Abstract
Background & Aims
The degree of cholestasis is an important disease driver in alcoholic hepatitis, a severe clinical condition that needs new biomarkers and targeted therapies. The mechanisms and biomarkers linked to cholestasis are largely unknown.
Methods
Here, we analyzed a well characterized cohort of patients with alcoholic hepatitis and correlated clinical and histological parameters and outcomes with serum bile acids and fibroblast growth factor 19 (FGF19), a major regulator of bile acids synthesis.
Results
We found that total and conjugated bile acids were significantly increased in patients with alcoholic hepatitis compared with controls. Serum FGF19 levels were strongly increased and gene expression of FGF19 was induced in biliary epithelial cells and ductular cells of patients with alcoholic hepatitis. De novo bile acid synthesis (CYP7A1 gene expression and C4 serum levels) was significantly decreased in alcoholic hepatitis patients. Importantly, total and conjugated bile acids correlated positively with FGF19 and with disease severity (MELD). FGF19 correlated best with conjugated cholic acid (CA), and MELD best with taurine-conjugated chenodeoxycholic acid (T-CDCA). Univariate analysis demonstrated significant associations between FGF19 and bilirubin as well as gamma-glutamyl-transferase (GGT), and negative correlations between FGF19 and fibrosis stage as well as polymorphonuclear leukocyte (PMN) infiltration in all alcoholic hepatitis patients.
Conclusion
Serum FGF19 and bile acids are significantly increased in patients with alcoholic hepatitis, while de novo bile acid synthesis is suppressed. Modulation of bile acid metabolism or signaling could represent a promising target for treatment of alcoholic hepatitis in humans.
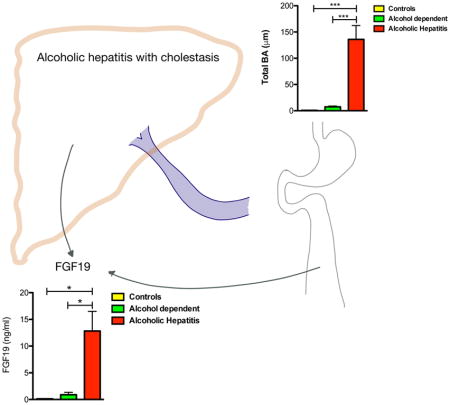
Understanding underlying mechanisms that drive alcoholic hepatitis is important for the development of new biomarkers and targeted therapies. Here we describe a molecule that is increased in patients with alcoholic hepatitis and modulating this pathway might lead to promising targets for the treatment of alcoholic hepatitis.
Keywords: FGF19, bile acids, microbiome
Graphical Abstract
Introduction
Alcohol abuse is the most important cause for liver disease worldwide 1. The most severe form of alcoholic liver disease is alcoholic hepatitis with mortality rates of 20–40% at 1–6 months, and a 90-day mortality rate of up to 75% in severe alcoholic hepatitis 2–4. Corticosteroids are the only effective medical therapy but failure in many patients has been reported 2. There is no cure for patients not responding to medical therapy, except for early liver transplantation that is offered in some centers to a highly selected group of patients 5.
Alcoholic hepatitis is a hepatocellular disease associated with cholestasis and accumulation of bile acids in the liver and the systemic circulation 6. Accumulation of bile acids in the liver can result in hepatocellular damage followed by inflammation and fibrosis 7. Early studies using high-performance liquid chromatography (HPLC) have identified elevated bile acid levels in patients with alcoholic hepatitis and a positive correlation with the alcoholic hepatitis score 8. The liver controls bile acid homeostasis via a complicated network of pathways regulated by nuclear receptors 7. One important regulator is fibroblast growth factor 19 (FGF19), a hormone that is secreted in the intestine and reaches the liver via the portal circulation. Binding of FGF19 to its receptor, fibroblast growth factor receptor 4 and β-Klotho, activates mitogen-activated protein kinase pathways, resulting in the downregulation of the rate-limiting enzyme of bile acid synthesis, cytochrome P450 family 7 subfamily A member 1 (CYP7A1), gene expression and inhibition of bile acid synthesis 9. Although FGF19 is not detectable in the liver under normal conditions, previous studies suggest that FGF19 is expressed under cholestatic conditions, such as in patients with primary biliary cholangitis. Plasma/serum elevated FGF19 levels are elevated in patients with various hepatic disorders including non-cirrhotic and cirrhotic primary biliary cholangitis and in patients with extrahepatic cholestasis 9–11. FGF19 mRNA expression was increased in the terminal ileum of actively drinking patients with cirrhosis when compared with patients with cirrhosis of nonalcoholic etiology 12.
In this study, we first determined serum bile acid levels and bile acid composition in patients with alcoholic hepatitis using mass spectrometry. We further investigated the relationship between bile acids, FGF19 and histopathological parameters and clinical outcomes in the setting of alcoholic hepatitis. The results of this study can help to understand the pathophysiology of alcoholic hepatitis as well as to evaluate the role of recombinant FGF19 preparations used in treatment.
Methods
Patient cohorts
132 patients with alcoholic hepatitis, nine patients with alcohol use disorder, and nine non-alcoholic control patients were enrolled from the InTeam Consortium from the 12 participating centers. Inclusion criteria were: 1. Age >18 years and ≤ 70 years, 2. Active alcohol abuse (>50 g/day for men and >40 g/day for women) in the last 3 months, 3. aspartate aminotransferase (AST) > alanine aminotransferase (ALT) and total bilirubin > 3 mg/dl in the past 3 months, 4. Liver biopsy and/or clinical picture consistent with alcoholic hepatitis. Exclusion criteria were: 1. Autoimmune liver disease (ANA > 1/320), 2. Chronic viral hepatitis, 3. Hepatocellular carcinoma, 4. Complete portal vein thrombosis, 5. Extrahepatic terminal disease, 6. Pregnancy, and 7. Lack of signed informed consent. The mortality was evaluated at 30 days. For the three patients who underwent liver transplantation, the transplantation date was considered as date of death. The protocol was approved by the Ethics Committee of each participating center and patients were enrolled after written informed consent was obtained from each patient. FGF19 levels, C4 levels, serum bile acid levels, other clinical laboratory parameters and clinical outcomes were measured and collected. The Model for End-stage Liver Disease (MELD) score was calculated from all patients from whom bilirubin level, international normalized ratio (INR), and creatinine level was available.
Bile acid measurements
Serum levels of total bile acids, conjugated and unconjugated as well as specific bile acids were measured in an Agilent series 6120 quadrupole mass detector as described previously 13. In brief, serum (20 μl) was protein precipitated with 80 μl of cold acetonitrile containing deuterated cholic acid as an internal standard, vortexed for 1 min and centrifuged at 10,000 rpm for 10 min at 4°C. Supernatant was evaporated under vacuum and reconstituted in assay mobile phase. Bile acid separation was achieved using an Acuity (Waters, Milford, MA) UPLC BEH C18 column (1.7microns 2.1×100mm) on a Nextera UPLC (Shimadzu, Tokyo, Japan); the temperature of the column and auto sampler was 65°C and 12°C, respectively. Sample injection volume was 1μl. The mobile phase consisted of 10% acetonitrile and 10% methanol in water containing 0.1% formic acid (Mobile Phase A) and 10% methanol in acetonitrile 0.1% formic acid (Mobile Phase B) delivered as a gradient: 0–5 min Mobile Phase B held at 22%; 5–12 min Mobile Phase B increased linearly to 60%, 12–15min Mobile Phase B increased linearly to 80% and 15–19 min Mobile Phase B constant at 80% at a flow rate of 0.5ml/min. The mass spectrometer (Q-Trap 5500; Sciex, Framingham, MA) was operated in negative electro-spray mode working in the multiple reaction mode (MRM). Operating parameters were: curtain gas 30psi; ion spray voltage 4500 V; temperature 550°C; ion source gas 1 60psi; ion source gas 2 65psi. Transition MRMs, declustering potential, entrance potentials and collision cell exit potentials were optimized using the Analyst software (Sciex, Framingham, MA). Dwell times were 25msec. Only bile acids with an overall contribution of >2% in healthy subjects were analyzed and depicted in figures and tables.
Quantification of C4 levels
7-alpha-Hydroxy-4-cholesten-3-one (C4) was purchased from Toronto Research Chemicals (Toronto, Canada). Cortisol-1,2-d2 was purchased from CDN Isotopes (Hornsby, NSW, Australia) and Charcoal Stripped Serum was purchased from Sigma-Aldrich (Castle Hill, NSW, Australia). Additional reagents and solvents were of HPLC grade. To 10μL of serum, 50μL of ice cold Acetonitrile containing 2% Formic Acid and 1.8μg of the assay Internal standard Cortisol-1,2-d2, was added. The mixture was vortexed for 1 minute and centrifuged at 10 000rpm for 10 minutes. The supernatant was collected and evaporated under vacuum at room temperature. The samples were then reconstituted in the assay mobile phase and transferred to a 96 well plate for analysis. Stock solution of C4 (1μM) was prepared in Charcoal Stripped Serum (CSS) and this was further diluted in CSS to give final concentrations of 0.01–1μM. The standards were then treated in the same way as the samples. A Nextera UPLC (SHIMADZU, Kyoto, Japan) system was used in combination with a Q-TRAP 5500 Mass Spectrometer (AB SCIEX, Toronto, Canada) with Analyst Software 1.6.2. Chromatographic separations were performed with an ACQUITY (WATERS, Milford, MA) UPLC BEH C18 column (1.7microns 2.1×100mm). The temperature of the column and auto sampler was 65°C and 12°C, respectively. Sample injection was 1μL. The mobile phase consisted of 10% Acetonitrile and 10% Methanol in water containing 0.1% Formic Acid (Mobile Phase A) and 10% Methanol in Acetonitrile 0.1% Formic Acid (Mobile Phase B) delivered as a gradient: 0–3-min Mobile Phase B 20%; 3–3.5-min Mobile Phase B 80%, 7–9min Mobile Phase B was a constant 80% at a flow rate of 0.5ml/min. The mass spectrometer was operated in positive electro-spray mode working in the multiple reaction mode (MRM). Transition MRMs for C4 and the Internal Standard Cortisol-1,2-d2 were 401.2→177.2 and 365.2→122.2 respectively. Operating parameters were: Curtain gas 30psi; Ion spray voltage 4500 V; Temperature 550C; Ion Source Gas 1 60psi; Ion Source Gas2 65psi. Declustering Potential, Entrance Potentials and Collision Cell Exit Potentials were optimized using the Analyst software.
FGF19 ELISA
FGF19 concentrations were measured from serum samples using an ELISA kit from Biovendor (Asheville, NC) according to the manufacturer’s protocol.
FGF19 immunohistochemistry
Immunohistochemistry was performed on 4 micron formalin-fixed paraffin-embedded liver core biopsies. The tissue sections were stained with an antibody to FGF19 (1:300) from Cell Signaling (Danvers, MA). Slides were stained on a Ventana Discovery Ultra (Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval was performed using CC1 for 92 minutes at 95°C. Due to the very low expression level of FGF19, IHC detection required the use of the HQ amplification system (Ventana) to detect the weakly expressing FGF19 cells within the liver and was followed by an HRP-conjugated antibody and DAB as a chromagen. IHC staining was followed by a hematoxylin counterstain. Slides were rinsed, dehydrated through alcohol and xylene and coverslipped.
RNA Sequencing
High-throughput transcriptome profiling by RNA sequencing (RNA-Seq) was performed using Illumina HiSeq2000 platform (San Diego, CA) with liver samples provided by the InTeam Consortium Human Biorepository Core (University of Pittsburgh, PA) as described (Manuscript in preparation; Argemi JM, Bataller R et al.). 51 patients with different phenotypes were selected: 12 patients with early alcoholic steatohepatitis, 11 patients with non-severe alcoholic hepatitis (with Maddrey discriminant function (MDF ≤32), 18 patients with severe alcoholic hepatitis (MDF>32), 10 explants from patients who underwent early liver transplantation for severe alcoholic hepatitis. Controls (n=10) were non-diseased livers obtained from biopsied single nodules, whose histology was without significant alterations.
Statistical analysis
Statistical analysis was conducted using GraphPad PRISM (La Jolla, USA) and R statistical software 14. Comparisons between three groups or more were conducted using One-Way ANOVA with Bonferroni post-test for multiple comparisons (for continuous variables) and Chi-squared tests (for categorical variables) using GraphPad PRISM. A comparison of two groups was performed using the unpaired student t-test. Univariate regression analysis was performed using R to determine associations of FGF19 and MELD with several biochemical markers and clinical outcomes. Cox univariate regression analysis was used to determine associations of FGF19 with 30-day mortality.
Results
Demographic and laboratory data
A total of 33 subjects were initially assessed: Nine non-alcoholic control subjects, nine patients with alcohol use disorder and 15 untreated patients with alcoholic hepatitis. Their demographic and laboratory data are shown in Table 1. Patients with alcoholic hepatitis had significantly higher levels of bilirubin, aspartate aminotransferase (AST), albumin and international normalized ratio (INR) compared with alcohol dependent and control subjects. Alanine aminotransferase (ALT) levels were significantly elevated in alcoholic hepatitis patients relative to controls. The mean Model for End-stage Liver Disease (MELD) value of patients with alcoholic hepatitis was 25.
Table 1.
Demographic and laboratory results of controls, alcohol dependent and patients with alcoholic hepatitis.a
| Variables | Controls (n=9) | Alcohol dependent (n=9) | Patients with alcoholic hepatitis (n=15)b |
|---|---|---|---|
| Gender (% male) | 100 | 100 | 60 |
| Age (years) | 59 (48–71) | 50 (44–57) | 54 (49–60) |
| Creatinine (mg/dl) | 1.1 (1.0–1.2) | 0.8 (0.6–1.0) | 1.5 (0.5–2.5) |
| Bilirubin (mg/dl) | 0.8 (0.6–1.0)b | 0.9 (0.6–1.1)c | 15.7 (10.7–20.7)b,c |
| AST (IU/L) | 20 (17–24)b | 78 (43–113)c | 201 (141–262)b,c |
| ALT (IU/L) | 23 (20–27)b,c | 64 (28–100)b | 76 (58–94)c |
| Albumin (g/dl) | 3.9 (3.6–4.3)b | 3.7 (3.1–4.3)c | 2.6 (2.3–2.9)b,c |
| INR | 1.0 (0.9–1.1)b | 1.0 (0.9–1.1)c | 1.7 (1.5–1.9)b,c |
| MELD score | NA | NA | 25 (20–30) |
Values are presented as mean with 95% Confidence Interval.
The fraction of missing values for each parameter is presented in Supplemental Table 1.
All patients with alcoholic hepatitis were untreated
One-Way ANOVA with Bonferroni post-test (for continuous variables) and Chi-squared tests (for categorical variables). For each comparison, identical letters (b, c) indicate the groups between which the statistical difference was significant (p-value < 0.05).
ALT, alanine aminotransferase; AST, aspartate aminotransferase; INR, international normalized ratio; MELD, Model for End-stage Liver Disease.
Serum bile acid levels and bile acid composition
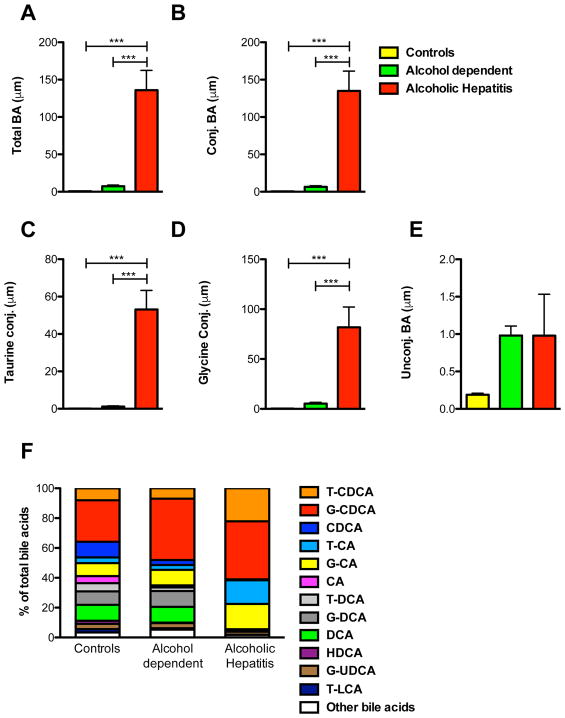
To assess alterations in bile acid levels and bile acid composition, serum samples were analyzed from patients with alcoholic hepatitis at the time of inclusion, patients with alcohol use disorder and controls using mass spectrometry. The results of the bile acid measurements are depicted in Fig. 1. Total and conjugated bile acids were significantly elevated in alcoholic hepatitis patients compared with controls and patients with alcohol use disorder (Fig. 1A and B). In addition, the individual taurine- and glycine-conjugates were significantly increased in patients with alcoholic hepatitis in relation to patients with alcohol use disorder and controls (Fig. 1C and D). In contrast, there was no significant difference in the unconjugated bile acids comparing the three groups in absolute concentrations (Fig. 1E).
Figure 1. Patients with alcoholic hepatitis have significantly higher serum levels of total and conjugated bile acids than controls and patients with alcohol use disorder.
Sera of 15 patients with alcoholic hepatitis, 9 patients with alcohol use disorder and 9 control subjects were analyzed. (A) Total bile acids. (B) Conjugated bile acids. (C) Taurine-conjugated bile acids. (D) Glycine-conjugated bile acids. (E) Unconjugated bile acids. (F) Bile acid composition. Only bile acids with contribution of > 2% in healthy subjects are depicted. One-Way ANOVA with Bonferroni post-test.
***P < 0.001. BA, bile acid; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; G, glycine; HDCA, hyodeoxycholic acid; T, taurine; UDCA, ursodeoxycholic acid; LCA, lithocholic acid.
Bile acid composition analysis of the different patient populations revealed significantly elevated glyco-cholic acid (G-CA), tauro-cholic acid (T-CA) and tauro-chenodeoxycholic acid (T-CDCA) in alcoholic hepatitis patients compared with alcohol dependent patients and controls. In contrast, patients with alcoholic hepatitis showed significantly lower levels of chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), glyco-deoxycholic acid (G-DCA) and cholic acid (CA) as well as lower proportional contributions of total unconjugated bile acids compared with control patients and patients with alcohol use disorder (Fig. 1F). Hyodeoxycholic acid (HDCA), tauro-deoxycholic acid (T-DCA) and tauro-lithocholic acid (T-LCA) were also significantly lower in patients with alcoholic hepatitis compared with controls. Conjugated CDCA and conjugated CA were significantly higher in alcoholic hepatitis patients when compared with controls (61.0% vs 35.8%, and 32.8% vs 12.6%, respectively) and conjugated CA were also significantly higher in alcoholic hepatitis patients compared with alcohol dependent patients (32.8% vs. 13.6%). In contrast, conjugated DCA were decreased in patients with alcoholic hepatitis compared with alcohol dependent patients and controls (1.4% vs 13.0% and 14.5%) (Fig. 1F).
Regulation of bile acid synthesis
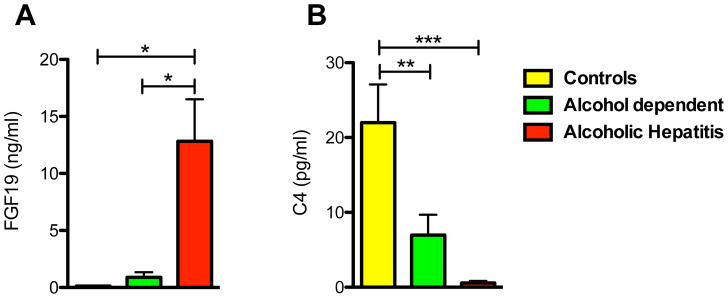
To understand the regulation and metabolism of bile acids in alcoholic hepatitis, serum 7-alpha-hydroxy-4-cholesten-3-one (C4) levels and FGF19 levels were determined. C4, a bile acid synthesis intermediate serves as a marker for de novo bile acid synthesis while FGF19 represents an important negative feedback regulator of bile acid synthesis. FGF19 levels were significantly elevated in patients with alcoholic hepatitis relative to patients with alcohol use disorder and controls (Fig. 2A). In line with these results, serum C4 levels were significantly decreased in patients with alcoholic hepatitis compared with controls (Fig. 2B). These results indicate that FGF19 regulates de novo bile acid synthesis, but total serum bile acid levels are not affected.
Figure 2. Patients with alcoholic hepatitis show significantly elevated serum levels of FGF19 and significantly suppressed de novo bile acid synthesis relative to controls and patients with alcohol use disorder.
Sera of 15 patients with alcoholic hepatitis, 8–9 patients with alcohol use disorder and 9 control subjects were analyzed. (A) FGF19 concentrations. (B) C4 levels, a marker for de novo bile acid synthesis. One-Way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, ***P < 0.001. C4, 7-alpha-hydroxy-4-cholesten-3-one; FGF19, fibroblast growth factor 19.
Analysis of hepatic FGF19 gene expression in different patient populations as well as of hepatic genes related to bile acid uptake, synthesis and efflux
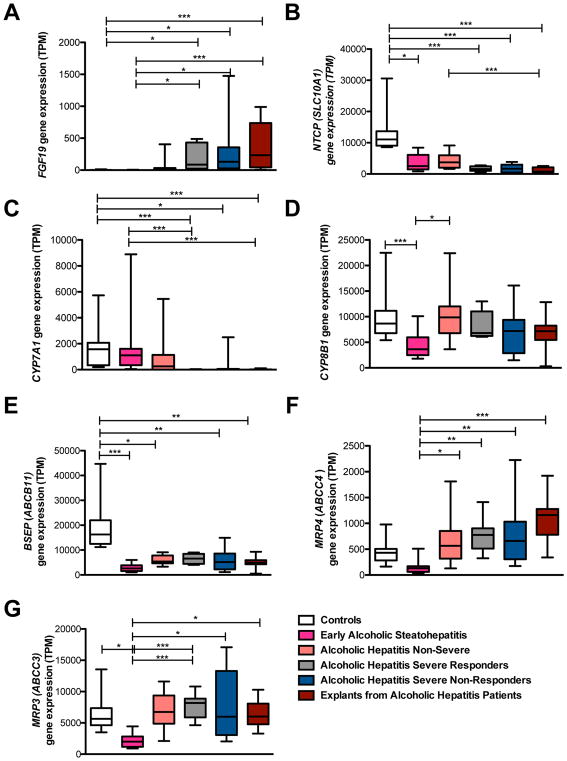
FGF19 serum levels measured in patients with alcoholic hepatitis were markedly higher (by ~100 fold) compared with FGF19 levels detected in patients with non-alcoholic steatohepatitis (NASH) or non-alcoholic fatty liver (NAFL) whose serum levels were similar to control subjects 15. In addition, as intrahepatic cholestasis is associated with hepatic expression of FGF19 in conditions such as primary biliary cholangitis 10, we wanted to determine whether hepatic FGF19 production was also induced in alcoholic hepatitis. Hepatic FGF19 gene expression was hence quantitated in multiple patient groups with different liver pathologies. Fig. 3 depicts mRNA expression levels in patients with normal liver histology (control subjects), early alcoholic steatohepatitis, non-severe alcoholic hepatitis, severe alcoholic hepatitis responders and non-responders as well as explants from alcoholic hepatitis patients. Hepatic FGF19 expression was essentially undetectable in controls and in patients with early alcoholic steatohepatitis, but its levels increased with severity of liver pathology, and significantly elevated hepatic FGF19 gene expression was detected in patients with severe alcoholic hepatitis (responders as well as non-responders) and in explants from alcoholic hepatitis patients compared with control subjects (Fig. 3A). In addition, hepatic expression levels of genes involved in bile acid uptake, synthesis, and excretion were analyzed in the aforementioned patient groups. Patients with increasing severity of liver pathology showed a lower gene expression of Na+-taurocholate co-transporting polypeptide (NTCP; SLC10A1) that is involved in bile acid uptake into hepatocytes (Fig. 3B). Worsening liver disease was also associated with a reduction of CYP7A1 expression and hence impaired bile acid synthesis (Fig. 3C), which is consistent with low serum C4 levels. The expression of another gene involved in bile acid synthesis, cytochrome P450 family 8 subfamily B member 1 (CYP8B1), was relatively stable and independent of the degree of liver disease (Fig. 3D). The expression of the bile salt export pump (BSEP; ABCB11) important for bile acid excretion into bile canaliculi was significantly decreased in patients with different severity of alcoholic hepatitis when compared with control subjects (Fig. 3E). The hepatic gene expression of transporters important for bile acid efflux from the basolateral membrane into the systemic circulation multidrug resistance-associated protein 4 (MRP4; ABCC4) and MRP3 (ABCC3) correlated with worsening alcoholic hepatitis (Fig. 3F and G).
Figure 3. Severe forms of alcoholic hepatitis are associated with marked hepatic FGF19 production, down-regulated hepatic bile acid uptake and synthesis, as well as induced bile acid efflux.
Hepatic gene expression was quantitated in 12 patients with early alcoholic steatohepatitis, 11 patients with non-severe alcoholic hepatitis, 18 patients with severe alcoholic hepatitis (responders and non-responders), 10 with “terminal” alcoholic hepatitis (with liver failure and early transplantation), and 10 controls with non-diseased livers. (A) FGF19. (B) Bile acid uptake transporter NTCP (SLC10A1). (C) CYP7A1. (D) CYP8B1. (E) BSEP (ABCB11) important for bile acid excretion into bile canaliculi. (F – G) Transporters for bile acid efflux MRP4 (ABCC4; F) and MRP3 (ABCC3; G). One-Way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, ***P < 0.001. ABCB11, ATP-binding cassette, sub-family B member 11; ABCC3/4, ATP-binding cassette, sub-family C member 3/4; BSEP, bile salt export pump; CYP7A1, cytochrome P450 family 7 subfamily A member 1; CYP8B1, cytochrome P450 family 8 subfamily B member 1; FGF19, fibroblast growth factor 19; MRP3/4, multidrug resistance-associated protein 3/4; NTCP, Na+-taurocholate co-transporting polypeptide; SLC10A1, solute carrier family 10 member 1; TPM, transcripts per million.
Biliary epithelial cells and ductular cells express FGF19 in the liver of patients with alcoholic hepatitis
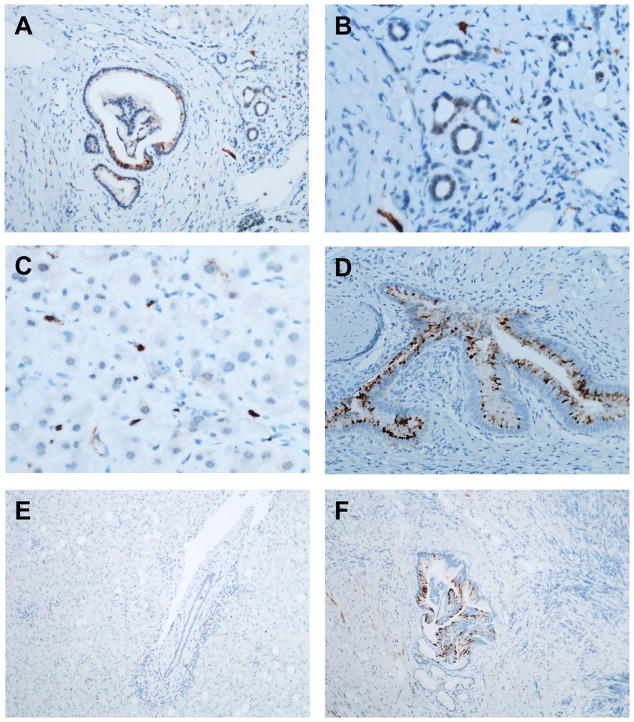
Given few reports about hepatic FGF19 synthesis in different cells such as hepatocytes in primary biliary cholangitis 10 or hepatic cancer stem cells in hepatocellular carcinoma (HCC) 16, we determined the cellular source of FGF19 production, after establishing that alcoholic hepatitis patients show induced FGF19 mRNA in their liver (Fig. 3A). Cholangiocytes in larger bile ducts and (to a lesser degree) ductular cells of smaller ductules were FGF19-positive in patients with alcoholic hepatitis (Fig. 4A and B). A few non-parenchymal cells (most likely endothelial cells) also stained positive for FGF19 (Fig. 4C). Gallbladder epithelial cells from a resected gall bladder served as positive staining control (Fig. 4D). Non-neoplastic liver (adjacent to a resected HCC) with portal tracts was negative for FGF19 protein (Fig. 4E), although large caliber bile ducts are weakly positive for FGF19 (Fig. 4F).
Figure 4. FGF19 immunohistochemistry in liver biopsies from patients with alcoholic hepatitis.
(A – C) Alcoholic hepatitis. (A) Intense staining in a large bile duct and moderate staining in bile ductules (magnification ×200). (B) Moderate staining in bile ductules (magnification ×400). (C) Intense staining in non-parenchymal cells in the sinusoids (magnification ×400). (D) Gallbladder specimen as positive control (magnification ×200). (E) Non-neoplastic liver tissue adjacent to resected HCC as negative control (magnification ×200). (F) Large caliber bile ducts in normal liver tissue adjacent to resected HCC stained positive for FGF19 (magnification ×100). FGF19, fibroblast growth factor 19; HCC, hepatocellular carcinoma.
Specific bile acids associated with serum FGF19, MELD, and ALT levels
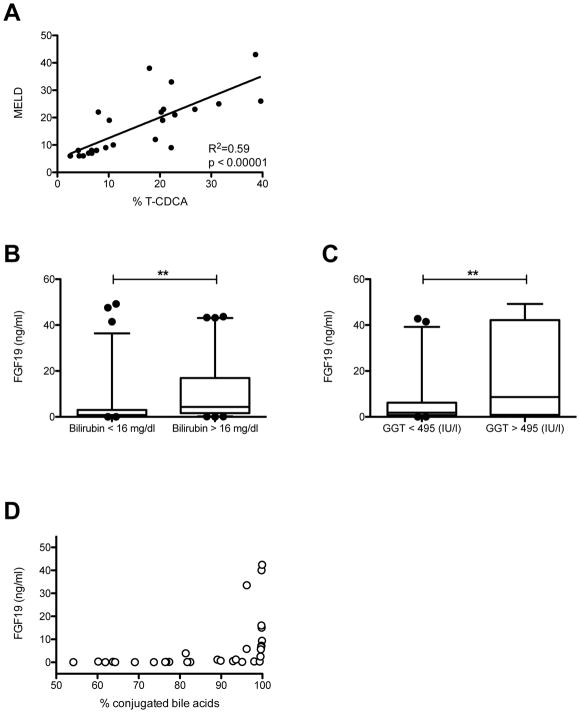
As bile acid levels were much higher in alcoholic hepatitis patients than in controls and patients with alcohol use disorder (Fig. 1), we proceeded to perform univariate regression analysis of the most abundant bile acids with MELD and serum FGF19 levels. Our analysis showed significant positive associations between total bile acids, absolute conjugated bile acids, proportional conjugated bile acids with MELD or FGF19 levels (Table 2, 3). The unconjugated bile acids CDCA, CA, DCA as well as conjugated DCA negatively correlated with MELD and FGF19 levels (Table 2, 3). MELD was also significantly associated with T-BA, T-CA and conjugated CA but even more significantly with T-CDCA (Table 2 and Fig. 5A). In addition to MELD, bilirubin values were analyzed and associated with various bile acids. Univariate regression analysis yielded similar results to those obtained with MELD and bile acids (Supplementary Table 1). Furthermore, the conjugated-to-unconjugated bile acid ratio, G-CA, T-CA, and conjugated CA were highly significantly correlated – all positively – with serum FGF19 (Table 3).
Table 2.
Univariable linear regression analysis for bile acids associated with MELD
| Simple linear regression with predictor variables | ||||
|---|---|---|---|---|
|
| ||||
| Variablea | B | SE | p-valueb | R2 |
| Total BA (abs.) | 0.045 | 0.020 | 0.036 | 0.177 |
|
| ||||
| Conj. BA (abs.) | 0.045 | 0.020 | 0.036 | 0.177 |
| Unconj. BA (abs.) | 0.294 | 1.343 | 0.829 | 0.002 |
|
| ||||
| Conj. BA | 48.590 | 11.96 | <0.001 | 0.418 |
| Unconj. BA | −48.597 | 11.96 | <0.001 | 0.418 |
| Conj./Unconj. BA | 0.0102 | 0.006 | 0.101 | 0.113 |
|
| ||||
| T-BA | 39.505 | 8.986 | <0.001 | 0.457 |
| G-BA | −12.60 | 13.35 | 0.355 | 0.037 |
|
| ||||
| CDCA | −120.454 | 43.061 | 0.010 | 0.254 |
| CA | −278.023 | 92.378 | 0.006 | 0.283 |
| DCA | −77.900 | 22.904 | 0.002 | 0.335 |
| HDCA | −288.234 | 141.664 | 0.054 | 0.153 |
|
| ||||
| T-CDCA | 75.666 | 13.064 | <0.00001 | 0.593 |
| T-CA | 54.89 | 17.90 | 0.005 | 0.290 |
| T-DCA | −168.010 | 77.169 | 0.040 | 0.171 |
| T-LCA | −199.300 | 110.167 | 0.084 | 0.125 |
|
| ||||
| G-CDCA | 1.292 | 14.936 | 0.932 | 0.000 |
| G-CA | 52.551 | 34.941 | 0.146 | 0.090 |
| G-DCA | −119.764 | 28.048 | <0.001 | 0.000 |
| G-UDCA | −54.546 | 54.630 | 0.328 | 0.042 |
|
| ||||
| Conj. CDCA | 32.7728 | 11.375 | 0.008 | 0.265 |
| Conj. CA | 36.234 | 12.971 | 0.010 | 0.253 |
| Conj. DCA | −89.155 | 21.852 | <0.001 | 0.420 |
Notes:
Bile acids (BA) are expressed as percent from total BA, except for “Total BA (abs.)”, “Conj. BA (abs.)” and “Unconj. BA (abs.)”; these are expressed as absolute (abs.) values.
Significance is indicated in bold. Univariable regression analysis.
Abbreviations:
B=non-standardized regression coefficient, SE=Standard Error, R2=coefficient of determination
BA, bile acid; CA, cholic acid; CDCA, chenodeoxycholic acid; conj., conjugated; DCA, deoxycholic acid G-BA, glycine-conjugated bile acid; G-CA, glycocholic acid; G-CDCA, glyco-chenodeoxycholic acid; G-DCA, glyco-deoxycholic acid; GUDCA, glyco-ursodeoxycholic acid, HDCA, hyodeoxycholic acid; MELD, Model for End-stage Liver Disease; T-BA, taurine-conjugated bile acid; T-CA, tauro-cholic acid; T-CDCA, tauro-chenodeoxycholic acid; T-DCA, tauro-deoxycholic acid; T-LCA, tauro-lithocholic acid; unconj., unconjugated.
Table 3.
Univariable linear regression analysis for bile acids associated with FGF19
| Simple linear regression with predictor variables | ||||
|---|---|---|---|---|
|
| ||||
| Variablea | B | SE | p-valueb | R2 |
| Total BA (abs.) | 0.061 | 0.019 | 0.003 | 0.259 |
|
| ||||
| Conj. BA (abs.) | 0.061 | 0.019 | 0.003 | 0.261 |
| Unconj. BA (abs.) | −0.848 | 1.392 | 0.547 | 0.012 |
|
| ||||
| Conj. BA | 35.750 | 12.770 | 0.009 | 0.207 |
| Unconj. BA | −35.754 | 12.773 | 0.009 | 0.207 |
| Conj./Unconj. BA | 0.024 | 0.005 | <0.0001 | 0.451 |
|
| ||||
| T-BA | 36.228 | 10.532 | 0.002 | 0.283 |
| G-BA | −10.642 | 13.124 | 0.424 | 0.022 |
|
| ||||
| CDCA | −96.023 | 38.762 | 0.019 | 0.170 |
| CA | −190.010 | 82.475 | 0.028 | 0.150 |
| DCA | −56.021 | 25.532 | 0.036 | 0.138 |
| HDCA | −196.596 | 141.172 | 0.174 | 0.061 |
|
| ||||
| T-CDCA | 34.599 | 19.657 | 0.089 | 0.094 |
| T-CA | 90.690 | 13.551 | <0.00001 | 0.599 |
| T-DCA | −139.161 | 68.300 | 0.051 | 0.122 |
| T-LCA | −121.791 | 105.929 | 0.259 | 0.042 |
|
| ||||
| G-CDCA | −16.468 | 14.243 | 0.257 | 0.043 |
| G-CA | 111.008 | 23.197 | <0.0001 | 0.433 |
| G-DCA | −81.897 | 32.877 | 0.019 | 0.171 |
| G-UDCA | −97.082 | 54.427 | 0.085 | 0.096 |
|
| ||||
| Conj. CDCA | 0.559 | 12.741 | 0.965 | 0.000 |
| Conj. CA | 63.651 | 8.183 | <0.00001 | 0.669 |
| Conj. DCA | −63.427 | 24.055 | 0.013 | 0.188 |
Notes:
Bile acids (BA) are expressed as percent from total BA, except for “Total BA (abs.)”, “Conj. BA (abs.)” and “Unconj. BA (abs.)”. These are expressed as absolute (abs.) values.
Significance is indicated in bold. Univariable regression analysis.
Abbreviations:
B=non-standardized regression coefficient, SE=Standard Error, R2=coefficient of determination
BA, bile acid; CA, cholic acid; CDCA, chenodeoxycholic acid; conj., conjugated; DCA, deoxycholic acid G-BA, glycine-conjugated bile acid; G-CA, glycocholic acid; G-CDCA, glyco-chenodeoxycholic acid; G-DCA, glyco-deoxycholic acid; GUDCA, glyco-ursodeoxycholic acid, HDCA, hyodeoxycholic acid; MELD, Model for End-stage Liver Disease; T-BA, taurine-conjugated bile acid; T-CA, tauro-cholic acid; T-CDCA, tauro-chenodeoxycholic acid; T-DCA, tauro-deoxycholic acid; T-LCA, tauro-lithocholic acid; unconj., unconjugated.
Figure 5. Association of MELD score and FGF19 with several variables or clinical parameters.
(A) MELD score and T-CDCA serum levels are similarly elevated in alcoholic hepatitis, and have a strong positive correlation. MELD scores and T-CDCA serum concentrations were analyzed from all patients in our cohorts with determinable MELD scores and T-CDCA serum concentrations (n=25). (B) Serum FGF19 levels were significantly higher in alcoholic hepatitis patients with above average serum bilirubin than in alcoholic hepatitis patients with below average serum bilirubin (mean was 16 mg/dl, n=128). (C) Serum FGF19 is significantly higher in alcoholic hepatitis patients with above average serum GGT (mean was 495 mg/dl, n= 61). (D) Serum FGF19 increases distinctly when the contribution of conjugated bile acids to the total bile acid content approaches 100%. FGF19 values from 15 patients with alcoholic hepatitis, 8 patients with alcohol use disorder and 9 control subjects, and their respective % conjugated bile acids were analyzed. FGF19, fibroblast growth factor 19; GGT, gamma-glutamyl-transferase; MELD, Model for End-stage Liver Disease; T-CDCA, taurine-conjugated chenodesoxycholic acid. **P < 0.01.
Laboratory and clinical parameters associated with FGF19
As FGF19 levels were highest in patients with severe alcoholic hepatitis (Fig. 2A), we next determined the role of serum FGF19 as potential biomarker in alcoholic hepatitis, and correlated it with multiple laboratory and clinical parameters. For this analysis, a larger cohort of patients with alcoholic hepatitis (treated and untreated at the time of inclusion) was analyzed. Demographic, clinical and biochemical parameters of patients with alcoholic hepatitis are presented in Table 4 (n=132). Univariate regression analysis revealed a significant positive correlation of FGF19 with bilirubin levels and gamma-glutamyl-transferase (GGT) (Table 5). A significant negative correlation was detected between FGF19 and the patients’ fibrosis stage as well as their level of polymorphonuclear cell (PMN) infiltration (Table 5).
Table 4.
Baseline characteristics of patients with alcoholic hepatitis (n=132).a
| Parameter | Mean (95% CI) or n (%) |
|---|---|
| Clinical and epidemiological variables | |
| Gender (male) | 88/132 (67%) |
| Age (years) | 51 (49–53) |
| Biochemical parameters at admission | |
| Creatinine (mg/dl) | 1.1 (0.9–1.3) |
| Bilirubin (mg/dl) | 15.5 (13.9 –17.1) |
| AST (IU/L) | 175 (145–205) |
| ALT (IU/L) | 59 (51–66) |
| GGT (IU/L) | 495 (315–675) |
| Albumin (g/dl) | 2.5 (2.4–2.6) |
| INR | 1.9 (1.8–2.0) |
| Scoring systems at admission | |
| ABIC score | 8.16 (7.91–8.41) |
| MELD score | 25 (24–26) |
| MELD > 21 | 91(72%) |
| MELDNa score | 27 (26–28) |
| Maddrey’s DF | 77.2 (67.7–86.7) |
| Maddrey’s DF > 32 | 99/107 (93%) |
| Treatment at admission | |
| Steroids | 39/129 (30%) |
| Pentoxifylline | 9/115 (8%) |
| Biopsy | |
| Liver biopsy available | 68/130 (52%) |
Data are shown as mean with 95% Confidence Interval or % (n).
The fraction of missing values for each parameter is presented in Supplementary Figure 2.
ABIC, ‘Age, serum Bilirubin, INR and serum Creatinine’; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma glutamyl transferase; INR, international normalized ratio; Maddrey’s DF, Maddrey’s discriminant function; MELD, Model for End-stage Liver Disease.
Table 5.
Univariable regression analysis of laboratory and clinical parameters associated with FGF19
| Variables | B | SE | p-valuea | R2 |
|---|---|---|---|---|
| Bilirubin (mg/dl) | 0.134 | 0.066 | 0.043 | 0.033 |
| GGT (IU/L) | 13.90 | 6.059 | 0.025 | 0.082 |
| ALT (IU/L) | 0.135 | 0.316 | 0.669 | 0.002 |
| AST (IU/L) | −0.681 | 1.250 | 0.587 | 0.002 |
| INR | −0.006 | 0.004 | 0.113 | 0.020 |
| Fibrosis stageb | −0.017 | 0.008 | 0.031 | 0.070 |
| PMN infiltrationc | −0.018 | 0.006 | 0.005 | 0.118 |
Notes:
Significance is indicated in bold. Univariable regression analysis.
Fibrosis stages were classified as 0 = no fibrosis, 1 = portal fibrosis, 2 = expansive periportal fibrosis, 3 = bridging fibrosis, 4 = cirrhosis.
PMN infiltration was classified as 0 = No PMN infiltration, 1 = mild PMN infiltration, 2 = severe PMN infiltration.
Abbreviations:
B=non-standardized regression coefficient, SE=Standard Error, R2=coefficient of determination.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; FGF19, fibroblast growth factor 19; GGT, gamma glutamyl transferase; INR, international normalized ratio; PMN, polymorphonuclear cells.
Similarly, when comparing FGF19 serum levels of alcoholic hepatitis patients with above average bilirubin levels with alcoholic hepatitis patients with below average bilirubin levels, significantly different FGF19 levels can be detected (Fig. 5B). The same holds true for the comparison of FGF19 and GGT. Significantly higher FGF19 are noted in patients with higher GGT than in alcoholic hepatitis patients with lower GGT levels (Fig. 5C).
We decided to examine a subgroup of patients with more severe alcoholic hepatitis more closely (MELD ≥ 29, >75th percentile) as FGF19 started to rise more markedly when the contribution of conjugated bile acids to the total bile acid content was close to 100% (Fig. 5D), indicating a more distinct increase with more severe cholestasis and alcoholic hepatitis. Univariate Cox regression analysis revealed FGF19 correlates significantly with 30-day mortality in patients with more severe alcoholic hepatitis (Table 6). However, the hazard ratio was fairly modest.
Table 6.
FGF19 is associated with 30-Day Mortality in patients with MELD score ≥29 (n=24).
| Univariate Cox Regression | |||
|---|---|---|---|
| Variables | HR | 95% CI | p-valuea |
| FGF 19 | 1.048 | 1.00–1.10 | 0.031 |
Notes:
Logrank test
Abbreviations:
HR=Hazard Ratio, CI=Confidence Interval.
FGF19, fibroblast growth factor 19; MELD, Model for End-stage Liver Disease
However, 30-day mortality was not significantly correlated with FGF19 when all alcoholic hepatitis patients were analyzed. Also, Cox multivariate analysis revealed no independent prognostic value of FGF19 in combination with MELD (data not shown).
Discussion
This study investigated the relationship between bile acids, FGF19 and clinical outcomes in patients with alcoholic hepatitis. Patients with alcoholic hepatitis had significantly increased total, and absolute and proportional conjugated bile acid levels in relation to controls and patients with alcohol use disorder. In particular the proportional contribution of G-/T-CDCA and G-/T-CA was significantly higher and that of G-/T-DCA was significantly lower in alcoholic hepatitis patients than in controls and patients with alcohol use disorder. The increase in the conjugated primary bile acids G-/T-CDCA and G-/T-CA in alcoholic hepatitis patients can in part be explained by the presence of cholestasis in alcoholic hepatitis 6, also demonstrated by increased bilirubin and GGT levels in alcoholic hepatitis patients in our study. Given cholestasis, significantly lower amounts of primary bile acids are secreted into the intestine. Normally, these primary bile acids are metabolized by bacterial 7-alpha dehydroxylase to secondary bile acids (CA to DCA, and CDCA to LCA) 17. However, as lower amounts of CA reach the intestine in cholestasis, less DCA will be produced by the intestinal bacteria and hence be absorbed in the ileum into the systemic circulation in form of conjugated DCA in alcoholic hepatitis patients (Fig. 1F).
Serum FGF19 levels were markedly increased in patients with alcoholic hepatitis by about 100 times in relation to controls. FGF19 inhibits hepatic CYP7A1, the rate-limiting enzyme for bile acid synthesis 18. Serum C4, a marker for de novo bile acid synthesis, was hence significantly decreased in alcoholic hepatitis patients compared with controls. However, total serum bile acid levels were not affected by the suppression of bile acid synthesis in alcoholic hepatitis patients suggesting a prominent role of cholestasis. As serum FGF19 levels were markedly increased in alcoholic hepatitis patients, the hepatic gene expression of FGF19 was quantitated. Hepatic gene expression increased with increasing severity of alcoholic hepatitis. This correlated with decreasing CYP7A1 in more severe forms of alcohol hepatitis. Interestingly, CYP8B1 was not affected by the degree of liver disease. As seen in obstructive cholestasis 19, bile acid uptake (NTCP) and bile acid transport into the biliary canaliculi (BSEP) were significantly decreased in alcoholic hepatitis patients in relation to controls. Similarly, bile acid efflux into the systemic circulation by MRP3 and MRP4 increased with worsening alcoholic hepatitis, as described previously in obstructive cholestasis 20,21. These changes in expression of bile acid transporters can be interpreted as a protective mechanism of the body to decrease the concentration of bile acids in the liver.
FGF19 is classically produced and secreted in the ileum, and travels to the liver to suppress CYP7A1 10. FGF19 has been shown to be produced by epithelial cells of the gallbladder, hepatocytes, and hepatic stem cells in various diseases 10,16. FGF19 gene expression increased in the small and large intestine in alcoholic cirrhotics in relation to controls 12,22. In the current study, we are the first to show that FGF19 gene expression is also significantly increased in the liver in severe alcoholic hepatitis, and that predominantly cholangiocytes and ductular cells from smaller ductules (progenitor cells) produce FGF19, which is consistent with high FGF19 expression in bile duct cells from a human liver with biliary cirrhosis 23. Although a de novo induction of hepatic FGF19 is the likely cause for the dramatic increase of serum FGF19 in patients with alcoholic hepatitis, we cannot exclude the possibility that other organs might contribute to this systemic increase as well. FGF19 gene expression was not significantly increased in patients with early alcoholic steatohepatitis compared with controls, which indicates that patients with alcohol use disorder do not induce FGF19 gene expression in the liver (Fig. 2A).
Serum FGF19 was not significantly different in patients with NASH compared with healthy controls despite significantly increased C4 levels and increased total bile acid levels in the feces 15. This is in contrast to alcoholic hepatitis where FGF19 serum levels are ~100 times higher than in controls (Fig. 2A) whose levels were similar to NASH patients. Alcoholic hepatitis patients have suppressed serum C4 levels likely in response to high FGF19 levels, possibly as a protective mechanism of the body to decrease overall bile acid levels in face of significantly elevated systemic bile acids.
The MELD score correlated positively with several serum bile acids and with total bile acid levels, in particular with the contribution of conjugated bile acids in percent, taurine-conjugated bile acids, and markedly with T-CDCA. There was also a significant negative correlation with G-DCA and conjugated DCA. In alcoholic cirrhotics, total fecal bile acids, LCA, and DCA among others were negatively correlated with the MELD score, but CA or CDCA were not significantly correlated with the MELD score 22. These findings appear in line with our findings to a certain extent with developing cholestasis in worsening liver disease, which results in less secretion of bile acids into the intestine and hence less secondary bile acids such as DCA after metabolism by intestinal bacteria.
FGF19 correlates positively with bilirubin and GGT in alcoholic hepatitis patients, and hence with cholestasis. It is also associated with fibrosis stage and PMN infiltration in a negative fashion in patients with alcoholic hepatitis. A negative correlation between FGF19 serum levels and hepatic fibrosis was also seen in pediatric onset intestinal failure 24. Injections with FGF19 reversed hepatic fibrosis in mice, supporting its beneficial effects 25. An increase in FGF19 could hence be viewed as an advantageous reaction of the body. Severe PMN infiltration was associated with improved survival in an alcoholic hepatitis cohort 6. Therefore, a negative correlation of PMN infiltration with FGF19 appears deleterious in our alcoholic hepatitis group. As FGF19 tends to increase with significant cholestasis (Table 5, Fig. 5D), we looked at a subgroup of patients with more severe alcoholic hepatitis (with MELD ≥ 29, >75th percentile). In this subgroup FGF19 correlated positively with mortality (Table 6). Whether this is causative or rather an expression of disease activity remains to be determined. Given the beneficial anti-fibrotic and anti-cholestatic effects 25, advantageous metabolic effects 26 and hepato-regenerative effects of FGF19 27, a compensatory up-regulation of FGF19 by the body in response to worsening liver health appears more likely.
This study has several limitations. First and foremost, this study involves many centers and therefore there are differences in sampling and patient management. Second, bile acids and C4 levels were only available for the exploratory patient cohort with alcoholic hepatitis who was untreated. Third, and in contrast to that, 30% of our larger cohort of 132 patients with alcoholic hepatitis were treated with steroids and 8% were treated with pentoxifylline, which might result in some heterogeneity.
Taken together, FGF19 correlates with disease activity in alcoholic liver disease. Serum FGF19 levels were positively associated with total bile acids, conjugated bile acids, conjugated/unconjugated bile acid ratio as well as T-CA, G-CA and conjugated CA. Given its (highly) significant correlation with several bile acids and ability to be induced by the hepatic FXR 18, therapeutic modulation of the master regulator of the bile acid metabolism, FXR, might present a therapeutic opportunity in alcoholic hepatitis. Obeticholic acid, an FXR agonist, has been shown to improve alkaline phosphatase and total bilirubin levels in primary biliary cholangitis in clinical trials 28. The FLINT trial demonstrated that obeticholic acid also improves ALT levels and histologic features of NASH 29. The FXR agonist obeticholic acid is currently being investigated in patients with moderately severe alcoholic hepatitis (TREAT, NCT02039219 on ClinicalTrials.gov).
The FGF19 analogue NGM282 is currently being investigated in patients with primary biliary cholangitis (NCT02135536 on ClinicalTrials.gov). NGM282 reduced non-invasive fibrosis markers and fat liver content in patients with NASH 30. Given convincing results after interventions with FGF19 or analogues in experimental cholestatic and alcoholic liver disease 25,26, human trials with FGF19 or derivatives in alcoholic hepatitis could be considered.
Supplementary Material
Supplementary Figure 1: Percentage of data that was not available for the indicated variable. Patient group indicates controls (n=9), patients with alcohol use disorder (n=9) and patients with untreated alcoholic hepatitis (n=15). INR, international normalized ratio.
Supplementary Figure 2: Percentage of data that was not available for the indicated variable. Patient group indicates patients with alcoholic hepatitis, treated and untreated (n=132). ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl-transferase; INR, international normalized ratio; PMN, polymorphonuclear cells.
Serum FGF19 and bile acids are significantly increased in patients with alcoholic hepatitis.
Total and conjugated bile acids correlated positively with FGF19 and with disease severity (MELD).
Modulation of bile acid metabolism or signaling could represent a promising target for treatment of alcoholic hepatitis in humans.
Acknowledgments
The manuscript was supported in part by NIH grants R01 AA020703, U01 AA021856, U01 AA24726 (to BS), by a laboratory service agreement from NGM Bio (to BS) and by Award Number I01BX002213 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to BS). RB was supported by NIH grant U01AA021908.
Abbreviations
- ALD
alcoholic liver disease
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BA
bile acid
- C4
7-alpha-hydroxy-4-cholesten-3-one
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- FGF
fibroblast growth factor
- FXR
Farnesoid X receptor
- INR
international normalized ratio
- LCA
lithocholic acid
- MELD
Model for End-stage Liver Disease
Footnotes
Conflict of interest: L.L., S.J.R. and A.M.D. are employees and stockholders of NGM Bio. B.S. reports a laboratory service agreement with NGM Bio. J.G.A. reports lecture fees from Lupin Pharma, consulting fees from Theravance, outside the submitted work.
Authors contributions: K.B. and P.H. were responsible for data analysis and interpretation of data, and writing of the manuscript; L.J.J. and D.P.P. were responsible for immunohistochemistry; J.A., M.V., and R.B. performed and analyzed RNA sequencing; S.C. and C.L. performed bile acid and C4 analysis; L.L., S.J.R. and A.M.D. measured FGF19; R.L., W.Z.M. assisted with data analysis; D.E.F. assisted with study design and manuscript editing; M.L., F.B., P.M., A.L., G.G., E.C.V., J.G.A., R.S.B., V.V., J.A., J.C., D.S., P.S., S.B.H. and R.B. were responsible for collection of human samples; B.S. was responsible for the study concept and design, editing the manuscript, and study supervision.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chacko KR, Reinus J. Spectrum of Alcoholic Liver Disease. Clin Liver Dis. 2016;20(3):419–427. doi: 10.1016/j.cld.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Thursz MR, Forrest EH, Ryder S investigators S. Prednisolone or Pentoxifylline for Alcoholic Hepatitis. N Engl J Med. 2015;373(3):282–283. doi: 10.1056/NEJMc1506342. [DOI] [PubMed] [Google Scholar]
- 3.Maddrey WC, Boitnott JK, Bedine MS, Weber FL, Jr, Mezey E, White RI., Jr Corticosteroid therapy of alcoholic hepatitis. Gastroenterology. 1978;75(2):193–199. [PubMed] [Google Scholar]
- 4.Dominguez M, Rincon D, Abraldes JG, Miquel R, Colmenero J, Bellot P, et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol. 2008;103(11):2747–2756. doi: 10.1111/j.1572-0241.2008.02104.x. [DOI] [PubMed] [Google Scholar]
- 5.Mathurin P, Lucey MR. Management of alcoholic hepatitis. J Hepatol. 2012;56(Suppl):S39–45. doi: 10.1016/S0168-8278(12)60005-1. [DOI] [PubMed] [Google Scholar]
- 6.Altamirano J, Miquel R, Katoonizadeh A, Abraldes JG, Duarte-Rojo A, Louvet A, et al. A histologic scoring system for prognosis of patients with alcoholic hepatitis. Gastroenterology. 2014;146(5):1231–1239. e1231–1236. doi: 10.1053/j.gastro.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halilbasic E, Baghdasaryan A, Trauner M. Nuclear receptors as drug targets in cholestatic liver diseases. Clin Liver Dis. 2013;17(2):161–189. doi: 10.1016/j.cld.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinchet JC, Gerhardt MF, Balkau B, Munz C, Poupon RE. Serum bile acids and cholestasis in alcoholic hepatitis. Relationship with usual liver tests and histological features. J Hepatol. 1994;21(2):235–240. doi: 10.1016/s0168-8278(05)80401-5. [DOI] [PubMed] [Google Scholar]
- 9.Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49(4):1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- 10.Wunsch E, Milkiewicz M, Wasik U, Trottier J, Kempinska-Podhorodecka A, Elias E, et al. Expression of hepatic Fibroblast Growth Factor 19 is enhanced in Primary Biliary Cirrhosis and correlates with severity of the disease. Sci Rep. 2015;5:13462. doi: 10.1038/srep13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Lin B, Lin G, Wu Y, Jie Y, Li X, et al. Circulating FGF19 closely correlates with bile acid synthesis and cholestasis in patients with primary biliary cirrhosis. PLoS One. 2017;12(6):e0178580. doi: 10.1371/journal.pone.0178580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajaj JS, Kakiyama G, Zhao D, Takei H, Fagan A, Hylemon P, et al. Continued Alcohol Misuse in Human Cirrhosis is Associated with an Impaired Gut-Liver Axis. Alcohol Clin Exp Res. 2017 doi: 10.1111/acer.13498. [DOI] [PubMed] [Google Scholar]
- 13.Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. 2015;21(2):159–165. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R: A language and environment for statistical computing [computer program] http://www.R-project.org/2014.
- 15.Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One. 2016;11(5):e0151829. doi: 10.1371/journal.pone.0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Zhang W, Doughtie A, Cui G, Li X, Pandit H, et al. Up-regulation of fibroblast growth factor 19 and its receptor associates with progression from fatty liver to hepatocellular carcinoma. Oncotarget. 2016;7(32):52329–52339. doi: 10.18632/oncotarget.10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Apte U. Bile Acid Metabolism and Signaling in Cholestasis, Inflammation, and Cancer. Adv Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49(1):297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JM, Trauner M, Soroka CJ, Stieger B, Meier PJ, Boyer JL. Expression of the bile salt export pump is maintained after chronic cholestasis in the rat. Gastroenterology. 2000;118(1):163–172. doi: 10.1016/s0016-5085(00)70425-2. [DOI] [PubMed] [Google Scholar]
- 20.Denk GU, Soroka CJ, Takeyama Y, Chen WS, Schuetz JD, Boyer JL. Multidrug resistance-associated protein 4 is up-regulated in liver but down-regulated in kidney in obstructive cholestasis in the rat. J Hepatol. 2004;40(4):585–591. doi: 10.1016/j.jhep.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3(Abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem. 2003;278(38):36688–36698. doi: 10.1074/jbc.M304011200. [DOI] [PubMed] [Google Scholar]
- 22.Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2014;306(11):G929–937. doi: 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naugler WE, Tarlow BD, Fedorov LM, Taylor M, Pelz C, Li B, et al. Fibroblast Growth Factor Signaling Controls Liver Size in Mice With Humanized Livers. Gastroenterology. 2015;149(3):728–740. e715. doi: 10.1053/j.gastro.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutanen A, Lohi J, Heikkila P, Jalanko H, Pakarinen MP. Loss of ileum decreases serum fibroblast growth factor 19 in relation to liver inflammation and fibrosis in pediatric onset intestinal failure. J Hepatol. 2015;62(6):1391–1397. doi: 10.1016/j.jhep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M, Learned RM, Rossi SJ, DePaoli AM, Tian H, Ling L. Engineered fibroblast growth factor 19 reduces liver injury and resolves sclerosing cholangitis in Mdr2-deficient mice. Hepatology. 2016;63(3):914–929. doi: 10.1002/hep.28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C, et al. Modulation of the intestinal bile acid-FXR-FGF15 axis improves alcoholic liver disease in mice. Hepatology. 2017 doi: 10.1002/hep.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez-Sola G, Uriarte I, Latasa MU, Jimenez M, Barcena-Varela M, Santamaria E, et al. Engineered fibroblast growth factor 19 protects from acetaminophen-induced liver injury and stimulates aged liver regeneration in mice. Cell Death Dis. 2017;8(10):e3083. doi: 10.1038/cddis.2017.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevens F, Andreone P, Mazzella G, Strasser SI, Bowlus C, Invernizzi P, et al. A Placebo-Controlled Trial of Obeticholic Acid in Primary Biliary Cholangitis. N Engl J Med. 2016;375(7):631–643. doi: 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- 29.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385(9972):956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison SA, Rinella ME, Abdelmalek MF, Trotter JF, Paredes AH, Arnold HL, et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2018 doi: 10.1016/S0140-6736(18)30474-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Percentage of data that was not available for the indicated variable. Patient group indicates controls (n=9), patients with alcohol use disorder (n=9) and patients with untreated alcoholic hepatitis (n=15). INR, international normalized ratio.
Supplementary Figure 2: Percentage of data that was not available for the indicated variable. Patient group indicates patients with alcoholic hepatitis, treated and untreated (n=132). ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl-transferase; INR, international normalized ratio; PMN, polymorphonuclear cells.