Abstract
Background
Grain size, which is determined by grain length, grain width, and grain thickness, is an important determinant for grain yield in rice. Identification and characterization of new genes that are associated with grain size will be helpful for the improvement of grain yield in rice.
Results
We characterized the grain size mutant, larger grain size 1 (lgs1), derived from rice activation-tagged T-DNA insertion lines. Histological analysis showed that increased cell numbers in the longitudinal direction of spikelet hulls was responsible for the grain mutant phenotype in lgs1. Quantitative real-time PCR (qRT-PCR) analysis further showed that the expression levels of genes associated with the cell cycle in the young panicles of the lgs1 were higher than those in the wild type (WT), which might result in the increased cell numbers in lgs1 spikelet hulls. Insertion site analysis together with transgenic experiments confirmed that the lgs1 phenotype was caused by enhanced expression of truncated OsbHLH107, corresponding to the nucleotide (nt) 331–846 region (i.e., the transcriptional activation region of OsbHLH107) of the OsbHLH107 coding sequence (CDS). OsbHLH107 is a nucleus-localized bHLH transcription factor, which can form a homodimer with itself. Phylogenetic analysis showed that OsbHLH107 belonged to the same subfamily as OsPILs. OsPIL13 (OsPIL1) and OsPIL16 (APG) were reported to regulate grain size in rice. By transgenic experiments, we found that OsPIL11 could also regulate grain size.
Conclusion
We concluded that OsbHLH107 and its homologs are important regulators of grain size development and might be useful for grain yield improvement in rice.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0237-y) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Cell proliferation, bHLH, PILs
Background
Rice is an important crop for both human food and research. Genetic and environmental factors determine grain yield. Genetic factors determine potential grain yield, and environmental factors determine how much of that potential can be captured. Almost half a century has passed since the ‘Green Revolution’, which was primarily based on semidwarf stature to ensure crop yield potential (Asano et al., 2011).
Grain yield in rice is determined by grain number per panicle, panicle number per plant, and grain weight (Zhou et al., 2017). When grain number per panicle and panicle number per plant reach optimum levels, improvement in grain weight becomes important to further increase grain yield in breeding programs (Jiao et al., 2010; Miura et al., 2010). Grain weight is determined by grain size, which comprises grain length, width, and thickness, which in turn involve cell number and size (Fukada and Kubo, 2015; Jia et al., 2016). GW2 encodes a RING-type E3 ubiquitin ligase and negatively regulates cell numbers in spikelet hulls (Song et al., 2007); GW5 is a grain width regulator and acts in the brassinosteroid signaling pathway to regulate grain width by promoting cell division (Shomura et al., 2008; Weng et al., 2008; Duan et al., 2017; Liu et al., 2017); GW7 and GW8 have very important roles in grain width regulation, and GW8 can inhibit the expression of GW7 to negatively regulate grain size (Wang et al., 2012, 2015a, b). Many genes or QTLs associated with grain length have been reported. GS3 and qGL3/qGL3.1 are major QTLs that modulate grain length by controlling cell numbers in the glumes (Fan et al., 2006; Mao et al., 2010; Zhang et al., 2012). GW6a encodes a histone H4 acetyltransferase, OsglHAT1. Elevated GW6a expression could enhance grain length and weight by increasing cell numbers in spikelet hulls (Song et al., 2015). As a negative regulator of grain length, TGW6 encodes an IAA-glucose hydrolase protein. Loss of function of TGW6 will lift restrictions on IAA supply, resulting in increased cell numbers in spikelet hulls (Ishimaru et al., 2013). SRS3 encodes a protein that contains a kinesin motor domain and a coiled-coil structure. The grain length of srs3 is shorter than that of WT, due to a reduction in the cell length of spikelet hulls in the longitudinal direction (Kitagawa et al., 2010; Wu et al., 2014). SRS5 encodes an alpha tubulin protein and has a similar role as SRS3 in regulation of grain length (Segami et al., 2012).
The basic helix-loop-helix (bHLH) proteins are a group of transcription factors that play various roles in plant development, and 167 bHLH proteins have been identified in the rice genome (Li et al., 2006). The bHLH motif contains two functionally distinctive regions: the basic (b) region for DNA-binding and the helix-loop-helix (HLH) region for protein dimerization (Massari and Murre, 2000). Based on DNA-binding ability, proteins that can bind DNA are called DNA-binding bHLH (typical bHLH), and the others are called non-DNA-binding bHLH (HLH or atypical bHLH) (Li et al., 2006). Recent studies have revealed that atypical bHLH proteins undergo heterodimerization with typical bHLH proteins through the bHLH domain and function antagonistically (Toledo-Ortiz, 2003). For example, Increased Leaf Inclination (ILL1) and ILI1 binding bHLH (OsIBH1) act antagonistically to control cell length in the lamina joint (Zhang et al., 2010). PIFs/PILs (Phytochrome-Interacting Factors/Phytochrome-Interacting factors-Like) are a subgroup within the bHLH transcription factor family. In Arabidopsis, PIFs interact with PHYs (Phytochromes) to regulate perception of light signals and affect heading (Leivar and Monte, 2014; Luo et al., 2014; Sakuraba et al., 2014; Soy et al., 2014). Six OsPILs (OsPIL11-OsPIL16) were identified by searching rice genomic database. Each of them encodes a protein with a PIL motif that is highly homologous to PIFs in Arabidopsis thaliana (Nakamura et al., 2007). Whether these OsPILs can interact with OsPHYs to regulate perception of light signals and induce heading in rice has not been determined. OsPIL13 (also named OsPIL1) and OsPIL16 (also named APG), but not OsPIL15, are regulators of grain length, but the functions of OsPIL11, OsPIL12, and OsPIL14 have not been reported. OsPIL13 is a positive regulator of grain length (Todaka et al., 2012). Three OsbHLH genes with antagonistic effects on grain size were identified: PGL1, PGL2, and APG (OsPIL16). PGL1 and PGL2 are positive grain length regulators, and APG belongs to the OsPIL family, which has functionally antagonistic effects on PGL1 and PGL2 as a negative grain length regulator (Heang and Sassa, 2012a, b, c).
In this study, we isolated and characterized a rice grain size mutant, lgs1, which has enhanced expression of OsbHLH107, a member of the bHLH transcription factor family that regulates grain size by influencing cell numbers in the longitudinal direction of spikelet hulls. We also found that OsPIL11, a homolog of OsbHLH107, regulates grain size.
Results
Characterization of lgs1
As part of our continuing efforts to investigate the molecular mechanisms regulating grain size, we searched for mutants with altered grain size from a collection of activation-tagged T-DNA insertion lines (Jeong et al., 2002, 2006). One mutant, named lgs1 on a japonica cultivar (cv.) ‘Dongjin’ (WT) background, displayed increased grain size compared to WT. At the heading stage, lgs1 showed almost the same plant architecture as WT (Additional file 1: Figure S1a-c). Statistical analysis showed no significant differences in plant height, heading date, tiller number per plant, primary branch number per panicle, spikelet number per panicle, and fully filled grain number per panicle between WT and lgs1 (Additional file 1: Figure S1d-i). However, grain size, especially grain length, of lgs1 appeared larger than that of WT (Fig. 1a). Statistical analysis showed that grain length and 1000-grain weight were significantly increased in lgs1 (Fig. 1b, d, e and g), but grain width was not significantly changed (Fig. 1c and f). To characterize the lgs1 phenotype in detail, we performed time-course analysis of grain development from 9 days after fertilization (DAF) to 30 DAF. Notably, lgs1 developed larger grains than WT early at 9 DAF (Fig. 1h and i).
Fig. 1.
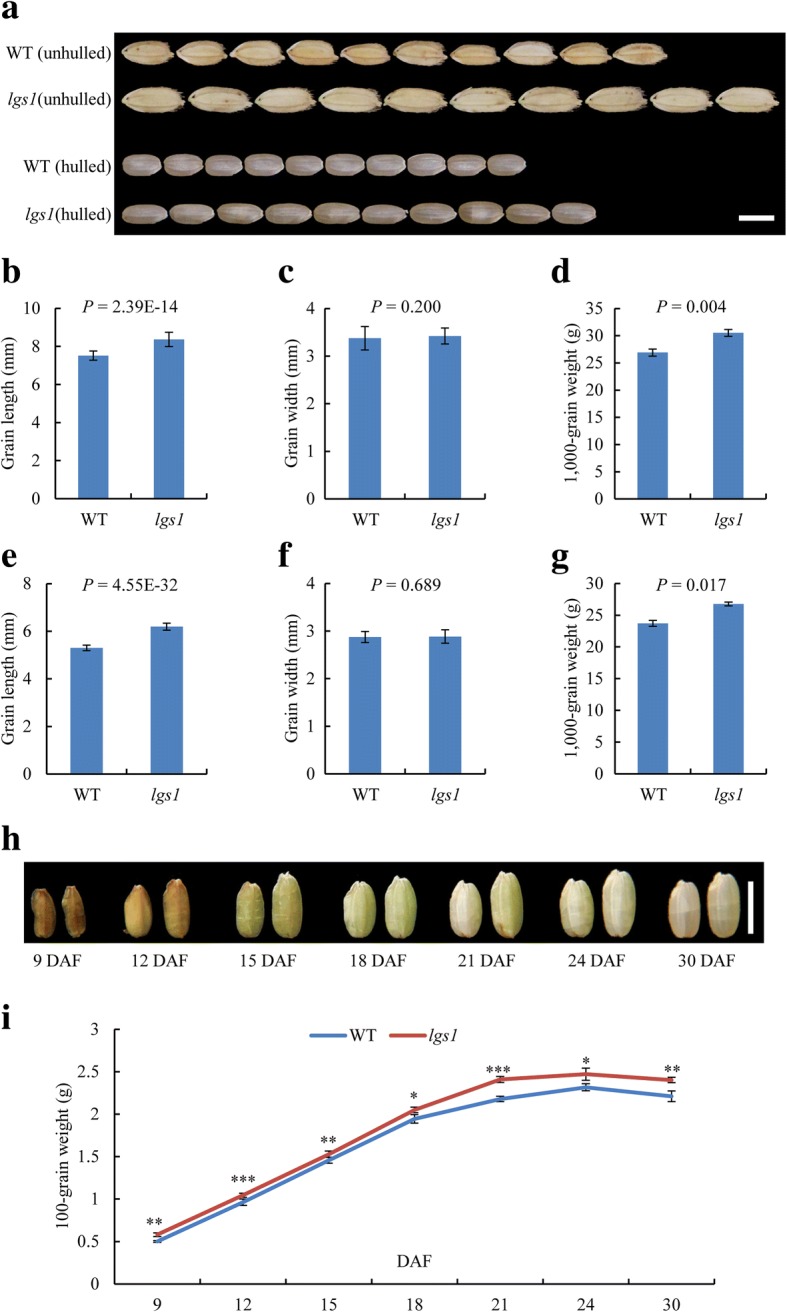
Comparisons of grain size between ‘Dongjin’ (WT) and lgs1. a Comparison of WT and lgs1 seeds (unhulled and hulled). Scale bar, 5 mm. b-d Comparisons of grain length (b, n = 30), grain width (c, n = 30), and 1000-grain weight (d, n = 3) of unhulled seeds between WT and lgs1. e-g Comparisons of grain length (e, n = 30), grain width (f, n = 30), and 1000-grain weight (g, n = 3) of hulled seeds between WT and lgs1. h Grain appearance of WT (left) and lgs1 (right) at a series of developmental stages as indicated. Scale bar, 5 mm. i Grain-filling process of WT and lgs1 (n = 5). Data are the mean ± SD. Student’s t-tests were used to generate the P values. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
Cell number increased in lgs1 spikelet hulls
To further clarify the causes of the longer grain in lgs1, we examined both cell size and cell number of spikelet hulls of WT and lgs1. Histological observations and statistical analysis showed that the average length of the inner epidermal cells of spikelet hulls in the longitudinal direction was not significantly changed in lgs1 compared with WT (Fig. 2a-d), while the cell number was significantly increased (Fig. 2e). Consistent with this finding, the relative expression levels of many genes involved in the cell cycle pathway were considerably higher in lgs1 than the WT (Fig. 2f, Additional file 1: Table S2), suggesting that the increased cell numbers in spikelet hulls of lgs1 might be due to increased expression levels of genes known to promote cell proliferation. These results indicated that lgs1 controls grain size most likely by influencing cell numbers in the longitudinal direction of spikelet hulls.
Fig. 2.
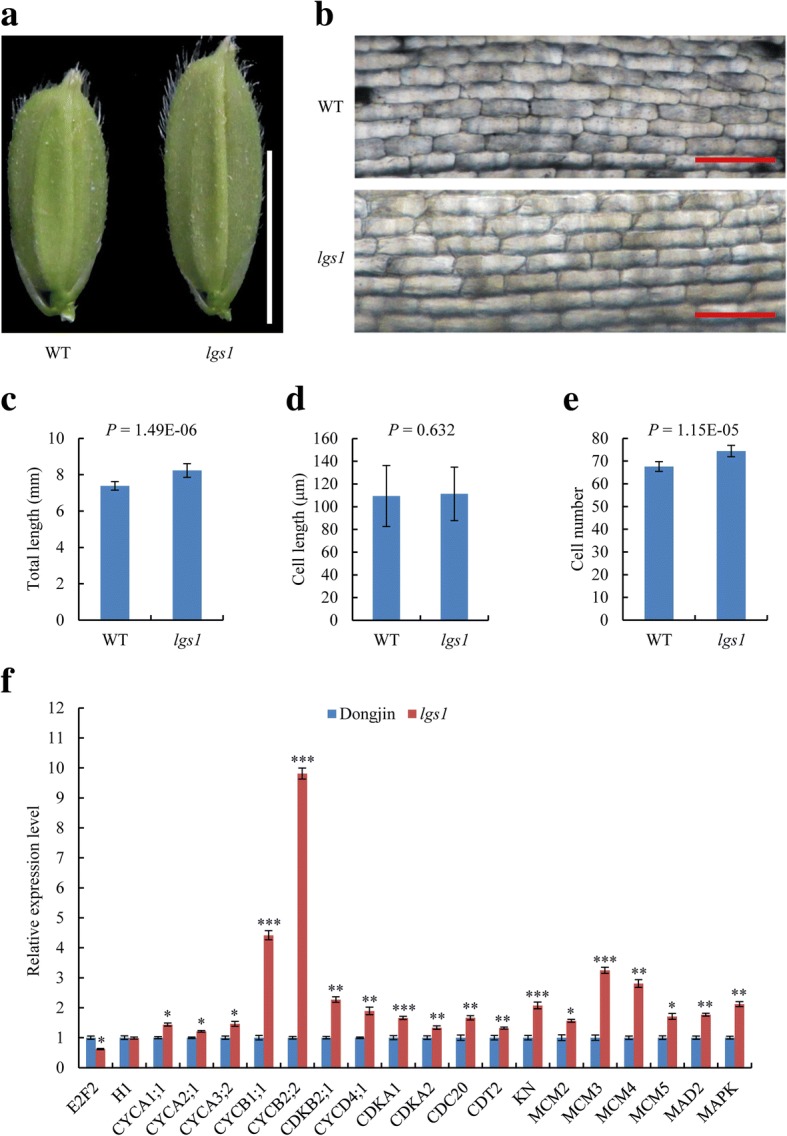
Increased cell number of spikelet hulls in the longitudinal direction determines lgs1 phenotype. a Young spikelet hulls of ‘Dongjin’ (WT) and lgs1. Scale bar, 5 mm. b Electron microscopic observation of inner epidermal cells. Scale bar, 100 μm. c-e Total length (c, n = 10), cell length (d, n = 100), and cell number (e, n = 10) of inner epidermal cells. f Relative expression levels of genes associated with the cell cycle. Relative expression levels of each gene were determined by qRT-PCR with RNA isolated from 5 cm panicles of WT and lgs1. OsActin1 was used as the internal control, and the values of the relative expression levels in the WT were set to one (n = 3). Data are the mean ± SD. Student’s t-tests were used to generate the P values. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
Enhanced expression of OsbHLH107 caused a large grain phenotype
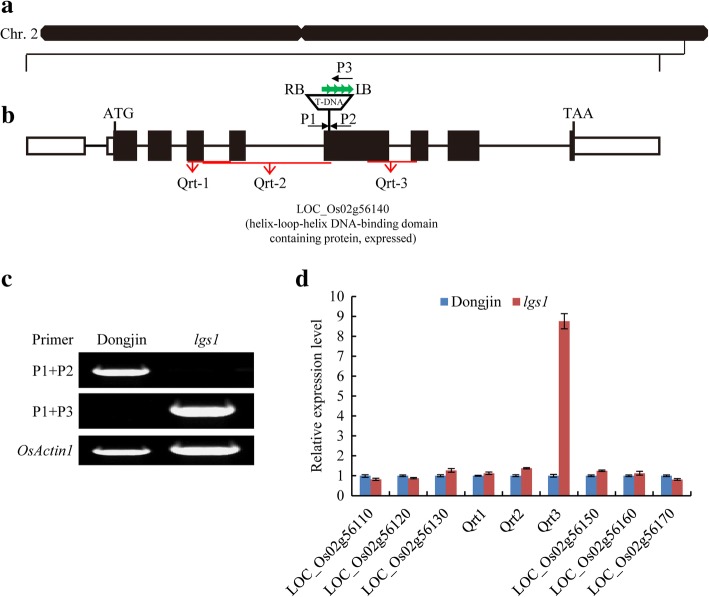
To isolate the gene responsible for the lgs1 (line 1B-04807) phenotype, we obtained the genomic flanking sequence from the T-DNA insertion database OryGenesDB (http://orygenesdb.cirad.fr/). pGA2715, an activation-tagged T-DNA vector with four copies of 35S enhancers on the left border (LB), was integrated into the 5th exon of LOC_Os02g56140, which encodes OsbHLH107, a member of the bHLH transcription factor family (Fig. 3b). PCR analysis further confirmed the T-DNA insertion site in lgs1 (Fig. 3c). A BLAST search (http://rice.plantbiology.msu.edu/) showed that OsbHLH107 is located at the end of rice chromosome 2 (Fig. 3a), with the genes LOC_Os02g56110, LOC_Os02g56120, and LOC_Os02g56130 upstream and LOC_Os02g56150, LOC_Os02g56160, and LOC_Os02g56170 downstream. To determine how the transcription of LOC_Os02g56140 was affected by the T-DNA insertion, we performed a 5′ rapid amplification of cDNA ends (5’ RACE) experiment. The 5’ RACE experiment showed that transcription of the region after the T-DNA insertion site was not disrupted, and at least two truncated transcripts were obtained (Additional file 1: Figure S2). Nine primer pairs were designed to examine whether the four 35S enhancers in the pGA2715 T-DNA vector had an influence on the expression levels of different regions of the LOC_Os02g56140 mRNA, and the genes upstream or downstream of it. qRT-PCR analysis showed that only the region after the T-DNA insertion site (primer pair, Qrt3) exhibited higher expression levels in lgs1 than WT (Fig. 3d). These results indicated that elevated expression of the truncated OsbHLH107 transcripts in lgs1 might be responsible for the mutant phenotype.
Fig. 3.
Identification of the T-DNA insertion site in lgs1. a OsbHLH107 (LOC_Os02g56140) located at the end of rice chromosome 2. Black lines, black rectangles, and white rectangles in the genomic region represent introns, exons, and UTRs, respectively. b The T-DNA insertion site in lgs1. ATG and TAA represent the start and stop codons in the CDS, respectively. The four copies of 35S enhancers in the T-DNA insertion sequence are indicated with green arrows. RB and LB indicate the T-DNA vector right and left borders, respectively. P1, P2, and P3 are primers used for identifying the T-DNA insertion site. Qrt1, Qrt2, and Qrt3 are primer pairs used for qRT-PCR analysis of different regions of LOC_Os02g56140. c Identification of the T-DNA insertion by PCR analysis. d qRT-PCR analysis of different regions of LOC_Os02g56140 and the genes upstream or downstream of it, with RNA isolated from leaves. OsActin1 was used as the internal control, and the relative expression levels in the WT were set to one (n = 3). Data are the mean ± SD
The OsbHLH107 gene was predicted to encode a protein composed of 281 amino acid (aa) residues that harbors a putative bHLH domain in the N-terminal and middle region (aa 51–110, Additional file 1: Figure S3). To test our assumption that overexpression of the truncated OsbHLH107 transcripts in lgs1 might be responsible for the mutant phenotype, we overexpressed (OE) nt 331–846 (i.e., aa 111–281) region, which contains the full-length sequence of altered transcripts after the T-DNA insertion site, under the control of the maize Ubi promoter (pUbi::CDSOsbHLH107 (nt 331–846), OE1) on a WT background. Statistical analysis showed that grain length of the transgenic plants was significantly increased compared to that of the WT (Fig. 4a and e; Additional file 1: Figure S4a). We also overexpressed full-length CDS (pUbi::CDSOsbHLH107 (nt 1–846), OE2) and nt 1–330 (pUbi::CDSOsbHLH107 (nt 1–330), OE3) of OsbHLH107. Interestingly, only transgenic plants overexpressing the full-length OsbHLH107 gene displayed significantly increased grain length (Fig. 4b, c, f and g; Additional file 1: Figure S4b and c). These results suggested that OsbHLH107 might positively regulate grain development in rice. To further confirm this, we generated OsbHLH107 knock-out and knock-down transgenic plants using CRISPR/Cas9 (CR) and RNA interference (RNAi) technologies. As expected, loss of function of OsbHLH107 on a WT background substantially reduced the grain length (Fig. 4d and h; Additional file 1: Figure S4d and Figure S12), and the same results were also obtained through RNAi experiments (Additional file 1: Figure S5). Collectively, these data suggested that the T-DNA insertion in lgs1 is responsible for the elevated expression of nucleotides after the T-DNA insertion site, which enhances grain size.
Fig. 4.
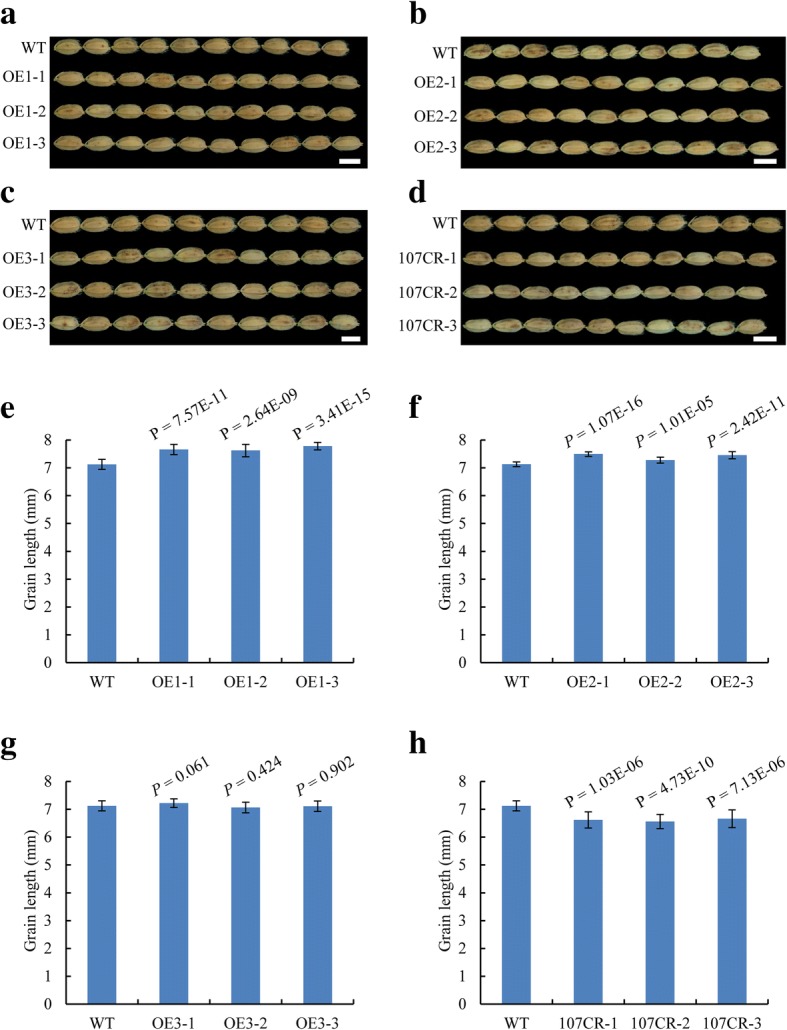
OsbHLH107 acts as a positive regulator in controlling grain development. a-d Grain length appearance of OE1 (a), OE2 (b), OE3 (c), and OsbHLH107-CRISPR/Cas9 (d, 107CR) transgenic plants on a ‘Dongjin’ (WT) background. Scale bar, 5 mm. e-h Statistical analysis of grain length shown in (a-d), respectively (n = 20). Data are the mean ± SD. Student’s t-tests were used to generate the P values
OsbHLH107 encodes a nucleus-localized protein that can form homodimers
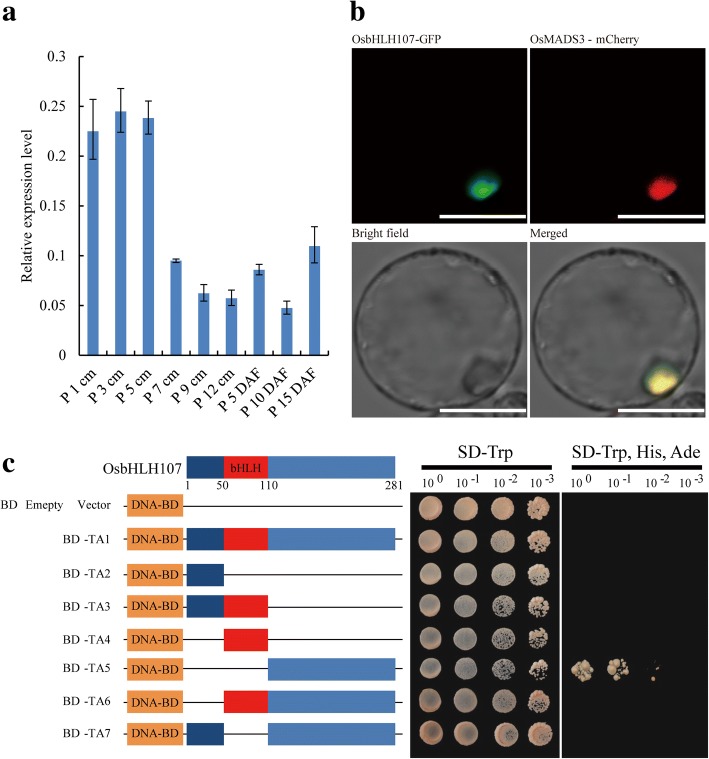
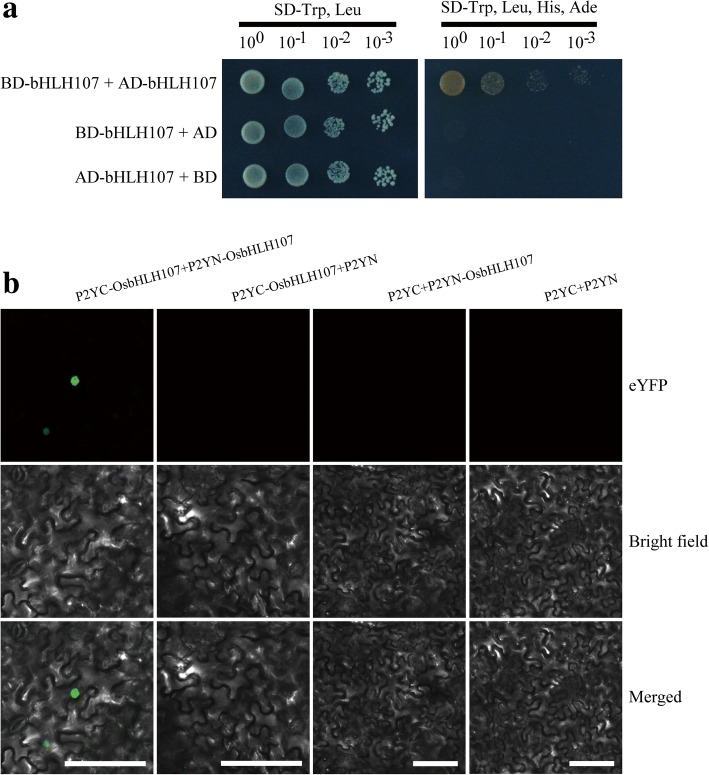
A search of the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp/) showed that OsbHLH107 is expressed throughout plant development (Additional file 1: Figure S6). To further examine the expression profile of OsbHLH107 at various developmental stages of the panicle in more detail, we performed qRT-PCR analysis and found that OsbHLH107 showed the highest expression in young panicles of 1 cm, 3 cm, and 5 cm in length, and then, the expression level gradually declined (Fig. 5a). Together, these data suggested that OsbHLH107 is broadly expressed in various tissues, with the highest expression in the early stages of panicle development. bHLH domain-containing proteins represent a group of transcription factors that putatively function in the nucleus (Li et al., 2006). To verify the subcellular-localization pattern of OsbHLH107, we transiently expressed the p35S::OsbHLH107-GFP vector in rice protoplasts. As expected, the OsbHLH107-GFP fusion protein was exclusively co-localized with the nucleus marker (OsMADS3-mCherry) (Li et al., 2011), indicating that OsbHLH107 indeed functions in the nucleus (Fig. 5b). We further fused full-length and various domain deletion variants of OsbHLH107 with the GAL4 DNA-binding domain to map its transcriptional activation region in yeast. As shown in Fig. 5c, only the C-terminal 171 amino acid residues could activate the reporter gene expression. This observation suggested that the activation domain is located in the C-terminus of OsbHLH107, which corresponds to the region responsible for the mutant phenotype (Fig. 4). Generally, bHLH transcription factors form protein dimerization (Massari and Murre, 2000; Li et al., 2006). A yeast two-hybrid (Y2H) assay showed that OsbHLH107 indeed interacted with itself (Fig. 6a). In addition, a bimolecular fluorescence complementation (BiFC) experiment confirmed the in vivo interaction of OsbHLH107 with itself in the nucleus of leaf epidermal cells of Nicotiana benthamiana (Fig. 6b). We also found that both aa 1–110 and aa 111–281, the truncated forms of the OsbHLH107 protein, were able to localize in the nucleus (Additional file 1: Figure S7) and had the ability to form homodimers (Additional file 1: Figure S8).
Fig. 5.
Expression patterns, subcellular-localization, and transcriptional activity analysis of OsbHLH107. a Relative expression levels of OsbHLH107 in various developmental stages of the panicles. P, panicles. OsActin1 was used as the internal control (n = 3). Data are the mean ± SD. b The OsbHLH107-GFP fusion protein was located in the nucleus in rice protoplasts. Scale bar, 20 μm. c Transcriptional activity assays of OsbHLH107 and its series of domain deletion variants in the yeast GAL4 system. Numbers below the schematic represent amino acid positions of OsbHLH107. DNA-BD represents the GAL4 DNA-binding domain. Empty pGBKT7 vector was used as the negative control
Fig. 6.
OsbHLH107 protein can form homodimers in vitro and in vivo. a Y2H assays showing that OsbHLH107 physically interacts with itself. AD, active domain. BD, binding domain. b BiFC assay showing that OsbHLH107 interacts with itself in leaf cells of Nicotiana benthamiana. YFP, yellow fluorescent protein. Scale bar, 100 μm
Phylogenetic analysis of OsbHLH107 and function analysis of OsPIL11
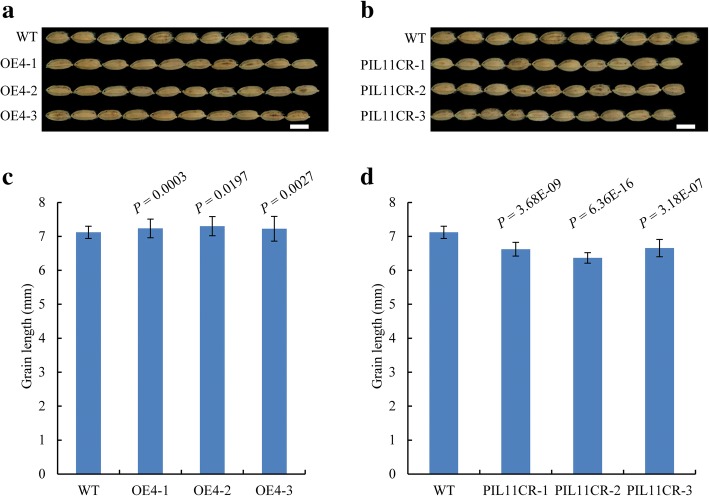
OsbHLH107 was reported to belong to the same phylogenetic subfamily as OsPILs (Li et al., 2006). We constructed a bHLH family phylogenetic analysis using 27 reported OsbHLH family proteins (Additional file 1: Table S3, China Rice Data Center, http://www.ricedata.cn/).The phylogenetic analysis showed that the amino acid sequences of OsbHLH107 and OsPILs are in highly similarity (Additional file 1: Figure S9). A BLAST search showed that OsbHLH107 harbors only one bHLH domain in the N-terminal and middle region (aa 51–110; Additional file 1: Figure S3). Therefore, OsbHLH107 might not belong to the OsPILs family due to the lack of the PIL motif. OsPIL13 (OsPIL1) and OsPIL16 (APG), but not OsPIL15, are known regulators of grain size, but the functions of OsPIL11, OsPIL12, and OsPIL14 have not been reported (Todaka et al., 2012; Heang and Sassa, 2012a, b). To clarify the functions of OsPIL11 and OsPIL14 (we could not obtain the transcript of OsPIL12), we overexpressed their full-length CDSs, and those of knock-out mutants were also obtained by the CRISPR/Cas9 technology. Overexpression of OsPIL11 significantly increased grain length, whereas the OsPIL11-CRISPR/Cas9 knock-out plants had short grains (Fig. 7; Additional file 1: Figure S10 and Figure S12), similar to the transgenic experiment results of OsbHLH107. These results indicated that OsbHLH107 and its homologs may play roles in regulation of grain size.
Fig. 7.
OsPIL11 is also a grain size regulator. Grain length appearance of pUbi::CDSOsPIL11 (a, OE4) and OsPIL11-CRISPR/Cas9 (b, PIL11CR) transgenic plants on a ‘Dongjin’ (WT) background. Scale bar, 5 mm. c and d Statistical analysis of grain length shown in (a and b), respectively (n = 20). Data are the mean ± SD. Student’s t-tests were used to generate the P values
Discussion
In this study, we proved that OsbHLH107 regulates grain size by influencing cell numbers in the longitudinal direction of spikelet hulls. Transgenic experiments confirmed that overexpression of either the full-length or the nt 331–846 region of the OsbHLH107 CDS could enhance grain size, which are similar to lgs1. On the other hand, knock-out (CRISPR/Cas9) or knock-down (RNAi) of OsbHLH107 reduced the grain length. However, we found that regardless of whether the full-length OsbHLH107 CDS or the nt 331–846 region was used, the effect on grain length did not reach the level of lgs1. The present study only simulated the mutation at the transcriptional level, and how these changes in the transcript sequence after the T-DNA insertion site affect gene function at the protein level requires further study. Moreover, we could not rule out the possibility that some other mutations induced by tissue culture also partly conferred to the enlarged grain size.
OsbHLH107 is a typical bHLH (with a DNA-binding element known as a G-box). In theory, because of the lack of the bHLH domain, this truncated protein (aa 111–281) of OsbHLH107 may not bind DNA and interact with its partners. However, through our experiments, we found that this truncated protein is also located in the nucleus and can form homodimers. These results indicated that this truncated protein may interact with other proteins in the nucleus to regulate the expression of downstream genes. Unfortunately, we have not yet identified proteins that can interact with OsbHLH107 to prove our hypothesis; this will be the focus of future research on the function of OsbHLH107. In addition, although aa 1–110 of OsbHLH107 was also located in the nucleus and could form homodimers, it did not have the ability to regulate grain size directly, possibly because of a lack of the transcriptional activation ability. This further indicated that the transcriptional activation ability of aa 111–281 was important for the OsbHLH107 function.
Phylogenetic analysis showed that OsbHLH107 has a tight evolutionary linkage with OsPILs. To investigate whether there are functional correlations between OsbHLH107 and its homologous genes, we conducted qRT-PCR analysis and found that there were no obvious differences at the expression levels between WT and lgs1 (Additional file 1: Figure S11). Transgenic experiments showed that overexpression of OsPIL11 significantly increased grain length and OsPIL11-CRISPR/Cas9 knock-out plants had short grains. In addition, although nucleotide changes were in 5’ UTR region of OsPIL11-CRISPR/Cas9 mutants, we think that these changes may affect the stability and/or expression of OsPIL11 mRNA, which leads to the mutant phenotype, and similar effects of UTR changes on mRNA stability and/or expression have been reported in several previous studies (Hughes, 2006; Srivastava et al., 2017). All these results indicated that OsbHLH107 and its homologs (OsPIL1/13, OsPIL11, OsPIL16) play roles in regulation of grain size but may act independently.
Plant organ size is determined by cell division and cell expansion (Sugimoto-Shirasu and Roberts, 2003). Many OsbHLH genes participate in regulation of plant organ size. For example, An-1 encodes a bHLH protein that regulates awn development, grain size, and grain number in rice (Luo et al., 2013). Histological observations indicated that increased cell numbers in the longitudinal direction of spikelet hulls determined the grain length phenotype in lgs1. Consistent with this finding, qRT-PCR analysis showed that relative expression levels of genes involved in the cell cycle mechanisms were up-regulated in the young panicles of lgs1, suggesting that the increased cell numbers might result from increased expression levels of genes promoting cell proliferation. Although we found that G-box elements were located in the promoter regions of some up-regulated cell cycle genes, we obtained no evidence to show that OsbHLH107 could bind to these G-boxes and regulate expression of these cell cycle genes, directly (data not shown). Because of the suggested grain size regulator role for OsbHLH107, we examined the relative expression levels of several genes that were previously reported to act as grain size regulators. There were no significant differences in the transcript levels of these genes between WT and lgs1 (data not shown), indicating that OsbHLH107 may act in a new way to regulate grain size.
Conclusions
OsbHLH107 is a member of the bHLH transcription factor family, which regulates grain size by influencing cell numbers in the longitudinal direction of spikelet hulls. We found that OsPIL11, a homolog of OsbHLH107, also regulates grain size. Our results revealed that OsbHLH107 and its homologs may play important roles in regulation of grain size in rice.
Methods
Plant materials and growth conditions
lgs1 (1B-04807) was isolated from a set of activation-tagged T-DNA insertion rice lines (Jeong et al., 2002, 2006) and was kindly provided by Prof. Gynheung An (Department of Plant Systems Biotech, Kyung Hee University, Yongin, South Korea). All plants were grown in the field at two sites, Shunyi in Beijing and Sanya in Hainan province.
Statistical analysis and histological observations
Agronomic traits were measured at the time of harvest. Harvested seeds were air-dried, and fully developed grains were measured for grain length, width, and weight. Fresh young spikelet hulls were collected and observed using an electron microscope (LEICA DM500 B). ImageJ software was used to measure the size of the inner epidermal cells. Total length of the spikelet hulls divided by the average length of the cells to obtain an approximate cell number.
Primer design, RNA extraction, qRT-PCR, and 5’ RACE experiment
Primer Premier 5.0 software was used to design primers, and all the primers used in this study are listed in Additional file 1: Table S1. Total RNA was isolated from tissues using a RNAprep Pure Plant Kit (Tiangen Biotech Co., Ltd., Beijing). First-strand cDNA was synthesized using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian). Synthesized cDNAs were utilized for qRT-PCR using a SYBR Premix Ex Taq kit (TaKaRa) on an ABI 7500 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Primers were designed using the GenScript real-time PCR (TaqMan) primer design tool (https://www.genscript.com/tools/real-time-pcr-tagman-primer-design-tool). The rice Actin1 gene (LOC_Os03g13170) was used to normalize the cDNA quantity. The 5’ RACE experiment was conducted using a SMARTer RACE 5’/3’ Kit (Clontech Laboratories, Inc. Mountain View, CA, USA).
Gene cloning, transgenic constructs, multiple sequence alignment analysis, and phylogenetic analysis
DNA and protein sequences of the genes mentioned in this study were obtained from the MSU database (http://rice.plantbiology.msu.edu/). The full-length or truncated CDS of OsbHLH107, OsPIL11, and OsPIL14 were amplified from rice variety ‘Dongjin’ and were cloned into the pCUbi1390 vector under the control of the maize Ubi promoter to create overexpression constructs. Gene specific fragment of OsbHLH107 was cloned, in sense and antisense orientations, into the LH-FAD2-1390RNAi vector (Li et al., 2013) for RNAi analysis of OsbHLH107. CRISPR/Cas9 technology was used to obtain knock-out mutants of OsbHLH107, OsPIL11, and OsPIL14 (Ma et al., 2017). sgRNA sequences were obtained by using online tool (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Plasmids were introduced into ‘Dongjin’ by Agrobacterium tumefactions-mediated transformation (strain EHA105). Multiple sequence alignment analysis was conducted using the software DNAMAN. A phylogenic tree was constructed using the software MEGA 5.0 based on the neighbor joining method as described previously (Wang et al., 2017).
Subcellular-localization and BiFC assays
The full-length or truncated amino acid sequences of OsbHLH107 were fused to the GFP protein under the control of the 2 × CaMV35S promoter in the transient expression vector pAN580-GFP. Rice protoplast preparation and transformation were performed as previously described (Chen et al., 2006). OsMADS3 (LOC_Os01g10504) (Li et al., 2011) and D53 (LOC_Os11g01330) (Zhou et al., 2013) were used as nucleus-localization markers. For BiFC assays, the CDS of OsbHLH107 was amplified and cloned into the p2YN and p2YC vectors to form the P2YN-OsbHLH107 and P2YC-OsbHLH107 plasmids, respectively. N. benthamiana leaf transformation was performed as described previously (Waadt and Kudla, 2008). Fluorescence was observed using a confocal laser-scanning microscope (Zeiss LSM 780).
Y2H assays
The CDS of OsbHLH107 and various truncated derivatives were amplified. The PCR products were inserted into the pGBKT7 and pGADT7 vectors, and these constructs were co-transformed into yeast strain AH109 as described in the Matchmaker™ Gold Yeast Two-Hybrid System User Manual (Clontech Laboratories). The yeast liquid culture was diluted to an absorbance of 0.5 at 600 nm (A600).
Additional file
Figure S1. Characterization of ‘Dongjin’ (WT) and lgs1 plants. Figure S2. Identification of transcripts after the T-DNA insertion site by 5’ RACE. Figure S3. Multiple sequence alignment of OsbHLH107 and its homologs. Figure S4. Relative expression analysis and molecular identification of OsbHLH107 overexpression and CRISPR/Cas9 transgenic plants. Figure S5. Characterization of OsbHLH107-RNAi seeds on a ‘Dongjin’ (WT) background. Figure S6. OsbHLH107 is broadly expressed in various tissues. Figure S7. Subcellular-localization of the truncated forms of OsbHLH107. Figure S8. Y2H assays showed that the truncated form of OsbHLH107 physically interacts with itself. Figure S9. Phylogenetic analysis of the reported OsbHLHs. Figure S10. Identification of OsPIL11 transgenic plants. Figure S11. Relative expression levels of OsPILs in ‘Dongjin’ and lgs1. Figure S12. Statistical analysis of grain length of OsbHLH109-CRISPR/Cas9 (107CR) and OsPIL11-CRSPR/Cas9 (PIL11CR) T1 generation plants. Table S1. Primers used in this study. Table S2. Information regarding cell cycle genes used in this study. Table S3. Information regarding bHLH genes used in this study. (DOCX 5206 kb)
Acknowledgements
We owe special thanks to Prof. Gynheung An (Department of Plant Systems Biotech, Kyung Hee University, Yongin, South Korea) for providing lgs1.
Funding
This research was supported by the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in the Mid-lower Yangtze River, Ministry of Agriculture, P.R. China; Jiangsu Collaborative Innovation Center for Modern Crop Production; the Yangtze River Valley Hybrid Rice Collaboration Innovation Center; grants from the National Key Research and Development Program of China (2016YFD0100101–08, 2017YFD0100305), National Transformation Science and Technology Program (2014ZX08001004–002), Jiangsu Science and Technology Development Program (BE2017368), and Agricultural Science and Technology Innovation Fund project of Jiangsu Province (CX(16)1029).
Availability of data and materials
All data supporting the conclusions of this article are provided within the article (and its Additional file 1).
Abbreviations
- 5’ RACE
5’ rapid amplification of cDNA ends
- aa
Amino acid
- AD
Active domain
- BD
Binding domain
- BiFC
Bimolecular fluorescence complementation
- CDS
Coding sequence
- CR
CRISPR/Cas9
- cv.
Cultivar
- DAF
Days after fertilization
- LB
Left border
- nt
Nucleotide
- OE
Overexpressed/Overexpression
- P
Panicles
- qRT-PCR
Quantitative real-time PCR
- RB
Right border
- RNAi
RNA interference
- WT
Wild type
- Y2H
Yeast two-hybrid
Authors’ contributions
JW, LJ and YR designed this study. XY, YR and LJ wrote the manuscript. XY, YC, MN and RJ performed the experiments. XY, ZF, CM, XL and LX managed the experimental materials. XZ, FW and XG produced the transgenic experiments. YR and LJ modified the manuscript. All authors read and approved the final manuscript.
Authors’ information
Correspondence and requests for materials should be addressed to Ling Jiang (jiangling@njau.edu.cn) and Jianmin Wan (wanjm@njau.edu.cn or wanjm@caas.cn).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Xiaoming Yang and Yulong Ren contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0237-y) contains supplementary material, which is available to authorized users.
Contributor Information
Ling Jiang, Phone: +86-25-84399061, Email: jiangling@njau.edu.cn.
Jianmin Wan, Phone: +86-25-84396516, Email: wanjm@njau.edu.cn, Email: wanjianmin@caas.cn.
References
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, Innan H, Kitano H, Ashikari M, Matsuoka M. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci U S A. 2011;108:11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang G. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol. 2006;7:417–427. doi: 10.1111/j.1364-3703.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Duan P, Xu J, Zeng D, Zhang B, Geng M, Zhang G, Huang K, Huang L, Xu R, Ge S, Qian Q, Li Y. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant. 2017;10:685–694. doi: 10.1016/j.molp.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Fukada F, Kubo Y. Colletotrichum orbiculare regulates cell cycle G1/S progression via a two-component GAP and a GTPase to establish plant infection. Plant Cell. 2015;27:2530–2544. doi: 10.1105/tpc.15.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed Sci. 2012;62:133–141. doi: 10.1270/jsbbs.62.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H. Overexpression of a basic helix-loop-helix gene antagonist of PGL1 (APG) decreases grain length of rice. Plant Biotechnol. 2012;29:65–69. doi: 10.5511/plantbiotechnology.12.0117a. [DOI] [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- Jeong D, An S, Kang H, Moon S, Han J, Park S, Lee HS, An K, An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D, An S, Park S, Kang H, Park G, Kim S, Sim J, Kim Y, Kim M, Kim S, Kim J, Shin M, Jung M, An G. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Jia N, Liu X, Gao H. A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 2016;171:318–333. doi: 10.1104/pp.16.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y. A novel kinesin 13 protein regulating Rice seed length. Plant Cell Physiol. 2010;51:1315–1329. doi: 10.1093/pcp/pcq092. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E. PIFs: Systems integrators in plant development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang L, Youn JH, Sun W, Cheng Z, Jin T, Ma X, Guo X, Wang J, Zhang X, Wu F, Wu C, Kim SK, Wan J. A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa) New Phytol. 2013;200:1076–1088. doi: 10.1111/nph.12427. [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D. Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell. 2011;23:2536–2552. doi: 10.1105/tpc.111.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, Zhang X, Guo X, Wang J, Wang H, Wan J. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y, Zhao Q, Wang A, Wang Z, Sang T, Wang Z, Han B. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell. 2013;25:3360–3376. doi: 10.1105/tpc.113.113589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell. 2014;26:2441–2456. doi: 10.1105/tpc.113.121657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Wu F, Sheng P, Wang X, Zhang Z, Zhou K, Zhang H, Hu J, Lin Q, Cheng Z, Wang J, Zhu S, Zhang X, Guo X, Wang H, Wu C, Zhai H, Wan J. The LBD12-1 transcription factor suppresses apical meristem size by repressing Argonaute 10 expression. Plant Physiol. 2017;173:801–811. doi: 10.1104/pp.16.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci U S A. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song X, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci Biotechnol Biochem. 2007;71:1183–1191. doi: 10.1271/bbb.60643. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang M, Kim J, Paek N, Choi G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- Segami S, Kono I, Ando T, Yano M, Kitano H, Miura K, Iwasaki Y. Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice. 2012;5:1–10. doi: 10.1186/1939-8433-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Song X, Huang W, Shi M, Zhu M, Lin H. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Song X, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J, Kitano H, Sakakibara H, Jacobsen SE, Ashikari M. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci U S A. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, Monte E. PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J Exp Bot. 2014;65:2925–2936. doi: 10.1093/jxb/ert465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2017;23:248–259. doi: 10.1016/j.tplants.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, Kojima M, Sakakibara H, Shinozaki K, Yamaguchi-Shinozaki K. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci U S A. 2012;109:15947–15952. doi: 10.1073/pnas.1207324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). Cold Spring Harb Protoc 2008:t4995 [DOI] [PubMed]
- Wang D, Wang Y, Long W, Niu M, Zhao Z, Teng X, Zhu X, Zhu J, Hao Y, Wang Y, Liu Y, Jiang L, Wang Y, Wan J. SGD1, a key enzyme in tocopherol biosynthesis, is essential for plant development and cold tolerance in rice. Plant Sci. 2017;260:90–100. doi: 10.1016/j.plantsci.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet. 2015;47:949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X, Wang J, Jiang L, Zhai H, Wan J. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C, Wang J, Jiang L, Wan J. Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice. Plant Cell Rep. 2014;33:235–244. doi: 10.1007/s00299-013-1524-0. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, Yang H, Xu Y, Kim SH, Fujioka S, Lin WH, Chong K, Lu T, Wang ZY. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2010;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, Zhang H. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci U S A. 2012;109:21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W, Gao H, Chen J, Yang C, Wang D, Tan J, Zhang X, Guo X, Wang J, Jiang L, Liu X, Chen W, Chu J, Yan C, Ueno K, Ito S, Asami T, Cheng Z, Wang J, Lei C, Zhai H, Wu C, Wang H, Zheng N, Wan J. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wang X, Zhou D, Ouyang Y, Yao J. Overexpression of the 16-kDa α-amylase/trypsin inhibitor RAG2 improves grain yield and quality of rice. Plant Biotechnol J. 2017;15:568–580. doi: 10.1111/pbi.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of ‘Dongjin’ (WT) and lgs1 plants. Figure S2. Identification of transcripts after the T-DNA insertion site by 5’ RACE. Figure S3. Multiple sequence alignment of OsbHLH107 and its homologs. Figure S4. Relative expression analysis and molecular identification of OsbHLH107 overexpression and CRISPR/Cas9 transgenic plants. Figure S5. Characterization of OsbHLH107-RNAi seeds on a ‘Dongjin’ (WT) background. Figure S6. OsbHLH107 is broadly expressed in various tissues. Figure S7. Subcellular-localization of the truncated forms of OsbHLH107. Figure S8. Y2H assays showed that the truncated form of OsbHLH107 physically interacts with itself. Figure S9. Phylogenetic analysis of the reported OsbHLHs. Figure S10. Identification of OsPIL11 transgenic plants. Figure S11. Relative expression levels of OsPILs in ‘Dongjin’ and lgs1. Figure S12. Statistical analysis of grain length of OsbHLH109-CRISPR/Cas9 (107CR) and OsPIL11-CRSPR/Cas9 (PIL11CR) T1 generation plants. Table S1. Primers used in this study. Table S2. Information regarding cell cycle genes used in this study. Table S3. Information regarding bHLH genes used in this study. (DOCX 5206 kb)
Data Availability Statement
All data supporting the conclusions of this article are provided within the article (and its Additional file 1).