Abstract
The zebrafish larva is a powerful tool for the study of dietary triglyceride (TG) digestion and how fatty acids (FA) derived from dietary lipids are absorbed, metabolized and distributed to the body. While fluorescent FA analogues have enabled visualization of FA metabolism, methods for specifically assaying TG digestion are badly needed. Here we present a novel High Performance Liquid Chromatography (HPLC) method that quantitatively differentiates TG and phospholipid (PL) molecules with one or two fluorescent FA analogues. We show how this tool may be used to discriminate between undigested and digested TG or phosphatidylcholine (PC), and also the products of TG or PC that have been digested, absorbed and re-synthesized into new lipid molecules. Using this approach, we explored the dietary requirement of zebrafish larvae for phospholipids. Here we demonstrate that dietary TG is digested and absorbed in the intestinal epithelium, but without dietary PC, TG accumulates and is not transported out of the enterocytes. Consequently, intestinal ER stress increases and the ingested lipid is not available support the energy and metabolic needs of other tissues. In TG diets with PC, TG is readily transported from the intestine and subsequently metabolized.
Keywords: Lipase/Digestive, Lipid droplets, Nutrition, Phospholipids/Absorption, Triglycerides, Zebrafish
1. Introduction
High postprandial plasma lipid levels have proven to be a major risk factor associated with cardiovascular disease [1,2], necessitating a better understanding of lipid digestion and subsequent export from the intestinal epithelium to the circulation. The zebrafish larva is a powerful tool for the study of the cell biology and metabolism of lipids, because it allows for the visualization of digestive processes at both the whole organ and subcellular levels, which can be coupled with tractable biochemical assays of lipid metabolism [3,4]. Recently the later larval stages, as well as the adult zebrafish, have come more into focus as many organ functions have been found analogous to their mammalian equivalents [4,5]. Additionally, gene expression and regulation in the intestinal epithelial cells is highly conserved between zebrafish and mammals, including functional specialization along the length of the digestive tract [6]. At five to seven days post fertilization (dpf), the optically clear zebrafish larva is ready for its first exogenous meal providing opportunities for live physiological studies not possible in mammals [3]. Furthermore, the minimally convoluted digestive tract is ideal for microscopy to explore the function of specific dietary lipids in digestion and fat absorption. While the small size and transparency of the zebrafish larva facilitates live imaging of lipid processing and transport in larvae fed fluorescently labeled lipids, complementary biochemical studies employing either fluorescent or radiolabeled lipids as metabolic tracers allow quantification of lipid processing [3,7,8]. Feeding fish larvae 14C-labeled lipids before placement in a metabolic chamber allows for the quantification of lipid that is evacuated in the feces, used in beta oxidation, and deposited as complex lipids (e.g. neutral/polar lipids)[9], and both fluorescent and radioactive lipids may be fed to larvae for quantitation of complex lipid products by chromatographic methods [10,11]. However, due to the small size of the zebrafish larva it is not practical to separate the contents of the intestinal lumen from the intestinal tissue before extraction of lipids for biochemical experiments. Because of this, the distinction between undigested lipid in the intestinal lumen and lipid contained within intestinal tissue has not been possible. When studying hydrolysis, absorption and transport of dietary lipids from the intestine to the body, it is crucial to be able to quantify these processes. Here we present such a method, broadening the utility of the zebrafish model system.
Lipids are digested in the vertebrate intestinal lumen by specific lipases; secretory phospholipase A2 group IB (sPLA2 IB) hydrolyzes dietary phospholipids (PL) to one free fatty acid (FFA) and one lyso-phospholipid (lyso-PL) [7,12]. Dietary triglyceride (TG) digestion varies among species: in mammals this process is initiated by gastric lipase (LIPF), belonging to the highly homologous family of acidic lipases [13]. Even though a putative LIPF gene is present in the zebrafish genome, there is no evidence of gastric lipase activity in any teleost fish [14]. While in most vertebrates TG are only partly digested to two FFA and monoacylglycerol in the intestinal lumen due to the hydrolytic activity of pancreatic lipase in combination with colipase [15], dietary triglycerides are processed differently in some species of fish. For example, there is a complete digestion of TG to three FFA and one glycerol molecule by bile-activated lipase in the Atlantic cod (Gadus morhua), and the pancreatic lipase is not predicted to be functional due to mutations within the domain known to be responsible for its lipolytic activity [16]. Here we present an analysis of putative lipase genes in the zebrafish genome, providing evidence that TG digestion to FFA in larval zebrafish is not stereospecific, but random.
We set out to develop tools allowing us to investigate how nutrients (e.g. PL) influence the digestion, absorption and re-synthesis of dietary TG, not only qualitatively but also quantitatively. We also investigated whether these tools could be used to explore the effect of pharmaceuticals on lipid digestion.
2. Materials and Methods
2.1. Fluorescent Lipid Synthesis
11-(dipyrrometheneboron difluoride) undecanoic acid (C11 TopFluor® fatty acid): C11 TopFluor® fatty acid was synthesized according to published methods [17]. 1H NMR (CDCl3, 400 MHz): δ 6.04 (S, 2H), 2.94(m, 2H), 2.51 (S, 6H), 2.41 (S, 6H), 2.35 (t, J; 6.3Hz, 2H), 1.63 (m, 4H), 1.49 (m, 2H), 1.30 (m, 10H). ESI-MS: [M-H]- 431.50; [M+Cl]- 467.60. 1,2-dioleoyl-3-[11-(dipyrrometheneboron)undecanoyl]-sn-glycerol (18:1-18:1-C11 TopFluor® TG): 18:1-18:1-C11 TopFluor® TG was prepared with EDCI coupling of 1,2-dioleoyl-sn-glycerol (Avanti Polar Lipids) and C11 TopFluor® acid in anhydrous dichloromethane at room temperature. 1H NMR (CDCl3, 400 MHz): δ 6.05 (S, 2H), 5.34 (m, 4H), 5.25 (m, 1H), 4.29 (dd, J, 2.5, 7.5 Hz; 2H), 4.14 (dd, J, 5.0, 7.5 Hz; 2H), 2.92 (m, 2H), 2.51 (S, 6H), 2.41 (S, 6H), 2.32 (m, 6H), 2.01 (m, 8H), 1.61 (m, 8H), 1.50 (m, 2H), 1.30 (m, 50H), 0.90 (t, J, 5.0 Hz, 6H). ESI-MS: [M+H]+ 1053.70.
1-oleoyl-2,3-di[11-(dipyrrometheneboron)undecanoyl]-sn-glycerol (18:1- C11 TopFluor®-C11 TopFluor® TG): 18:1- C11 TopFluor®-C11 TopFluor® TG was prepared with EDCI coupling of 1-oleoyl-sn-glycerol (Avanti Polar Lipids) and C11 TopFluor® acid in anhydrous dichloromethane at room temperature. 1H NMR (CDCl3, 400 MHz): δ 6.05 (S, 4H), 5.34 (m, 2H), 5.25 (m, 1H), 4.29 (dd, J, 2.5, 7.5 Hz; 2H), 4.14 (dd, J, 5.0, 7.5 Hz; 2H), 2.92 (m, 4H), 2.51 (S, 12H), 2.41 (S, 12H), 2.32 (m, 6H), 2.02 (m, 4H), 1.61 (m, 8H), 1.50 (m, 4H), 1.30 (m, 42H), 0.90 (t, J, 5.0 Hz, 3H). ESI-MS: [M+NH4]+ 1202.7, [M+NH3+Na]+ 1224.6, [M+NaCl+H]+ 1244.7.
1-palmitoyl-2-[11-(dipyrrometheneboron difluoride)undecanoyl]-sn-glycerol-3-phosphocholine (TopFluor® PC, Avanti Polar Lipids): 1H NMR (CDCl3:CD3OD 8:2, 400 MHz): δ 6.07 (S, 2H), 5.22 (m, 1H), 4.41 (m; 1H), 4.24 (m, 2H), 4.15 (m; 1H), 4.02 (m; 2H), 3.57 (m; 2H), 3.21 (s; 9H),2.95 (m, 2H), 2.50 (S, 6H), 2.43 (S, 6H), 2.31 (m, 4H), 1.61 (m, 6H), 1.48 (m, 2H), 1.26 (m, 30H), 0.90 (t, J, 5.0 Hz, 3H). ESI-MS: [M+H]+ 911.10, [M+Na]+ 933.10, [M+NaCl+H]+ 969.20. 1,2-di[11-(dipyrrometheneboron difluoride)undecanoyl]-sn-glycerol-3-phosphocholine (Bis-C11 TopFluor® PC): Bis-C11 TopFluor® PC was prepared with DCC coupling of sn-glycerol-3-phosphocholine and C11 TopFluor® acid in anhydrous chloroform at room temperature. 1H NMR (CDCl3, 400 MHz): δ 6.05 (S, 4H), 5.22 (m, 1H), 4.40 (m; 1H), 4.34 (m, 2H), 4.15 (m; 1H), 4.00 (m; 2H), 3.76 (m; 2H), 3.34 (s; 9H), 2.92 (m, 4H), 2.51 (S, 12H), 2.41 (S, 12H), 2.29 (m, 4H), 1.61 (m, 8H), 1.48 (m, 4H), 1.27 (m, 20H), 0.90 (t, J, 5.0 Hz, 3H). ESI-MS: [M+H]+ 1087.20.
2.2. Diets
The diet emulsions consisted of 20.6 µM total lipid. The 100 % TG (100TG) emulsions comprised 14.2 µM unlabeled lipids (canola oil, Bunnpris, Norway) and 6.4 µM TopFluor® labeled lipids. In emulsion diets including PC (60TG), mixed L-α-Phosphatidylcholine from chicken egg yolk (Sigma-Aldrich, Germany) was added. See Table 1 for the lipid compositions of the different diets. Lipids dissolved in chloroform were flushed with N2 until chloroform was totally evaporated. Lipids were then dissolved in 5 µL ethanol per mL of emulsion. To aid emulsification of diets, 1.5 µL Tween 20 was added per mL emulsion. ddH2O was then added to the desired volume and the oil/water mix was vortexed before sonication. Double labeled fluorescent TG or PC (TopFluor®, Avanti Polar Lipids, USA) was added at 6.4 µM to both diets. TopFluor is a boron-dipyrromethene (BODIPY)-like fluorescent dye. The synthesis of BODIPY was first described by Loudet et al., in 2007 [18].
Table 1.
The effect of dietary lipid composition on expression of genes in several pathways involved in lipid metabolism was measured by qRT-PCR.
| Gene | Accession # | Gene name | Primer sequence | Amp. size |
Pathway |
|---|---|---|---|---|---|
| mttp | NM_212970.1 | Microsomal Triglyceride Transfer Protein | F: GGACACCTGCCACATGAGAT | 186 | Chylomicron assembly |
| R: ATCCACGGTTTCTGCCATGA | |||||
|
| |||||
| apoA-1a | NM_131128.1 | Apolipoprotein AIa | F: TAAGCTGACCGAGCGTCTTG | 179 | Chylomicron assembly |
| R: TCTGTGCGAATGTGGTCCTC | |||||
|
| |||||
| apoA-IV | NM_001079861.1 | Apolipoprotein AIV | F: ACTGTGACCCAAATCACACTTCT | 132 | Chylomicron assembly |
| R: GCATCTGGTGGGATGGCTAA | |||||
|
| |||||
| apoB | XM_689735.8 | Apolipoprotein B | F: CTCTTTGGAGAGCGCTTGGA | 123 | Chylomicron assembly |
| R: AGCGTGGAACGAAGACCATT | |||||
|
| |||||
| sar1b | NM_001024377.2 | Secretion Associated Ras Related GTPase 1b | F: CAGACGAGACCATCGGCAAT | 124 | Chylomicron assembly |
| R: CCCTTTCCTGTCGTCTGACC | |||||
|
| |||||
| edem1 | NM_201189.2 | ER Degradation Enhancing Alpha-Mannosidase Like Protein 1 | F: GGCACCCGTTTGGAGATGTA | 115 | ER stress |
| R: GGGATGCCTGTGCTAGTGTT | |||||
|
| |||||
| edem2 | NM_001305614.1 | ER Degradation Enhancing Alpha-Mannosidase Like Protein 2 | F: GACAGAATCAGCCGGGTCAA | 165 | ER stress |
| R: CTCAAACTCTGTGCCCGTGT | |||||
|
| |||||
| hsp70 | NM_001113589.1 | Heat Shock Protein 70 | F: GACCTCTTCAGGGGAACACTA | 101 | ER stress |
| R: GTTGATCCCCCAACCAGCAC | |||||
|
| |||||
| plin3 | XM_005166931.3 | Perilipin 3 | F: ATGGTGCGTGGTTTGAGTGA | 174 | Lipid droplet |
| R: CTGCAGTAAGAGGACGGGTG | |||||
|
| |||||
| pnpla2 | XM_005166931.3 | Patatin Like Phospholipase Domain Containing 2 | F: CACCGGAGCCTGTTTAGGAG | 135 | Lipid droplet |
| R: ATCACGCGGTGTAAGTGTGT | |||||
|
| |||||
| cpt1 | XM_005166473.3 | Carnitine Palmitoyltransferase 1 | F: GCTGTACAGGAGGAAGCTGG | 131 | Betaoxidation |
| R: AGTCCCTCTCCTTTCCTGGG | |||||
|
| |||||
| Agpat4 | NM_212992.1 | 1-acylglycerol-3-phosphate O-acyltransferase 4 | F: ATATTCCGCTCGGTGCCATC | 167 | PA synthesis |
| R: TCCTTTTGTCCCTGTCGTGG | |||||
|
| |||||
| Agpat9-like | NM_001099450 | 1-acylglycerol-3-phosphate O-acyltransferase 9, like | F: GACCTGGTTGGTAATCGGCA | 166 | PA synthesis |
| R: GCCGCCTTTCTTAGGTCTGT | |||||
|
| |||||
| Gpat3 | NM_001002685 | glycerol-3-phosphate acyltransferase 3 | F: GCAAGGTGAAGAACTGGCTC | 149 | LPA synthesis |
| R: AATCGGGGAAGTGTGGTTGG | |||||
|
| |||||
| Gpat4 | NM_001040249 | glycerol-3-phosphate acyltransferase 4 | F:GTCTCACGATGCTCCAATATACA | 112 | LPA synthesis |
| R: TCACATCAAGGTCCACAGCC | |||||
|
| |||||
| Mogat2 | NM_001326700.1 | monoacylglycerol O-acyltransferase 2 | F: CGAGCATGAGAGTGGGAAGC | 148 | DAG synthesis |
| R: GCCAGGAATGAGAAGACCCA | |||||
|
| |||||
| Mogat3b | NM_001008626.3 | monoacylglycerol O-acyltransferase 3b | F: GGGGACAGGTCACCATGGAA | 181 | DAG synthesis |
| R: TCTCAGGTGTGTGCCAATCA | |||||
|
| |||||
| Dgat1a | NM_199730.1 | diacylglycerol O-acyltransferase 1a | F: CCAAAGCTCGAACCCTGTCT | 107 | TG synthesis |
| R: TCTGTGTGTGAGGTTTCCCG | |||||
|
| |||||
| Dgat1b | NM_001002458.1 | diacylglycerol O-acyltransferase 1b | F: GAGTTGCCACAAGTTGCAGG | 112 | TG synthesis |
| R: GAGACGTGCGTTGCTCAAAA | |||||
|
| |||||
| Dgat2 | NM_001030196.1 | diacylglycerol O-acyltransferase 2 | F: TTCCGGTGTCAAAAAGGGCT | 169 | TG synthesis |
| R: CAGCAGCAAAGAGCAAGCAA | |||||
2.3. Larval Zebrafish & Feeding Trials
The wild-type AB zebrafish strain was used for the experiments. Eggs were incubated in E3 medium for 6 days post fertilization at 28 °C, with 14 hours light exposure and 10 hours darkness per day.
This study was carried out within the Norwegian animal welfare act guidelines, in accordance with the Animal Welfare Act of 20th December 1974, amended 19th June, 2009 and approved by FDU, FOTS ID 8131.
Feeding was carried out in 24-well plates with 15 larvae in each well for 60 or 120 min, and each treatment was done in triplicate for the HPLC analysis. Orlistat (GlaxoSmithKline, UK) [19] was used to block neutral lipase activity in the intestinal lumen of the larvae, and 2,4-dibromoacetophenone (Sigma-Aldrich, Germany) was used to block luminal phospholipase activity [20]. As both of these drugs are water-insoluble, they were ground to a fine powder that was suspended in E3 medium at 1 mg/mL. This treatment protocol was validated, and larvae exposed to either drug showed significantly reduced lipase activity (Supplementary Fig. 1). In feeding trials, the larvae swam in the drug-saturated medium for one hour before feeding in addition to inclusion of either Orilstat or 2,4-dibromoacetophenone in the emulsion diets. For imaging to demonstrate emulsification of intestinal contents and/or accumulation of lipid in intestinal enterocytes, six larvae from each drug treatment group were fed the lipid diet emulsions described above. For live imaging, larvae were anaesthetized with metacaine (100 mg/L, MS-222, Norsk medisinaldepot AS, Bergen, Norway). For images demonstrating accumulation of lipid in enterocytes, larvae were kept in E3 medium post-feeding to evacuate excess gut content before being euthanized with an overdose of metacaine (300 mg/L) and fixed overnight in 4 % paraformaldehyde in PBS (pH 7.2) before transfer to 30 % sucrose. At the end of the feeding experiments for HPLC analysis, the larvae were euthanized with an overdose of metacaine (300 mg/L) and rinsed in ddH2O.
2.4. HPLC
To extract lipids for HPLC, larvae were added to 750 µl of a 2:1 methanol:chloroform mix and sonicated until completely homogenized, then 250 µl chloroform and 250 µl ddH2O were added and vortexed thoroughly[21]. After centrifugation (14000 rpm, 5 min at 4 °C), the lipid fraction was collected. Total lipid extracted from the zebrafish was then separated by HPLC (Dionex UltiMate 3000 LC, Thermo Fisher Scientific Inc.) and quantified with a fluorescence detector (Dionex UltiMate 3000 RS). For optimal separation of the lipid classes, separate runs were performed for neutral and polar lipids. Neutral lipids were separated on a silica column (Kromasil 100-3.5-SIL 2.1×150 mm, Eka Chemicals AB., Bohus, Sweden). Each run takes 34 min with a flow of 0.4 ml/min, starting with a gradient of heptane with 0.5% to 10% of isopropanol for 21 min followed by 14 min reconditioning with 0.5% isopropanol in heptane. Polar lipids were separated on a silica column (Pinnacle DB Silica 150 × 2.1 mm, 3µm, Restek, Bellfonte, PA, USA) in a 12 min isocratic run, with a 0.4 ml/min flow of 52% isopropanol, 40% heptane and 8% ddH2O. Standards mixed with unlabeled lipids and total lipids from samples were dissolved in chloroform as the injection solvent. The detector was set to λ Ex = 489 nm and Em = 509 nm.
2.5. Microscopy
Larvae were photographed to demonstrate emulsification of intestinal content with an AZ100 makroskope (Nikon, Japan) and DS Fi1 camera (Nikon, Japan). For images demonstrating accumulation of lipid in intestinal membrane, larvae were mounted in methylcellulose. Imaging was done on a Ti-E inverted microscope (Nikon, Japan) with a CFI Super Plan Fluor ELWD ADM 20 X C PH-1 objective, numerical aperture 0.45 (Nikon, Japan), and a C2+ confocal scanner (Nikon, Japan). The confocal scanner settings were; first filter cube: 447/60, second filter cube: 525/50 595 LP, laser wavelength: 488, laser power: 4.8 and pinhole size: 30 µm2.
2.6. RNA extraction and qPCR
Euthanized larvae were homogenized on Quiazol (Qiagen) using a Precellys 24 (Bertin Technologies). Total RNA was extracted from the whole fish on a Bio Robot EZ1 using the EZ1 RNA Universal Tissue Kit with the RNase-free DNase Set (Qiagen), according to the manufacturer's instructions. The quantification and purity of RNA was assessed with the NanoDrop ND-1000 UV–Vis Spectrophotometer (NanoDrop Technologies). For all total RNA samples, the optical density ratio at 260/280 nm ranged between 1.70 and 1.98. RNA integrity numbers [22,23] were between 8.2 and 9.7.
RT reactions and qPCR were run according to published methods [24]. Genes coding for proteins involved in assembly and transport of chylomicrons, ER-stress, cytosolic lipid droplet metabolism and mitochondrial ß-oxidation were analyzed (Table 1). Results were calculated as the arithmetic mean using ubiquitin and beta-actin as reference genes based on the study of Sæle et al [24]. Normalization was performed using the geNorm Visual Basic for Applications applet for Microsoft as previously described by Vandesompele et al [25]. Accession number, primer sequences and product size for each quantitative PCR assay are given together with the corresponding biological pathway in Table 1.
2.7. Data mining & Statistics
Blast searches were done in the Zebrafish genome (GRCz10, ensemble genome browser 90). Pancreatic lipase (pnlip) has been annotated Pancreatic lipase related protein 2 (pnliprp2) in fish; therefore, pnliprp2 was used when searching the zebrafish genome for pancreatic lipase. Verified pancreatic lipase and colipase sequences from cave fish (Astyanax mexicanus) [pnliprp2 (ENSG00000266200) and CLPS (ENSAMXG00000007696)], spotted gar (Lepisosteus oculatus) [pnliprp2 (ENSLOCG00000009574) and CLPS (ENSLOCG00000011147)] and Xenopus (Xenopus tropicalis) [pnliprp2 (ENSXETG00000004181) and clps (ENSXETG00000032290)] were used as templates for the searches. Synteny maps of the colipase (clps) gene locus and the pancreatic lipase (pnlip) gene locus in human and zebrafish were generated in Ensembl release 90 - August 2017.
Potential differences in fluorescence between the feeding groups (60TG and 100TG) after 60 and 120 min were tested with a factorial ANOVA. The Tukey HSD post hoc test was used to identify significant differences between group means. For the analysis of gene expression levels, a Student’s t-test was used. Differences and effects were considered significant at p < 0.05, and all significant differences were annotated with unlike letters in figures. ANOVA, post hoc analyses and t-tests were performed on Statistica 12.0 (StatSoft, Inc.). Data are presented as means and standard deviations.
3. Results
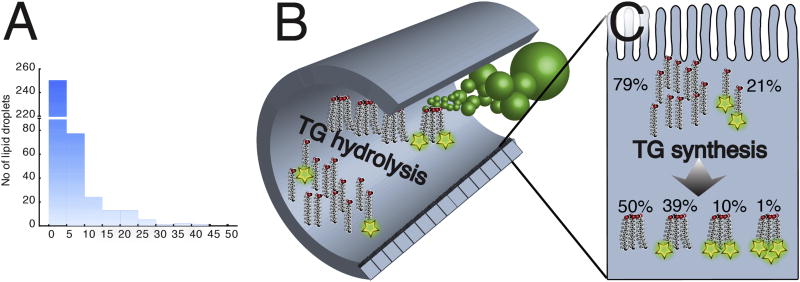
To investigate dietary triglyceride lipolysis and transport by the intestinal epithelium, larval zebrafish were immersed in emulsified liquid defined diets containing varying amounts of PL and TG. We synthesized a novel triglyceride containing two fluorescently labeled acyl chains to track both lipolysis and transport through the digestive organs of transparent larvae. The fluorescence enables tracing of labeled lipids using live imaging. However, to enable quantitative separation between luminal undigested lipid and digested, absorbed and re-synthesized lipid, feeding larvae the dual acyl chain label in the TG in combination with non-labeled TG presented a solution. Our novel HPLC method allows for separation between TG with single and dual acyl chain labels. Feeding the larvae large quantities of non-labeled TG together with the dual acyl chain label TG ensures that all single labeled TG can only originate from digested and absorbed lipid (Fig. 1). Dietary PL play an important role in absorption and transport of TG [26–28]. Based on this we hypothesized that a shortage of dietary PL could limit lipoprotein synthesis and subsequent TG transport from enterocytes, the absorptive cells of the intestine [29]. To test the effect of PL on TG digestion and transport, we produced one emulsion with 100% TG (100TG) and one with 60% TG and 40% phosphatidylcholine (60TG) by mass, both containing the fluorescent TG described above. Zebrafish larvae (6 dpf) were fed the fluorescently labeled 100TG and 60TG diets and then whole–larva lipid extracts were analyzed by HPLC with fluorescent detection.
Fig.1.
A) Fish larvae swim in the emulsion of 20.6 µM lipid of which 6.4 µM is fluorescently labeled with TopFluor. The graph shows the size distribution (area) of dietary lipid particles. B) The emulsion diet enters the intestine and lipids are hydrolyzed. C) Absorbed free fatty acids inside the enterocyte are incorporated into TG molecules. The theoretical distribution of TopFluor-labeled FAs is given for the 100TG diet.
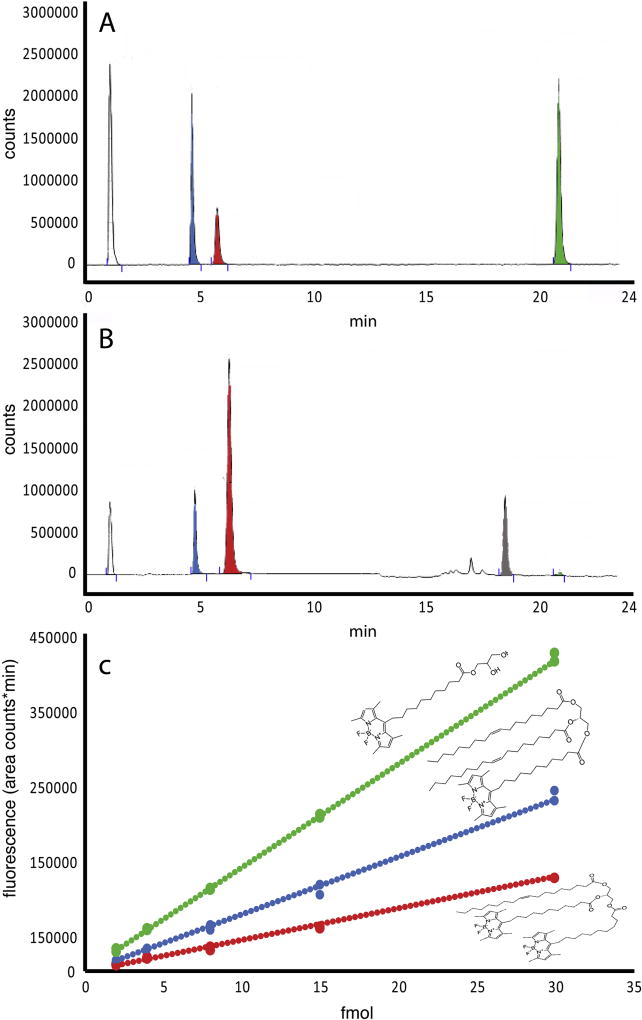
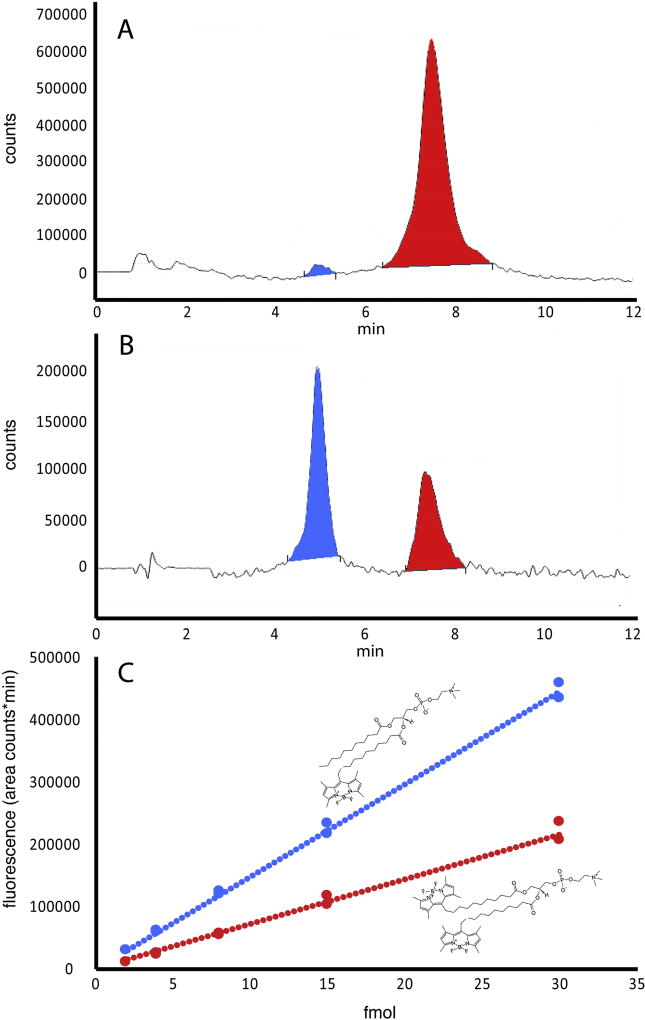
To predict whether or not larval zebrafish would express the enzymes necessary for complete or partial dietary triglyceride digestion, we searched for orthologs of genes encoding human digestive enzymes in the zebrafish genome (GRCz10, ensemble genome browser 90). A BLAST search did not locate any orthologs of human pancreatic lipase (pnlin), pnlin 1 and 2, or colipase (clps) in zebrafish. Aligning a 702.61 Kb part of the human genome, comprising the loci for pnlin and flanked by known neighboring genes such as gfra1 and ENO4 with the corresponding grfa1 and eno4 containing region of the zebrafish genome show the absence of pnlin in the zebrafish (Supplementary fig. 2). For closer investigation of the presence of colipase a 151.24 Kb region of the human genome, comprising the loci for clps, with known neighboring genes such as lhfpl5 and srpk1 with the corresponding lhfpl5 and srpk1 containing region of the zebrafish genome show the absence of clps in the zebrafish (see Supplementary Fig. 2). However, the digestive enzyme Carboxyl ester lipase (cel), sometimes referred to as Bile-activated lipase (bal), is present. Zebrafish have two cel paralogs, cel1 (ENSDARG00000017490) and cel2 (ENSDARG00000029822). This supports the hypothesis that zebrafish, like the Atlantic cod, should be able to digest TG completely to FFA and glycerol in the intestinal lumen. Assuming that complete TG digestion takes place and the rates of hydrolysis of fluorescently labeled and unlabeled TG are equal, the probability of finding a double labeled TG resynthesized in the intestinal tissue was 10%, TG with one labeled fatty acid was 39%, unlabeled TG was 50%, and TG with all three fatty acids labeled was less than 1 % for larvae fed the 100TG diet. The theoretical distribution of lipid types in larvae fed the other test diets is given in Table 2. HPLC peaks that were present in lipid extracts from larvae were identified as single and double labeled TG and labeled FFA using standards (Fig. 2A). When lipid extracts from fish fed the 100TG diet were run additional peaks appeared at 16 and 18 min, in the area where diacyl- and monoacylglycerols are expected. A minimal quantity of labeled FFA was detected (Fig. 2B). Standard curves prepared by HPLC quantitation of known amounts of purchased fluorescent lipid standards show that the level of fluorescence increases twice as fast (2.1× for PC and 1.8× for TG) in the single labeled lipid molecule when compared to the double labeled counterpart (see Figs 2 and 3). This is the result of the quenching effect of the proximity of two fluorophores, resulting from the fluorescence resonance energy transfer (FRET) in the double labeled lipids [30,31] (Fig. 2C, 3C). HPLC of polar lipids from larvae fed labeled PC resolved peaks corresponding to single and double labeled PC standards (Fig. 3).
Table 2.
Top panel: concentration of lipids in diets. The average molecular weight of canola oil was calculated based on the fatty acid profile. Lower panel: theoretical distribution of TopFluor labeled fatty acids in resynthesized TGs and PCs in intestinal tissue after digestion and absorption of the trial diets. Calculations are based on full hydrolysis of TG (three FFAs) and partial hydrolysis of PC (one lysoPC and one FFA).
| Diets | 100TG | 60TG | PC60TG |
|---|---|---|---|
| Canola oil | 14.2 µM | 8.52 µM | 8.52 µM |
| PC | 5.68 µM | 5.68 µM | |
| bis-TopFluor TG | 6.4 µM | 6.4 µM | |
| bis-TopFluor PC | 6.4 µM | ||
| Theoretical products | |||
| 0 TopFluor TG | 49.8 % | 30.1 % | 23.6 % |
| 1 TopFluor TG | 39.1 % | 30.7 % | 14.5 % |
| 2 TopFluor TG | 10.2 % | 10.4 % | 3.0 % |
| 3 TopFluor TG | 0.9 % | 1.2 % | 0.2 % |
| 0 TopFluor PC | 20.6 % | 24.3 % | |
| 1 TopFluor PC | 7.0 % | 29.3 % | |
| 2 TopFluor PC | 5.0 % |
Fig. 2.
Chromatograms of a standard mix (A) of double (red) and single (blue) labeled TG and labeled free fatty acid (green). B) Separation of a sample from zebrafish larvae fed the 100TF diet. The large peak of double-labeled TG originates from undigested luminal lipid. The peaks at 16 and 18 minutes appear where diacylglycerol and monoacylglycerol are expected to be. C) Standard curves for double labeled TG (y = 4201.7×, R2 = 0.99), single labeled TG (y = 7617.9×, R2 = 0.99) and labeled FA (y = 13853×, R2 = 0.99) from 4 to 30 fmol.
Fig. 3.
A) Chromatogram of a standard mix of double (red) and single (blue) labeled PC. B) Separation of a sample from zebrafish larvae fed the 60% TG and 40% PC diet containing labeled PC. C) Standard curves for double labeled PC (red, y = 7219.8×, R2 = 0.99) and single labeled PC (blue, y = 14850×, R2 = 0.99) from 2 to 30 fmol.
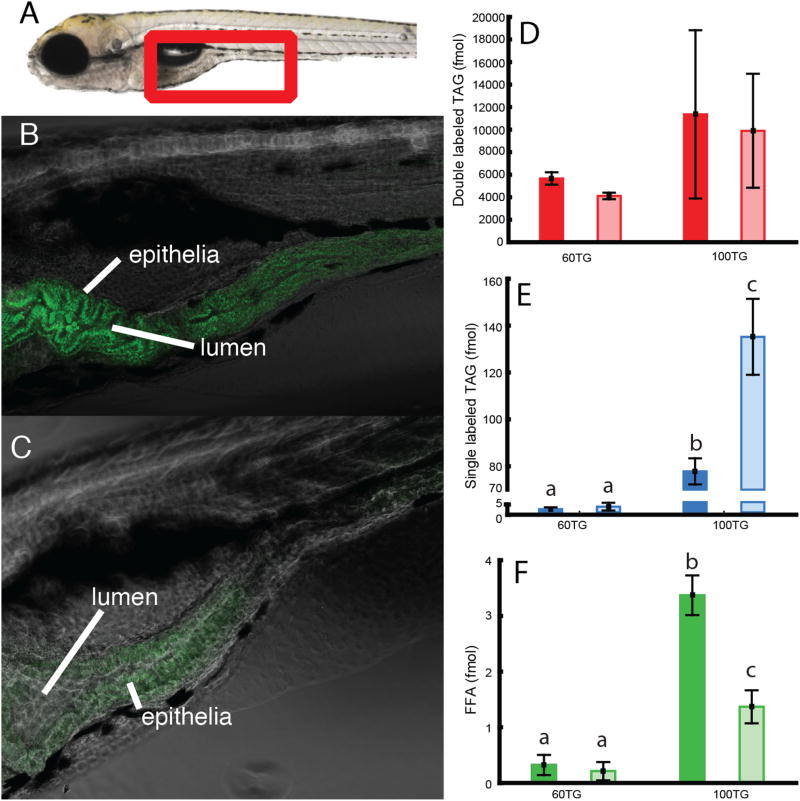
HPLC analysis revealed major metabolic differences in response to the two diets. Specifically, analysis of the lipid extracts from animals exposed to the diet lacking PL (100TG) revealed an accumulation of single labeled TG indicating that the double labeled substrate was subject to lipolysis and resynthesis/remodeling. Confocal microscopy performed to determine the subcellular localization of this TG revealed fluorescence within enterocytes, likely in cytosolic lipid droplets (CLD) (Fig. 4). Stored single labeled TG in the larvae given the 60TG diet was low; 0.46 (0.24 SD) fmol per larva after 60 min and 0.79 (0.55 SD) fmol per larva after 120 min. This is in contrast to when PC was excluded from the emulsion diet (100TG), significantly more TG was stored; 18.46 (1.32 SD) fmol per larva after 60 min (p = 0.0002) increasing to 32.09 (3.85 SD) fmol after 120 min (p = 0.0002). Interestingly, there was a trend that more double labeled TG was found in zebrafish larvae fed the 100TG diet compared to those fed 60TG, but this was not statistically significant. There was more fluorescently labeled FFA in the larvae fed the 100TG diet compared to the 60TG diet group, but this decreased with time.
Fig. 4.
Left panel: anterior intestine of zebrafish larvae (see red frame in A). Larvae in panel B were fed a 100% TG diet and larvae in panel C were fed 60 % TG and 40 % PC diet. Both diets were labeled with double labeled TG. The pure TG diet is digested and absorbed, but accumulates in intestinal epithelia (B). When PC is added to the diet, products of digested fluorescent TG do not accumulate in intestinal tissue (C). Images are representative of 6 larvae per group. Right panel: D, E and F show quantification by HPLC of labeled lipid per larva after 60 min feeding (dark colored column) and 120 min (light colored columns).
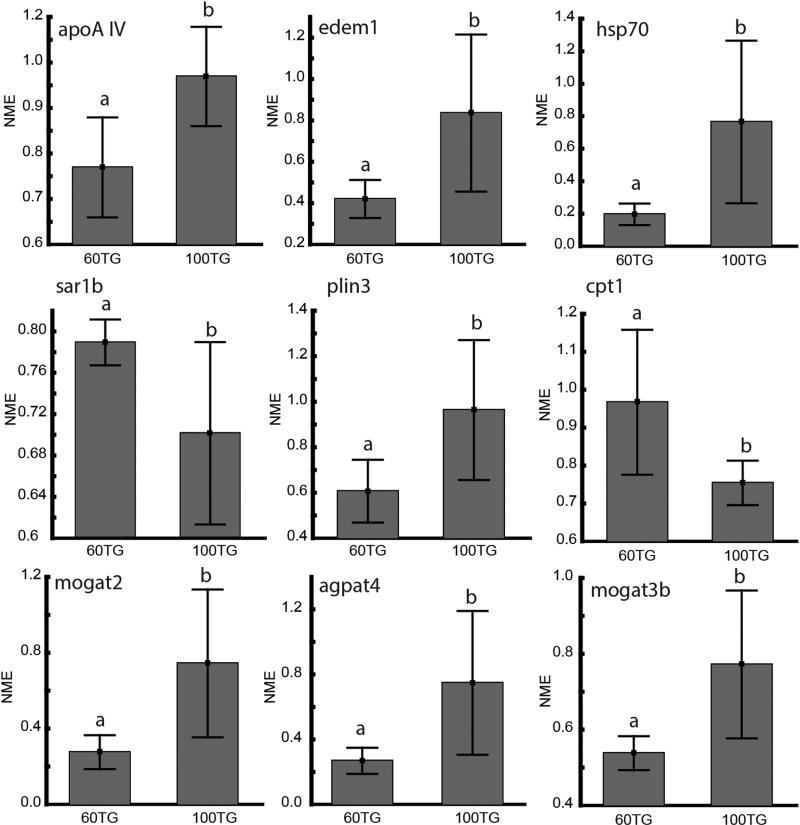
Expression of genes related to lipid transport and synthesis, chylomicron assembly, beta-oxidation, ER stress and cytosolic lipid droplet metabolism was measured by qRT-PCR in larval zebrafish. The expression of the sar1b (Secretion-Associated Ras-Related GTPase 1B) gene, important for transferring the nascent chylomicron from the ER to the Golgi apparatus [32], was higher in larvae fed the 60TG and 100TG diets (Fig. 5). On the other hand, edem1 (ER Degradation Enhancing Alpha-Mannosidase-Like Protein 1), an important component in ER-associated degradation (ERAD) [33] and hsp70 (Heat Shock Protein 70), a chaperone involved in stabilization of apolipoproteins [34], were significantly up-regulated in the 100TG group. Expression of apoA-IV (Apolipoprotein A-IV), an apolipoprotein involved in the assembly of ApoB-containing lipoproteins in enterocytes [35,36], was up-regulated in the 100TG group.
Fig. 5.
Transcriptional levels of genes associated with lipid metabolism expressed as the mean normalized expression (NME). Larvae were fed a diet composed of 60 % TG and 40 % PC (60TG) or 100 % TG (100TG). Statistically different gene expression levels are indicated with different letters. (p<0.05, Student’s t-test).
Expression of cpt1 (Carnitine Palmitoyltransferase 1), a controlling factor of mitochondrial beta-oxidation, was also down-regulated in the 100TG group (Fig. 5). Out of the two genes associated with CLD metabolism, plin3 (Perilipin 3) was up regulated in the 100TG group (Fig. 5), and Patatin-Like Phospholipase Domain-Containing 2 (pnpla2), was not regulated in response to the diet (Supplementary Fig. 3). Mogat2 and mogat3b (Monoacylglycerol O-acyltransferase 2 and 3b), coding for enzymes synthesizing diacylglycerol (DAG) from 2-monoacylglycerol (2-MAG) and fatty acyl-CoA in the intestine, were up-regulated in the 100TG group (Fig 5), while the genes coding for the enzyme required for the following step in TG synthesis, dgat1a, dgat1b and dgat2 (Diacylglycerol O-acyltransferase 1a, 1b and 2), were not regulated in response to the diet (Supplementary Fig. 3). Out of the genes involved in de novo synthesis of TG (the Kennedy pathway) only agpat4 (1-acylglycerol-3-phosphate O-acyltransferase 4), which synthesizes phosphatidic acid (PA) from lyso-phosphatidic acid (LPA), was regulated according to diet, with higher expression in the 100TG group.
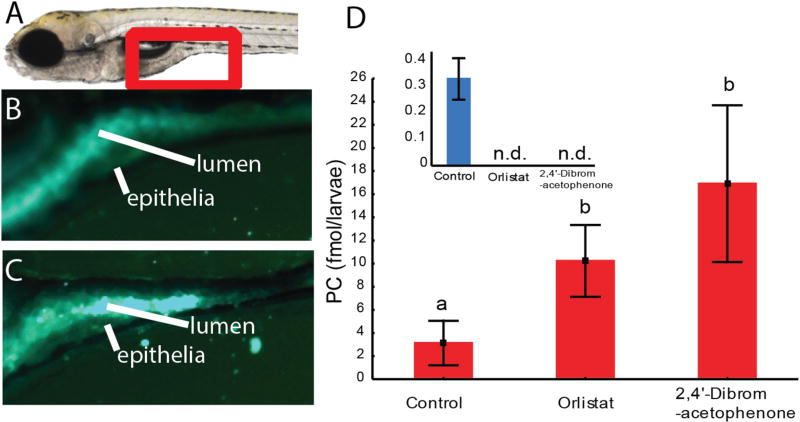
To investigate if feeding double labeled lipids in an emulsion to zebrafish larvae may be a suitable tool to investigate pharmaceutical impact on lipid digestion, digestion of both PL and TG was challenged with two known lipase inhibitors. To inhibit neutral lipase activity, Orlistat (GlaxoSmithKline, UK) [19] was added to the medium of one treatment group. For the inhibition of luminal sPLA2 activity, 2,4-dibromoacetophenone (Sigma-Aldrich, Germany) [20] was added to the second treatment group. Together with a control treatment without any lipase inhibitors, the three groups were fed PC60TG diet, with double labeled PC. Microscopy of the intestines of fed fish larvae reveals that the intestinal lumens of the control group exhibit diffuse fluorescence, a result of advanced emulsification facilitated by FFA produced from lipase activity (Fig. 6B). When TG hydrolysis is blocked with Orlistat, the larger water-dispersed lipid globules from the emulsified feed remain intact, producing an intense fluorescence within the intestinal lumen (Fig. 6C).
Fig. 6.
A) Left panel: anterior intestine of zebrafish larvae (see red frame). B) and C) Larvae were fed a diet composed of 60 % TG and 40 % PC. Both diets contained fluorescent double labeled PC. B) Fluorescence in the intestinal lumen and epithelium is diffuse, indicating strong emulsification. C) Larvae were treated with Orlistat, a drug blocking neutral lipase activity. Emulsification of dietary lipid was inhibited, visualized as strong globular fluorescence in the lumen. Images represent results from 6 larvae per treatment group. D) More double labeled PC is present in zebrafish treated with lipase inhibitors. Red is double labeled and blue (graph in inset) is single labeled PC.
The HPLC analysis showed that the control group contained an average of 0.34 (0.08 SD) fmol single labeled PC per larvae, but none could be detected in the two lipase inhibitor treatment groups. Significantly more double labeled PC was found in the groups given lipase blockers (p=0.02). This indicates that both lipase blockers prevented uptake of double-labeled PC or its labeled FFA or lyso-PC products into enterocytes, where they would have been used to synthesize single-labeled PC (among other products not detected by the assay).
Discussion
Feeding larval zebrafish a meal containing only triglyceride (100TG diet) led to dietary TG accumulation in the enterocytes when compared with larvae fed phospholipid along with triglyderide (60TG diet). Similar results have been observed by electron microscopy in another fish species, where the lack of dietary PL led to increased CLD size and numbers [37], but the use of double-labeled fluorescent lipids in the larval zebrafish reveals the contribution of dietary lipid to this accumulation of TG in enterocytes. We hypothesize that neutral lipid accumulates when the supply of PL is insufficient to form chylomicron/VLDL particles for export at a high enough rate to keep up with dietary lipid absorption. Both transport (chylomicron/VLDL type particles) and storage (CLD) particles contain a PL monolayer surrounding the more hydrophobic lipids contained within their cores. Despite this PL requirement for both particles, the surface area to volume ratio is typically much lower in CLDs than in chylomicrons and VLDL. A large VLDL particle may have a diameter of 60 to 200 nm whereas an enterocytic CLD may be several microns [38]. As concluded by Penno et al., [39] “The amount of phospholipid required for lipid droplet monolayer expansion is remarkably small.” It has also been demonstrated that compromised PC biosynthesis by RNAi inactivation of Phosphate Cytidylyltransferase 1 and 2, Choline alpha leads to fewer but larger lipid droplets [40].
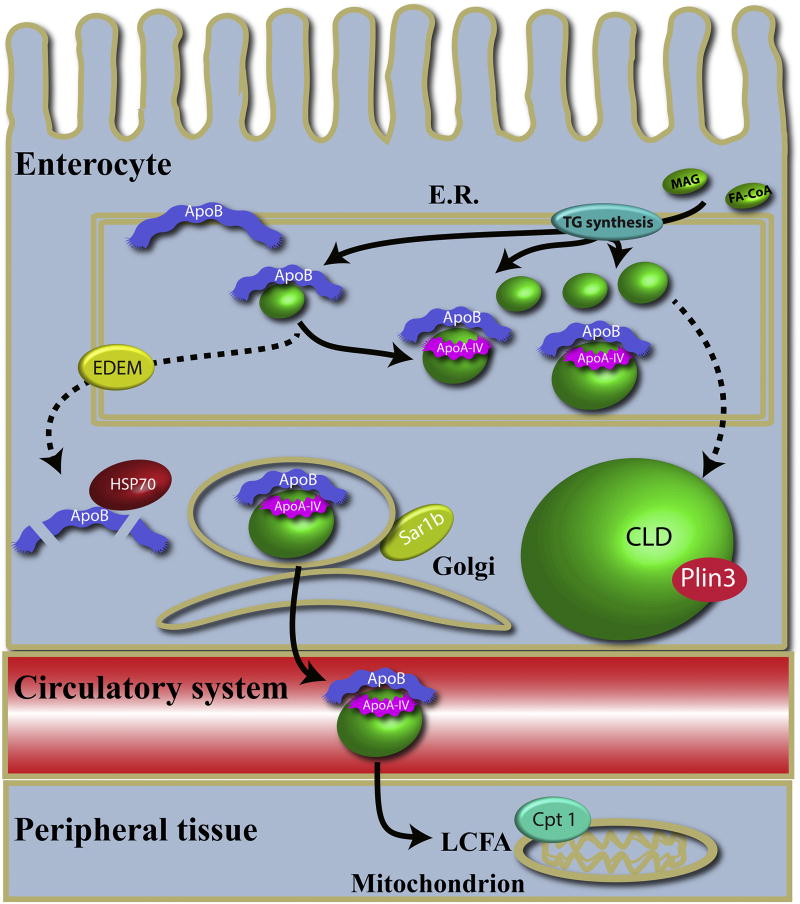
One of the first steps in chylomicron formation is when ApoB, chaperoned by MTTP, forms a complex with a dense particle (Fig. 7). The dense particle contains mostly PLs and cholesterol [36]. If the ApoB molecule does not merge with a dense particle, ApoB becomes unstable and is degraded via the endoplasmic-reticulum associated protein degradation (ERAD) and autophagy pathways. Edem1 and Hsp70 are essential proteins in these systems, respectively [41]. Depriving the larvae of dietary PL should lead to dramatically lower dense particles in the enterocyte ER increasing the pressure on the ERAD and autophagy pathways, which may explain the increase in edem1 and hsp70 expression in larvae fed the 100TG diet. As expected, expression of plin3 (a CLD coating protein) increased along with enterocyte lipid droplet-associated fluorescence in larvae fed the 100TG diet, while expression of pnpla2, a lipase responsible for hydrolyzing TG from CLD [42], was unchanged (Fig. 7). Genes associated with re-synthesis of TG were also upregulated in the 100TG group. The observed increase in agpat4, coding for one of the enzymes synthesizing TG from acyl-CoA, was expected. However, the reason for up-regulation of mogat2 and mogat3b in larvae fed only TG is less clear. Since zebrafish lack both colipase and pancreatic lipase, the only known enzymes that produce MAG in the vertebrate intestinal lumen [43], increases in luminal FFA and glycerol but not MAG were expected in larvae fed the 100TG diet. It remains to be determined whether MAG increases in response to the 100TG diet by another mechanism, or if there is another process driving up-regulation of mogat2 and mogat3b.
Fig. 7.
Biogenesis of chylomicrons. TG is synthesized de novo via gpat, agpat, pap, and dgat, or remodeled via mogat and dgat. MTTP chaperones ApoB in the ER where it forms a complex with a dense lipid particle, of which the major component is PC. We hypothesize that without dietary PC the dense lipid particle level is low, and the ApoB-lipid particle complex is not formed. This will destabilize ApoB and it will be degraded, a process in which edem1 and hsp70 are central. ApoAI-V merges with the ApoB-lipid complex mediating expansion of the particle with TG [47], and the particle is transported to the Golgi facilitated by the coatomer complex COPII of which Sar1b is a part [36]. Fatty acids are absorbed by peripheral tissue in the body and a large part of the FAs originating from TG is ß-oxidized in the mitochondria. However, if dietary PC is low the dense lipid particles will not be formed and a large excess of TG will accumulate in the ER. These will be incorporated into CLDs (stippled arrows).
Kalogeris et al., [44] demonstrated that the secretion of apoA-IV in the lymph of mice depended upon lipid transport by chylomicrons. It was later demonstrated that ingestion of lipids upregulates the transcription of apoA-IV in zebrafish [45]. Since both diets used in the present study were 100 % lipid with the only difference being the lipid class composition, our results demonstrate that this mechanism is linked to TG and not PL. Since PLs are only partially hydrolyzed by sPLA2 to one FFA and one lyso-PL, the 100TG experimental diet yields more FFA than the 60TG diet. Our results therefore suggest that the transcription of apoA-IV is associated with TG synthesis.
We hypothesized that the absence of dietary PC would obstruct lipid transport to peripheral tissues where TG may be used for energy, with the result of increasing levels of digested and resynthesized TG (single labeled TG) trapped in the enterocyte CLDs. This hypothesis was supported by the difference in levels of single labeled TG found in larvae fed a PC and TG mix (60TG) versus larvae only fed TG (100TG). As expected, digested and absorbed TG (single labeled TG) did not accumulate in the 60TG group, probably due to ß-oxidation. However, when dietary PC was not administered, digested and absorbed TG accumulated in the larval enterocytes. This is further supported by the down-regulation of cpt1 mRNA in the 100TG group which is characteristic for decreased conversion of FA into energy [46]. In the group of larvae fed PC and TG, the level of single labeled TG is low, and does not increase over time. We have shown in previous work that most ingested neutral lipid was used in β-oxidation in larvae of Atlantic cod [9]. We demonstrated that cod larvae of 9.75 (SD 0.63) mm catabolized 100% of absorbed dietary PL up to 2 µg (0.04% of body weight). For dietary TG the cod larvae had to absorb more than 3 µg (0.07% of body weight) of lipid before any TG was retained in the body tissue [9].
To evaluate the role of intestinal lipases in the digestion, absorption and remodeling of PLs, lipase inhibitors were added to the double TopFluor PC labeled 60TG diet. As expected, the phospholipase inhibitor 2,4-dibromoacetophenone blocked digestion of PL. Surprisingly, the neutral lipase blocking agent Orlistat had the same effect. As the fluorescent dietary PC had both fatty acids labeled, the only way PC with only one labeled fatty acid may be produced in the larva is by hydrolyzing one of the labeled fatty acids (digestion) and absorbing the lyso-PC from the intestinal lumen into the enterocyte where lysophosphatidylcholine acyltransferase attaches an unlabeled fatty acid, forming a single labeled PC [48]. The single labeled PC, being the verification of digestion, absorption and re-modeling, could only be detected in larvae fed the control diet.
We interpret the fact that Orlistat could prevent digestion of PL as a demonstration of the importance of the emulsifying effect of products from intestinal TG digestion. The emulsifying effect of TG hydrolysis has been shown in the human stomach [49]. Most of the ingested lipid in the human stomach is emulsified by partial hydrolysis by gastric lipase one-hour post ingestion. Partial gastric hydrolysis of TG produced lipid droplets with the same average droplet size as those found in the duodenum, where bile is present. In contrast, larval zebrafish do not have a stomach or gastric lipase but hydrolyze neutral lipids primarily with bile activated lipase in the intestine [14]. However, the products of both enzymatic processes are FFAs, and here we demonstrate the importance of TG hydrolysis for emulsification of the zebrafish meal. The lack of emulsion with consequently poor hydrolysis and absorption may also be reflected in the amount of double labeled lipids (luminal lipids). When luminal lipases are inhibited, ingested luminal lipids accumulate (Fig. 6D). Similarly, in the fish larvae given the emulsion diet without PC (100TG) there was a trend that more undigested luminal TG was found. This is consistent with earlier findings in which emulsions with PC were hydrolyzed and absorbed at a faster rate compared to emulsions made with Tween 80 [50,51]. An alternate explanation is that zebrafish intestinal TG lipases, like mammalian CEL [52,53], have phospholipase activity that is directly inhibited by Orlistat.
We have demonstrated how labeling two fatty acids with a BODIPY-like fluorophore (TopFluor) in a single lipid molecule like TG or PC enables us to trace digestion and absorption into the enterocyte and further metabolism of lipids in the zebrafish larva in a way not previously possible. This gives the opportunity for qualitative analysis by microscopy as well as quantitative analysis by HPLC using the same tracer. Using this system we revealed a requirement for dietary PL for efficient TG processing by the intestine and that when PC is lacking, the components of luminal TG end up being resynthesized and stored within the cytoplasm of enterocytes. The translucent zebrafish larva, now available with an increasingly larger number of transgenic lines, offers in combination with the methods presented here valuable tools in research on lipid digestion and metabolism. This system may also present itself as a valuable tool in the search for pharmaceuticals affecting lipid digestion and transport.
Supplementary Material
Highlights.
-
-
Fluorescent lipids allow biochemical and visual examination of dietary lipid metabolism.
-
-
Dietary double-labeled fluorescent TG and PC allows quantitation of digestion and absorption.
-
-
Dietary phospholipid is necessary for export of lipid from enterocytes.
Acknowledgments
The work was funded by the Research Council of Norway (CODE knowledge platform project; Grant no. 199482/S40). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Support was also provided by the National Institutes of Health (NIDDK), RO1DK093399 (S.A.F.), and RO1GM63904 (The Zebrafish Functional Genomics Consortium: PI Stephen Ekker and Co-PI S.A.F). This content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Additional support for this work was provided by the Carnegie Institution for Science endowment and the G. Harold and Leila Y. Mathers Charitable Foundation (S.A.F). We thank Nini Sissener for assistance with calculations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Freiberg JJ, Tybjaerg-Hansen A, Jensen JS, Nordestgaard BG. Nonfasting Triglycerides and Risk of Ischemic Stroke in the General Population. Circulation. 2008;118:S756–S756. doi: 10.1001/jama.2008.621. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of- Function Mutations in APOC3 and Risk of Ischemic Vascular Disease. New England Journal of Medicine. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 3.Carten JD, Farber SA. A new model system swims into focus: using the zebrafish to visualize intestinal lipid metabolism. Clinical Lipidology. 2009;4:501–515. doi: 10.2217/clp.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz-Garcia L, Schlegel A. Lxr-driven enterocyte lipid droplet formation delays transport of ingested lipids. Journal of Lipid Research. 2014 doi: 10.1194/jlr.M052845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadler KC, Rawls JF, Farber SA. Getting the Inside Tract: New Frontiers in Zebrafish Digestive System Biology. Zebrafish. 2013;10:129–131. doi: 10.1089/zeb.2013.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lickwar CR, Camp JG, Weiser M, Cocchiaro JL, Kingsley DM, Furey TS, et al. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017;15:e2002054. doi: 10.1371/journal.pbio.2002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farber SA, Pack M, Ho SY, Johnson LD, Wagner DS, Dosch R, et al. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 8.Otis JP, Farber SA. Imaging vertebrate digestive function and lipid metabolism in vivo. Drug Discovery Today: Disease Models. 2013;10:e11–e16. doi: 10.1016/j.ddmod.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamre K, Lukram IM, Ronnestad I, Nordgreen A, Sæle Ø. Pre-digestion of dietary lipids has only minor effects on absorption, retention and metabolism in larval stages of Atlantic cod (Gadus morhua) British Journal of Nutrition. 2011;105:846–856. doi: 10.1017/S0007114510004459. [DOI] [PubMed] [Google Scholar]
- 10.Quinlivan VH, Wilson MH, Ruzicka J, Farber SA. An HPLC-CAD/fluorescence lipidomics platform using fluorescent fatty acids as metabolic tracers. Journal of Lipid Research. 2017;58:1008–1020. doi: 10.1194/jlr.D072918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Developmental Biology. 2011;360:276–285. doi: 10.1016/j.ydbio.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saele O, Nordgreen A, Olsvik PA, Hamre K. Characterisation and expression of secretory phospholipase A(2) group IB during ontogeny of Atlantic cod (Gadus morhua) . Br. J. Nutr. 2011;105:228–237. doi: 10.1017/S0007114510003466. [DOI] [PubMed] [Google Scholar]
- 13.Lohse P, Chahrokh-Zadeh S, Seidel D. The acid lipase gene family: three enzymes, one highly conserved gene structure. Journal of Lipid Research. 1997:880–891. [PubMed] [Google Scholar]
- 14.Ronnestad I, Yufera M, Ueberschaer B, Ribeiro L, Sæle Ø, Boglione C. Feeding behaviour and digestive physiology in larval fish: current knowledge, and gaps and bottlenecks in research. Reviews in Aquaculture. 2013;5:S59–S98. doi: 10.1111/raq.12010. [DOI] [Google Scholar]
- 15.Sternby B, Larsson A, Borgström B. Evolutionary studies on pancreatic colipase. Biochimica Et Biophysica Acta (BBA) - Lipids and Lipid Metabolism. 1983;750:340–345. doi: 10.1016/0005-2760(83)90038-3. [DOI] [PubMed] [Google Scholar]
- 16.Sæle Ø, nordgreen A, Olsvik PA, Hamre K. Characterization and expression of digestive neutral lipases during ontogeny of Atlantic cod (Gadus morhua) Comparative Biochemistry and Physiology Part a: Molecular & Integrative Physiology. 2010;157:252–259. doi: 10.1016/j.cbpa.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Mintzer E, Bittman R. First synthesis of free cholesterol-BODIPY conjugates. J. Org. Chem. 2006;71:1718–1721. doi: 10.1021/jo052029x. [DOI] [PubMed] [Google Scholar]
- 18.Loudet A, Burgess K. BODIPY dyes and their derivatives: syntheses and spectroscopic properties. Chem. Rev. 2007;107:4891–4932. doi: 10.1021/cr078381n. [DOI] [PubMed] [Google Scholar]
- 19.Sternby B, Hartmann D, Borgstrom B, Nilsson A. Degree of in vivo inhibition of human gastric and pancreatic lipases by Orlistat (Tetrahydrolipstatin, THL) in the stomach and small intestine. Clinical Nutrition. 2002;21:395–402. doi: 10.1054/clnu.2002.0565. [DOI] [PubMed] [Google Scholar]
- 20.Markova M, Koratkar RA, Silverman KA, Sollars VE, Mac-Phee-Pellini M, Walters R, et al. Diversity in secreted PLA(2)-IIA activity among inbred mouse strains that are resistant or susceptible to Apc(Min/+) tumorigenesis. Oncogene. 2005;24:6450–6458. doi: 10.1038/sj.onc.1208791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959 doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Imbeaud S. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Research. 2005;33:e56–e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller O, Hahnenberger K, Dittmann M, Yee H, Dubrow R, Nagle R, et al. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis. 2000;21:128–134. doi: 10.1002/(SICI)1522-2683(20000101)21:1<128::AID-ELPS128>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 24.Sæle Ø, Nordgreen A, Hamre K, Olsvik PA. Evaluation of candidate reference genes in Q-PCR studies of Atlantic cod (Gadus morhua) ontogeny, with emphasis on the gastrointestinal tract. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 2009;152:94–101. doi: 10.1016/j.cbpb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Doherty PJA, Kakis G, Kuksis A. Role of luminal lecithin in intestinal fat absorption. Lipids. 1973;8:249–255. doi: 10.1007/BF02531899. [DOI] [PubMed] [Google Scholar]
- 27.Tso P, Kendrick H, Balint JA, Simmonds WJ. Role of biliary phosphatidylcholine in the absorption and transport of dietary triolein in the rat. Gastroenterology. 1981;80:60–65. doi: 10.5555/uri:pii:0016508581901918. [DOI] [PubMed] [Google Scholar]
- 28.Mansbach CM, 2, Arnold A, Cox MA. Factors influencing triacylglycerol delivery into mesenteric lymph. Am. J. Physiol. 1985;249:G642–G648. doi: 10.1152/ajpgi.1985.249.5.G642. [DOI] [PubMed] [Google Scholar]
- 29.Hui DY, Cope MJ, Labonté ED, Chang H-T, Shao J, Goka E, et al. The phospholipase A(2) inhibitor methyl indoxam suppresses diet-induced obesity and glucose intolerance in mice. Br. J. Pharmacol. 2009;157:1263–1269. doi: 10.1111/j.1476-5381.2009.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahim M, Mizuno NK, Li X-M, Momsen WE, Momsen MM, Brockman HL. Physical and photophysical characterization of a BODIPY phosphatidylcholine as a membrane probe. Biophys. J. 2002;83:1511–1524. doi: 10.1016/S0006-3495(02)73921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 32.Mansbach CM, Siddiqi SA. The biogenesis of chylomicrons. Annu. Rev. Physiol. 2010;72:315–333. doi: 10.1146/annurev-physiol-021909-135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das I, Png CW, Oancea I, Hasnain SZ, Lourie R, Proctor M, et al. Glucocorticoids alleviate intestinal ER stress by enhancing protein folding and degradation of misfolded proteins. Journal of Experimental Medicine. 2013;210:1201–1216. doi: 10.1084/jem.20121268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL. The Hsp110 Molecular Chaperone Stabilizes Apolipoprotein B from Endoplasmic Reticulum-associated Degradation (ERAD) J. Biol. Chem. 2007;282:32665–32675. doi: 10.1074/jbc.M705216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberg RB, Gallagher JW, Fabritius MA, Shelness GS. ApoA-IV modulates the secretory trafficking of apoB and the size of triglyceride-rich lipoproteins. Journal of Lipid Research. 2012;53:736–743. doi: 10.1194/jlr.M019992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansbach CMI, Gorelick F. Development and Physiological Regulation of Intestinal Lipid Absorption. II. Dietary lipid absorption, complex lipid synthesis, and the intracellular packaging and secretion of chylomicrons. Am. J. Physiol. 2007;293:G645–G650. doi: 10.1152/ajpgi.00299.2007. [DOI] [PubMed] [Google Scholar]
- 37.Olsen RE, Dragnes BT, Myklebust R, Ringø E. Effect of soybean oil and soybean lecithin on intestinal lipid composition and lipid droplet accumulation of rainbow trout, Oncorhynchus mykiss Walbaum. Fish Physiol. Biochem. 2003;29:181–192. doi: 10.1023/B:FISH.0000045708.67760.43. [DOI] [Google Scholar]
- 38.Wojczynski MK, Glasser SP, Oberman A, Kabagambe EK, Hopkins PN, Tsai MY, et al. High-fat meal effect on LDL, HDL, and VLDL particle size and number in the Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN): an interventional study. Lipids Health Dis. 2011;10:181. doi: 10.1186/1476-511X-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochim. Biophys. Acta. 2013;1831:589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao C, Hsieh J, Adeli K, Lewis GF. Gut-liver interaction in triglyceride-rich lipoprotein metabolism. American Journal of Physiology - Endocrinology and Metabolism. 2011;301:E429–E446. doi: 10.1152/ajpendo.00178.2011. [DOI] [PubMed] [Google Scholar]
- 42.Beilstein F, Corriere V, Leturque A, Demignot S. Characteristics and functions of lipid droplets and associated proteins in enterocytes. Exp. Cell Res. 2016;340:172–179. doi: 10.1016/j.yexcr.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 43.Lowe M. The triglyceride lipases of the pancreas. Journal of Lipid Research. 2002 doi: 10.1194/jlr.r200012-jlr200. [DOI] [PubMed] [Google Scholar]
- 44.Kalogeris TJ, Monroe F, Demichele SJ, TSO P. Intestinal synthesis and lymphatic secretion of apolipoprotein A-IV vary with chain length of intestinally infused fatty acids in rats. J. Nutr. 1996;126:2720–2729. doi: 10.1093/jn/126.11.2720. [DOI] [PubMed] [Google Scholar]
- 45.Otis JP, Zeituni EM, Thierer JH, Anderson JL, Brown AC, Boehm ED, et al. Zebrafish as a model for apolipoprotein biology: comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Disease Models and Mechanisms. 2015;8:295–309. doi: 10.1242/dmm.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial betaoxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11:380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 47.Cheng D, Xu X, Simon T, Boudyguina E, Deng Z, VerHague M, et al. Very Low Density Lipoprotein Assembly Is Required for cAMP-responsive Element-binding Protein H Processing and Hepatic Apolipoprotein A-IV Expression. J. Biol. Chem. 2016;291:23793–23803. doi: 10.1074/jbc.M116.749283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lands W, Hart P. Metabolism of glycerolipids. J. Biol. Chem. 1960 [PubMed] [Google Scholar]
- 49.Armand M, Borel P, Dubois C, Senft M, Peyrot J, Salducci J, et al. Characterization of Emulsions and Lipolysis of Dietary Lipids in the Human Stomach. Am. J. Physiol. 1994;266:G372–G381. doi: 10.1152/ajpgi.1994.266.3.G372. [DOI] [PubMed] [Google Scholar]
- 50.Couëdelo L, Amara S, Lecomte M, Meugnier E, Monteil J, Fonseca L, et al. Impact of various emulsifiers on ALA bioavailability and chylomicron synthesis through changes in gastrointestinal lipolysis. Food Funct. 2015;6:1726–1735. doi: 10.1039/C5FO00070J. [DOI] [PubMed] [Google Scholar]
- 51.Vors C, Capolino P, Guerin C, Meugnier E, Pesenti S, Chauvin M-A, et al. Coupling in vitro gastrointestinal lipolysis and Caco-2 cell cultures for testing the absorption of different food emulsions. Food Funct. 2012;3:537–546. doi: 10.1039/c2fo10248j. [DOI] [PubMed] [Google Scholar]
- 52.Duan RD, Borgstrom B. Is there a specific lysophospholipase in human pancreatic juice? Biochim. Biophys. Acta. 1993;1167:326–330. doi: 10.1016/0005-2760(93)90236-3. [DOI] [PubMed] [Google Scholar]
- 53.Mackay K, Starr JR, Lawn RM, Ellsworth JL. Phosphatidylcholine hydrolysis is required for pancreatic cholesterol esterase- and phospholipase A2-facilitated cholesterol uptake into intestinal Caco-2 cells. J. Biol. Chem. 1997;272:13380–13389. doi: 10.1074/jbc.272.20.13380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.