Abstract
Purpose of review
Recent concerns regarding the clinical utilization of clustered regularly interspaced short palindromic repeats (CRISPR) involve uncertainties about the potential detrimental effects that many arise due to unintended genetic changes, as in off-target mutagenesis, during CRISPR genome surgery. This review gives an overview of off-targeting detection methods and CRISPR’s place in the clinical setting, specifically in the field of ophthalmology.
Recent findings
As CRISPR utilization in the laboratory setting has increased, as well as the knowledge regarding CRISPR mechanisms including its off-target effects. Although a perfect method for achieving 100% specificity has yet to be determined, the last few years has seen many developments in off-targeting detection and in increasing CRISPR tools efficacy.
Summary
The CRISPR system has high potential to be an invaluable therapeutic tool as it has the ability to modify and repair pathogenic retinal lesions. Although it is not yet a perfect system, with further efforts to improve its specificity and efficacy along with careful screening of off-target mutations, CRISPR-mediated genome surgery potential can become maximized and applied to patients.
Keywords: CRISPR, genome surgery, off-targeting, precision medicine, retina
Introduction
Numerous recent studies have pointed towards both potential promises and perils of clustered regularly interspaced short palindromic repeats (CRISPR) genome surgery in the clinical setting1–10. Of the potential perils of the CRISPR genome surgery system, one study raised questions about utilizing CRISPR in the clinical setting without further study of off-target effects1, 11. While this report raises warranted concern, it should also be considered that the study, though in vivo, the CRISPR genome surgery was performed on zygotes, not adult retinae11. As such, the off-target effects reported1 may not necessarily be reflective of therapeutic interventions that may typically occur in postnatal or post-mitotic retinas. Interestingly, the heated debate regarding off-target effects was raised when nine clinical trials utilizing CRISPR to treat various malignancies had already begun (Table 1). Of note, the first patient recruitment for a CRISPR clinical trial occurred in the past year in October 2016 (NCT027933856). This debate and public concern raise the question of whether these clinical trials began too early without proper consideration of safety1. Yet, while the debate is a necessary and important consideration, it is also worth noting that all the CRISPR clinical trials that had begun were in phase I or phase II, evaluating safety (Table 1).
Table 1.
CRISPR Clinical Trials at time of debate
| NCT ID | Phase | Intervention | Conditions Targeted | Study Sponsor |
|---|---|---|---|---|
| NCT03057912 | I | TALEN- or CRISPR/Cas9- mediated disruption of HPV E6/E7 DNA | Human Papillomavirus-Related Malignant Neoplasm | First Affiliated Hospital, Sun Yat-Sen University |
| NCT0381715 | II | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Esophageal Cancer | Hangzhou Cancer Hospital |
| NCT03044743 | I/II | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Stage IV Gastric Carcinoma; Stage IV Nasopharyngeal Carcinoma; T-Cell Lymphoma Stage IV; Stage IV Adult Hodgkin Lymphoma; Stage IV Diffuse Large B-Cell Lymphoma | Yang Yang, The Affiliated Nanjing Drum Tower Hospital of Nanjing University Medical School |
| NCT02793856 | I | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Metastatic Non-small Cell Lung Cancer | Sichuan University |
| NCT02867332 | I | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Metastatic Renal Cell Carcinoma | Peking University |
| NCT02867345 | I | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Hormone Refractory Prostate Cancer | Peking University |
| NCT02863913 | I | Autologous CRISPR/Cas9-engineered PD-1 knockout-T cells | Invasive Bladder Cancer Stage IV | Peking University |
| NCT03164135 | – | CRISPR/Cas9 CCR5 gene modification of CD34+ hematopoietic stem cells | HIV-1-infection Hematological Malignancies | Affiliated Hospital to Academy of Military Medical Sciences |
| NCT03166878 | I/II | Autologous CRISPR/Cas9-engineered universal CD19-specific CAR-T cells | B Cell Leukemia; B Cell Lymphoma | Chinese PLA General Hospital |
It is also important to note that off-targeting effects, unintended mutations that arise from CRISPR-engineering, are not new to the world of genetics, and methods for detecting such changes do exist and are in further development12–15. Unintended changes due to DNA engineering in ophthalmology has certainly been encountered before. In 2008, Kleinman et al. in 2008 presented that while choroidal neovascularization (CNV) could be inhibited by small interfering RNAs (siRNAs), the siRNAs may elicit immune effects as the mechanism of action was in suppressing toll-like receptor 3 (TLR3)16. This review seeks to address the questions raised by many on the efficacy of CRISPR, safety considerations, and its place in the clinical setting. This review in particular focuses in part on CRISPR-mediated ophthalmic genome surgery.
Brief CRISPR Mechanism
The CRISPR system is a genome engineering tool derived from bacteria and archaea immune systems17–19 considered to be highly specific7, 17. The CRISPR system has been adapted to serve many purposes in genome engineering including DNA modification and repair, transcription modification, and directed evolution7, 8, 20–23. Such diverse applications of CRISPR have allowed further study and understanding of CRISPR. As such, much is known about CRISPR and how it can be directed to specific DNA strands15, 17, 24. The most commonly used version of CRISPR is comprised of single guide RNA (sgRNA)19, denoting a chimera of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), and CRISPR associated protein Cas9 endonuclease7, 17, 19. The sgRNA binds to the Cas9 which activates the endonuclease24. In the actual target DNA, another component is necessary for Cas9 to recognize and cleave at the target location: a protospacer adjacent motif (PAM) adjacent to the target site24–26. The required PAM sequence for spCas9, the most commonly used Cas9 orthologue derived from Streptococcus pyogenes3, is “NGG,” with N denoting any of the four nucleotides. Once Cas9 makes a double strand break (DSB) at the target location, the DSB can be repaired in a variety of ways which determines the application of CRISPR; an example homology-directed repair (HDR) utilizes a template strand25, 27 which results in DNA repair of a pathogenic allele to wild-type (WT), and non-homologous end joining (NHEJ) without a template strand introduces insertions and deletions (INDELs) which results in gene knockout of pathogenic allele by frameshift caused by the non-specific INDELs7, 8, 17, 20. Figure 1 provides a simplified visual schematic.
Figure 1.
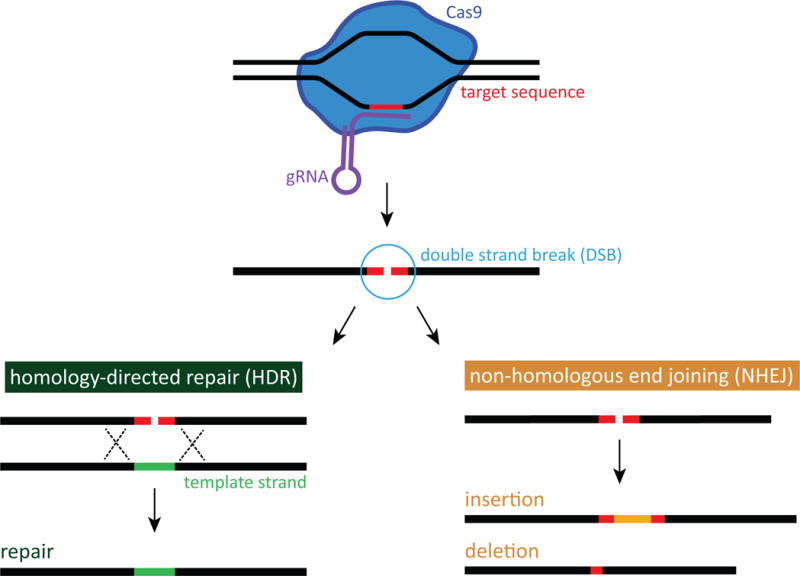
CRISPR-mediated genome surgery simplified schematic. Homology-directed repair (HDR) and non-homologous end joining (NHEJ). Cas9 endonuclease creates a double strand break (DSB) at the target location. DSB repair by HDR (left) utilizes a wild-type template strand resulting in DNA repair. DSB repair by NHEJ (right) introduces insertions and deletions (INDELs).
Along with the development of different CRISPR techniques, studies have also shown that during CRISPR/Cas9 mediated genome engineering, changes to the DNA can also occur at unintended locations, as in off-target sites15, 28–32. This type of off-targeting may arise due to a variety of reasons. One such reason may be that though the lack of the presence of a PAM sequence can become a limiting factor in targeting specific sequences in some cases, the opposite case in which the presence of a PAM sequence in unintended locations may also become targeted by Cas9 seems possible. Or perhaps the fact that Cas9 is directed by RNA33, and inside the eukaryotic cell many non-coding RNAs (ncRNAs) are present playing regulatory roles34; it is possible that these ncRNA may direct Cas9 to unintended sites. Thus, many efforts to characterize and predict off-target effects of CRISPR exist5, 6, 9, 15, 35. The ultimate goal of these studies is to not only identify off-targeting but determine how CRISPR’s specificity can be maximized with minimal off-target effects5, 9, 15.
Brief Overview of Current Off-Targeting Detection Methods
In recent years, many methods of genome engineering off-targeting detection have been developed. A brief overview of different methods follows. Figure 2 provides a simplified visual overview. A number of reviews are recommended for a more comprehensive overview5, 15, 36–38. Although diverse variations of off-targeting detection methods exist, they can be summarized into the following categories: in silico algorithm predictions, in vitro selection, and genome-wide assays.
Figure 2.

Off-target detection methods simplified overview. (A) in silico algorithm prediction. Computer algorithms are utilized to predict off-target cleavage sites based on sequence similarities. The predicted sites are checked for off-target cleavage that may have occurred but does not allow detection of novel or random off-target sites. (B) in vitro selection. Partially randomized concatemeric DNA libraries are created via rolling-circle amplification of potential cleavage sites, which are checked for off-target cleavage. While a large library is created, the sequences are selected for by sequence similarities and may miss some off-target sites (C) Genome-wide assay. High-throughput sequencing of the entire genome is performed, essentially being an unbiased method as the entire genome is checked for off-target cleavage, although mosaicism may be missed.
A common method utilized when selecting a sgRNA and checking for off-target mutagenesis is algorithm-based prediction of potential cleavage sites9, 13, 31, 39. The algorithms predict off-target cleavage based on sequence similarity to the on-target site. When utilizing in silico prediction methods, the predicted sites are checked for INDELs that may have arisen from NHEJ or nucleic acid bulges that occur due to mismatch or gaps in DNA and sgRNA base pairing31, 32. Studies of off-targeting using in silico prediction have revealed many aspects of off-targeting. Mismatched sites can have high-frequency mutagenesis with five or fewer mismatches31, 39. It seems that 10-12 base-pairs proximal to 5′ of the PAM is more determinate of Cas9 specificity than more distal portions39. Interestingly, although the PAM sequence is a required component for cleavage, when tested for spCas9 cleavage of sequences with a 5′-NAG-3′ PAM sequence, cleavage was still achieved at 20% efficiency of that of 5′-NGG-3′ PAM sequence39. In consideration of the current knowledge, in silico prediction methods have shown that the sequence of the sgRNA is perhaps most indicative of mismatches and off-targeting9, 13. Current knowledge has allowed for further development in prediction algorithms including online tools such as CRISPR Design Tool39, E-CRISP40 and Cas-OFFinder13. But perhaps the biggest consideration of in silico prediction methods is that by nature, it is a biased review of off-target sites as the method is based on checking for what is predicted, but not for novel or random potential off-targeting sites that are not yet understood38.
Another method of detecting off-target mutagenesis is in vitro selection combined with high-throughput sequencing; this method was developed in order to more comprehensively characterize spCas9 specificity30, 41. This method by Pattanayak et al. utilizes partially randomized concatemeric DNA libraries created by rolling-circle amplification of potential cleavage sites30. The libraries are cleaved by CRISPR/Cas9, ligation-tagged, amplified by PCR, then analyzed by high-throughput sequencing computational analysis. Pattanayak et al. reported this method as a comprehensive method; because each selection library includes ~1012 target sequence variants, it is theoretically large enough to have tenfold coverage of all sequences with eight or fewer mutations relative to the target sequence. Pattanayak et al. were able to generate over 1012 potential off-target sites for the target sequences tested, better defining cleavage characteristics. However, because the randomized oligonucleotide libraries are created to resemble the target sequence, in vitro selection may still be a biased method of defining off-target sites15.
In order to have a true comprehensive understanding of CRISPR/Cas9 actions, especially in off-target sites, an unbiased method is necessary. A number of genome-wide assay methods have thus been developed15, 36, 38. In regards to a truly unbiased method, whole-genome sequencing (WGS) could identify even single nucleotide polymorphisms (SNPs) and small INDELs42. However, WGS is costly and time-consuming36. Another concern with WGS is that although the screening would be accurate for single-cell clones42, mosaicism is seen in experimental studies of in vivo CRISPR/Cas9 editing11, 43, 44. It would not be feasible to definitively utilize WGS on every cell in mosaics, but that would be required in order to truly comprehensively identify off-targeting45. Thus, even WGS is still an insensitive method of detecting off-target cleavage. Some groups have adapted using whole-exome sequencing (WES) as a more cost-efficient alternative46 while still detecting changes in the protein-coding regions47. However, WES does not detect changes in introns or regulatory regions and is an even more insensitive measure than WGS36. Other methods of genome-wide analyses such as integrase-defective lentiviral vector (IDLV) capture, genome-wide unbiased identification of DSBs enabled by sequencing (GUIDE-seq), high-throughput genome-wide translocation sequencing (HTGTS), breaks labelling, enrichment on streptavidin and next-generation sequencing (BLESS), and digested genome sequencing (Digenome-seq) have been developed in recent years to meet the need for better off-target mutagenesis detection35, 48–51.
Of these, methods that can capture off-target events in living cells is of particular interest as therapeutic applications of genome engineering will need to be applied in living cells. Gabriel et al. developed IDLV capture in order to show in vivo action of endonucleases capture transient DSBs48. IDLV capture is a method that detects for DSBs in the genome by essentially tagging DSBs with IDLV that become integrated, or captured, during NHEJ. The sites of integration are amplified by linear amplification-mediated polymerase chain reaction (LAM-PCR) then mapped by high-throughput sequencing. A limitation of IDLV capture however is that IDLVs sometimes randomly integrate into the DNA without endonuclease action. Thus, although sites of high frequency integration can be better identified, and the sites of IDLV capture are not biased by preselection, IDLV capture could show non-specific events. GUIDE-seq, developed by Tsai et al., is a similar method to IDLV capture but integrates double-stranded oligodeoxynucleotide (dsODN) by NHEJ35. Another difference is that GUIDE-seq utilizes single-tail adapter/tag (STAT)-PCR, which amplifies only the sequences with integrated dsODN which allows for correction of background and PCR bias. Although GUIDE-seq has high sensitivity with detection of sites with frequencies as low as 0.1% in off-target DSBs in living cells, the detection relies on dsODN insertion, which may not always occur efficiently and is limited by sequencing reads. Another off-targeting detection tool that can be used in living cells is HTGTS, a modified LAM-PCR method by Frock et al.49. HTGTS utilizes the detection of translocation events between DSBs created by the endonuclease and off-target DSBs to identify off-target mutagenesis and chromosomal damage49, 52, 53. HTGTS does not require any components other than the CRISPR/Cas9 and sgRNA and therefore has the advantage of not introducing other foreign material49. However, the rate at which translocations occur is low and are biased towards cleavage sites that are closer in proximity.
Other genome-wide methods cannot be completed in living cells, but offer other advantages. Crosetto et al. developed BLESS which detects telomere ends and endonuclease-induced DSBs for visualization of a genome-wide DSB landscape50. BLESS is able to capture transient DSBs by in situ labeling of DSBs by ligation of biotinylated linkers. Although BLESS is able to detect DSBs after CRISPR/Cas9 delivery in vivo and is a high-resolution method, BLESS requires cell fixation and lysis for analysis. Another approach developed by Kim et al. specifically for human cells, Digenome-seq, is an in vitro approach51. Digenome-seq is performed by isolating then digesting the genome in vitro using Cas9 ribonucleoprotein (RNP), preassembled Cas9 protein and sgRNA. The digested genome is analyzed by WGS. In vitro delivery allows the DSB events to be isolated from other in vivo factors such as cell conditions and chromatin accessibility. The preassembled Cas9 RNP also allows for maximization of cleavage events identifying off-target sites with frequency of 0.1% or lower. Other than it being an in vitro approach, Digenome-seq is also limited by WGS background information, as well as potential inefficient sequencing.
Some Characteristics of Off-Targeting
As described above, no current method of off-target detection is completely sensitive or specific, and more developments are needed. Still, despite limitations of off-targeting detection methods, the development of these methods has given a better understanding of Cas9-induced DSBs and off-targeting events. A finding of greatest concern is that some off-target mutagenesis occurs at frequencies equivalent to or higher than the target site31, 35 which further emphasizes the need to better define off-targeting. Current understanding of how the CRISPR/Cas9 system determines DSB locations has led to studies focusing on sgRNA’s effects on off-targeting5, 9. O’Geen et al. report based on a review of in silico predictions, in vitro screenings, and genome-wide detections that what determines CRISPR/Cas9 specificity most is the sgRNA and suggest that it is not Cas9 that should be categorized as specific or nonspecific but rather the sgRNA9. Though efforts toward better defining sgRNA sequence for higher specificity exist, further study is needed54, 55. In regards to the targeted genome, some studies have shown a correlation between off-targeting and chromatin accessibility and non-methylated regions56, 57. Another characteristic that has been observed by genome-wide assays is up to six mismatches in the PAM and non-canonical PAM sequences seem to be tolerated by Cas915. It has also been shown that Cas9 will cleave DNA sequences in the presence of DNA or RNA bulges created by mismatched target DNA and sgRNA32.
CRISPR as a Therapeutic and Safety Considerations
In light of current understanding of off-target mutagenesis, safety considerations for CRISPR/Cas9 as a therapeutic tool is critical. Wu et al.’s study “CRISPR Repair Reveals Causative Mutation in a Preclinical Model of Retinits Pigmentosa” is a relevant and important preclinical model of CRISPR utilization for therapeutic purposes11. Wu et al.’s success in disease model rescue following CRISPR-mediated correction of a pathogenic point mutation is an example of CRISPR’s potency as a clinical tool. In recent years, CRISPR has been applied to many different species and to a variety of diseases5, 58 and the successful applications of CRISPR in disease model rescue has built much anticipation for the benefits of CRISPR7, 17. Furthering the anticipation is Ma et al.’s recent report in August 2017 of CRISPR/Cas9-mediated correction of MYPC3 in human preimplantation embryos59. Yet, some respond that correction as described by Ma et al. to be unlikely, but rather what likely occurred is deletion of the gene of interest60, which raises uncertainty and consideration of safety. In evaluating CRISPR as a therapeutic tool, this review focuses on the retina as Wu et al.’s experiment is a retinal disease study, as the vast volume of recent CRISPR-mediated genome engineering does not allow for a comprehensive overview.
The retina is of particular importance as one of the most feared illnesses in America is blindness61. This is unsurprising as blindness or low vision can cause impairment in mobility and activities of daily living, and has been linked to depression and anxiety61. Retinal pigment epithelium (RPE) disorders, which affect more than 10 million Americans62, lead to retinal degenerations causing irreversible blindness, as the RPE has little regenerative potential63. At this time, retinal degenerations with genetic cause have no curative treatments available. It is hoped that these previously unapproachable genetic retinal dystrophies may be treated by gene and cell-based therapies63. An example of such an approach currently undergoing clinical trials is gene therapy. Gene therapy uses viral vectors to insert therapeutic (or wild-type) genes64; clinical trials of this therapeutic model have had varying degrees of success63, 65–72. The variability of success has been theorized to be attributable in part to the timing of gene therapy; beyond a certain degree of disease progression there may be a “point of no return” due to cell death65. Another great limitation of gene therapy is its inability to treat dominant mutations; only recessive conditions and haplo-insufficiency can be approached through gene therapy as it is a gene addition method63. Thus, a different treatment approach is needed. Figure 3 demonstrates this need. The therapeutic method that is expected to have the most potential for success in treating inherited disorders at this time is the CRISPR system8, 63, 73.
Figure 3.

Therapeutic options for recessive and dominant conditions. (A) Diseases resulting from recessive mutations can be corrected by the addition of a wild-type gene, as in gene therapy. (B) Diseases resulting from dominant mutations cannot be corrected by the addition of a wild-type gene. Correction of the pathogenic dominant mutation by DNA repair, e.g. CRISPR-mediated genome surgery, is needed.
In the laboratory setting, CRISPR/Cas9 has proven to be a useful genome engineering tool in preclinical models, especially in ophthalmology (see Table 2, Table 3). The advantage of CRISPR is that it has the potential to directly correct the genetic defect in induced pluripotent stem cells (iPSCs)74, which allows corrections for both dominant and recessive genetic mutations. These corrected iPSCs could be transplanted into the patient’s eye as a cell-based therapy potentially curing blindness63, 75. In 2015, Bassuk et al. were the first to report correction of a retinal dystrophy causative point mutation in patient-derived iPSCs using CRISPR/Cas974. Since then, a number of groups have reported successful correction of pathogenic mutations in patient-derived iPSCs (see Table 3).
Table 2.
CRISPR/Cas9 Applications for the Retina in Non-human Preclinical Models
| Gene of Interest | Associated Human Disease | CRISPR method | Purpose | Model Used or Generated | Reference |
|---|---|---|---|---|---|
| Reep6 | arRP | CRISPR/Cas9 Reep6L135P/L135P knock-in | Disease model generation | Reep6 knock-in mouse model | 107 |
| Reep6 | arRP | CRISPR/Cas9 Reep6 knock-out | Disease model generation | Reep6 knock-out mouse model | 108 |
| Mertk | arRP | CRISPR/Cas9 mediated HITI | New application and Gene rescue | RCS rat | 109 |
| Pde6b | arRP | CRISPR/Cas9 HDR | Discern causative mutation in rd1 mouse model and gene rescue | rd1 mouse | 11 |
| Nrl | – | CRISPR/Cas9 knock-down | Improve rod survival in multiple mouse models of retinal degeneration | Rho−/− mouse, RHO-P347S transgenic mouse, Rd10 mouse | 110, 111 |
| Rho | adRP | CRISPR/Cas9 ablation of pathogenic mutation | Disease model rescue | Transgenic S334ter rat | 112 |
| RHO | adRP | CRISPR/Cas9-induced knock-down of P23H-mutant RHO | Feasibility of knock-down in vivo | P23H RHO transgenic mouse | 113 |
| Kcnj13 | LCA | CRISPR/Cas9-induced mosaicism of Kcnj13 function | Disease model generation | Kcnj13-related LCA mouse model | 44 |
| Mfrp | Multiple including: nanophthalmia, hyperopia, RPE atrophy | CRISPR/Cas9-induced mutagenesis | Disease model generation | Mfrp zebrafish | 114 |
| Vegfa | Multiple including: CNV, AMD | Cas9 RNP-mediated gene inactivation | Feasibility of gene editing therapy for a non-genetic CNV disease model | laser-induced CNV mouse | 115 |
| Pax6 | – | CRISPR/Cas9 knock-out | Expand understanding of eye development | Pax6 knock-out mouse model | 43 |
abbreviations: arRP, autosomal recessive retinitis pigmentosa; adRP, autosomal dominant retinitis pigmentosa; LCA, Leber congenital amaurosis; RPE, retinal pigment epithelium; CNV, choroidal neovascularization; AMD, age-related macular degeneration; HITI, homology-independent targeted integration; HDR, homology-directed recombination; RNP, ribonucleoprotein
Table 3.
CRISPR/Cas9 Applications for the Retina in Human Cell Line Preclinical Models
| Gene of Interest | Associated Human Disease | CRISPR method | Purpose | Cell Type Used | Reference |
|---|---|---|---|---|---|
| MAK | arRP | CRISPR/Cas9 HDR | Gene rescue | Patient-derived iPSCs | 108 |
| RPGR | XLRP | CRISPR/Cas9 HDR | Gene rescue | Patient-derived iPSCs | 71 |
| CEP290 | LCA | Targeted genomic deletion with self-limiting CRISPR/Cas9 | Gene rescue | HEK293FT cells with IVS26 mutation in CEP290 | 75 |
| CEP 290 | LCA | CRISPR/Cas9 NHEJ of splice site mutation IVS26 | Feasibility of protein expression restoration via splice site correction | Patient-derived iPSCs | 108 |
| PROM1 | STGD4 | CRISPR/Cas9 PROM1 knock-out | Expand understanding of disease mechanism | ARPE-19 cells (human RPE cell line) | 109 |
| VEGF-A | Multiple including: CNV, AMD | CRISPR/Cas9-mediated INDEL formation | Feasibility of gene editing therapy in human RPE cells | ARPE-19 cells (human RPE cell line) | 110 |
abbreviations: arRP, autosomal recessive retinitis pigmentosa; XLRP, X-linked retinitis pigmentosa; LCA, Leber congenital amaurosis; STGD4, Stargardt-like macular dystrophy; CNV, choroidal neovascularization; AMD, age-related macular degeneration; HDR, homology-directed recombination; NHEJ, Non-homologous end joining; INDEL, insertions or deletions; iPSCS, induced pluripotent stem cells; RPE, retinal pigment epithelium
Of the numerous CRISPR system applications, Leber congenital amaurosis (LCA) is of particular interest to the current discussion of CRISPR as a therapeutic in the retina, as Editas and Allergan announced in March 2017 plans to develop CRISPR-mediated treatment of LCA10 and begin testing the treatment in 2018. LCA is also important to the discussion of CRISPR correction in general as affected individuals show symptoms within a few months from birth, and the disease-causative mutations are genetically heterogeneous76, 77. Of these, LCA10 is caused by the most frequently detected amongst the affected individuals: an intronic mutation in CEP290 (c.2991+1655A>G). CEP290 is challenging to target with CRISPR due to its size exceeding adeno-associated virus (AAV) delivery capacity78, 79. A dual AAV approach of pAAV-SpCas9 and pAAV-sgRNA has been shown to circumvent the AAV carrying capacity limitation78. Another approach which utilized a smaller S. aureus-derived Cas9 with two guide RNAs has also been shown to be effective in circumventing the AAV carrying capacity limitation79. These successful corrections are demonstrative of the accelerated developments for potential application that the field has seen in recent years.
Further, expectations for advances in CRISPR applications in ophthalmology is greater than in other fields as the human retina is one of the simplest areas to evaluate experimental gene and cell therapies because of its relative immune privilege by the blood-retina barrier and easy accessibility for monitoring without invasive techniques80. That is to say, one of the safest platforms for evaluating CRISPR’s safety in clinical trials may be the eye.
Even still, as discussed above, CRISPR has yet to be developed into a perfect system and there are many considerations especially for clinicians to navigate, including lack of accurate and specific off-target mutagenesis detection tools15. When safety is not properly assessed, even in a more ideal platform such as the eye, adverse events can occur. Related to the use of CRISPR is the use of stem cells as a therapeutic as mentioned above. Unfortunately, public perception of stem cells, driven in part by deceptive advertising, has led patients to seek stem-cell therapy under unregulated conditions leading to deleterious effects81. In such a case, a patient with an inherited retinal dystrophy who was expected to maintain useful vision for many years sought unregulated stem cell treatment and returned with sudden central vision loss in the treated eye82. Additionally, recent article “Immunity to CRISPR Cas9 and Cas12a therapeutics” highlights the potential of the CRISPR system to mount a host immune response, via innate, cellular, and humoral immunity2. Further, it is noted that CRISPR correction that introduces new protein products, while therapeutic for the treated disease, may introduce immunologically foreign proteins to the host2. Thus, in applying CRISPR to patient-derived iPSCs for autologous transplantation, and all other applications of CRISPR as a therapeutic, all of the benefits and risks must be considered. Without careful consideration of treatments and all of their effects, clinicians may cause more harm than good. For such reasons, efforts to improve CRISPR efficacy, reduce off-targeting, and better understand CRISPR mechanism and its effects are in progress.
Safer CRISPR?
A brief overview of experimental methods of increasing CRISPR/Cas9 specificity is given. For a more complete mechanism of individual methods, studies which report the use of these methods are recommended4, 29, 83–93. As discussed above, the specificity of Cas9 is dependent on the sgRNA sequence. Thus, some methods to improve Cas9 specificity utilize the sgRNA sequence54. One such example is truncated gRNA (tru-gRNA) which is 2-3 nucleotides shorter than traditional sgRNAs83, 94. Tru-gRNA has been shown to reduce off-target mutagenesis in comparison to traditional sgRNA by two to fivefold, but does not reduce off-target mutagenesis to undetectable levels35. Another method uses the opposite approach and elongates the sgRNA by two additional guanine nucleotides to the 5′ end29. Although the extended gRNA has been shown to reduce off-targeting, it has also been shown to reduce actual on-target site activity as well. Thus, though these two methods of sgRNA manipulation seem to have some success, these methods alone do not seem to be enough for the specificity desired.
Additionally, some groups have modified the Cas9 protein itself in order to improve CRISPR/Cas9 specificity. One method utilizes paired Cas9 nickases (Cas9n) which create single-strand breaks instead of DSBs84, 85. This is achieved by deactivating one of the two nuclease domains of the Cas9. Two separate offset nicks, or single-strand breaks, are made on each strand of DNA, guided by two different sgRNAs. This method has been shown to reduce off-target mutagenesis by 50 to 1500-fold in human cells. It is not yet clear whether the usage of a second sgRNA may cause off-target mutagenesis at other locations. Cas9n has also been observed to have an increased frequency of point mutations, which are more difficult to detect than INDELs15, 87. Another variation of the Cas9 protein is dimeric RNA-guided FokI-dCas9nuclease (RFN) which has been created to reduce the off-target effects of Cas9n’s activity as a monomer86–88. The variation is created by fusing catalytically inactive Cas9, or dead Cas9 (dCas9) to dimerization-dependent FokI nuclease domain95, meaning two co-localized RFN are necessary for cleavage. This dimerization dependent activity also allows extended double-length target sites to be recognized for cleavage. RFN in combination with tru-gRNA, also known as tru-RFNs, has been shown to especially reduce off-target monomer activity86. The requirements of the dimerization however also limit the target range of the endonuclease.
Another variation or approach is engineered variants of SpCas9. SpCas9 high-fidelity variant 1 (SpCas9-HF1)4 and enhanced specificity SpCas9 version 1.1 (eSpCas9 1.1)89 have alanine substitutions at the predicted Cas9 protein residue that makes contact with the target DNA. This method disrupts the non-specific DNA contact points and diminishes off-target events. SpCas9-HF1 and eSpCas9 1.1 both show target site cleavage activity at rates comparable to SpCas9. More recently, another interesting variation of Cas9 has been reported: an expanded PAM SpCas9 variant (xCas9), engineered by phage-assisted continuous evolution (PACE)96. xCas9 is unique in that it recognizes a broader range of PAM sequences including NG, GAA, and GAT. The wider PAM recognition widens the scope of genetic sequences accessible to CRISPR. Current experimental data shows that though PAM compatibility is broadened, xCas9 actually yields lower rates of off-target activity with increased specificity. Still, while the engineered variants show great specificity with great reduction in off-target events, some off-targeting is observed.
Other methods of improving Cas9 specificity limits the duration of Cas9 activity to reduce the likelihood of off-target events. These methods include utilization of ribonucleoproteins (RNPs) which have been shown to be degraded within 24 hours versus several days of traditional Cas9 delivery90. Inducible mechanisms also exist including split Cas9 which is induced to dimerize in the presence of rapamycin for activation of enzymatic activity91. Another inducible mechanism represses the enzymatic activity of Cas9 by intein insertion92. This particular insertion is cleaved in the presence of 4-hydroxytamoxifen (4-HT), inducing Cas9 activity. Even a photoactivatable Cas9 (paCas9) exists93. PaCas9 is composed of split Cas9 fragments that are fused to dimerization domains called Magnets which dimerize upon blue light irradiation, activating endonuclease activity. Optogenetic control allows an inducible system that is also reversible. A very recent advancement in limiting Cas9 activity duration is the use of anti-CRISPR protein, AcrIIA4, or anti-CRISPR DNA mimic97. AcrIIA4 mimics DNA and binds to Cas9-sgRNA complexes, interfering with PAM recognition. AcrIIA4 could be administered after a specified time in order to stop Cas9 activity. Overall, reducing the duration of Cas9 activity does reduce off-target events, but still does not yield perfect specificity.
Another CRISPR approach that is in development as an alternative to HDR-dependent repair is the use of base editing98–101. Base editing systems are unique as they substitute nucleotides, unlike endonuclease cleavage systems which make DSBs. A third-generation base editor (BE3), which can permanently covert C:G base pairs to T:A base pairs, was introduced in the past year99. The BE3 mechanism is similar to a traditional CRISPR system in that a gRNA targets Cas9 to a specific locus. The difference lies in Cas9 engineering; BE3 utilizes inactive Cas9 tethered to a cytidine deaminase enzyme99, 100, 102, 103. Upon binding of the engineered Cas9 complex to DNA, a small window of DNA is exposed allowing for any cytidine to be deaminated to uracil. Through a series of steps, the mutated U:G is converted to a T:A104. Base editing has been applied to induction of STOP codons (iSTOP), which works by converting codons CAA, CAG, CGA, and TGG into STOP codons, TAA, TAG, and TGA105. iSTOP is advantageous as it could potentially treat dominant negative disorders without the need for HDR or NHEJ. Another application of base editing is CRISPR-X, which pairs dCas9 with activation-induced cytidine deaminase (AID), resulting in a diverse library of point mutations106. While base editing methods are promising, there are still drawbacks including: conversion of base pairs to T:A at undesired locations, limitation to the C:G to T:A conversion, and varied efficiency depending on the target. Recently, fourth-generation base editing (BE4) has been unveiled which increases editing efficiency and specificity104. BE4 is a fusion of BE3 and Gam, a bacteriophage Mu protein which binds ends of double-stranded DNA. This fusion seems to reduce INDEL formation leading to greater efficiency and specificity, but is still confined to some of the limitations of BE3, specifically the inability to make edits beyond the C:G to T:A conversion.
Conclusions
In recent years, immense research has been completed to improve genome engineering techniques and better understand the mechanisms and effects of genome engineering. Despite great successes and progress, limitations to genome engineering and our understanding of it exist. These limitations bring safety concerns to the usage of CRISPR-mediated genome surgery in the clinical setting. Yet, the CRISPR system has the potential to cure illnesses previously unapproachable. Further efforts to improve CRISPR/Cas9 specificity and efficacy and better define and understand off-target mutagenesis should be made in order to maximize the potentials of the CRISPR system and apply to clinical therapeutics. There is still great anticipation and expectation for CRISPR to be utilized as a therapeutic tool, given that it is with warranted judicious care.
Summary Statement.
This review gives an overview of off-targeting detection methods and CRISPR’s place in the clinical setting, specifically in the field of ophthalmology.
Acknowledgments
Funding
Funding/Support: VBM and AGB are supported by NIH grants [R01EY026682, R01EY024665, R01EY025225, R01EY024698 and R21AG050437], and Research to Prevent Blindness (RPB), New York, NY. The Barbara & Donald Jonas Laboratory of Regenerative Medicine and Bernard & Shirlee Brown Glaucoma Laboratory are supported by the National Institute of Health [5P30EY019007, R01EY018213, R01EY024698, R21AG050437], National Cancer Institute Core [5P30CA013696], the Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, NY, USA. SHT is a member of the RD-CURE Consortium and is supported by the Tistou and Charlotte Kerstan Foundation, the Schneeweiss Stem Cell Fund, New York State [C029572], the Foundation Fighting Blindness New York Regional Research Center Grant [C-NY05-0705-0312], the Joel Hoffman Fund, the Professor Gertrude Rothschild Stem Cell Foundation, and the Gebroe Family Foundation.
Role of the Sponsor: The funding organizations had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions
G.Y.C. performed the literature searches and composed the manuscript. K.A.S. assisted in the manuscript composition. V.B.M., A.G.B., S.H.T. oversaw all aspects of the manuscript preparation and hold final responsibility for contained information.
Conflict of Interest Disclosures
None reported.
References
- 1.Schaefer KA, et al. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 2017;14:547–548. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chew WL. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip Rev Syst Biol Med. 2018;10 doi: 10.1002/wsbm.1408. [DOI] [PubMed] [Google Scholar]
- 3.Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamal M, et al. Keeping CRISPR/Cas on-Target. Curr Issues Mol Biol. 2016;20:1–12. [PubMed] [Google Scholar]
- 6.Zhang XH, et al. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 8.Cho GY, et al. CRISPR-mediated Ophthalmic Genome Surgery. Curr Ophthalmol Rep. 2017;5:199–206. doi: 10.1007/s40135-017-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Geen H, Yu AS, Segal DJ. How specific is CRISPR/Cas9 really? Curr Opin Chem Biol. 2015;29:72–78. doi: 10.1016/j.cbpa.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanik M, et al. In vivo genome editing as a potential treatment strategy for inherited retinal dystrophies. Prog Retin Eye Res. 2017;56:1–18. doi: 10.1016/j.preteyeres.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Wu WH, et al. CRISPR Repair Reveals Causative Mutation in a Preclinical Model of Retinitis Pigmentosa. Mol Ther. 2016;24:1388–1394. doi: 10.1038/mt.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo T, Lee J, Kim JS. Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol Cells. 2015;38:475–481. doi: 10.14348/molcells.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakurai T, et al. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 2014;14:69. doi: 10.1186/1472-6750-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 21.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabral T, et al. CRISPR applications in ophthalmologic genome surgery. Curr Opin Ophthalmol. 2017 doi: 10.1097/ICU.0000000000000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 35.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv. 2017;35:95–104. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Lee CM, et al. Nuclease Target Site Selection for Maximizing On-target Activity and Minimizing Off-target Effects in Genome Editing. Mol Ther. 2016;24:475–487. doi: 10.1038/mt.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin F, et al. Biased and Unbiased Methods for the Detection of Off-Target Cleavage by CRISPR/Cas9: An Overview. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17091507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 41.Pattanayak V, et al. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasue A, et al. Relationship between somatic mosaicism of Pax6 mutation and variable developmental eye abnormalities-an analysis of CRISPR genome-edited mouse embryos. Sci Rep. 2017;7:53. doi: 10.1038/s41598-017-00088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong H, et al. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 2015;5:8366. doi: 10.1038/srep08366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai SQ, Joung JK. What’s changed with genome editing? Cell Stem Cell. 2014;15:3–4. doi: 10.1016/j.stem.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 46.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–280. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 48.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 49.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 231 p following 243. [DOI] [PubMed] [Google Scholar]
- 52.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuscu C, et al. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Niu Y, Ji W. Genome editing in nonhuman primates: approach to generating human disease models. J Intern Med. 2016;280:246–251. doi: 10.1111/joim.12469. [DOI] [PubMed] [Google Scholar]
- 59.Ma H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 60.Egli D, et al. Inter-homologue repair in fertilized human eggs? bioRxiv. 2017 doi: 10.1038/s41586-018-0379-5. [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg EA, Sperazza LC. The visually impaired patient. Am Fam Physician. 2008;77:1431–1436. [PubMed] [Google Scholar]
- 62.Friedman DS, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 63.Sengillo JD, et al. Gene and cell-based therapies for inherited retinal disorders: An update. Am J Med Genet C Semin Med Genet. 2016;172:349–366. doi: 10.1002/ajmg.c.31534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sundaram V, et al. Retinal dystrophies and gene therapy. Eur J Pediatr. 2012;171:757–765. doi: 10.1007/s00431-011-1615-2. [DOI] [PubMed] [Google Scholar]
- 65.Cepko CL, Vandenberghe LH. Retinal gene therapy coming of age. Hum Gene Ther. 2013;24:242–244. doi: 10.1089/hum.2013.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bennett J, et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med. 2012;4:120ra115. doi: 10.1126/scitranslmed.3002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bennett J, et al. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacLaren RE, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conlon TJ, et al. Preclinical potency and safety studies of an AAV2-mediated gene therapy vector for the treatment of MERTK associated retinitis pigmentosa. Hum Gene Ther Clin Dev. 2013;24:23–28. doi: 10.1089/humc.2013.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghazi NG, et al. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum Genet. 2016;135:327–343. doi: 10.1007/s00439-016-1637-y. [DOI] [PubMed] [Google Scholar]
- 71.Feuer WJ, et al. Gene Therapy for Leber Hereditary Optic Neuropathy: Initial Results. Ophthalmology. 2016;123:558–570. doi: 10.1016/j.ophtha.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan X, et al. Efficacy and Safety of rAAV2-ND4 Treatment for Leber’s Hereditary Optic Neuropathy. Sci Rep. 2016;6:21587. doi: 10.1038/srep21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DiCarlo JE, et al. CRISPR-Cas Genome Surgery in Ophthalmology. Transl Vis Sci Technol. 2017;6:13. doi: 10.1167/tvst.6.3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bassuk AG, et al. Precision Medicine: Genetic Repair of Retinitis Pigmentosa in Patient-Derived Stem Cells. Sci Rep. 2016;6:19969. doi: 10.1038/srep19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cramer AO, MacLaren RE. Translating induced pluripotent stem cells from bench to bedside: application to retinal diseases. Curr Gene Ther. 2013;13:139–151. doi: 10.2174/1566523211313020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.den Hollander AI, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanein S, et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23:306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 78.Ruan GX, et al. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol Ther. 2017;25:331–341. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maeder ML, et al. Therapeutic Correction of an LCA-Causing Splice Defect in the CEP290 Gene by CRISPR/Cas-Mediated Gene Editing. Molecular Therapy. 2016;24(Supplement 1):S51–S52. 124. [Google Scholar]
- 80.Yang T, et al. BEST1: the Best Target for Gene and Cell Therapies. Mol Ther. 2015;23:1805–1809. doi: 10.1038/mt.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor-Weiner H, Graff Zivin J. Medicine’s Wild West–Unlicensed Stem-Cell Clinics in the United States. N Engl J Med. 2015;373:985–987. doi: 10.1056/NEJMp1504560. [DOI] [PubMed] [Google Scholar]
- 82.Boudreault K, et al. Complication of Autologous Stem Cell Transplantation in Retinitis Pigmentosa. JAMA Ophthalmol. 2016;134:711–712. doi: 10.1001/jamaophthalmol.2016.0803. [DOI] [PubMed] [Google Scholar]
- 83.Fu Y, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wyvekens N, et al. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim S, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davis KM, et al. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nihongaki Y, et al. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 94.Fu Y, Reyon D, Joung JK. Targeted genome editing in human cells using CRISPR/Cas nucleases and truncated guide RNAs. Methods Enzymol. 2014;546:21–45. doi: 10.1016/B978-0-12-801185-0.00002-7. [DOI] [PubMed] [Google Scholar]
- 95.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J Mol Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 96.JH Hu, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018 doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shin J, et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv. 2017;3:e1701620. doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rees HA, et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hess GT, et al. Methods and Applications of CRISPR-Mediated Base Editing in Eukaryotic Genomes. Mol Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim YB, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nishida K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 104.Komor AC, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Billon P, et al. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol Cell. 2017;67:1068–1079 e1064. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hess GT, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arno G, et al. Mutations in REEP6 Cause Autosomal-Recessive Retinitis Pigmentosa. Am J Hum Genet. 2016;99:1305–1315. doi: 10.1016/j.ajhg.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Agrawal SA, et al. REEP6 Deficiency Leads to Retinal Degeneration through Disruption of ER Homeostasis and Protein Trafficking. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Suzuki K, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu W, et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu J, et al. Gene and mutation independent therapy via CRISPR-Cas9 mediated cellular reprogramming in rod photoreceptors. Cell Res. 2017;27:830–833. doi: 10.1038/cr.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bakondi B, et al. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Latella MC, et al. In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina. Mol Ther Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Collery RF, et al. Loss of Zebrafish Mfrp Causes Nanophthalmia, Hyperopia, and Accumulation of Subretinal Macrophages. Invest Ophthalmol Vis Sci. 2016;57:6805–6814. doi: 10.1167/iovs.16-19593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim K, et al. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017 doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burnight ER, et al. Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhattacharya S, et al. Prominin-1 Is a Novel Regulator of Autophagy in the Human Retinal Pigment Epithelium. Invest Ophthalmol Vis Sci. 2017;58:2366–2387. doi: 10.1167/iovs.16-21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yiu G, et al. Genomic Disruption of VEGF-A Expression in Human Retinal Pigment Epithelial Cells Using CRISPR-Cas9 Endonuclease. Invest Ophthalmol Vis Sci. 2016;57:5490–5497. doi: 10.1167/iovs.16-20296. [DOI] [PMC free article] [PubMed] [Google Scholar]


