNA
- Super-enhancer
An enhancer region with high density of TF binding sites
- Writer
An enzyme that adds a covalent modification to DNA or histones
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aapola U, Liiv I, Peterson P. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Research. 2002;30:3602–3608. doi: 10.1093/nar/gkf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Wakabayashi S, Soma A, Sato S, Nakatake Y, Oda M, Murakami M, Sakota M, Chikazawa-Nohtomi N, Ko SB, Ko MS. Transient ectopic expression of the histone demethylase JMJD3 accelerates the differentiation of human pluripotent stem cells. Development. 2016;143:3674–3685. doi: 10.1242/dev.139360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, Puri PL. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I, Ajioka I, Xu B, Zhang J, Chen X, Benavente C, Finkelstein D, Johnson D, Akiyama J, Pennacchio LA, Dyer MA. Brg1 coordinates multiple processes during retinogenesis and is a tumor suppressor in retinoblastoma. Development (Cambridge, England) 2015;142:4092–4106. doi: 10.1242/dev.124800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I, Moore KB, Hutcheson DA, Zhang J, Vetter ML. Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/beta-catenin signaling. Development. 2013;140:2867–2878. doi: 10.1242/dev.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I, Xu B, Wang L, Chen X, Hiler D, Griffiths L, Valentine M, Shirinifard A, Thiagarajan S, Sablauer A, Barabas ME, Zhang J, Johnson D, Frase S, Zhou X, Easton J, Zhang J, Mardis ER, Wilson RK, Downing JR, Dyer MA St Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome P. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron. 2017;94:550–568. e510. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Anderson KW, Chen J, Wang M, Mast N, Pikuleva IA, Turko IV. Quantification of histone deacetylase isoforms in human frontal cortex, human retina, and mouse brain. PLoS One. 2015;10:e0126592. doi: 10.1371/journal.pone.0126592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D, Chan CC. Aging is not a disease: distinguishing age-related macular degeneration from aging. Prog Retin Eye Res. 2013;37:68–89. doi: 10.1016/j.preteyeres.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilano SR, Malik D, Chwa M, Cáceres-Del-Carpio J, Nesburn AB, Boyer DS, Kuppermann BD, Jazwinski SM, Miceli MV, Wallace DC, Udar N, Kenney MC. Mitochondrial DNA variants can mediate methylation status of inflammation, angiogenesis and signaling genes. Hum Mol Genet. 2015;24:4491–4503. doi: 10.1093/hmg/ddv173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad GK, Tomar RS. Proteolytic clipping of histone tails: the emerging role of histone proteases in regulation of various biological processes. Molecular Biology Reports. 2014;41:2717–2730. doi: 10.1007/s11033-014-3181-y. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. The Journal of Biological Chemistry. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Barabino A, Plamondon V, Abdouh M, Chatoo W, Flamier A, Hanna R, Zhou S, Motoyama N, Hebert M, Lavoie J, Bernier G. Loss of Bmi1 causes anomalies in retinal development and degeneration of cone photoreceptors. Development. 2016;143:1571–1584. doi: 10.1242/dev.125351. [DOI] [PubMed] [Google Scholar]
- Barres R, Zierath JR. DNA methylation in metabolic disorders. Am J Clin Nutr. 2011;93:897S–900. doi: 10.3945/ajcn.110.001933. [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Sassone-Corsi P. Metabolic Signaling to Chromatin. Cold Spring Harb Perspect Biol. 2016:8. doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet. 1999;21:355–356. doi: 10.1038/7678. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Chaum E, Johnson DA, Johnson LR. Age-related susceptibility to apoptosis in human retinal pigment epithelial cells is triggered by disruption of p53-Mdm2 association. Invest Ophthalmol Vis Sci. 2012;53:8350–8366. doi: 10.1167/iovs.12-10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, Oshimura M, Elowitz MB. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Molecular Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright JH, Nickerson JM, Borst DE. Site-specific DNA hypomethylation permits expression of the IRBP gene. Brain Res. 2000;887:211–221. doi: 10.1016/s0006-8993(00)02990-5. [DOI] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnel S, Mohand-Said S, Sahel JA. The aging of the retina. Exp Gerontol. 2003;38:825–831. doi: 10.1016/s0531-5565(03)00093-7. [DOI] [PubMed] [Google Scholar]
- Booij JC, Baas DC, Beisekeeva J, Gorgels TG, Bergen AA. The dynamic nature of Bruch’s membrane. Prog Retin Eye Res. 2010;29:1–18. doi: 10.1016/j.preteyeres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet. 2015;47:479–485. doi: 10.1038/ng.3272. [DOI] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Reh TA. Photoreceptor cell fate specification in vertebrates. Development. 2015;142:3263–3273. doi: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Fields MA, Hoshino R, Priore LV. Effects of aging and anatomic location on gene expression in human retina. Front Aging Neurosci. 2012;4:8. doi: 10.3389/fnagi.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cao Y, Feng B, Chen S, Chu Y, Chakrabarti S. Mechanisms of endothelial to mesenchymal transition in the retina in diabetes. Invest Ophthalmol Vis Sci. 2014;55:7321–7331. doi: 10.1167/iovs.14-15167. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Cavallotti C, Artico M, Pescosolido N, Leali FMT, Feher J. Age-related changes in the human retina. Canadian Journal of Ophthalmology Journal Canadien D’ophtalmologie. 2004;39:61–68. doi: 10.1016/s0008-4182(04)80054-1. [DOI] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nature Reviews Neuroscience. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chader GJ, Taylor A. Preface: The aging eye: normal changes, age-related diseases, and sight-saving approaches. Invest Ophthalmol Vis Sci. 2013;54:ORSF1–4. doi: 10.1167/iovs.13-12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitankar V, Karakulah G, Ratnapriya R, Giuste FO, Brooks MJ, Swaroop A. Next generation sequencing technology and genomewide data analysis: Perspectives for retinal research. Prog Retin Eye Res. 2016;55:1–31. doi: 10.1016/j.preteyeres.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N, He S, Spee CK, Ishikawa K, Hinton DR. Attenuation of choroidal neovascularization by histone deacetylase inhibitor. PLoS One. 2015;10:e0120587. doi: 10.1371/journal.pone.0120587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatoo W, Abdouh M, Duparc RH, Bernier G. Bmi1 distinguishes immature retinal progenitor/stem cells from the main progenitor cell population and is required for normal retinal development. Stem Cells. 2010;28:1412–1423. doi: 10.1002/stem.462. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Stockwell PA, Rodger EJ, Morison IM. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res. 2012;40:e79. doi: 10.1093/nar/gks150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Cepko CL. Requirement of histone deacetylase activity for the expression of critical photoreceptor genes. BMC Dev Biol. 2007;7:78. doi: 10.1186/1471-213X-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Kaya KD, Dong L, Swaroop A. Three-dimensional retinal organoids from mouse pluripotent stem cells mimic in vivo development with enhanced stratification and rod photoreceptor differentiation. Mol Vis. 2016;22:1077–1094. [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khan NW, Roger JE, Swaroop A. Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum Mol Genet. 2011;20:4102–4115. doi: 10.1093/hmg/ddr334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, Akbarian S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchore Y, Begaj T, Wu D, Drokhlyansky E, Cepko CL. Glycolytic reliance promotes anabolism in photoreceptors. Elife. 2017:6. doi: 10.7554/eLife.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm NC, Henderson ML, Selvamani A, Park MJ, Dindot S, Miranda RC, Sohrabji F. Histone methylation patterns in astrocytes are influenced by age following ischemia. Epigenetics. 2015;10:142–152. doi: 10.1080/15592294.2014.1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Heo K, Byun HM, An W, Lu W, Yang AS. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Research. 2011;39:104–118. doi: 10.1093/nar/gkq774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AV, James J, Bhattacharya S, Imbalzano AN, Antony ML, Hegde G, Zhao X, Mallya K, Ahmad F, Knudsen E, Ahmad I. SWI/SNF chromatin remodeling ATPase Brm regulates the differentiation of early retinal stem cells/progenitors by influencing Brn3b expression and Notch signaling. J Biol Chem. 2007;282:35187–35201. doi: 10.1074/jbc.M706742200. [DOI] [PubMed] [Google Scholar]
- Das AV, Mallya KB, Zhao X, Ahmad F, Bhattacharya S, Thoreson WB, Hegde GV, Ahmad I. Neural stem cell properties of Muller glia in the mammalian retina: regulation by Notch and Wnt signaling. Dev Biol. 2006;299:283–302. doi: 10.1016/j.ydbio.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Derrien T, Estelle J, Marco Sola S, Knowles DG, Raineri E, Guigo R, Ribeca P. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. The Journal of Biological Chemistry. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schubeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte DA, Rosales MAB, Papadimitriou A, Silva KC, Amancio VHO, Mendonça JN, Lopes NP, de Faria JBL, de Faria JML. Polyphenol-enriched cocoa protects the diabetic retina from glial reaction through the sirtuin pathway. The Journal of Nutritional Biochemistry. 2015;26:64–74. doi: 10.1016/j.jnutbio.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Eberhart A, Feodorova Y, Song C, Wanner G, Kiseleva E, Furukawa T, Kimura H, Schotta G, Leonhardt H, Joffe B, Solovei I. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 2013;21:535–554. doi: 10.1007/s10577-013-9375-7. [DOI] [PubMed] [Google Scholar]
- Elsner VR, Lovatel GA, Moysés F, Bertoldi K, Spindler C, Cechinel LR, Muotri AR, Siqueira IR. Exercise induces age-dependent changes on epigenetic parameters in rat hippocampus: a preliminary study. Exp Gerontol. 2013;48:136–139. doi: 10.1016/j.exger.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nature Structural & Molecular Biology. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, Sun YE. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schonegger A, Klughammer J, Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Ferreira RC, Popova EY, James J, Briones MR, Zhang SS, Barnstable CJ. Histone Deacetylase 1 Is Essential for Rod Photoreceptor Differentiation by Regulating Acetylation at Histone H3 Lysine 9 and Histone H4 Lysine 12 in the Mouse Retina. J Biol Chem. 2017;292:2422–2440. doi: 10.1074/jbc.M116.756643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Muller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247–252. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- Fischle W. One, two, three: how histone methylation is read. Epigenomics. 2012;4:641–653. doi: 10.2217/epi.12.56. [DOI] [PubMed] [Google Scholar]
- Frith MC, Mori R, Asai K. A mostly traditional approach improves alignment of bisulfite-converted DNA. Nucleic Acids Res. 2012;40:e100. doi: 10.1093/nar/gks275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Said S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanche H, Deleuze JF, Igo RP, Jr, Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA, Jr, Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Leveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N, Kuzelova A, Ebert A, Strnad H, Lachova J, Machon O, Busslinger M, Kozmik Z. Polycomb repression complex 2 is required for the maintenance of retinal progenitor cells and balanced retinal differentiation. Dev Biol. 2018;433:47–60. doi: 10.1016/j.ydbio.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genetics. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. The EMBO journal. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Research. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani R, Lai WW, Sum R, McGhee SM, Chan CW, Hedley AJ, Wong D. The incidental findings of age-related macular degeneration during diabetic retinopathy screening. Graefes Arch Clin Exp Ophthalmol. 2014;252:723–729. doi: 10.1007/s00417-013-2530-1. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Sankpal UT, Robertson AK, Zhao Y, Zhao Y, Robertson KD. DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochemical and Biophysical Research Communications. 2004;318:544–555. doi: 10.1016/j.bbrc.2004.04.058. [DOI] [PubMed] [Google Scholar]
- Gemenetzi M, Lotery AJ. The role of epigenetics in age-related macular degeneration. Eye (Lond) 2014;28:1407–1417. doi: 10.1038/eye.2014.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestaneh N, Chu Y, Cheng SK, Cao H, Poliakov E, Berinstein DM. Repressed SIRT1/PGC-1α pathway and mitochondrial disintegration in iPSC-derived RPE disease model of age-related macular degeneration. Journal of Translational Medicine. 2016;14:344. doi: 10.1186/s12967-016-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Gong H, Qian H, Ertl R, Astle CM, Wang GG, Harrison DE, Xu X. Histone modifications change with age, dietary restriction and rapamycin treatment in mouse brain. Oncotarget. 2015;6:15882–15890. doi: 10.18632/oncotarget.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan G, Mathews S, Srinivasan K, Ramasamy K, Periasamy S. Establishment of human retinal mitoscriptome gene expression signature for diabetic retinopathy using cadaver eyes. Mitochondrion. 2017 doi: 10.1016/j.mito.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans CY, Wallace VA, Gregory-Evans K. Gene networks: dissecting pathways in retinal development and disease. Prog Retin Eye Res. 2013;33:40–66. doi: 10.1016/j.preteyeres.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Groman-Lupa S, Adewumi J, Park KU, Brzezinski JA., IV The Transcription Factor Prdm16 Marks a Single Retinal Ganglion Cell Subtype in the Mouse Retina. Invest Ophthalmol Vis Sci. 2017;58:5421–5433. doi: 10.1167/iovs.17-22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhu P, Guo F, Li X, Wu X, Fan X, Wen L, Tang F. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc. 2015a;10:645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011a;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011b;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang SB, Xu H, Ribic A, Mohns EJ, Zhou Y, Zhu X, Biederer T, Crair MC, Chen B. A short N-terminal domain of HDAC4 preserves photoreceptors and restores visual function in retinitis pigmentosa. Nat Commun. 2015b;6:8005. doi: 10.1038/ncomms9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O, Gronke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, Andrews S, Zhang Q, Wakelam MJ, Beyer A, Reik W, Partridge L. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18:56. doi: 10.1186/s13059-017-1187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Han D, Yan Z, Boyd-Kirkup JD, Green CD, Khaitovich P, Han JD. Stress-associated H3K4 methylation accumulates during postnatal development and aging of rhesus macaque brain. Aging Cell. 2012;11:1055–1064. doi: 10.1111/acel.12007. [DOI] [PubMed] [Google Scholar]
- Hao H, Kim DS, Klocke B, Johnson KR, Cui K, Gotoh N, Zang C, Gregorski J, Gieser L, Peng W, Fann Y, Seifert M, Zhao K, Swaroop A. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Tummala P, Guzman E, Mali RS, Gregorski J, Swaroop A, Mitton KP. The transcription factor neural retina leucine zipper (NRL) controls photoreceptor-specific expression of myocyte enhancer factor Mef2c from an alternative promoter. J Biol Chem. 2011;286:34893–34902. doi: 10.1074/jbc.M111.271072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EY, Ponts N, Le Roch KG, Lonardi S. BRAT-BW: efficient and accurate mapping of bisulfite-treated reads. Bioinformatics. 2012;28:1795–1796. doi: 10.1093/bioinformatics/bts264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner W, Pevny L. Eye development and retinogenesis. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, Devys D. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Smith MM. Histone variants and epigenetics. Cold Spring Harb Perspect Biol. 2015;7:a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. PLoS One. 2013;8:e69721. doi: 10.1371/journal.pone.0069721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiler D, Chen X, Hazen J, Kupriyanov S, Carroll PA, Qu C, Xu B, Johnson D, Griffiths L, Frase S, Rodriguez AR, Martin G, Zhang J, Jeon J, Fan Y, Finkelstein D, Eisenman RN, Baldwin K, Dyer MA. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell Stem Cell. 2015;17:101–115. doi: 10.1016/j.stem.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS computational biology. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, Eszes M, Faull RLM, Curtis MA, Waldvogel HJ, Choi OW, Tung S, Vinters HV, Coppola G, Yang XW. Huntington’s disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 2016;8:1485–1512. doi: 10.18632/aging.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, Bates O, Walton A, Bermingham-McDonogh O, Glass IA, Wong ROL, Swaroop A, Reh TA. Molecular Anatomy of the Developing Human Retina. Dev Cell. 2017;43:763–779. doi: 10.1016/j.devcel.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, Xia S, Liu S, Lyu H, Ming GL, Wade H, Song H, Qian J, Zhu H. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6:73–88. doi: 10.2217/epi.13.72. [DOI] [PubMed] [Google Scholar]
- Hughes AE, Enright JM, Myers CA, Shen SQ, Corbo JC. Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors. Sci Rep. 2017;7:43184. doi: 10.1038/srep43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Spechler PA, Cwanger A, Song Y, Zhang Z, Ying GS, Hunter AK, Dezoeten E, Dunaief JL. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53:2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JB, Lindsay KJ, Du J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J Neurosci Res. 2015;93:1079–1092. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Fagerness J, Kirby A, Reynolds R, Zak A, Gimelbrant A, Plenge R, Daly M, Chess A, Seddon JM. (Epi)Genetic analyses of age-related macular degeneration: case-control and discordant twin studies. Hum Hered. 2014;78:59–72. doi: 10.1159/000362814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine. 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Baba Y, Satoh S, Mochizuki Y, Nakauchi H, Furukawa T, Koseki H, Murakami A, Watanabe S. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Dev Neurobiol. 2015;75:947–960. doi: 10.1002/dneu.22261. [DOI] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Kuribayashi H, Satoh S, Mochizuki Y, Baba Y, Nakauchi H, Furukawa T, Koseki H, Murakami A, Watanabe S. Histone demethylase Jmjd3 is required for the development of subsets of retinal bipolar cells. Proc Natl Acad Sci U S A. 2014;111:3751–3756. doi: 10.1073/pnas.1311480111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson GR, Owsley C, Curcio CA. Photoreceptor degeneration and dysfunction in aging and age-related maculopathy. Ageing Res Rev. 2002;1:381–396. doi: 10.1016/s1568-1637(02)00007-7. [DOI] [PubMed] [Google Scholar]
- Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S, Nellen W, Schaefer M, Helm M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2016:1–16. doi: 10.1080/15476286.2016.1191737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science (New York, NY) 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA. Stimulation of functional neuronal regeneration from Muller glia in adult mice. Nature. 2017;548:103–107. doi: 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CC, Atan D, Ng D, Ploder L, Ross SE, Klein M, Birch DG, Diez E, McInnes RR. Transcription factor PRDM8 is required for rod bipolar and type 2 OFF-cone bipolar cell survival and amacrine subtype identity. Proc Natl Acad Sci U S A. 2015;112:E3010–3019. doi: 10.1073/pnas.1505870112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala CS, Zheng L, Du Y, Yohannes E, Kao HY, Miyagi M, Kern TS. Acetylation of retinal histones in diabetes increases inflammatory proteins: effects of minocycline and manipulation of histone acetyltransferase (HAT) and histone deacetylase (HDAC) J Biol Chem. 2012;287:25869–25880. doi: 10.1074/jbc.M112.375204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells. 2015;33:3504–3518. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Friedman JS, Nishiguchi KM, Swaroop A. Retinopathy mutations in the bZIP protein NRL alter phosphorylation and transcriptional activity. Hum Mutat. 2007;28:589–598. doi: 10.1002/humu.20488. [DOI] [PubMed] [Google Scholar]
- Kandpal RP, Rajasimha HK, Brooks MJ, Nellissery J, Wan J, Qian J, Kern TS, Swaroop A. Transcriptome analysis using next generation sequencing reveals molecular signatures of diabetic retinopathy and efficacy of candidate drugs. Mol Vis. 2012;18:1123–1146. [PMC free article] [PubMed] [Google Scholar]
- Karimzadeh M, Ernst C, Kundaje A, Hoffman MM. Umap and Bismap: Quantifying genome and methylome mappability. bioRxiv. 2016 doi: 10.1093/nar/gky677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Yamazaki R, Onishi A, Sanuki R, Furukawa T. G9a histone methyltransferase activity in retinal progenitors is essential for proper differentiation and survival of mouse retinal cells. J Neurosci. 2012;32:17658–17670. doi: 10.1523/JNEUROSCI.1869-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Chen S, Xin X, Chiu J, Khan ZA, Chakrabarti S. Diabetes-induced extracellular matrix protein expression is mediated by transcription coactivator p300. Diabetes. 2006;55:3104–3111. doi: 10.2337/db06-0519. [DOI] [PubMed] [Google Scholar]
- Kawashima M, Ozawa Y, Shinmura K, Inaba T, Nakamura S, Kawakita T, Watanabe M, Tsubota K. Calorie restriction (CR) and CR mimetics for the prevention and treatment of age-related eye disorders. Exp Gerontol. 2013;48:1096–1100. doi: 10.1016/j.exger.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Kim CH, An MJ, Kim DH, Kim JW. Histone deacetylase 1 (HDAC1) regulates retinal development through a PAX6-dependent pathway. Biochem Biophys Res Commun. 2017;482:735–741. doi: 10.1016/j.bbrc.2016.11.103. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kim JW, Jang SM, An JH, Song KH, Choi KH. Transcriptional activity of paired homeobox Pax6 is enhanced by histone acetyltransferase Tip60 during mouse retina development. Biochem Biophys Res Commun. 2012a;424:427–432. doi: 10.1016/j.bbrc.2012.06.126. [DOI] [PubMed] [Google Scholar]
- Kim CY, Kuehn MH, Clark AF, Kwon YH. Gene expression profile of the adult human retinal ganglion cell layer. Mol Vis. 2006;12:1640–1648. [PubMed] [Google Scholar]
- Kim DI, Park MJ, Choi JH, Kim IS, Han HJ, Yoon KC, Park SW, Lee MY, Oh KS, Park SH. PRMT1 and PRMT4 Regulate Oxidative Stress-Induced Retinal Pigment Epithelial Cell Damage in SIRT1-Dependent and SIRT1-Independent Manners. Oxidative Medicine and Cellular Longevity. 2015;2015:617919. doi: 10.1155/2015/617919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-i, Park M-j, Lim S-k, Choi J-h, Kim J-c, Han H-j, Kundu TK, Park J-i, Yoon K-c, Park S-w, Park J-s, Heo Y-r, Park S-h. High-glucose-induced CARM1 expression regulates apoptosis of human retinal pigment epithelial cells via histone 3 arginine 17 dimethylation: role in diabetic retinopathy. Archives of Biochemistry and Biophysics. 2014;560:36–43. doi: 10.1016/j.abb.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Kim JW, Jang SM, Kim CH, An JH, Choi KH. Transcriptional activity of neural retina leucine zipper (Nrl) is regulated by c-Jun N-terminal kinase and Tip60 during retina development. Mol Cell Biol. 2012b;32:1720–1732. doi: 10.1128/MCB.06440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Yang HJ, Brooks MJ, Zelinger L, Karakulah G, Gotoh N, Boleda A, Gieser L, Giuste F, Whitaker DT, Walton A, Villasmil R, Barb JJ, Munson PJ, Kaya KD, Chaitankar V, Cogliati T, Swaroop A. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Rep. 2016a;17:2460–2473. doi: 10.1016/j.celrep.2016.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Yang HJ, Oel AP, Brooks MJ, Jia L, Plachetzki DC, Li W, Allison WT, Swaroop A. Recruitment of Rod Photoreceptors from Short-Wavelength-Sensitive Cones during the Evolution of Nocturnal Vision in Mammals. Dev Cell. 2016b;37:520–532. doi: 10.1016/j.devcel.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Ju WK, Neufeld AH. Neuronal susceptibility to damage: comparison of the retinas of young, old and old/caloric restricted rats before and after transient ischemia. Neurobiol Aging. 2004;25:491–500. doi: 10.1016/j.neurobiolaging.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci U S A. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AD, Huang K, Rubbi L, Liu S, Wang CY, Wang Y, Pellegrini M, Fan G. Reversible Regulation of Promoter and Enhancer Histone Landscape by DNA Methylation in Mouse Embryonic Stem Cells. Cell Rep. 2016;17:289–302. doi: 10.1016/j.celrep.2016.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilyaprak C, Spehner D, Devys D, Schultz P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PloS one. 2010;5:e11039. doi: 10.1371/journal.pone.0011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilyaprak C, Spehner D, Devys D, Schultz P. The linker histone H1C contributes to the SCA7 nuclear phenotype. Nucleus. 2011;2:444–454. doi: 10.4161/nucl.2.5.17843. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YX, van Bergen N, Bui BV, Chrysostomou V, Vingrys AJ, Trounce IA, Crowston JG. Impact of aging and diet restriction on retinal function during and after acute intraocular pressure injury. Neurobiol Aging. 2012;33:1126e1115–1125. doi: 10.1016/j.neurobiolaging.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kooragayala K, Gotoh N, Cogliati T, Nellissery J, Kaden TR, French S, Balaban R, Li W, Covian R, Swaroop A. Quantification of Oxygen Consumption in Retina Ex Vivo Demonstrates Limited Reserve Capacity of Photoreceptor Mitochondria. Invest Ophthalmol Vis Sci. 2015;56:8428–8436. doi: 10.1167/iovs.15-17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kowluru RA. Diabetic retinopathy, metabolic memory and epigenetic modifications. Vision Res. 2017 doi: 10.1016/j.visres.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Kowluru RA, Kowluru A, Mishra M, Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog Retin Eye Res. 2015;48:40–61. doi: 10.1016/j.preteyeres.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhevnikova OS, Korbolina EE, Ershov NI, Kolosova NG. Rat retinal transcriptome: effects of aging and AMD-like retinopathy. Cell Cycle. 2013;12:1745–1761. doi: 10.4161/cc.24825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba DA, Hayes S, Karl MO, Reh T. Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev Dyn. 2008;237:3016–3023. doi: 10.1002/dvdy.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoke F, Shaw S, Yuan J, Ananth S, Duncan M, Martin P, Bartoli M. Increased Oxidative and Nitrative Stress Accelerates Aging of the Retinal Vasculature in the Diabetic Retina. PLoS One. 2015;10:e0139664. doi: 10.1371/journal.pone.0139664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010;Chapter 11(Unit 11):17. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, Bartlett JJ, Pavlenok M, Gundlach JH. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci U S A. 2013;110:18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SJ, Kenta N. An integrative approach for efficient analysis of whole genome bisulfite sequencing data. BMC Genomics. 2015;16(Suppl 12):S14. doi: 10.1186/1471-2164-16-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AHFM. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Current biology: CB. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY) 2015;7:1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Daniel M, Tollefsbol TO. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011;9:98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GI, Wang M, Matragoon S. Timing of interphotoreceptor retinoid-binding protein (IRBP) gene expression and hypomethylation in developing mouse retina. Dev Biol. 1994;161:345–356. doi: 10.1006/dbio.1994.1036. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247. e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi PM, Yan Q, Wajapeyee N. DNA Bisulfite Sequencing for Single-Nucleotide-Resolution DNA Methylation Detection. Cold Spring Harb Protoc. 2017;2017 doi: 10.1101/pdb.prot094839. pdb prot094839. [DOI] [PubMed] [Google Scholar]
- Lluch-Senar M, Luong K, Llorens-Rico V, Delgado J, Fang G, Spittle K, Clark TA, Schadt E, Turner SW, Korlach J, Serrano L. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 2013;9:e1003191. doi: 10.1371/journal.pgen.1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341:1110–1112. doi: 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- Lu AT, Hannon E, Levine ME, Crimmins EM, Lunnon K, Mill J, Geschwind DH, Horvath S. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nature Communications. 2017;8:15353. doi: 10.1038/ncomms15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AK, Zhang P, Cardoso MC. Modifiers and Readers of DNA Modifications and Their Impact on Genome Structure, Expression, and Stability in Disease. Front Genet. 2016;7:115. doi: 10.3389/fgene.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, Jelinek J, Colman RJ, Issa JJ. Caloric restriction delays age-related methylation drift. Nat Commun. 2017;8:539. doi: 10.1038/s41467-017-00607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold CA, Masser DR, Stanford DR, Bixler GV, Pisupati A, Giles CB, Wren JD, Ford MM, Sonntag WE, Freeman WM. CNS-wide Sexually Dimorphic Induction of the Major Histocompatibility Complex 1 Pathway With Aging. The Journals of Gerontology Series A, Biological Sciences and Medical Sciences. 2017;72:16–29. doi: 10.1093/gerona/glv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 2012;22:1334–1349. doi: 10.1101/gr.127191.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K, Schroeder DI, LaSalle JM, Lalande M, Xu RH. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7:71–82. doi: 10.4161/epi.7.1.18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The neuronal organization of the retina. Neuron. 2012;76:266–280. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbs SL, Khan MA, Hackler L, Jr, Oliver VF, Wan J, Qian J, Zack DJ. Cell-specific DNA methylation patterns of retina-specific genes. PloS one. 2012;7:e32602. doi: 10.1371/journal.pone.0032602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura T, Kaji Y, Noma H, Funatsu H, Okamoto S. The role of SIRT1 in ocular aging. Exp Eye Res. 2013;116:17–26. doi: 10.1016/j.exer.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Mitteldorf J. Is programmed aging a cause for optimism? Curr Aging Sci. 2015;8:69–75. doi: 10.2174/1874609808666150422112826. [DOI] [PubMed] [Google Scholar]
- Mo A, Luo C, Davis FP, Mukamel EA, Henry GL, Nery JR, Urich MA, Picard S, Lister R, Eddy SR, Beer MA, Ecker JR, Nathans J. Epigenomic landscapes of retinal rods and cones. Elife. 2016;5:e11613. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, Washburn MP, Workman JL, Abmayr SM. Loss of Drosophila Ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev. 2014;28:259–272. doi: 10.1101/gad.225151.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- Morse SJ, Butler AA, Davis RL, Soller IJ, Lubin FD. Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology. 2015;4:298–313. doi: 10.3390/biology4020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortuza R, Chen S, Feng B, Sen S, Chakrabarti S. High glucose induced alteration of SIRTs in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS One. 2013;8:e54514. doi: 10.1371/journal.pone.0054514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortuza R, Feng B, Chakrabarti S. SIRT1 reduction causes renal and retinal injury in diabetes through endothelin 1 and transforming growth factor β1. Journal of Cellular and Molecular Medicine. 2015;19:1857–1867. doi: 10.1111/jcmm.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes & Development. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney DL. Epigenetic Control Systems. Proc Natl Acad Sci U S A. 1958;44:712–717. doi: 10.1073/pnas.44.7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasonkin IO, Lazo K, Hambright D, Brooks M, Fariss R, Swaroop A. Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina. J Comp Neurol. 2011;519:1914–1930. doi: 10.1002/cne.22613. [DOI] [PubMed] [Google Scholar]
- Nasonkin IO, Merbs SL, Lazo K, Oliver VF, Brooks M, Patel K, Enke RA, Nellissery J, Jamrich M, Le YZ, Bharti K, Fariss RN, Rachel RA, Zack DJ, Rodriguez-Boulan EJ, Swaroop A. Conditional knockdown of DNA methyltransferase 1 reveals a key role of retinal pigment epithelium integrity in photoreceptor outer segment morphogenesis. Development. 2013;140:1330–1341. doi: 10.1242/dev.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Lu A, Swaroop A, Sharlin DS, Swaroop A, Forrest D. Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J Neurosci. 2011;31:11118–11125. doi: 10.1523/JNEUROSCI.1709-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Oh EC, Cheng H, Hao H, Jia L, Khan NW, Swaroop A. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 2008;1236:16–29. doi: 10.1016/j.brainres.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliver VF, Franchina M, Jaffe AE, Branham KE, Othman M, Heckenlively JR, Swaroop A, Campochiaro B, Vote BJ, Craig JE, Saffery R, Mackey DA, Qian J, Zack DJ, Hewitt AW, Merbs SL. Hypomethylation of the IL17RC promoter in peripheral blood leukocytes is not a hallmark of age-related macular degeneration. Cell Rep. 2013a;5:1527–1535. doi: 10.1016/j.celrep.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver VF, Jaffe AE, Song J, Wang G, Zhang P, Branham KE, Swaroop A, Eberhart CG, Zack DJ, Qian J, Merbs SL. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics. 2015;10:698–707. doi: 10.1080/15592294.2015.1060388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver VF, Wan J, Agarwal S, Zack DJ, Qian J, Merbs SL. A novel methyl-binding domain protein enrichment method for identifying genome-wide tissue-specific DNA methylation from nanogram DNA samples. Epigenetics Chromatin. 2013b;6:17. doi: 10.1186/1756-8935-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkhov-Mitsel E, Bapat B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 2012;1:237–260. doi: 10.1002/cam4.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Kubo S, Kon T, Furuhashi M, Narita H, Kominami T, Ueno A, Tsutsumi R, Chaya T, Yamamoto H, Suetake I, Ueno S, Koseki H, Nakagawa A, Furukawa T. Samd7 is a cell type-specific PRC1 component essential for establishing retinal rod photoreceptor identity. Proc Natl Acad Sci U S A. 2017;114:E8264–E8273. doi: 10.1073/pnas.1707021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Kasinathan S, Zentner GE, Henikoff S, Ahmad K. Mapping regulatory factors by immunoprecipitation from native chromatin. Curr Protoc Mol Biol. 2015;110:21 31 21–25. doi: 10.1002/0471142727.mb2131s110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsley C. Vision and Aging. Annual Review of Vision Science. 2016;2:255–271. doi: 10.1146/annurev-vision-111815-114550. [DOI] [PubMed] [Google Scholar]
- Owsley C, Huisingh C, Jackson GR, Curcio CA, Szalai AJ, Dashti N, Clark M, Rookard K, McCrory MA, Wright TT, Callahan MA, Kline LB, Witherspoon CD, McGwin G. Associations between abnormal rod-mediated dark adaptation and health and functioning in older adults with normal macular health. Invest Ophthalmol Vis Sci. 2014;55:4776–4789. doi: 10.1167/iovs.14-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2:e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parapuram SK, Cojocaru RI, Chang JR, Khanna R, Brooks M, Othman M, Zareparsi S, Khan NW, Gotoh N, Cogliati T, Swaroop A. Distinct signature of altered homeostasis in aging rod photoreceptors: implications for retinal diseases. PLoS One. 2010;5:e13885. doi: 10.1371/journal.pone.0013885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B, Hsieh TF, Ibarra C, Fischer RL. MethylCoder: software pipeline for bisulfite-treated sequences. Bioinformatics. 2011;27:2435–2436. doi: 10.1093/bioinformatics/btr394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pelzel HR, Schlamp CL, Nickells RW. Histone H4 deacetylation plays a critical role in early gene silencing during neuronal apoptosis. BMC neuroscience. 2010;11:62. doi: 10.1186/1471-2202-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CH, Chang YL, Kao CL, Tseng LM, Wu CC, Chen YC, Tsai CY, Woung LC, Liu JH, Chiou SH, Chen SJ. SirT1--a sensor for monitoring self-renewal and aging process in retinal stem cells. Sensors (Basel, Switzerland) 2010;10:6172–6194. doi: 10.3390/s100606172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CH, Cherng JY, Chiou GY, Chen YC, Chien CH, Kao CL, Chang YL, Chien Y, Chen LK, Liu J-h, Chen SJ, Chiou SH. Delivery of Oct4 and SirT1 with cationic polyurethanes-short branch PEI to aged retinal pigment epithelium. Biomaterials. 2011;32:9077–9088. doi: 10.1016/j.biomaterials.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A, Eisen D, Wagner M, Laube SK, Kunzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, Muller M, Biel M, Carell T, Michalakis S. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Pervouchine DD, Djebali S, Breschi A, Davis CA, Barja PP, Dobin A, Tanzer A, Lagarde J, Zaleski C, See LH, Fastuca M, Drenkow J, Wang H, Bussotti G, Pei B, Balasubramanian S, Monlong J, Harmanci A, Gerstein M, Beer MA, Notredame C, Guigo R, Gingeras TR. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat Commun. 2015;6:5903. doi: 10.1038/ncomms6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova TD, Seigel GM, Otteson DC. A role for DNA methylation in regulation of EphA5 receptor expression in the mouse retina. Vision Res. 2011;51:260–268. doi: 10.1016/j.visres.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna A, Zinellu A, Tendas D, Blasetti F, Carru C, Castiglia P. Plasma Homocysteine and Asymmetrical Dimethyl-l-Arginine (ADMA) and Whole Blood DNA Methylation in Early and Neovascular Age-Related Macular Degeneration: A Pilot Study. Current Eye Research. 2016;41:88–96. doi: 10.3109/02713683.2014.1002044. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. The Necessity of Chromatin: A View in Perspective. Cold Spring Harb Perspect Biol. 2016;8:a019547. doi: 10.1101/cshperspect.a019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Grigoryev SA, Fan Y, Skoultchi AI, Zhang SS, Barnstable CJ. Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem. 2013;288:17895–17907. doi: 10.1074/jbc.M113.452144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Pinzon-Guzman C, Salzberg AC, Zhang SS, Barnstable CJ. LSD1-Mediated Demethylation of H3K4me2 Is Required for the Transition from Late Progenitor to Differentiated Mouse Rod Photoreceptor. Mol Neurobiol. 2016;53:4563–4581. doi: 10.1007/s12035-015-9395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Xu X, DeWan AT, Salzberg AC, Berg A, Hoh J, Zhang SS, Barnstable CJ. Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One. 2012;7:e46867. doi: 10.1371/journal.pone.0046867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Elsaeidi F, Goldman D. Injury-dependent Muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J Neurosci. 2012;32:1096–1109. doi: 10.1523/JNEUROSCI.5603-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, Jones DA. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, Ramsahoye BH, Whitelaw E, Greally JM, Adams IR, Bickmore WA, Meehan RR. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE, Keeley PW. Genomic control of neuronal demographics in the retina. Prog Retin Eye Res. 2016;55:246–259. doi: 10.1016/j.preteyeres.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KD, Yu J, Zhao CY, Fan G, Yang XJ. Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival. Cell Death Dis. 2012;3:e427. doi: 10.1038/cddis.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer S, Patel S, Sockanathan S, Ulanski LJ, 2nd, Miller L, Podella C. Resveratrol based oral nutritional supplement produces long-term beneficial effects on structure and visual function in human patients. Nutrients. 2014;6:4404–4420. doi: 10.3390/nu6104404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer S, Stiles W, Ulanski L, Carroll D, Podella C. Observation of human retinal remodeling in octogenarians with a resveratrol based nutritional supplement. Nutrients. 2013;5:1989–2005. doi: 10.3390/nu5061989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Strbenac D, Stirzaker C, Statham AL, Song J, Speed TP, Clark SJ. Copy-number-aware differential analysis of quantitative DNA sequencing data. Genome Res. 2012;22:2489–2496. doi: 10.1101/gr.139055.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues HF, Souza TA, Ghiraldini FG, Mello ML, Moraes AS. Increased age is associated with epigenetic and structural changes in chromatin from neuronal nuclei. J Cell Biochem. 2014;115:659–665. doi: 10.1002/jcb.24705. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler R, Smallwood SA, Veenvliet JV, Pechlivanoglou P, Peng SP, Chakrabarty K, Groot-Koerkamp MJ, Pasterkamp RJ, Wesseling E, Kelsey G, Boddeke E, Smidt MP, Copray S. Detailed analysis of the genetic and epigenetic signatures of iPSC-derived mesodiencephalic dopaminergic neurons. Stem Cell Reports. 2014;2:520–533. doi: 10.1016/j.stemcr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis J, Hu L, Cohen S, Lin Y, Harrar D, McInnes RR, Greenberg ME. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genetics. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Ruiz MA, Feng B, Chakrabarti S. Polycomb repressive complex 2 regulates MiR-200b in retinal endothelial cells: potential relevance in diabetic retinopathy. PLoS One. 2015;10:e0123987. doi: 10.1371/journal.pone.0123987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Ehninger D. Bison: bisulfite alignment on nodes of a cluster. BMC Bioinformatics. 2014;15:337. doi: 10.1186/1471-2105-15-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SH, Kang K, Yoo T, Joe CO, Chung JH. Transcriptional repression of repeat-derived transcripts correlates with histone hypoacetylation at repetitive DNA elements in aged mice brain. Experimental Gerontology. 2011;46:811–818. doi: 10.1016/j.exger.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Sajgo S, Ghinia MG, Brooks M, Kretschmer F, Chuang K, Hiriyanna S, Wu Z, Popescu O, Badea TC. Molecular codes for cell type specification in Brn3 retinal ganglion cells. Proc Natl Acad Sci U S A. 2017;114:E3974–E3983. doi: 10.1073/pnas.1618551114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biology. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16:716–726. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor Perspectives in Biology. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Duan X, Zhang Y, Liu XS. Computational methodology for ChIP-seq analysis. Quant Biol. 2013;1:54–70. doi: 10.1007/s40484-013-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, Roska B. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15:487–495. S481–482. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Sifuentes CJ, Kim JW, Swaroop A, Raymond PA. Rapid, Dynamic Activation of Muller Glial Stem Cell Responses in Zebrafish. Invest Ophthalmol Vis Sci. 2016;57:5148–5160. doi: 10.1167/iovs.16-19973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman DM, Ross K, Sande PH, Kubota S, Ramaswamy S, Apte RS, Mostoslavsky R. SIRT6 is required for normal retinal function. PLoS One. 2014;9:e98831. doi: 10.1371/journal.pone.0098831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Mallela RK, Hayes A, Dunham NR, Hedden ME, Enke RA, Fariss RN, Sternberg H, West MD, Nasonkin IO. Dnmt1, Dnmt3a and Dnmt3b cooperate in photoreceptor and outer plexiform layer development in the mammalian retina. Exp Eye Res. 2017;159:132–146. doi: 10.1016/j.exer.2016.11.014. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017:6. doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–4454. doi: 10.1242/dev.01302. [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Snigdha S, Prieto GA, Petrosyan A, Loertscher BM, Dieskau AP, Overman LE, Cotman CW. H3K9me3 Inhibition Improves Memory, Promotes Spine Formation, and Increases BDNF Levels in the Aged Hippocampus. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2016;36:3611–3622. doi: 10.1523/JNEUROSCI.2693-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrin L, Seddon JM. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev AA, Josefowicz SZ, Allis CD. Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome. Mol Cell. 2016;62:681–694. doi: 10.1016/j.molcel.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stelzer Y, Shivalila CS, Soldner F, Markoulaki S, Jaenisch R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015;163:218–229. doi: 10.1016/j.cell.2015.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storvall H, Ramskold D, Sandberg R. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PLoS One. 2013;8:e53822. doi: 10.1371/journal.pone.0053822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Stubbs TM, Bonder MJ, Stark AK, Krueger F, Team BIAC, von Meyenn F, Stegle O, Reik W. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017;18:68. doi: 10.1186/s13059-017-1203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syreeni A, El-Osta A, Forsblom C, Sandholm N, Parkkonen M, Tarnow L, Parving HH, McKnight AJ, Maxwell AP, Cooper ME, Groop PH FinnDiane Study G. Genetic examination of SETD7 and SUV39H1/H2 methyltransferases and the risk of diabetes complications in patients with type 1 diabetes. Diabetes. 2011;60:3073–3080. doi: 10.2337/db11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG. “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623. doi: 10.1016/j.neuron.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu KL, Vucic EA, Kennett JY, Heryet C, Brown CJ, Lam WL, Wilson IM. Methylated DNA immunoprecipitation. J Vis Exp. 2009 doi: 10.3791/935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Iwagawa T, Kuribayashi H, Baba Y, Nakauchi H, Murakami A, Nagasaki M, Suzuki Y, Watanabe S. Transition of differential histone H3 methylation in photoreceptors and other retinal cells during retinal differentiation. Sci Rep. 2016;6:29264. doi: 10.1038/srep29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Iwagawa T, Ochiai G, Koso H, Nakauchi H, Nagasaki M, Suzuki Y, Watanabe S. Analysis of Muller glia specific genes and their histone modification using Hes1-promoter driven EGFP expressing mouse. Sci Rep. 2017;7:3578. doi: 10.1038/s41598-017-03874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui A, Iwagawa T, Mochizuki Y, Iida A, Wegner M, Murakami A, Watanabe S. Expression of Sox4 and Sox11 is regulated by multiple mechanisms during retinal development. FEBS Lett. 2013;587:358–363. doi: 10.1016/j.febslet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Molecular Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Van Schil K, Karlstetter M, Aslanidis A, Dannhausen K, Azam M, Qamar R, Leroy BP, Depasse F, Langmann T, De Baere E. Autosomal recessive retinitis pigmentosa with homozygous rhodopsin mutation E150K and non-coding cis-regulatory variants in CRX-binding regions of SAMD7. Sci Rep. 2016;6:21307. doi: 10.1038/srep21307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury WJ, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkner M, Zschatzsch M, Rostovskaya M, Overall RW, Busskamp V, Anastassiadis K, Karl MO. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Reports. 2016;6:525–538. doi: 10.1016/j.stemcr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. Organisers and genes. The Cambridge University Press; 1940. [Google Scholar]
- Waddington C. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Wallace DM, Donovan M, Cotter TG. Histone deacetylase activity regulates apaf-1 and caspase 3 expression in the developing mouse retina. Invest Ophthalmol Vis Sci. 2006;47:2765–2772. doi: 10.1167/iovs.05-1383. [DOI] [PubMed] [Google Scholar]
- Wan J, Oliver VF, Wang G, Zhu H, Zack DJ, Merbs SL, Qian J. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genomics. 2015;16:49. doi: 10.1186/s12864-015-1271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Oliver VF, Zhu H, Zack DJ, Qian J, Merbs SL. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013;41:8503–8514. doi: 10.1093/nar/gkt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jia ST, Jia S. New Insights into the Regulation of Heterochromatin. Trends in genetics: TIG. 2016;32:284–294. doi: 10.1016/j.tig.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603. doi: 10.1523/JNEUROSCI.4488-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Method to detect differentially methylated loci with case-control designs using Illumina arrays. Genet Epidemiol. 2011;35:686–694. doi: 10.1002/gepi.20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sidoli S, Zhang W, Wang Q, Wang L, Jensen ON, Guo L, Zhao X, Zheng L. Abnormal levels of histone methylation in the retinas of diabetic rats are reversed by minocycline treatment. Sci Rep. 2017;7:45103. doi: 10.1038/srep45103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Bloch W. Histone methylation and acetylation indicates epigenetic change in the aged cochlea of mice. Eur Arch Otorhinolaryngol. 2013;270:1823–1830. doi: 10.1007/s00405-012-2222-1. [DOI] [PubMed] [Google Scholar]
- Wei L, Liu B, Tuo J, Shen D, Chen P, Li Z, Liu X, Ni J, Dagur P, Sen HN, Jawad S, Ling D, Park S, Chakrabarty S, Meyerle C, Agron E, Ferris FL, Chew EY, McCoy JP, Blum E, Francis PJ, Klein ML, Guymer RH, Baird PN, Chan CC, Nussenblatt RB. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012;2:1151–1158. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TY, Cheung CMG, Larsen M, Sharma S, Simó R. Diabetic retinopathy. Nature Reviews Disease Primers. 2016;2:16012. doi: 10.1038/nrdp.2016.12. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, Tackett A, Wang G, Hon LS, Fang G, Swenberg JA, Xiao AZ. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Tian J, Luo Y, Wei L, Lin S, Tang S. Effects of 5-aza-2′-deoxycytidine and trichostatin A on high glucose- and interleukin-1β-induced secretory mediators from human retinal endothelial cells and retinal pigment epithelial cells. Mol Vis. 2014;20:1411–1421. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, Masai I. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Yan N, Cheng L, Cho K, Malik MT, Xiao L, Guo C, Yu H, Zhu R, Rao RC, Chen DF. Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1. Sci Rep. 2016;6:33887. doi: 10.1038/srep33887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Ratnapriya R, Cogliati T, Kim JW, Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog Retin Eye Res. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Ohashi A, Nishida S, Kamiya T, Suwa T, Hara H, Takeda J, Itoh Y, Adachi T. Exendin-4 induces extracellular-superoxide dismutase through histone H3 acetylation in human retinal endothelial cells. Journal of Clinical Biochemistry and Nutrition. 2016;59:174–181. doi: 10.3164/jcbn.16-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Yashar BM, Hiriyanna S, Swaroop A. Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci. 2002;43:2554–2560. [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, Oshlack A, Majewski IJ. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–7427. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 1985;353:229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelinger L, Swaroop A. RNA Biology in Retinal Development and Disease. Trends in Genetics. 2018 doi: 10.1016/j.tig.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yang K. Sirtuin 1 participates in the process of age-related retinal degeneration. Biochem Biophys Res Commun. 2015;468:167–172. doi: 10.1016/j.bbrc.2015.10.139. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yang K, Wang F, Zhou L, Hu Y, Tang M, Zhang S, Jin S, Zhang J, Wang J, Li W, Lu L, Xu GT. The glucagon like peptide 1 analogue, exendin-4, attenuates oxidative stress-induced retinal cell death in early diabetic rats through promoting Sirt1 and Sirt3 expression. Exp Eye Res. 2016;151:203–211. doi: 10.1016/j.exer.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- Zhang J, Taylor RJ, La Torre A, Wilken MS, Cox KE, Reh TA, Vetter ML. Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation. Dev Biol. 2015a;403:128–138. doi: 10.1016/j.ydbio.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen XA, Wang Y, Lin L, Chen L, Jin P, Wu GY, Chen G. Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell. 2015b;17:735–747. doi: 10.1016/j.stem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015c;16:1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Research. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, Su J, Li X, Wu Q, Wang F, Cui Y. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res. 2011;39:e58. doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Li J, Wang N, Zheng B, Li T, Gu Q, Xu X, Zheng Z. Fenofibrate suppresses cellular metabolic memory of high glucose in diabetic retinopathy via a sirtuin 1-dependent signalling pathway. Molecular Medicine Reports. 2015;12:6112–6118. doi: 10.3892/mmr.2015.4164. [DOI] [PubMed] [Google Scholar]
- Zhao S, Li T, Li J, Lu Q, Han C, Wang N, Qiu Q, Cao H, Xu X, Chen H, Zheng Z. miR-23b-3p induces the cellular metabolic memory of high glucose in diabetic retinopathy through a SIRT1-dependent signalling pathway. Diabetologia. 2016;59:644–654. doi: 10.1007/s00125-015-3832-0. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harbor Perspectives in Biology. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110:1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–565. doi: 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Widschwendter M, Teschendorff AE. A comparison of feature selection and classification methods in DNA methylation studies using the Illumina Infinium platform. BMC Bioinformatics. 2012;13:59. doi: 10.1186/1471-2105-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuge CC, Xu JY, Zhang J, Li W, Li P, Li Z, Chen L, Liu X, Shang P, Xu H, Lu Y, Wang F, Lu L, Xu GT. Fullerenol protects retinal pigment epithelial cells from oxidative stress-induced premature senescence via activating SIRT1. Invest Ophthalmol Vis Sci. 2014;55:4628–4638. doi: 10.1167/iovs.13-13732. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, Gnirke A, Meissner A. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla-Zubilete MA, Yeste A, Quintana FJ, Toiber D, Mostoslavsky R, Silberman DM. Epigenetic control of early neurodegenerative events in diabetic retinopathy by the histone deacetylase SIRT6. J Neurochem. 2018;144:128–138. doi: 10.1111/jnc.14243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.