Abstract
Previously, we have succeeded in converting induced pluripotent stem cells (iPSCs) into cancer stem cells (CSCs) by treating the iPSCs with conditioned medium of Lewis lung carcinoma (LLC) cells. The converted CSCs, named miPS-LLCcm cells, exhibited the self-renewal, differentiation potential, and potential to form malignant tumors with metastasis. In this study, we further characterized miPS-LLCcm cells both in vivo and in vitro. The tumors formed by subcutaneous injection showed the structures with pathophysiological features consisting of undifferentiated and malignant phenotypes generally found in adenocarcinoma. Metastasis in the lung was also observed as nodule structures. Excising from the tumors, primary cultured cells from the tumor and the nodule showed self-renewal, differentiation potential as well as tumor forming ability, which are the essential characters of CSCs. We then characterized the epigenetic regulation occurring in the CSCs. By comparing the DNA methylation level of CG rich regions, the differentially methylated regions (DMRs) were evaluated in all stages of CSCs when compared with the parental iPSCs. In DMRs, hypomethylation was found superior to hypermethylation in the miPS-LLCcm cells and its derivatives. The hypo- and hypermethylated genes were used to nominate KEGG pathways related with CSC. As a result, several categories were defined in the KEGG pathways from which most related with cancers, significant and high expression of components was PI3K-AKT signaling pathway. Simultaneously, the AKT activation was also confirmed in the CSCs. The PI3K-Akt signaling pathway should be an important pathway for the CSCs established by the treatment with conditioned medium of LLC cells.
Introduction
Tumors are composed of heterogeneous cancer cells with distinct morphological and functional profiles. This heterogeneity could be partially explained by the classical cancer initiation theory; evolutional accumulation of one or more mutations in a single or a few cells resulting in uncontrolled growth [1]. Another explanation is based on the presence of stem cells in tumors. Even in a few number the stem cells are considered to comprise the whole tumor in patient [2], [3], [4]. The heterogeneous population in cancer tissues are thought to be the progeny of stem cell population resulting from self-renewal and differentiation. Coupled with the malignant tumorigenic potential, the stem cells have been termed as cancer stem cells (CSCs) generating a functionally hierarchical structure in the tumor. Leukemic stem cells were first to be identified with the cell-surface markers, CD34+ CD38- differentiating in vivo into leukemic blasts [2]. This particular approach led to the emergence of new studies which described tissue-specific markers for solid tumors. The phenotype associated with cancers including motility, invasion and chemo-/radio-resistance could be traced to CSCs. Metastasis through the activation of the CXCR4 receptor was previously demonstrated by the migration of invasive CSCs defined with CD133 in pancreatic cancer [5]. On the other hand, other studies attributed their metastatic potential to the epithelial-mesenchymal transition (EMT) events showing that differentiated cancer cells could turn into CSC-like mesenchymal cells [6].
Recent reports determined that epigenetic mechanisms, such as DNA methylation, can be involved in stem-cell maintenance and in the regulation of differentiation of stem cells [7], [8]. Transcriptional silencing of tumor suppressor genes by promoter DNA hypermethylation is frequently found in human carcinogenesis [9]. However, hypomethylation was thought to be the epigenetic changes found in the early stages of carcinogenesis. Hepatocellular carcinoma lines that had been transformed with chemical carcinogens showed an aberrant hypomethylation of endogenous DNA during neoplastic transformation [10]. The global methylation difference between normal tissues and tumors showed that overall global hypomethylation is involved in oncogenesis or tumor progression [11].
Adult somatic cells have been successfully reprogrammed to pluripotent stem cells (iPSCs) with the transduction of four transcription factors [12]. The differentiation potential of iPSCs is largely expected to develop multiple potential avenues for the regenerative therapy. Immune rejection of embryonic stem cells can be avoided by replacing these with iPSCs. However, the risks of potential tumor development and other unpredictable biological changes during transplantation are still unresolved. The cellular interaction of transplanted cells in the microenvironment has been reported important to obtain successful results in regeneration therapy [13]. Recently, we demonstrated that the microenvironment of cancer cells can affect stem cells to convert iPSCs into CSC-like cells, with tumorigenic capacity as well as self-renewal and differentiation potential. In the present study, we tried to prove our hypothesis that epigenetic alterations can induce CSCs from normal stem cells in the tumor microenvironment.
Methods
Cell Culture
Mouse Lewis Lung Carcinoma cell lines (LLC) were purchased from ATCC (USA) and maintained in DMEM (D5796 Sigma) medium containing 10% fetal bovine serum (FBS, Gibco, NY) and 100 U/ml penicillin/streptomycin (Wako, Japan). Mouse induced pluripotent stem cells (miPS, iPS-MEF-Ng-20D-17; Lot No.012, Fiken Cell Bank, Japan) were cultured in DMEM containing 15% FBS, 0.1 mM NEAA (100X NEAA, Gibco, NY), 2mM L-Glutamine (Nacalai Tesque, Japan), 50 U/ml of penicillin/streptomycin (P/S), 0.1 mM 2-mercaptoethanol (Sigma) and 1000 U/ml of Leukemia Inhibitory Factor (LIF, Millipore, MA) on feeder layers of mitomycin treated mouse embryonic fibroblast (MEF) cells (Reprocell, Japan). In the case of feeder-less, the miPS cells were cultured on gelation (0.1%) coated dishes.
To prepare the primary culture, the mouse allografts were excised and cut into small pieces (approximately 1 mm3) and washed in the HBSS for three times. These pieces were transferred into a 15-ml tube with 4 ml of dissociation buffer prepared in PBS containing 0.25% trypsin, 0.1% collagenase, 20% KnockOut™ Serum Replacement (Gibco, NY), 1 mM of CaCl2 and incubated at 37°C for 40 mins. To terminate the digestion, 5 ml of DMEM containing 10% FBS was then added. The cellular suspension transferred into the new tubes and centrifuged at 1000 rpm for 5 mins. The cell pellet was resuspended in 5 ml HBSS, and centrifuged at 1000 rpm for 5 min. The cell pellet was then placed into an appropriate volume of miPS medium without LIF and the cells were seeded into a dish at a density of 5×105/ml. The primary cells derived from mouse allografts treated with puromycin for 24 hours to remove the host cells. The metastatic nodules of lung were separated and also placed into primary culture. The preparation was done like we prepared in the primary culture of tumor allografts. The expression of GFP and cell morphology was observed and photographed using Olympus IX81 microscope equipped with a light fluorescence device (Olympus, Japan).
Conversion of miPSCs Into the CSC-Like Cells
According to the methods reported by Chen L and Kasai T et al., we cultured the miPSCs in the presence of conditioned medium of LLC cells [14]. Nanog-GFP reporter expression was used in miPSC cells and the expression of GFP reflects the maintenance of stemness [15]. LLC cells were cultured and prior to collecting conditioned medium, the cells were changed into 5% serum medium at 70-80% confluency. After 48hrs incubation, the conditioned medium(CM) from LLC cells was collected and filtered through a 0.22 μm filter (Millipore, Ireland). The miPSCs were treated with the CM for 4 weeks and miPSCs were cultured in the presence or absence of LIF as controls. The miPSCs cultured under feeder-less conditions were treated with the CM in 1:1 ratio of miPS medium and CM for four weeks.
Sphere Formation Assay
To generate the spheroids, serum free medium (DMEM 97.5%, NEAA 1%, L-Glutamine 1%, 100X Pen/Strep 0.5%, 0.1 mM 2-mercaptoethanol, and Insulin-transferrin-selenium-X 1/100 v/v (ITS-X, life technologies, CA) was used and single cells were plated on ultra-low attachment dishes (Corning incorporated, NY) at cell density of 1x104 cells/ml [16].
Animal Experiments
The plan of animal experiments was reviewed and approved by the ethics committee for animal experiments of Okayama University under the IDs OKU-2013252, OKU-2014157, OKU-2014429 and OKU-2016078. All experiments were performed according to the Policy on the Care and Use of the Laboratory Animals, Okayama University. Nude mice (Balb/c-nu/nu, female, 4 weeks) were purchased CharlesRiver, Japan. Cells at 1x106 were suspended in 200 μl of HBSS (Gibco, NY) and were subcutaneously transplanted into nude mice.
RNA Extraction, cDNA Synthesis and qPCR mRNA Expression Analysis
Total RNA was extracted using RNAeasy Mini kit (QUIAGEN, Germany) according to the manufacturer's instructions and 1 μg of RNA was reverse transcribed using Superscript First strand kit (Invitrogen, CA). Quantitative real time PCR was performed with cycler 480 SYBR green I Master Mix (Roche, Switzerland) according to manufacturer’s instructions. Primers used for qPCR are listed in Table S1.
Histological Analysis and Immunohistochemistry (IHC)
Paraffin embedded tumor sections (5μm) were stained with Hematoxylin (Sigma Aldrich,USA;0.5%) and Eosin Y (Sigma Aldrich, USA) (HE) for histological analysis. Primary antibodies and dilutions used for IHC were used as follows; anti-GFP antibody 1:200 (#2956, Cell Signaling, USA), anti-MUC1 antibody 1:100 (Abcam/ab15481, UK), anti-ALDH1 antibody 1:200 (Abcam/ab52492, UK) and Anti-CD44 antibody 1:200 (Abcam/ab24504, UK).
Western Blotting
Proteins following the SDS-PAGE were transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Germany) and probed with Antibodies against anit-Akt and anit-p-Akt (Ser473), anit-p110γ (D55D5), anit-p101 (D32A5) (Cell Signaling Technology, USA) at a 1:1000 dilution followed by horseradish peroxidase-conjugated secondary antibodies (1: 2,000-1: 5,000, Cell signaling Technology). In -LIF experiment, miPSCs were cultured in miPSC medium without LIF for 6 hours before protein extraction.
RRBS DNA Methylation Analysis
The RRBS methylation analysis was performed by the BGI (BGI Japan, Kobe, Japan). RRBS libraries were sequenced on Illumina HiSeq 4000 using the PE100 sequencing strategy. The clean reads were mapped to the reference sequence (mm10) using BSMAP in which the mapping rate and bisulfite conversion rate of each sample is calculated [17].
Methylation level (Rm) was determined by the reads which covered in cytosine, which was also to equal the mC/C ratio of each reference cytosine [18], [19]. The formula was as following;
(Nm represents the reads number of mC, while Nnm represents the reads number of non-methylation reads.)
Putative DMRs were identified by comparison of the sample1 and sample 2 methylomes using windows that contained at least 5 CpG (CHG or CHH) sites with a 2-fold change in methylation level and Fisher test P value <.05. After iteratively merging interdependent DMRs, the final dataset of DMRs was made up of those that were independent from each other [20]. The DMR related genes was annotated with UCSC table browser tool [21], [22].
KEGG Pathway Enrichment
Pathway-based analysis assists to further define gene biological functions. DAVID bioinformatics resources was used to analyze the pathway enrichment of DMR -related genes [23], [24], [25]. This analysis identifies significantly enriched metabolic pathways or signal transduction pathways in DMR-related genes compared with the target regions background.
Statistical Analysis
The data were analyzed using two-tailed student’s t-test and are presented as the mean ± standard deviation (SD) at least three-time determinations. A P-value less than .05 was considered to be statically significant, while less than .01 was highly significant.
Results
Conversion of miPSCs Into CSC-like Cells With Tumorigenic and Metastatic Potential
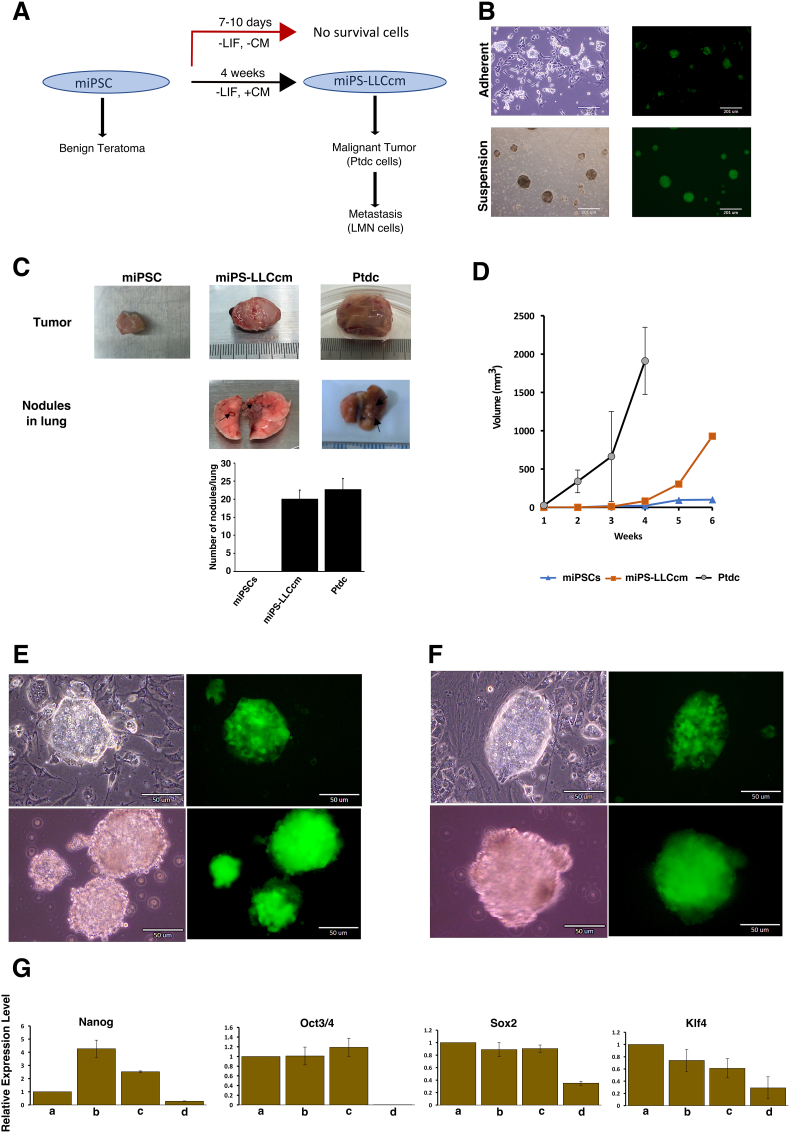
We have reported a model of CSC-like cells converted from miPSCs after exposure of miPSCs to the conditioned medium (CM) from various cancer cell lines including LLC cells [14], [26], [27]. The miPSCs used in the studies had a GFP reporter gene under the control of Nanog promoter wherein undifferentiated stem cells exhibit strong GFP expression and differentiated cells lose green fluorescence. miPSCs were found to be viable in the presence of conditioned medium even when differentiation was allowed but failed to survive beyond 10 days without LLC-CM. This scheme is briefly summarized in Figure 1A. After 4 weeks of treatment with LLC-CM, the population of miPSCs that survived and remained undifferentiated cells and were expressing GFP were miPS-LLCcm cells. The self-renewal of miPS-LLCcm cells, which is a specific characteristic of undifferentiated stem cells was confirmed by sphere-forming assay as well as differentiation potential which was evident by the appearance of highly adhesive fibroblast-like cells which had lost GFP expression and which may have undergone EMT (Figure 1B). The subcutaneous transplantation of miPS-LLCcm cells into immunocompromised Balb/c nude mice generated malignant tumors together with the metastatic-like nodule structures in the lung (Figure 1C). The number of nodules per lung was calculated for each of the three cells and the metastatic incidence was shown in Supplementary Table S2. These primary tumors and lung nodules were subjected to cell culture to isolate GFP expressing cells. The primary cultured cells that were derived from tumors were classified as Ptdc cells and the cells that were derived from lung nodules was classified as LMN cells. The Ptdc cells were subsequently transplanted into Balb/c nude mice to generate the secondary tumors. In the subcutaneous injection of 106 cells, the growth of tumors was compared (Figure 1D). The Ptdc cells showed the most rapid growth when compared to miPS-LLCcm cells and miPSCs. Primary cultures derived from the tumors were positive for GFP signal, confirming they had originated from the miPS-LLCcm cells and not from host cells and displayed sphere-forming activity suggesting they preserve stem-like characteristics (Figure 1, E and F). miPS-LLCcm cells, Ptdc cells and LMN cells maintained the expression of endogeneous stemness markers, such as Nanog, Oct3/4, Sox2 and Klf4 without LIF similar with the untreated miPSCs cultured in the presence of LIF (Figure 1G). Therefore, stemness can be maintained in the presence of LLC-CM and this conversion supports the establishment of CSC-like features.
Figure 1.
Characterization of miPS-LLCcm cells converted from miPSCs. (A) Summarized scheme of conversion of miPSCs. (B) Converted miPSCs in adherent culture (top) and in suspension culture (bottom). (C) Tumors and metastatic nodules in lungs generated by subcutaneously transplanted miPSCs, miPS-LLCcm cells and Ptdc cells into the Balb/c-nu mouse. Arrows in lung indicate the positions of nodules. The histogram showed the average number of lung nodules for each of the three cells. (D) The sizes of tumors growing in 4-6 weeks. (E) Ptdc cells in adherent culture (top) and sphere formation in suspension culture (bottom) with the expression of GFP. (F) LMN cells in adherent culture (top) and sphere formation in suspension culture (bottom) with the expression of GFP. (G) Comparison of the expression levels of stemness markers by rt-qPCR. a, miPSCs; b, miPS-LLCcm cells; c, Ptdc cells; d, LMN cells.
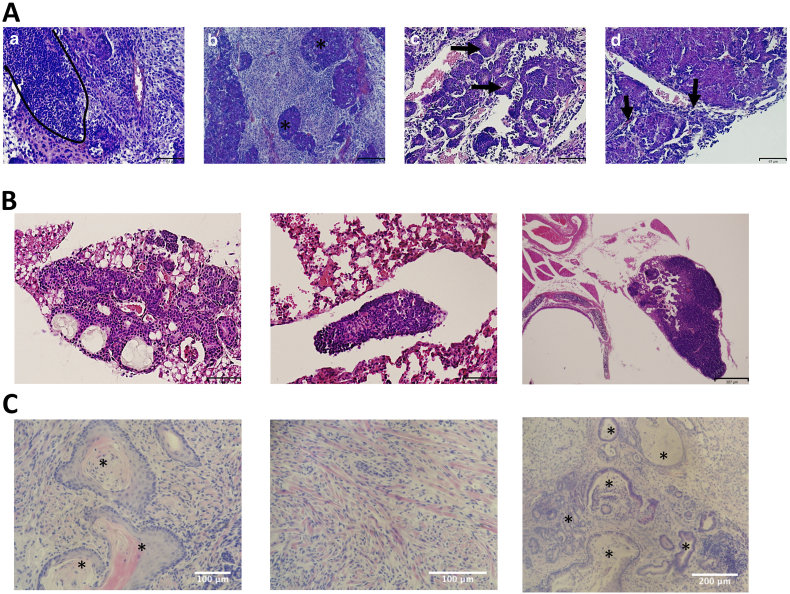
Histological analysis of tumors derived from miPS-LLCcm cells showed poorly differentiated phenotype, high nuclear to cytoplasmic ratio, micrometastasis and some epithelial ductal-like structure, which are the signs of malignancy (Figure 2A). Metastatic node-like structures were observed in the lung of the mice (Figure 2B). Untreated miPSCs tumors had a teratoma like phenotype with various germ layers (Figure 2C). These observations confirmed the tumorigenic and metastatic potential of miPS-LLCcm cells.
Figure 2.
Histological analyses of allografts of miPS-LLCcm cells. (A) High nuclear to cytoplasmic ratio (a) and with the mass of undifferentiated cells (asterisks in (b)), granular epithelial structure (arrows in (c)) and micrometastasis (arrows in (d)). (B) Metastatic nodules-like tumor in the lung pulmonary tissue (left), in the lung tissue (middle), and in the chest (right). (C) Teratoma from untreated miPSCs showing the three germ layers: squamous epithelial tissue (asterisks) in ectoderm (left), muscle tissue in mesoderm (middle) and gland-like structures (asterisks) in endoderm (right). (A-C) H&E staining.
Characterization of miPS-LLCcm, Ptdc and LMN Cells
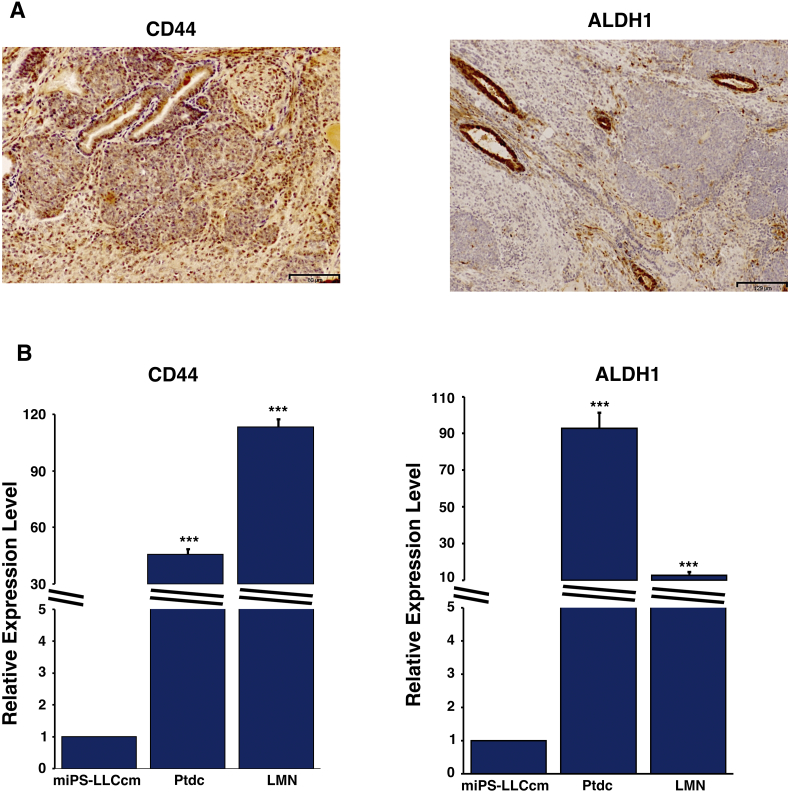
In order to further confirm the acquisition of CSC-like phenotype, we assessed the expression of the commonly known CSC markers, CD44 and ALDH1 by rt-qPCR as well as immuno-histochemical analysis. (Figure 3). The LMN and Ptdc cells showed the significantly higher expression of ALHD1 and CD44 than the miPSCs. The miPS-LLCcm derived tumor also showed the expression of these two markers.
Figure 3.
The localization and expression of CSC markers. (A) IHC analysis showing the expression of CD44 and ALDH1 in the tumor of the miPS-LLCcm cells. (B) The comparison of the expression of CD44 and ALDH1 the Ptdc and LMN cells with miPS-LLCcm by rt-qPCR analysis. ***P < .001, **P < .01, *P < .05.
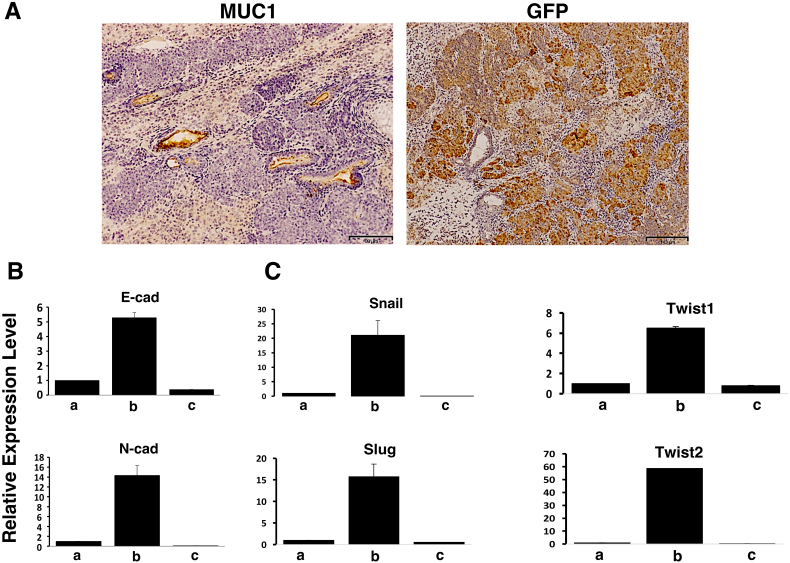
MUC1 plays a crucial role in cancer progression and is considered as a suitable marker for the adenocarcinoma tumor phenotype [28]. Immuno-histochemical analysis showed that tumor associated MUC1 expression was detected in tumor of miPS-LLCcm (Figure 4A). Mass of highly proliferating cells showed the expression of GFP as population of cells that are maintaining self-renewal ability. Ptdc cells showed the up-regulation of both E-cadherin, an epithelial marker, and N-cadherin, a mesenchymal marker, as compared to miPS-LLCcm and LMN cells (Figure 4B). The expression of Snail, Slug, Twist1 and Twist2 were also up-regulated in Ptdc cells suggesting the potential of partial and metastable epithelial-to-mesenchymal transition (EMT) phenotype in Ptdc cells.
Figure 4.
The localization and expression of EMT markers in miPS-LLCcm derived tumors. (A) Immuno-histochemical (IHC) analysis showed that ductal epithelial structure expressed MUC1 (left) and the undifferentiated mass of cells expressed the GFP (right). Comparison of the expression of (B) epithelial and mesenchymal markers (E-cadherin and N-cadherin) and (C) EMT markers in a, miPS-LLCcm cells; b, Ptdc cells and c, LMN cells.
DNA Methylation in the Conversion of iPSCs Into CSCs
Epigenetic alterations have been attributed to play an important role in carcinogenesis. Methylation and demethylation is generally considered to silence and activate gene expression, respectively. However, the epigenetic changes in early stages of cancer development in this research has not yet been assessed. Since the 4-week treatment of iPSCs with LLC-CM appears to be the main source of conferring the potential of CSCs to generate miPS-LLCcm, we hypothesized that altered epigenetic regulation rather than gene mutations, may perform an essential role in this conversion process. To quantify the DNA methylation profile in miPS-LLCcm cells, PtDC cells and LMN cells, we performed a reduced representation bisulfite sequencing (RRBS) on those genes covering most of promoters and CpG islands (Table S3). Sliding-window approach identified differentially methylated regions (DMRs), which contains at least five CG sites. The methylation levels were significantly different between the samples when assessed by Fisher test (P < .05).
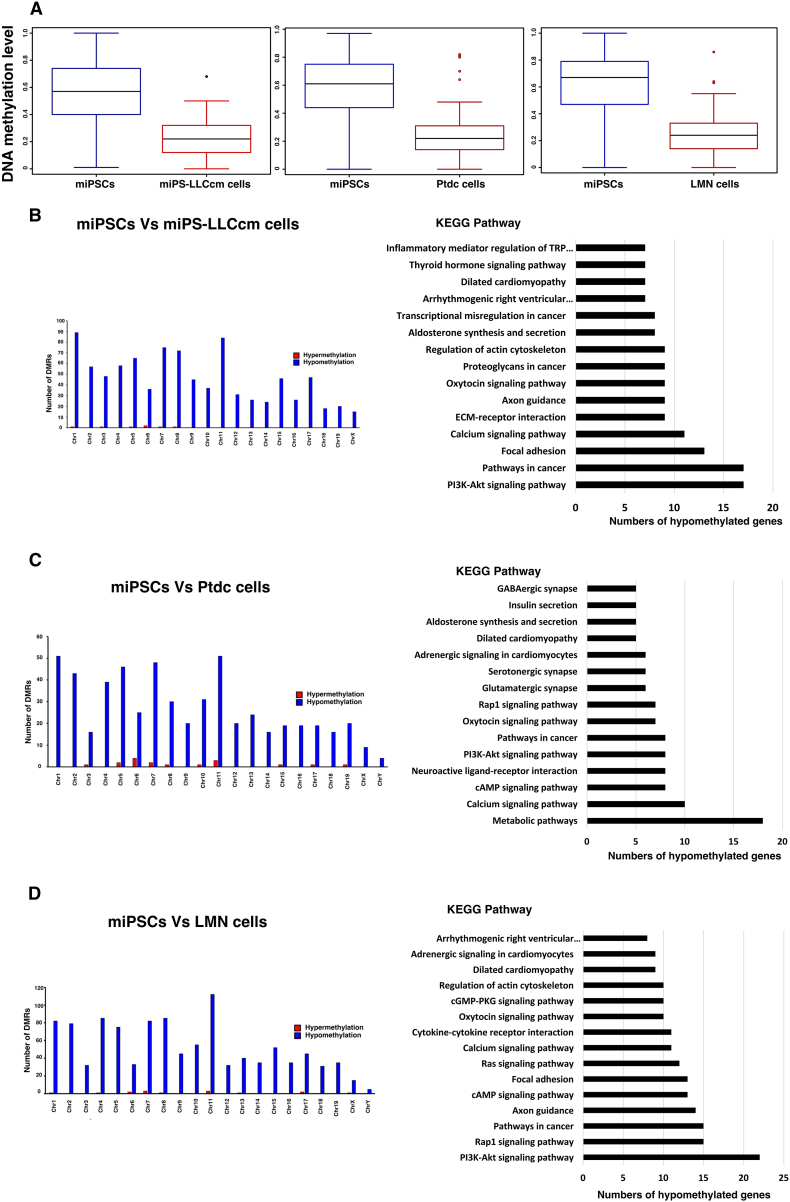
We compared the methylation in miPS-LLCcm, LMN and Ptdc cells with that in miPSC by three sets; (1) miPSCs vs. miPS-LLCcm cells, (2) miPSCs vs. Ptdc cells and (3) miPSCs vs. LMN cells. All comparisons between the different cell populations were found to exhibit hypomethylation as compared to miPSCs (Figure 5A) and 926, 583 and 1105 DMRs were identified respectively from the comparisons (Figure 5, B–D and Dataset S1A,B,C). DMRs-associated genes were further identified and segregated into hypo- and hyper-methylated genes categories (Dataset S2A,B, S3A,B, S4A,B). To validate whether the genes with DMRs were enriched for certain pathways, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the hypomethylated genes in miPS-LLCcm, Ptdc and LMN cells. According the numbers of hypomethylated genes involved in the pathway, the top 15 KEGG pathways were nominated (Figure 5, B–D and Dataset S2C, S3C, S4C). We then selected the pathways most relevant for carcinogenesis including Focal adhesion, PI3K-Akt signaling pathway, Calcium signaling pathway, Pathway in cancer and Transcriptional misregulation in cancer.
Figure 5.
Results of hypomethylation analyses. Identification of DMRs and KEGG pathway analysis. (A) Relative DNA methylation levels of miPS-LLCcm cells, Ptdc cells and LMN cells compared to miPSCs. miPSCs vs. miPS-LLCcm cells (left), miPSCs vs. Ptdc cells (middle) and miPSCs vs. LMN cells (right). (B-D) Numbers of DMRs hypo- and hyper-methylated in each chromosome are depicted and KEGG pathways nominated with the number of hypomethylated genes. (B) miPSCs vs. miPS-LLCcm cells, (C) miPSCs vs. Ptdc cells and (D) miPSCs vs. LMN cells.
Overexpression of PI3K-Gamma Candidates in the Model were Relating to Oncogenic Potential
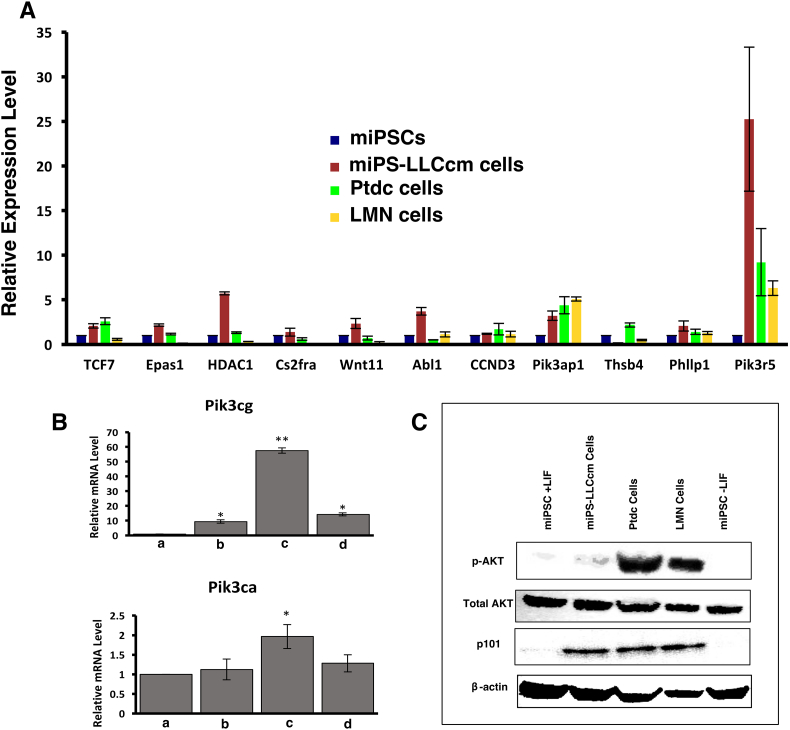
To evaluate the up-regulated expression of the hypomethylated genes in the pathway selected above, we performed rt-qPCR analysis on the miPS-LLCcm cells together with the Ptdc and LMN cells. As the results, the expression of 11 genes were found up-regulated in miPS-LLCcm cells when compared to miPSCs. Among the candidates, Pik3r5 (p101) showed significantly high expression in all the cells (Figure 6A). Recent reports have suggested that the overexpression of the catalytic subunit Pik3cg (p110γ) or the regulatory subunit Pik3r5 leads to oncogenic cellular transformation and malignancy [29]. These two subunits are class IB PI3Ks and considered to make a heterodimer of PI3K-gamma. We assessed the expression levels of Pik3cg and Pik3ca, which were class IB and class IA PI3Ks, in the miPS-LLCcm, Ptdc and LMN cells. As the result, Pik3cg showed significantly higher expression (P < .01) than that of Pik3ca (Figure 6B).
Figure 6.
Evaluation of the candidate signaling pathway nominated by the KEGG analysis. (A) rt-qPCR analyses of the candidate hypomethylated genes; (B) rt-qPCR analyses of Pik3cg (top) and Pik3ca (bottom); ***P < .001, **P < .01, *P < .05. (a, miPSCs; b, miPS-LLCcm cells; c, Ptdc cells; d, LMN cells). (C) Immunoblotting analysis of AKT activation and the expression of Pik3r5.
PI3K-Akt Activation Drives iPSC-CSCs Model
The up-regulated expression of Pik3r5 should induce the activation of Akt resulting in the onset of tumorigenicity and metastatic potential of miPS-LLCcm cells and Ptdc cells. Based on the evaluation in rt-qPCR, the constitutive expression of Pik3r5 was detected by immunoblotting with anti-p101 antibody in miPS-LLCcm and its derivatives (Figure 6C). Akt activation was simultaneously assessed by Western blotting with anti-phosphorylated Akt (p-Akt) antibody. We found Akt was constitutively activated in miPS-LLCcm, Ptdc and LMN cells (Figure 6C). In miPSCs, Akt was weakly activated in the presence of LIF but not in the absence of LIF. Taking these into consideration, the hypomethylation should lead to overexpress PI3K-gamma to enhance PI3K-Akt pathway in CSC-like cells without mutations in the open reading frames of DNA (Figure S1). These findings are consistent with the PI3K-Akt pathway as recognized one of the most frequent signaling pathway enhanced in human cancers [30].
Discussion
The current study focused on the DNA methylation in the CSC model converted from iPSCs by the treatment with conditioned medium of cancer cells. By the treatment, miPSCs obtained the ability of unlimited growth and the capacity to maintain their stemness, while they were allowed to differentiate without LIF. The subcutaneous transplantation of the survived cells into the mouse formed malignant tumors and metastasized into lung tissues. Thus, we concluded that miPSCs should be converted into CSCs without any intended genetic manipulation. The immunohistochemical analysis revealed the heterogeneity of the tumors, in which miPS-LLCcm cells should on one hand maintain the undifferentiated population expressing GFP and differentiate on the other into adenocarcinoma phenotype expressing MUC1 while miPSCs developed benign teratoma showing the three germ-layers differentiation and the loss of undifferentiated phenotype. Furthermore, the cells from benign teratoma cannot be maintained but the cells from tumors at primary site and metastatic nodules can be maintained in vitro.
The heterogeneous phenotypes characterized by the expression of E-cadherin and N-cadherin in the Ptdc cells should be implying the progression of cancer and metastatic potential of the tumor maintaining the plasticity of the transition between epithelial and mesenchymal states. Further study is required to confirm the cells are undergoing the EMT being involved in maintenance of stemness, invasiveness and metastasis of tumor cells.
Recently our group generated CSC models converted from iPSCs with the aid of CM of various cancer cell lines such as human pancreatic carcinoma cell lines PK-8 cells and KLM-1 cells, human breast cancer cell lines T47D cells and BT549 cells, mouse carcinoma cell lines LLC cells, P19 cells, B16 cells and MC.E12 cells [14], [26], [27]. In the conversion of pancreatic duct like adenocarcinoma (PDAC) like CSC model, there was no evidence relating to single point mutations even in Kras oncogene and its xenografts tumors showed the features of acinoductal metaplasia, pancreatic intraepithelial neoplasia and PDAC lesions [26]. We postulated that CSCs may be induced by epigenetic changes without any known mutation(s).
Premature termination of reprogramming were reported to result in tumor development in various tissues with undifferentiated dysplastic cells exhibiting global changes in DNA methylation at H19 DMRs identifying IGF-2 expression up-regulated in the tumor initiating cells [31]. Since the changes in DNA methylation was considered responsible for the conversion of iPSCs into CSCs, the patterns of DNA methylation were compared between the converted cells (miPS-LLCcm cells), tumor derived cells (Ptdc cells and LMN cells) and miPSCs. As the results of bisulfite sequencing, we evaluated the list of epigenetically affected genes regarding to the DMRs in the miPS-LLCcm cells and Ptdc cells and LMN cells. Hypo- and hypermethylated genes were identified and hypomethylation was found overall superior to hypermethylation in all CSCs when compared to the parental cell line miPSCs.
The analysis of KEGG pathways relating to hypomethylated genes revealed the several notable pathways important in cancers. Checking the expression of genes associated with these pathways, the expression of hypomethylated genes relating to PI3K-Akt pathway was found significantly high among those of the other genes. PI3K-Akt-mTOR signaling pathway has previously been reported as a key driver of carcinogenesis in several cancer types [32], [33]. In this study, we found Pik3r5 (p101), which is a regulatory subunit of Pik3cg enzyme, as a hypomethylated and highly up-regulated gene relating to PI3K-Akt pathway. In the recent report, the evidence of PIK3CG as a potential oncogene were evaluated by analyzing the differential role each unit of PIK3CG, of which overexpression of the catalytic subunit PIK3CG (p110γ) or the regulatory subunit PIK3R5 (p101) leads to oncogenic cellular transformation and malignancy [34]. Therefore, the hypomethylation of Pik3r5 gene leading to the up-regulation is closely related to the activation/phosphorylation of AKT that is the downstream target molecule and Pik3cg should play a key role in carcinogenesis. In fact, the multiple myeloma cells derived from patients, the up-regulation of PI3K components, in which PIK3CG has been proved to be a main regulator of cells adhesion and migration [35]. The PIK3CA gene has been reported to be hypomethylated in esophageal cancer cases when compared to the adjacent normal tissues [36]. On the other hand, both Pik3r5 and Pik3cg were overexpressed resulting in the up-regulation of PI3K-gamma in the class IB PI3Ks, but not the PI3K-alpha in the class IA in our CSCs. Collectively, the activation of PI3K-Akt signaling pathway should significantly be relating with the conversion of miPSC into miPS-LLCcm cells resulting in the constitutive activation of Akt in Ptdc and LMN cells.
According to the recent reports, the tumor cells produced a variety of molecules such as growth factors, cytokines and chemokines, which exhibited various effects such as on tumor growth and angiogenesis, providing them with various microenvironments [37], [38]. In our study, we have successfully demonstrated the CSCs generated from iPSCs by the treatment with CM from cancer derived cells acquired the DNA hypomethylation.
Conclusions
Significant overall DNA hypomethylation during the conversion should lead to the activation of certain proto-oncogene, which represent the malignant conversion even without mutations. In this context, the hypomethylation might be considered to contribute to the progression and metastasis of the cancer stem cells.
The following are the supplementary data related to this article.
Trace data of codons in Pik3ca and Pten cDNA from miPSCs, miPS-LLCcm cells and Ptdc cells and LMN cells. The sequencing was carried out to cover the positions of codons that are frequently found mutated in human cancers, of the genes related to PI3K-Akt signaling pathway. (A) The sequencing of Pik3ca cDNA showed that there is no point mutation at the codons of 542E, 545E and 1047H in all cells. (B) The sequencing of Pten cDNA showed that there is no point mutation at the codons of 130R, 173R and 233R in all cells. (DNA sequencing and position of the codons are shown at the top. Names of the cells are indicated on the left.)
List of primers used in the experiments.
Metastatic incidence in vivo transplantation.
Summary of RRBS analysis showing mapping efficiencies, proportion of mCG in promoters and CGIs.
Identification of DMRs from three sets of comparison (A; miPSCs vs. miPS-LLCcm cells, B; miPSCs vs. Ptdc cells, C; miPSCs vs. LMN cells).
DMR related genes between miPSCs vs. miPS-LLCcm cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).
DMR related genes between miPSCs vs. Ptdc cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).
DMR related genes between miPSCs vs. LMN cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).
Funding
This research was supported by the Grant-in-Aid for Scientific Research (A) No. 25242045 (M.S.); the Grant-in-Aid for Challenging Exploratory Research No. 26640079 (M.S.), Grant-in-Aid for Scientific Research (C) No. 16K07116 (Y.I.); the Japan Science and Technology Agency, Matching Planner Program-Tansaku Shiken-Grant (T.K.) and the Natural Science Foundation of Tianjin, China, No. 16JCYBJC27000 (L.C.).
Authors Contribution
A.K.K.O. and T.K. conceived and designed the study. A.K.K.O, A.S.C., and A.V conducted the experiments. H.B.M., Y.J. and N.N. helped in performing some experiments. A.S.C., H.B.M. and H.M., executed the IHC and Western Blotting experiments. A.M.J., A.C., A.S., and D.J., and H.M. gave advice in designing the experiments. T.K., and L.C., helped in analyzing the histopathological images. M.S. and T.K. directed the research project and wrote the manuscript together with A.K.K.O. All authors were involved in the final version of the manuscript.
Contributor Information
Aung Ko Ko Oo, Email: kokooo.aung@gmail.com.
Anna Sanchez Calle, Email: annasc.nit@gmail.com.
Yoshiaki Iwasaki, Email: yiwasaki@okayama-u.ac.jp.
Ling Chen, Email: chenlingdoctor@126.com.
Masaharu Seno, Email: mseno@okayama-u.ac.jp.
References
- 1.Rozhok AI, Salstrom JL, DeGregori J. Stochastic modeling indicates that aging and somatic evolution in the hematopoietic system are driven by non-cell-autonomous processes. Aging (Albany NY) 2014 doi: 10.18632/aging.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997 doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006 doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 5.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007 doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz-sánchez E. Vol. 1. 2014. Overview : Epigenetic Regulation in Cancer Stem Cells by Methylation; pp. 3–5. [Google Scholar]
- 8.Toh TB, Lim JJ, Chow EK-H. Epigenetics in cancer stem cells. Mol Cancer. 2017 doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG, Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000 doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 10.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979 doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich M, Jiang G, Fiala E, Dome JS, Yu MC, Long TI, Youn B, Sohn O, Widschwendter M, Tomlinson GE. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002 doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Wan P-X. Importance of the stem cell microenvironment for ophthalmological cell-based therapy. World J Stem Cells. 2015 doi: 10.4252/wjsc.v7.i2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M, Vaidyanath A, Mizutani A, Satoh A, Kudoh T. A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One. 2012 doi: 10.1371/journal.pone.0033544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007 doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 16.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem / progenitor cells. Genes Dev. 2003 doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009 doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y. Single base–resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 2010 doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 19.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park I, Yu J. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009 doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008 doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karolchik D. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. UCSC genome browser tutorial. Genomics. 2008 doi: 10.1016/j.ygeno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S., Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008 doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calle AS, Nair N, Oo AKK, Prieto-Vila M, Koga M, Khayrani AC, Zahra MH, Hurley L, Vaidyanath A, Seno A. A new PDAC mouse model originated from iPSCs-converted pancreatic cancer stem cells (CSCcm) Am J Cancer Res. 2016;6:2799–2815. [PMC free article] [PubMed] [Google Scholar]
- 27.Nair N, Calle AS, Zahra MH, Prieto-Vila M, Oo AKK, Hurley L, Vaidyanath A, Seno A, Masuda J, Iwasaki Y. A cancer stem cell model as the point of origin of cancer-associated fibroblasts in tumor microenvironment. Sci Rep. 2017 doi: 10.1038/s41598-017-07144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nath S, Mukherjee P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014 doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson C, Marriott SJ, Levy LS. Overexpression of p101 activates PI3Kgamma signaling in T cells and contributes to cell survival. Oncogene. 2007 doi: 10.1038/sj.onc.1210504. [DOI] [PubMed] [Google Scholar]
- 30.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2014 doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K., Osafune K, Arioka Y, Maeda T. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014 doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Chang L, Graham PH, Ni J, Hao J, Bucci J, Cozzi PJ, Li Y. Targeting PI3K/Akt/mTOR signaling pathway in the treatment of prostate cancer radioresistance. Crit Rev Oncol Hematol. 2015 doi: 10.1016/j.critrevonc.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Papadimitrakopoulou V. Development of PI3K/AKT/mTOR Pathway Inhibitors and Their Application in Personalized Therapy for Non-Small-Cell Lung Cancer. J Thorac Oncol. 2012 doi: 10.1097/JTO.0b013e31825493eb. [DOI] [PubMed] [Google Scholar]
- 34.Brazzatti JA, Klingler-Hoffmann M, Haylock-Jacobs S, Harata-Lee Y, Niu M, Higgins MD, Kochetkova M, Hoffmann P, McColl SR. Differential roles for the p101 and p84 regulatory subunits of PI3Kγ in tumor growth and metastasis. Oncogene. 2012 doi: 10.1038/onc.2011.414. [DOI] [PubMed] [Google Scholar]
- 35.Piddock RE, Loughran N, Marlein CR, Robinson SD, Edwards DR, Yu S, Pillinger GE, Zhou Z, Zaitseva L, Auger MJ. PI3Kδ and PI3Kγ isoforms have distinct functions in regulating pro-tumoural signalling in the multiple myeloma microenvironment. Blood Cancer J. 2017 doi: 10.1038/bcj.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Xie Y, Zhou Z, Qin Z, Wu J, He J. PIK3CA hypomethylation plays a key role in activation of the PI3K/AKT pathway in esophageal cancer in Chinese patients. Acta Pharmacol Sin. 2013 doi: 10.1038/aps.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuyama T, Ichiki Y, Yamada S, Shigematsu Y, Baba T, Nagata Y, Mizukami M, Sugaya M, Takenoyama M, Hanagiri T. Cytokine production of lung cancer cell lines: Correlation between their production and the inflammatory/immunological responses both in vivo and in vitro. Cancer Sci. 2007 doi: 10.1111/j.1349-7006.2007.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasmin R, Siraj S, Hassan A, Khan AR, Abbasi R, Ahmad N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediat Inflamm. 2015 doi: 10.1155/2015/201703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trace data of codons in Pik3ca and Pten cDNA from miPSCs, miPS-LLCcm cells and Ptdc cells and LMN cells. The sequencing was carried out to cover the positions of codons that are frequently found mutated in human cancers, of the genes related to PI3K-Akt signaling pathway. (A) The sequencing of Pik3ca cDNA showed that there is no point mutation at the codons of 542E, 545E and 1047H in all cells. (B) The sequencing of Pten cDNA showed that there is no point mutation at the codons of 130R, 173R and 233R in all cells. (DNA sequencing and position of the codons are shown at the top. Names of the cells are indicated on the left.)
List of primers used in the experiments.
Metastatic incidence in vivo transplantation.
Summary of RRBS analysis showing mapping efficiencies, proportion of mCG in promoters and CGIs.
Identification of DMRs from three sets of comparison (A; miPSCs vs. miPS-LLCcm cells, B; miPSCs vs. Ptdc cells, C; miPSCs vs. LMN cells).
DMR related genes between miPSCs vs. miPS-LLCcm cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).
DMR related genes between miPSCs vs. Ptdc cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).
DMR related genes between miPSCs vs. LMN cells (A; Hypomethylated genes, B; Hypermethylated genes, C; Nominated KEGG pathways).